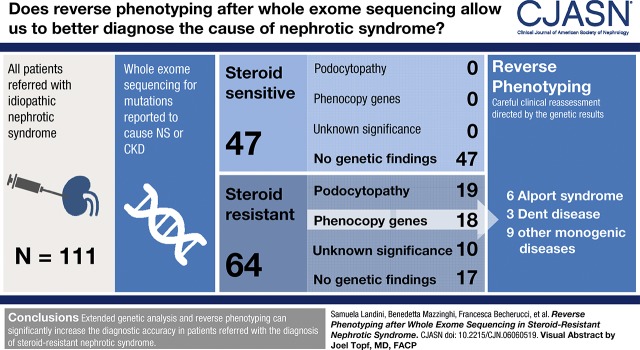
Visual Abstract
Keywords: nephrotic syndrome, chronic kidney disease, whole-exome sequencing, genetic testing, humans, United States, retrospective studies, workflow, whole exome sequencing, medical genetics, phenotype, steroids, chronic renal insufficiency, multiple drug resistance
Abstract
Background and objectives
Nephrotic syndrome is a typical presentation of genetic podocytopathies but occasionally other genetic nephropathies can present as clinically indistinguishable phenocopies. We hypothesized that extended genetic testing followed by reverse phenotyping would increase the diagnostic rate for these patients.
Design, setting, participants, & measurements
All patients diagnosed with nephrotic syndrome and referred to our center between 2000 and 2018 were assessed in this retrospective study. When indicated, whole-exome sequencing and in silico filtering of 298 genes related to CKD were combined with subsequent reverse phenotyping in patients and families. Pathogenic variants were defined according to current guidelines of the American College of Medical Genetics.
Results
A total of 111 patients (64 steroid-resistant and 47 steroid-sensitive) were included in the study. Not a single pathogenic variant was detected in the steroid-sensitive group. Overall, 30% (19 out of 64) of steroid-resistant patients had pathogenic variants in podocytopathy genes, whereas a substantial number of variants were identified in other genes, not commonly associated with isolated nephrotic syndrome. Reverse phenotyping, on the basis of a personalized diagnostic workflow, permitted to identify previously unrecognized clinical signs of an unexpected underlying genetic nephropathy in a further 28% (18 out of 64) of patients. These patients showed similar multidrug resistance, but different long-term outcome, when compared with genetic podocytopathies.
Conclusions
Reverse phenotyping increased the diagnostic accuracy in patients referred with the diagnosis of steroid-resistant nephrotic syndrome.
Introduction
Isolated nephrotic syndrome is classified, according to the response to steroids, as steroid-sensitive or steroid-resistant nephrotic syndrome (1,2). Although steroid-sensitive nephrotic syndrome usually has a favorable prognosis, steroid-resistant nephrotic syndrome can progress to ESKD (3–7). Indeed, despite some patients’ response to immunosuppression in terms of proteinuria reduction, many are multidrug resistant (6). Genetic testing using extended panels of podocytopathy genes has become a valuable diagnostic tool to identify monogenic podocytopathies, which account for about 30% of patients affected by steroid-resistant nephrotic syndrome (5,8–12). In addition, a recent report suggested that steroid-sensitive nephrotic syndrome may also occasionally be of genetic origin (13). All other cases of isolated nephrotic syndrome are usually assumed to be of nongenetic cause. Genetic nephropathies outside the podocytopathy spectrum (e.g., Alport syndrome, Dent disease, or Fabry disease) are usually recognized upon standard diagnostic work-up. However, steroid-resistant nephrotic syndrome can rarely be the only evident clinical sign, at disease onset or even later (14–18). When this happens, these genetic nephropathies are often misclassified and treated as isolated steroid-resistant nephrotic syndrome when genetic testing for commonly reported disease-causing genes (i.e., podocytopathy genes) proves to be negative. These conditions are referred to as phenocopies of monogenic podocytopathies (19–25). A phenocopy is defined as “a phenotypic trait or disease that resembles the trait expressed by a particular genotype, but in an individual who is not a carrier of that genotype” (26). Recently, by applying whole-exome sequencing, Warejko et al. (27) reported the diagnosis of a phenocopy in 5% of patients with steroid-resistant nephrotic syndrome. We hypothesized that establishing standardized criteria to define podocytopathy versus phenocopy genes, and adding re-evaluation of patients and their family (i.e., “reverse phenotyping”) after genetic testing, could correctly segregate the identified genetic variants to previously unrecognized clinical symptoms and increase the sensitivity of the genetic analysis.
Materials and Methods
Patients
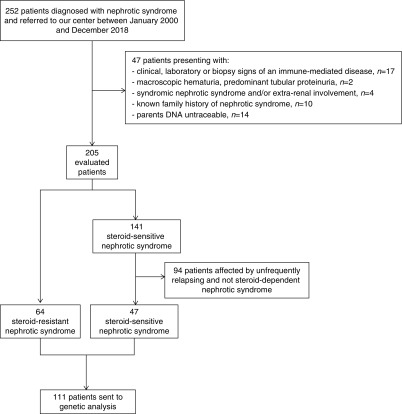
All consecutive patients diagnosed with nephrotic syndrome and referred to the Nephrology Unit of the Meyer Children’s University Hospital of Florence from January 2000 to December 2018 were assessed for inclusion in this retrospective study (Figure 1, Supplemental Table 1). Participants were followed from the day of referral until 31 December 2018, with no loss to follow-up. Demographic, clinical, and laboratory data were retrospectively collected from direct interview of patients, families, and referring nephrologists and from medical records. Inclusion criteria were onset of symptoms before 30 years of age and a clinical diagnosis of nephrotic syndrome (e.g., nephrotic range proteinuria, hypoalbuminemia, edema) or nephrotic range proteinuria with kidney histology of FSGS, minimal change disease, or diffuse mesangial sclerosis. Exclusion criteria were (1) evidence of clinical, laboratory, or kidney biopsy signs of an immune-mediated disease; (2) macroscopic hematuria or predominantly tubular proteinuria (low-molecular-weight proteins >50% according to urine protein electrophoresis) (28); (3) syndromic nephrotic syndrome and/or presence of extrarenal signs or symptoms (e.g., sensorineural hearing loss, ocular abnormalities); (4) known family history of nephrotic syndrome; and (5) untraceable parents’ DNA (Figure 1). The remaining patients were then subclassified in steroid-resistant or steroid-sensitive nephrotic syndrome. The majority of patients with steroid-resistant nephrotic syndrome were diagnosed in another nephrology center and subsequently referred to our hospital, therefore detailed laboratory information at disease onset was not always available. Those affected by congenital nephrotic syndrome, or with a histologic diagnosis of diffuse mesangial sclerosis, or advanced kidney failure at diagnosis were considered comparable with patients who were steroid-resistant, although not treated with steroids. Among patients who were steroid-sensitive, only frequently relapsing or steroid-dependent patients were selected for genetic testing and inclusion in the study. First-degree relatives were either included before the study or asked to participate after the identification of potentially causative gene variants in the patient. The local Ethics Committee of the Meyer Children’s University Hospital of Florence approved the study. The study was conducted according to the Declaration of Helsinki. A clinical geneticist counseled all patients and their families regarding the whole-exome sequencing procedure, and all participants or their legal guardians gave written informed consent.
Figure 1.
Flowchart for the selection of 111 patients included in the study.
Sequencing and Bioinformatic Analysis
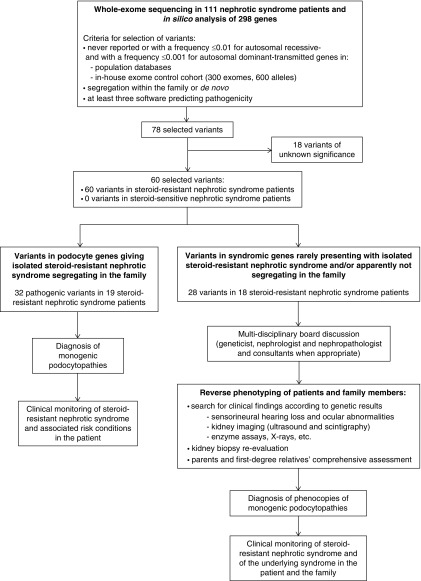
We evaluated whole-exome sequencing data for causative mutations in 298 genes reported as even rare cause of steroid-resistant nephrotic syndrome or CKD (Supplemental Table 2). Genomic DNA was isolated from blood lymphocytes and sequenced with the platform NextSeq500 (Illumina Inc., San Diego, CA); reads were aligned with the human reference hg19 genome using Burrows–Wheeler Aligner (29), mapped and analyzed with the Integrative Genome Viewer software (2013 Broad Institute) (30). Downstream alignment processing (i.e., alignment sorting, indexing, deduplication, and base quality score recalibration) was performed with the Genome Analysis Toolkit Unified Genotyper Module (GATK) (31), SAMtools (32), and Picard Tools (http://picard.sourceforge.net/). The GATK Unified Genotyper was used to obtain a set of single nucleotide variants and indel calls. We performed a hard filtering of called variants on the basis of four criteria: (1) the variant confidence (QUAL), that is, the Phred-scaled posterior probability of the variant to be homozygous reference; (2) the quality by depth (QD), that is, the QUAL divided by the unfiltered depth (DP); (3) the DP, that is, the number of reads spanning the variant site; and (4) the strand bias (FS), measured as the Phred-scaled P value, using Fisher exact test to detect FS (the variation being seen on only either the forward or the reverse strand) in the reads. We excluded the variant calls for which at least one of these filters was true: QD<5.0, DP<5, FS>60.0, QUAL<30.0. Variants were annotated using Annovar tool (33) to obtain information such as variants frequency in different populations. Variants were bioinformatically prioritized by using the diagnostic workflow shown in Figure 2 and detailed in Supplemental Appendix 1, in accordance with the American College of Medical Genetics and Genomics guidelines (34). Probable effect on the function of the encoded protein was assessed using Polyphen-2, SIFT, Mutation Taster, Mutation Assessor, FATHMM, and FATHMM MKL. Mutations were designated as pathogenic on the basis of the criteria reported in Figure 2 and Supplemental Table 3. We used manual inspection for the P.[Arg229Gln] variant in NPHS2 when segregating with variants localized in exon 7 and 8 (35) and for APOL1 G1 and G2 risk alleles (36). To estimate copy number variations we used a normalized read count approach as detailed in Supplemental Appendix 1. Synonymous variants and intronic variants that were not located within splice site regions were excluded. Variants were confirmed in the patients’ and families’ DNA by Sanger sequencing.
Figure 2.
Reverse phenotyping after genetic testing doubled diagnostic rate in steroid-resistant nephrotic syndrome patients.
Definition of Podocytopathy and Phenocopy Genes
We systematically defined podocytopathy versus phenocopy genes on the basis of Online Mendelian Inheritance in Man (OMIM; https://www.omim.org). Accordingly, genes identified in OMIM as causing “nephrotic syndrome” or “FSGS” were considered “podocytopathy genes.” In contrast, genes identified in OMIM to cause a syndromic disorder, with nephrotic syndrome being only one among many other clinical signs or even not mentioned at all, were considered “phenocopy genes.” Recently published literature indicating a causative pathogenic role for a gene in nephrotic syndrome was also considered to stratify cases as podocytopathy versus phenocopy when not yet reported in OMIM (37). Patients carrying pathogenic variants in phenocopy genes and their families underwent reverse phenotyping.
Reverse Phenotyping
To perform reverse phenotyping, we established a multidisciplinary team composed of at least one geneticist, one nephrologist and one nephropathologist, discussing clinical cases of patients undergoing whole-exome sequencing during regularly scheduled meetings. Patients with mutations in phenocopy genes and their families were thoroughly revised, also with the help of external consultants, according to the clinical suspicion raised by the genetic test. As a general procedure, the geneticist illustrated the suspicious variant(s) and the related existing literature. The nephrologist proposed a list of clinical exams and specialist consultations to detect overlooked signs or symptoms of the syndromic genetic disorder suggested by DNA analysis. In addition, the nephropathologist proposed if and how to re-evaluate the kidney biopsy looking for specific signs of the suspected genetic nephropathy, eventually including further staining.
Statistical Analyses
Statistical analysis was performed using SPSS software (SPSS, Inc., Evanston, IL). All continuous data are expressed as median and interquartile range.
Kaplan–Meier estimates were used to generate an overall survival curve for the development of ESKD and differences among groups were assessed by log-rank test. A P value <0.05 was considered statistically significant. Length of follow-up was compared between groups using one-way ANOVA.
Results
Whole-Exome Sequencing Identifies Heterogeneous Genotypes in Patients with Idiopathic Steroid-Resistant Nephrotic Syndrome
A total of 252 consecutive patients were assessed for inclusion. A total of 111 patients affected by isolated nephrotic syndrome were selected for final analysis (Figure 1, Supplemental Table 1, Table 1). The baseline characteristics of 64 patients with steroid-resistant nephrotic syndrome and 47 patients with steroid-sensitive nephrotic syndrome are shown in Table 1 and detailed in Supplemental Table 1. After the diagnostic workflow shown in Figure 2, we identified 32 pathogenic variants in 19 out of 64 (30%) patients with steroid-resistant nephrotic syndrome in “podocytopathy genes,” identified in OMIM as causing “nephrotic syndrome” or “FSGS.” In addition, 28 variants in genes that did not apparently fit the clinical phenotype were identified in 18 out of 64 (28%) patients. These genes included those already indicated as “phenocopy genes” by Warejko et al. (27), as well as all other genes identified in OMIM with the name of the underlying disease, but where nephrotic syndrome is rare and/or represents only one of the many possible symptoms of a syndromic disorder. Moreover, ten out of 64 (16%) patients with steroid-resistant nephrotic syndrome carried variants of unknown significance (Supplemental Table 4). Because this group potentially included unrecognized genetic patients, it was considered separately being referred as “undefined.” Only 17 out of 64 (26%) patients with steroid-resistant nephrotic syndrome were negative upon genetic analysis. Finally, none of the patients with steroid-sensitive nephrotic syndrome showed pathogenic variants at genetic analysis. Genetic profiles of patients showing pathogenic variants are detailed in Supplemental Table 5.
Table 1.
Baseline characteristics of the study population
| Characteristics | Steroid-Sensitive Nephrotic Syndrome (n=47) | Steroid-Resistant Nephrotic Syndrome (n=64) | Total (n=111) |
|---|---|---|---|
| Age at onset, yr | 4 [3–8] | 5 [3–9] | 5 [3–8] |
| Sex (male) | 30/47 (64%) | 32/64 (50%) | 62/111 (56%) |
| Ethnicity (European) | 34/47 (72%) | 60/64 (94%) | 94/111 (85%) |
| Clinical onset | |||
| Nephrotic-range proteinuria | 45/45 (100%) | 53/53 (100%) | 98/98 (100%) |
| Serum albumin, g/dl | 2.0 [1.6–2.4] | 2.2 [1.9–2.7] | 2.1 [1.8–2.6] |
| Hypoalbuminemia, ≤3g/dl | 41/45 (91%) | 42/48 (87%) | 83/93 (89%) |
| Dyslipidemia | 28/30 (93%) | 36/44 (81%) | 64/74 (86%) |
| Edema | 31/36 (86%) | 29/54 (54%) | 60/90 (67%) |
| Histopathological findings | |||
| FSGS | 1/5 (20%) | 43/59 (73%) | 44/64 (69%) |
| Minimal change disease | 4/5 (80%) | 14/59 (24%) | 18/64 (28%) |
| Diffuse mesangial sclerosis | 0/5 (0%) | 2/59 (3%) | 2/64 (3%) |
| Length of follow-up, yr | 7 [5–11] | 7 [4–11] | 7 [5–11] |
Continuous variables are presented as median [interquartile range] and categorical variables are presented as n (%). Dyslipidemia was defined as increased cholesterol and/or triglycerides levels according to age-adjusted reference values of each laboratory.
Reverse Phenotyping
Reverse phenotyping was performed after obtaining the genetic testing results, which was on average 5±1 years after disease onset. Clinical reassessment of the 18 patients with unexpected genetic findings and their families led to the identification of minor or overlooked signs referable to the genetic diseases suggested by whole-exome sequencing, as summarized in Table 2. Those signs and symptoms were either present at the time of referral or appeared later on, during follow-up. Reverse phenotyping allowed correct identification of six patients with Alport syndrome, three patients with Dent disease, two patients with Papillo-renal syndrome, one patient with cystinosis, one patient with Fabry disease, and five patients with other rare monogenic diseases (Table 2) who had been misdiagnosed and treated for isolated steroid-resistant nephrotic syndrome. Clinical reassessment of all 19 patients with pathogenic variants in podocytopathy genes was unremarkable instead, as expected (Supplemental Tables 3 and 5, Table 3).
Table 2.
Summary of the results of whole-exome sequencing and reverse phenotyping in phenocopy patients
| Patient | Sex | Ethnicity | Pre Whole-Exome Sequencing Diagnosis/Biopsy Results | Gene | Variant | Reverse Phenotyping Diagnosis | Post Whole-Exome Sequencing Diagnosis | Phenotype OMIM Number (#) |
|---|---|---|---|---|---|---|---|---|
| Case 1 | F | Senegalese | SRNS/FSGS | COL4A4 APOL1 | Hom Comp het | - SNHL not present at onset and diagnosed by rechecking WES results | Alport syndrome | 203780 |
| - Father with mild proteinuria and microscopic hematuria | ||||||||
| - Mother with microscopic hematuria | ||||||||
| Confirmed Alport syndrome | ||||||||
| Case 2 | F | European | SRNS/FSGS | COL4A4 | Comp het | - Parents with microscopic hematuria | Alport syndrome | 203780 |
| Confirmed Alport syndrome | ||||||||
| Case 3 | F | European | SRNS/MCD | COL4A3 | Comp het | - Parents with microscopic hematuria | Alport syndrome | 203780 |
| Confirmed Alport syndrome | ||||||||
| Case 4 | F | European | SRNS/FSGS | COL4A5 | Het | - Mother affected by SNHL | Alport syndrome | 301050 |
| Confirmed Alport syndrome | ||||||||
| Case 5 | M | European | SRNS/MCD | COL4A5 | Hem | - SNHL not present at onset and diagnosed by rechecking WES results | Alport syndrome | 301050 |
| - Mother with microscopic hematuria | ||||||||
| Confirmed Alport syndrome | ||||||||
| Case 6 | F | European | SRNS/FSGS | COL4A5 | Het | - Mother developed nephrotic syndrome during pregnancy (regressed after delivery) | Alport syndrome | 301050 |
| Confirmed Alport syndrome | ||||||||
| Case 7 | M | European | SRNS/FSGS | LAMB2 | Comp het | - Sister with chronic glomerulopathy of unknown etiology | Pierson syndrome | 609049 |
| Confirmed Pierson syndrome | ||||||||
| Case 8 | F | European | SRNS/FSGS | GLA | Het | - Podocyte vacuolization and microscopic lamellar bodies on EM | Fabry disease | 301500 |
| - Father affected by SNHL | ||||||||
| Confirmed Fabry disease | ||||||||
| Case 9 | M | European | SRNS/FSGS | FAT1 | Hom | - Cluster headache | FAT1-related glomerulotubular nephropathy | — |
| - Tubular atrophy | ||||||||
| Confirmed FAT1-related glomerulotubular nephropathy | ||||||||
| Case 10 | F | European | SRNS/- | FAT4 | Comp het | - Left kidney hypodysplasia at kidney scintigraphy (normal US scanning) | Van Maldergem syndrome 2 | 615546 |
| Confirmed kidney involvement in Van Maldergem syndrome 2 | ||||||||
| Case 11 | F | European | SRNS/FSGS | PAX2 | Het | - No ocular abnormalities | Papillo-renal syndrome/FSGS | 120330 |
| Confirmed FSGS in Papillo-renal syndrome | ||||||||
| Case 12 | F | Latino | SRNS/- | PAX2 | Het | - No ocular abnormalities | Papillo-renal syndrome/FSGS | 120330 |
| Confirmed FSGS in Papillo-renal syndrome | ||||||||
| Case 13 | M | European | SRNS/FSGS | CLCN5 | Hem | - Growth failure | Dent disease | 300009 |
| Confirmed Dent disease | ||||||||
| Case 14 | M | European | SRNS/MCD | CLCN5 | Hem | - Late-onset hypercalciuria | Dent disease | 300009 |
| Confirmed Dent disease | ||||||||
| Case 15 | M | European | SRNS/FSGS | CLCN5 | Hem | - Microlithiasis | Dent disease | 300009 |
| Confirmed Dent disease | ||||||||
| Case 16 | M | European | SRNS/MCD | CTNS | Comp het | - Corneal crystals at slit-lamp examination | Cystinosis | 219800 |
| - Multinucleated podocytes on EM on kidney biopsy | ||||||||
| - Intraleukocytes cystin levels 2 nmol/mg protein (normal <0.3) | ||||||||
| Confirmed cystinosis | ||||||||
| Case 17 | M | Tunisian | SRNS/FSGS | LMX1B | Het | - Lack of ossification nuclei of the radium (bilateral) | Nail-patella syndrome | 161200 |
| - Severe growth failure | ||||||||
| - Absence of thumbs lunulae | ||||||||
| - Father with microscopic hematuria and mild proteinuria | ||||||||
| Confirmed Nail-patella syndrome | ||||||||
| Case 18 | M | European | SRNS/MCD | KANK1 | Comp het | - Multidrug resistant epilepsy | Cerebral palsy | 612900 |
| Confirmed cerebral palsy |
All postwhole-exome sequencing diagnoses are defined according to OMIM nomenclature (https://www.omim.org) with the exception of FAT1-related glomerulotubular nephropathy (37). OMIM, Online Mendelian Inheritance in Man; F, female; SRNS, steroid-resistant nephrotic syndrome; Hom, homozygous; Comp het, compound heterozygous; SNHL, sensorineural hearing loss; MCD, minimal change disease; Het, heterozygous; M, male; Hem, hemizygous; WES, whole-exome sequencing; EM, electron microscopy; -, biopsy not performed; US, ultrasound.
Table 3.
Summary of the results of whole-exome sequencing in podocytopathy patients
| Patient | Sex | Ethnicity | Pre Whole-Exome Sequencing Diagnosis/Biopsy Results | Gene | Variant | Post Whole-Exome Sequencing Diagnosis | Phenotype OMIM Number (#) |
|---|---|---|---|---|---|---|---|
| Case 19 | M | European | SRNS/FSGS | NPHS2 | Comp het | Nephrotic syndrome, type 2 | 600995 |
| Case 20 | M | European | SRNS/FSGS | NPHS2 | Comp het | Nephrotic syndrome, type 2 | 600995 |
| Case 21 | F | European | SRNS/FSGS | NPHS2 | Comp het | Nephrotic syndrome, type 2 | 600995 |
| Case 22 | M | European | SRNS/MCD | NPHS2 | Comp het | Nephrotic syndrome, type 2 | 600995 |
| Case 23 | M | European | SRNS/FSGS | NPHS2 | Comp het | Nephrotic syndrome, type 2 | 600995 |
| Case 24 | M | European | SRNS/FSGS | NPHS2 | Hom | Nephrotic syndrome, type 2 | 600995 |
| Case 25 | F | European | SRNS/- | NPHS2 | Hom | Nephrotic syndrome, type 2 | 600995 |
| Case 26 | M | European | SRNS/FSGS | NPHS2 | Comp het | Nephrotic syndrome, type 2 | 600995 |
| Case 27 | F | European | SRNS/FSGS | NPHS2 | Comp het | Nephrotic syndrome, type 2 | 600995 |
| Case 28 | M | European | SRNS/MCD | NPHS2 | Comp het | Nephrotic syndrome, type 2 | 600995 |
| Case 29 | F | European | SRNS/- | NPHS1 | Comp het | Nephrotic syndrome, type 1 | 256300 |
| Case 30 | M | European | SRNS/FSGS | ANLN | Het | FSGS 8 | 616032 |
| Case 31 | M | European | SRNS/FSGS | PLCE1 | Comp het | Nephrotic syndrome, type 3 | 610725 |
| Case 32 | F | European | SRNS/DMS | PLCE1 | Comp het | Nephrotic syndrome, type 3 | 610725 |
| Case 33 | F | European | SRNS/FSGS | ACTN4 | Het | Glomerulosclerosis, focal segmental, 1 | 603278 |
| Case 34 | F | European | SRNS/FSGS | ACTN4 | Het | Glomerulosclerosis, focal segmental, 1 | 603278 |
| Case 35 | F | European | SRNS/FSGS | WT1 | Het | Nephrotic syndrome, type 4 | 256370 |
| Case 36 | F | European | SRNS/- | WT1 | Het | Nephrotic syndrome, type 4 | 256370 |
| Case 37 | M | European | SRNS/FSGS | WT1 | Het | Nephrotic syndrome, type 4 | 256370 |
All postwhole-exome sequencing diagnoses are defined according to OMIM nomenclature (https://www.omim.org). OMIM, Online Mendelian Inheritance in Man; M, male; SRNS, steroid-resistant nephrotic syndrome; Comp het, compound heterozygous; F, female; MCD, minimal change disease; Hom, homozygous; -, biopsy not performed; Het, heterozygous; DMS, diffuse mesangial sclerosis.
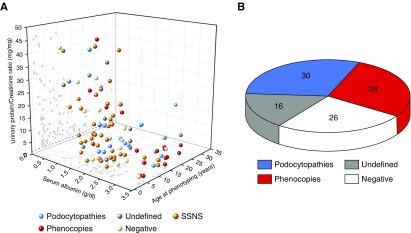
Plotting age, proteinuria, and blood albumin levels at the time of referral to our hospital resulted in a large overlap that made single patients indistinguishable (Figure 3A). Overall, 84% of patients with steroid-resistant nephrotic syndrome had been diagnosed and treated in at least another (pediatric) nephrology unit distributed all over Europe before referral to our hospital. All 18 out of 64 phenocopy patients showed specific extrarenal involvement in the patient (cases 1, 5, and 8–18, Table 2) or in first-degree relatives (cases 1–8 and 17, Table 2), thus confirming the diagnosis upon reverse phenotyping (Figure 3B). FSGS was the most common histologic pattern in all groups (Supplemental Table 1), making kidney biopsy unremarkable in distinguishing patients belonging to different genetic groups.
Figure 3.
Clinical and genetic findings of the patients included in the study. (A) Age, urinary protein-to-creatinine ratio, and serum albumin levels at referral for all of the patients enrolled in the study. Each point represents a single patient. (B) Conclusive diagnosis in patients with steroid-resistant nephrotic syndrome: podocytopathies (10 NPHS2, 1 NPHS1, 1 ANLN, 2 PLCE1, 2 ACTN4, 3 WT1) and phenocopies (2 COL4A4, 1 COL4A3, 3 COL4A5, 1 LAMB2, 1 GLA, 1 FAT1, 1 FAT4, 2 PAX2, 3 CLCN5, 1 CTNS, 1 LMX1B, 1 KANK1). SSNS, steroid-sensitive nephrotic syndrome.
Genetic Patients Are Multidrug Resistant and do Not Relapse after Transplantation
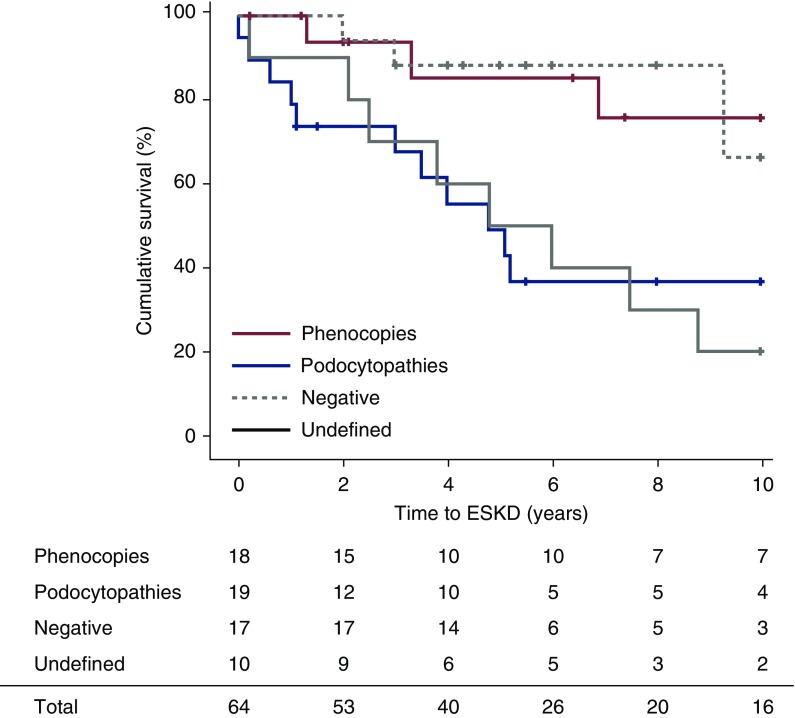
We next analyzed how treatment response and long-term kidney outcome related to the genetic diagnosis in patients with steroid-resistant nephrotic syndrome. Strikingly, complete remission occurred exclusively in negative patients (eight out of 17, 47%; Table 4) and had been induced by calcineurin inhibitors in five out of ten negative patients treated (50%; Table 4). By contrast, 13 out of 14 (93%) phenocopies and podocytopathies were resistant to calcineurin inhibitor treatment, and only one patient showed partial response (Table 4). At Kaplan–Meier analysis, patients affected by monogenic podocytopathies had a 10-year kidney survival rate of 38% (Figure 4). Undefined patients also showed a poor outcome, with a 10-year kidney survival rate of only 20%. By contrast, negative patients and phenocopy patients showed a 10-year survival rate of 66% and 75%, respectively (Figure 4). Finally, one major problem of patients with steroid-resistant nephrotic syndrome is relapse of the disease after kidney transplantation (38). In our cohort, none of the patients with a clear genetic diagnosis developed disease recurrence after transplantation (zero out of 11 patients, zero out of 15 grafts). By contrast, approximately 40% of negative and undefined patients developed disease recurrence (four out of ten patients, five out of 11 grafts). Together, phenocopy patients, as well as podocytopathies, do not benefit from immunosuppressive treatment.
Table 4.
Summary of the clinical and pathologic characteristics of the patients with steroid-resistant nephrotic syndrome included in the study, divided by genetic group
| Characteristics | Podocytopathies (n=19) | Phenocopies (n=18) | Negative (n=17) | Undefined (n=10) |
|---|---|---|---|---|
| Age at onset, yr | 4 [1–7] | 5 [3–10] | 5 [3–8] | 5 [3–13] |
| Sex (male) | 10/19 (53%) | 9/18 (50%) | 10/17 (59%) | 3/10 (30%) |
| Ethnicity (European) | 19/19 (100%) | 15/18 (83%) | 17/17 (100%) | 9/10 (90%) |
| Histopathological findings (FSGS) | 13/16 (81%) | 11/16 (69%) | 11/17 (65%) | 8/10 (80%) |
| Remission | ||||
| Complete | 0/19 (0%) | 0/18 (0%) | 8/17 (47%) | 0/0 (0%) |
| Partial | 1/19 (5%) | 11/18 (61%) | 5/17 (29%) | 0/0 (0%) |
| None | 18/19 (95%) | 7/18 (39%) | 4/17 (24%) | 10/10 (0%) |
| Response to CNIs | 1/8 (13%) | 0/6 (0%) | 5/10 (50%) | 0/6 (0%) |
| Response to RASi | 1/15 (7%) | 10/18 (56%) | 7/15 (47%) | 0/8 (0%) |
| ESKD at 10 yr | 11/15 (73%) | 3/10 (30%) | 3/6 (50%) | 8/10 (80%) |
| Post-transplant recurrence | 0/11 (0%) | 0/0 (0%) | 1/3 (33%) | 3/7 (43%) |
| Length of follow-up, yr | 8 [5–12] | 7 [2–10] | 6 [5–10] | 8 [4–12] |
Continuous variables are presented as median [interquartile range] and categorical variables are presented as n (%). CNIs, calcineurin inhibitors; RASi, inhibitors of the renin-angiotensin-aldosterone system.
Figure 4.
Kidney survival of patients with steroid-resistant nephrotic syndrome according to the genetic diagnosis. Kaplan–Meier kidney survival analysis over a period of 10 years of follow-up. Patients with steroid-resistant nephrotic syndrome are stratified according to whole-exome sequencing results. Length of follow-up was similar between groups (ANOVA P=0.74). (+) Censored observations. Survival rates were compared between groups by log-rank test (significant values): podocytopathies versus phenocopies (log-rank 5.47; P=0.02), podocytopathies versus negative (log-rank 5.77; P=0.02), undefined versus phenocopies (log-rank 6.15; P=0.01), and undefined versus negative (log-rank 6.12; P=0.01).
Discussion
We hypothesized that among patients with steroid-resistant nephrotic syndrome, particularly those who are multidrug resistant, phenocopies of monogenic podocytopathies may be more frequent than previously thought and that whole-exome sequencing for an extended panel of nephropathy-related genes plus reverse phenotyping may help identifying them. In this study, we show that (1) by combining a whole-exome sequencing–based diagnostic workflow for an extended panel of nephropathy-related genes with reverse phenotyping of the patients and their families, we double the current diagnostic rate for genetic diagnosis underlying steroid-resistant nephrotic syndrome from 30% to 60%; (2) phenocopies comprise a significant subgroup within patients with steroid-resistant nephrotic syndrome, involving subtle phenotypes of syndromic genetic disorders; and (3) phenocopies are usually multidrug resistant. Recent studies have applied genetic screening using whole-exome sequencing and bioinformatic filtering of gene panels to patients with different CKDs (39–41) including nephrotic syndrome (11,27). By using a filtering strategy on the basis of 140 genes, Bullich et al. (40), increased the rate of diagnosis in previously undiagnosed patients but could rescue wrong diagnoses in only 2% of patients with inherited glomerular disorders. By adding the further step of reverse phenotyping, we increased the rate of rescued diagnoses to 28% thanks to the identification of other genetic disorders appearing as phenocopies of monogenic podocytopathies.
To optimize the identification of phenocopies, we systematically considered genes identified in OMIM as causing “nephrotic syndrome” or “FSGS” as “podocytopathy genes” and genes identified as causing a syndromic disorder as “phenocopy genes” (see Materials and Methods). On the basis of this, we performed reverse phenotyping every time that phenocopy genes were identified by whole-exome sequencing. In none of the patients we describe, the true diagnosis had been suspected from clinical phenotype and biopsy results. Indeed, when specific signs of the underlying disease are absent at onset, only whole-exome sequencing may suggest the correct diagnosis. However, even with whole-exome sequencing the diagnosis can be missed, especially when (1) filtering criteria for in silico analysis of variants are set very tight to increase specificity at the expense of sensitivity; (2) interpretation of whole-exome sequencing results does not include analysis of segregation within the family, which is essential to validate the presence and significance of pathogenic variants occurring in unexpected genes; and (3) genetic testing is performed without the opportunity of clinical re-evaluation of the patients and the family members for subtle and previously unrecognized clinical signs, especially when unexpected findings occur. This is particularly relevant for patients referred for genetic testing from peripheral centers, when only the DNA is shipped and there is no independent clinical re-evaluation.
The results of this study demonstrate the power of “reverse phenotyping” in the interpretation of whole-exome sequencing data directly in the clinical setting with the awareness that, for certain genetic diseases, syndromic features may be nonpenetrant in the patient as well as in the family or may become evident only with age. In these patients, extrarenal involvement and syndromic features may be subtle and need to be specifically assessed to reach conclusive diagnoses. For these reasons, we suggest that for phenocopy genes, reverse phenotyping of the patient and relatives should always be part of the diagnostic workup with geneticist, nephrologist, and nephropathologist working closely to avoid misdiagnosis. These results suggest that phenocopies of monogenic podocytopathies may be underestimated. Indeed, cases like the ones we describe in this study are difficult to recognize in registry-based studies, suggesting the limitations of this kind of analyses in moving forward our daily clinical practice. Identification of phenocopies permits to avoid unnecessary multiple immunosuppressive treatments and inform treatment, in particular when a specific therapy is available (e.g., cystinosis). In addition, a correct identification of phenocopies would make it possible to accurately enroll them in clinical trials in larger cohorts and better establish their prognosis on a case-by-case basis, determined by the underlying disorder. Moreover, our study suggests that phenocopies show a better outcome compared with genetic podocytopathies. This may reflect the fact that although in monogenic podocytopathies the podocyte is primarily affected, in phenocopies podocyte damage may be, at least partially, secondary to nephron loss, justifying the longer time needed for kidney function loss.
The high number of analyzed genes led to the identification of many patients carrying variants of unknown clinical significance. These “undefined” patients had a significantly worse outcome compared with negative ones, likely because they included unrecognized genetic causes, as well as disorders possibly related to permeability factor(s) (42,43). Consistently, these patients also showed a high risk of nephrotic syndrome recurrence after kidney transplant (43,44). By contrast, patients with genetic forms showed a negligible risk of post-transplantation disease recurrence, in agreement with previous studies (11,12). Finally, in case of a misdiagnosed genetic phenocopy, kidney donation may be detrimental not only for the recipient, but also for the affected parent.
The generalizability of these results is limited by the low number of patients with a long follow-up that could be analyzed. For this reason, independent replications and meta-analysis efforts are needed and can be addressed in future research.
Taken together, our results suggest that in steroid-resistant nephrotic syndrome a workflow including analysis of an extended panel of genes, multidisciplinary meetings and reverse phenotyping increased the diagnostic rate from 30% to 60%. Relevant clinical implications deriving from this approach include: (1) the identification of the underlying cause of the disease; (2) the definition of appropriate diagnostic work-up, with referral and screening for previously unrecognized extrarenal features; (3) the possibility to guide therapy and identify potential tailored treatments; (4) a more accurate prognostic evaluation; and (5) genetic counseling, cascade screening of at-risk relatives, and donor selection for transplantation.
Disclosures
The authors have nothing to disclose.
Funding
This article is supported by the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (grant agreement 648274), the Tuscan Region to the Meyer Children’s Hospital “Programma attuativo regionale Fas-FSC,” and from the Tuscan Association for Childhood Renal Diseases.
Supplementary Material
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
See related editorial, ““It’s In Your Genes”: Exome Sequencing Enables Precision Diagnostics in Proteinuric Kidney Diseases,” on pages 10–12.
Supplemental Material
This article contains the following supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.06060519/-/DCSupplemental.
Supplemental Table 1. Clinical profile of all of the patients included in the study.
Supplemental Table 2. Panel of 298 genes analyzed with whole-exome sequencing.
Supplemental Table 3. List of pathogenic variants on the basis of the American College of Medical Genetics and Genomics score.
Supplemental Table 4. List of variants of unknown significance on the basis of the ACMG score.
Supplemental Table 5. Genetic profile of patients showing pathogenic variants.
Supplemental Appendix 1. DNA extraction, DNA library preparation, assembly, variant calling, variant interpretation strategy, statistical analysis, web resources, case reports, and references.
References
- 1.Kidney Disease Improving global outcomes (KDIGO) Glomerulonephritis Work Group : KDIGO clinical practice guidelines for glomerulonephritis. Kidney Int Suppl (2011) 2: 139–274, 2012 [Google Scholar]
- 2.Noone DG, Iijima K, Parekh R: Idiopathic nephrotic syndrome in children. Lancet 392: 61–74, 2018 [DOI] [PubMed] [Google Scholar]
- 3.Gipson DS, Chin H, Presler TP, Jennette C, Ferris ME, Massengill S, Gibson K, Thomas DB: Differential risk of remission and ESRD in childhood FSGS. Pediatr Nephrol 21: 344–349, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Samuel SM, Flynn R, Zappitelli M, Dart A, Parekh R, Pinsk M, Mammen C, Wade A, Scott SD; Canadian Childhood Nephrotic Syndrome Project Team* : Factors influencing practice variation in the management of nephrotic syndrome: A qualitative study of pediatric nephrology care providers. CMAJ Open 5: E424–E430, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Büscher AK, Beck BB, Melk A, Hoefele J, Kranz B, Bamborschke D, Baig S, Lange-Sperandio B, Jungraithmayr T, Weber LT, Kemper MJ, Tönshoff B, Hoyer PF, Konrad M, Weber S; German Pediatric Nephrology Association (GPN) : Rapid response to cyclosporin a and favorable renal outcome in nongenetic versus genetic steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 11: 245–253, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trautmann A, Schnaidt S, Lipska-Ziętkiewicz BS, Bodria M, Ozaltin F, Emma F, Anarat A, Melk A, Azocar M, Oh J, Saeed B, Gheisari A, Caliskan S, Gellermann J, Higuita LMS, Jankauskiene A, Drozdz D, Mir S, Balat A, Szczepanska M, Paripovic D, Zurowska A, Bogdanovic R, Yilmaz A, Ranchin B, Baskin E, Erdogan O, Remuzzi G, Firszt-Adamczyk A, Kuzma-Mroczkowska E, Litwin M, Murer L, Tkaczyk M, Jardim H, Wasilewska A, Printza N, Fidan K, Simkova E, Borzecka H, Staude H, Hees K, Schaefer F; PodoNet Consortium : Long-term outcome of steroid-resistant nephrotic syndrome in children. J Am Soc Nephrol 28: 3055–3065, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romagnani P, Remuzzi G, Glassock R, Levin A, Jager KJ, Tonelli M, Massy Z, Wanner C, Anders HJ: Chronic kidney disease. Nat Rev Dis Primers 3: 17088, 2017 [DOI] [PubMed] [Google Scholar]
- 8.Giglio S, Provenzano A, Mazzinghi B, Becherucci F, Giunti L, Sansavini G, Ravaglia F, Roperto RM, Farsetti S, Benetti E, Rotondi M, Murer L, Lazzeri E, Lasagni L, Materassi M, Romagnani P: Heterogeneous genetic alterations in sporadic nephrotic syndrome associate with resistance to immunosuppression. J Am Soc Nephrol 26: 230–236, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, Engelmann S, Vega-Warner V, Fang H, Halbritter J, Somers MJ, Tan W, Shril S, Fessi I, Lifton RP, Bockenhauer D, El-Desoky S, Kari JA, Zenker M, Kemper MJ, Mueller D, Fathy HM, Soliman NA, Hildebrandt F; SRNS Study Group : A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 26: 1279–1289, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Zhang Y, Mao J, Yu Z, Yi Z, Yu L, Sun J, Wei X, Ding F, Zhang H, Xiao H, Yao Y, Tan W, Lovric S, Ding J, Hildebrandt F: Spectrum of mutations in Chinese children with steroid-resistant nephrotic syndrome. Pediatr Nephrol 32: 1181–1192, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bierzynska A, McCarthy HJ, Soderquest K, Sen ES, Colby E, Ding WY, Nabhan MM, Kerecuk L, Hegde S, Hughes D, Marks S, Feather S, Jones C, Webb NJ, Ognjanovic M, Christian M, Gilbert RD, Sinha MD, Lord GM, Simpson M, Koziell AB, Welsh GI, Saleem MA: Genomic and clinical profiling of a national nephrotic syndrome cohort advocates a precision medicine approach to disease management. Kidney Int 91: 937–947, 2017 [DOI] [PubMed] [Google Scholar]
- 12.Trautmann A, Bodria M, Ozaltin F, Gheisari A, Melk A, Azocar M, Anarat A, Caliskan S, Emma F, Gellermann J, Oh J, Baskin E, Ksiazek J, Remuzzi G, Erdogan O, Akman S, Dusek J, Davitaia T, Özkaya O, Papachristou F, Firszt-Adamczyk A, Urasinski T, Testa S, Krmar RT, Hyla-Klekot L, Pasini A, Özcakar ZB, Sallay P, Cakar N, Galanti M, Terzic J, Aoun B, Caldas Afonso A, Szymanik-Grzelak H, Lipska BS, Schnaidt S, Schaefer F; PodoNet Consortium : Spectrum of steroid-resistant and congenital nephrotic syndrome in children: The PodoNet registry cohort. Clin J Am Soc Nephrol 10: 592–600, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashraf S, Kudo H, Rao J, Kikuchi A, Widmeier E, Lawson JA, Tan W, Hermle T, Warejko JK, Shril S, Airik M, Jobst-Schwan T, Lovric S, Braun DA, Gee HY, Schapiro D, Majmundar AJ, Sadowski CE, Pabst WL, Daga A, van der Ven AT, Schmidt JM, Low BC, Gupta AB, Tripathi BK, Wong J, Campbell K, Metcalfe K, Schanze D, Niihori T, Kaito H, Nozu K, Tsukaguchi H, Tanaka R, Hamahira K, Kobayashi Y, Takizawa T, Funayama R, Nakayama K, Aoki Y, Kumagai N, Iijima K, Fehrenbach H, Kari JA, El Desoky S, Jalalah S, Bogdanovic R, Stajić N, Zappel H, Rakhmetova A, Wassmer SR, Jungraithmayr T, Strehlau J, Kumar AS, Bagga A, Soliman NA, Mane SM, Kaufman L, Lowy DR, Jairajpuri MA, Lifton RP, Pei Y, Zenker M, Kure S, Hildebrandt F: Mutations in six nephrosis genes delineate a pathogenic pathway amenable to treatment. Nat Commun 9: 1960, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gast C, Pengelly RJ, Lyon M, Bunyan DJ, Seaby EG, Graham N, Venkat-Raman G, Ennis S: Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol Dial Transplant 31: 961–970, 2016 [DOI] [PubMed] [Google Scholar]
- 15.Pierides A, Voskarides K, Athanasiou Y, Ioannou K, Damianou L, Arsali M, Zavros M, Pierides M, Vargemezis V, Patsias C, Zouvani I, Elia A, Kyriacou K, Deltas C: Clinico-pathological correlations in 127 patients in 11 large pedigrees, segregating one of three heterozygous mutations in the COL4A3/ COL4A4 genes associated with familial haematuria and significant late progression to proteinuria and chronic kidney disease from focal segmental glomerulosclerosis. Nephrol Dial Transplant 24: 2721–2729, 2009 [DOI] [PubMed] [Google Scholar]
- 16.Fogo AB: The spectrum of FSGS: Does pathology matter? Nephrol Dial Transplant 25: 1034–1036, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Saida K, Kamijo Y, Matsuoka D, Noda S, Hidaka Y, Mori T, Shimojo H, Ehara T, Miura K, Takita J, Sekine T, Igarashi T, Koike K: A case of adult Dent disease in Japan with advanced chronic kidney disease. CEN Case Rep 3: 132–138, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fervenza FC: A patient with nephrotic-range proteinuria and focal global glomerulosclerosis. Clin J Am Soc Nephrol 8: 1979–1987, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malone AF, Phelan PJ, Hall G, Cetincelik U, Homstad A, Alonso AS, Jiang R, Lindsey TB, Wu G, Sparks MA, Smith SR, Webb NJ, Kalra PA, Adeyemo AA, Shaw AS, Conlon PJ, Jennette JC, Howell DN, Winn MP, Gbadegesin RA: Rare hereditary COL4A3/COL4A4 variants may be mistaken for familial focal segmental glomerulosclerosis. Kidney Int 86: 1253–1259, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vallés P, Peralta M, Carrizo L, Martin L, Principi I, Gonzalez A, Manucha W: Follow-up of steroid-resistant nephrotic syndrome: Tubular proteinuria and enzymuria. Pediatr Nephrol 15: 252–258, 2000 [DOI] [PubMed] [Google Scholar]
- 21.He G, Zhang H, Cao S, Xiao H, Yao Y: Dent’s disease complicated by nephrotic syndrome: A case report. Intractable Rare Dis Res 5: 297–300, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Anglani F, Beara-Lasic L, Mehta AJ, Vaughan LE, Herrera Hernandez L, Cogal A, Scheinman SJ, Ariceta G, Isom R, Copelovitch L, Enders FT, Del Prete D, Vezzoli G, Paglialonga F, Harris PC, Lieske JC; Investigators of the Rare Kidney Stone Consortium : Glomerular pathology in dent disease and its association with kidney function. Clin J Am Soc Nephrol 11: 2168–2176, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koulousios K, Stylianou K, Pateinakis P, Zamanakou M, Loules G, Manou E, Kyriklidou P, Katsinas C, Ouzouni A, Kyriazis J, Speletas M, Germenis AE: Fabry disease due to D313Y and novel GLA mutations. BMJ Open 7: e017098, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.du Moulin M, Koehn AF, Golsari A, Dulz S, Atiskova Y, Patten M, Münch J, Avanesov M, Ullrich K, Muschol N: The mutation p.D313Y is associated with organ manifestation in Fabry disease. Clin Genet 92: 528–533, 2017 [DOI] [PubMed] [Google Scholar]
- 25.Köping M, Shehata-Dieler W, Cebulla M, Rak K, Oder D, Müntze J, Nordbeck P, Wanner C, Hagen R, Schraven S: Cardiac and renal dysfunction is associated with progressive hearing loss in patients with Fabry disease. PLoS One 12: e0188103, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Cancer Institute. Available at : https://www.cancer.gov/publications/dictionaries/genetics-dictionary/def/phenocopy. Accessed July 15, 2019
- 27.Warejko JK, Tan W, Daga A, Schapiro D, Lawson JA, Shril S, Lovric S, Ashraf S, Rao J, Hermle T, Jobst-Schwan T, Widmeier E, Majmundar AJ, Schneider R, Gee HY, Schmidt JM, Vivante A, van der Ven AT, Ityel H, Chen J, Sadowski CE, Kohl S, Pabst WL, Nakayama M, Somers MJG, Rodig NM, Daouk G, Baum M, Stein DR, Ferguson MA, Traum AZ, Soliman NA, Kari JA, El Desoky S, Fathy H, Zenker M, Bakkaloglu SA, Müller D, Noyan A, Ozaltin F, Cadnapaphornchai MA, Hashmi S, Hopcian J, Kopp JB, Benador N, Bockenhauer D, Bogdanovic R, Stajić N, Chernin G, Ettenger R, Fehrenbach H, Kemper M, Munarriz RL, Podracka L, Büscher R, Serdaroglu E, Tasic V, Mane S, Lifton RP, Braun DA, Hildebrandt F: Whole exome sequencing of patients with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 13: 53–62, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Wang F, Xiao H, Yao Y: The ratio of urinary α1-microglobulin to microalbumin can be used as a diagnostic criterion for tubuloproteinuria. Intractable Rare Dis Res 7: 46–50, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorvaldsdóttir H, Robinson JT, Mesirov JP: Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA: The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup : The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, Li M, Hakonarson H: ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee : Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tory K, Menyhárd DK, Woerner S, Nevo F, Gribouval O, Kerti A, Stráner P, Arrondel C, Huynh Cong E, Tulassay T, Mollet G, Perczel A, Antignac C: Mutation-dependent recessive inheritance of NPHS2-associated steroid-resistant nephrotic syndrome. Nat Genet 46: 299–304, 2014 [DOI] [PubMed] [Google Scholar]
- 36.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gee HY, Sadowski CE, Aggarwal PK, Porath JD, Yakulov TA, Schueler M, Lovric S, Ashraf S, Braun DA, Halbritter J, Fang H, Airik R, Vega-Warner V, Cho KJ, Chan TA, Morris LG, ffrench-Constant C, Allen N, McNeill H, Büscher R, Kyrieleis H, Wallot M, Gaspert A, Kistler T, Milford DV, Saleem MA, Keng WT, Alexander SI, Valentini RP, Licht C, Teh JC, Bogdanovic R, Koziell A, Bierzynska A, Soliman NA, Otto EA, Lifton RP, Holzman LB, Sibinga NE, Walz G, Tufro A, Hildebrandt F: FAT1 mutations cause a glomerulotubular nephropathy. Nat Commun 7: 10822, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fine RN: Recurrence of nephrotic syndrome/focal segmental glomerulosclerosis following renal transplantation in children. Pediatr Nephrol 22: 496–502, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Groopman EE, Marasa M, Cameron-Christie S, Petrovski S, Aggarwal VS, Milo-Rasouly H, Li Y, Zhang J, Nestor J, Krithivasan P, Lam WY, Mitrotti A, Piva S, Kil BH, Chatterjee D, Reingold R, Bradbury D, DiVecchia M, Snyder H, Mu X, Mehl K, Balderes O, Fasel DA, Weng C, Radhakrishnan J, Canetta P, Appel GB, Bomback AS, Ahn W, Uy NS, Alam S, Cohen DJ, Crew RJ, Dube GK, Rao MK, Kamalakaran S, Copeland B, Ren Z, Bridgers J, Malone CD, Mebane CM, Dagaonkar N, Fellström BC, Haefliger C, Mohan S, Sanna-Cherchi S, Kiryluk K, Fleckner J, March R, Platt A, Goldstein DB, Gharavi AG: Diagnostic utility of exome sequencing for kidney disease. N Engl J Med 380: 142–151, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bullich G, Domingo-Gallego A, Vargas I, Ruiz P, Lorente-Grandoso L, Furlano M, Fraga G, Madrid Á, Ariceta G, Borregán M, Piñero-Fernández JA, Rodríguez-Peña L, Ballesta-Martínez MJ, Llano-Rivas I, Meñica MA, Ballarín J, Torrents D, Torra R, Ars E: A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int 94: 363–371, 2018 [DOI] [PubMed] [Google Scholar]
- 41.Yao T, Udwan K, John R, Rana A, Haghighi A, Xu L, Hack S, Reich HN, Hladunewich MA, Cattran DC, Paterson AD, Pei Y, Barua M: Integration of genetic testing and pathology for the diagnosis of adults with FSGS [published online ahead of print January 15, 2019]. Clin J Am Soc Nephrol 10.2215/CJN.08750718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Agati VD, Kaskel FJ, Falk RJ: Focal segmental glomerulosclerosis. N Engl J Med 365: 2398–2411, 2011 [DOI] [PubMed] [Google Scholar]
- 43.Gallon L, Leventhal J, Skaro A, Kanwar Y, Alvarado A: Resolution of recurrent focal segmental glomerulosclerosis after retransplantation. N Engl J Med 366: 1648–1649, 2012 [DOI] [PubMed] [Google Scholar]
- 44.Francis A, Trnka P, McTaggart SJ: Long-term outcome of kidney transplantation in recipients with focal segmental glomerulosclerosis. Clin J Am Soc Nephrol 11: 2041–2046, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.