Supplemental Digital Content is available in the text.
Keywords: adult, genetics, humans, immune system, infection
Objective:
HDL (high-density lipoprotein) cholesterol (HDL-C) and LDL (low-density lipoprotein) cholesterol (LDL-C) are inversely associated with infectious hospitalizations. Whether these represent causal relationships is unknown.
Approach and Results:
Adults of 40 to 69 years of age were recruited from across the United Kingdom between 2006 and 2010 and followed until March 31, 2016, as part of the UK Biobank. We determined HDL-C, LDL-C, and triglyceride polygenic scores for UK Biobank participants of British white ancestry (n=407 558). We examined the association of lipid levels and polygenic scores with infectious hospitalizations, antibiotic usage, and 28-day sepsis survival using Cox proportional hazards or logistic regression models. Measured levels of HDL-C and LDL-C were inversely associated with risk of infectious hospitalizations, while triglycerides displayed a positive association. A 1-mmol/L increase in genetically determined levels of HDL-C associated with a hazard ratio for infectious disease of 0.84 ([95% CI, 0.75–0.95]; P=0.004). Mendelian randomization using genetic variants associated with HDL-C as an instrumental variable was consistent with a causal relationship between elevated HDL-C and reduced risk of infectious hospitalizations (inverse weighted variance method, P=0.001). Furthermore, of 3222 participants who experienced an index episode of sepsis, there was a significant inverse association between continuous HDL-C polygenic score and 28-day mortality (adjusted hazard ratio, 0.37 [95% CI, 0.14–0.96] per 1 mmol/L increase; P=0.04). LDL-C and triglyceride polygenic scores were not significantly associated with hospitalization for infection, antibiotic use, or sepsis mortality.
Conclusions:
Our results provide causal inference for an inverse relationship between HDL-C, but not LDL-C or triglycerides, and risk of an infectious hospitalization.
Highlights.
Elevated levels of high-density lipoprotein cholesterol and low-density lipoprotein cholesterol were observationally associated with reduced risk of infectious disease hospitalizations.
Alternatively, elevated levels of triglycerides were observationally associated with increased risk of infectious disease hospitalizations.
For genetically determined lipid levels, only high-density lipoprotein cholesterol was significantly associated with reduced risk of hospitalizations for infectious disease, lower odds of outpatient antibiotic usage, and reduced risk of mortality from sepsis.
Mendelian randomization analysis suggested that the observational relationship between higher levels of high-density lipoprotein cholesterol and reduced risk of hospitalization for infectious disease could be causal in nature.
Despite the epidemiological evidence for a strong association between low levels of HDL (high-density lipoprotein) cholesterol (HDL-C) and atherosclerotic cardiovascular disease, genetic studies and clinical trials have failed to demonstrate that this relationship is causal.1,2 However, there is interest in the role of HDL particles as an important component of the innate immune system and protecting against infection.3–8
See accompanying editorial on page 5
HDL is able to sequester pathogen-associated lipids, such as lipopolysaccharide and lipoteichoic acid, and mitigate host inflammation during sepsis.9–11 These findings are consistent with preclinical studies that show that HDL-based interventions reduce inflammation and improve survival in rodent models of experimental sepsis.6,12,13 Furthermore, low levels of HDL-C and LDL (low-density lipoprotein) cholesterol (LDL-C) are associated with poor clinical outcomes and increased risk of infectious disease, respectively, in sepsis cohorts and large epidemiological studies.3,5,14–16 However, these epidemiological associations are subject to confounding and reverse causation, and it remains unclear whether they represent causal relationships. Indeed, recent studies have suggested that the associations between low levels of LDL-C and sepsis mortality are likely due to confounding rather than representing a causal relationship.17,18 It is unknown whether the same is true for HDL-C.
The objective of this study was to assess the influence of common variants associated with HDL-C, LDL-C, and triglyceride levels on infectious disease risk among the general population. Using genetic variants as risk factors can circumvent many issues of confounding and reverse causation and has the potential to provide causal inference regarding the effect of genetically determined levels of HDL-C on susceptibility to infectious disease. We hypothesized that individuals with elevated levels of genetically determined HDL-C, but not LDL-C or triglycerides, would have reduced risk of hospitalization for infectious disease hospitalizations and improved sepsis survival.
Methods
Data Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.
UK Biobank Cohort
This study was approved by the UK Biobank (application identification: 42857) and by the Clinical Research Ethics Board of the University of British Columbia (H18-02181).
Participants from the UK Biobank prospective population study who consented to genotyping analysis and were of British white ancestry were included in this analysis.19,20 A dataset of genotyped and imputed variants was available from the UK Biobank. Genotypes of interest were extracted from bgen files using the rbgen R package. Samples that had a mismatch between reported sex and genetic sex or missing >3 of the 223 single-nucleotide variants (SNVs) of interest were excluded from analyses.
Biochemical measurements, physical exam measurements, and medical histories were assessed at the time of study enrollment (Methods in the online-only Data Supplement; Table I in the online-only Data Supplement).
Index infectious disease hospitalization events were grouped into categories based on primary or secondary International Classification of Diseases, version 10, diagnosis codes (Table II in the online-only Data Supplement). Time-to-first index events were assessed between the date of participant enrollment (2006–2010) to March 31, 2016. Baseline medications considered to be oral or injectable antibiotics are detailed in Table II in the online-only Data Supplement.
Polygenic Scores for HDL-C, LDL-C, and Triglyceride
Weighted HDL-C, LDL-C, and triglyceride polygenic scores were calculated using the standardized effect sizes (in units of SD) for the association between lipid traits and 223 SNVs described by Klarin et al.21 The effect sizes published by Klarin et al21 represent the association between each SNV with measured levels of HDL-C, LDL-C, and triglycerides after adjustment for age, age squared, and study-specific covariates, including principal components to account for population structure before transformation using the inverse normal distribution.22 The exome genotyping array data from the Global Lipids Genetics Consortium are displayed in Table IV in the online-only Data Supplement.21–23 Weighted polygenic scores were calculated for UK Biobank participants using the formula Σ [β1 × SNV1]+…+[β223 × SNV223], where SNV is the number of lipid trait increasing alleles (0, 1, or 2) and β is the positive effect size (β-coefficient) for the lipid trait of interest for each of the 223 SNVs. HDL-C, LDL-C, and triglyceride polygenic scores are positively associated with measured levels of HDL-C, LDL-C, and triglycerides.
Statistical Analyses
Analyses were performed using R, version 3.5.1. Measured lipid levels and polygenic scores were grouped into quintiles or above and below the median for visualization and description of cohort characteristics at recruitment. χ2 tests were used for contingency analyses. Normally distributed data were analyzed with 1-way ANOVA tests, whereas non-normal data were analyzed with Kruskal-Wallis tests. Linear regression was used to assess the correlation between lipid levels and polygenic scores and was adjusted for age, sex, genotyping array/batch, and the first 4 principal components of ancestry.
Odds ratios (ORs) and hazard ratios (HRs) were calculated using logistic regressions and Cox regression models adjusted for age, sex, genotyping array/batch, and the first 4 principal components of ancestry, respectively. Adjustments were made for age and sex to account for variabilities in lipid levels and risk of infection that are well-established demographic covariates, genotyping array and batch to control for batch effects of genotyping, and the first 4 principal components of genetic ancestry to mitigate the risk of confounding from population substructure.24 Adjusting for these covariates is common practice in population genetics.21,25,26 Hazards ratios for time-to-first infectious event used the time post–study enrollment as a time scale and were prematurely censored if a loss to follow-up or death event occurred. In individuals who experienced an index sepsis episode, Cox regression models were used to assess the risk of mortality using time post-hospitalization as a time scale with censoring occurring at 28 days post-hospitalization or at the date in which a loss to follow-up event occurred. A log-rank test was used to assess whether there were significant differences in the risk of mortality between polygenic scores above versus below the median.
A 2-step Mendelian randomization (MR) analysis was performed using the inverse weighted variance and MR Egger methods for HDL-C.27 A set of 57 SNVs of the 223 SNVs included in the lipid trait polygenic scores were chosen based on their genome-wide significant association with HDL-C levels in the original Global Lipid Genetics Consortium study.23 The effect sizes and SEs for the exposure of HDL-C are reported in Table V in the online-only Data Supplement,21–23 while individual SNV effect sizes and SEs for HRs of outcomes were determined in the UK Biobank. To further address the pleiotropic nature of SNVs associated with lipid traits, we performed a multivariable weighted linear regression Mendelian randomization analysis using the 57 SNVs and their associations with both infectious hospitalization and HDL-C, LDL-C, and triglycerides.28,29 Data were analyzed using the MendelianRandomization v0.3.0 package in R, version 3.5.1.30
Statistical significance was claimed when 2-sided P values were ≤0.05.
Results
Baseline Characteristics of Study Participants
The baseline characteristics of UK Biobank participants stratified by quintiles of measured levels of HDL-C (n=357 202), LDL-C (n=389 564), and triglycerides (n=389 971) are displayed in Tables VI through VIII in the online-only Data Supplement, respectively. There was a significantly greater prevalence of male sex, diabetes mellitus, and other cardiovascular comorbidities among individuals in lower quintiles of measured levels of HDL-C.
Association of HDL-C, LDL-C, and Triglyceride Levels With Risk of Infectious Disease
We first sought to determine the associations between levels of HDL-C, LDL-C and triglycerides with risk of infectious disease–related hospitalizations.3 Using measured levels of HDL-C as a continuous variable, the HRs for a 1-mmol/L increase in HDL-C for infections classified by site were 0.57 for sepsis ([95% CI, 0.50–0.64]; P<0.0001), 0.62 for pneumonia ([95% CI, 0.57–0.67]; P<0.0001), 0.60 for gastroenteritis ([95% CI, 0.56–0.65]; P<0.0001), 0.54 for urinary tract infections ([95% CI, 0.50–0.58]; P<0.0001), 0.53 for skin infections ([95% CI, 0.49–0.59]; P<0.0001), and 0.62 for any infectious disease ([95% CI, 0.59–0.64]; P<0.0001; Figure I in the online-only Data Supplement). There was a dose-dependent relationship between ascending quintiles of measured HDL-C and reduced risk of any infectious disease–related hospitalizations (Figure 1). Specifically, a 1-mmol/L increase in HDL-C levels was associated with significantly reduced risk of infections that were diagnosed as bacterial (HR, 0.58 [95% CI, 0.54–0.62]; P<0.0001), viral (HR, 0.57 [95% CI, 0.50–0.65]; P<0.0001), or fungal in pathogenesis (HR, 0.75 [95% CI, 0.65–0.86]; P<0.0001; Figure II in the online-only Data Supplement).
Figure 1.

Elevated levels of HDL (high-density lipoprotein) cholesterol (HDL-C) and LDL (low-density lipoprotein) cholesterol (LDL-C) associate with reduced risk of hospitalization for infectious disease, whereas elevated levels of triglycerides associate with increased risk of hospitalization for infectious disease. Cox proportional hazards models and the associated hazard ratios (HRs) with 95% CIs are displayed for all infectious disease hospitalizations vs quintiles of measured levels of (A) HDL-C, (B) LDL-C (direct measurement), and (C) triglycerides. All models were adjusted for age, sex, genotyping array, and the first 4 principal components of genetic ancestry.
Similar to HDL-C levels, LDL-C levels displayed significant inverse associations with risk of infection when classified by either site of infection (Figure I in the online-only Data Supplement) or suspected pathogenesis of infection (Figure II in the online-only Data Supplement). The HR for any infectious hospitalization was 0.83 for a 1-mmol/L increase in LDL-C levels ([95% CI, 0.82–0.84]; P<0.0001; Figure 1). In contrast, triglyceride levels displayed a significant positive association with risk of infection when classified by site of infection (Figure I in the online-only Data Supplement) and infections suspected to be of bacterial pathogenesis (Figure II in the online-only Data Supplement). The adjusted HR for any infectious hospitalization was 1.07 for a 1-mmol/L increase in triglyceride levels ([95% CI, 1.06–1.09]; P<0.0001; Figure 1).
To test for a potential pleiotropic effect of triglycerides influencing the association between measured levels of HDL-C and risk of any infectious hospitalization, we performed analyses in which the HRs for measured levels of HDL-C were adjusted for triglycerides and the HRs for measured levels of triglycerides were adjusted for HDL-C. The positive association between a 1-mmol/L increase in triglycerides and risk of any infectious hospitalization was lost when triglyceride levels were adjusted for HDL-C levels (HR, 1.01 [95% CI, 1.00–1.02]; P=0.18; Figure III in the online-only Data Supplement). Alternatively, the inverse association between a 1-mmol/L increase in HDL-C levels and risk of any infectious hospitalization was unchanged by adjustment for triglyceride levels (HR, 0.62 [95% CI, 0.60–0.65]; P<0.0001; Figure III in the online-only Data Supplement).
The risk of any infectious hospitalization was significantly greater for men than women (HR, 1.15 [95% CI, 1.12–1.18]; P<0.0001; Figure IV in the online-only Data Supplement). This difference became nonsignificant when adjusted for measured levels of HDL-C (HR, 0.99 [95% CI, 0.97–1.02]; P=0.71; Figure IV in the online-only Data Supplement).
Polygenic Scores Are Associated With Measured Lipid Levels
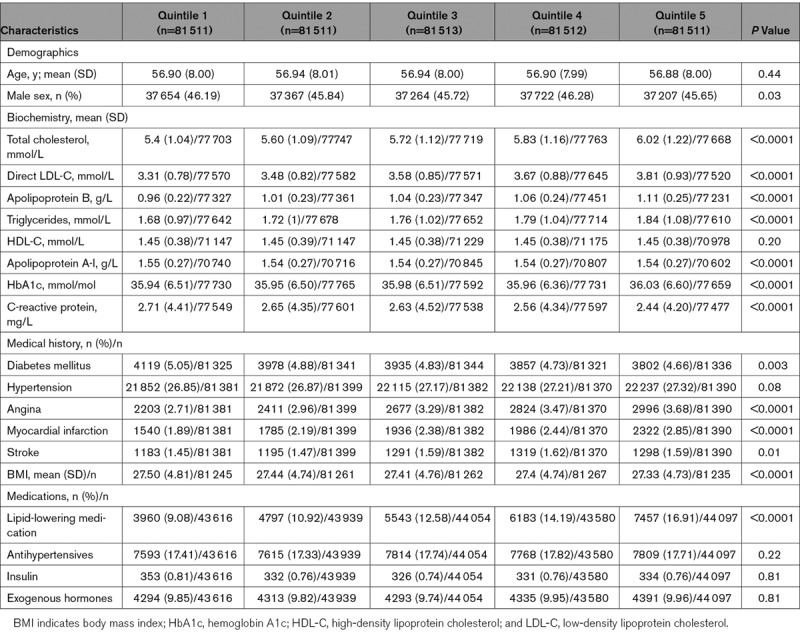
We next constructed polygenic scores for HDL-C, LDL-C, and triglycerides. The baseline characteristics of 407 558 individuals stratified by quintiles of HDL-C, LDL-C, and triglyceride polygenic scores are shown in Tables 1 and 2 and Table IX in the online-only Data Supplement, respectively. The mean HDL-C levels for quintiles of HDL-C polygenic score were 1.32, 1.40, 1.45, 1.50, and 1.60 mmol/L. The mean direct LDL-C levels for quintiles of LDL-C polygenic score were 3.31, 3.48, 3.58, 3.67, and 3.81 mmol/L. The mean triglyceride levels for quintiles of triglyceride polygenic score were 1.46, 1.62, 1.74, 1.86, and 2.12 mmol/L. There were nominal, but significant, differences in the prevalence of hypertension, diabetes mellitus, angina, and myocardial infarction, with greater prevalence among lower quintiles of HDL-C polygenic scores (Table 1). As expected, there was significantly greater prevalence of angina and myocardial infarction among higher quintiles of LDL-C and triglyceride polygenic scores (Table 2; Table IX in the online-only Data Supplement, respectively).
Table 1.
Baseline Characteristics of UK Biobank Participants With British White Genetic Ancestry Stratified by Quintiles of HDL-C Polygenic Score
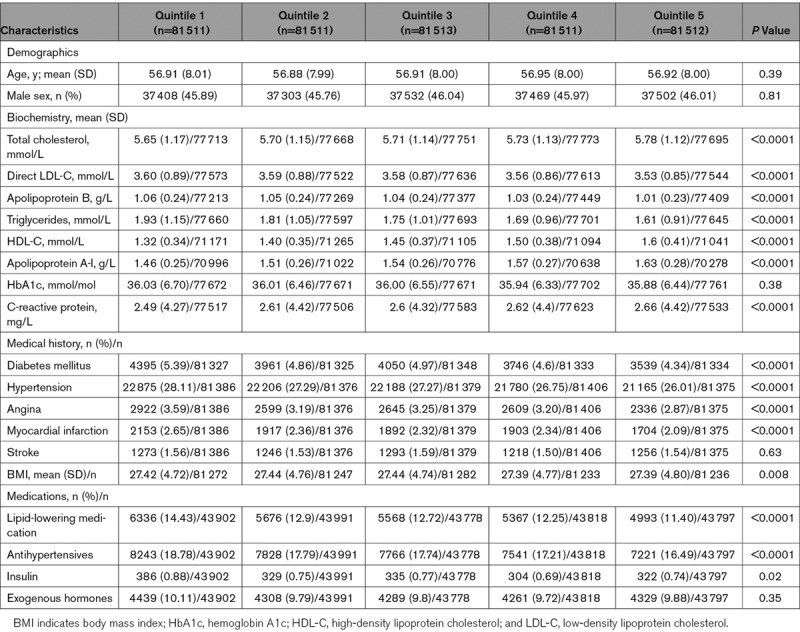
Table 2.
Baseline Characteristics of UK Biobank Participants With British White Genetic Ancestry Stratified by Quintiles of LDL-C Polygenic Score
HDL-C, LDL-C, and triglyceride polygenic scores were normally distributed. Measured levels of HDL-C, LDL-C, and triglycerides were significantly correlated with HDL-C (R2=0.24; P<0.0001), LDL-C (R2=0.05; P<0.0001), and triglyceride (R2=0.10; P<0.0001) polygenic scores when adjusted for age, sex, genotyping array/batch, and the first 4 principal components of ancestry, respectively (Figure V in the online-only Data Supplement).
HDL-C Polygenic Score Is Associated With Reduced Risk of Infectious Disease–Related Hospitalizations, Whereas LDL-C and Triglyceride Polygenic Scores Are Not
Increasing HDL-C polygenic score trended toward reduced risk of hospitalizations for infections classified by specific site (Figure VI in the online-only Data Supplement). In addition, HDL-C polygenic score was inversely associated with risk of hospitalization for bacterial and viral infections but not fungal infections (Figure VII in the online-only Data Supplement). There was a general dose-dependent relationship between ascending quintiles of HDL-C polygenic score and reduced risk of any infectious disease–related hospitalizations (Figure 2). The HR for any infectious hospitalization was 0.94 for a 1-SD-unit increase in HDL-C polygenic score ([95% CI, 0.91–0.98]; P=0.004). This translated to a HR for any infectious hospitalization of 0.84 for a 1-mmol/L increase in genetically predicted levels of HDL-C ([95% CI, 0.75–0.95]; P=0.004; Figure 2). These results remained significant even when additionally adjusted for physician diagnosis of diabetes mellitus at baseline (HR, 0.96 per 1 SD unit increase in HDL-C polygenic score [95% CI, 0.92–0.99]; P=0.03) or when individuals with known diabetes mellitus at enrollment were excluded (n=387 864; HR, 0.95 per 1 SD unit increase in HDL-C polygenic score [95% CI, 0.91–0.99]; P=0.03).
Figure 2.
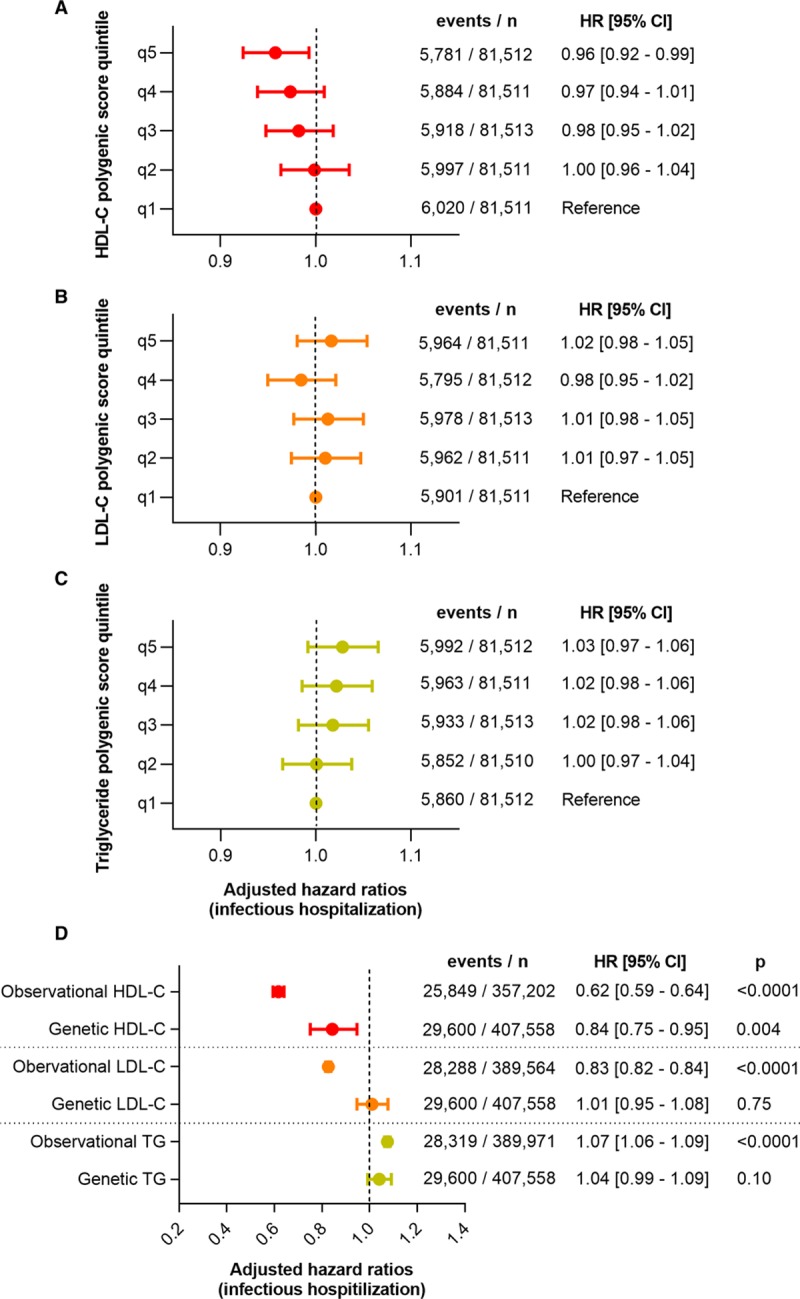
Elevated HDL (high-density lipoprotein) cholesterol (HDL-C) polygenic score, but not LDL (low-density lipoprotein) cholesterol (LDL-C) or triglyceride (TG) polygenic score, associates with reduced risk of hospitalization for infectious disease. Cox proportional hazards models and the associated hazard ratios (HRs) with 95% CIs are displayed for any infectious disease hospitalizations vs quintiles of (A) HDL-C, (B) LDL-C, and (C) triglyceride polygenic scores (SD units). D, The HRs and 95% CIs for any infectious disease hospitalization are depicted for a 1-mmol/L increase in observational or genetically predicted lipid levels. All models were adjusted for age, sex, genotyping array, and the first 4 principal components of genetic ancestry.
In contrast, continuous LDL-C polygenic score (HR, 1.01 [95% CI, 0.97–1.05]; P=0.75) and continuous triglyceride polygenic score (HR, 1.04 [95% CI, 0.99–1.09]; P=0.10) did not display significant associations with risk of hospitalization for any infectious disease (Figure 2; Figure VI in the online-only Data Supplement). The HRs for any infectious hospitalization were 1.01 and 1.04 for a 1-mmol/L increase in genetically predicted levels of LDL-C ([95% CI, 0.95–1.08]; P=0.75) and triglycerides ([95% CI, 0.99–1.09]; P=0.10), respectively. There were no statistically significant relationships between LDL-C polygenic score or triglyceride polygenic score and microbial pathogenesis of infectious hospitalizations (Figure VII in the online-only Data Supplement).
Given that elevated HDL-C polygenic score was associated with reduced risk of infections requiring hospitalization, we next investigated whether elevated HDL-C polygenic scores would also be associated with fewer episodes of minor-to-moderate infections, as reflected by outpatient antibiotic usage. The OR for any antibiotic usage was 0.82 and 0.86 for a 1-mmol/L increase in measured levels of HDL-C ([95% CI, 0.75–0.89]; P<0.0001) and LDL-C ([95% CI, 0.83–0.89]; P<0.0001), respectively (Figure VIII in the online-only Data Supplement). In contrast, the OR for any antibiotic usage was 1.03 for a 1-mmol/L increase in measured levels of triglycerides ([95% CI, 1.00–1.06]; P=0.04; Figure VIII in the online-only Data Supplement). Like the observations between antibiotic usage and measured levels of HDL-C, the OR for any antibiotic usage was 0.90 for a 1-SD-unit increase in HDL-C polygenic score ([95% CI, 0.83–0.99]; P=0.03; Figure IX in the online-only Data Supplement). This translated to an OR for any antibiotic usage of 0.74 for a 1-mmol/L increase in genetically predicted levels of HDL-C (95% CI, 0.57–0.97). There were no significant associations between antibiotic usage and LDL-C polygenic score (OR, 0.92 [95% CI, 0.83–1.01]; P=0.07) or triglyceride polygenic score (OR, 0.96 [95% CI, 0.87–1.07]; P=0.47; Figure IX in the online-only Data Supplement).
Causal Inference for the Effect of HDL-C Polygenic Score on Infectious Disease–Related Hospitalizations
The availability of genetic variants associated with HDL-C levels provides the opportunity to determine whether the epidemiologically observed association between HDL-C and hospitalizations for overall infectious diseases represents a causal relationship. To investigate this, we performed a 2-step Mendelian randomization analysis.31,32 The first step used the effect sizes of 57 of the 223 SNVs used in the polygenic score that were found to be significantly associated with HDL-C in the genome-wide association study performed by the Global Lipid Genomics Consortium used to construct the HDL-C polygenic score.23 In the second step, we used the estimates of SNV effects on HDL-C levels as instrumental variables to investigate whether genetically elevated HDL-C had a significant causal estimate for decreased risk of overall infectious disease–related hospitalization. We observed a 0.061 decrease in the logHR for infectious disease–related hospitalization per 1 SD increase in genetically determined levels of HDL-C using the inverse weighted variance method (95% CI, −0.105 to −0.017; P=0.006). These results were consistent with the MR Egger method, which demonstrated a 0.082 decrease in the logHR for infectious disease–related hospitalizations per a 1-SD increase in genetically determined levels of HDL-C ([95% CI, −0.146 to −0.018]; P=0.01; Figure 3). Furthermore, the estimate of the intercept for the MR Egger method was 0.002 ([95% CI, −0.002 to 0.005]; P=0.39) with a nonsignificant test statistic for heterogeneity of 54.13 (55 df; P=0.51), suggesting that the results were not influenced by unbalanced horizontal pleiotropy. These results were confirmed with a multivariable Mendelian randomization analysis that included HDL-C, LDL-C, and triglycerides as risk factors for infectious hospitalization. Only HDL-C displayed a significant association with reduced risk infectious hospitalization (logHR—HDL-C: −0.060 [95% CI, −0.113 to −0.006], P=0.028; triglycerides: 0.006 [95% CI, −0.067 to 0.080], P=0.86; LDL-C: −0.012 [95% CI, −0.110 to 0.086], P=0.81). The heterogeneity test statistic for this model was nonsignificant (54.80 on 54 df; P=0.44).
Figure 3.
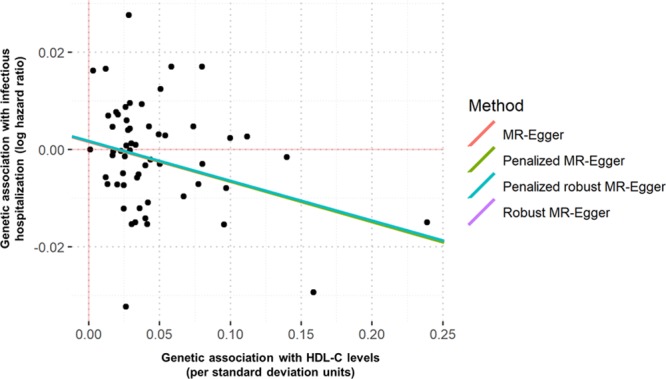
Mendelian randomization (MR) analysis of genetically determined HDL (high-density lipoprotein) cholesterol (HDL-C) levels and risk of infectious hospitalization. Genetic associations with outcomes (log hazard ratios) are displayed against genetic associations with SD units of HDL-C. Lines represent causal estimates from the different methods.
Elevated HDL-C Polygenic Score Associates With Reduced Sepsis Mortality, Whereas LDL-C and Triglyceride Polygenic Scores Do Not
When an infection triggers a dysregulated inflammatory response, it can lead to sepsis—a condition associated with a high mortality rate. Previous work has identified genetic variants in HDL-related genes that influence survival in sepsis.4 We examined the effect of HDL-C, LDL-C, and triglyceride polygenic scores on survival in 3222 participants in the UK Biobank who were hospitalized for sepsis. We observed a significant increase in 28-day survival among participants with an HDL-C polygenic score greater than the median relative to participants with an HDL-C polygenic score below the median (log-rank test: P=0.007; Figure 4A). When HDL-C polygenic score was assessed as a continuous variable, the HR for 28-day sepsis mortality was 0.71 for a 1-SD-unit increase in HDL-C polygenic score ([95% CI, 0.52–0.99]; P=0.04; Figure 4B). This translated to an HR for sepsis mortality of 0.37 per 1 mmol/L increase in genetically predicted levels of HDL-C (95% CI, 0.14–0.96). In contrast, there were no significant differences in 28-day sepsis survival between participants stratified by LDL-C or triglyceride polygenic score (Figure 4C through 4F). The HRs for sepsis mortality for LDL-C polygenic score and triglyceride polygenic score were 0.86 ([95% CI, 0.62–0.1.20]; P=0.38) and 1.19 ([95% CI, 0.83–1.70]; P=0.34) for a 1-SD-unit increase in polygenic score, respectively (Figure 4).
Figure 4.
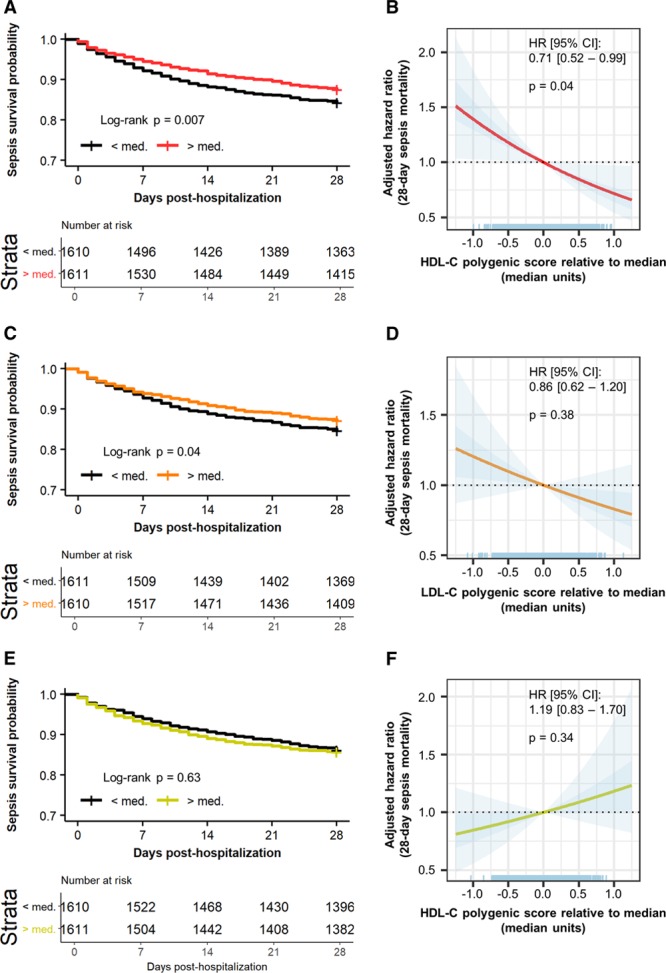
HDL (high-density lipoprotein) cholesterol (HDL-C) polygenic score, but not LDL (low-density lipoprotein) cholesterol (LDL-C) or triglyceride polygenic score, is associated with improved acute survival from sepsis. Twenty-eight–day survival curves for UK Biobank participants stratified by a polygenic score above or below the median for (A) HDL-C, (C) LDL-C, and (E) triglycerides. Cox proportional hazards models for 28-d sepsis mortality vs (B) HDL-C, (D) LDL-C, and (F) triglyceride polygenic score were adjusted for age, sex, genotyping array, and the first 4 principal components of genetic ancestry. Polygenic scores are scaled as median units. The darker blue depicts the SE, while light blue depicts the 95% CI. HR indicates hazard ratio.
Discussion
Here, we report that genetically determined levels of HDL-C have a modest but significant influence on the risk of hospitalization for infectious diseases. The strength of this study is the demonstration that higher HDL-C polygenic scores, which are associated with relatively small increases in HDL-C levels, are associated with lower risk of infectious disease–related hospitalizations, less outpatient antibiotic usage, and reduced short-term mortality from sepsis. Furthermore, Mendelian randomization suggested that the inverse associations between genetically determined levels of HDL-C and risk of hospitalization for infectious disease events could be causal in nature. In contrast, there were no statistically significant associations between genetically determined levels of LDL-C or triglycerides and risk of infectious disease. These findings provide evidence to support that HDL-C levels are causally related to risk of clinical infectious disease among the general population.
Previous work by Madsen et al3 demonstrated that low levels of HDL-C associate with increased risk of infectious disease in the large Copenhagen City Heart and the Copenhagen General Population Study epidemiological studies. We were able to confirm their finding of an increased risk of infections in individuals with lower levels of HDL-C and LDL-C in an even larger population cohort. Our findings also extend that previous work by inferring the causal nature of this relationship. Unlike Madsen et al, we did not observe an increased risk of infectious disease hospitalizations at higher extremes of HDL-C levels (a U-shaped relationship between HDL-C and infection risk). This discrepancy could be due to the overall healthier status of individuals participating in UK Biobank33 and our study’s focus on individuals of British white ancestry. Importantly, neither our study nor the Copenhagen City Heart and the Copenhagen General Population Study had HDL-C levels measured at the time of infectious disease events, and, therefore, these levels may differ considerably from the pre-illness measurements available.34
HDL particles have several properties relevant to modulating the immune system and infectious disease. Of all lipoproteins, HDLs have the greatest affinity to bind pathogen-associated lipids (eg, lipopolysaccharide, lipoteichoic acid) that mediate the excessive immune activation in sepsis.9–11 HDL also has immunomodulatory,35,36 antithrombotic,37 and antioxidant effects that could provide important clues to why genetically determined levels of HDL-C, but not LDL-C,17,18 confer a causal protective effect against infectious disease. In particular, HDL-related apolipoproteins, such as apolipoprotein A1 and apolipoprotein M, interact with lipid rafts on cellular membranes that are enriched in immune cell receptors such as Toll-like receptors on macrophages,38–40 T-cell receptors,41 and B-cell receptors42 to modulate immune responses.41 In this large epidemiological study, we were only able to use HDL-C as a surrogate for HDL particle function and were not able to assess how differences in the lipid and protein components of HDL particles influence infectious risk. Further studies will be needed to differentiate which of the proposed mechanisms are most important to conferring the anti-infectious properties of HDL.
Our observations support the concept of HDL-C being a potential therapeutic target in clinical infectious diseases such as sepsis. We have previously shown that the CETP (cholesteryl ester transfer protein) gain-of-function variant, rs1800777, is associated with significant reductions in HDL-C levels during sepsis, increased risk of acute kidney injury, and increased mortality in clinical sepsis cohorts.4,44 This study’s findings demonstrate another genetic mechanism by which lower levels of genetically determined HDL-C associate with increased sepsis mortality. These findings raise the intriguing possibility that drugs that raise HDL-C levels, such as inhibitors of CETP, may be beneficial to treat or reduce the risk of infectious diseases.39 However, it is important to point out that in some studies of HDL-C–raising therapies, including ILLUMINATE (Investigation of Lipid Level Management to Understand its Impact in Atherosclerotic Events)45 of torcetrapib and HSP2-THRIVE (Heart Protection Study 2–Treatment of HDL to Reduce the Incidence of Vascular Events)46 of niacin and laropiprant, an increased risk of infectious disease events was observed in those allocated to HDL-C raising therapies. However, this risk was not observed in trials of other CETP inhibitors,1,47,48 and it will be critical for future clinical trials to specifically assess this hypothesis in an infectious disease setting and to be carefully monitored for safety and efficacy.
The genetic approach that we used in this study reduces the risk of confounding and reverse causation that can occur in observational studies.3,15 However, an important assumption of Mendelian randomization is that genetic instruments are not associated with confounding factor that could influence the outcome of interest.49 In this regard, it is important to note that we observed a small, but significant, increase in the prevalence of reported diabetes mellitus among participants in the lower quintiles of HDL-C polygenic score. This was unexpected, as a previous Mendelian randomization study found that genetically lowered HDL-C is not a causal risk for the development of diabetes mellitus.50 We hypothesize that the higher prevalence of type 2 diabetes mellitus diagnoses in participants with genetically low HDL-C may reflect more intensive screening of these individuals because of their perceived higher cardiovascular risk, as determined by commonly used risk calculators.51–53 This hypothesis is supported by the finding that there was no significant difference in hemoglobin A1c levels among lower compared with higher quintiles of HDL-C polygenic score.
This study has some limitations that should be considered. Firstly, the potential effect of HDL-C polygenic score on the dynamic changes to HDL-C during the infection or sepsis event remains unknown. Secondly, we used diagnosis codes to identify hospitalization and mortality events of interest. This method sacrifices the depth of participant phenotyping for increased sample size and these classifications may not be reflective of the most recent guidelines for some conditions (eg, sepsis definitions).54 However, the observed ≈10% mortality suggests our approach reasonably classified patients with clinically suspected sepsis. Third, we focused our analyses on participants of British white genetic ancestry to minimize the risk of confounding from population stratification, to match the ancestry of most participants from the Global Lipid Genetics Consortium, and because this population reflects the majority of UK Biobank participants. It will be critical to assess the generalizability of these findings in other large population studies of more diverse ancestral groups and perform secondary replication of these findings in another epidemiological study.
In summary, we report that elevated genetically determined levels of HDL-C are associated with reduced risk of hospitalization for infectious disease, reduced outpatient antibiotic usage, and reduced mortality from sepsis. These results provide new insight into the potential causal role of HDL in infectious diseases and support the concept that raising HDL-C levels in individuals with certain infectious disease such as sepsis could be a viable therapeutic target.
Acknowledgments
We acknowledge the participants and individuals involved in the UK Biobank project, Compute Canada, and Alexandra Sull for assistance with data curation.
Sources of Funding
This project was supported by the Providence Health Care Research Institute, Vanier Canada Graduate Scholarship (M. Trinder), Canadian Institutes of Health Research Foundation Grant (K.R. Walley), Michael Smith Foundation for Health Research Scholar Award, and Canada Research Chair in Precision Cardiovascular Disease Prevention (L.R. Brunham).
Disclosures
J.H. Boyd and K.R. Walley have a patent Methods and Uses for Proprotein Convertase Subtilisin Kexin 9 Inhibitors (PCT/CA2013/000488 issued). The other authors report no conflicts.
Supplementary Material
Nonstandard Abbreviations and Acronyms
- CETP
- cholesteryl ester transfer protein
- HDL
- high-density lipoprotein
- HDL-C
- high-density lipoprotein cholesterol
- HR
- hazard ratio
- LDL
- low-density lipoprotein
- LDL-C
- low-density lipoprotein cholesterol
- OR
- odds ratio
- SNV
- single-nucleotide variant
For Sources of Funding and Disclosures, see page 277.
The online-only Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.119.313381.
References
- 1.Bowman L, Hopewell JC, Chen F, Wallendszus K, Stevens W, Collins R, Wiviott SD, Cannon CP, Braunwald E, Sammons E, et al. HPS3/TIMI55–REVEAL Collaborative Group. Effects of anacetrapib in patients with atherosclerotic vascular disease. N Engl J Med. 2017;377:1217–1227. doi: 10.1056/NEJMoa1706444. doi: 10.1056/NEJMoa1706444. [DOI] [PubMed] [Google Scholar]
- 2.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madsen CM, Varbo A, Tybjærg-Hansen A, Frikke-Schmidt R, Nordestgaard BG. U-shaped relationship of HDL and risk of infectious disease: two prospective population-based cohort studies. Eur Heart J. 2018;39:1181–1190. doi: 10.1093/eurheartj/ehx665. doi: 10.1093/eurheartj/ehx665. [DOI] [PubMed] [Google Scholar]
- 4.Trinder M, Genga KR, Kong HJ, Blauw LL, Lo C, Li X, Cirstea M, Wang Y, Rensen PCN, Russell JA, et al. Cholesteryl ester transfer protein influences high-density lipoprotein levels and survival in sepsis. Am J Respir Crit Care Med. 2019;199:854–862. doi: 10.1164/rccm.201806-1157OC. doi: 10.1164/rccm.201806-1157OC. [DOI] [PubMed] [Google Scholar]
- 5.Kaysen GA, Ye X, Raimann JG, Wang Y, Topping A, Usvyat LA, Stuard S, Canaud B, van der Sande FM, Kooman JP, et al. Monitoring Dialysis Outcomes (MONDO) Initiative. Lipid levels are inversely associated with infectious and all-cause mortality: international MONDO study results. J Lipid Res. 2018;59:1519–1528. doi: 10.1194/jlr.P084277. doi: 10.1194/jlr.P084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morin EE, Guo L, Schwendeman A, Li XA. HDL in sepsis - risk factor and therapeutic approach. Front Pharmacol. 2015;6:244. doi: 10.3389/fphar.2015.00244. doi: 10.3389/fphar.2015.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feingold KR, Grunfeld C. The effect of inflammation and infection on lipids and lipoproteins. In: Feingold KR, Anawalt B, Boyce A, Chrousos G, Dungan K, Grossman A, Hershman JM, Kaltsas G, et al., editors. In: Endotext. South Dartmouth, MA: MDText.com Inc; 2000. [Google Scholar]
- 9.Levels JH, Abraham PR, van Barreveld EP, Meijers JC, van Deventer SJ. Distribution and kinetics of lipoprotein-bound lipoteichoic acid. Infect Immun. 2003;71:3280–3284. doi: 10.1128/IAI.71.6.3280-3284.2003. doi: 10.1128/iai.71.6.3280-3284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levels JH, Abraham PR, van den Ende A, van Deventer SJ. Distribution and kinetics of lipoprotein-bound endotoxin. Infect Immun. 2001;69:2821–2828. doi: 10.1128/IAI.69.5.2821-2828.2001. doi: 10.1128/IAI.69.5.2821-2828.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levels JH, Marquart JA, Abraham PR, van den Ende AE, Molhuizen HO, van Deventer SJ, Meijers JC. Lipopolysaccharide is transferred from high-density to low-density lipoproteins by lipopolysaccharide-binding protein and phospholipid transfer protein. Infect Immun. 2005;73:2321–2326. doi: 10.1128/IAI.73.4.2321-2326.2005. doi: 10.1128/IAI.73.4.2321-2326.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine DM, Parker TS, Donnelly TM, Walsh A, Rubin AL. In vivo protection against endotoxin by plasma high density lipoprotein. Proc Natl Acad Sci USA. 1993;90:12040–12044. doi: 10.1073/pnas.90.24.12040. doi: 10.1073/pnas.90.24.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pirillo A, Catapano AL, Norata GD. HDL in infectious diseases and sepsis. In: von Eckardstein A, Kardassis D, editors. In: High Density Lipoproteins. Handbook of Experimental Pharmacology. Cham, Switzerland: Springer; 2015. pp. 483–508. [DOI] [PubMed] [Google Scholar]
- 14.Cirstea M, Walley KR, Russell JA, Brunham LR, Genga KR, Boyd JH. Decreased high-density lipoprotein cholesterol level is an early prognostic marker for organ dysfunction and death in patients with suspected sepsis. J Crit Care. 2017;38:289–294. doi: 10.1016/j.jcrc.2016.11.041. doi: 10.1016/j.jcrc.2016.11.041. [DOI] [PubMed] [Google Scholar]
- 15.Chien YF, Chen CY, Hsu CL, Chen KY, Yu CJ. Decreased serum level of lipoprotein cholesterol is a poor prognostic factor for patients with severe community-acquired pneumonia that required intensive care unit admission. J Crit Care. 2015;30:506–510. doi: 10.1016/j.jcrc.2015.01.001. doi: 10.1016/j.jcrc.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Lekkou A, Mouzaki A, Siagris D, Ravani I, Gogos CA. Serum lipid profile, cytokine production, and clinical outcome in patients with severe sepsis. J Crit Care. 2014;29:723–727. doi: 10.1016/j.jcrc.2014.04.018. doi: 10.1016/j.jcrc.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Walley KR, Boyd JH, Kong HJ, Russell JA. Low low-density lipoprotein levels are associated with, but do not causally contribute to, increased mortality in sepsis. Crit Care Med. 2019;47:463–466. doi: 10.1097/CCM.0000000000003551. doi: 10.1097/CCM.0000000000003551. [DOI] [PubMed] [Google Scholar]
- 18.Feng Q, Wei WQ, Chaugai S, Leon BGC, Mosley JD, Leon DAC, Jiang L, Ihegword A, Shaffer CM, Linton MF, et al. Association between low-density lipoprotein cholesterol levels and risk for sepsis among patients admitted to the hospital with infection. JAMA Netw Open. 2019;2:e187223. doi: 10.1001/jamanetworkopen.2018.7223. doi: 10.1001/jamanetworkopen.2018.7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Band G, Marchini J. BGEN: a binary file format for imputed genotype and haplotype data. bioRxiv. 2018:1–6. doi:10.1101/308296. [Google Scholar]
- 21.Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, Gagnon DR, DuVall SL, Li J, Peloso GM, et al. Global Lipids Genetics Consortium; Myocardial Infarction Genetics (MIGen) Consortium; Geisinger-Regeneron DiscovEHR Collaboration; VA Million Veteran Program. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu X, Peloso GM, Liu DJ, Wu Y, Zhang H, Zhou W, Li J, Tang CS, Dorajoo R, Li H, et al. GLGC Consortium. Exome chip meta-analysis identifies novel loci and East Asian-specific coding variants that contribute to lipid levels and coronary artery disease. Nat Genet. 2017;49:1722–1730. doi: 10.1038/ng.3978. doi: 10.1038/ng.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, Leed A, Weale ME, Spencer CCA, Aguet F, et al. CARDIoGRAMplusC4D Consortium. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet. 2017;49:1392–1397. doi: 10.1038/ng.3914. doi: 10.1038/ng.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35:1880–1906. doi: 10.1002/sim.6835. doi: 10.1002/sim.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–260. doi: 10.1093/aje/kwu283. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson KE, Siewert KM, Klarin D, Damrauer SM, Chang KM, Tsao PS, Assimes TL, Maxwell KN, Voight BF VA Million Veteran Program. Assessing a causal relationship between circulating lipids and breast cancer risk: Mendelian randomization study. bioRxiv. 2019:1–19. doi: 10.1371/journal.pmed.1003302. doi: 10.1101/794594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26:2333–2355. doi: 10.1177/0962280215597579. doi: 10.1177/0962280215597579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017;14:577–590. doi: 10.1038/nrcardio.2017.78. doi: 10.1038/nrcardio.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, Collins R, Allen NE. Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–1034. doi: 10.1093/aje/kwx246. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madsen CM, Varbo A, Nordestgaard BG. Extreme high high-density lipoprotein cholesterol is paradoxically associated with high mortality in men and women: two prospective cohort studies. Eur Heart J. 2017;38:2478–2486. doi: 10.1093/eurheartj/ehx163. doi: 10.1093/eurheartj/ehx163. [DOI] [PubMed] [Google Scholar]
- 35.De Nardo D, Labzin LI, Kono H, Seki R, Schmidt SV, Beyer M, Xu D, Zimmer S, Lahrmann C, Schildberg FA, et al. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat Immunol. 2014;15:152–160. doi: 10.1038/ni.2784. doi: 10.1038/ni.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pajkrt D, Doran JE, Koster F, Lerch PG, Arnet B, van der Poll T, ten Cate JW, van Deventer SJ. Antiinflammatory effects of reconstituted high-density lipoprotein during human endotoxemia. J Exp Med. 1996;184:1601–1608. doi: 10.1084/jem.184.5.1601. doi: 10.1084/jem.184.5.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birjmohun RS, van Leuven SI, Levels JH, van ‘t Veer C, Kuivenhoven JA, Meijers JC, Levi M, Kastelein JJ, van der Poll T, Stroes ES. High-density lipoprotein attenuates inflammation and coagulation response on endotoxin challenge in humans. Arterioscler Thromb Vasc Biol. 2007;27:1153–1158. doi: 10.1161/ATVBAHA.106.136325. doi: 10.1161/ATVBAHA.106.136325. [DOI] [PubMed] [Google Scholar]
- 38.Fessler MB, Parks JS. Intracellular lipid flux and membrane microdomains as organizing principles in inflammatory cell signaling. J Immunol. 2011;187:1529–1535. doi: 10.4049/jimmunol.1100253. doi: 10.4049/jimmunol.1100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yvan-Charvet L, Kling J, Pagler T, Li H, Hubbard B, Fisher T, Sparrow CP, Taggart AK, Tall AR. Cholesterol efflux potential and antiinflammatory properties of high-density lipoprotein after treatment with niacin or anacetrapib. Arterioscler Thromb Vasc Biol. 2010;30:1430–1438. doi: 10.1161/ATVBAHA.110.207142. doi: 10.1161/ATVBAHA.110.207142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki M, Pritchard DK, Becker L, Hoofnagle AN, Tanimura N, Bammler TK, Beyer RP, Bumgarner R, Vaisar T, de Beer MC, et al. High-density lipoprotein suppresses the type I interferon response, a family of potent antiviral immunoregulators, in macrophages challenged with lipopolysaccharide. Circulation. 2010;122:1919–1927. doi: 10.1161/CIRCULATIONAHA.110.961193. doi: 10.1161/CIRCULATIONAHA.110.961193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabouridis PS, Jury EC. Lipid rafts and T-lymphocyte function: implications for autoimmunity. FEBS Lett. 2008;582:3711–3718. doi: 10.1016/j.febslet.2008.10.006. doi: 10.1016/j.febslet.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta N, DeFranco AL. Lipid rafts and B cell signaling. Semin Cell Dev Biol. 2007;18:616–626. doi: 10.1016/j.semcdb.2007.07.009. doi: 10.1016/j.semcdb.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Vorst EPC, Theodorou K, Wu Y, Hoeksema MA, Goossens P, Bursill CA, Aliyev T, Huitema LFA, Tas SW, Wolfs IMJ, et al. High-density lipoproteins exert pro-inflammatory effects on macrophages via passive cholesterol depletion and PKC-NF-κB/STAT1-IRF1 signaling. Cell Metab. 2017;25:197–207. doi: 10.1016/j.cmet.2016.10.013. doi: 10.1016/j.cmet.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 44.Genga KR, Trinder M, Kong HJ, Li X, Leung AKK, Shimada T, Walley KR, Russell JA, Francis GA, Brunham LR, et al. CETP genetic variant rs1800777 (allele A) is associated with abnormally low HDL-C levels and increased risk of AKI during sepsis. Sci Rep. 2018;8:16764. doi: 10.1038/s41598-018-35261-2. doi: 10.1038/s41598-018-35261-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, Lopez-Sendon J, Mosca L, Tardif JC, Waters DD, et al. ILLUMINATE Investigators. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 46.Landray MJ, Haynes R, Hopewell JC, Parish S, Aung T, Tomson J, Wallendszus K, Craig M, Jiang L, Collins R, et al. HPS2-THRIVE Collaborative Group. Effects of extended-release niacin with laropiprant in high-risk patients. N Engl J Med. 2014;371:203–212. doi: 10.1056/NEJMoa1300955. doi: 10.1056/NEJMoa1300955. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, Chaitman BR, Holme IM, Kallend D, Leiter LA, et al. dal-OUTCOMES Investigators. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med. 2012;367:2089–2099. doi: 10.1056/NEJMoa1206797. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 48.Lincoff AM, Nicholls SJ, Riesmeyer JS, Barter PJ, Brewer HB, Fox KAA, Gibson CM, Granger C, Menon V, Montalescot G, et al. ACCELERATE Investigators. Evacetrapib and cardiovascular outcomes in high-risk vascular disease. N Engl J Med. 2017;376:1933–1942. doi: 10.1056/NEJMoa1609581. doi: 10.1056/NEJMoa1609581. [DOI] [PubMed] [Google Scholar]
- 49.Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318:1925–1926. doi: 10.1001/jama.2017.17219. doi: 10.1001/jama.2017.17219. [DOI] [PubMed] [Google Scholar]
- 50.Haase CL, Tybjærg-Hansen A, Nordestgaard BG, Frikke-Schmidt R. HDL cholesterol and risk of type 2 diabetes: a Mendelian randomization study. Diabetes. 2015;64:3328–3333. doi: 10.2337/db14-1603. doi: 10.2337/db14-1603. [DOI] [PubMed] [Google Scholar]
- 51.Anderson TJ, Grégoire J, Pearson GJ, Barry AR, Couture P, Dawes M, Francis GA, Genest J, Jr, Grover S, Gupta M, et al. 2016 Canadian Cardiovascular Society guidelines for the management of dyslipidemia for the prevention of cardiovascular disease in the adult. Can J Cardiol. 2016;32:1263–1282. doi: 10.1016/j.cjca.2016.07.510. doi: 10.1016/j.cjca.2016.07.510. [DOI] [PubMed] [Google Scholar]
- 52.Piepoli MF, Hoes AW, Agewall S, Albus C, Brotons C, Catapano AL, Cooney MT, Corrà U, Cosyns B, Deaton C, et al. ESC Scientific Document Group. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts)developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR). Eur Heart J. 2016;37:2315–2381. doi: 10.1093/eurheartj/ehw106. doi: 10.1093/eurheartj/ehw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Catapano AL, Graham I, De Backer G, Wiklund O, Chapman MJ, Drexel H, Hoes AW, Jennings CS, Landmesser U, Pedersen TR, et al. ESC Scientific Document Group. 2016 ESC/EAS guidelines for the management of dyslipidaemias. Eur Heart J. 2016;37:2999–3058. doi: 10.1093/eurheartj/ehw272. doi: 10.1093/eurheartj/ehw272. [DOI] [PubMed] [Google Scholar]
- 54.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]


