Abstract
Myasthenia gravis (MG) is an autoimmune neuromuscular junction disorders mediated by various autoantibodies. Although most patients with MG require chronic immunosuppressive treatment to control disease activity, appropriate surveillance biomarkers that monitor disease activity or potential toxicity of immunosuppressants are yet to be developed. Herein, we investigated quantitative distribution of peripheral blood B cell subsets and transcriptional profiles of memory B cells (CD19+ CD27+) in several subgroups of MG patients classified according to the Myasthenia Gravis Foundation of America (MGFA) Clinical Classification. This study suggests potential immunologic B-cell markers that may guide treatment decision in future clinical settings.
Keywords: Myasthenia gravis, B-lymphocytes, Biomarkers, Flow cytometry, Transcriptome
INTRODUCTION
Myasthenia gravis (MG) is an autoantibody-mediated post-synaptic neuromuscular junction disorder. Anti-acetylcholine receptor antibodies are found in about 80% of patients with MG [1], and numerous auto-antibodies have been recently discovered; muscle-specific tyrosine kinase (MuSK) [2], low-density lipoprotein receptor-related protein 4 (LRP4) [3], agrin [4], voltage-gated K+ channel Kv1.4 [5], ryanodine receptor [6] and cortactin [7].
Chronic immunosuppressive treatment (ISTX), such as tacrolimus, azathioprine, mycophenolate mofetil and prednisolone, is required in most patients at some point in their courses, in order to maintain disease stability. Nevertheless, appropriate biomarkers that reflect disease activity or toxicity of ISTX lack [8]. Herein, we immunophenotyped peripheral blood mononuclear cells (PBMC) from participants and tested if B cell subsets were altered by disease activity or ISTX. Furthermore, given that MG is an apparently antibody-specific autoimmune disease, we analyzed transcriptional profiles of memory B cells (CD19+ CD27+) in order to develop appropriate biomarkers to monitor disease status. For transcriptome study, we used Nanostring analysis which is a digital multiplexed mRNA assay that provides highly reproducible data even in small-sized samples, by detecting native RNAs directly, without reverse transcription or amplification [9].
MATERIALS AND METHODS
Study subjects
A total of 21 patients with MG and 10 healthy controls were recruited from December 2015 to August 2019 in Seoul Metropolitan Government Boramae Medical Center. Written informed consent was obtained from all participants. This study was approved by the local institutional review board (IRB no. 20151016/16-2015-147/111).
All participants provided peripheral blood samples. MGFA classification class and the titer of anti-acetylcholine receptor antibody (AchR-Ab) at the time of sample were identified [10]. For those on ISTX, the treatment regimen at the time of sample were also investigated.
Patients with MG were classified into 3 groups according to the disease activity and whether they received immunosuppressive treatment at the sample date; group 1 (complete stable remission or minimal manifestation without immunosuppressive treatment), group 2 (pharmacologic remission or minimal manifestation with immunosuppressive agents), and group 4 (drug naïve MG). Healthy participants were designated as group 3.
Fluorescence-activated cell sorting (FACS) analysis
PBMCs were isolated by centrifugation for 30min at 100g in 4°C. The whitish buffy coat between histopaque and medium was aspirated and was washed twice with 10 ml of sterile PBS. Frozen PBMC samples w thawed and were immunolabelled with following anti-human monoclonal antibodies: PerCP-conjugated anti-CD45 (Biolegend, San Diego, CA, USA), FITC-conjugated anti-CD3 (Biolegend, San Diego, CA, USA), APC-Cy7-conjugated anti-CD19 (BD Biosciences, San Jose, CA, USA), APC-conjugated anti-CD27 (Biolegend, San Diego, CA, USA) and PE-conjugated anti-IgD (BD Biosciences, San Jose, CA, USA). For memory B-cell transcriptome study using Nanostring assay, lymphocytes positive for CD19 and CD27 were sorted in 1.5 ml tubes. After 30 minutes of incubation in refrigerator at 4°C, samples were read by BD LSR Fortessa (BD Biosciences, San Jose, CA, USA).
Nanostring assay
Prior to the Nanostring assay, 100% Buffer RLT (RNeasy Lysis Buffer, QIAGEN, Mississauga, Canada) was diluted to 1/3 diluted RLT in nuclease free water. Then 5ul of the diluted RLT buffer was added to each tube and lysis process was performed by pipetting up and down 15 times per sample while avoiding bubble formation. After lysis, lysates were centrifuged at 12,000g for 2 min, 4°C, then transferred to a PCR tube, and were placed on ice to use immediately.
Memory B cells sorted from PBMC samples of 3 patients per group were gathered into 1 pool. A total of 4 memory B cell pools, 1 per group were used in Nanostring analysis with Human Immunology Panel Kit (Nanostring Technologies). Hybridizations were carried out by combining 5 ul of each cell lysates with 8 ul of nCounter Reporter probes in hybridization buffer and 2 ul of nCounter Capture probes (for a total reaction volume of 15 ul) overnight at 65°C for 18 hrs. Excess probes were removed using two-step magnetic bead-based purification on the nCounter Prep Station (Nanostring Technologies). Abundances of specific target molecules were quantified on the nCounter Digital Analyzer by counting the individual fluorescent barcodes and assessing the target molecules. For each assay, a high-density scan encompassing 555 fields of view was performed. The data was collected using the nCounter Digital Analyzer after taking images of the immobilized fluorescent reporters in the sample cartridge with a CCD camera.
nSolver software analysis and pathway analysis
mRNA data analysis was performed using the nSolver software analysis. Threshold count value was set to 20, so that nonspecific counts would be excluded in further analysis. After background thresholding, positive control normalization and codeset control normalization was conducted using geometric mean of 6 synthetic positive control counts and 15 housekeeping gene counts, respectively. This procedure reduces several sources of error such as pipetting errors, instrument scan resolution, lot-to-lot variation in probes, and sample input variability [9]. Based on the normalized gene expression data, relatively over- or under-expressed genes in group 2 and 4 in contrast to group 1 were identified in term of fold-changes.
The Database for Annotation Visualization and Integrated Discovery (DAVID v.6.8) was used for functional enrichment analysis [11]. 4 lists of differentially expressed genes with absolute fold-change over 2 (either up- or down-regulated in two pairs of intergroup comparison) were submitted on the website.
Statistical analysis
R 3.6.1 was used to analyze data, which is described as the mean±standard deviation. Comparison between study groups were performed using ANOVA test or Kruskal-Wallis test. p-value <0.05 was regarded statistically significant. Benjamini-Hochberg procedure was used for identification of functional annotation clusters in order to reduce type 1 errors from multiple-comparison.
Data availability
The raw data of this study are available from the corresponding author upon reasonable request.
RESULTS
Demographic and clinical characteristics
Table 1 summarizes the full dataset of demographic, clinical, serological characteristics and MGFA classification of individual participants. Disease duration, AchR-Ab titer, and the presence of thymoma or thymic hyperplasia did not show any correlation with B cell subsets.
Table 1.
Demographic, clinical, serological characteristics and MGFA classification of individual participants
| Participant (Group-number) | Sex | Age | Naïve | UM | SM | DN | MGFA | Disease duration (months) | AchR-Ab titer (nmol/L) | Thymoma or thymic hyperplasia | Used in Nanostring analysis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-1 | F | 71 | 65.6 | 5.4 | 23.5 | 5.5 | CSR | 10 | 4.9 | Yes | Yes |
| 1-2 | M | 59 | 64.2 | 4.3 | 26.4 | 5.2 | CSR | 23 | 9.77 | Yes | Yes |
| 1-3 | M | 65 | 93.1 | 2.1 | 3.6 | 1.2 | CSR | 8 | 9 | Yes | No |
| 1-4 | F | 48 | 81.8 | 5.2 | 9.5 | 3.5 | CSR | 6.5 | 18 | No | No |
| 1-5 | M | 69 | 78.1 | 4.3 | 14.9 | 2.7 | CSR | 23 | 3.85 | Yes | Yes |
| 1-6 | F | 85 | 53.9 | 9.4 | 24.3 | 12.4 | CSR | 11 | 0.23 | No | No |
| 1-7 | M | 36 | 71.5 | 7.5 | 16.5 | 4.5 | CSR | 5 | 0.67 | No | No |
| 1-8 | M | 75 | 83.7 | 5.1 | 7.5 | 3.7 | CSR | 5 | 4.04 | No | No |
| 2-1 | M | 43 | 66.8 | 7.2 | 17.1 | 9.0 | PR | 8 | (−) | No | No |
| 2-2 | M | 80 | 84.4 | 4.2 | 8.2 | 3.2 | PR | 2 | 0.76 | No | No |
| 2-3 | F | 41 | 66.5 | 4.4 | 20.3 | 8.8 | PR | 21 | 0.24 | No | No |
| 2-4 | M | 62 | 63.7 | 8.3 | 23.0 | 5.0 | PR | 6 | (−) | No | No |
| 2-5 | F | 80 | 21.2 | 5.2 | 46.7 | 26.9 | PR | 5 | 7.88 | No | No |
| 2-6 | F | 45 | 55.6 | 11.7 | 25.0 | 7.7 | PR | 1.5 | 11.87 | Yes | No |
| 2-7 | M | 60 | 40.4 | 6.2 | 25.7 | 27.7 | PR | 3.5 | 7.09 | Yes | No |
| 2-8 | F | 42 | 82.3 | 2.4 | 12.3 | 3.0 | MM | 1 | 16.05 | No | Yes |
| 2-9 | M | 72 | 30.4 | 6.1 | 38.9 | 24.5 | MM | 12 | 17.65 | No | Yes |
| 2-10 | F | 56 | 57.3 | 2.0 | 23.8 | 16.9 | MM | 0.5 | 6.36 | Yes | Yes |
| 2-11 | F | 79 | 82.2 | 2.6 | 12.7 | 2.5 | PR | 3 | 1.91 | No | No |
| 2-12 | M | 77 | 28.5 | 8.4 | 42.0 | 21.1 | PR | 4 | 7.44 | No | No |
| 2-13 | M | 50 | 56.9 | 2.5 | 24.8 | 15.7 | PR | 11 | 4.66 | Yes | No |
| 3-1 | M | 54 | 47.1 | 6.2 | 40.2 | 6.5 | N/A | N/A | N/A | N/A | Yes |
| 3-2 | F | 67 | 78.8 | 5.6 | 12.4 | 3.2 | N/A | N/A | N/A | N/A | No |
| 3-3 | F | 69 | 88.0 | 3.2 | 5.1 | 3.8 | N/A | N/A | N/A | N/A | No |
| 3-4 | F | 78 | 84.3 | 2.7 | 9.6 | 3.4 | N/A | N/A | N/A | N/A | No |
| 3-5 | F | 61 | 86.2 | 1.9 | 7.3 | 4.6 | N/A | N/A | N/A | N/A | No |
| 3-6 | M | 53 | 61.7 | 9.2 | 20.3 | 8.9 | N/A | N/A | N/A | N/A | No |
| 3-7 | M | 54 | 69.5 | 4.5 | 21.7 | 4.3 | N/A | N/A | N/A | N/A | No |
| 3-8 | M | 60 | 60.3 | 6.0 | 30.4 | 3.3 | N/A | N/A | N/A | N/A | Yes |
| 3-9 | M | 67 | 42.5 | 9.3 | 42.5 | 5.6 | N/A | N/A | N/A | N/A | Yes |
| 3-10 | F | 70 | 76.0 | 3.5 | 13.0 | 7.6 | N/A | N/A | N/A | N/A | No |
| 4-1 | F | 67 | 59.8 | 7.3 | 20.4 | 12.5 | V | 0.5 | 6.7 | Yes | Yes |
| 4-2 | F | 46 | 74.9 | 6.8 | 13.0 | 5.3 | IIa | 10 | 9.76 | No | Yes |
| 4-3 | M | 46 | 72.3 | 4.5 | 17.5 | 5.7 | IIb | 3.5 | 2.45 | Yes | Yes |
Every peripheral blood sample was analyzed by flow cytometry for B cell immunophenotyping. Memory B cells sorted from 3 patients per group were gathered into each pool and were submitted for Nanostring assay. UM, unswitched memory B cells; SM, switched memory B cells; DN, double-negative B cells; MGFA, Myasthenia Gravis Foundation of America; AchR-Ab, anti-acetylcholine receptor antibody; F, female; M, male; CSR, chronic stable remission; PR, pharmacologic remission; MM, minimal manifestation; N/A, not applicable.
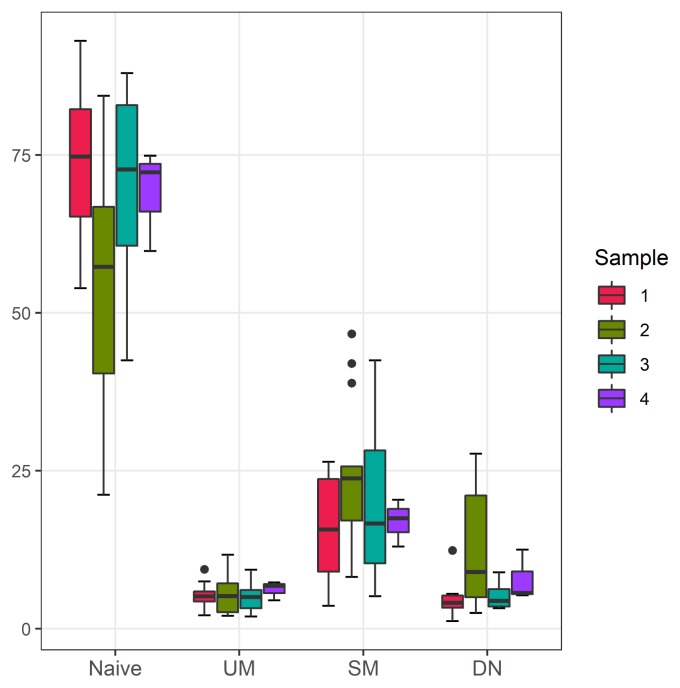
Double-negative B cells are increased in group 2
B-cells were classified into 4 types depending on the presence of IgD and/or CD27; naïve B cells (IgD+CD27−), unswitched memory B cells (IgD+CD27+), switched memory B cells (IgD−CD27+), and double-negative B cells (IgD−CD27−). The frequencies of 4 B cell subsets were analyzed in each group (Table 2, Fig. 1).
Table 2.
Frequencies of B cell subsets in 4 groups (naïve: IgD+CD27−, double-negative: IgD−CD27−, unswitched memory: IgD+CD27+, switched memory: IgD−CD27+)
| Group | 1 | 2 | 3 | 4 | p-value |
|---|---|---|---|---|---|
| Naïve | 73.99±12.61 | 56.63±21.08 | 69.44±16.14 | 69.00±8.07 | 0.2466 |
| UM | 5.40±2.19 | 5.48±2.90 | 5.19±2.57 | 6.20±1.5 | 0.8475 |
| SM | 15.78±8.47 | 24.65±11.68 | 20.26±13.42 | 16.97±3.73 | 0.4078 |
| DN | 4.83±3.36 | 13.23±9.40 | 5.11±1.98 | 7.81±4.06 | 0.0828 |
Although none of 4 subsets revealed statistically significant difference between 4 groups, group 2 showed marked increase of double-negative B cells and relative decrease of naïve B cells. Data were presented as mean±standard deviation. UM, unswitched memory B cells; SM, switched memory B cells; DN, double-negative B cells.
Fig. 1.
Box plot representing B cell subset frequencies in 4 groups.
Group 2 exhibited marked increase in double-negative B cell subset in contrast to the other groups, although statistically not significant (p=0.0828). Although double-negative B cells tended to be slightly higher in group 4 than group 1 and 3, given the small gap and number of samples from group 4, this tendency was not considered significant. Moreover, marked decrease in naïve B cells was noted in group 2 (p=0.2466). Other B cell subsets did not reveal any between-group differences.
Memory B cell transcriptional profiles
We initially assumed that transcripts of memory B cells would impact the disease status, and those differentially expressed between group 4 (drug naïve) and 1 (complete stable remission or minimal manifestation without ISTX) reflect disease activity. This is because while both groups did not receive ISTX in common, patients in group 4 were in active status. In a similar way, differential transcription in group 2 (pharmacologic remission or minimal manifestation with ISTX) compared to group 1 was regarded as epigenetic change in memory B cells induced by ISTX. We investigated which genes in nCounter Human Immunology V2 Panel showed inter-group difference in RNA counts. The lists of differentially expressed genes were summarized in Table 3, sorted in descending order by the fold change (complete data in supplemental file S1). We identified 9 and 26 genes up- and down-regulated, respectively, in the patient group of active disease compared to the group of complete stable remission. In the comparison to evaluate the effect of immunosuppressive treatments, 10 and 5 genes were identified to be up- and down-regulated in the pharmacologic remission group compared to the complete stable remission group.
Table 3.
Transcripts differentially expressed in group 4, 2 versus 1 and their fold changes
| Disease activity (group 4 vs 1) | ISTX effect (group 2 vs 1) | |||
|---|---|---|---|---|
|
|
|
|||
| Up-regulated | Down-regulated | Up-regulated | Down-regulated | |
| CCR2 (4.71) | CD83 (5.55) | IL10RA (2.74) | CD45RB (3.88) | DUSP4 (2.96) |
| CD45RB (4.4) | DUSP4 (5.54) | ICAM1 (2.67) | EGR1 (3.1) | LGALS3 (2.44) |
| LAIR (2.53) | NFKBIA (4.97) | ICOSLG (2.52) | IFIT2 (2.44) | NOD2 (2.2) |
| SLAMF6 (2.4) | SOCS3 (4.68) | NFKBIZ (2.42) | LILRB2 (2.3) | GBP5 (2.11) |
| BCL6 (2.34) | CXCR4 (4.51) | PRDM1 (2.36) | LAIR1 (2.25) | PRDM1 (2.05) |
| TGFBI (2.34) | BCL3 (4.37) | CCND3 (2.33) | EBI3 (2.19) | |
| IL7 (2.33) | CD96 (3.66) | CDKN1A (2.23) | EGR2 (2.16) | |
| TLR4 (2.2) | BCL2L11 (3.19) | CCL5 (2.21) | BCL6 (2.16) | |
| HRE (2) | FKBP5 (2.89) | HLA-DRB1 (2.17) | NFIL3 (2.08) | |
| TNFAIP3 (2.89) | TRAF4 (2.16) | HRE (2.08) | ||
| IRF1 (2.83) | GBP5 (2.11) | |||
| CCR7 (2.76) | IL13RA1 (2.07) | |||
| SOCS1 (2.74) | NOD2 (2.05) | |||
Among the 4 lists of differentially regulated genes, only those down-regulated in group 4 compared to group 1 revealed significant functional annotation clusters, shown in Table 4.
Next, we performed functional enrichment analysis using DAVID v6.8 in order to clarify which immunological pathways were significantly altered in patients with active disease or in those receiving ISTX. 3 functional annotation clusters were identified from the list of genes down-regulated in group 4 compared to group 1. Among genes included in each cluster, those from KEGG pathway database are selectively demonstrated in Table 4, as they provide pathophysiological relevance.
Table 4.
KEGG pathways which functional annotation clusters of down-regulated genes in group 4 versus 1 revealed
| KEGG pathway | p-value | Benjamini | Genes |
|---|---|---|---|
| TNF signaling pathway | 5.5E-7 | 4.5E-5 | BCL3, CCL5, NFKBIA, TNFAIP3, ICAM1, NOD2, SOCS3 |
| NOD-like receptor signaling pathway | 6.1E-4 | 1.6E-2 | CCL5, NKFBIA, TNFAIP3, NOD2 |
| Epstein-Barr virus infection | 4.5E-4 | 1.8E-2 | NFKBIA, TNFAIP3, CDKN1A, ICAM1, HLA-DRB1 |
| Influenza | 1.7E-3 | 2.7E-2 | CCL5, NFKBIA, ICAM1, HLA-DRB1, SOCS3 |
| HTLV-1 infection | 6.6E-3 | 6.5E-2 | NFKBIA, CCND3, CDKN1A, ICAM1, HLA-DRB1 |
| Cytokine-cytokine receptor interaction | 5.7E-3 | 6.4E-2 | CCL5, CCR7, CXCR4, IL10RA, IL13RA1 |
| Chemokine signaling pathway | 1.8E-2 | 1.2E-1 | CCL5, CCR7, CXCR4, NFKBIA |
Most of them are associated with proinflammatory cascades, of which down-regulation might suppress disease activity of MG.
DISCUSSION
We performed flow cytometry and Nanostring analysis to investigate B cell immunophenotype and transcriptional profile of memory B cell populations in peripheral blood samples of MG patients. Immunophenotyping of B cells suggested a marked increase of double-negative B cells (IgD−CD27−) and a decrease of naïve B cells in group 2. mRNA quantification of memory B cell lysates revealed that several genes were differentially regulated within groups. Pathway analysis using DAVID v6.8 revealed 3 functional annotation clusters in genes under-expressed in group 4 (drug naïve MG) compared to 1, and most of them were associated with proinflammatory cascades.
A double-negative B lymphocyte is a recently recognized B cell entity of which exact role and origin remain to be demonstrated. However, increasing evidences suggest that they belong to memory B cell category, but are incapable of antigen presentation or replication, so-called “exhausted” or “senescent” memory B cells [12, 13]. Expansion of DN B lymphocytes has been reported in elderly population or various chronic inflammatory conditions; Alzheimer’s disease (AD) [14], rheumatoid arthritis (RA) [15], systemic lupus arthritis (SLE) [16], and chronic Human Immunodeficiency Virus (HIV) [17]. Therefore, it might be assumed that time-enduring antigenic stimulation or immune dysregulation may accumulate double-negative B cells, while deleting naïve or functional memory B cells. Our result might imply that ISTX suppresses B cell-mediated antigen presentation leading to reduction in MG activity. Given positive correlation with double-negative B cell expansion and disease activity in SLE, double-negative B cells in active versus inactive MG patients need to be further investigated with larger number of samples to clarify potential association with this B cell subset and MG activity.
Based on our hypothesis that genes differentially expressed in memory B cells of group 4 and 2 versus 1 reflect disease activity and ISTX-induced transcriptional alterations respectively, Nanostring analysis revealed potential memory B cell biomarkers, some of which also addressed in our previous report [18]. Functional annotation cluster analysis revealed several proinflammatory pathways down-regulated in group 4 in contrast to in group 1: tumor necrosis factor (TNF) signaling pathway, NOD-like receptor (NLR) signaling pathway, cytokine-cytokine receptor interaction, and chemokine signaling pathway. TNF signaling mediates B cell maturation and differentiation, dysregulation of which can cause autoimmunity or neoplasia [19]. NLR pathway was recently discovered to mediate B cell activation without T cell signaling [20]. Interestingly, the other 3 KEGG pathways suggested that HLA-DRB1 is associated with active versus inactive MG, of which polymorphism has been posited to be associated with MG in several previous studies [21–23]. Clinical relevance as potential biomarkers should be further investigated in future studies with larger number of patients.
This study has several limitations. First, number of samples analyzed in flow cytometry was too small to reach statistical significance. Especially, only 3 samples from group 4 were not enough to show the pattern of B cell subset alteration. Similarly, statistical analysis was not available in Nanostring analysis, because only single pool per group had been used. Hence, some “differentially expressed genes (DEGs)” in this study may reflect individual variations, rather than disease status. Given that a pool contains memory B cells of 3 patients per group, these “DEGs” worth further analysis to confirm their role as biomarkers.
Moreover, our assumptions on group-group comparison have potential errors. Because group 1 and 4 show substantial difference in disease duration (mean 11.44 months in group 1 and 4.67 months in group 4), chronological changes not directly related to disease activity might have affected their transcriptional profiles. Some patients in group 2 may aggravate when ISTX is discontinued, implying relatively higher disease activity in group 2 than 1. Thus, transcriptional profile of group 2 compared to group 1 owe not only to ISTX, but also to disease activity in some parts.
As we had initially assumed that memory B cells have a major impact on disease status, we only sorted memory B cells from flow cytometry for Nanostring analysis and did not sort out double-negative subset for transcript profiling. Given the unexpected increase of double-negative B cells in group 2, however, transcriptome analysis of this subset in various MG status might also be promising. Because double-negative B cells are rare in entire B cell population, each pool needs samples from more participants, requiring another study with larger number.
Despite these limitations, this study suggests a novel mean to investigate detailed immunologic evaluation of autoimmune diseases. A follow-up study with well controlled design and larger number of samples will suggest practical biomarkers that guide treatment decision in future clinical settings.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by grants from the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (2017R1D-1A1B03030569), and funded by the Astellas Pharma Korea Inc.
REFERENCES
- 1.Lindstrom JM, Seybold ME, Lennon VA, Whittingham S, Duane DD. Antibody to acetylcholine receptor in myasthenia gravis. Prevalence, clinical correlates, and diagnostic value. Neurology. 1976;26:1054–1059. doi: 10.1212/WNL.26.11.1054. [DOI] [PubMed] [Google Scholar]
- 2.Hoch W, McConville J, Helms S, Newsom-Davis J, Melms A, Vincent A. Auto-antibodies to the receptor tyrosine kinase MuSK in patients with myasthenia gravis without acetylcholine receptor antibodies. Nat Med. 2001;7:365–368. doi: 10.1038/85520. [DOI] [PubMed] [Google Scholar]
- 3.Higuchi O, Hamuro J, Motomura M, Yamanashi Y. Autoantibodies to low-density lipoprotein receptor-related protein 4 in myasthenia gravis. Ann Neurol. 2011;69:418–422. doi: 10.1002/ana.22312. [DOI] [PubMed] [Google Scholar]
- 4.Gasperi C, Melms A, Schoser B, Zhang Y, Meltoranta J, Risson V, Schaeffer L, Schalke B, Kröger S. Anti-agrin autoantibodies in myasthenia gravis. Neurology. 2014;82:1976–1983. doi: 10.1212/WNL.0000000000000478. [DOI] [PubMed] [Google Scholar]
- 5.Romi F, Suzuki S, Suzuki N, Petzold A, Plant GT, Gilhus NE. Anti-voltage-gated potassium channel Kv1.4 antibodies in myasthenia gravis. J Neurol. 2012;259:1312–1316. doi: 10.1007/s00415-011-6344-y. [DOI] [PubMed] [Google Scholar]
- 6.Mygland A, Tysnes OB, Matre R, Volpe P, Aarli JA, Gilhus NE. Ryanodine receptor autoantibodies in myasthenia gravis patients with a thymoma. Ann Neurol. 1992;32:589–591. doi: 10.1002/ana.410320419. 1992. [DOI] [PubMed] [Google Scholar]
- 7.Gallardo E, Martínez-Hernández E, Titulaer MJ, Huijbers MG, Martínez MA, Ramos A, Querol L, Díaz-Manera J, Rojas-García R, Hayworth CR, Verschuuren JJ, Balice-Gordon R, Dalmau J, Illa I. Cortactin autoantibodies in myasthenia gravis. Autoimmun Rev. 2014;13:1003–1007. doi: 10.1016/j.autrev.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 8.Heldal AT, Eide GE, Romi F, Owe JF, Gilhus NE. Repeated acetylcholine receptor antibody-concentrations and association to clinical myasthenia gravis development. PLoS One. 2014;9:e114060. doi: 10.1371/journal.pone.0114060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol. 2011;Chapter 25(Unit25B.10) doi: 10.1002/0471142727.mb25b10s94. [DOI] [PubMed] [Google Scholar]
- 10.Jaretzki A, 3rd, Barohn RJ, Ernstoff RM, Kaminski HJ, Keesey JC, Penn AS, Sanders DB. Myasthenia gravis: recommendations for clinical research standards. Task Force of the Medical Scientific Advisory Board of the Myasthenia Gravis Foundation of America. Neurology. 2000;55:16–23. doi: 10.1212/WNL.55.1.16. [DOI] [PubMed] [Google Scholar]
- 11.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colonna-Romano G, Bulati M, Aquino A, Pellicanò M, Vitello S, Lio D, Candore G, Caruso C. A double-negative (IgD−CD27−) B cell population is increased in the peripheral blood of elderly people. Mech Ageing Dev. 2009;130:681–690. doi: 10.1016/j.mad.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Martorana A, Balistreri CR, Bulati M, Buffa S, Azzarello DM, Camarda C, Monastero R, Caruso C, Colonna-Romano G. Double negative (CD19+IgG+IgD−CD27−) B lymphocytes: a new insight from telomerase in healthy elderly, in centenarian offspring and in Alzheimer’s disease patients. Immunol Lett. 2014;162:303–309. doi: 10.1016/j.imlet.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Bulati M, Buffa S, Martorana A, Gervasi F, Camarda C, Azzarello DM, Monastero R, Caruso C, Colonna-Romano G. Double negative (IgG+IgD−CD27−) B cells are increased in a cohort of moderate-severe Alzheimer’s disease patients and show a pro-inflammatory trafficking receptor phenotype. J Alzheimers Dis. 2015;44:1241–1251. doi: 10.3233/JAD-142412. [DOI] [PubMed] [Google Scholar]
- 15.Mahmood Z, Muhammad K, Schmalzing M, Roll P, Dörner T, Tony HP. CD27−IgD− memory B cells are modulated by in vivo interleukin-6 receptor (IL-6R) blockade in rheumatoid arthritis. Arthritis Res Ther. 2015;17:61. doi: 10.1186/s13075-015-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei C, Anolik J, Cappione A, Zheng B, Pugh-Bernard A, Brooks J, Lee EH, Milner EC, Sanz I. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J Immunol. 2007;178:6624–6633. doi: 10.4049/jimmunol.178.10.6624. [DOI] [PubMed] [Google Scholar]
- 17.Bamford A, Hart M, Lyall H, Goldblatt D, Kelleher P, Kampmann B. The influence of paediatric HIV infection on circulating B cell subsets and CXCR5(+) T helper cells. Clin Exp Immunol. 2015;181:110–117. doi: 10.1111/cei.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park KH, Jung J, Lee JH, Hong YH. Blood transcriptome profiling in myasthenia gravis patients to assess disease activity: a pilot RNA-seq study. Exp Neurobiol. 2016;25:40–47. doi: 10.5607/en.2016.25.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rickert RC, Jellusova J, Miletic AV. Signaling by the tumor necrosis factor receptor superfamily in B-cell biology and disease. Immunol Rev. 2011;244:115–133. doi: 10.1111/j.1600-065X.2011.01067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petterson T, Jendholm J, Månsson A, Bjartell A, Riesbeck K, Cardell LO. Effects of NOD-like receptors in human B lymphocytes and crosstalk between NOD1/NOD2 and Toll-like receptors. J Leukoc Biol. 2011;89:177–187. doi: 10.1189/jlb.0210061. [DOI] [PubMed] [Google Scholar]
- 21.Xie YC, Qu Y, Sun L, Li HF, Zhang H, Shi HJ, Jiang B, Zhao Y, Qiao SS, Wang SH, Wang DX. Association between HLA-DRB1 and myasthenia gravis in a Northern Han Chinese population. J Clin Neurosci. 2011;18:1524–1527. doi: 10.1016/j.jocn.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Maniaol AH, Elsais A, Lorentzen ÅR, Owe JF, Viken MK, Sæther H, Flåm ST, Bråthen G, Kampman MT, Midgard R, Christensen M, Rognerud A, Kerty E, Gilhus NE, Tallaksen CM, Lie BA, Harbo HF. Late onset myasthenia gravis is associated with HLA DRB1*15:01 in the Norwegian population. PLoS One. 2012;7:e36603. doi: 10.1371/journal.pone.0036603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos E, Bettencourt A, da Silva AM, Boleixa D, Lopes D, Brás S, Costa PPE, Lopes C, Gonçalves G, Leite MI, da Silva BM. HLA and age of onset in myasthenia gravis. Neuromuscul Disord. 2017;27:650–654. doi: 10.1016/j.nmd.2017.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of this study are available from the corresponding author upon reasonable request.