Abstract
More than 60 years after the first description of differentiation in cell culture and 40 years after the synthesis of 5-azacytidine,epigenetic therapies have been added to the anticancer armamentarium. DNA methyltransferase (DNMT) inhibitors such as 5-aza-2’-deoxycytidine or 5-azacytidine have been approved in myelodysplastic syndrome (MDS) and acute myelogenous leukemia (AML), whereas the histone deacetylase inhibitors (HDIs) including vorinostat, romidepsin, panobinostat, belinostat, and entinostat have been shown to be active in cutaneous and peripheral T-cell lymphoma. Although the range of malignancies in which monotherapy with DNMT inhibitors or HDIs are effective has been limited to date, the possibility remains that a broader spectrum of activity will be identified as combination studies are completed. Meanwhile, basic science has provided a steadily increasing understanding of the complexity of the epigenome, including the histone code and triggers for aberrant methylation, and their contribution to oncogenesis. As our basic understanding of the epigenetics of cancer increases, the number of potential therapeutic targets will also increase, offering more hope in the quest to treat cancer by normalizing the epigenome. This issue of CCR Focus is dedicated to understanding the clinical and translational aspects of epigenetics research.
The inclusion of epigenetics in the National Institutes of Health (NIH) roadmap has highlighted the need for research in both epigenetic mechanisms of oncogenesis and in epigenetic therapies. This decade has seen the addition of several epigenetic therapies to the anticancer armamentarium. The Food and Drug Administration (FDA) has approved three epigenetic targeting agents for oncology: Vidaza (Celgene, Summit, NJ, azacitidine, 2004), Dacogen (SuperGen, Inc., Dublin, CA, decitabine, 5-aza-2’-deoxycytidine, 2006), and Zolinza (Merck & Co., Inc., Whitehouse Station, NJ, vorinostat, 2006); and many more are in clinical and preclinical development (Table 1). Yet, we still do not fully understand the best schedule, the ideal dosing, or true mechanism of action of these agents. Their enzymatic targets are known; however, the downstream effectors have not been elucidated. This issue of CCR Focus addresses our current understanding of these matters and addresses the arrival of epigenetic therapies as conventional anticancer therapies.
Table 1.
Drugs with epigenetic targets either approved or in development
| Drug target | Generic or trade name |
Development name |
Chemical class |
Pharmaceutical sponsor |
Status |
|---|---|---|---|---|---|
| DNMT inhibitor | Azacitidine Vidaza | 5-azacytidine | Nucleoside analog | Celgene (Summit, NJ) | FDA approved, 2004 EMEA approved, 2008 |
| DNMT inhibitor | Decitabine Dacogen | 2’-deoxy-5-azacytidine | Nucleoside analog | Eisai Co., Ltd. (Tokyo, Japan) | FDA approved, 2006 |
| DNMT inhibitor | Zebularine | Nucleoside analog | |||
| DNMT inhibitor | SGI-110 | Nucleoside analog | SuperGen (Dublin, CA) | ||
| DNMT inhibitor | RG108 | Active site inhibitor | |||
| DNMT inhibitor | SGI-1036 | Quinoline | SuperGen | ||
| EZH2 antagonist | DZnep 3-deazaneplanocin A | Nucleoside analog | |||
| HDAC inhibitor | Sodium phenylbutyrate Buphenyl Ammonaps | Small chain fatty acid | Ucyclyd Pharma (Scottsdale, AZ) Orphan International (Stockholm, Sweden) |
FDA approved EMEA approved (urea cycle disorders) | |
| HDAC inhibitor | Valproic acid Depakote, (others) | Small chain fatty acid | Abbott Laboratories (Abbott Park, IL) | FDA approved (seizure disorders, others) | |
| HDAC inhibitor | Vorinostat Zolinza | SAHA | Hydroxamic acid Merck & Co. | FDA approved, 2006 | |
| HDAC inhibitor | Panobinostat | LBH589 | Hydroxamic acid | Novartis Pharmaceuticals (East Hanover, NJ) | |
| HDAC inhibitor | Belinostat | PXD101 | Hydroxamic acid | TopoTarget (Rockaway, NJ) | |
| HDAC inhibitor | JNJ-26481585 | Hydroxamic acid | Johnson & Johnson (Langhorne, PA) | ||
| HDAC inhibitor | Romidepsin | Depsipeptide, FK228 | Cyclic peptide | Gloucester Pharmaceuticals (Cambridge, MA) | New drug application filed with FDA |
| HDAC inhibitor | Entinostat | MS275, SNDX-275 | Benzamide | Syndax Pharmaceuticals (Waltham, MA) | |
| HDAC inhibitor | MGCD-0103 | Benzamide | MethylGene (Montreal, Quebec, Canada) | ||
Abbreviation: EMEA, European Medicines Agency.
Differentiating Agents: Epigenetic Targets
One interesting aspect of epigenetic therapies is their historical origin as differentiating agents. The spontaneous differentiation of leukemic cells was first noted in cell culture systems in the 1940s (1). Over time, it became clear that many different agents and culture conditions could promote such differentiation, although there were model-specific differences (2). Agents found to induce differentiation in leukemic cells included antifolates, anthracyclines, camptothecins, retinoids, phorbol esters, vitamins D3 and B12, and various cytokines. Among these agents were also compounds later understood to alter chromatin structure: hypomethylating agents and histone deacetylase inhibitors (HDIs). In 1977, Constantinides reported that 5-azacytidine was able to induce striated muscle cells from murine embryonic 10T1/2 cells and in 1979 this work was extended to show that pluripotency in these cells following exposure to 5-azacytidine could produce chondrocytes, adipocytes, and muscle cells (3, 4). Jones and Taylor then reported in 1980 that these differentiating effects were associated with alterations in DNA methylation (5). DNA hypomethylation was soon linked to activation of gene expression resulting in phenotypic cell differentiation in murine Friend erythroleukemia cells by observation of the tight association between the activities following treatment with 5-azacytidine or its analog 5-aza-2’-deoxycytidine (6). In independent studies, erythroid differentiation following the addition of sodium butyrate was observed at the phenotypic level in the human erythroleukemic cell line, K562 (7, 8). A review of sodium butyrate in 1982 observed that the agent induced histone hyperacetylation and connected this property with the inhibition of cell growth and the differentiated phenotype in certain model systems, notably in erythroleukemia cells (9).
These observations launched the search for compounds that could be used clinically in the differentiation therapy of hematologic disorders and malignancies, despite the differing mechanisms of action. Case reports described the first clinical use of 5-azacytidine to induce fetal hemoglobin in a patient with beta-thalassemia in 1982 (10) and in a patient with sickle cell anemia in 1983 (11). The approval of 5-azacytidine in 2004 for myelodysplastic syndrome (MDS) marked the end of a 40-year quest to find a clinical application for a compound originally synthesized in 1964 (12). Clinical experience with sodium butyrate was first reported with a partial response in a patient with acute myelogenous leukemia (AML) in 1983 (13), reviewed by Gore and Carducci (14). Although butyrate was never approved as an anticancer therapy, its evaluation and that of other short chain fatty acids led directly to the development of hydroxamic acids, notably vorinostat, now approved for the treatment of cutaneous T-cell lymphoma (CTCL) (ref. 15).
DNA Methylation as a Therapeutic Target
Recognition of the extent of aberrant methylation found in human cancer has led to the understanding that DNA methylation, if not a first hit in oncogenesis, is at least a mediator of oncologic progression. In distilling the literature surrounding this topic for this issue of CCR Focus, McCabe, Brandes, and Vertino note that cancer cells exhibit widespread loss of intergenic DNA methylation and gain of DNA methylation in promoter-associated CpG islands, defined as clusters of the CpG dinucleotide, that are normally found in an unmethylated state (16). These methylated CpG islands are often found associated with the promoters of tumor suppressor genes, such as the retinoblastoma (Rb) gene (17) or p21 (18).
As new methods for examining genome-wide methylation patterns have become available, the extent to which aberrant and varying methylation patterns exist in human cancer has begun to emerge. Neither the trigger, nor the initiating event, nor the sequence of events that follow and give rise to hypermethylation, nor the mechanisms underlying the selection of a high prevalence of tumor suppressor genes for methylation have been determined. McCabe and colleagues illustrate the multiple pathways involved in hypermethylation (16), leading to the sobering realization that, although we have been successful with one strategy for reducing hypermethylation with the nucleoside analogs 5-aza-2’-deoxycytidine or 5-azacytidine (now understood to function as DNA methyltransferase (DNMT) inhibitors following their incorporation into DNA), we are still far from understanding how to normalize DNA methylation in cancer. However, the progress made in basic science has allowed the identification of multiple genes in which methylation has potential clinical application in diagnosis, early detection, prognosis, or therapy outcome.
Issa and Kantarjian offer a more sanguine outlook in the translation of these two agents to the clinic in the therapy of MDS (19). The authors describe the earlier clinical failure of agents that had clear ability to demethylate and reactivate genes in preclinical models, followed by the slow recognition that there was a biphasic dose response curve in which high doses of the nucleoside analogs inhibited DNA synthesis whereas hypomethylation requires DNA synthesis. At lower doses there would be DNA incorporation followed by DNMT inhibition. This meant a prolonged dosing period for MDS, eventually yielding response rates in the 60% to 70% range for both 5-aza-2’-deoxycytidine and 5-azacytidine. Having an effective agent offers the opportunity to develop biomarkers of response. Issa summarizes these efforts, noting that whereas reduced 5-methyl cytosine content can be detected in correlative studies, change in gene expression has correlated better with response than has alteration in global methylation. This observation leads to the important issues of determining mechanisms of resistance to DNMT inhibitors (20) and how to improve on the clinical results already obtained.
Histone Deacetylase as a Therapeutic Target
HDIs were found to be remarkable and effective cytotoxic agents in vitro. Whereas the traditional mechanism of action underlying the antineoplastic activity of HDIs was considered the increased acetylation of lysine residues that form the octomeric histone core of chromatin, in recent studies a remarkable array of potential mechanisms has been proposed, suggesting the mechanisms underlying HDAC activity are pleiotropic and likely to be cell context-dependent. In this issue of CCR Focus, Schrump discusses the varied mechanisms of action proposed to underlie the activity of HDIs in vitro (21). These mechanisms include:
Histone acetylation with alterations in gene expression that effect cell cycle arrest and limit cell growth, including up-regulation of genes such as p21, p27, and other genetic markers of differentiation; and down-regulation of genes involved in growth such as cyclin D (22 –24);
Acetylation of nonhistone proteins such as p53, HIF-1alpha, pRb, STAT-3, Rel A/p65, or estrogen receptor that may impair their function and thereby influence cell growth or survival (25 – 27);
Acetylation of Hsp90, with its attendant loss of ability to chaperone client proteins resulting in their ubiquitinylation and proteasomal degradation (28, 29);
A prometaphase cell cycle arrest that results from reduced premitotic phosphorylation of pericentromeric histone H3 and disruption of kinetochore assembly (30);
An antiangiogenic effect potentially mediated by impairing HIF-1a stability (31);
Direct activation of apoptotic pathways through reduction of antiapoptotic proteins such as Bcl-2 and increased expression of proapoptotic proteins such as BAX and BAK (32, 33);
Enhanced production of reactive oxygen species (ROS) (refs. 34, 35);
Disruption of aggresome formation through acetylation of tubulin (36);
Enhanced antitumor immunity through enhancement of TRAIL or up-regulation of antigen expression that could facilitate cancer cell recognition (37 – 40);
Disruption of DNA repair through acetylation or downregulation of proteins such as Ku70, Ku86, BRCA1, and RAD51 (41 – 43).
One question that such a wide-ranging list raises is whether histone acetylation is actually an indispensable component of HDI activity. If differentiation is important, then histone acetylation per se may not be needed, as other agents induce differentiation. On the other hand, any one of the activities above could be critical in a given cell type. One of the complicating aspects of the laboratory evaluations that elucidated the multiple mechanisms of action itemized above is the use of different cell lines and the focus on only one or two aspects within each study. It is important that, going forward, in vitro and in vivo studies of HDIs embrace the possibility that multiple activities can occur concurrently and that these are assessed simultaneously. Furthermore, some consideration for evaluating these various mechanisms should be incorporated into clinical trials. Some investigators have championed one effect over another as being more important in limiting cell growth or inducing cytotoxicity. On the other hand, the multiplicity of these activities has raised the question of whether more specific HDIs should be developed. The current HDIs target class I (HDACs 1, 2, 3, and 8), class II (HDAC 4, 5, 6, 7, 9, and 10), and HDAC 11 (generally considered class IV) enzymes to varying degrees. It is quite possible that different HDIs may have a spectrum of activities much as we have already observed for different agents that belong to a general drug class such as anthracyclines, vinca alkaloids, or platinums.
Although, as noted above, the HDIs have a range of pathways by which to inhibit cell growth or trigger cell death, the clinical activity to date has been largely confined to hematologic malignancies, and particularly T-cell lymphomas. Prince and coauthors examine the clinical experience with HDIs to date, including a systematic review of results from the early phase trials for HDIs in development, including both outcome and toxicities (44). The authors note the dramatic activity in T-cell lymphoma, both peripheral T-cell lymphoma (PTCL) and CTCL, as a class effect observed with several HDIs and then go on to describe examples of activity in patients with B-cell lymphoma, Hodgkin lymphoma, and multiple myeloma. They note dose-and schedule-dependent activity in AML, suggesting that in the absence of the ability to markedly increase dosing of the HDI, effective combinations will be needed to exploit the observed activity.
HDAC Inhibitors in T-Cell Lymphomas
The demonstration that HDIs are active in T-cell lymphoma, and the approval of vorinostat for CTCL has raised a number of questions:.
What is the mechanism of the marked efficacy of HDIs in T-cell lymphoma? With what seems to be a class effect, is there a basis for differences among the HDIs? And finally, with the approval of vorinostat, should other HDIs be developed?
The marked efficacy of HDIs in T-cell lymphoma is not understood. It is tempting to speculate that the responsive subset of T-cell lymphomas has its origin in an as-yet unknown chromosomal rearrangement that recruits the class 1 HDACs to the promoter of a gene normally limiting cell proliferation. However, chromosomal alterations such as those described in AML have not been described in T-cell lymphoma. Further, the lymphomas in general have proven to be a tumor type distinctly susceptible to different therapeutic interventions. It seems equally likely that HDIs trigger apoptosis in an apoptosis-prone environment.
Differences between the structurally very different HDIs currently in development include schedule, potency, and pharmacology. Examples of scheduling differences include vorinostat administered on a daily oral schedule; panobinostat administered orally three times weekly; romidepsin with an intravenous day 1, 8, and 15 schedule; and belinostat administered by short intravenous infusion on days 1 through 5. There are marked differences in potency, when observed from an in vitro standpoint. Romidepsin is active at nanomolar concentrations whereas vorinostat is active at micromolar concentrations. In pharmacokinetics, romidepsin has been found to have a 2.5-hour half-life, only slightly longer than that of vorinostat. In contrast, panobinostat has a longer half-life at 8 hours, and entinostat a much longer half-life of more than 30 hours (45, 46). Differences in pharmacology are incompletely understood. One mechanism of protection from normal tissue toxicity is the efflux of compounds by P-glycoprotein. As a substrate for P-glycoprotein – mediated drug efflux, romidepsin would likely not penetrate into the central nervous system, whereas other HDAC inhibitors may have central nervous system effects (47, 48). As one example, Depakote (Abbott Laboratories, Abbott Park, IL, valproic acid), is in clinical use as an antiseizure medication and not thought to be susceptible to P-glycoprotein-or ABCG2 – mediated drug efflux, important components in the blood brain barrier (49). With variations in structure, schedule, and pharmacology, differences in spectrum of activity are likely to emerge over time.
Recognizing that the authors have a bias about the answer to the last question above, on the basis of a decade of effort in the development of romidepsin, we would argue in the affirmative. We have noted dramatic, durable activity of romidepsin in the National Cancer Institute multicenter trial in patients with T-cell lymphoma (Fig. 1) (ref. 50). As discussed elsewhere in this issue of CCR Focus, a comparable level of activity was noted in an independent registration trial (44), and romidepsin is currently under review by the FDA for this indication. One need only think of current antineoplastic agents to conclude that agents with identical targets may have very different antineoplastic activities. Daunorubicin and doxorubicin have very similar structures, differing only by a hydroxyl group, and have a very different spectrum of activity, in leukemias and solid tumors, respectively. The most recent addition to the microtubule targeting armamentarium, the epothilones, and their predecessors the taxanes, provide an example of drugs with differing structures, but identical targets.
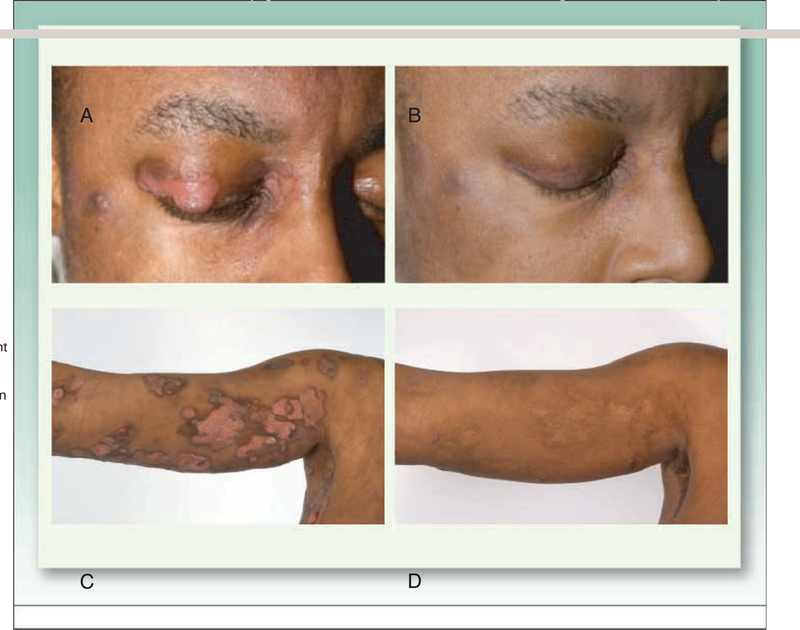
Fig. 1.
This patient with tumor stage CTCL was enrolled on a phase II trial of romidepsin for patients with CTCL or PTCL and treated at 14 mg/m2 on a day 1,8, and 15 schedule every 28 d (50). A, As shown in the upper panel,a lesion next to the eye and on the eyelid showed significant improvement, B, after the first cycle of therapy. Lesions in other areas of the body also showed significant response,as shown in the photograph, C, of her right arm prior to and, D, 5 mo after starting therapy.This partial response lasted 15 mo.
Understanding the Restricted Clinical Efficacy
One observation is clear—the extraordinary activity of DNMT inhibitors and HDIs in vitro has not translated to clinical efficacy in solid tumors.
As regards the DNMT inhibitors, Issa and Kantarjian address the question of why hypomethylating agents were able to succeed in MDS and yet have failed to benefit patients with other tumors, at least to date (19). One possible answer lies in the high doses of the drug used in earlier studies, typically the maximally tolerated dose, likely resulting in inhibition of DNA synthesis rather than DNMT inhibition and hypomethylation. Alternatively, the agents may be susceptible to the drug resistance mechanisms prevalent in the patient populations enrolled in earlier studies, suggesting that a patient population without a significant degree of prior therapy should be tested (19, 20). Improved clinical outcomes in nonhematologic malignancies maybe observed if lower doses of demethylating agents are used, and if rational combination therapies are studied.
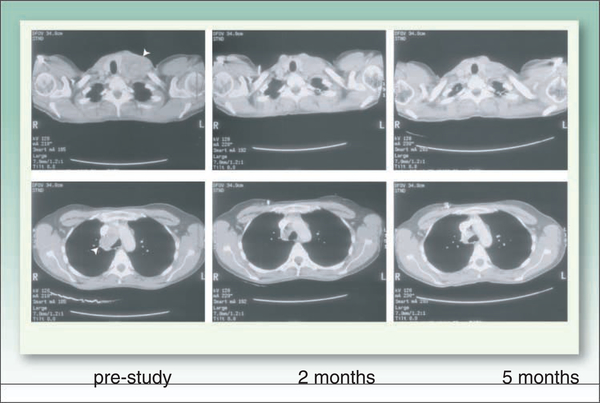
For currently available HDIs this raises the general question of whether the lack of activityin solid tumors is due to incorrect dosing or a lack of potency. An earlier assumption that resistance to HDIs could result simply from the slower growth rate of solid tumors has been discounted to some extent. In vitro studies have shown sensitivity to HDAC inhibition in culture conditions in which growth is inhibited, such as in tumor spheroids or in serum starvation, in which resistance to classical cytotoxic agents is observed (51, 52). This stands in contrast to DNMT inhibitors, which require DNA synthesis. Despite the in vitro data arguing against drug resistance due to slow growth, the fact remains that responses to HDIs in solid tumors in monotherapy trials have been rare, almost individually reportable. Figure 2 shows CT scans obtained in a patient with renal cell carcinoma who enrolled in a phase I study of romidepsin (53) and experienced a partial remission lasting 5 months following therapy with romidepsin. Unfortunately, a follow-up phase II trial failed to confirm activity in renal cancer (54).
Fig. 2.
CT scans obtained in a patient with renal cell carcinomawhowas enrolled on a phase I trial of romidepsin and treated at escalating doses from 9.1to17.8 mg/m2 on a day 1and 5 schedule every 21d (53). A significant responsewas noted after 2 cycles of therapy withmarked shrinkage of mediastinal adenopathy and a supraclavicular lesion also palpable on physical examination (arrowheads). This met the criteria for a partial response, however,t he patient experienced progression of disease in the liver after 6 cycles of therapy,wh ile the remainder of the disease was in control.
One explanation for the drug resistance and the trademark failure of HDI activity in adult solid tumors may be that the changes in gene expression, as mentioned above, are associated with phenotypic and differentiation effects in solid tumors that may promote survival, rather than apoptosis. Several studies have suggested that cells that undergo p21-mediated growth arrest are rescued from the more cytotoxic effects of the HDIs (24, 55). Similarly, HDIs may activate nuclear factor-nB (NF-κB), which is believed to contribute to resistance to HDAC-induced apoptosis through its prosurvival activities. Prevention of NF-κB activation through siRNA knockdown or proteasome inhibition sensitizes head and neck cancer cells to HDIs (56). Finally, in addition to direct activation of apoptotic pathways following HDAC exposure, autophagy has been noted in cells treated with HDIs (57). Although autophagy can enhance apoptosis, it may also be a cell survival mechanism (58). Although these activities can be seen as a mechanism of drug resistance or an adaptive process as found following exposure to anticancer agents, it seems more likely at this writing that they are among the many downstream effects of the HDIs.
Another possible explanation for the difference in sensitivity to HDAC inhibition between solid tumors and hematologic malignancies is that the latter may have fewer mutations and more intact apoptotic pathways. One lesson derived from the genomic sequencing of breast, colon, and pancreatic cancer is that the number of mutations in these solid tumors is quite large and often not overlapping between cancers. Individual tumors contained an average of 90 mutant genes, 11 of which seemed to be oncogenic, among the 13,023 genes sequenced in 11 breast and 11 colorectal cancers (59). Perhaps such an explanation underlies sensitivity to epigenetic therapies in general as well as the more successful outcome of classical cancer chemotherapies in lymphomas and other hematologic malignancies. If solid tumors have intrinsic mechanisms of resistance, then increasing the dose or adding a second agent that exploits any of the molecular effects of the HDI might help overcome resistance in solid tumors.
Combination Therapy
Combination therapies employing DNMT inhibitors and HDIs together or with other agents are being pursued clinically. The possibility to optimally re-express methylated genes following treatment with a DNMT inhibitor followed by an HDI has been the basis for combined epigenetic therapies. Given the in vitro evidence for synergy in such combinations, this remains an area of active study, with initial trials focusing on hematologic malignancies (44, 60). Eventually, randomized trials will be required to establish whether the clinical outcomes correlate with preclinical synergy for these agents. Whether combination epigenetic therapy will improve efficacy in solid tumors remains to be determined.
In addition to combined epigenetic therapies, regimens using HDIs in combination with established agents are actively being pursued. Rationally conceived combination therapies with HDIs can be considered to fit one of several strategies: (1) to counter the molecular effects of HDIs that abrogate their efficacy, (2) to extend the molecular effects of the HDI, or (3) to exploit the molecular effects of the HDI. Bots and Johnstone elaborate on the development of combination therapies (60). An example of the first strategy is the addition of flavopiridol to prevent the induction of p21, which causes cell cycle arrest and to some extent limits the activity of the HDI (61, 62). An example of the second type of combination is to add HDIs to death receptor ligands such as TRAIL that can synergize to increase apoptosis (63). Several examples of the third strategy involve the depletion of Hsp90 client proteins that follows acetylation of Hsp90 and impairment of its chaperone function. Hsp90 acetylation results in polyubiquitinylation of client proteins, such as ABL, Bcr-Abl, EGFR, ErbB2, cRaf, c-kit, FLT3, ER, and AKT, and their degradation by the proteasome (64, 65).
One could predict that this last strategy will be the most promising, allowing selection of tumor type based on pre-existing dysregulation of client proteins. Thus, combining an HDI with trastuzumab should generate a synergistic anticancer effect in HER2 over-expressing breast cancer, a concept that has been confirmed in vitro and is undergoing clinical testing in breast cancer (66, 67). A similar concept should hold true for EGFR inhibitors in lung cancer, in which gefitinib and erlotinib effectively block signaling from mutant EGFR, but from which resistance eventually emerges (68). Other tyrosine kinase inhibitors could synergize with HDIs by this mechanism: imatinib, nilotinib and dasatinib could be potentiated in chronic myelogenous leukemia by loss of Bcr-Abl (69). The list of potentially active combinations grows verylong employing this strategy.
In vitro data also suggest synergy with traditional cytotoxic chemotherapeutics (60). It has been previously noted that HDIs sensitize cancer cells to topoisomerase targeted therapies (70). Initially this was related to a direct interaction of HDAC 1 and 2 and topoisomerase II enzymes (70), but it was later noted that HDIs also increase expression of topoisomerase II (71). Moreover, it was found that acetylation of Ku70 (such as that following treatment of cells with an HDI) results in a functional impairment of DNA repair leading to an increased sensitivity to DNA damaging agents (72). An alternate effect of the acetylation of the cytoplasmic form of Ku70 is the release of Bax, a pro-apoptotic protein, which when released from Ku70 translocates to the mitochondria and triggers apoptosis through the release of cytochrome c and caspase-activation (73).
Caveats about Combination Therapy
An important question is whether combination studies can succeed in which there has been no single agent activity. Few examples of anticancer therapy can be identified in which an agent has failed as monotherapy but has succeeded in combination. However, DNMT inhibitors and HDIs may be different in that a molecular effect that is insufficient to cause cell death could nonetheless set up an environment in which another agent could exert a more potent effect. Thus, the critical point for development of these agents in combination therapy will be to obtain clinical material to test for the molecular effect that is to be exploited in the combination. Only with this information can the results of combination trials be correctly interpreted.
Arguing for combination therapy as a means to fully exploit the activity of epigenetic therapy leads to a related issue, that of scheduling: determining how best to schedule the combined therapy. A chemotherapeutic-inducing cell cycle arrest could not be combined with a DNMT inhibitor. Similarly, the p21 induction and cell cycle arrest that results from HDAC inhibition could lead to resistance with most conventional cytotoxic agents that require cell division for activity. Theoretical rationale suggests that there could be synergy from an HDI combination with platinum due to the increased accessibilityof DNA for the platinum binding. However, this actual mechanism has not been confirmed in any model. Published data confirm sequence dependence, with some agreement that the HDI should not be administered first (74, 75), although in one report greater sensitivityis onlyobserved when the HDI is given first (76). These results may again be cell-type specific.
Finally, when considering combination therapy with conventional chemotherapy agents, it should be recalled that HDIs have direct effects on the promoters of ABC transporters. MDR-1/ABCB1, which encodes P-glycoprotein, is uniformly increased at the promoter level following HDI exposure (47, 77, 78). ABCG2, a drug efflux transporter with a differing spectrum of activity, is also frequently up-regulated following HDI exposure in in vitro models (79). To the extent these multidrug transporters can reduce intracellular drug concentrations in vivo, then efficacy of a chemotherapy substrate could be reduced in combination with the HDI.
Epigenetic Therapies Reach Main Street
Although the cancer communityhas become acutelyaware of epigenetic aberrations in cancer, and therapies aimed at normalizing the epigenetic profile of the cancer cell are now available and in the clinic, much is yet to be discovered. Although we know the cellular target for current epigenetic therapy, we do not really understand how these targets play a role in cancer, or what we could actuallyexpect if we had a drug that fullynormalized the epigenetic profile. Would this in effect be the differentiation therapy that was imagined from the 1970s onward? Would we, in fact, completely reverse the malignant phenotype, especially in solid tumors? It seems doubtful given the array of oncogenic mutations now understood to be involved in human cancer.
It is interesting to note that both of our clinically active classes of epigenetic therapies involve inhibition of enzymes that work to silence DNA. As enzyme inhibitors, the target is clear; only their specificity for a given HDAC or DNMT remains to be worked out. However, the critical downstream events that follow enzyme inhibition and determine cell death are not elucidated, with HDIs even less so than the DNMT inhibitors. Both hypomethylating agents and HDIs invoke multiple changes in cells that sometimes cause cell death and sometimes promote cell survival. Downstream events resulting from these therapies may provoke disparate results. Both agents have numerous potential avenues of synergy in treating cancer. One of the most compelling strategies is to use the epigenetic therapies as radiation sensitizers, as we consider DNA to be a target of both (80).
It must be recognized that any discussion of epigenetic therapies for the future must go beyond DNA methylation and histone deacetylase inhibition. Because epigenetic modifications have many components, it is exceedingly likely that other as yet unidentified therapeutic targets exist. It is critical that investigators evaluate such targets, develop active compounds, and then establish proof of concept clinical trials in a much shorter time frame than it took to develop our current epigenetic therapies. Potential “other” epigenetic therapies include histone acetyltransferase (HAT) inhibitors (recall that as many genes are down-regulated following HDAC inhibition as up-regulated), inhibitors of HDAC class III enzymes such as sirtuin inhibitors, EZH2 antagonists to prevent perpetuation of DNA methylation, and histone methyltransferase inhibitors (81–83). Although HAT activity provides the acetyl groups that allow gene expression following inhibition of HDACs, it is interesting to note that in some cancer models increased HAT activity has been associated with tumor progression and inhibition with an anticancer effect (81).
Determining and exploiting clinically beneficial effects is a major goal for the future. The pleiotropic nature of the molecular effects of chromatin modifying agents is perhaps not surprising, given the ubiquitous nature of the methyl transferase and histone deacetylase enzymes they target. It remains to be determined if more specific agents will be more or less beneficial, or enjoy a greater therapeutic window. We do know that most of our successful anticancer agents have multiple mechanisms of action.
Further progress also requires the development of better methods to detect the epigenetic modifications and a clearer understanding of factors that drive these changes. The inclusion of epigenetics in the NIH roadmap indicates recognition of its role in human disease including but certainly not limited to malignancy. At least two large consortia have taken the lead in attempting to map the epigenetic modifications in detail over a defined region of the human genome. The American Association of Cancer Research has launched the Human Epigenome Task Force, whereas the Human Epigenome Project (HEP) has its origins in the United Kingdom. HEP has focused initially on DNA methylation profiling whereas the Human Epigenome Task Force will also examine histone modifications associated with histone acetylation patterns in addition to DNA methylation. In 2006, HEP released data including 1.9 million CpG methylation values, obtained from the analysis of 2,524 amplicons across chromosomes 6, 20, and 22 in 43 samples (84). This work is in its infancy but has the potential to give us many more clues to the role of epigenetics in cancer, beyond a lifetime of individual gene promoter studies.
With the approval of three epigenetic agents and additional ones in the pipeline, we have the beginnings of a toolbox for manipulating the epigenome. Welcome to Main Street.
AcknowledEgments
The authors would like to acknowledge the expert assistance of Dr. Robert Robey in preparing this manuscript.
Footnotes
Epigenetics—the study of stable genetic modifications that result in changes in gene expression and function without a corresponding alteration in DNA sequence. The epigenome is a catalog of the epigenetic modifications that occur in the genome. Epigenetic changes have been associated with disease, but further progress requires the development of better methods to detect the modifications and a clearer understanding of factors that drive these changes. (National Institutes of Health Roadmap)
Disclosure of Potential Conflicts of Interest
The authors have received research support from Gloucester Pharmaceuticals through a Cooperative Research and Development Agreement (CRADA) between the National Cancer Institute and Gloucester Pharmaceuticals.
References
- 1.Nowell PC. Differentiation of human leukemic leukocytes in tissue culture. Exp Cell Res 1960;19:267–77. PubMed doi: 10.1016/0014-4827(60)90007-0. [DOI] [PubMed] [Google Scholar]
- 2.Tsiftsoglou AS, Pappas IS, Vizirianakis IS. Mechanisms involved in the induced differentiation of leukemia cells. Pharmacol Ther 2003;100:257–90. PubMed doi: 10.1016/jpharmthera.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Constantinides PG, Jones PA, GeversW. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature 1977;267: 364–6. PubMed doi: 10.1038/267364a0. [DOI] [PubMed] [Google Scholar]
- 4.Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 1979;17:771–9. PubMed doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA,Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell 1980;20:85–93. PubMed doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 6.Creusot F, Acs G, Christman JK. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2’-deoxycytidine. J Biol Chem 1982;257:2041–8. PubMed. [PubMed] [Google Scholar]
- 7.Hoffman R, Murnane M, Benz EJ, et al. Induction of erythropoietic colonies in a human chronic myelogenous leukemia cell line. Blood 1979;54:1182–7. PubMed. [PubMed] [Google Scholar]
- 8.Lozzio CB, Lozzio BB, Machado EA, Fuhr JE, Lair SV, Bamberger EG. Effects of sodium butyrate on human chronic myelogenous leukaemia cell line K562. Nature 1979;281:709–10. PubMed doi: 10.1038/281709b0. [DOI] [PubMed] [Google Scholar]
- 9.Kruh J. Effects of sodium butyrate, a new pharmacological agent, on cells in culture. Mol Cell Biochem 1982;42:65–82. PubMed. [DOI] [PubMed] [Google Scholar]
- 10.Ley TJ, DeSimone J, Anagnou NP, et al. 5-azacytidine selectively increases gamma-globin synthesis in a patient with beta+ thalassemia. N Engl J Med 1982; 307:1469–75. PubMed. [DOI] [PubMed] [Google Scholar]
- 11.Charache S, Dover G, Smith K,Talbot CJ, Moyer M, Boyer S. Treatment of sickle cell anemia with 5-azacytidine results in increased fetal hemoglobin production and is associated with nonrandom hypomethylation of DNA around the gamma-delta-beta-globin gene complex. Proc Natl Acad Sci U S A 1983;80:4842–6. PubMed doi: 10.1073/pnas.80.15.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaminskas E, Farrell A, Abraham S, et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res 2005;11:3604–8. PubMed doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- 13.NovogrodskyA DvirA, Ravid A, et al. Effect of polar organic compounds on leukemic cells. Butyrate-induced partial remission of acute myelogenous leukemia in a child. Cancer 1983;51:9–14. PubMed doi:. [DOI] [PubMed] [Google Scholar]
- 14.Gore SD, Carducci MA. Modifying histones to tame cancer: clinical development of sodium phenylbutyrate and other histone deacetylase inhibitors. Expert Opin Investig Drugs 2000;9:2923–34. PubMed doi: 10.1517/13543784.9.12.2923. [DOI] [PubMed] [Google Scholar]
- 15.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol 2007;25:84–90. PubMed doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 16.McCabe M, Brandes J, Vertino P. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res 2009;15:3927–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai T,Toguchida J, Ohtani N,Yandell D, Rapaport J, DryjaT. Allele-specific hypermethylation of the retinoblastoma tumor-suppressor gene. Am J Hum Genet 1991;48:880–8. PubMed. [PMC free article] [PubMed] [Google Scholar]
- 18.Roman-Gomez J, Castillejo JA, Jimenez A, et al. 5’ CpG island hypermethylation is associated with transcriptional silencing of the p21 (CIP1/WAF1/SDI1) gene and confers poor prognosis in acute lymphoblastic leukemia. [comment] Blood 2002;99:2291–6. PubMed doi: 10.1182/bloodV99.7.2291. [DOI] [PubMed] [Google Scholar]
- 19.Issa J, Kantarjian H. Targeting DNA methylation. Clin Cancer Res 2009;15:3938–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin T, Jelinek J, Si J, Shu J, Issa J. Mechanisms of resistance to 5-aza-2’-deoxycytidine in human cancer cell lines. Blood 2009;113:659–67. PubMed doi: 10.1182/blood-2008-02-140038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrump D. Cytotoxicity mediated by histone deacetylase inhibitors: mechanisms and potential clinical implications. Clin Cancer Res 2009;15:3947–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gui CY Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A 2004;101:1241–6. PubMed doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci U S A 2000; 97:10014–9. PubMed doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandor V, Senderowicz A, Mertins S, et al. P21-dependent g(1)arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br J Cancer 2000;83:817–25. PubMed doi: 10.1054/bjoc.2000.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Lu S,Wu L, et al. Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21(Waf1/Cip1). Mol Cell Biol 2006;26:2782–90. PubMed doi: 10.1128/MCB.26.7.2782-2790.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai Y, Rahmani M, Dent P, Grant S. Blockade of histone deacetylase inhibitor-induced RelA/p65 acetylation and NF-kappaB activation potentiates apoptosis in leukemia cells through a process mediated by oxidative damage, XIAP downregulation, and c-Jun N-terminal kinase 1 activation. Mol Cell Biol 2005;25: 5429–44. PubMed doi: 10.1128/MCB.25.13.5429-5444.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spange S, WagnerT, Heinzel T, Krämer O. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol 2009;41: 185–98. PubMed doi: 10.1016/jbiocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y,Wang S, Zhang X, et al. FK228 inhibits Hsp90 chaperone function in K562 cells via hyperacetylation of Hsp70. Biochem Biophys Res Commun 2007;356:998–1003. PubMed doi: 10.1016/jbbrc.2007.03.076. [DOI] [PubMed] [Google Scholar]
- 29.Nishioka C, Ikezoe T, Yang J, Takeuchi S, Koeffler H, Yokoyama A. MS-275, a novel histone deacetylase inhibitor with selectivity against HDAC1, induces degradation of FLT3 via inhibition of chaperone function of heat shock protein 90 in AML cells. Leuk Res 2008;32:1382–92. PubMed doi: 10.1016/jleukres.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 30.Robbins AR, Jablonski SA,Yen TJ, et al. Inhibitors of histone deacetylases alter kinetochore assembly by disrupting pericentromeric heterochromatin. Cell Cycle 2005;4:717–26. PubMed. [DOI] [PubMed] [Google Scholar]
- 31.Kim SH, Jeong JW, Park JA, et al. Regulation of the HIF-1alpha stability by histone deacetylases. Oncol Rep 2007;17:647–51. PubMed. [PubMed] [Google Scholar]
- 32.Suzuki T, Yokozaki H, Kuniyasu H, et al. Effect of trichostatin A on cell growth and expression of cell cycle-and apoptosis-related molecules in human gastric and oral carcinoma cell lines. Int J Cancer 2000;88:992–7. PubMed doi:. [DOI] [PubMed] [Google Scholar]
- 33.Dong G,Wang L,Wang C,Yang T, Kumar M, Dong Z. Induction of apoptosis in renal tubular cells by histone deacetylase inhibitors, a family of anticancer agents. J Pharmacol ExpTher 2008;325:978–84. PubMed doi: 10.1124/jpet.108.137398. [DOI] [PubMed] [Google Scholar]
- 34.Rosato RR, Almenara JA, Grant S. The histone deacetylase inhibitor MS-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21CIP1/WAF1 1. Cancer Res 2003;63:3637–45. PubMed. [PubMed] [Google Scholar]
- 35.Martirosyan A, Leonard S, Shi X, Griffith B, Gannett P, Strobl J. Actions of a histone deacetylase inhibitor NSC3852 (5-nitroso-8-quinolinol) link reactive oxygen species to cell differentiation and apoptosis in MCF-7 human mammary tumor cells. J Pharmacol ExpTher 2006;317:546–52. PubMed doi: 10.1124/jpet.105.096891. [DOI] [PubMed] [Google Scholar]
- 36.Catley L, Weisberg E, Kiziltepe T, et al. Aggresome induction by proteasome inhibitor bortezomib and alpha-tubulin hyperacetylation by tubulin deacetylase (TDAC) inhibitor LBH589 are synergistic in myeloma cells. Blood 2006;108:3441–9. PubMed doi: 10.1182/blood-2006-04-016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue H, Shiraki K, Ohmori S, et al. Histone deacetylase inhibitors sensitize human colonic adenocarcinoma cell lines toTNF-related apoptosis inducing ligandmediated apoptosis. Int J Mol Med 2002;9:521–5. PubMed. [PubMed] [Google Scholar]
- 38.Guo F, Sigua C,Tao J, et al. Cotreatment with histone deacetylase inhibitor LAQ824 enhances Apo-2L/tumor necrosis factor-related apoptosis inducing ligand-induced death inducing signaling complex activity and apoptosis of human acute leukemia cells. Cancer Res 2004;64:2580–9. PubMed doi: 10.1158/0008-5472.CAN-03-2629. [DOI] [PubMed] [Google Scholar]
- 39.Weiser TS, Ohnmacht GA, Guo ZS, et al. Induction of MAGE-3 expression in lung and esophageal cancer cells. AnnThorac Surg 2001;71:295–301; discussion 301 −2. [DOI] [PubMed] [Google Scholar]
- 40.Weiser TS, Guo ZS, Ohnmacht GA, et al. Sequential 5-Aza-2’-deoxycytidine-depsipeptide FR901228 treatment induces apoptosis preferentially in cancer cells and facilitates their recognition by cytolytic T lymphocytes specific for NY-ESO-1. J Immunother (1991) 2001;24:151–61. doi: 10.1097/00002371-200103000-00010. [DOI] [PubMed] [Google Scholar]
- 41.Munshi A, Kurland J, Nishikawa T, et al. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin Cancer Res 2005;11:4912–22. PubMed doi: 10.1158/1078-0432.CCR-04-2088. [DOI] [PubMed] [Google Scholar]
- 42.Rosato RR, Almenara JA, Maggio SC, et al. Role of histone deacetylase inhibitor-induced reactive oxygen species and DNA damage in LAQ-824/fludarabine antileukemic interactions. Mol Cancer Ther 2008;7: 3285–97. PubMed doi: 10.1158/1535-7163.MCT-08-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geng L, Cuneo K, Fu A, Tu T, Atadja P, Hallahan D. Histone deacetylase (HDAC) inhibitor LBH589 increases duration of gamma-H2AX foci and confines HDAC4 to the cytoplasm in irradiated non-small cell lung cancer. Cancer Res 2006;66: 11298–304. PubMed doi: 10.1158/0008-5472.CAN-06-0049. [DOI] [PubMed] [Google Scholar]
- 44.Prince H, Bishton M, Harrison S. Clinical studies of histone deacetylase inhibitors. Clin Cancer Res. In press 2009. [DOI] [PubMed] [Google Scholar]
- 45.Kummar S, Gutierrez M, Gardner ER, et al. Phase I trial of MS-275, a histone deacetylase inhibitor, administered weekly in refractory solid tumors and lymphoid malignancies. Clin Cancer Res 2007;13:5411–7 PubMed doi: 10.1158/1078-0432.CCR-07-0791. [DOI] [PubMed] [Google Scholar]
- 46.Gojo I, Jiemjit A,Trepel J, et al. Phase 1 and pharmacologic study of MS-275, a histone deacetylase inhibitor, in adults with refractory and relapsed acute leukemias. Blood 2007;109:2781–90. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robey RW, Zhan Z, Piekarz RL, Kayastha GL, Fojo T, Bates SE. Increased MDR1 expression in normal and malignant peripheral blood mononuclear cells obtained from patients receiving depsipeptide (FR901228, FK228, NSC630176). Clin Cancer Res 2006;12:1547–55. PubMed doi: 10.1158/1078-0432.CCR-05-1423. [DOI] [PubMed] [Google Scholar]
- 48.Berg SL, Stone J, Xiao JJ, et al. Plasma and cerebrospinal fluid pharmacokinetics of depsipeptide (FR901228) in nonhuman primates. Cancer Chemother Pharmacol 2004;54:85–8. http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids=15042312&dopt=AbstractPubMeddoi:10.1007/s00280-004-0766-5. [DOI] [PubMed] [Google Scholar]
- 49.LÖscher W, Potschka H. Role of multidrug transporters in pharmacoresistance to antiepileptic drugs. J Pharmacol ExpTher 2002;301:7–14. PubMed doi: 10.1124/jpet.301.1.7. [DOI] [PubMed] [Google Scholar]
- 50.Piekarz RL, Frye R,Turner M, et al. A multi-institutional phase II trial of the HDAC inhibitor romidepsin as monotherapy for patients with cutaneousT-cell lymphoma. J Clin Oncol. In press 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steadman K, Stein W, Litman T, et al. PolyHEMA spheroids are an inadequate model for the drug resistance of the intractable solid tumors. Cell Cycle 2008; 7:818–29. [DOI] [PubMed] [Google Scholar]
- 52.Burgess A, Ruefli A, Beamish H, et al. Histone deacetylase inhibitors specifically kill nonproliferating tumour cells. Oncogene 2004;23:6693–701. PubMed doi: 10.1038/sjonc.1207893. [DOI] [PubMed] [Google Scholar]
- 53.Sandor V,Bakke S,Robey RW,et al. Phase I trialofthe histonedeacetylase inhibitor,depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin Cancer Res 2002;8:718–28. [PubMed] [Google Scholar]
- 54.Stadler WM, Margolin K, Ferber S, McCulloch W, Thompson JA. A phase II study of depsipeptide in refractory metastatic renal cell cancer. Clin Genitourin Cancer 2006;5:57–60. PubMed doi: 10.3816/CGC.2006.n.018. [DOI] [PubMed] [Google Scholar]
- 55.Rosato RR, Almenara JA,Yu C, Grant S. Evidence of a functional role for p21WAF1/CIP1 down-regulation in synergistic antileukemic interactions between the histone deacetylase inhibitor sodium butyrate and flavopiridol. Mol Pharmacol 2004;65:571–81. PubMed doi: 10.1124/mol.65.3.571. [DOI] [PubMed] [Google Scholar]
- 56.Duan J, Friedman J, Nottingham L, Chen Z, Ara G,Van Waes C. Nuclear factor-kappaB p65 small interfering RNA or proteasome inhibitor bortezomib sensitizes head and neck squamous cell carcinomas to classic histone deacetylase inhibitors and novel histone deacetylase inhibitor PXD101. Mol Cancer Ther 2007;6:37–50. PubMed doi: 10.1158/1535-7163.MCT-05-0285. [DOI] [PubMed] [Google Scholar]
- 57.Shao Y, Gao Z, Marks P, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA 2004;101: 18030–5. PubMed doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amaravadi RK,Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res 2007;13:7271–9. PubMed doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 59.SjÖblom T, Jones S, Wood L, et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006;314:268–74. PubMed doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 60.Bots M, Johnstone R. Rational combinations using HDAC inhibitors. Clin Cancer Res 2009;15:3970–7. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen DM, Schrump WD, Chen GA, et al. Abrogation of p21 expression by flavopiridol enhances depsipeptide-mediated apoptosis in malignant pleural mesothelioma cells. Clin Cancer Res 2004;10:1813–25 PubMed doi: 10.1158/1078-0432.CCR-0901-3. [DOI] [PubMed] [Google Scholar]
- 62.Rosato RR, Almenara JA, Cartee L, BettsV, Chellappan S, Grant S. The cyclin-dependent kinase inhibitor flavopiridol disrupts sodium butyrate-induced p21WAF1/CIP1 expression and maturation while reciprocally potentiating apoptosis in human leukemia cells. Mol CancerTher 2002;1:253–66. [PubMed] [Google Scholar]
- 63.Glaser KB, Li J, Pease LJ, et al. Differential protein acetylation induced by novel histone deacetylase inhibitors. Biochem Biophys Res Commun 2004;325:683–90. PubMed doi: 10.1016/jbbrc.2004.10.082. [DOI] [PubMed] [Google Scholar]
- 64.Banerji U. Heat shock protein 90 as a drug target: some like it hot. Clin Cancer Res 2009;15:9–14. PubMed doi: 10.1158/1078-0432.CCR-08-0132. [DOI] [PubMed] [Google Scholar]
- 65.Bali P, Pranpat M, Swaby R, et al. Activity of suberoylanilide hydroxamic acid against human breast cancer cells with amplification of her-2. Clin Cancer Res 2005;11:6382–9. PubMed doi: 10.1158/1078-0432.CCR-05-0344. [DOI] [PubMed] [Google Scholar]
- 66.Drummond DC, Marx C, Guo Z, et al. Enhanced pharmacodynamic and antitumor properties of a histone deacetylase inhibitor encapsulated in liposomes or ErbB2-targeted immunoliposomes. Clin Cancer Res 2005;11:3392–401. PubMed doi: 10.1158/1078-0432.CCR-04-2445. [DOI] [PubMed] [Google Scholar]
- 67.Fuino L, Bali P, Wittmann S, et al. Histone deacetylase inhibitor LAQ824 down-regulates Her-2 and sensitizes human breast cancer cells to trastuzumab, taxotere, gemcitabine, and epothilone B. Mol Cancer Ther 2003;2:971–84. [PubMed] [Google Scholar]
- 68.Witta SE, Gemmill RM, Hirsch FR, et al. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res 2006;66:944–50. PubMed doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- 69.Fiskus W, Pranpat M, Bali P, et al. Combined effects of novel tyrosine kinase inhibitor AMN107 and histone deacetylase inhibitor LBH589 against Bcr-Abl-expressing human leukemia cells. Blood 2006;108:645–52. PubMed doi: 10.1182/blood-2005-11-4639. [DOI] [PubMed] [Google Scholar]
- 70.Tsai SC, Valkov N, Yang WM, Gump J, Sullivan D, Seto E. Histone deacetylase interacts directly with DNA topoisomerase II. Nat Genet 2000;26:349–53. PubMed doi: 10.1038/81671. [DOI] [PubMed] [Google Scholar]
- 71.Kurz EU,Wilson SE, Leader KB, et al. The histone deacetylase inhibitor sodium butyrate induces DNA topoisomerase II alpha expression and confers hypersensitivity to etoposide in human leukemic cell lines. Mol CancerTher 2001;1:121–31. [PubMed] [Google Scholar]
- 72.Chen CS,Wang YC,Yang HC, et al. Histone deacetylase inhibitors sensitize prostate cancer cells to agents that produce DNA double-strand breaks by targeting Ku70 acetylation. Cancer Res 2007;67:5318–27. PubMed doi: 10.1158/0008-5472.CAN-06-3996. [DOI] [PubMed] [Google Scholar]
- 73.Subramanian C, Opipari AJ, Bian X, Castle V, Kwok R. Ku70 acetylation mediates neuroblastoma cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci U S A 2005;102:4842–7. PubMed doi: 10.1073/pnas.0408351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bevins RL, Zimmer SG. It’s about time: scheduling alters effect of histone deacetylase inhibitors on camptothecin-treated cells. Cancer Res 2005;65: 6957–66. PubMed doi: 10.1158/0008-5472.CAN-05-0836. [DOI] [PubMed] [Google Scholar]
- 75.Rosato RR, Maggio SC, Almenara JA, et al. The histone deacetylase inhibitor LAQ824 induces human leukemia cell death through a process involving XIAP down-regulation, oxidative injury, and the acid sphingomyelinase-dependent generation of ceramide. Mol Pharmacol 2006;69:216–25. [DOI] [PubMed] [Google Scholar]
- 76.Kim MS, Blake M, Baek JH, Kohlhagen G, Pommier Y, Carrier F. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res 2003;63:7291–300. [PubMed] [Google Scholar]
- 77.Jin S, Scotto KW. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol 1998; 18: 4377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tabe Y, Konopleva M, Contractor R, et al. Up-regulation of MDR1 and induction of doxorubicin resistance by histone deacetylase inhibitor depsipeptide (FK228) and ATRA in acute promyelocytic leukemia cells. Blood 2006;107:1546–54. PubMed doi: 10.1182/blood-2004-10-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.To KK, Polgar O, Huff LM, Morisaki K, Bates SE. Histone modifications at the ABCG2 promoter following treatment with histone deacetylase inhibitor mirror those in multidrug-resistant cells. Mol Cancer Res 2008;6:151–64. PubMed doi: 10.1158/1541-7786.MCR-07-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Schutter H, Nuyts S. Radiosensitizing potential of epigenetic anticancer drugs. Anticancer Agents Med Chem 2009;9:99–108. [DOI] [PubMed] [Google Scholar]
- 81.Chandregowda V, Kush A, Reddy G. Synthesis of benzamide derivatives of anacardic acid and their cytotoxic activity. Eur J Med Chem 2009;44:2711–9. PubMed doi: 10.1016/jejmech.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 82.Lara E, Mai A, Calvanese V, et al. Salermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene 2009;28:781–91. PubMed doi: 10.1038/onc.2008.436. [DOI] [PubMed] [Google Scholar]
- 83.Wang H, Huang Z, Xia L, et al. Methylation of histone H4at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 2001;293: 853–7. PubMed doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 84.Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 2006;38:1378–85. PubMed doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]