Abstract
p97 is a hexameric AAA+ adenosine triphosphatase (ATPase) that is an attractive target for cancer drug development. We report cryo–electron microscopy (cryo-EM) structures for adenosine diphosphate (ADP)–bound, full-length, hexameric wild-type p97 in the presence and absence of an allosteric inhibitor at resolutions of 2.3 and 2.4 angstroms, respectively. We also report cryo-EM structures (at resolutions of ~3.3, 3.2, and 3.3 angstroms, respectively) for three distinct, coexisting functional states of p97 with occupancies of zero, one, or two molecules of adenosine 5′-O-(3-thiotriphosphate) (ATPγS) per protomer. A large corkscrew-like change in molecular architecture, coupled with upward displacement of the N-terminal domain, is observed only when ATPγS is bound to both the D1 and D2 domains of the protomer. These cryo-EM structures establish the sequence of nucleotide-driven structural changes in p97 at atomic resolution. They also enable elucidation of the binding mode of an allosteric small-molecule inhibitor to p97 and illustrate how inhibitor binding at the interface between the D1 and D2 domains prevents propagation of the conformational changes necessary for p97 function.
The super family of AAA+ proteins(ATPases associated with diverse cellular activities) consists of molecular chaperones that serve major roles in cellular protein quality control; DNA replication, transcription, recombination, and repair; membrane fusion; movement of intracellular cargo; and cell cycle regulation (1, 2). These oligomeric ring-like enzymes, which typically form hexamers, contain common functional features including one or more conserved AAA+ ATPase cassettes in each protomer. These cassettes bind and hydrolyze ATP at the interface between adjacent subunits. The acquired energy from binding and catalysis of nucleotides induces a series of conformational changes to enable these enzymes to modulate their substrates. p97, a well-characterized member of this protein family, binds multiple proteins including deubiquitylating enzymes as well as ubiquitin-binding adaptors and ligases (3–7). The central role that p97 plays in protein quality control makes it an attractive target for cancer chemotherapy, as disruption of both of these processes hinders cancer cell survival (8). Up-regulation of p97 expression in canscer cells supports this premise (9), as do reports of numerous structurally diverse inhibitor types (10–14), including a potent inhibitor that entered a phase I clinical trial in 2015 (15).
Structurally, each p97 protomer has a predicted molecular weight of ~90 kDa and comprises an N-terminal domain (henceforth N domain), two tandem ATPase domains (designated D1 and D2) that pack as hexameric rings, and a short C-terminal domain. Structural studies of full-length p97 by x-ray crystallography have so far been limited to medium resolution (3.5 Å to 4.7 Å) (16), although higher-resolution structures of subcomplexes that contain D1 and the N domain or the D2 domain have revealed further architectural insights in these regions of the protein (16–20). Determination of the atomic-resolution structure of native, full-length p97 in different conformational states and in complex with inhibitors is therefore of considerable biological and clinical interest.
Using cryo-EM, we first performed structural analysis of p97 without addition of exogenous nucleotides. This structure, determined at an overall resolution of 2.4 Å, shows the expected architecture of the hexameric complex (fig. S1, A and B). ADP occupies both the D1 and D2 domain nucleotide-binding pockets (fig. S1, C and D). Analysis of the conformational variants that emerge during three-dimensional classification suggests that subtle conformational heterogeneity in the ADP-bound D2 hexamer (fig. S2) is a likely reason for the lower resolutions achieved in x-ray crystallographic analyses of full-length p97. Overall, the cryo-EM–derived structure of the D2 domain in the full-length hexamer is comparable to that determined for full-length p97 at 3.5 Å by x-ray crystallography, except for some differences in the relative disposition of the N domain (16) and better definition of the C-terminal a-helical subdomain that spans amino acid residues 645 to 763.
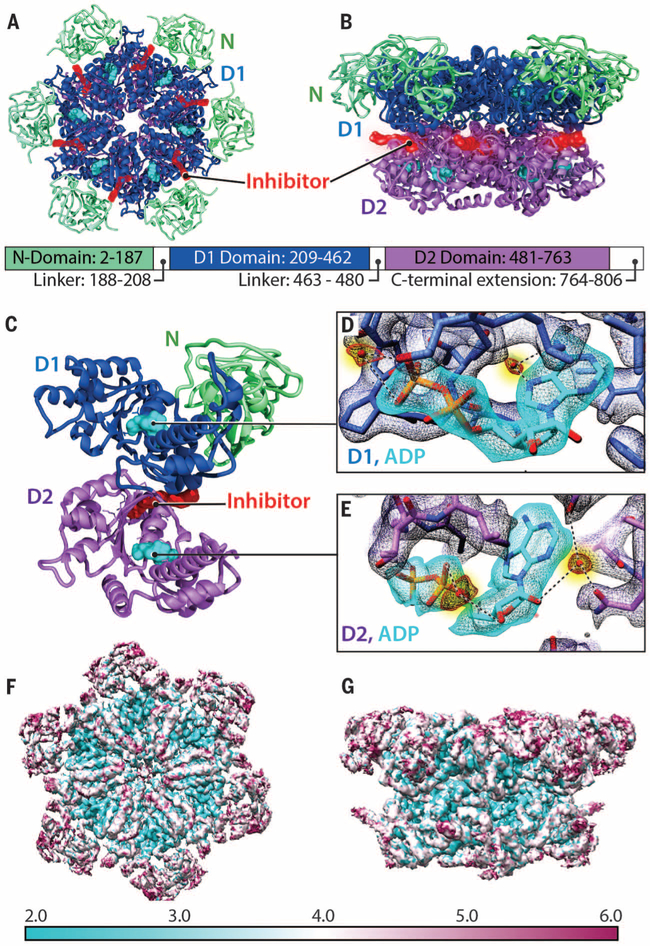
Several classes of inhibitors, including those that act in an allosteric or competitive manner, have been identified that impair p97 ATPase activity (10–14, 21–27). Determination of the cryo-EM structure at 2.3 Å resolution of p97 in complex with UPCDC30245, a phenyl indole derivative (fig. S3) that is structurally related to a recently described series of allosteric inhibitors (14), shows that this compound (half-maximal inhibitory concentration ~27 nM) binds at the junction between the D1 and D2 domains (Fig. 1, A and B). A detailed view of the structure of a single p97 protomer shows the location of the inhibitor and bound ADP (Fig. 1, C to E), including features such as the interaction of the ADP β-phosphate with a water molecule in the D1 domain (Fig. 1D) and hydrogen bonding of a water molecule to the α-phosphate moiety and to the hydroxyl group on the ribose ring proximal to the α-phosphate group (Fig. 1E). Furthermore, the distal hydroxyl group of the ribose ring interacts with a water molecule hydrogen-bonded to the side-chain oxygen atom of Asn660 and the main-chain carbonyl atom of residue Asp478. Local map resolution measured by ResMap (28) indicates that the density is weakest in the N domain, consistent with plots of B-factor profiles of the refined structure, which show that the average B-factor is highest in the N domain and in the D1-D2 connector region and lowest in the core of the D1 and D2 domains (Fig. 1, F and G, and fig. S4). The most ordered regions are at the p97 core, with a clear gradient toward higher B-factors at the periphery (movie S1).
Fig. 1. Atomic-resolution model derived from cryo-EM structure of p97 in the presence of bound inhibitor.
(A and B) Top and side views, respectively, of the cryo-EM structure of the p97 hexamer presented as a ribbon diagram, showing the N (green), D1 (blue), and D2 (purple) domains. The ADP molecule is colored cyan. The inhibitor (red) is bound at the junction between the D1 and D2 domains. The relative position of each domain in the primary sequence is indicated. (C) Ribbon diagram of a p97 protomer highlighting the location of the bound inhibitor (red) relative to the two bound nucleotides (cyan) in D1 and D2 domains. (D and E) Density maps for bound nucleotides, establishing that ADP is bound to both D1 and D2 domains, and visualization of densities for tightly bound water molecules (colored red, highlighted in yellow) at the nucleotide-binding sites. (F and G) Top and side views, respectively, of the structure (uncorrected density map), color-coded to represent variation in resolution across the protein as determined using ResMap (28).
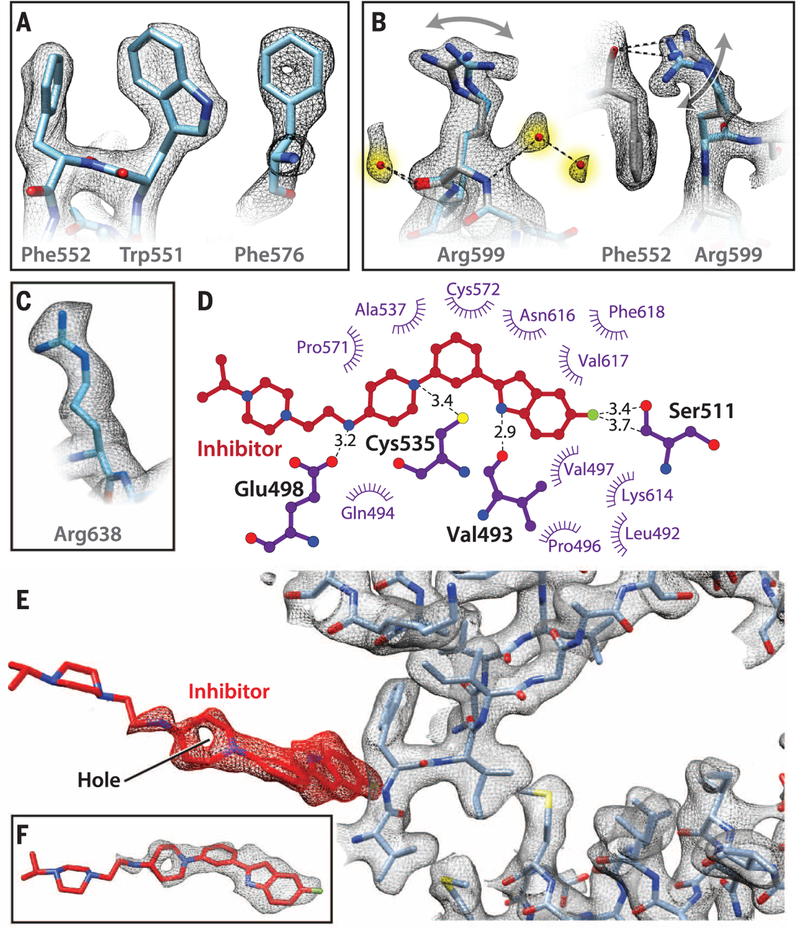
Selected examples of the quality of the cryo-EM map in the D2 domain (residues 481 to 763) are presented in Fig. 2, A to C. It is conceivable that the two discrete conformations of the Arg599 side chain (Fig. 2B) are relevant to the ability of p97 to locally adapt to structural variations in bound substrates because this residue has been implicated in substrate interactions in the pore region surrounded by the D2 hexamer. UPCDC30245 binding did not result in significant changes in conformation of the polypeptide backbone, including at the nucleotide-binding sites (fig. S5). Visualization of density for the bound inhibitor in the map enabled determination of its conformation in the bound state and the local interactions involved in its binding at the D1-D2 interface. The inhibitor conformation is most ordered in the half that includes the indole ring that nestles into a binding pocket (fig. S6A). This inhibitor-binding site is sandwiched by the C-terminal regions of two α-helical segments that range from residues 483 to 498 and 523 to 534 (Fig. 2D). The binding tunnel is capped at the end where the fluorinated indole ring is docked between two loop elements (amino acid residues 506 to 512 and 610 to 618) that lead into two parallel b strands. The other end of the binding cleft is open and solvent-exposed (Fig. 2, E and F), with the piperazine ring of the inhibitor protruding outward into the bulk solvent. Residues that interact with ADP in the D1 and D2 sites are summarized in fig. S6, B and C, respectively.
Fig. 2. Depiction of cryo-EM map quality and inhibitor interactions.
(A) Illustration of hydrophobic packing between the aromatic side chains of Trp551 and Phe552, and the presence of a hole in the aromatic ring of Phe576. (B) Density for water molecules hydrogen-bonded to the Arg599 backbone and evidence for alternate conformations of the terminal guanidinium group, shown in orthogonal orientations. The main-chain oxygen atom of Phe552 shown in (A) is hydrogen-bonded to the Arg599 guanidinium moiety. (C) Density for Arg638, part of the “arginine finger” motif at the outer surface of the D2 domain. (D) LIGPLOT representation showing residues in p97 that are in close proximity to the inhibitor, highlighting key H-bond interactions between the nitrogen on the indole ring and the backbone carbonyl oxygen atom of Val493 and side chain of Glu498 with the nitrogen atom in the linker of the inhibitor. The interaction between the fluorine atom present on the indole ring of the inhibitor and the main-chain carbonyl atom of Ser511 is also indicated. (E and F) Close-up view of the conformation of UPCDC30245 bound to p97 shown along with selected residues in its vicinity. There is strong density for the bound inhibitor for the segment encompassing the indole group to the piperidine ring and virtually no density at the other extremity, where the molecule is expected to be highly flexible because of minimal interactions with the protein.
The key interactions that anchor the inhibitor arise from the main-chain carbonyl oxygen of Val493 hydrogen-bonded to the N-H of the indole ring (2.9 Å), weak interaction of the fluorine atom present on the indole ring with the main-chain carbonyl oxygen atom of Ser511, hydrogen bonding of the thiol group of Cys535 to the nitrogen atom in the piperidine ring (3.4 Å), and the side chain of Glu498 hydrogen-bonded to the nitrogen atom in the linker between the piperidine and piperazine groups (3.2 Å). The rest of the binding environment is largely hydrophobic in nature, stemming from interactions with residues Phe618, Pro496, Pro510, Pro571, Ala537, and Val497 (Fig. 2D). Interactions at the indole ring end are central to the binding of UPCDC30245 to p97, with a potential edge-to-face, weak π-π interaction between Phe618 and the indole ring. Consistent with these findings, recent functional studies show that substitutions at the 5-position of the indole ring (where the fluorine atom is attached) have profound effects on activity, with both steric and electronic factors being important (14). The interactions at the piperazine ring end are minimal, suggesting high conformational flexibility in this region, although the highly negatively charged outer surface of the binding tunnel (fig. S6A) may enable better anchoring to the surface of p97. Mutagenesis studies have shown that the D1-D2 interface is important for p97 function (11, 12) and provide support for the structural studies we report here.
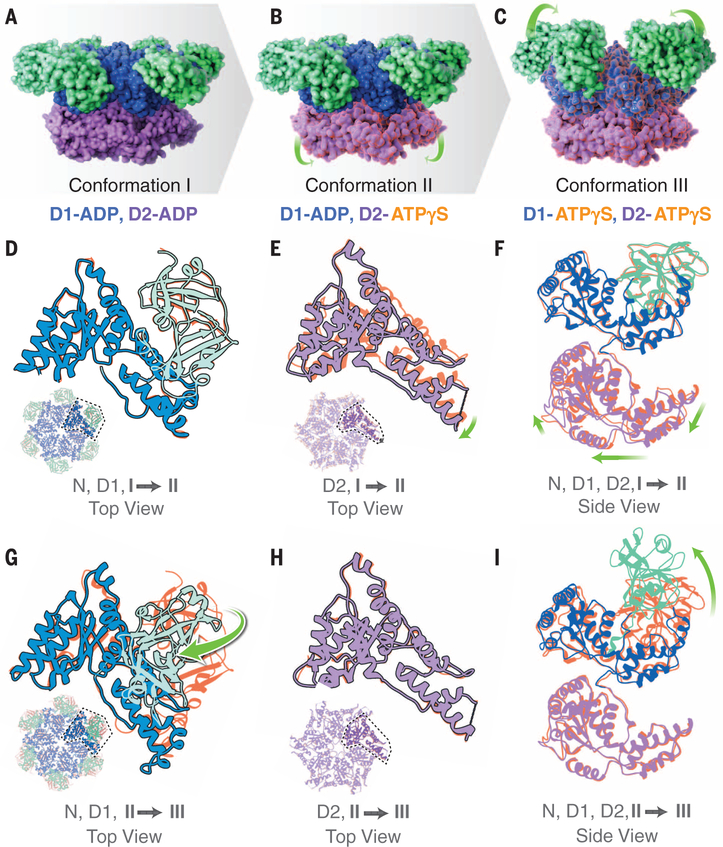
To unravel the intra- and interprotomer conformational changes that occur with ATP binding, we performed structural analysis of p97 in the presence of ATPγS at physiologically relevant concentrations. Earlier crystallographic studies of p97 with ATP analogs revealed only modest structural changes (16), raising the concern that crystal packing constraints may have prevented capturing functionally relevant protein conformational changes. Cryo-EM methods have been applied previously to study p97 conformational changes; however, the resulting structures were at very low resolutions [~20 to 30 Å (29–31)] and did not result in definitive identification of the structural changes relative to the ADP-bound conformation. Here, we report separation of three well-defined, coexisting conformational states at near-atomic resolution (~3.2 to 3.3 Å) that are simultaneously populated when ATPγS is added to p97 (Fig. 3 and figs. S7 and S8). In conformation I, ADP is bound in both D1 and D2 nucleotide-binding domains; in conformation II, ADP is bound in the D1 domain while ATPγS is bound in the D2 domain; and in conformation III, ATPγS is bound in the nucleotide-binding regions of both D1 and D2.
Fig. 3. Cryo-EM structures at ~3.3 Å, ~3.2 Å, and ~3.3 Å resolution, respectively, of three distinct p97 conformational states populated upon addition of ATPγS.
(A to C) Side views of molecular surface models of the three states, color-coded to show the N, D1, and D2 domains in green, blue, and purple, respectively. The green arrows indicate the motion of the D2 domain in the transition from conformation I to II (B) and the motion of the N domain in the transition from conformation II to III (C). (D to F) Superposition of the polypeptide backbones of conformations I and II in ribbon representation to illustrate that the N and D1 domains display similar conformations but that there are substantial differences in the D2 domain, as indicated by the green arrows. The color scheme for conformation II is as in (A) to (C), with conformation I shown in orange. (G to I) Superposition of the polypeptide backbones of conformations II and III in ribbon representation to illustrate that the D2 domain is similar but that there are substantial differences in conformation of the N and D1 domains, as indicated by the green arrows. The color scheme for conformation III is as in (A) to (C), with conformation II now shown in orange. The insets in (D) and (G) show top views of the D1 domain; the insets in (E) and (H) show top views of the D2 domain, providing context for the superpositions shown in the main panels.
The differences in structure among the three states reveal an unambiguous and stepwise evolution of the conformational change with binding of ATPγS (Fig. 3, A to C). Conformation II differs from I only at the D2 domain, while the N and D1 domains remain essentially unchanged (Fig. 3, D to F). The changes in the D2 domain occur largely proximal to the nucleotide-binding region and result from the exchange of the bound ADP with ATPγS, as verified by direct visualization of density for the additional phosphate moiety in the binding site (fig. S8). Within each protomer, there is a ~10° to 15° rotational twist of the D2 domain with respect to the D1 domain, with the D1-D2 linker region serving as a hinge. This relative rotational displacement of the D2 ring with respect to the D1 ring is consistent with atomic force microscopy studies that have reported ATP-dependent rotation in p97 (32).
The difference between conformation II and III is primarily in the N and D1 domains, with minimal changes in the D2 domain (Fig. 3, G to I). Here again, the change in conformation is directly associated with the exchange of ADP for ATPgS, with the density map demonstrating density for the additional phosphate moiety present in the D1 nucleotide-binding site (fig. S8). The most striking feature of this transition is the large-scale motion of the N domain away from the plane of the D1 hexamer. The structure of the N and D1 domains in conformation III is essentially the same as that reported from x-ray crystallographic analysis of an isolated N-D1 fragment derived from a mutant p97 (33). The changes include rearrangements at the peripheral helix in D1 and its interface with the D2 domain, and movement of the N domain from its original isoplanar or “down” position to an “up” position, where it is rotated by ~75° and displaced by ~14 Å. This out-of-plane conformation of the N-D1 domain and the double occupancy of both nucleotide-binding regions with ATPγS have not been reported in any of the full-length p97 structures solved by x-ray crystallography (16). Additionally, the presence of ATPγS in conformations II and III results in the stabilization of the C-terminal helix of the D2 domain and extension of the traceable density from residue 763 to residue 768 (fig. S8, G to I).
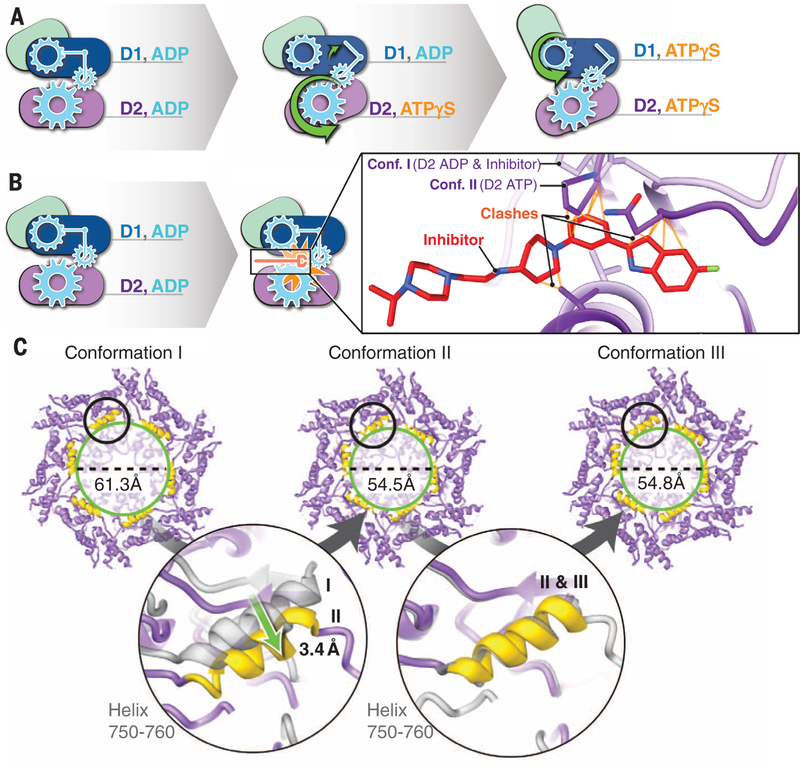
Our structural analyses thus show that the effect of ATPγS binding is a two-step, sequential conformational change in the D2 and D1 hexameric layers, respectively (Fig. 4A). Analysis of the images without imposition of six-fold symmetry shows that the majority of the molecules are in one of the three conformations obtained with the use of symmetry, indicating that the structural differences among conformations I, II, and III are cooperative in nature (fig. S9). The location of the inhibitor-binding site at the interface of domains D1 and D2 suggests that inhibitor binding prevents the nucleotide-driven conformational changes at the interface that are pivotal for p97 activity. To explore this further, we superimposed the structure of the p97-UPCDC30245 complex onto that of the ATPγS-bound p97 conformation II. The superposed models (Fig. 4B and fig. S8J) suggest that there would be steric clashes between the indole ring of the inhibitor and residues in the D2 loop (amino acid residues 611 to 616). Even if there is some flexibility in the D2 domain that enables some level of binding, key interactions that enable the binding of UPCDC30245 in the p97-ADP complex, such as that between the terminal fluorine atom and the backbone carbonyl Ser511 (Fig. 2D), cannot be achieved in the ATPγS-bound state. These observations indicate that UPCDC30245 is a conformation-selective inhibitor that preferentially binds the ADP state to act as a “wrench in the works.”
Fig. 4. Mechanism of ATP-driven conformational change.
(A) Schematic of the two-step sequence involved in p97 activation resulting from the sequential binding of ATPγS first to D2 and then to D1, resulting in a large-scale movement of the N domain. (B) Close-up view of the location of the bound inhibitor when ADP is present in both D1 and D2 domains, indicating that there is a steric clash with the structure adopted by the D2 polypeptide upon ATPγS binding and that binding of the inhibitor blocks the conformational changes. The steric clashes between conformation II and the bound inhibitor are marked. (C) View of the D2 domain in the three conformational states, illustrating narrowing of the pore diameter (green circles) upon transition from conformation I to II, with minimal further changes upon transition to conformation III. The regions marked by the black circles are highlighted in magnified form in the lower part of the figure. They show views of the protein in the vicinity of residues 750 to 760 in one protomer and illustrate the extent of the helix movement in narrowing of the pore.
The pivot-like movement of the D2 hexamer with ATPγS binding also affects pore dimensions (Fig. 4C). Thus, with ATPγS binding, the diameter of the cytoplasmic face of the pore in the center of the D2 hexamer (measured by the α carbon of residue 755 in the 750–760 helix) contracts from ~61 Å to ~54 Å (movie S2), whereas the average outer (~28 Å) and inner (~6 Å) diameters of D1 are similar in the three conformations. Many of the residues that line the central pore are negatively charged and could be important in the context of binding of ubiquitylated substrates. The cavity at the center is large enough to accommodate substrates destined for degradation, but given that the changes in size with nucleotide binding are relatively small, our results are consistent with the suggestion (16) that substrates degraded by p97 may not be threaded through the central axis across the length of the barrel.
The ability to determine atomic-resolution structures for multiple conformational states that are present simultaneously in a dynamic molecular machine such as p97 is likely to be an increasingly common signature of the application of cryo-EM methods in structural biology. p97 mediates its function in the cell by interacting with a large array of effector proteins, most of which bind at the N domain, which undergoes a large movement with ATP binding, as our present work and previous structural studies demonstrate (5, 18, 34–36). Cryo-EM studies could therefore be useful for a more detailed understanding of the structural basis of these interactions in normal cells and in diseases such as cancer. Further, the delineation of the inhibitor-bound p97 structure provides another example [besides β-galactosidase (37)] of a regulatory metabolic enzyme where cryo-EM at ~2 Å resolution enables visualization of structural detail such as hydrogen-bonded water molecules, with features comparable to those seen in model-based 2Fobs − Fcalc maps at similar reported resolutions obtained using x-ray crystallography (fig. S10). The discovery of the specific mode of binding of a compound that blocks p97 activity provides new insights into the interactions and residues that are critical for function, and may enable the design of clinically useful inhibitors.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by funds from the Center for Cancer Research, National Cancer Institute, and with federal funds from the National Cancer Institute under Chemical Biology Consortium Contract No. HHSN261200800001E Agreement No. 29XS127TO15. R.J.D. is an investigator of the Howard Hughes Medical Institute. This work used the computational resources of the NIH High-Performance Computing Biowulf cluster (http://hpc.nih.gov). We thank K. Moynihan, R. Mueller, and J. Cometa for technical assistance with electron microscopy; F. Ulmer, P. Mooney, and C. Booth for advice and assistance with optimizing K2 detector performance; and M. G. Laporte for contributions to the medicinal chemistry program. The University of Pittsburgh has filed a provisional patent application that covers the inhibitor compound described in this manuscript. R.J.D. is on the Scientific Advisory Board of Cleave Biosciences. The density maps and refined atomic models have been deposited in the Electron Microscopy Data Bank with accession numbers EMD-3295, 3296, 3297, 3298, and 3299, and in the Protein Data Bank with matching accession numbers of PDB-5FTJ, 5FTK, 5FTL, 5FTM, and 5FTN, respectively, for native p97 in the presence and absence of bound inhibitor, and for p97 conformational states I, II, and III observed in the presence of ATPγS.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/351/6275/871/suppl/DC1
Materials and Methods
Figs. S1 to S10
Movies S1 and S2
REFERENCES AND NOTES
- 1.Hanson PI, Whiteheart SW, Nat. Rev. Mol. Cell Biol 6, 519–529 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Ogura T, Wilkinson AJ, Genes Cells 6, 575–597 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Barthelme D, Sauer RT, J. Mol. Biol 10.1016/j.jmb.2015.11.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otter-Nilsson M, Hendriks R, Pecheur-Huet EI, Hoekstra D,Nilsson T, EMBO J 18, 2074–2083 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer H, Weihl CC, J. Cell Sci 127, 3877–3883 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avci D, Lemberg MK, Trends Cell Biol 25, 611–622 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Fang L et al. , Front. Cell. Neurosci 9, 16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshaies RJ, BMC Biol 12, 94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valle CW et al. , PLOS ONE 6, e29073 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou TF et al. , Proc. Natl. Acad. Sci. U.S.A 108, 4834–4839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou TF, Li K, Frankowski KJ, Schoenen FJ, Deshaies RJ, ChemMedChem 8, 297–312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magnaghi P et al. , Nat. Chem. Biol 9, 548–556 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Polucci P et al. , J. Med. Chem 56, 437–450 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Alverez C et al. , ACS Med. Chem. Lett 6, 1225–1230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson DJ et al. , “Inhibition of the AAA-ATPase p97 with the first in class inhibitor CB-5083 as a novel approach to treat cancer” Paper presented at the American Association for Cancer Research annual meeting, Philadelphia, 18 to 22 April 2015. [Google Scholar]
- 16.Davies JM, Brünger AT, Weis WI, Structure 16, 715–726 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Kim SJ et al. , J. Biol. Chem 289, 12264–12274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hänzelmann P, Buchberger A, Schindelin H, Structure 19, 833–843 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Dreveny I et al. , EMBO J 23, 1030–1039 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang WK, Xia D, J. Biol. Chem 288, 36624–36635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman E, Maksim N, de la Cruz F, La Clair JJ, Molecules 20, 3027–3049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang CJ et al. , ChemMedChem 10, 52–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou TF, Deshaies RJ, Autophagy 7, 1091–1092 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao S et al. , ACS Chem. Biol 10, 1916–1924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi P et al. , Mol. Cancer Ther 11, 2610–2620 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Brown SJ et al. , in Probe Reports from the NIH Molecular Libraries Program (National Center for Biotechnology Information, Bethesda, MD, 2010). [PubMed] [Google Scholar]
- 27.Chimenti MS et al. , J. Biomol. Screen 20, 788–800 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Kucukelbir A, Sigworth FJ, Tagare HD, Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X et al. , Mol. Cell 6, 1473–1484 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Niwa H et al. , J. Biol. Chem 287, 8561–8570 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeung HO et al. , Open Biol 4, 130142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noi K et al. , Structure 21, 1992–2002 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Tang WK et al. , EMBO J 29, 2217–2229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chia WS, Chia DX, Rao F, Bar Nun S, Geifman Shochat S, PLOS ONE 7, e50490 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beuron F et al. , EMBO J 25, 1967–1976 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almeida B et al. , Biochim. Biophys. Acta 1852, 1950–1959 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Bartesaghi A et al. , Science 348, 1147–1151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.