Abstract
Background:
Accumulated evidence has indicated the associations between single-nucleotide polymorphisms (SNPs) in microRNAs (miRNAs) and the susceptibility to diabetes mellitus (DM), but the conclusions remain controversial. This study was to investigate the true contribution of miRNA SNPs to the risk of DM by using a meta-analysis of all the published studies.
Methods:
Relevant studies were identified in the databases of PubMed and the Cochrane Library databases. The strength of associations between miRNA polymorphisms and DM risk was assessed by odds ratios (ORs) and 95% confidence intervals (95% CIs) under five genetic models using the STATA software.
Results:
Six studies, containing 2773 cases and 2632 controls, were enrolled, 5 of which evaluated miR-146a (rs2910164), 4 for miR-27a (rs895819), and 3 for miR-124 (rs531564) and 2 for miR-375 (rs6715345), miR-128a (rs11888095), miR-194a (rs3820455). The meta-analysis indicated that the G allele or GG genotype of miR-146a rs2910164 was associated with a significantly increased risk for DM compared with C allele or GC/CC genotype in Latin American population; CC genotype of miR-27a rs895819 polymorphism was associated with a significantly decreased risk for DM in Asian population compared with the TT genotype; patients carrying with CC genotype of miR-124 rs531564 had a lower probability to develop DM regardless of ethnicity; no associations were identified between polymorphisms in miR-375, miR-128a, miR-194a and the susceptibility to DM.
Conclusion:
Our study suggests that miR-146a/miR-27a and miR-124 polymorphisms may be ethnicity-dependent or -independent susceptibility factors to DM, respectively.
Keywords: diabetes mellitus, microRNAs, meta-analysis, polymorphism, single nucleotide
1. Introduction
Diabetes mellitus (DM), characterized by progressive hyperglycemia due to insulin resistance or insulin deficiency, has been a common metabolic disorder worldwide. It was estimated that 366 million people suffered from DM in 2013, and this number will rise to 592 million by 2035.[1] Patients with DM are at higher risk of developing various microvascular complications (such as nephropathy,[2] retinopathy,[3] neuropathy,[4] diabetic foot,[5] hypertension, dyslipidemia,[6] and atherosclerosis[7]) which lead to the disability and even death of patients. Thus, prediction of the population at a higher risk of DM is essential in order to reduce the incidence of DM and its related complications.
Although DM is a multifactorial disease, accumulating evidence has shown genetics, especially the single nucleotide polymorphism (SNP), is a strong contributor.[8] MicroRNAs (miRNAs) are a group of short, non-coding RNAs of approximately 25 nucleotides in length, which involve in various physiological and pathophysiological processes (such as inflammation, insulin secretion) by negatively controlling the expression of target genes via complementarily binding to their 3’-untranslated region (3’-UTR).[9,10] Thus, SNPs in miRNAs that change the expressions and functions of miRNAs may be underlying biomarkers for the prediction of DM risk. This hypothesis has been reported previously. miR-146a was found to be downregulated in DM patients[11] and model rats[12] compared with normal controls. The relative expression level of miR-146a was significantly lower in diabetic patients with the rs2910164 CC genotypes than that in GG carriers.[13] Therefore, miR-146a CC genotype may increase the risk of developing type 2 DM (T2DM), which has been validated in the study of Li et al[14] and Alipoor et al[15]. However, the controversial results were also observed recently. For example, Wang et al found that there was no association between the individuals carrying the variant genotype of the miR-146a and DM risk.[16] Assmann et al identified CC and GC genotypes were associated with protection against DM.[17] Furthermore, there was also no consensus on the association between other miRNA polymorphisms (such as rs895819 of hsa-miR-27a,[18,19] rs531564 in hsa-miR-124a[16,19]) and susceptibility to DM. This discrepancy may be partially attributed to small sample size and ethnicity of the patients.
The goal of our present study was to conduct a meta-analysis to combine all the available studies and to investigate the true contribution of all the studied miRNA-SNPs to the risk of DM, which, to our knowledge, has not been reported.
2. Materials and methods
2.1. Publication search
This meta-analysis was conducted using the Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA). All results were collected from published studies; thus, ethical approval and patient consent were waived.
The PubMed and the Cochrane Library databases were searched by two independent authors to obtain relevant studies published before October, 2018. The used search terms included: [“diabetes” OR diabetic OR T2DM AND (“microRNA” OR “miRNA” OR “miR”) AND (“polymorphism” OR “polymorphisms” OR “SNP” OR “mutation” OR “variant”)]. Citations of retrieved articles were also manually searched to identify additional literatures.
2.2. Inclusion and exclusion criteria
Studies were suggested to be eligible if they:
-
1.
investigated the associations between miRNA polymorphisms and the risk of DM by at least two studies;
-
2.
were in a case-control design;
-
3.
provided genotype frequencies for cases and controls to calculate the odds ratio (OR) and 95% confidence interval (CI);
-
4.
had genotype distribution of controls in accordance with Hardy-Weinberg equilibrium (HWE); and
-
5.
were published in the English language.
The papers were excluded if they were:
-
1.
not related with miRNA-SNPs;
-
2.
lack of healthy controls;
-
3.
animal model research;
-
4.
genotype frequencies unavailable;
-
5.
duplicated publications; and
-
6.
abstracts, case reports/series, comments, editorial articles, summary, reviews or meta-analysis.
2.3. Data extraction
Two authors independently extracted the following data from the included studies: first author, year of publication, country, ethnicity, genotyping method, source of controls, DM type, HWE for controls, age/sex/genotype distribution for cases and controls. Discrepancies were resolved by consensus.
2.4. Quality assessment
The quality of all case-control studies was assessed using a nine-point scoring system of the Newcastle–Ottawa Scale (NOS) [20] by 2 independent authors. A study with a score of ≥ 5 stars was defined as high quality. If discrepancies existed, consensus would be finally reached on discussion.
2.5. Statistical analysis
The crude OR with 95%CI was used to estimate the strength of the association between miRNA polymorphisms and the risk of DM. The significance of pooled ORs was determined using the Z test and P value < .05 was considered to be significant. Meta-analysis was performed using a fixed-effect model if no heterogeneity was present among studies (P value > .10 for the Q test and I2 < 50% for I2 statistic); otherwise, a random-effect model was employed. If there was contradiction for P value and I2 to define the heterogeneity, the Q test result was dependent. The subgroup analysis was also performed to investigate the source of heterogeneity according to ethnicity, DM type, sample size, genotyping method and source of controls. Publication bias was assessed using the Egger linear regression test. If obvious publication bias was detected (P value < .05), trim and fill method was used to adjust for the effect of publication bias.[21] Sensitivity analysis was performed by eliminating studies one by one to evaluate the stability of the results. All these statistical tests were performed using STATA software (version 13.0; STATA Corporation, College Station, TX).
3. Results
3.1. Study characteristics
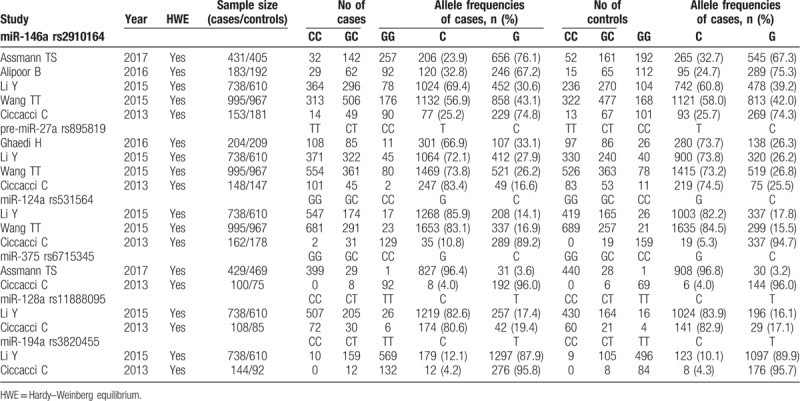
A total of 797 studies were yielded after preliminary database search. Based on the inclusion and exclusion criteria (Fig. 1), 6 studies containing 2773 cases and 2632 controls were finally included in this meta-analysis.[14–19] As shown in Table 1, the eligible studies were published between 2010 and 2018. Most of the included studies (5/6, 83.3%) investigated the association with T2DM. Four studies were conducted in Asians populations (2 in China and 2 in Iran), and the other 2 were performed for European (Italy) and Latin America (Brazil) populations, respectively. Three papers were population-based case-control studies, while the other three were a hospital-based case-control study. Genotyping methods included polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) in two, TaqMan in 2 and sequencing in two studies. Of the 6 studies, 5 evaluated the association between the association of miR-146a polymorphism (rs2910164) with DM risk, four explored pre-miR-27a (rs895819), 3 analyzed miR-124a (rs531564) and 2 focused on miR-375 (rs6715345), miR-128a (rs11888095) and miR-194a (rs3820455) (Table 2). The genotype frequencies of the controls in all studies conformed to HWE. All the studies scored equal or more than 5 on the NOS, which indicated they were of high quality (Table 1).
Figure 1.
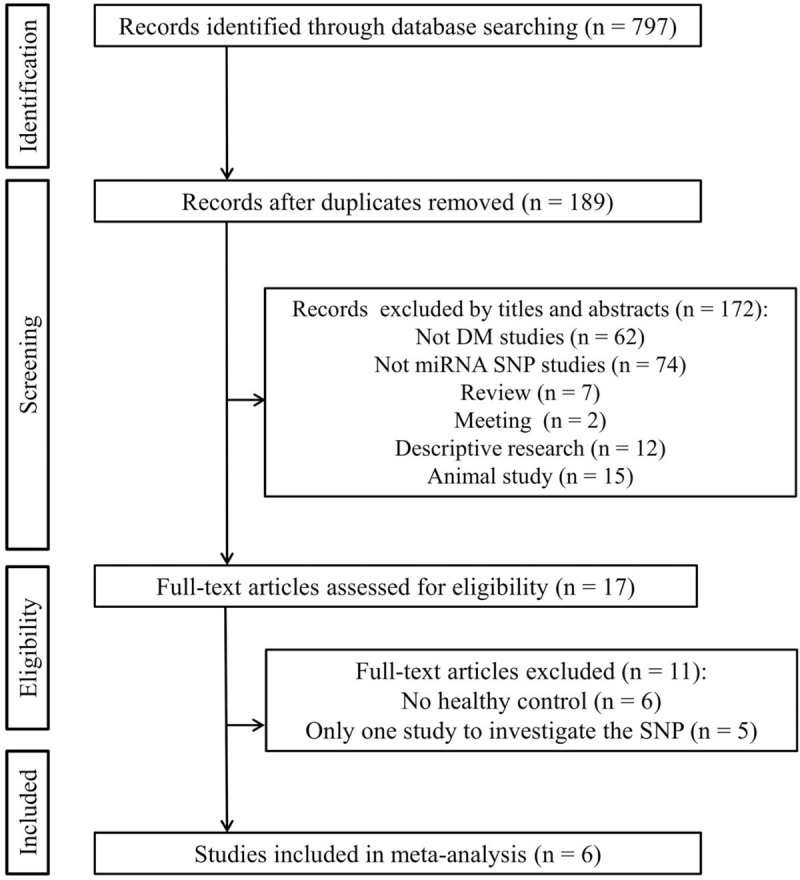
Flow diagram of study identification.
Table 1.
Characteristics of included studies.
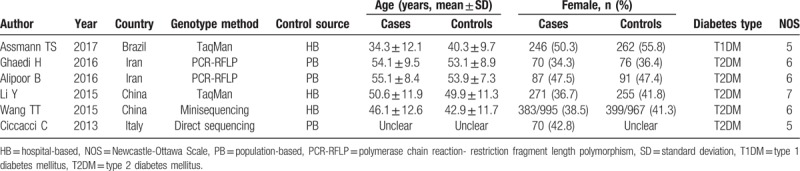
Table 2.
Genotype distributions of miRNA polymorphisms.
3.2. miR-146a polymorphism and the risk of DM
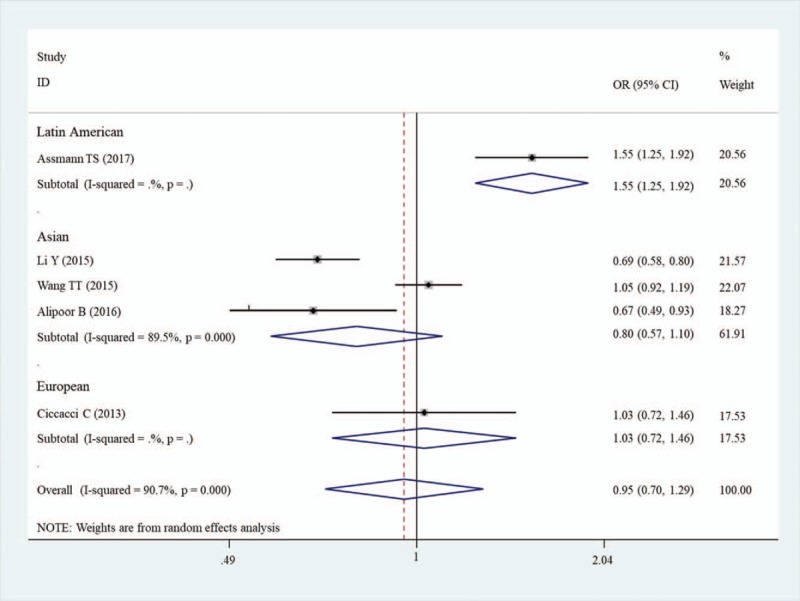
Overall analysis showed there was no significant association between miR-146a rs2910164 polymorphism and the risk of DM under five genetic models (Table 3). Due to the presence of significant heterogeneity in the overall analysis, subgroup analyses were performed. The results showed the G allele or GG carriers seemed to have a significantly higher risk for DM only in Latin American population with T1DM (G vs C, OR = 1.55, 95%CI = 1.25–1.92, P < .001; GG vs CC, OR = 2.18, 95%CI = 1.35–3.51, P = .001; GG vs GC, OR = 1.52, 95%CI = 1.13–2.03, P = .005; GG + GC vs CC, OR = 1.84, 95%CI = 1.16–2.92, P = .01; GC + CC vs GG, OR = 0.61, 95%CI = 0.46–0.80, P < .001) (Table 4; Fig. 2), but this conclusion was only obtained in one study and thus the validity needed further confirmation.
Table 3.
Overall meta-analysis of the association between DM and miRNA polymorphisms.
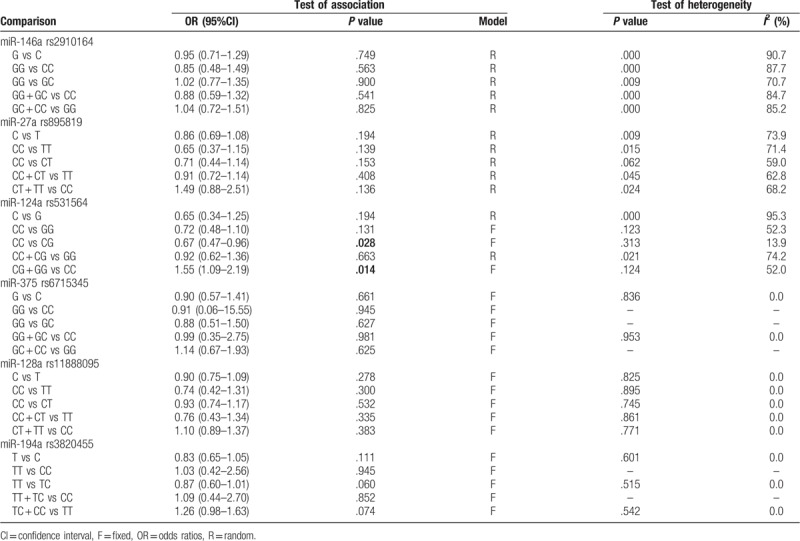
Table 4.
Subgroup meta-analysis of the association between DM and miR-146a rs2910164.
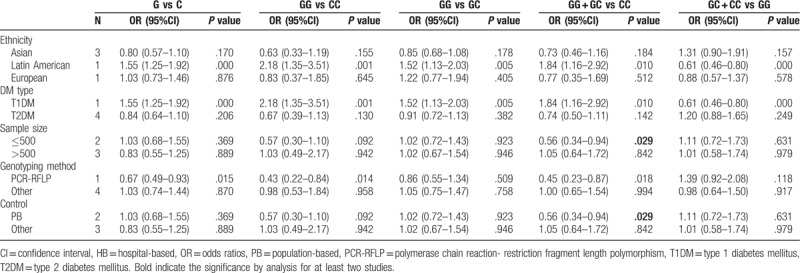
Figure 2.
Forest plots of the association of miR-146a rs2910164 and susceptibility to diabetes mellitus under G vs C model. CI = confidence intervals, OR = odds ratio.
3.3. miR-27a polymorphism and the risk of DM
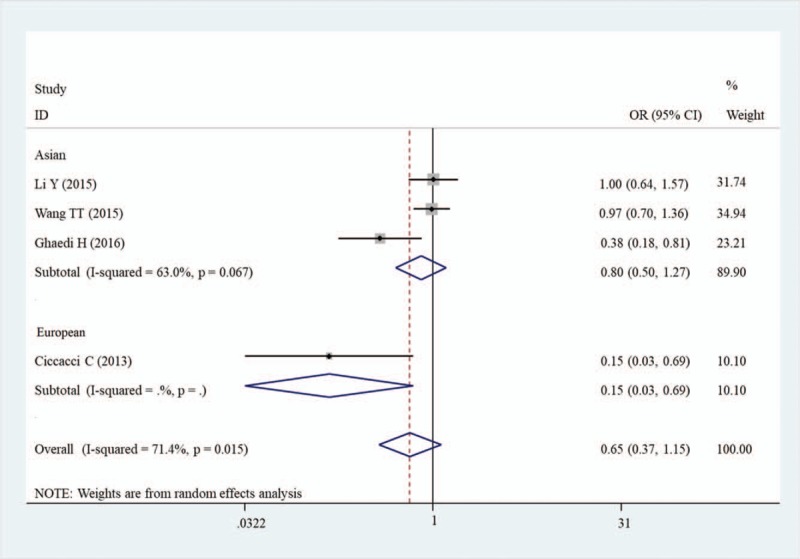
No significant correlation was also found in the overall analyses of the association between miR-27a rs895819 polymorphism and the risk of DM under five genetic models (Table 3). However, the stratified analysis results revealed CC genotype was significantly associated with a decreased risk of DM in the Asian population (OR = 0.80, 95%CI = 0.51–1.27, P = .015) compared with the TT genotype (Table 5; Fig. 3).
Table 5.
Subgroup meta-analysis of the association between DM and miR-27a rs895819.
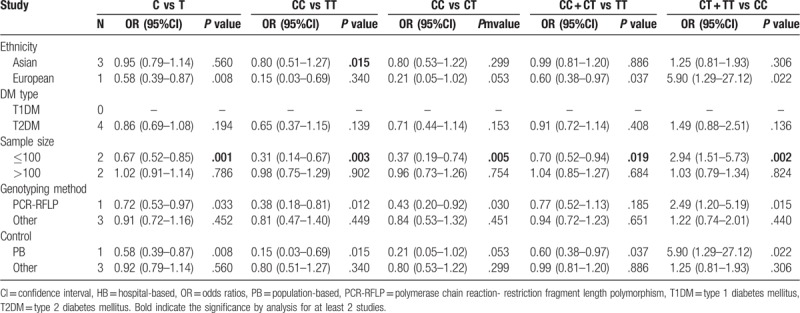
Figure 3.
Forest plots of the association of miR-27a rs895819 and susceptibility to diabetes mellitus under CC vs TT model. CI = confidence intervals, OR = odds ratio.
3.4. miR-124a polymorphism and the risk of DM
The overall analysis showed that patients carrying with CC genotype of miR-124 rs531564 had a lower probability to develop DM (CC vs CG, OR = 0.67, 95% CI = 0.47–0.96, P = .028; CG + GG vs CC, OR = 1.55, 95% CI = 1.09–2.19; P = .014) (Table 3; Fig. 4). No obvious heterogeneity between the studies was found in overall analysis and the number of studies was smaller. Thus, the subgroup analysis was not performed.
Figure 4.
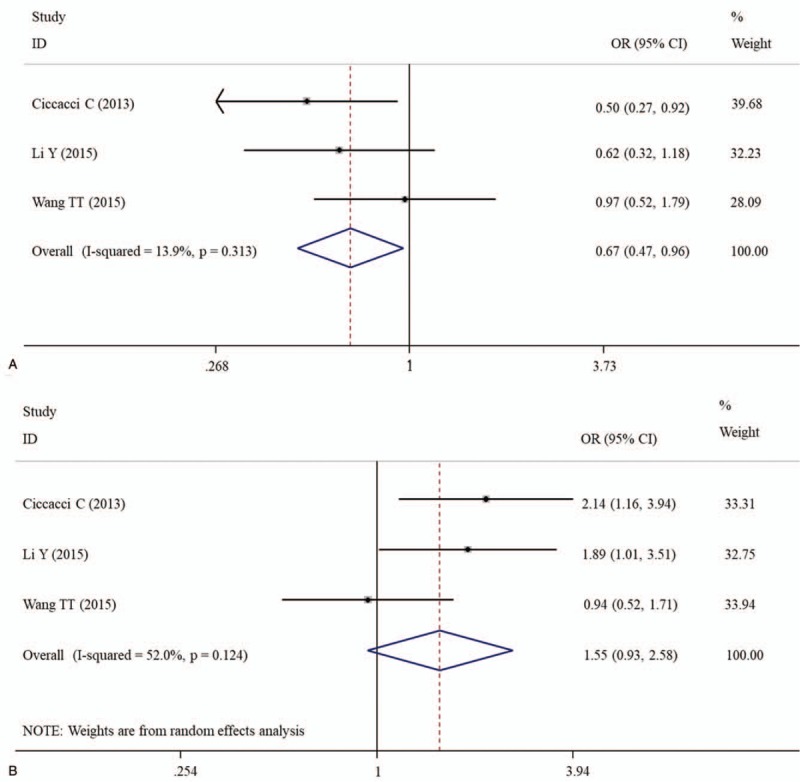
Forest plots of the association of miR-124a rs531564 and susceptibility to diabetes mellitus under CC vs CG (A) and CG + GG vs CC (B) models. CI = confidence intervals, OR = odds ratio.
3.5. miR-375, miR-128a, miR-194a polymorphisms and the risk of DM
No significant associations were observed between miR-375 rs6715345/miR-128a rs11888095 polymorphisms and DM susceptibility (Table 3).
There was a marginal correlation between miR-194a rs3820455 polymorphism and the susceptibility to DM, with TT genotype as a protective factor compared with TC (TT vs TC, OR = 0.87, 95% CI = 0.60–1.01, P = .060) and TC + CC (TC + CC vs TT, OR = 1.26, 95% CI = 0.98–1.63, P = .074) (Table 3). Subgroup analysis was also not performed because only 2 studies were included for them.
3.6. Publication bias and sensitivity analysis
Egger linear regression test showed the intercept did not pass through the origin (that is, asymmetry) in association analysis of miR-146a rs2910164 (G v C, P = .017; GG vs GC, P = .015; GG + GC vs CC, P = .006) and miR–27a rs895819 (CC vs CT, P = .034; CT + TT vs CC, P = .006), indicating the presence of publication bias (Fig. 5A). Subsequently, trim and fill method was used to further adjust for the publication bias. The results showed the association remained not changed after adjusting for publication bias, implying our results were statistically robust (Fig. 5B). No evidence of publication bias was detected for others polymorphisms (data not shown).
Figure 5.
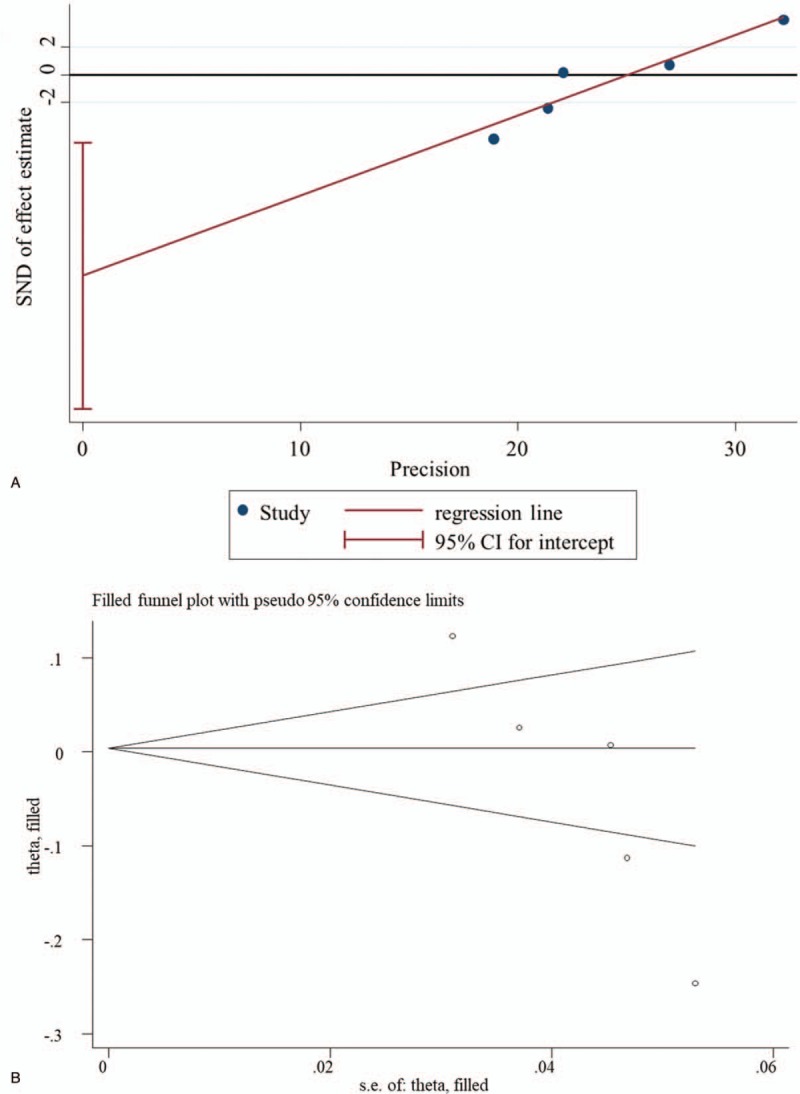
Potential publication bias. A. Egger funnel plot for the assessment of rs2910164 (G v C); B. Trim and fill for adjusting publication bias. CI = confidence intervals, SND = standard normal deviation.
Sensitivity analysis also showed no obvious change in the pooled results after omitting each individual study, suggesting the results were unstable (Fig. 6).
Figure 6.
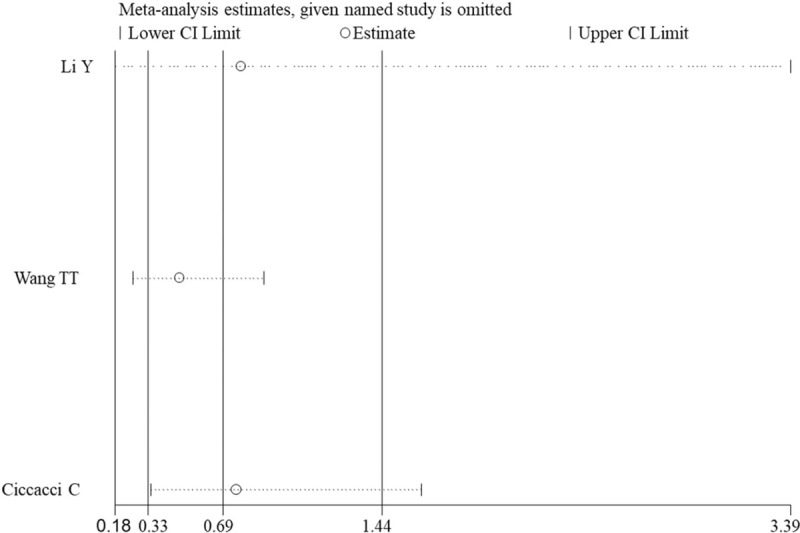
Sensitivity analysis for the assessment of influence of each study for miR-124a polymorphism (rs531564) and diabetes mellitus under CC vs CG model. CI = confidence intervals.
4. Discussion
In the present study, an association between 6 SNPs in miRNAs and DM risk was, for the first time, assessed by the pooled results from 6 case–control studies. The results demonstrated that G allele or GG genotype of miR-146a rs2910164 was associated with a significantly increased risk for DM compared with C allele or GC/CC genotype in Latin American population; CC genotype of miR-27a rs895819 polymorphism was associated with a significantly decreased risk for DM in Asian population compared with the TT genotype; patients carrying with CC genotype of miR-124 rs531564 had a lower probability to develop DM regardless of ethnicity; no associations were identified between polymorphisms in miR-375, miR-128a, miR-194a and the susceptibility to DM.
Previous studies have demonstrated that inflammation and oxidative stress are important mechanisms for the development of DM.[12,22] Xie et al observed the expression of miR-146a was significantly downregulated in DM model rats and its expression was negatively correlated with the expression of inflammatory mediators (COX-2, TNF-α, IL-1β) and oxidative stress indicators (MDA and p22phox).[12] Subsequent in vitro and in vivo transgenic studies proved overexpression of miR-146a inhibited diabetes-induced generation of reactive oxygen species and upregulation of NF-κB downstream pro-inflammatory genes,[23,24] followed by improving neurological,[25] renal or retinal functional defects.[24,26] Luciferase reporter assay indicated miR-146a may regulate the endothelial inflammation via targeted inhibition of interleukin-1 receptor-associated kinase-1 and tumor necrosis factor -associated factor 6.[9,27] These findings imply SNPs that cause the lower expression of the miR-146a and higher expression of its target genes may contribute to increased DM risk. There had a study to reveal colorectal cancer cells with miR-146a GG genotype possessed significantly lower expression of miR-146a compared with those with the GC/CC genotype.[28] The study of Wang et al showed that chronic pancreatitis individuals carrying the G allele conferred a lower expression level of mature miR-146a.[29] Pan et al observed the G allele of rs2910164 was significantly associated with increased abundance of TRAF6.[30] Hereby, patients with GG genotype of miR-146a rs2910164 may have a higher risk for the development of DM, which was confirmed in Latin American population of our study.
High glucose was also reported to stimulate miR-27a expression which, in turn, suppressed peroxisome proliferator-activated receptor γ and then activated β-catenin signaling as evidenced by upregulation of β-catenin target genes, snail1 and α-smooth muscle actin (α-SMA) and downregulation of podocyte-specific markers podocin and synaptopodin. These changes caused disrupted podocyte architectural integrity and increased podocyte apoptosis, ultimately deteriorating diabetic nephropathy.[31,32] Furthermore, miR-27a was also revealed to be up-regulated in embryos and cultured neural stem cells exposed to diabetes, which subsequently down-regulated nuclear factor erythroid 2-related factor 2 and nuclear factor erythroid 2-related factor 2-controlled antioxidant enzymes and led to diabetic embryopathy.[33] Therefore, SNPs that cause the higher expression of the miR-27a may be potential risk factor for DM. Song et al observed the expression level of total pri- and pre-miR27a was significantly higher in rs895819 CT and TT groups compared with CC group,[34] indicating CC genotype may be a protective factor for DM, which was confirmed in Asian population of our study.
miRNA microarray and real-time PCR analysis demonstrated level of miR-124 was significantly increased during the progress of diabetic retinopathy.[35] Administration of antisense RNA oligonucleotide for miR-124 for 2 weeks significantly reduced urinary podocytic nephrin, podocin and albumin excretion and up-regulated integrin α3 expression, thus achieving the treatment goal for idiabetic retinopathy.[36] Curcumin was also reported to prevent against podocytic adhesive capacity damage under mechanical stress by inhibiting miR-124.[37] It was also reported the serum miR-124 was significantly upregulated in the distal tibia metaphyseal fracture patients with GG genotype of rs531564 than those with GC and CC genotypes.[38] Accordingly, rs531564 CC variant in miR-124 might prevent the genetic predisposition of DM by inhibiting the production of miR-124. In line with these findings, we also proved patients carrying with CC genotype of miR-124 rs531564 had a lower probability to develop DM regardless of ethnicity.
Although no controversial conclusions were reported in the studies on miR-375, miR-128a, miR-194a, our study seemed to further indicate the necessity of meta-analysis because the association of miR-194a rs3820455 seemed to be of marginal significance (P < .1) when two studies were combined, not P > .1 as described in individual studies.[14,19]
Some limitations in this meta-analysis should be acknowledged. First, the sample size was relatively small, which may be an underlying cause to result in negative associations between SNPs in miRNAs (miR-375, miR-128a, miR-194a) and DM risk. Second, our pooled outcomes were based on the initial data and were not adjusted by environmental factors due to lack of related information. Third, only published studies in English or Chinese were enrolled in this analysis, and a potential language bias may exist.
5. Conclusion
Our study suggests that miR-146a/miR-27a and miR-124 polymorphisms may be ethnicity-dependent or -independent susceptibility factors to DM, respectively.
Author contributions
Conceptualization: Xi Chen, Wenjing Wang, Lei Gao.
Data curation: Xi Chen, Wenjing Wang.
Formal analysis: Xi Chen, Wenjing Wang.
Investigation: Ruien Li.
Methodology: Jing Yu.
Software: Ruien Li.
Supervision: Lei Gao.
Visualization: Jing Yu.
Writing – original draft: Xi Chen, Wenjing Wang.
Writing – review & editing: Lei Gao.
Footnotes
Abbreviations: CI = confidence interval, DM = diabetes mellitus, HWE = Hardy-Weinberg equilibrium, NOS = Newcastle-Ottawa Scale, OR = odds ratio, PRISMA = Preferred Reporting Items for Systematic Review and Meta-analysis, SNP = single nucleotide polymorphism, 3’-UTR = 3’-untranslated region.
How to cite this article: Chen X, Wang W, Li R, Yu J, Gao L. Association between polymorphisms in microRNAs and susceptibility to diabetes mellitus. Medicine. 2019;98:44(e17519).
XC and WW contributed equally to this work.
The authors have no funding and conflicts of interest to disclose.
All data in this meta-analysis can be seen in previous published studies.
References
- [1].Guariguata L, Whiting DR, Hambleton I, et al. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract 2014;103:137–49. [DOI] [PubMed] [Google Scholar]
- [2].Cunjie L, Ting L. Rs12976445 polymorphism is associated with risk of diabetic nephropathy through modulating expression of MicroRNA-125 and Interleukin-6R. Med Sci Monit 2015;21:3490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mcauley AK, Mohamed D, Jin WJ, et al. A genetic variant regulating miR-126 is associated with sight threatening diabetic retinopathy. Diab Vasc Dis Res 2015;12:133–8. [DOI] [PubMed] [Google Scholar]
- [4].Ciccacci C, Latini A, Greco C, et al. Association between a MIR499A polymorphism and diabetic neuropathy in type 2 diabetes. J Diabetes Complications 2018;32:11–7. [DOI] [PubMed] [Google Scholar]
- [5].Mir KA, Pugazhendhi S, Paul MJ, et al. Heat-shock protein 70 gene polymorphism is associated with the severity of diabetic foot ulcer and the outcome of surgical treatment. Br J Surg 2010;96:1205–9. [DOI] [PubMed] [Google Scholar]
- [6].Abbas S, Raza S, Mir S, et al. Role of variants rs5030717 and rs5030718 of TLR4 in the risk prediction of nephropathy, hypertension and dyslipidaemia in type 2 diabetes mellitus. Br J Biomed Sci 2018;75:163–8. [DOI] [PubMed] [Google Scholar]
- [7].Shen J, Zhang M, Sun M, et al. The relationship of miR-146a gene polymorphism with carotid atherosclerosis in Chinese patients with type 2 diabetes mellitus. Thromb Res 2015;136:1149–55. [DOI] [PubMed] [Google Scholar]
- [8].Nadeem A, Mumtaz S, Naveed AK, et al. Association of IL-6 C-174G (rs 1800795) single nucleotide polymorphism with type 2 diabetes mellitus in Pakistani population. J Pak Med Assoc 2017;67:428–33. [PubMed] [Google Scholar]
- [9].Lo WY, Peng CT, Wang HJ. MicroRNA-146a-5p mediates high glucose-induced endothelial inflammation via targeting interleukin-1 receptor-associated kinase 1 Expression. Front Physiol 2017;8:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sebastiani G, Po A, Miele E, et al. MicroRNA-124a is hyperexpressed in type 2 diabetic human pancreatic islets and negatively regulates insulin secretion. Acta Diabetol 2015;52:523–30. [DOI] [PubMed] [Google Scholar]
- [11].Alipoor B, Ghaedi H, Meshkani R, et al. Association of MiR-146a expression and Type 2 diabetes mellitus: a meta-analysis. Int J Mol Cell Med 2017;6:156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Xie Y, Chu A, Feng Y, et al. MicroRNA-146a: a comprehensive indicator of inflammation and oxidative stress status induced in the brain of chronic T2DM rats. Front Pharmacol 2018;9:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Alipoor B, Ghaedi H, Meshkani R, et al. The rs2910164 variant is associated with reduced miR-146a expression but not cytokine levels in patients with type 2 diabetes. J Endocrinol Invest 2018;41:557–66. [DOI] [PubMed] [Google Scholar]
- [14].Yiping L, Yu Z, Xianli L, et al. Association study of polymorphisms in miRNAs with T2DM in Chinese population. Int J Med Sci 2015;12:875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Alipoor B, Meshkani R, Ghaedi H, et al. Association of miR-146a rs2910164 and miR-149 rs2292832 Variants with Susceptibility to Type 2 Diabetes. Clin Lab 2016;62:1553–61. [DOI] [PubMed] [Google Scholar]
- [16].Wang TT, Chen YJ, Sun LL, et al. Affection of single-nucleotide polymorphisms in miR-27a, miR-124a, and miR-146a on susceptibility to type 2 diabetes mellitus in Chinese Han people. Chin Med J 2015;128:533–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Assmann TS, Duarte GCK, Brondani LA, et al. Polymorphisms in genes encoding miR-155 and miR-146a are associated with protection to type 1 diabetes mellitus. Acta Diabetol 2017;54:433–41. [DOI] [PubMed] [Google Scholar]
- [18].Ghaedi H, Tabasinezhad M, Alipoor B, et al. The pre-mir-27a variant rs895819 may contribute to type 2 diabetes mellitus susceptibility in an Iranian cohort. J Endocrinol Invest 2016;39:1187–93. [DOI] [PubMed] [Google Scholar]
- [19].Ciccacci C, Fusco DD, Cacciotti L, et al. MicroRNA genetic variations: association with type 2 diabetes. Acta Diabetol 2013;50:867–72. [DOI] [PubMed] [Google Scholar]
- [20].Andreas S. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [21].Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000;56:455. [DOI] [PubMed] [Google Scholar]
- [22].Domingueti CP, Dusse LM, Md C, et al. Diabetes mellitus: the linkage between oxidative stress, inflammation, hypercoagulability and vascular complications. J Diabetes Complications 2016;30:738–45. [DOI] [PubMed] [Google Scholar]
- [23].Wan RJ, Li YH. MicroRNA-146a/NAPDH oxidase4 decreases reactive oxygen species generation and inflammation in a diabetic nephropathy model. Mol Med Rep 2018;17:4759–66. [DOI] [PubMed] [Google Scholar]
- [24].Chen S, Feng B, Thomas AA, et al. miR-146a regulates glucose induced upregulation of inflammatory cytokines extracellular matrix proteins in the retina and kidney in diabetes. PloS One 2017;12:e0173918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu XS, Fan B, Szalad A, et al. MicroRNA-146a mimics reduce the peripheral neuropathy in Type II diabetic mice. Diabetes 2017;66:3111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhuang P, Muraleedharan CK, Xu S. Intraocular delivery of miR-146 inhibits diabetes-induced retinal functional defects in diabetic rat model. Invest Ophthalmol Vis Sci 2017;58:1646–55. [DOI] [PubMed] [Google Scholar]
- [27].Zhong X, Liao Y, Chen L, et al. The MicroRNAs in the pathogenesis of metabolic memory. Endocrinology 2015;156:3157–68. [DOI] [PubMed] [Google Scholar]
- [28].Iguchi T, Nambara S, Masuda T, et al. miR-146a Polymorphism (rs2910164) predicts colorectal cancer patients’ susceptibility to liver metastasis. PloS One 2016;11:e0165912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang Z, Li D, Jin J, et al. Association between microRNA polymorphisms and chronic pancreatitis. Pancreatology 2016;16:244–8. [DOI] [PubMed] [Google Scholar]
- [30].Pan Y, Li D, Lou S, et al. A functional polymorphism in the pre-miR-146a gene is associated with the risk of non-syndromic orofacial cleft. Hum Mutat 2018;39:742–50. [DOI] [PubMed] [Google Scholar]
- [31].Hou X, Tian J, Geng J, et al. MicroRNA-27a promotes renal tubulointerstitial fibrosis via suppressing PPARÎ3 pathway in diabetic nephropathy. Oncotarget 2016;7:47760–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhou Z, Wan J, Hou X, et al. MicroRNA-27a promotes podocyte injury via PPARγ-mediated β-catenin activation in diabetic nephropathy. Cell Death Dis 2017;8:e2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhao Y, Dong D, Reece EA, et al. Oxidative stress-induced miR-27a targets the redox gene Nrf2 in diabetic embryopathy. Am J Obstet Gynecol 2017;112:71–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Song B, Yan G, Hao H, et al. rs11671784 G/A and rs895819 A/G polymorphisms inversely affect gastric cancer susceptibility and miR-27a expression in a Chinese population. Med Sci Monit 2014;20:2318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wu JH, Gao Y, Ren AJ, et al. Altered microRNA expression profiles in retinas with diabetic retinopathy. Ophthalmic Res 2012;47:195–201. [DOI] [PubMed] [Google Scholar]
- [36].Dong L, Zhenyu L, Junya J, et al. MiR-124 is related to podocytic adhesive capacity damage in STZ-induced uninephrectomized diabetic rats. Kidney Blood Press Res 2013;37:422–31. [DOI] [PubMed] [Google Scholar]
- [37].Li D, Lu Z, Jia J, et al. Curcumin ameliorates Podocytic adhesive capacity damage under mechanical stress by inhibiting miR-124 expression. Kidney Blood Press Res 2013;38:61–71. [DOI] [PubMed] [Google Scholar]
- [38].Zou L, Zhang G, Liu L, et al. A MicroRNA-124 polymorphism is associated with fracture healing via modulating BMP6 expression. J Cell Biochem 2017;41:2161–70. [DOI] [PubMed] [Google Scholar]


