Abstract
Tropomyosin 1 (TPM1) is a protein that constitutes the sarcomere filaments and is encoded by the TPM1 gene. The aim of the present study is to investigate the correlation between the 3′ untranslated region (3′UTR) single nucleotide polymorphisms (SNPs) of the TPM1 gene and dilated cardiomyopathy (DCM).
A total of 245 patients with DCM and 245 healthy controls were recruited with 5 ml of venous blood. Genomic DNA was extracted to analyze the TPM1 gene rs12148828, rs11558748, rs707602, rs6738, rs7178040 loci genotypes, and the plasma miR-21 level was analyzed by reverse transcription-PCR (RT-PCR).
The risk of DCM development in the rs6738 locus G allele carriers were 1.69 times more than A allele carriers (95% CI: 1.22-2.33, P = .001). Age and gender had no effect on the association of TPM1 gene SNPs with DCM risk (P > .05). The plasma miR-21 level of TPM1 gene rs6738 locus AA carriers was significantly higher than that of the AG and GG genotypes (P < .001).
The SNPs of TPM1 gene rs6738 locus is associated with the risk of DCM, which may be related to the abnormal increase of miR-21 level in DCM patients, but further research is needed to prove the causal relationship between miR-21 level and DCM risk.
Keywords: dilated cardiomyopathy, miR-21, single nucleotide polymorphism, tropomyosin, 3’UTR
1. Introduction
Dilated cardiomyopathy (DCM) is an undetermined myocardial disease with ventricular dilatation and left ventricular systolic dysfunction, which might result in progressive heart failure, malignant arrhythmia, thromboembolism, and sudden death.[1,2] It was reported that DCM patients have a survival rate of only 50% for more than 5-year disease course.[3,4]
In many cases, DCM is found to be hereditary, also known as familial DCM.[5,6] Indeed, The familial DCM may account for 20% to 48% among all DCM cases.[5] In order to better prevent and improve the clinical outcomes of this population, it is necessary to investigate the underlying mechanisms of this disease more widely.
Mutations in multiple sarcomeric genes were thought to be associated with the development of DCM, such as Tropomyosin 1 (TPM1),[7]desmin,[8] and cardiac actin.[8] TPM1 protein is a key component of sarcomere, which stabilizes thin filament and promotes the interaction of actin with myosin during muscle contraction.[9] In the genetic analysis of DCM patients, it was documented that the missense mutations of TPM1 gene Glu40Lys and Glu54Lys result in autosomal dominant inheritance of DCM.[10] In recent years, it has been found that a variety of TPM1 gene mutations are associated with DCM.[11] In addition, Chinese investigators observed that the rs1071646 SNP in the exon of TPM1 is related to the DCM in a Kazakh population from Xinjiang province; however, it is not related to the DCM in the Han population.[12]
In the present study, we analyzed the association of TPM1 gene SNPs with the risk of DCM from the perspective of changes in TPM1 gene expression level. Prior study demonstrated that there is a miR-21 binding site in the 3′ untranslated region (3′UTR) region of the TPM1 gene, moreover, miR-21 could down-regulate the expression of TPM1 gene, and inhibit tumor growth through it binding to the 3’UTR region of the TPM1 gene.[13] Here, we select five SNPs of the TPM1 gene according to the following 2 criteria: the minor allele frequency (MAF) >0.05 (NCBI Variation at https://www.ncbi.nlm.nih.gov/variation/view/) and located in the 3’UTR region of the TPM1 gene. These SNP loci are rs12148828, rs11558748, rs707602, rs6738, and rs7178040. We analyzed the correlation between DCM risk and the five SNPs.
2. Materials and methods
2.1. Ethical aspects
This study was approved by the corresponding ethics committees of the Zhejiang Chinese Medicine and Western Medicine Integrated Hospital/Hangzhou Red Cross Hospital, and Tongde Hospital of Zhejiang Province. All the participants signed their informed written consent. This study conforms to the principles outlined in the World Medical Association Declaration of Helsinki.
2.2. Participants
A cohort of 245 DCM patients who fulfilled the modified version of standardized diagnostic criteria for DCM,[3] was recruited from the Zhejiang Chinese Medicine and Western Medicine Integrated Hospital/Hangzhou Red Cross Hospital, and Tongde Hospital of Zhejiang Province in this case-control study, including 135 men and 110 women, aged 32 to 85 years old, with an average of 52.6 ± 11.2 years old. Another cohort of 245 “age- and gender-matched” healthy control was also recruited, aged 33 to 85 years, with a mean 53.0 ± 11.5 years old. The exclusion criteria include: patients with hypertension, coronary heart disease, diabetes, valvulopathy, acute viral myocarditis, autoimmune systemic disease, and family history of DCM. There is no family tie among all participants. Clinical features of all participants were collected, including end-diastolic inter ventricular septal diameter (IVSd), end-diastolic left ventricular posterior wall diameter (LVPWd), left ventricular end-diastolic diameter (LVEDd), left ventricular end-systolic diameter (LVESd), left atrium (LA) and left ventricular ejection fraction (LVEF).
2.3. Genetic analyses
We collected 5 ml of venous blood from all participants with limosis, and extracted genomic DNA from peripheral blood lymphocytes using the TIANamp Blood DNA Kit (Tiangen Biotect; Beijing, China). Polymerase Chain Reaction (PCR) was conducted using the nucleotide sequence of primers for different SNPs as shown in Table 1. PCR was conducted in a total volume of 20 μL, which contains 2 μL of 10 × PCR buffer, 0.5 μL of 10 mM dNTPs, 1 μL of 10 μmol/L of Forward and Reverse primers, 0.2 μL of Taq enzyme, 100 ng of genomic DNA, and add sterile water to a final volume of 20 μL. The PCR conditions were: 95 °C, 30 seconds; 95 °C, 5 seconds; 60 °C, 30 seconds; 72 °C, 30 seconds; a total of 30 cycles followed by 72 °C, 2 minutes, and 4 °C, 1 minute. After PCR, Sanger sequencing was used to analyze the genotype of SNPs, which was performed by GENEWIZ. The sequencing results of SNPs heterozygotes are shown in Figure 1.
Table 1.
PCR primer sequences for TPM1 gene SNPs.

Figure 1.
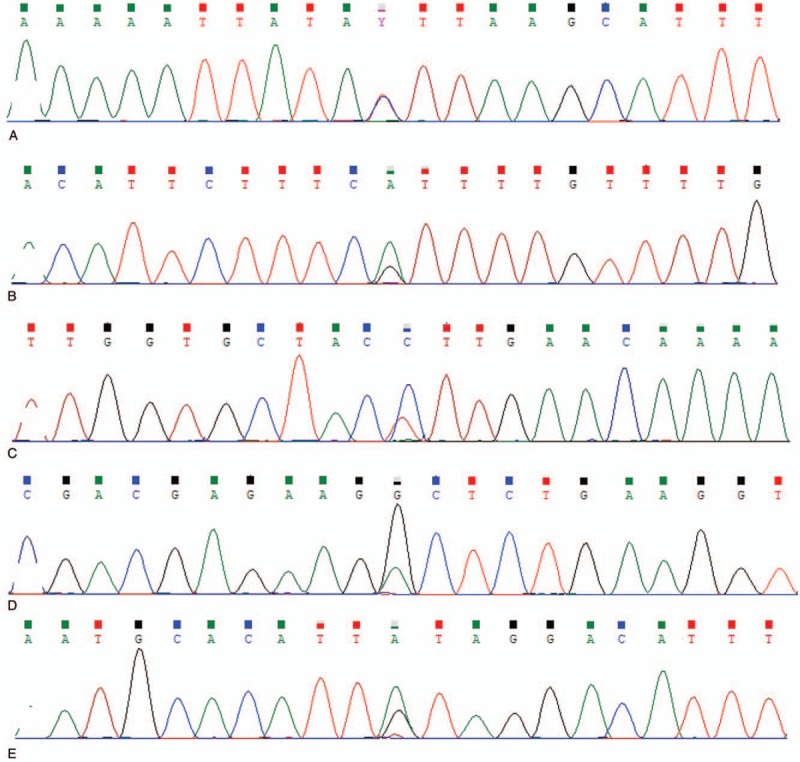
Sanger sequencing results of TPM1 gene SNPs. (A) The TC genotype of rs12148828 locus. (B) The GA genotype of rs11558748 locus. (C) The TC genotype of rs707602 locus. (D) The AG genotype of rs6738 locus. (E) The AG genotype of rs7178040 locus.
2.4. Real-time PCR (RT-PCR)
Total RNA was extracted from plasma using TRIzol reagent (Life Technologies), and cDNA was synthesized using Universal cDNA Synthesis Kit (Exiqon, Copenhagen, Denmark). The relative level of miR-21 was detected using SYBR GreenMaster Mix KitTM (Exiqon, Copenhagen, Denmark), with primer sequence: 5’-TGC GCT AGC TTA TCA GAC TGA-3 ’(forward), 5’-CCA GTG CAG GGT CCG AGG TAT T-3’ (reverse). The level of glyceraldehyde-3 -phosphate dehydrogenase (GAPDH) was set as a control, with primer sequence: 5′-GAG CCC GCA GCC TCC CGC TT-3′ (forward), 5′-CCC GCG GCC ATC ACG CCA CAG-3′ (reverse). The 2-ΔΔCt method was used to compute the relative expression level of miR-21, and all assays were repeated for 3 times.
2.5. Statistical analysis
The SPSS 21.0 (IBM Inc.; Armonk, NY) was used for statistical analysis. The Pearson chi-squared (χ2) and unpaired student t tests were used to test the significance for categorical and continuous variables, respectively. For allele association analysis, a 2 × 2 contingency table evaluated by Pearson's χ2 test was applied to compare the frequency difference of the minor alleles of SNPs between the DCM patients and the control group. The odds ratio (OR), and the corresponding 95% confidence interval (95% CI) were calculated. A 2 × 3 contingency table evaluated by the Pearson χ2 test was adopted for genotype association analysis under three genetic models (additive, dominant, recessive). Multiple logistic regression analysis was used to adjust covariates, such as gender and age. P < .05 was considered as statistically significant difference.
3. Results
3.1. Clinical characteristics of participants
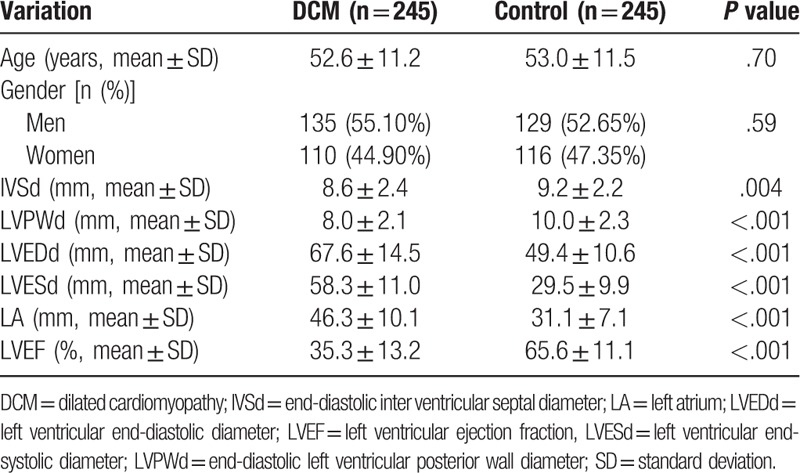
Basically, clinical characteristics of both DCM patients and healthy controls are shown in Table 2. There was no significant difference in age and gender between DCM and control groups (P > .05). Compared with the control group, the IVSd, LVPWd, and LVEF were lower in DCM patients, while the LVEDd, LVESd, and LA were higher, and these differences were statistically significant (P < .05). All DCM patients were curing according to the medical guidelines for heart failure.
Table 2.
Basic characteristics of DCM patients and healthy controls.
3.2. Genotype and allele frequency of TPM1 gene SNPs in DCM and control groups
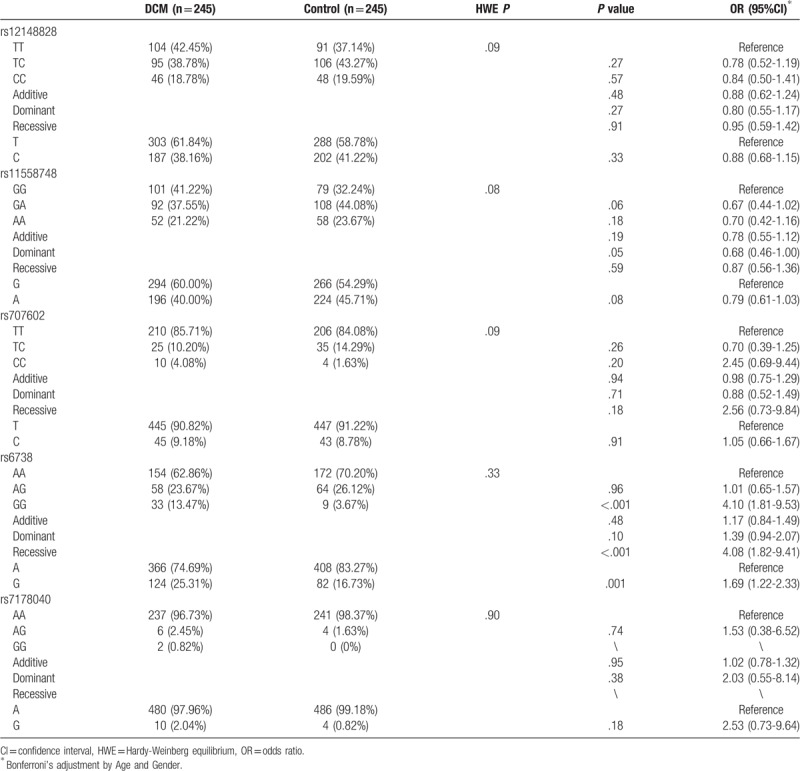
We investigated the genotypes and allele frequencies of the rs12148828, rs11558748, rs707602, rs6738, and rs7178040 loci of TPM1 gene, and found that the genotype distributions of these five SNPs of TPM1 gene were in the Hardy-Weinberg equilibrium for both DCM and control groups (P > .05; Table 3). The genotype frequencies of TPM1 gene rs12148828, rs11558748, rs707602, rs7178040 were not significantly different between DCM group and control group (P > .05). However, the DCM risks were significantly increased (OR = 4.10, 95% CI: 1.81-9.53, P < .001; OR = 4.08, 95% CI: 1.82–9.41, P < .001) in subjects with rs6738 locus homozygous and/or recessive model, and the risk of DCM was 1.69 times higher in subjects carrying G allele than A allele carriers (95% CI: 1.22-2.33, P = .001).
Table 3.
TMP1 gene SNPs genotypes in DCM and control groups and its relationship with DCM risk.
3.3. Correlation between TPM1 gene SNP and clinical characteristics of participants
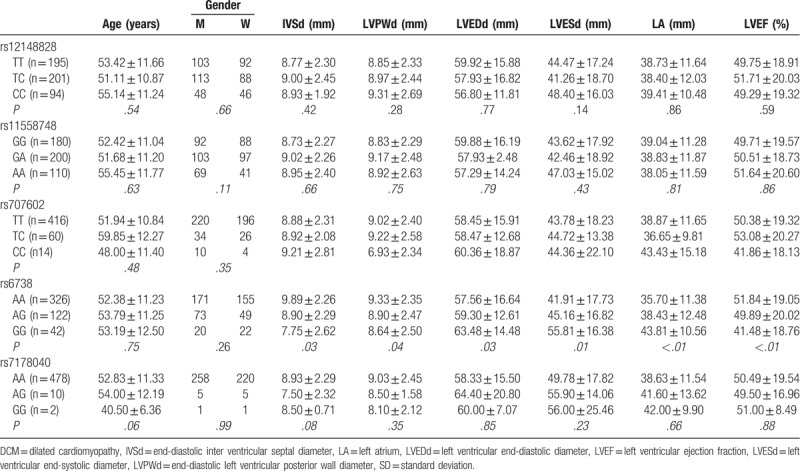
We found that there were significant differences in IVSD, LVPWD, LVEDd, LVESD, LA, LVEF and other characteristics between different genotypes of rs6738 locus (P < .05), rs12148828, rs11558748. And there were no significant differences in IVSD, LVPWD, LVEDd, LVESD, LA, LVEF and other characteristics between different genotypes of rs707602 and rs7178040 loci (P > .05). Additionally, no differences were detected in age and sex of the subjects between different genotypes of each SNPs (P > .05; Table 4).
Table 4.
Comparison of clinical characteristic between subjects with different genotypes of TPM1 gene SNP loci.
3.4. Multiple comparisons for correlation between TPM1 gene and the risk for DCM
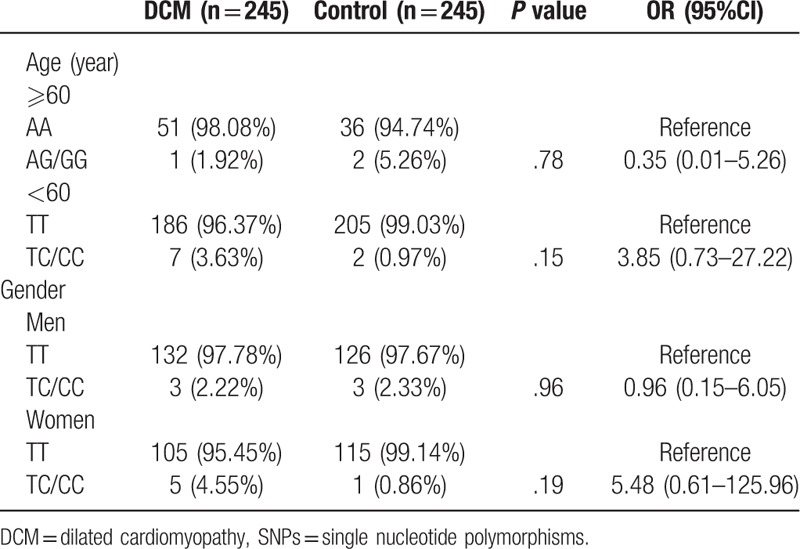
Further, we analyzed the correlation between TMP1 gene SNPs and DCM risk in different ages (≥60 years and <60 years) and gender subjects, and observed that both age and gender had no effect on the association of TPM1 gene SNPs with DCM risk (P > .05; Table 5 ).
Table 5.
Multiple comparisons for correlation between TPM1 SNPs and DCM risk.
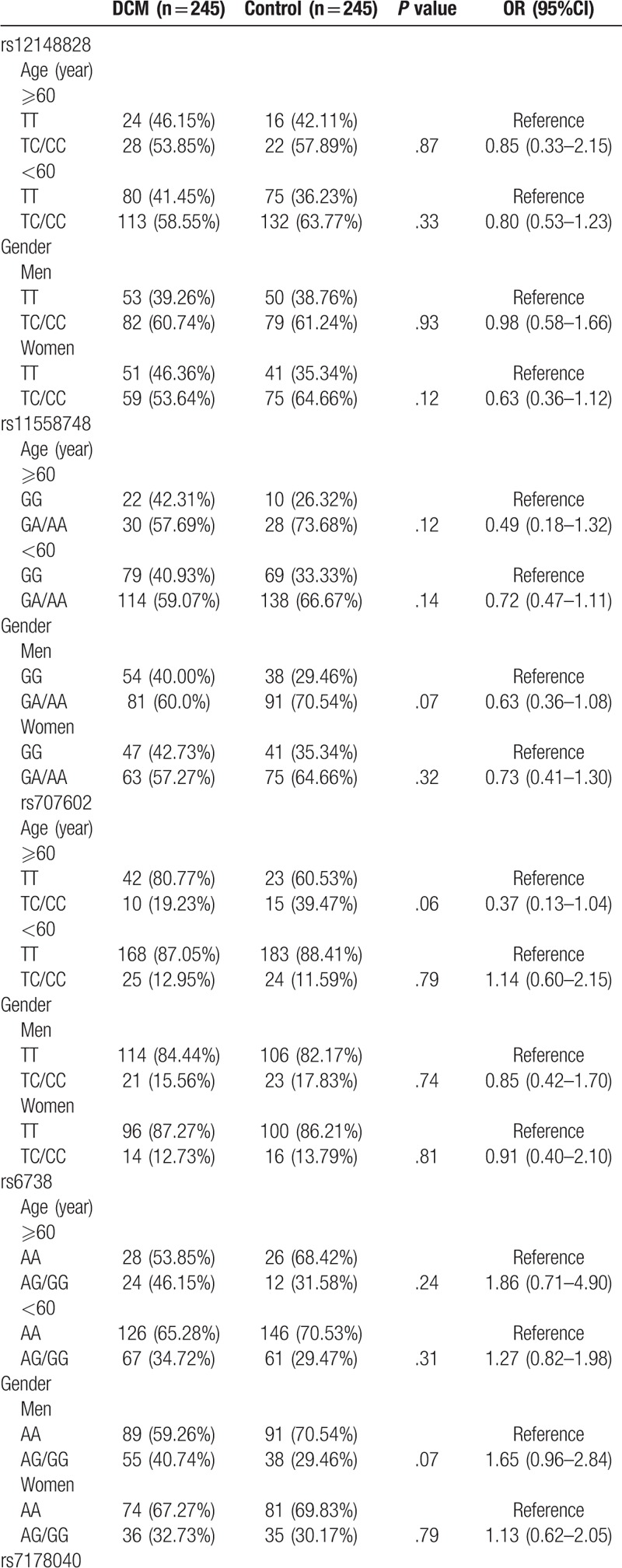
Table 5 (Continued).
Multiple comparisons for correlation between TPM1 SNPs and DCM risk.
3.5. Plasma miR-21 level
Finally, we detected plasma miR-21 level in both DCM and control subjects using RT-PCR, and found a significant higher plasma miR-21 level in DCM patients than that of the healthy control (P < .001; Fig. 2). Meanwhile, we also analyzed the plasma miR-21 levels in subjects with different SNPs of the TPM1 gene in the DCM (Fig. 3A1–E1) and the control group (Fig. 3A2–E2). We found that there was a statistically significant difference in plasma miR-21 levels in subjects with different genotypes of TPM1 gene rs6738 locus in both DCM and the control groups (P < .001). However, no difference in plasma miR-21 levels was observed in subjects with different genotypes of rs12148828, rs11558748, rs707602, and rs7178040 in both DCM and the control groups (P > .05).
Figure 2.
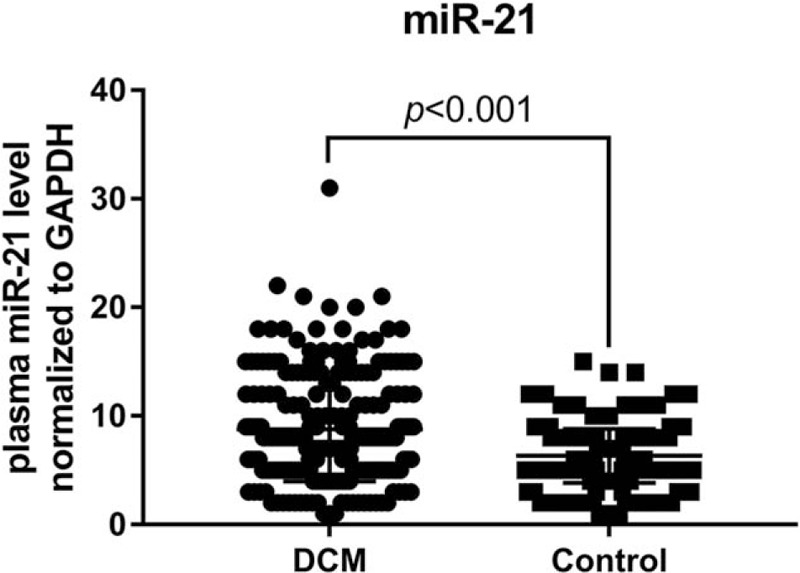
The plasma miR-21 levels in DCM and control groups.
Figure 3.
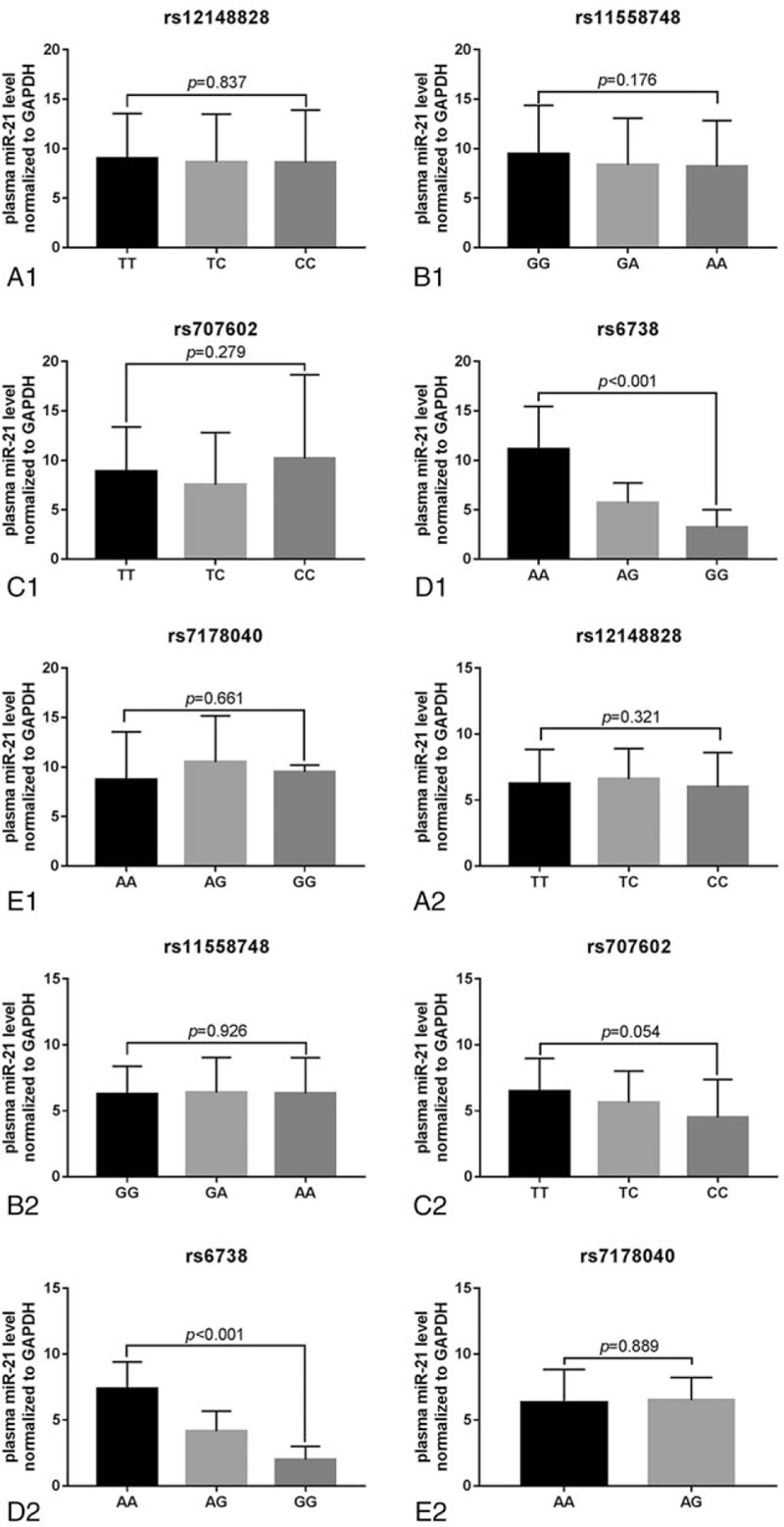
The plasma miR-21 levels in subjects with different genotypes of TPM1 gene SNPs in DCM and control groups. (A1-E1) DCM patients. (A2-E2) healthy control.
4. Discussion
In the present study, we analyzed the relationship between 5 SNPs with MAF > 0.05 in the 3′UTR region of TPM1 gene and the risk for DCM. We showed that SNPs of TPM1 gene rs6738 locus were associated with DCM risk. Further, we found that rs6738 SNPs were related to plasma miR-21 levels in subjects. Hence, it is likely that the higher risk of DCM was associated higher levels of miR-21 in DCM patients; however, further researches are still necessary to confirm this notion.
TPM1 is a parallel double-stranded α-helical coiled-coil protein that binds to the long groove of actin, which is essential for actin regulation and stability.[14] Furthermore, its function is also associated with troponin complex in response to Ca2+ signaling.[15] The gene encoding for TPM1 protein is located on 15q22.2, and the two isoforms of the TPM1 gene, TPM1α and TPM1κ, with the difference in containing exon 2b or 2a, are expressed equally in fetal and adult heart.[16–18] Up to date, most studies which investigate the relationship between TPM1 gene mutation and disease are focused on its coding region. For instance, the T230 N mutation of TPM1 gene results in a decline in the flexibility of its C-terminus via the alteration of helical structure of the protein, which thereby decreases the flexibility of the TPM1 overlap and impairs its ability to regulate contraction.[7] However, there are few studies focusing on the relationship between the non-coding region mutations of the TPM1 gene and disease, in particularly rarely focused on the 3’UTR region.
As an exploratory study, we screened 5 mutation-prone SNPs in the 3′UTR region of the TPM1 gene, rs12148828, rs11558748, rs707602, rs6738, rs7178040. The rs12148828 locus was found to be unrelated to non-chemotaxis orbital fissure (NSOC) according to Qian et al.[19] However, Weymouth et al found that this site is associated with clubfoot in non-Hispanic whites.[20] There is no research evidence showing that rs11558748 is related to disease, and rs707602 is not associated with myocardial infarction (MI) according to Shiffman et al.[21] It was also found that the rs6738 and rs7178040 loci were not associated with the occurrence and prognosis of triple negative breast cancer (TNBC) according to Wang et al.[22] In the current study, we found that rs6738 locus SNPs were associated with DCM risk. By comparing between 245 DCM patients and 245 healthy controls, a TMP1 gene rs6738 G allele carrier was 1.69 times higher DCM risk that A allele carrier, and in the recessive model it was as high as 4.08 times. Further, we showed that that the level of miR-21 in the plasma was significantly lower in subjects with rs6738 locus GG genotype than that of AG and AA genotypes, and there is a significant correlation between plasma miR-21 level and rs6738 locus genotype. After comparing the rs6738 locus sequence and miR-21 sequence (Fig. 4), it is speculated that the binding site of miR-21 to TPM1 gene is likely to be located at the rs6738 locus, and the binding efficiency is higher in A allele that G allele. As a consequence, the level of TPM1 gene expression in the G allele carrier is relatively lower.
Figure 4.
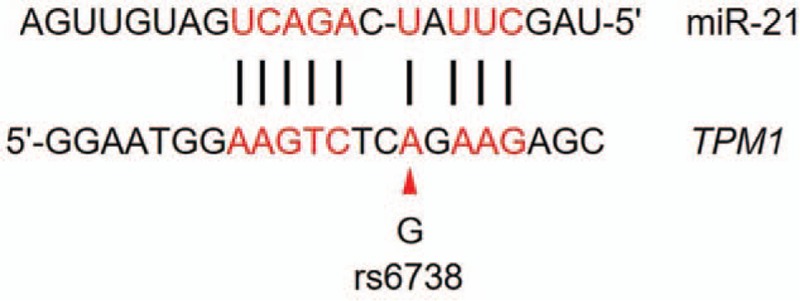
The comparison of partial sequence between 3′UTR region of TPM1 gene and miR-21. The red arrow indicates the base of the rs6738 locus mutation.
It is well known that miRNAs influence a variety of cellular functions by regulating the expression of multiple target genes.[23] The reason for investigating miR-21 in the present study is that the TPM1 gene is target gene of miR-21, and there is a specific miR-21 binding site at the 3′UTR of the TPM1 variants V1 and V5. After cloning the 3′UTR of TPM1 into a luciferase reporter gene, it was found that although miR-21 down-regulates the luciferase activity, anti-miR-21 upregulates it.[13] This observation provides us with a new idea that SNPs located at miRNAs targets of the gene 3’UTR region may be associated with risk of disease, since SNPs at these loci may affect the regulating efficiency of miRNAs on gene expression.
Interestingly, we did not find that rs6738 locus SNPs were associated with DCM risk in different ages or genders. The explanation for this might be the limited sample size. Unfortunately, we could not recruit a sufficiently large sample for further verification in a limited time, and this is also a shortcoming of this study. In addition, we have no direct evidence that miR-21 regulates TPM1 gene expression through its binding to rs6738 locus, since we lack an animal model to verify our speculation. In addition, we did not analyze the correlation between follow-up results and SNPs, which needs to be supplemented in later studies. We believe that this is just the beginning, we will find more sensitive loci in genetic mechanisms underlying the pathogenesis of DCM, and provide novel targets for the prevention and treatment of DCM through gene-gene or gene-environment interaction analysis.
5. Conclusion
SNPs at rs6738 locus in the 3′UTR region of TPM1 gene are associated with DCM risk. G allele carriers have a higher risk of DCM, and plasma miR-21 level is related to SNPs at rs6738 locus. Therefore, it is speculated that abnormal elevation of miR-21 levels in carriers with G allele of rs6738 locus is a reason for higher risk of DCM, which needs further researches to verify.
Author contributions
Conceptualization: Tianjie Zhang.
Data curation: Qiang Yao, Wei Zhang.
Formal analysis: Qiang Yao, Wei Zhang.
Funding acquisition: Tianjie Zhang.
Investigation: Qiang Yao, Wei Zhang.
Methodology: Qiang Yao, Wei Zhang.
Supervision: Tianjie Zhang.
Validation: Wei Zhang.
Writing – original draft: Qiang Yao, Tianjie Zhang.
Writing – review & editing: Tianjie Zhang.
Footnotes
Abbreviations: DCM = dilated cardiomyopathy, IVSd = end-diastolic inter ventricular septal diameter, LA = left atrium, LVEDd = left ventricular end-diastolic diameter, LVEF = left ventricular ejection fraction, LVESd = left ventricular end-systolic diameter, LVPWd = end-diastolic left ventricular posterior wall diameter, MAF = minor allele frequency, OR = odds ratio, PCR = polymerase chain reactions, RT-PCR = reverse transcription-PCR, SNPs = single nucleotide polymorphisms, TPM1 = Tropomyosin 1, 3′UTR = 3′ untranslated region.
How to cite this article: Yao Q, Zhang W, Zhang T. Association of single nucleotide polymorphisms in the 3′UTR region of TPM1 gene with dilated cardiomyopathy: a case-control study. Medicine. 2019;98:44(e17710).
This work was supported by grants from Zhejiang Provincial Medicine Health Science and Technology Program (2008B152).
The authors have no conflicts of interests to disclose.
References
- [1].Dilaveris P, Antoniou CK, Gatzoulis KA. Arrhythmic risk stratification in non-ischemic dilated cardiomyopathy: where do we stand after DANISH? Trends Cardiovasc Med 2017;27:542–55. [DOI] [PubMed] [Google Scholar]
- [2].Jefferies JL, Towbin JA. Dilated cardiomyopathy. Lancet 2010;375:752–62. [DOI] [PubMed] [Google Scholar]
- [3].Kaski JP, Elliott P, Group ESCW. The classification concept of the ESC Working Group on myocardial and pericardial diseases for dilated cardiomyopathy. Herz 2007;32:446–51. [DOI] [PubMed] [Google Scholar]
- [4].Hershberger RE, et al. Genetic evaluation of cardiomyopathy–a Heart Failure Society of America practice guideline. J Card Fail 2009;15:83–97. [DOI] [PubMed] [Google Scholar]
- [5].Taylor MR, Carniel E, Mestroni L. Cardiomyopathy, familial dilated. Orphanet J Rare Dis 2006;1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhao CM, et al. TBX20 loss-of-function mutation associated with familial dilated cardiomyopathy. Clin Chem Lab Med 2016;54:325–32. [DOI] [PubMed] [Google Scholar]
- [7].Lynn ML, et al. The structural basis of alpha-tropomyosin linked (Asp230Asn) familial dilated cardiomyopathy. J Mol Cell Cardiol 2017;108:127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tesson F, et al. Epidemiology of Desmin and cardiac actin gene mutations in a European population of dilated cardiomyopathy. Eur Heart J 2000;21:1872–6. [DOI] [PubMed] [Google Scholar]
- [9].England J, et al. Tropomyosin 1: multiple roles in the developing heart and in the formation of congenital heart defects. J Mol Cell Cardiol 2017;106:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Olson TM, et al. Mutations that alter the surface charge of alpha-tropomyosin are associated with dilated cardiomyopathy. J Mol Cell Cardiol 2001;33:723–32. [DOI] [PubMed] [Google Scholar]
- [11].Redwood C, Robinson P. Alpha-tropomyosin mutations in inherited cardiomyopathies. J Muscle Res Cell Motil 2013;34:285–94. [DOI] [PubMed] [Google Scholar]
- [12].Ji Y, et al. TPM1 gene mutation is associated with dilated cardiomyopathy in Kazaks in Xinjiang. Zhonghua Xin Xue Guan Bing Za Zhi 2015;43:521–6. [PubMed] [Google Scholar]
- [13].Zhu S, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008;18:350–9. [DOI] [PubMed] [Google Scholar]
- [14].Brown JH, et al. Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proc Natl Acad Sci U S A 2005;102:18878–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jagatheesan G, et al. Striated muscle tropomyosin isoforms differentially regulate cardiac performance and myofilament calcium sensitivity. J Muscle Res Cell Motil 2010;31:227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dube S, et al. Expression of TPM1kappa, a novel sarcomeric isoform of the TPM1 gene, in mouse heart and skeletal muscle. Mol Biol Int 2014;2014:896068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang J, et al. Ectopic expression and dynamics of TPM1alpha and TPM1kappa in myofibrils of avian myotubes. Cell Motil Cytoskeleton 2007;64:767–76. [DOI] [PubMed] [Google Scholar]
- [18].Lohmeier-Vogel EM, Heeley DH. Biochemical comparison of Tpm1.1 (alpha) and Tpm2.2 (beta) tropomyosins from rabbit skeletal muscle. Biochemistry 2016;55:1418–27. [DOI] [PubMed] [Google Scholar]
- [19].Qian Y, et al. TPM1 polymorphisms and nonsyndromic orofacial clefts susceptibility in a Chinese Han population. Am J Med Genet A 2016;170A:1208–15. [DOI] [PubMed] [Google Scholar]
- [20].Weymouth KS, et al. Variants in genes that encode muscle contractile proteins influence risk for isolated clubfoot. Am J Med Genet A 2011;155A:2170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shiffman D, et al. Identification of four gene variants associated with myocardial infarction. Am J Hum Genet 2005;77:596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang W, et al. A single-nucleotide polymorphism in the 3’-UTR region of the adipocyte fatty acid binding protein 4 gene is associated with prognosis of triple-negative breast cancer. Oncotarget 2016;7:18984–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]


