Abstract
Objective:
PR interval (PR) is a heritable electrocardiographic measure of atrial and atrioventricular nodal conduction. Changes in PR duration may be associated with atrial fibrillation, heart failure, and all-cause mortality. Hispanic/Latino populations have high burdens of cardiovascular morbidity and mortality, are highly admixed, and represent exceptional opportunities for novel locus identification. However, they remain chronically understudied. We present the first genome-wide association study (GWAS) of PR in 14,756 participants of Hispanic/Latino ancestry from three studies.
Methods:
Study-specific summary results of the association between 1000 Genomes Phase 1 imputed SNPs and PR assumed an additive genetic model and were adjusted for global ancestry, study center/region, and clinical covariates. Results were combined using fixed-effects, inverse variance weighted meta-analysis. Sequential conditional analyses were used to identify independent signals. Replication of novel loci was performed in populations of Asian, African, and European descent. ENCODE and RoadMap data were used to annotate results.
Results:
We identified a novel genome-wide association (P<5×10−8) with PR at ID2 (rs6730558), which replicated in Asian and European populations (P<0.017). Additionally, we generalized ten previously identified PR loci to Hispanics/Latinos. Bioinformatics annotation provided evidence for regulatory function in cardiac tissue. Further, for six loci that generalized, the Hispanic/Latino index SNP was genome-wide significant and identical to (or in high linkage disequilibrium with) the previously identified GWAS lead SNP.
Conclusions:
Our results suggest that genetic determinants of PR are consistent across race/ethnicity, but extending studies to admixed populations can identify novel associations, underscoring the importance of conducting genetic studies in diverse populations.
Hispanic/Latino populations are chronically understudied in medical research, despite a growing population that will represent 30% of the U.S. population by 2060 [1]. Hispanic/Latino populations shoulder a higher burden of cardiovascular diseases (CVD), including myocardial infarction, stroke, and death from CVD, compared to European American populations [2, 3, 4]. Hispanic/Latino populations are highly admixed, tracing their origins to Europe, Africa, and the Americas [5]. Thus, Hispanic/Latino populations are uniquely positioned to inform on the genetic architecture of not just populations from the Americas, but also on populations of European, Asian, and African descent. Hispanic/Latino populations represent a unique genetic architecture in which to study the genetics of cardiovascular disease [6].
The PR interval (PR), a heritable measure of atrial depolarization and atrioventricular nodal conduction [7], is a promising yet unexplored candidate for study in Hispanic/Latino populations. PR is associated with atrial fibrillation, a risk factor for stroke, pacemaker implantation, heart failure, and all-cause mortality [8, 9, 10]. Previous genome-wide association studies (GWAS) in European, African American, and Asian populations have identified more than 15 genetic loci associated with PR, including SCN5A, SCN10A, CAV1-CAV2, TBX5-TBX3, and SOX5 [11, 12, 13, 14, 15, 16, 17, 18, 19].
Hispanic/Latino populations pose an interesting paradox. The prevalence of prolonged PR is higher in Hispanics/Latinos, whereas their risk for atrial fibrillation is lower compared to European descent populations [20]. As researchers begin to develop genetic risk scores for conditions such as atrial fibrillation, it is important to acknowledge evidence supporting population-specific variants underlying common complex traits such as atrial fibrillation [21]. Population-specific variants makes it critical that genetic risk scores used in Hispanic/Latino populations incorporate results from genetic studies in Hispanic/Latino populations rather than extending genetic risk scores developed in European descent populations. We present the first GWAS of PR in Hispanic/Latino populations, bringing together three large cohorts of Hispanics/Latinos, including the Hispanic Community Health Study/Study of Latinos (HCHS/SOL), the Women’s Health Initiative (WHI), and the Multi-Ethnic Study of Atherosclerosis (MESA).
MATERIALS AND METHODS
Study Populations
Three cohorts participated in this meta-analysis (N=14,756): the HCHS/SOL (N=11,703), the MESA (N=1,480), and the WHI (N=1,573; Table 1, Supplemental Text). Each study was approved by the Institutional Review Board at the respective sites and all participants provided written consent.
Table 1.
Descriptive characteristics of participants in three cohorts in Hispanic/Latino genome-wide association study
| Population | N | Mean Age in years (SD) |
Mean PR in ms (SD) |
% Female |
Mean Height in cm (SD) |
Mean BMI in kg/m2 (SD) |
Mean SBP in mmHg (SD) |
|---|---|---|---|---|---|---|---|
| HCHS/SOL | 11,703 | 46 (14) | 157 (21) | 59 | 162 (9) | 30 (6) | 122 (18) |
| MESA | 1,480 | 61 (10) | 162 (22) | 52 | 162 (9) | 29 (5) | 127 (22) |
| WHI | 1,573 | 60 (6) | 156 (21) | 100 | 157 (6) | 29 (5) | 127 (17) |
BMI, Body mass index; cm, Centimeters; HCHS/SOL, Hispanic Community Health Study/Study of Latinos; kg/m2, Kilograms per meter squared; MESA, Multi-Ethnic Study of Atherosclerosis; mmHG, Millimeters of mercury; ms, Milliseconds; N, Number of participants; PR, PR interval; SBP, Systolic blood pressure; SD, Standard deviation; WHI, Women’s Health Initiative
PR Interval Measurement
Resting, supine, or semi-recumbent ECGs were digitally recorded in each study at baseline by certified technicians using standard 12-lead ECGs using either Marquette MAC12 or MAC PC machines (GE Healthcare, Milwaukee, WI, USA; Supplemental Table 1). Comparable procedures were used for preparing participants, placing electrodes, recording, transmitting, processing, and controlling the quality of the ECGs. The PR interval was measured electronically using the Marquette 12SL algorithm.
Genotyping
Participants were genotyped on the Affymetrix Genome-Wide Human SNP Array 6.0 (MESA, WHI) or an Illumina custom array. Genotypes were imputed using the 1000 Genomes Phase 1 reference panel (Additional details in Supplemental Text; Supplemental Table 1).
Statistical Analysis
Genome-wide analyses were performed by each cohort independently across approximately 25 – 39 million SNPs assuming an additive genetic model, using linear regression (MESA, WHI) or linear mixed models (HCHS/SOL, Supplemental Text). All analyses were adjusted for age, sex, study center or region, height, body mass index, systolic blood pressure, heart rate, beta blocker use, and study principal components of ancestry to maintain consistency with prior PR GWAS [11, 12, 13, 14, 15, 16, 17, 18, 19]. Study specific results were corrected for genomic inflation (λ, Supplemental Figure 1) and combined with inverse-variance weighted meta-analysis. For all significant index SNPs, we performed conditional analyses by adjusting for the index SNPs and any subsequently significant SNPs until no remaining genome-wide significant SNPs were identified.
Generalization of Previously Reported PR SNPs
For SNPs previously associated with PR in published GWAS reports [11, 16], we examined evidence for generalization using the approach described by Sofer et al. [22]. The method assigns an r-value to every index SNP. The generalization null hypothesis testing generalization of the PR index SNPs to Hispanics/Latinos was rejected when the r-value was less than 0.05, controlling the false discovery rate of the generalization null hypothesis.
Replication of Novel Associations
For novel associations, we attempted to replicate our results in populations of East Asian [14], European, and African (WHI study, Supplemental Text) descent using exclusion criteria and analytic procedures that overlapped with previously described approaches.
Linkage Disequilibrium Analysis
LD was calculated with the r2 correlation statistic using 1000 Genomes meta-populations as reference (AMR, EUR, AFR, ASN). RFMix and ASAFE were used to estimated local ancestry and ancestry-specific allele frequencies (Supplemental Text).
Bioinformatic Annotation
For loci associated with PR at a genome-wide significance level in our Hispanic/Latino population, we used epigenetic data from the ENCODE and RoadMap projects to examine evidence of functional annotation. Results were restricted to annotation found in available heart tissue (fetal heart, right atrium, and right or left ventricle). Additionally, publically available data from genetic association studies of clinical phenotypes associated with PR were queried for associations with lead Hispanic/Latino SNPs (Supplemental Text).
RESULTS
Study Population Characteristics
A total of 14,756 participants from three cohorts (MESA, HCHS/SOL, WHI) contributed to this study (Table 1). The majority of participants were from HCHS/SOL (N=11,703), who were, on average, 14 years younger than MESA and WHI participants. In addition, participants were predominantly female (52%−100%) and obese (average BMI=29–30 kg/m2). After study-specific quality control and filtering by effective sample size, studies contributed between 8,217,098 (MESA) and 17,322,742 (HCHS/SOL) imputed single nucleotide polymorphisms (SNPs; Supplemental Table 1), which together represented 18,828,993 unique SNPs.
Novel Association at ID2
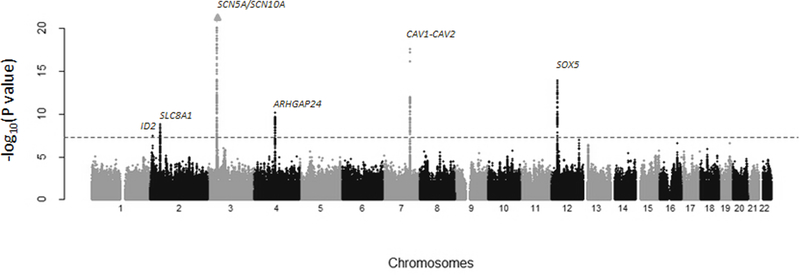
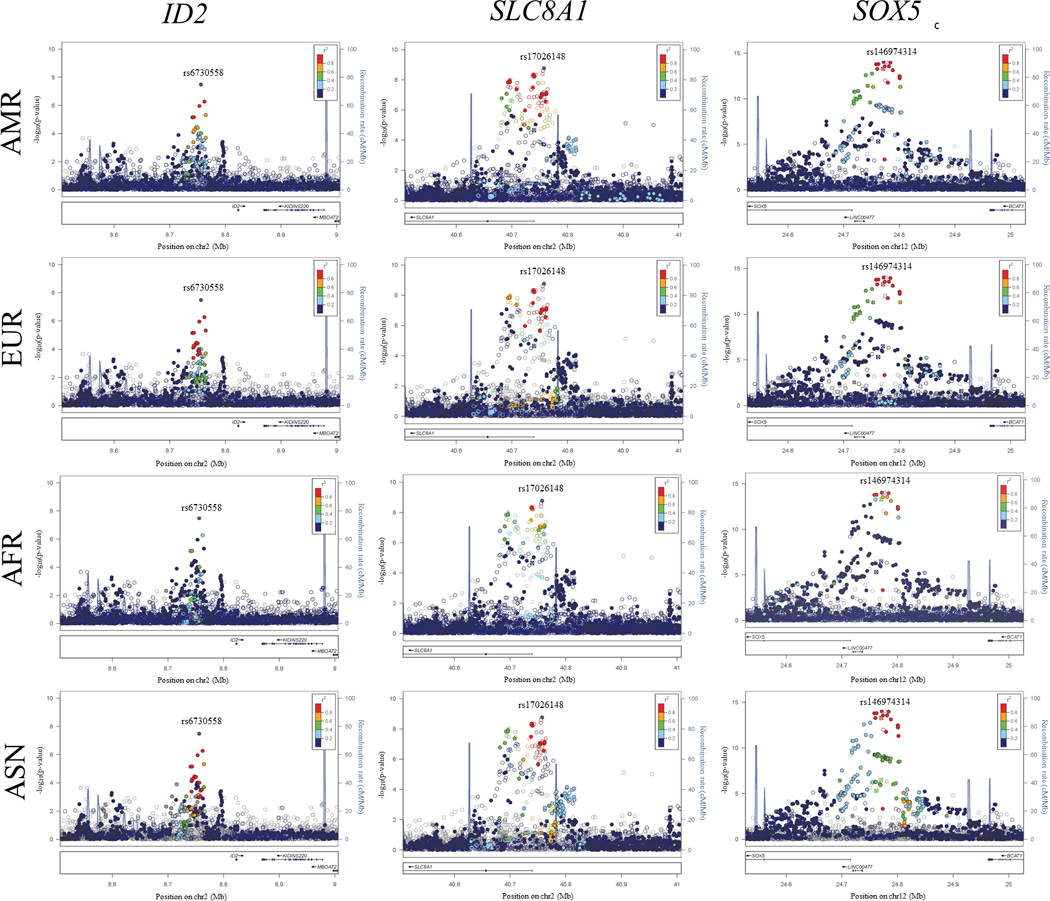
A total of 454 SNPs at seven loci were genome-wide significant (P < 5×10−8, Figure 1, Table 2), with no evidence of genomic inflation (study-specific λ range: 0.975 to 1.02; meta-analysis λ=1.004, Supplemental Figure 1). One of these seven loci located upstream of ID2 (inhibitor of DNA binding 2, HLH protein), a gene encoding a transcriptional regulator, was novel. The Hispanic/Latino lead SNP, rs6730558 (P=3×10−8; Table 2, Figure 1), was common in our population (coded allele frequency [CAF]=0.49) and had homogenous results (Phet=0.7). Rs6730558 also was common in 1000 Genomes meta-populations (African [AFR]: CAF=0.67, European [EUR]: CAF=0.39, and East Asian [ASN]: CAF=0.57; Table 3). Furthermore, when we calculated the local ancestry at each index SNP in the HCHS/SOL population and then calculated the CAF by ancestral haplotype, the T allele of rs6730558 was common across all three ancestral populations (European descent: 0.39, African descent: 0.70, and Native American: 0.56; Table 3). Examination of linkage disequilibrium (LD) patterns from four 1000 Genomes meta-populations (Admixed American [AMR], AFR, EUR, and ASN) showed that rs6730558 resides in a very narrow LD block, with particularly weak LD in AFR populations (Figure 2). Conditional analyses did not identify any independent SNPs at this locus.
Figure 1.
Manhattan plot of genome-wide association meta-analysis results in Hispanic/Latino population (N=14,756). Loci meeting genome-wide significance (P < 5×10−8, denoted with red line) are annotated with nearest gene name.
Table 2.
Common genetic variants (coded allele frequency > 0.05) associated with PR interval in Hispanic/Latino population (N=14,756) at genome-wide significance (P < 5×10−8)
| Locus | Position | Index SNP | CA | CAF | P-value | Effect Estimate (SE) |
Phet |
|---|---|---|---|---|---|---|---|
| ID2* | 2p25.1 | rs6730558 | T | 0.49 | 3×10−8 | 1.36 (0.25) | 0.7 |
| SLC8A1 | 2p22.1 | rs17026148 | A | 0.17 | 2×10−9 | 1.98 (0.33) | 0.4 |
| SCN5A | 3p21.0 | rs3922844 | T | 0.37 | 4×10−41 | −3.39 (0.25) | 0.09 |
| rs7374004** | A | 0.65 | 3×10−19 | 2.66 (0.30) | 0.3 | ||
| rs45567533*** | A | 0.87 | 1×10−11 | −2.49 (0.37) | 0.4 | ||
| SCN10A | 3p22.0 | rs6801957 | T | 0.38 | 1×10−55 | 3.90 (0.25) | 0.9 |
| ARHGAP24 | 4q22.1 | rs13105921 | A | 0.41 | 6×10−11 | 1.76 (0.27) | 0.06 |
| CAV1-CAV2 | 7q31.1 | rs3807989 | A | 0.41 | 2×10−18 | 2.18 (0.25) | 0.7 |
| SOX5 | 12p12.1 | rs146974314 | A | 0.14 | 9×10−15 | −2.66 (0.35) | 0.2 |
Novel association
Conditioned on rs3922844 and rs6801957
Conditioned on rs3922844, rs6801957, and rs7374004
CA, Coded allele; CAF, Coded allele frequency in our study population meta-analysis; Phet, P-value for heterogeneity calculated using Cochran’s Q statistic; SE, Standard error; SNP, Single nucleotide polymorphism
Table 3.
Comparison of coded allele frequencies in Hispanic/Latino population and in 1000 Genomes meta-populations at signals associated with PR interval in Hispanic/Latino population
| Locus | Index SNP (Coded Allele) |
Hispanic/Latino CAF |
CAF By Ancestral Haplotype* |
1000 Genomes Population CAF |
||||
|---|---|---|---|---|---|---|---|---|
| EU | AF | NA | EUR | AFR | ASN | |||
| ID2 | rs6730558 (T) | 0.49 | 0.39 | 0.70 | 0.56 | 0.39 | 0.67 | 0.57 |
| SLC8A1 | rs17026148 (A) | 0.17 | 0.03 | 0.14 | 0.70 | 0.03 | 0.16 | 0.43 |
| SCN5A | rs3922844 (T) | 0.37 | 0.32 | 0.69 | 0.30 | 0.29 | 0.67 | 0.14 |
| SCN10A | rs6801957 (T) | 0.38 | 0.44 | 0.11 | 0.40 | 0.42 | 0.14 | 0.22 |
| ARHGAP24 | rs13105921 (A) | 0.41 | 0.67 | 0.12 | 0.02 | 0.65 | 0.14 | 0.07 |
| CAV1-CAV2 | rs3807989 (A) | 0.41 | 0.42 | 0.72 | 0.23 | 0.40 | 0.73 | 0.31 |
| SOX5 | rs146974314 (A) | 0.14 | 0.17 | 0.001 | 0.17 | 0.15 | 0.01 | 0.13 |
CAF was calculated in the Hispanic Community Health Study/Study of Latinos after calculating local ancestry (EU, AF, or NA). From there, the CAF was calculated for each ancestral population at each separate chromosome region.
AF, African ancestry; AFR, 1000 Genomes African meta-population; ASN, 1000 Genomes East Asian meta-population; CAF, Coded allele frequency; EU, European ancestry; EUR, 1000 Genomes European meta-population; NA, Native American ancestry; SNP, Single nucleotide polymorphism
Figure 2.
LocusZoom plots of common (coded allele frequency > 0.05) single nucleotide polymorphisms at three select regions (ID2, SLC8A1, SOX5) associated with PR interval in Hispanic/Latino populations. Results from the genome-wide association study m
We attempted to replicate the association of rs6730558 with PR in independent European descent (N=4,296), African American (N=3,763), and Asian (N=6,805) populations given the admixed nature of Hispanic/Latino populations (Table 4). A Bonferroni-corrected significance level of 0.017 (i.e. 0.05/3 tests) was used to determine significance. For replication in Asian populations, we identified rs3856447 as a proxy in high LD with rs6730558 (r2 > 0.9), which was associated with PR at P=7×10-3. Rs6730558 was also associated with PR in the European descent replication population (P=0.002) but failed to replicate within the African American replication population (P=0.05). However, the direction of effect and magnitude of the effect size was consistent with the Hispanic/Latino lead SNP across all three populations (range: 1.07 – 1.60 ms).
Table 4.
Replication of ID2 Hispanics/Latinos Signal in Asian (N=6,805), African American (N=3,763), and European American (N=4,296) Populations
| Race/Ethnic Group |
Replicated SNP |
r2 between Replicated SNP and Hispanics/Latino Index SNP |
CA | CAF | P-value | Effect Estimate (SE) |
|---|---|---|---|---|---|---|
| Asian* | rs3856447 | 0.9 | A | 0.44§ | 0.007 | 1.07 (0.40) |
| African American** | rs6730558 | NA | T | 0.61 | 0.05 | 1.16 (0.60) |
| European** | rs6730558 | NA | T | 0.38§ | 0.002 | 1.60 (0.52) |
Abbreviations: CA, Coded Allele; CAF, Coded Allele Frequency; SE, Standard Error; SNP, Single Nucleotide Polymorphism
Asian results obtained from Hong et al. [14] The lead SNP in Hispanics/Latinos, rs6730558, was not present in the dataset. A proxy SNP in Asian populations was identified using the SNAP database and the r2 value reported in the table represents the linkage disequilibrium between rs3856447 and rs6730558 in the combined Han Chinese (CHB) and Japanese (JPT) 1000 Genomes populations.
African American and European results obtained using WHI populations. African American population from the SHARe study. European populations from the WHIMS study.
CAF not available in study dataset. Reported CAF obtained from 1000 Genomes meta-poulations, EAS (East Asian) and EUR (European).
The ID2 locus showed no associations among previously published genetic associations with atrial fibrillation (Supplemental Table 2).
Replication of Previously Identified PR Loci
Six loci previously associated with PR in other race/ethnicities were significantly associated with PR in Hispanics/Latinos (Table 2, Figure 1). Of the lead SNPs at these six loci in Hispanics/Latinos, three (rs3922844 at SCN5A, rs6801957 at SCN10A, and rs3807989 at CAV1-CAV2) were identical to previously reported index SNPs (Table 5). Furthermore, at ARGHAP24, the Hispanic/Latino lead SNP was in high LD with an index SNP previously identified in only European populations and a signal previously identified in only African American populations (r2=0.93 and 0.82, respectively).
Table 5.
Comparison of genetic loci associated with PR interval in Hispanic/Latino populations with results from previous genome-wide association studies in European [13, 16], African [11, 19], and Asian [14, 17, 18] descent populations
| Locus | Index SNP in Hispanics/Latinos |
Previously-Identified Index SNPs |
||
|---|---|---|---|---|
| Index SNP | Population | r2 | ||
| SLC8A1 | rs17026148 | rs17026114 | AS | 0.93 |
| rs17026156 | AS | 0.88 | ||
| SCN5A | rs3922844 | rs3922844 | EU, AA | -- |
| rs7638909 | AS | 0.008 | ||
| rs11708996 | EU | 0.06 | ||
| rs6599222 | AA | 0.0003 | ||
| rs6763048 | AA | 0.05 | ||
| SCN10A | rs6801957 | rs6801957 | EU, AA, AS | -- |
| rs6795970 | EU, AS | 0.79 | ||
| rs6800541 | EU, AS | 0.77 | ||
| rs6798015 | AA | 0.80 | ||
| rs6599257 | AS | 0.57 | ||
| ARHGAP24 | rs13105921 | rs7692808 | EU | 0.93 |
| rs7660702 | EU | 0.74 | ||
| rs11732231 | AA | 0.82 | ||
| CAV1-CAV2 | rs3807989 | rs3807989 | EU | -- |
| rs11773845 | EU, AA, AS | 0.99 | ||
| SOX5 | rs146974314 | rs11047543 | EU | 0.97 |
AA, African-descent population; AS, Asian-descent population; EU, European-descent population; r2, Correlation between previously identified index SNP and index SNP identified in Hispanic/Latino population using Hispanic/Latino linkage disequilibrium pattern; SNP, Single nucleotide polymorphism
The remaining two loci have only been previously described in a single race/ethnicity. The Hispanic/Latino index SNP was in high LD with the previously-described index signals at both the SOX5 (European populations) and SLC8A1 (Asian populations) loci (r2=0.93 and 0.97, respectively). The lead SNP at the SLC8A1 locus was rare in the EUR population (CAF=0.03) and also resided in a narrow LD block in the AFR population, although LD was similar across AMR, EUR, and ASN populations (Table 3, Figure 2). Rs17026148 at SLC8A1 was common in both the ASN population as well as in HCHS/SOL participants with Native American local ancestry at the rs17026148 chromosomal position (CAF=0.43 and 0.70, respectively; Table 3). Similarly, the Hispanic/Latino association at SOX5 was rare in the AFR population (CAF=0.01) and the Hispanic/Latino lead SNP had less LD in the ASN population than the EUR population, which showed similar LD patterns to the AMR population.
Conditional analyses identified independent signals at the SCN5A/SCN10A locus (rs7374004 and rs45567533) but did not identify independent signals at any other loci (Table 2). Rs6801957 was also identified as an secondary signal in African Americans [11] and was in high LD with previously published index SNPs rs6800541 (r2=0.93) [12, 14, 16], rs6795970 (r2=0.93) [12, 13], and rs6798015 (r2=0.83) [19] in HapMap CEU populations and rs6798015 (r2=0.87) in CHB+JPT populations (Supplemental Table 3). Secondary signal rs45567533 was not in high LD with any previously published index SNP (r2 < 0.8). There was little LD at this locus in YRI or AFR populations (Supplemental Figure 2).
Generalization
Additionally, we evaluated evidence of generalization for fifteen index SNPs reported by previous GWAS in Europeans (N=9) and African Americans (N=6) [11, 16]. Eight of the nine SNPs previously identified in the European population and all six SNPs previously identified in the African American population generalized to Hispanics/Latinos (r-value < 0.05) (Supplemental Table 4, Supplemental Figure 3). Notably, SNPs that generalized to Hispanics/Latinos included four SNPs at the MEIS1, ITGA9, TBX5/TBX3, and NKX2–5 loci in addition to six index SNPs at loci identified above as genome-wide significant in Hispanics/Latinos.
Bioinformatic Annotation of Associated Variants
Finally, we performed bioinformatic annotation of the lead SNPs associated with PR in our Hispanic/Latino population (Supplemental Table 5). The ID2 locus (rs6730558) showed evidence of enhancer activity and H3K4me1 and H3K27ac modification, also associated with enhancer activity, in heart tissues (left ventricle, right ventricle, right atrium, and fetal heart). The lead SNPs at SCN5A, SCN10A, and CAV1-CAV2 also showed evidence of cardiac enhancer activity. Our lead SNP at ARHGAP24 (rs13105921) showed evidence of promoter and transcriptional activity, including H3K4me3 and H3K9ac modification. Signals at SLC8A1 and SOX5 had no evidence of functional annotation.
DISCUSSION
In this study, the first PR GWAS in Hispanics/Latinos, we identified a novel association with PR at ID2 and generalized ten loci, including six at genome-wide significant levels, to Hispanics/Latinos. Additionally, we identified one population specific secondary signal at SCN5A and generalized another secondary signal at SCN5A. Finally, the results at SLC8A1, a locus previously only identified in East Asian populations [14, 17], demonstrated that evaluation of an admixed population can aid in improving characterization of loci that had not previously extended across populations. These results emphasize the need to conduct genetic studies in admixed populations.
We identified a novel association at rs6730558, located 54 kb downstream of ID2, a gene encoding a DNA binding inhibitor protein. ID2 has been implicated in the development of the cardiac conduction system of mice [23] but has yet to be associated with electrophysiology phenotypes in humans. However, other DNA regulatory proteins, such as TBX5/TBX3, have been implicated in the regulation of PR in both GWAS and animal models. Associations with regulatory genes indicate that transcriptional control can be involved in PR duration. The evidence of enhancer activity at the ID2 Hispanic/Latino lead SNP (rs6730558) also suggests that the underlying association may be affecting PR through the increased transcription of ID2, which in turn would affect PR duration.
There are several reasons ID2 may not have been identified in previous GWAS of PR. First, rs6730558 may be a population specific signal. However, it is more likely that the ID2 signal was poorly captured in previous analyses. While rs6730558 is common across genetic ancestries, it lies on a narrow LD block, suggesting that previous genotyping and imputation platforms may not have adequately captured the ID2 signal. In particular, African populations demonstrate extremely low LD with the lead SNP identified in this analysis, making it more difficult to characterize the region in African descent populations.
The admixed genomes in Hispanic/Latino populations enabled us to detect two previously population-specific associations. While the SLC8A1 and SOX5 loci had previously not been replicated in any other populations, we were able to successfully replicate the same signal at both of these two loci in our highly admixed Hispanic/Latino population. There are several possible explanations for the lack of findings at the SLC8A1, and SOX5 loci. Similar to ID2, the signals could have been inadequately genotyped or imputed in previous studies, particularly those using HapMap imputation panels in non-EU populations. Imputation accuracy is an existing question in admixed populations such as African Americans, which have large proportions of African ancestry and shorter LD blocks [24]. The narrow LD blocks found in our study also suggest that poor imputation anywhere in the narrow LD block would make it difficult to adequately capture the signal at that locus. Use of a broader range of populations along with the increasing use of whole genome sequencing (WGS) data, for both genetic association studies and imputation, will aid future genetic research in better capturing disease association signals.
However, another possible explanation is that the SLC8A1, SOX5, and ID2 signals lie on a European or Asian/Native American haplotypes. There are several lines of evidence that could support the hypothesis that these signals lie on population-specific haplotypes. For example, although our index signal at ID2 (rs6730558) is common across ancestral populations (EUR, AFR, AMR, and ASN), there is almost no LD at the ID2 locus in AFR populations. Conversely, the SLC8A1 and SOX5 loci both demonstrate more LD across all four ancestral populations. However, the index SNP at SLC8A1, rs17026148, is rare in European populations, and the index SNP at SOX5, rs146974314, is rare in African populations. Prior work on other disease states have found population-specific alleles and haplotypes associated with diseases such as schizophrenia [25], glaucoma [26], and asthma [27].
Furthermore, rare and population-specific variants are now being identified through whole genome sequencing, which continues to lead to the discovery of new population-specific signals. For example, sequencing of a Sardinian population found ~76,000 variants common among Sardinians but rare in the 1000 Genomes panel, including four novel loci associated with lipid levels and inflammatory markers [28]. Additionally, whole exome sequencing results have found low-frequency (1–5%) variants and rare variants (<5%) associated with asthma unique to either Hispanics/Latinos or African Americans [29]. Results from sequencing studies suggest that rare and low-frequency variants can be ethnic specific.
Despite this knowledge, genomics research remains dominated by studies in European ancestry populations. As of 2016, minority populations account for 20% of all populations in GWAS [30]. However, much of this is focused in populations of Asian and some African ancestry, leaving little research in populations of Hispanic ancestry. Minority populations are better represented in WGS efforts. However, even in the National Heart, Lung, and Blood Institute’s (NHLBI) Trans-Omics for Precision Medicine (TOPMed), African American ancestry participants account for 30% of the TOPMed population while Hispanics/Latinos remain chronically under sampled, accounting for just 10% of the TOPMed population [30]. Our study, along with those discussed herein, emphasize the need to include these minority populations in health research but work still needs to be done to adequately include these populations in future research.
Furthermore, clinical applications of genetic research will require diverse ancestral populations. Work discussed herein, as well as the results of this study, demonstrates that the genetic underpinnings of disease-related traits can vary by population. As precision medicine advances, it is imperative that genetic research expands to include diverse populations, such as Hispanic/Latinos. Current advances in precision medicine disproportionately benefit European descent populations which can exacerbate health disparities but work such as that presented herein ensures that advances in the knowledge of disease etiology and any subsequent clinical applications are applicable to relevant populations.
This study had several notable limitations. First, our sample size of Hispanics/Latinos was only half that of the largest study in EU populations [16]. However, our sample was comparable to previous work in African American ancestry populations [11] and twice that of previous Asian ancestry GWAS of PR [14, 17]. Furthermore, even with our reduced sample size, we were able to identify a novel association at ID2 and replicate all but one previous association, suggesting that our diverse population allowed us to better capture areas of the genome such as the ID2 locus. Our ability to examine differences among Hispanic/Latino ancestries was limited also by the composition of our cohorts. While our study included a broad range of genetic ancestries (Mexican, Cuban, Dominican, Puerto Rican, South American, Central American), Mexican ancestry accounted for more than double the proportion of our population than the next best represented ancestry. Finally, despite our broad range of ancestries, we were unable to further refine our populations of South or Central American ancestries. However, our study represents the first study of the genetics of PR in Hispanics/Latinos and includes a broad representation of Hispanic/Latino populations, improving on previous genetic studies of Hispanics/Latinos which were previously focused on Mexican Americans.
In conclusion, our study identified a novel association with PR near ID2 and generalized ten previously identified loci to our Hispanic/Latino population. Our results support consistency in multiple genetic determinants of PR across race/ethnicities and underscore the importance of conducting genetic studies in diverse populations. Genetic research seeking to elucidate the genetic architecture of PR, as well as research aiming to eliminate health disparities must make greater efforts to include diverse populations.
Supplementary Material
KEY MESSAGES.
What is already known about this subject?
The PR interval (PR) is a heritable measure of atrial depolarization and atrioventricular nodal conduction. Previous genome-wide association studies (GWAS) have identified more than 15 genetic loci associated with PR but much of the heritability remains unexplained. Hispanic/Latino populations are particularly understudied in genetic analyses, including those of PR.
What does this study add?
We have conducted the first GWAS of PR in Hispanic/Latino populations, identifying one novel locus associated with PR and generalizing a further six loci (which had previously been associated with PR in other race/ethnicities) to Hispanics/Latinos.
How might this impact on clinical practice?
Hispanic/Latino populations pose an interesting paradox, with a higher prevalence of prolonged PR but a lower risk of atrial fibrillation compared to European descent populations. Better understanding the genetic architecture of PR in Hispanics/Latinos can help better identify individuals at risk of negative health outcomes due to prolonged PR.
ACKNOWLEDGEMENTS
Hispanic Community Health Study/Study of Latinos (HCHS/SOL): We thank the participants and staff of the HCHS/SOL study for their contributions to this study. The baseline examination of HCHS/SOL was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Genotyping efforts were supported by NHLBI HSN 26220/20054C, NCATS CTSI grant UL1TR000124, and NIDDK Diabetes Research Center (DRC) grant DK063491. AAS was also supported by training grants T32HL7055 and T32HL07779. NS was supported by R01HL116747 and R01 HL111089.
Multi-Ethnic Study of Atherosclerosis (MESA): This research was supported by the Multi-Ethnic Study of Atherosclerosis (MESA) contracts HHSN2682015000031, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and by grants UL1-TR-000040, UL1-TR-001079, and UL1-RR-025005 from NCRR. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02-HL-6-4278. We also thank the other investigators, the staff, and the participants of MESA for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
Women’s Health Initiative Clinical Trial (WHI CT): The Women’s Health Initiative clinical trials were funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C. All contributors to WHI science are listed @ https://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf.
Footnotes
Conflict of Interest Statement
The authors have no conflict of interest to declare.
Contributor Information
Amanda A Seyerle, Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Division of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN, USA.
Henry J Lin, Institute for Translational Genomics and Populations Sciences, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA; Division of Medical Genetics, Department of Pediatrics, Harbor-UCLA Medical Center, Torrance, CA, USA.
Stephanie M Gogarten, Department of Biostatistics, University of Washington, Seattle, WA, USA.
Adrienne Stilp, Department of Biostatistics, University of Washington, Seattle, WA, USA.
Raul Méndez-Giraldez, Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Elsayed Z Soliman, Epidemiology Cardiology Research Center (EPICARE), Division of Public Health Sciences, Wake Forest School of Medicine, Winston-Salem, NC, USA; Department of Medicine, Section of Cardiology, Wake Forest School of Medicine, Winston-Salem, NC, USA.
Antoine Baldassari, Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Mariaelisa Graff, Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Susan R Heckbert, Department of Epidemiology, University of Washington, Seattle, WA, USA; Cardiovascular Health Research Unit, University of Washington, Seattle, WA, USA.
Kathleen F Kerr, Department of Biostatistics, University of Washington, Seattle, WA, USA.
Charles Kooperberg, Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Carlos J Rodriguez, Department of Medicine, Section of Cardiology, Wake Forest School of Medicine, Winston-Salem, NC, USA; Department of Epidemiology and Prevention, Division of Public Health Sciences, Wake Forest School of Medicine, Winston-Salem, NC, USA.
Xiuqing Guo, Institute for Translational Genomics and Populations Sciences, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA; Division of Medical Genetics, Department of Pediatrics, Harbor-UCLA Medical Center, Torrance, CA, USA.
Jie Yao, Institute for Translational Genomics and Populations Sciences, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA; Division of Medical Genetics, Department of Pediatrics, Harbor-UCLA Medical Center, Torrance, CA, USA.
Nona Sotoodehnia, Cardiovascular Health Research Unit, University of Washington, Seattle, WA, USA; Division of Cardiology, University of Washington, Seattle, WA, USA.
Kent D Taylor, Division of Genomic Outcomes and Institute for Translational Genomics and Population Sciences, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA.
Eric A Whitsel, Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Department of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Jerome I Rotter, Institute for Translational Genomics and Populations Sciences, Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA; Division of Medical Genetics, Department of Pediatrics, Harbor-UCLA Medical Center, Torrance, CA, USA.
Cathy C. Laurie, Department of Biostatistics, University of Washington, Seattle, WA, USA
Christy L Avery, Department of Epidemiology University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Carolina Population Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
REFERENCES
- 1.Colby SL, Ortman JM, U.S. Census Bureau. Projections of the Size and Composition of the U.S. Population: 2014 to 2060. Current Population Reports 2015:25–1143. [Google Scholar]
- 2.Goff DC, Nichaman MZ, Chan W, et al. Greater incidence of hospitalized myocardial infarction among Mexican Americans than non-Hispanic whites. The Corpus Christi Heart Project, 1988–1992. Circulation 1997;95:1433–40. [DOI] [PubMed] [Google Scholar]
- 3.Morgenstern LB, Smith MA, Lisabeth LD, et al. Excess stroke in Mexican Americans compared with non-Hispanic Whites: the Brain Attack Surveillance in Corpus Christi Project. American journal of epidemiology 2004;160:376–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey DK, Labarthe DR, Goff DC, et al. Community-wide coronary heart disease mortality in Mexican Americans equals or exceeds that in non-Hispanic whites: the Corpus Christi Heart Project. The American journal of medicine 2001;110:81–7. [DOI] [PubMed] [Google Scholar]
- 5.Manichaikul A, Palmas W, Rodriguez CJ, et al. Population Structure of Hispanics in the United States: The Multi-Ethnic Study of Atherosclerosis. PLoS Genet 2012;8:e1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nature reviews Genetics 2011;12:523–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson B, Tuna N, Bouchard T, et al. Genetic factors in the electrocardiogram and heart rate of twins reared apart and together. The American Journal of Cardiology 1989;63:606–9. [DOI] [PubMed] [Google Scholar]
- 8.Cheng S, Keyes MJ, Larson MG, et al. Long-term outcomes in individuals with prolonged PR interval or first-degree atrioventricular block. JAMA : the journal of the American Medical Association 2009;301:2571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy D, Talajic M, Dubuc M, et al. Atrial fibrillation and congestive heart failure. Current Opinion in Cardiology 2009;24:29–34 10.1097/HCO.0b013e32831c8c58. [DOI] [PubMed] [Google Scholar]
- 10.Soliman EZ, Prineas RJ, Case LD, et al. Ethnic Distribution of ECG Predictors of Atrial Fibrillation and Its Impact on Understanding the Ethnic Distribution of Ischemic Stroke in the Atherosclerosis Risk in Communities (ARIC) Study. Stroke 2009;40:1204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler AM, Yin X, Evans DS, et al. Novel loci associated with PR interval in a genome-wide association study of 10 African American cohorts. Circulation Cardiovascular genetics 2012;5:639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers JC, Zhao J, Terracciano CMN, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet 2010;42:149–52. [DOI] [PubMed] [Google Scholar]
- 13.Holm H, Gudbjartsson DF, Arnar DO, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet 2010;42:117–22. [DOI] [PubMed] [Google Scholar]
- 14.Hong KW, Lim JE, Kim JW, et al. Identification of three novel genetic variations associated with electrocardiographic traits (QRS duration and PR interval) in East Asians. Hum Mol Genet 2014;23:6659–67. [DOI] [PubMed] [Google Scholar]
- 15.Newton-Cheh C, Guo C-Y, Wang T, et al. Genome-wide association study of electrocardiographic and heart rate variability traits: the Framingham Heart Study. BMC Medical Genetics 2007;8:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet 2010;42:153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sano M, Kamitsuji S, Kamatani N, et al. Genome-wide association study of electrocardiographic parameters identifies a new association for PR interval and confirms previously reported associations. Hum Mol Genet 2014;23:6668–76. [DOI] [PubMed] [Google Scholar]
- 18.Smith JG, Lowe JK, Kovvali S, et al. Genome-wide association study of electrocardiographic conduction measures in an isolated founder population: Kosrae. Heart Rhythm 2009;6:634–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith JG, Magnani JW, Palmer C, et al. Genome-Wide Association Studies of the PR Interval in African Americans. PLoS Genet 2011;7:e1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rane S, Patton KK. Impact of Sex and Ethnicity on Arrhythmic Risk. Current Cardiology Reports 2015;17:50. [DOI] [PubMed] [Google Scholar]
- 21.Avery CL, Wassel CL, Richard MA, et al. Fine-mapping of QT regions in global populations refines previously identified QT loci and identifies signals unique to african and hispanic descent populations. Heart Rhythm 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sofer TH, Bogomolov M, Avery CL, et al. A Powerful Statistical Framework for Generalization Testing in GWAS, with Application to the HCHS/SOL. UW Biostatistics Working Paper Series 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moskowitz IPG, Kim JB, Moore ML, et al. A Molecular Pathway Including Id2, Tbx5, and Nkx2–5 Required for Cardiac Conduction System Development. Cell 2007;129:1365–76. [DOI] [PubMed] [Google Scholar]
- 24.Chanda P, Yuhki N, Li M, et al. Comprehensive evaluation of imputation performance in African Americans. Journal of human genetics 2012;57:411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Li M, Su B. GWAS‐identified schizophrenia risk SNPs at TSPAN18 are highly diverged between Europeans and East Asians. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 2016. [DOI] [PubMed] [Google Scholar]
- 26.Nakano M, Ikeda Y, Tokuda Y, et al. Novel common variants and susceptible haplotype for exfoliation glaucoma specific to Asian population. Scientific reports 2014;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torgerson D, Galanter JM, Roth LA, et al. Admixture Mapping From Existing Genome-Wide Association Data Identifies SMAD2 As A Population-Specific Risk Factor For Asthma In Latinos. C66 THE INTERACTION BETWEEN GENES, PROTEINS AND ENVIRONMENT: Am Thoracic Soc 2013:A6102-A. [Google Scholar]
- 28.Sidore C, Busonero F, Maschio A, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nature genetics 2015;47:1272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Igartua C, Myers RA, Mathias RA, et al. Ethnic-specific associations of rare and low-frequency DNA sequence variants with asthma. Nature communications 2015;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature 2016;538:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.