Abstract
Breast cancer is a frequent female malignant tumor with high mortality and poor prognosis. Peroxidasin like (PXDNL) has many biological functions, including characteristic activity of hormone biosynthesis, host defense, and cell motility. In addition, PXDNL is closely connected with the progression of breast cancer. In this study, we found that PXDNL may be an independent prognostic biomarker of breast cancer.
We tested the mRNA expression of PXDNL in breast cancer by detecting The Cancer Genome Atlas (TCGA) database. The chi-squared test was used to evaluate clinical correlation. The receiver operating characteristic (ROC) curves were drawn to evaluate diagnosis potential in breast cancer. Subsequently, survival analyses were performed to identify the relevance between the expression of PXDNL and the overall survival/relapse-free survival of patients with breast cancer. Univariate/multivariate Cox regression model was executed to detect risk factors affecting the prognosis of patients with breast cancer.
PXDNL is highly expressed in breast cancer tissues and is related to survival status of patients. The ROC curve showed that PXDNL had beneficial diagnostic ability in breast cancer. Survival analysis indicated that patients with breast cancer with high PXDNL expression generally had decreased overall survival/relapse-free survival. Univariate/multivariate Cox model analyses further suggested an association between PXDNL expression and prognosis of patients with breast cancer.
High PXDNL expression is a potential and independent prognostic biomarker in breast cancer.
Keywords: biomarker, breast cancer, peroxidasin like, prognosis
1. Introduction
Breast cancer has a high incidence and is a common malignant tumor among women.[1,2] In recent years, with the ongoing development of modern molecular diagnostic technology, the treatment of breast cancer has made progress.[3–5] However, the poor prognosis of breast cancer, and particularly late-stage, is still an urgent problem. Therefore, identifying a reliable and feasible prognostic biomarker of breast cancer has been a key field in breast cancer research.
peroxidasin like (PXDNL) is a member of the peroxidase gene family. This gene encodes for peroxidase-like protein, and its isomer PMR1 encodes nucleic acid endonucleases that selectively degrade certain target genes. The expression of human PXDNL stimulates cell motility in a similar way to Xenopus PMR1, thus providing clear evidence which links the downregulation of endonucleases with the regulation of cell motility.[6] In a cohort study, researchers found that the top-driving oncogenes, including PXDNL 7, had mutation prevalence of >5% in 6697 patients with breast cancer, but their role in breast cancer was unsharp.[7]
For further prove the clinical roles of PXDNL in breast cancer patients, we investigated the expression of PXDNL by detecting The Cancer Genome Atlas (TCGA). The chi-squared test was implemented to assess clinical relevance. Receiver operating characteristic (ROC) curves were performed to calculate the capability of diagnosis in patients. Survival analyses were executed to identify the effect of PXDNL on the overall survival/relapse-free survival of patients with breast cancer. Univariate/multivariate Cox regression model were used to find out independent risk factors for breast cancer prognosis.
2. Materials and methods
2.1. Data source
Using TCGA (https://cancergenome.nih.gov/) databases, we obtained currently RNAseq data and available clinical from breast cancer and normal tissues, respectively. All data used was from public datasets.
2.2. Data mining and statistical analyses
We included the full TCGA-BRCA cohort in this analysis, and “NA” values were removed when specific analyses (Fisher exact test, chi-squared test, and survival analysis) were executed. In order to evaluate the prognosis results more accurately, some clinical features of patients are used. We do data mining by using R (version 3.6.1).[8] The ggplot2 package[9] was used to plot the boxplot of clinical features. The pROC package[10] mapped ROC, which was used to assess diagnostic capabilities and the best OS cut-off values based on the Youden index, to separate the high PXDNL expression group from the low PXDNL expression group. The chi-squared test was used to test the possible clinical interdependency between clinical features and the expression of PXDNL. We also used a survival package[11,12] to plot survival curves, and then a logarithmic rank test to check survival bias. The univariate Cox model and multivariate Cox model were used to distinguish the independence of PXDNL expression from other clinical features.
3. Results
3.1. Data cohort characteristics
The PXDNL expression levels, as well as clinical features of patients, including age, sex, progesterone receptor (PR) status, human epidermal growth factor 2 (HER2) status, estrogen receptor (ER) status, cancer stage, new type, histologic grade, molecular subtype, menopause status, T/N/M classification, lymph node status, margin status, radiation therapy, neoadjuvant treatment, targeted molecular therapy, sample type, overall survival, relapse-free survival, and vital status, are shown in Table 1.
Table 1.
Correlation between the expression of peroxidasin like and the clinicopathologic characteristics in breast cancer.
3.2. PXDNL expression was correlated with breast cancer
Boxplots of PXDNL expression showed that higher PXDNL expression was obtained in breast cancer (P = 1.8 × 10−06; Fig. 1). In addition, PXDNL was also differentially expressed by molecular subtype (P = .015), menopause status (P = .00028), age (P = .00039), M classification (P = .0016), and vital status (P = .0034).
Figure 1.
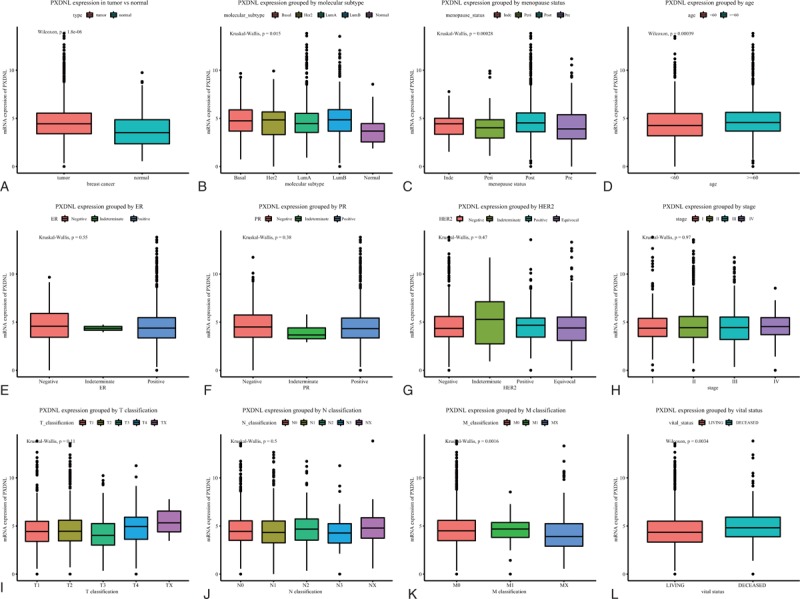
The different PXDNL expressions in the boxplot. The expression of PXDNL is grouped by type (A), molecular subtype (B), menopause status (C), age (D), ER (E), PR (F), HER2 (G), stage (H), T classification (I), N classification (J), and M classification (K), and vital status (L). ER = estrogen receptor, HER2 = human epidermal growth factor 2, PR = progesterone receptor, PXDNL = peroxidasin like.
3.3. PXDNL has substantial diagnostic capability
According to ROC analysis, we found that the area under the curve (AUC) was 0.636, suggesting that PXDNL may have considerable diagnostic ability. We reached the same results by analyzing the subgroups of different stages (AUC: 0.628, 0.637, 0.633, 0.627 for stage I, stage II, stage III, stage IV, respectively; Fig. 2).
Figure 2.
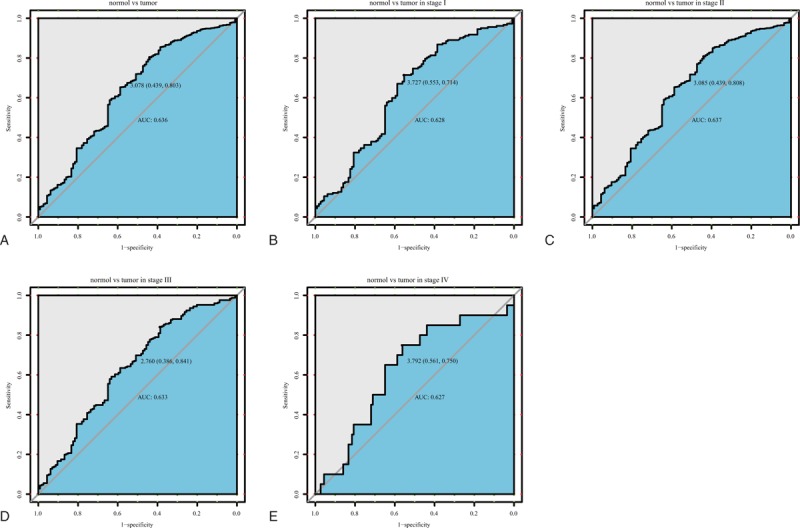
The ROC curve of peroxidasin like (PXDNL) in The Cancer Genome Atlas (TCGA) cohort. A, Nontumor sample and tumor sample. B, Nontumor sample and tumor sample of stage I. C, Nontumor sample and tumor sample of stage II. D, Nontumor sample and tumor sample of stage III. E, Nontumor sample and tumor sample of stage IV. AUC = area under the curve, ROC = receiver-operating characteristic curve.
3.4. Relationship between PXDNL and patients’ clinical features
As shown in Table 1, the expression of PXDNL was closely related to age (P = .032), ER status (P = .0003), T/N/M classification (P = .0001; P = 0; P = 0), margin status (P = .0002), stage (P = 0), and vital status (P = 0) of patients with breast cancer.
3.5. The increase in PXDNL expression was related to poor overall survival of breast cancer
We found that high PXDNL expression in patients with overall survival (P < .0001; Fig. 3), Subgroup analysis showed that PXDNL expression had significant prognostic value in infiltrating ductal carcinoma (P = .00097), infiltrating lobular carcinoma (P = .0073), ER-positive cancer (P < .0001), PR-negative cancer (P = .032), PR-positive cancer (P = 0.00013), HER2-negative cancer (P = .011), Lum A (P = .0015), and Lum B (P = .041).
Figure 3.
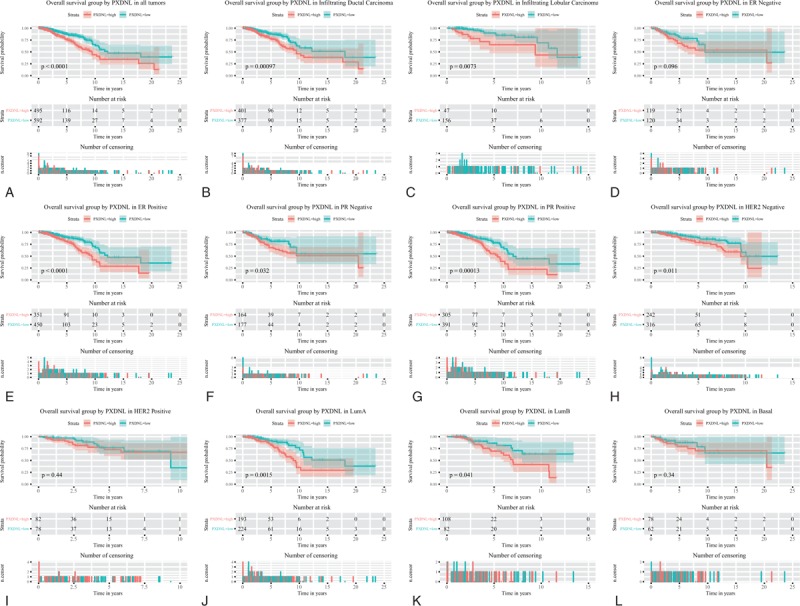
Survival analysis of PXDNL expression in terms of overall survival. Kaplan-Meier curves produced survival analysis (A) and subgroup analysis of infiltrating ductal carcinoma (B), infiltrating lobular carcinoma (C), ER positive (D), PR negative (E), PR positive (F), HER2 negative (G), Lum A (H), and Lum B (I). ER = estrogen receptor, HER2 = human epidermal growth factor 2, PR = progesterone receptor, PXDNL = peroxidasin like.
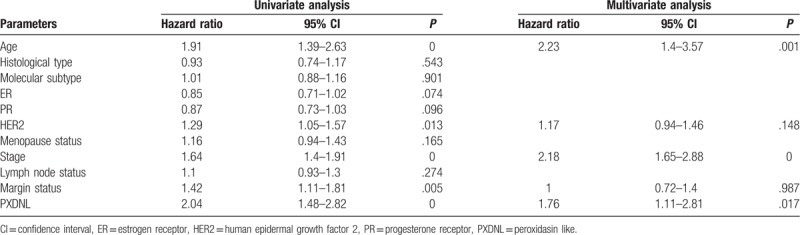
Univariate and multivariate Cox model analysis recommended that high expression of PXDNL may be an independent risk factor for overall survival in patients with breast cancer (hazard ratio [HR] = 1.76, P = .017, Table 2).
Table 2.
Summary of univariate and multivariate Cox regression analyses of overall survival duration.
3.6. The increase of PXDNL expression is related to poor relapse-free survival in patients with breast cancer
We found that high PXDNL expression in patients with shorter relapse-free survival time (P < .0059; Fig. 4). Subgroup analysis showed that PXDNL expression had significant prognostic value in infiltrating ductal carcinoma (P = .016), ER-positive cancer (P = .0035), Lum A (P = .0019), and PR-positive cancer (P = .012).
Figure 4.
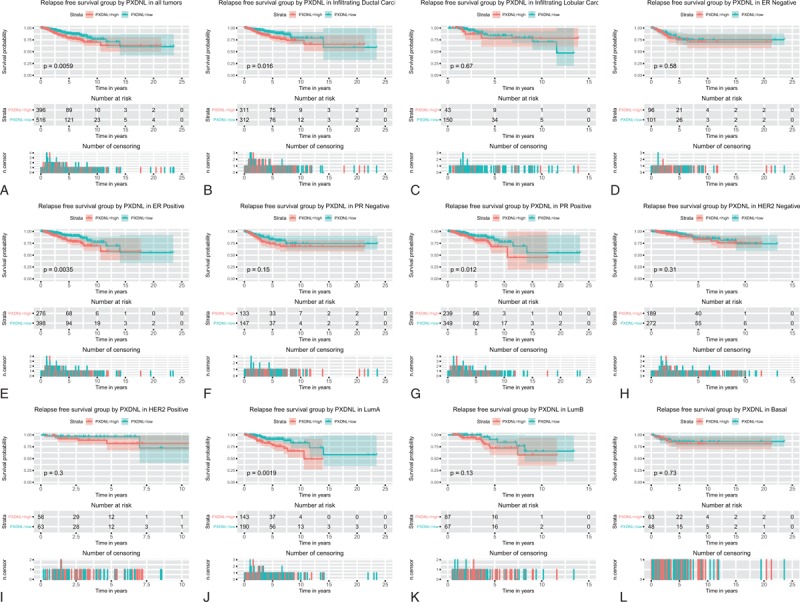
Survival analysis of PXDNL expression in terms of relapse-free survival. Kaplan-Meier curves produced survival analysis (A) and subgroup analysis of infiltrating ductal carcinoma (B), infiltrating lobular carcinoma (C), ER positive (D), PR negative (E), PR positive (F), HER2 negative (G), Lum A (H), and Lum B (I). ER = estrogen receptor, HER2 = human epidermal growth factor 2, PR = progesterone receptor, PXDNL = peroxidasin like.
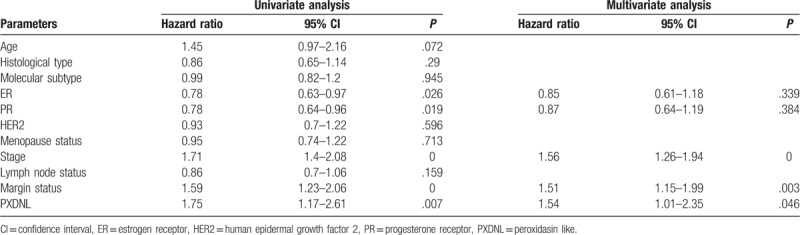
Univariate and multivariate Cox model analysis recommended that high expression of PXDNL may be an independent risk factor for the relapse-free survival in patients with breast cancer (HR = 1.54, P = .046; Table 3).
Table 3.
Summary of univariate and multivariate Cox regression analyses of relapse-free survival duration.
4. Discussion
Breast cancer is a fatal disease that particularly endangers women's health. With the development of modern social economy, various lifestyle changes such as premature sexual maturity, postponement of menopause, infertility, late childbearing, unhealthy lifestyle, increased intake of exogenous estrogen, and other environmental factors have increased in the incidence of breast cancer year by year.[13] In 2018, there were 209.96 million new cases of breast cancer worldwide, making it one of the most common causes of female cancerous deaths in developed and developing countries.[14] Although the clinical treatment of breast cancer has made progress, the prognosis of the disease remains poor. Most common treatments, such as radiotherapy or surgical resection, do not guarantee the survival of patients with advanced breast cancer.[15,16] Conversely, continued chemotherapy can lead to severe endothelial dysfunction, leading to complications such as heart attack, heart failure, stroke, or congestive decline.[17] Therefore, the field urgently requires a prognostic biomarker to guide clinicians to improve the prognosis of patients with breast cancer. Our team has been working for exploring novel biomarkers in various cancer.[18–24] In this study, we found that the high PXDNL expression may be a potential and independent risk factor for the prognosis of patients with breast cancer.
PMR1 endonuclease was found in the breasts of Xenopus laevis and identified as a member of a large and diverse peroxidase gene family. PXDNL was later identified as another member of this family.[25] PXDNL mutations have been found in 6697 patients with breast cancer and a system (http://www.g-2-o.com) was set up to quickly verify the mutations found in a large breast cancer cohort, which contributed greatly to breast cancer research.[7] We found that the expression of PXDNL was increased, suggesting that PXDNL played an important function in breast cancer progression. In addition, we also found increased expression of PXDNL in deceased patients, suggesting a link between high expression of PXDNL and poor prognosis in patients with breast cancer. Nonetheless, as the potential mechanism is still unclear, it is necessary to further explore the role played by PXDNL in patients with breast cancer.
PXDNL is closely related to the diagnosis and prognosis of cancer. Researchers have found that PXDNL is involved in many disease processes, such as neurodegenerative disorders[26] and severe depressive disorders.[27] However, the specific role of PXDNL in breast cancer has not yet been elucidated. In this research, we used ROC curves to show that that PXDNL has an excellent diagnostic ability in breast cancer. In addition, we found that when PXDNL was overexpressed, the overall/ relapse-free survival time of patients with breast cancer was shorter. By subgroup analysis, we found that PXDNL had statistical significance in the overall survival rate of fibroductal carcinoma and invasive lobular carcinoma, which have been identified as the main sources of breast cancer.[28,29] Interestingly, we also found that PXDNL had statistical significance in the overall survival analysis of HER2-negative molecular subtyping, which has further improved patient individualization and precise treatment.
In view of the correlation between endonuclease and metastasis mechanisms of malignant tumors, and the established role of PXDNL in endonucleases,[30] it seems that PXDNL take part in the metastasis of breast cancer. In fact, previous studies have confirmed that PXDNL expression is associated with the migration ability of MCF-7 cells.[31] Therefore, the association between high PXDNL expression and low survival rate in patients with breast cancer in this study may be due to the potential contribution of PXDNL to tumor metastasis.
As far as we know, this is the first study to investigate the prognostic value of PXDNL in breast cancer. With joint efforts from other studies of PXDNL, our study should provide utility for researchers to fully understand the effect of PXDNL in cancer progression and provide a greater possibility to offer patients prognosis prediction. However, in-depth molecular mechanisms have not been fully elucidated in this cancer. Therefore, in the future, we plan to obtain more tissue samples from clinical patients, and further detect and understand levels of PXDNL expression according to various treatments received by patients.
5. Conclusion
In a word, we generally payed close attention to the prognosis values of PXDNL in patients with breast cancer. High PXDNL expression has been shown to be a potential and independent prognosis biomarker in breast cancer. Consider noting the sample size of the patient subset in the TCGA, or that further in vitro/in vivo data are required to formally prove the correlation between high PXDNL expression and poor prognosis.
Author contributions
Conceptualization: Yanan Liu.
Data curation: Yanqing Li, Yanan Liu.
Formal analysis: Zhangping Luo, Yanan Liu.
Investigation: Yanan Liu.
Methodology: Yan Jiao, Yang Li.
Software: Zhangping Luo.
Visualization: Yang Li.
Writing – original draft: Yang Li, Yanan Liu.
Writing – review and editing: Yanqing Li, Yang Li, Yanan Liu.
Yang Li orcid: 0000-0003-4399-1505.
Footnotes
Abbreviations: AUC = area under the curve, ER = estrogen receptor, HER2 = human epidermal growth factor 2, PR = progesterone receptor, PXDNL = peroxidasin like, ROC = receiver operating characteristic, TCGA = The Cancer Genome Atlas.
How to cite this article: Li Y, Jiao Y, Luo Z, Li Y, Liu Y. High PXDNL expression is a potential and independent prognostic biomarker in breast cancer. Medicine. 2019;98:44(e17703).
Funding Statement: The project is funded by Bethune Program of Jilin University (2018A02), Jilin Provincial Department of Health (2018J061) and basic research fees.
Compliance with ethical standards: The manuscript has not yet been published or submitted for publication elsewhere. All authors contributed to and agreed with the contents of the manuscript. All participants obtained informed consent. In addition, there is no proprietary or commercial interest in any of the products mentioned or concepts discussed in this article. The TCGA database is publicly available. The study manuscript does not contain open clinical studies or patient data.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Radisky DC, Visscher DW, Frank RD, et al. Natural history of age-related lobular involution and impact on breast cancer risk. Breast Cancer Res Treat 2016;155:423–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kamel AM, Teama S, Fawzy A, et al. Plasma DNA integrity index as a potential molecular diagnostic marker for breast cancer. Tumour Biol 2016;37:7565–72. [DOI] [PubMed] [Google Scholar]
- [4].Marchio C, Balmativola D, Castiglione R, et al. Predictive diagnostic pathology in the target therapy era in breast cancer. Curr Drug Targets 2017;18:4–12. [DOI] [PubMed] [Google Scholar]
- [5].McGale P, Taylor C, Correa C, et al. Effect of radiotherapy after mastectomy and axillary surgery on 10-year recurrence and 20-year breast cancer mortality: meta-analysis of individual patient data for 8135 women in 22 randomised trials. Lancet 2014;383:2127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gu SQ, Bakthavachalu B, Han J, et al. Identification of the human PMR1 mRNA endonuclease as an alternatively processed product of the gene for peroxidasin-like protein. RNA 2012;18:1186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pongor L, Kormos M, Hatzis C, et al. A genome-wide approach to link genotype to clinical outcome by utilizing next generation sequencing and gene chip data of 6,697 breast cancer patients. Genome Med 2015;7:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].R Development Core Team. 2018. R: a language and environment for statistical computing. Version 3.5.2. Vienna: R Foundation for Statistical Computing. Available at https://www.R-project.org. [Google Scholar]
- [9].Wickham HJ. 2011. Ggplot2: elegant graphics for data analysis, vol. 174. New York: Springer-Verlag, 245–246. [Google Scholar]
- [10].Robin X, Turck N, Hainard A, et al. 2011. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77. [Google Scholar]
- [11].Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Vol. 97. New York: Springer; 2000. [Google Scholar]
- [12].Therneau TM. A Package for Survival Analysis in S. 2015. [Google Scholar]
- [13].Torre LA, Islami F, Siegel RL, et al. Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev 2017;26:444–57. [DOI] [PubMed] [Google Scholar]
- [14].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [15].Chan A, Bauwens A, Pontre S, et al. Efficacy of scalp cooling in reducing alopecia in early breast cancer patients receiving contemporary chemotherapy regimens. Breast (Edinburgh, Scotland) 2018;41:127–32. [DOI] [PubMed] [Google Scholar]
- [16].Maishman T, Cutress RI, Hernandez A, et al. Local recurrence and breast oncological surgery in young women with breast cancer: the POSH observational cohort study. Ann Surg 2017;266:165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bodai BI, Tuso P. Breast cancer survivorship: a comprehensive review of long-term medical issues and lifestyle recommendations. Perm J 2015;19:48–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hou L, Zhang X, Jiao Y, et al. ATP binding cassette subfamily B member 9 (ABCB9) is a prognostic indicator of overall survival in ovarian cancer. Medicine (Baltimore) 2019;98:e15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jiao Y, Fu Z, Li Y, et al. High EIF2B5 mRNA expression and its prognostic significance in liver cancer: a study based on the TCGA and GEO database. Cancer Manag Res 2018;10:6003–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jiao Y, Fu Z, Li Y, et al. Aberrant FAM64A mRNA expression is an independent predictor of poor survival in pancreatic cancer. PLoS One 2019;14:e0211291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jiao Y, Li Y, Jiang P, et al. PGM5: a novel diagnostic and prognostic biomarker for liver cancer. PeerJ 2019;7:e7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jiao Y, Li Y, Liu S, et al. ITGA3 serves as a diagnostic and prognostic biomarker for pancreatic cancer. Onco Targets Ther 2019;12:4141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jiao Y, Li Y, Lu Z, et al. High trophinin-associated protein expression is an independent predictor of poor survival in liver cancer. Dig Dis Sci 2019;64:137–43. [DOI] [PubMed] [Google Scholar]
- [24].Li Y, Jiao Y, Fu Z, et al. High miR-454-3p expression predicts poor prognosis in hepatocellular carcinoma. Cancer Manag Res 2019;11:2795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A 2002;99:16899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nikoletopoulou V, Tavernarakis N. The PMR1 pump in alpha-synuclein toxicity and neurodegeneration. Neurosci Lett 2018;663:66–71. [DOI] [PubMed] [Google Scholar]
- [27].Chen X, Long F, Cai B, et al. A novel relationship for schizophrenia, bipolar, and major depressive disorder. Part 8: a hint from chromosome 8 high density association screen. Mol Neurobiol 2017;54:5868–82. [DOI] [PubMed] [Google Scholar]
- [28].Christgen M, Langer F, Kreipe H. Histological grading of breast cancer [in German]. Pathologe 2016;37:328–36. [DOI] [PubMed] [Google Scholar]
- [29].Yamashita T, Kaneko S. Liver cancer. Jpn J Clin Pathol 2016;64:787–96. [PubMed] [Google Scholar]
- [30].Lu R, Wang Y, Xu X, et al. Establishment of a detection assay for DNA endonuclease activity and its application in the screening and prognosis of malignant lymphoma. BMC Biochem 2018;19:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gu SQ, Gallego-Perez D, McClory SP, et al. The human PMR1 endonuclease stimulates cell motility by down regulating miR-200 family microRNAs. Nucleic Acids Res 2016;44:5811–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


