Abstract
The present study aimed to detect the marker-trait association of a selected diverse panel of 127 mungbean genotypes against mungbean yellow mosaic India virus (MYMIV). Virus-specific primers pairs viz., AC-abut/AV-abut and BC-abut/BV-abut confirmed the involvement of MYMIV in yellow mosaic disease development and the same was validated through restriction digestion analysis. 256 genome-wide microsatellite markers were screened on a test panel in which 93 polymorphic markers were used in association studies. Population structure analysis led to formation of six distinct subpopulations. 1097 alleles were detected among 127 test genotypes whereas number of alleles ranged 2–22 and PIC values ranged 0.27–0.92%, indicating ample amount of variation at genome level. 15 microsatellite markers were detected as associated with MYMIV resistance, among them three microsatellites explained 11–14% phenotypic variation. The specific regions close to CEDG293, DMB-SSR008 and DMB-SSR059 associated with MYMIV resistance were detected, located on linkage group 2, 4 and 9 and may prove useful in marker-assisted mungbean improvement programme for enhancing MYMIV resistance.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-2035-7) contains supplementary material, which is available to authorized users.
Keywords: Marker-trait association, MYMIV, Simple sequence repeats, Vigna radiata, YMD
Introduction
Mungbean or green gram [Vigna radiata (L.) Wilczek] has a unique position in prominent cropping systems in the Indian subcontinent as well other countries of East and South-East Asia due to its shorter life cycle, high per day productivity and use in numerous food preparations (Singh et al. 2017). Development of disease resistant, short duration and photo-thermo insensitive varieties of mungbean during the last three decades has further helped in expanding its cultivation to newer areas and cropping systems in the world. However, several biotic and abiotic stresses such as drought, fluctuating temperatures and pest and disease problems continue to impart a challenge towards realizing the true yield potential of the released mungbean cultivars (Ali et al. 2010). Among them, viral diseases have emerged as the major threats to its cultivation (Singh et al. 2019). Yellow mosaic disease (YMD) caused by begomoviruses (family Geminiviridae, genus Begomovirus) is the most serious disease which has been reported to cause tremendous yield losses (Varma et al. 1992; Maiti et al. 2011) as well as deterioration in the seed quality in mungbean.
Three species of Begomoviruses, i.e., mungbean yellow mosaic India virus (MYMIV), mungbean yellow mosaic virus (MYMV) and horsegram yellow mosaic virus (HgYMV) are known to cause YMD in mungbean (Malathi and John 2008). Dolichos yellow mosaic virus (DoYMV) is also noticed as very close at genome level to these three viruses but its infection is reported limited to Dolichos bean only (Naimuddin et al. 2016). In Vigna, the YMD causing viruses are bipartite having DNA-A and DNA-B components, approx 2.7–2.8 kb in size (Pant et al. 2001; Borah and Dasgupta 2012) which are about 2723 and 2675 nucleotides long, respectively (Qazi et al. 2007). The DNA-A component of MYMIV and MYMV share about 82% identity in their sequences and therefore justify their separation into distinct species. The second group of DNA-B component is more related to MYMIV (about 92%) than to MYMV. The DNA-B group represents MYMIV, MYMV and HgYMV which cause similar disease symptoms and it is extremely difficult to differentiate them on the basis of morphological symptoms on the plant. However, genome characterization of these three described viruses helps in identifying the virus causing YMV diseases (Akaram et al. 2015).
The conventional breeding approach coupled with molecular marker technology has proven to be effective in introgressing disease resistance in food legumes such as chickpea (Varshney et al. 2014; Pratap et al. 2017), which can help in accelerating mungbean improvement programmes also. In mungbean, molecular breeding now sounds more practicable after the availability of whole genome sequence (Kang et al. 2014). Earlier, several workers utilized the cross-transferable microsatellite markers from related legumes in developing mungbean maps for various important traits (Isemura et al. 2012; Kitsanachandee et al. 2013; Chankaew et al. 2014), which indicated availability of reliable markers for marker-assisted selection (MAS) and identification of quantitative trait loci (QTLs) for desired traits. Developing high yielding varieties with enhanced resistance by tagging and mapping of loci/gene(s) governing the specific disease resistance is more effective and efficient approach. Based on published reports it is amply clear that MYMIV resistance in mungbean is governed by QTLs suggesting the quantitative inheritance of MYMIV. To date, seven QTLs have been reported for MYMIV resistance through bi-parental linkage analysis (Kitsanachandee et al. 2013; Alam et al. 2014). However, linkage mapping has its own limitations due to bi-parental segregation. In recent years, exploring QTLs by association analysis has been one of the effective approaches in quantitative genetics, which performs rapid and fine-mapping of the target locus. In the present investigation, we focused on identification of novel QTLs/associated markers for MYMIV resistance through SSR-based genome-wide association mapping approach.
Materials and methods
Mapping panel, field experimentation and disease scoring
The plant materials comprised 127 mungbean genotypes including released varieties, advanced breeding materials, elite lines, and local and exotic collections (Supplementary Table 1). The materials were grown under natural field conditions in six different environments viz., the tropical climate at ICAR-Indian Institute of Pulses Research, Kanpur, India (26.28° N, 80.24° E, 152.4 m above mean sea level) during Monsoon season (Kharif) of 2016 (E1), Summer season of 2017 (E2), Monsoon season of 2017 (E3), New Research Farm, Kanpur during Monsoon 2016 (E4), Summer season of 2017 (E5) and Monsoon season of 2017 (E6). In each environment, all test genotypes were grown in two rows of 4 m length. Four highly MYMIV susceptible genotypes viz., Co-5, Vamban-1, Barabanki local and DGGV-2 were grown after every five test genotypes for creating sufficient innoculum load to ensure field epiphytotic conditions. The recommended cultural practices without any plant protection measure such as chemical seed treatment, insecticidal spray, etc. were followed to raise the crop. Disease scoring was done on 0–9 scale (0 as highly resistant and 9 as highly susceptible) following Ahmed (1985) (Table 1). When infector genotypes recorded > 75% of yellow mosaic symptoms, which usually occurred at 40–45 days after sowing, severity of disease was calculated as per the following formula:
Table 1.
Disease scoring for categorization of yellow mosaic disease in mungbean
| Disease score | Disease reaction | Remarks |
|---|---|---|
| 0 | HR | No visible disease symptoms on leaves |
| 1 | R | < 5.0% disease symptoms on leaves |
| 3 | MR | 5.1–15.0% disease symptoms on leaves |
| 5 | MS | 15.1–30.0% disease symptoms on leaves |
| 7 | S | 30.1–75.0% disease symptoms on leaves |
| 9 | HS | > 75.1% disease symptoms on leaves and sometimes may be on pods |
HR Highly resistance, R resistance, MR moderately resistance, MS moderately susceptible, S susceptible, HS highly susceptible
Disease severity (DS) = {sum of all disease ratings/(total number of plants × maximum grade)} × 100.
Pathogen identification, validation and microsatellite markers
The genomic DNA extracted from symptomatic leaves showing yellow mosaic infestation was subjected to rolling circular amplification using RPLI-g kit (Qiagen, India) as per manufacturer’s instruction to enrich the copy of circular DNA. The amplified product was diluted @1:20 (DNA: H2O) and used for detection of virus(es). Six primer pairs each for DNA-A and DNA-B of MYMIV, MYMV and HgYMV were used for detection of presence of virus species (Table 2). The identified virus species leading to YMD development was validated through restriction digestion analysis. Initially, 256 microsatellite markers were screened on a test panel of 20 genotypes comprising highly resistant to highly susceptible genotypes to detect the polymorphic markers. The selected 93 genome-wide polymorphic microsatellite markers covering the whole genome were used to find out the strongly associated markers in the vicinity of loci conferring resistance to MYMIV (Supplementary table-2).
Table 2.
Primer pairs used for characterization of virus(es) species involved in disease development
| Primer code | Sequence | Remarks | References |
|---|---|---|---|
| AC-abut/AV-abut |
GTAAAGCTTTACGCATAATG AAAGCTTACATCCTCCAC |
MYMIV Full length DNA-A | Islam et al. (2012) |
| BV-abut/BC-abut |
CCAGGATCCAATGATGCCT ATTGGATCCTGGAGATTCA |
MYMIV Full length DNA-B | |
| MYMV-CP |
ATG GG (T/G) TCC GTT GTA TGC TTG GGC GTC ATT AGC ATA GGC AAT |
MYMV/DNA A | Naimuddin et al. (2011a, b, c) |
| MYMV-MP |
ATG GAG AAT TAT TCA GGC GCA TTA CAA CGC TTT GTT CAC ATT |
MYMV/DNA B | |
| HYMV-CP |
ATG CTT GCA ATT AAG TAC TTG CA TAG GCG TCA TTA GCA TAG GCA |
HgYMV/DNA A | |
| HYMV-MP |
ATG GAG CAT TAT TCC GGT GCA TTA CA(G/A) GGT TTT GTT TAC AGT |
HgYMV/DNA B |
DNA extraction and PCR conditioning
Total genomic DNA was extracted from 15-day-old seedlings of all mungbean genotypes following the Cetyl Trimethyl Ammonium Bromide (CTAB) method (Doyle and Doyle 1990). The quality of the extracted DNA was equated on 0.8% agarose gel and the quantity was determined using Nanodrop spectrophotometer ND 1000 (Nanodrop Technologies, DE, USA). DNA samples were normalized at a concentration of 50 ng/µl for PCR amplification. The PCR amplification was carried out in 20 µl reaction mixture containing 10× Taq buffer A with 15 mM MgCl2, 2 mM dNTPs and 1U of Taq DNA polymerase (Fermentas, Mumbai), 50 ng template DNA and 10 pmol each of forward and reverse primers (ILS, India) in a tetrad thermocycler (G-Strom, Somerset, UK). PCR conditions were programmed at initial denaturation for 3 min at 95 °C, followed by 35 cycles of denaturation for 1 min at 94 °C, annealing for 1 min at 45–55 °C (primer specific), extension at 72 °C for 1–3 min (primer specific) and final extension of 72 °C for 10 min. The PCR products thus obtained were resolved on 1–3% agarose gel (based on expected size of fragments) in 1× TAE buffer and stained with ethidium bromide. Gel images were taken using gel documentation system (Uvitech, Cambridge). Polymorphic microsatellite markers were identified on the basis of differences in amplicons by comparison with 100-bp DNA ladder.
Data generation and statistical analysis
The clear and fine bands were scored and their reproducibility was validated by repeated amplification with selected markers. The data were subjected to analysis using Power Marker version 3.25 (Liu and Muse 2005) to calculate the total number of alleles (NA), major allele frequency (MAF), gene diversity (GD) and polymorphic information content (PIC) value. Population genetic structure analysis was carried out using STRUCTURE version 2.3.4 (Pritchard et al. 2000). The number of presumed population (K) was set from 2 to 15 with admixture model. The Markov Chain Monte Carlo (MCMC) chains were run at 50,000 burn-in-period at fixed number of population. The final results were based on 1,00,000 and three iterations on this chain using a model allowing for admixture. The optimal value of K was determined by examining Delta K statistic and L (K) (Evanno et al. 2005) using Structure Harvester (Earl and Von-Holdt 2012) available free online (https://taylor0.biology.ucla.edu/structureHarvester).
The TASSEL software version 2.1 (https://www.maizegenetics.net) was used to establish marker-trait association between individual markers and MYMIV. Two approaches viz., general linear model (GLM) with Q (without consideration of kinship) and multiple linear model (MLM) with Q + K (with consideration of population structure and kinship) were adopted to extract information on association of the genome-wide microsatellite markers with MYMIV resistance.
Results
Identification and validation of YMD-causing virus species
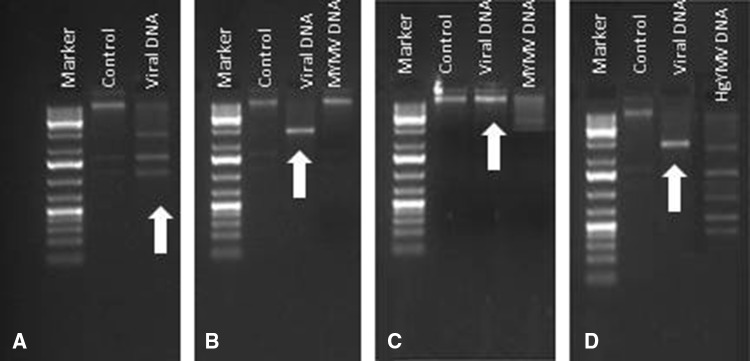
Rolling circular amplification (RCA) was performed to enrich the quantity of virus DNA followed by PCR analysis and restriction digestion with different enzymes for both DNA-A and DNA-B. PCR amplification was done with specific primers for MYMIV, MYMV and HgYMV to confirm the presence of virus(es) leading to disease development. The MYMIV specific markers, AC-abut/AV-abut (GTAAAGCTTTACGCATAATG/AAAGCTTACATCCTCCAC) and BC-abut/BV-abut (CCAGGATCCAATGATGCCT/ATTGGATCCTGGAGATTCA), the MYMIV specific primers, gave the expected fragment size of about 2.7 kb (Fig. 1a), which indicated the presence of MYMIV leading to YMD development in mungbean, whereas primer pairs MYMV-CP, MYMV-MP (Fig. 1b), HgYMV-CP and HgYMV-MP (Fig. 1c) did not amplify.
Fig. 1.
Identification and confirmation of virus species a MYMIV, b MYMV, c HgYMV in development of yellow mosaic disease
The presence of MYMIV in the infected samples was also validated through the analysis of restriction pattern of host DNA with specific enzymes (Fig. 2). Restriction digestion with XbaI (Fig. 2a) indicated the absence of DoYMV, but it could not rule out the presence of the other three viruses in the sample. To check the spectrum of other YMD-causing virus species, samples was further subjected to restriction digestion with PmeI (Fig. 2b), a cutter of MYMIV but non-cutter of MYMV, which confirmed that MYMV was not present in the samples. Likewise, the sample was again digested with non-cutter of MYMIV and one positive control of MYMV was digested with the cutter of MYMV, SmaI (Fig. 2c). The sample was further digested with HindIII (Fig. 2d), which is a single cutter of MYMIV and multiple cutter of HgYMV. Based on the digestion pattern with various restriction enzymes, it was validated that only MYMIV was present in the diseased leaf samples collected from different experimental sites.
Fig. 2.
Validation of involvement of MYMIV in developing YMD through restriction digestion a XbaI, b PmeI, c SmaI, d HindIII
MYMIV-phenotyping
MYMIV scores of test entries were recorded after reaching the disease severity > 75% in spreader rows. The sufficient number of white flies were also recorded (data not sown), whose spreaded the MYMIV over the population. Based on phenotypic score, the genotypes were grouped into six categories viz., HR, R, MR, MS, S and HS. The extent of disease severity of MYMIV was recorded upto 81.00% in E1, 88.89% in E2, 40.00% in E3, 88.33% in E4, 89.93% in E5, 93.89% in E6 and 81.02% in pooled over environments (Fig. 3). In all environments individually as well as in pooled over environments all the four susceptible genotypes exhibited more than 75% disease severity (data not shown). Accordingly, 37 (29.13%) mungbean genotypes were categorized in HR and 31 (24.41%) in R category, whereas rest of the 59 genotypes (46.10%) exhibited variable disease reactions ranging between moderately susceptible to highly susceptible.
Fig. 3.
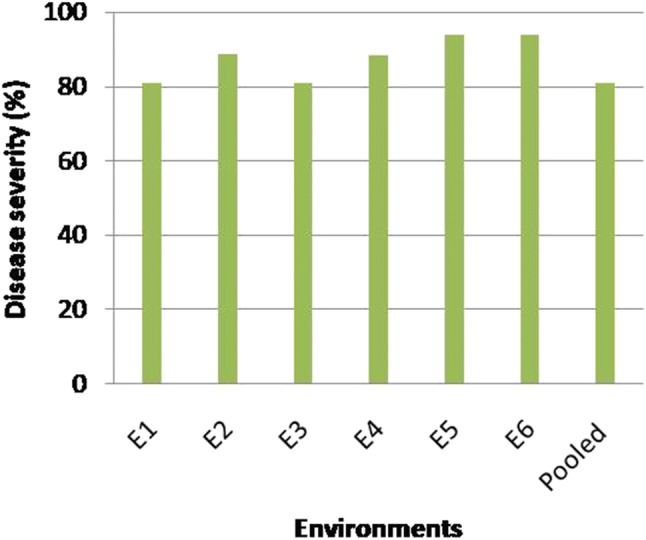
The maximum percent MYMIV-disease severity exhibited in different environments
Genetic and phenotypic variations
Ample amount of genetic variation was detected among the test genotypes in terms of major allele frequency (MAF), number of alleles (NA), gene diversity (GD) and polymorphic information content (PIC) presented in Table 3. Based on 93 polymorphic microsatellite loci, 1097 alleles were detected among the 127 test genotypes. Number of alleles ranged from 2 (CEDG244, cp08695) to 22 (CEDG254), with a mean of 11.72 alleles/locus. GD and PIC values ranged between 0.27 (cp05096) to 0.92 (CEDG100) and 0.25 (CDEG244) to 0.96 (CEDG097) with a mean value of 0.80 and 0.79, respectively. Locus CEDG097 was found most informative, whereas CDEG244 was the least. Any constant pattern of correlation was not observed between number of repeat units/number of alleles and GD or PIC. The structured data of population exhibited almost equal distribution of mungbean genotypes with differential MYMIV reactions.
Table 3.
Diversity statistics of microsatellite marker in mungbean population
| Marker | Major. Allele. Frquency | Allele No | Gene Diversity | PIC | Marker | Major. Allele. Frquency | Allele No | Gene Diversity | PIC |
|---|---|---|---|---|---|---|---|---|---|
| RGA 1F-CG/RGA 1R | 0.64 | 8 | 0.52 | 0.46 | CEDG 191 | 0.42 | 13 | 0.77 | 0.75 |
| CEDG 036 | 0.16 | 12 | 0.73 | 0.73 | CEDG 118 | 0.27 | 15 | 0.89 | 0.89 |
| CEDG 003 | 0.1 | 11 | 0.73 | 0.73 | CP1038 | 0.2 | 12 | 0.82 | 0.82 |
| CEDG 053 | 0.31 | 11 | 0.73 | 0.71 | CEDG 146 | 0.27 | 18 | 0.88 | 0.88 |
| CEDG 204 | 0.5 | 10 | 0.73 | 0.72 | CEDG 015 | 0.74 | 7 | 0.43 | 0.41 |
| CEDG 256 | 0.23 | 13 | 0.76 | 0.84 | GMES1823 | 0.14 | 13 | 0.81 | 0.81 |
| CEDG 128 | 0.75 | 14 | 0.44 | 0.43 | CP09781 | 0.15 | 13 | 0.84 | 0.84 |
| CDEG 220 | 0.45 | 11 | 0.78 | 0.77 | CEDG 225 | 0.16 | 11 | 0.82 | 0.82 |
| CEDG 254 | 0.13 | 13 | 0.83 | 0.73 | CDEG 244 | 0.85 | 2 | 0.27 | 0.25 |
| CEDG 041 | 0.16 | 11 | 0.9 | 0.79 | GMES 0211 | 0.09 | 16 | 0.86 | 0.86 |
| GMES 2320 | 0.42 | 14 | 0.78 | 0.77 | CEDG 064 | 0.15 | 15 | 0.82 | 0.82 |
| CEDG 048 | 0.14 | 11 | 0.83 | 0.82 | CEDG174 | 0.06 | 13 | 0.88 | 0.88 |
| CEDG 293 | 0.17 | 8 | 0.82 | 0.82 | MB-SSR 008 | 0.12 | 11 | 0.84 | 0.84 |
| CEDG 136 | 0.26 | 21 | 0.88 | 0.87 | CEDG186 | 0.33 | 11 | 0.86 | 0.85 |
| CEDG 297 | 0.55 | 14 | 0.67 | 0.65 | CEDG 218 | 0.39 | 11 | 0.81 | 0.8 |
| CEDG 100 | 0.15 | 12 | 0.92 | 0.92 | CEDG176 | 0.1 | 14 | 0.84 | 0.84 |
| CP02662 | 0.33 | 12 | 0.87 | 0.86 | CEDG 295 | 0.12 | 12 | 0.84 | 0.84 |
| CEDG 225 | 0.18 | 11 | 0.9 | 0.89 | CEDC 302 | 0.15 | 12 | 0.84 | 0.84 |
| CDEG 244 | 0.16 | 13 | 0.84 | 0.83 | CEDG 271 | 0.16 | 11 | 0.82 | 0.82 |
| CEDG 050 | 0.11 | 12 | 0.84 | 0.84 | CEDGAT 009 | 0.12 | 13 | 0.85 | 0.85 |
| GMES 0211 | 0.4 | 10 | 0.81 | 0.8 | CEDG 035 | 0.12 | 14 | 0.84 | 0.84 |
| CP00361 | 0.15 | 8 | 0.82 | 0.82 | VM 37 | 0.08 | 13 | 0.87 | 0.87 |
| CEDG 296 | 0.15 | 12 | 0.83 | 0.82 | CEDG 030 | 0.17 | 11 | 0.83 | 0.83 |
| BM 146 | 0.33 | 13 | 0.86 | 0.85 | CEDG 071 | 0.35 | 13 | 0.74 | 0.7 |
| DMB SSR035 | 0.09 | 15 | 0.86 | 0.86 | CEDG 247 | 0.66 | 8 | 0.54 | 0.51 |
| CEDG 305 | 0.07 | 13 | 0.86 | 0.86 | DMB-SSR 059 | 0.37 | 10 | 0.8 | 0.78 |
| CEDG 254 | 0.17 | 22 | 0.81 | 0.81 | CEDG 070 | 0.27 | 11 | 0.86 | 0.85 |
| CEDG 159 | 0.06 | 11 | 0.86 | 0.86 | CEDG 024 | 0.19 | 12 | 0.83 | 0.83 |
| CEDG 084 | 0.11 | 12 | 0.86 | 0.86 | CEDG 290 | 0.18 | 13 | 0.82 | 0.82 |
| CEDC 139 | 0.37 | 11 | 0.78 | 0.76 | VM 27 | 0.08 | 15 | 0.86 | 0.86 |
| CEDC 055 | 0.12 | 11 | 0.85 | 0.84 | CEDG 166 | 0.24 | 9 | 0.87 | 0.86 |
| GMES0162 | 0.1 | 10 | 0.83 | 0.83 | CEDG267 | 0.34 | 8 | 0.78 | 0.76 |
| CEDG 185 | 0.15 | 9 | 0.83 | 0.82 | BM 212 | 0.55 | 10 | 0.69 | 0.68 |
| CEDG 088 | 0.21 | 7 | 0.79 | 0.88 | CEDG 147 | 0.42 | 11 | 0.8 | 0.79 |
| CEDC 033 | 0.13 | 13 | 0.85 | 0.85 | CEDG113 | 0.12 | 10 | 0.83 | 0.83 |
| GMES 0035 | 0.42 | 11 | 0.79 | 0.78 | DMB-SSR043 | 0.19 | 13 | 0.9 | 0.89 |
| DMB SSR008 | 0.27 | 11 | 0.82 | 0.89 | CEDG 116 | 0.32 | 14 | 0.87 | 0.86 |
| DMB-SSR199 | 0.11 | 13 | 0.84 | 0.84 | CEDG097 | 0.11 | 11 | 0.86 | 0.96 |
| DMB-SSR080 | 0.12 | 15 | 0.84 | 0.84 | CEDG 150 | 0.33 | 14 | 0.8 | 0.78 |
| DMB SSR024 | 0.26 | 8 | 0.89 | 0.88 | CEDG 168 | 0.3 | 10 | 0.85 | 0.84 |
| CEDG 008 | 0.08 | 10 | 0.87 | 0.87 | CP00464 | 0.28 | 14 | 0.88 | 0.88 |
| CEDG 115 | 0.1 | 11 | 0.85 | 0.85 | CEDG 013 | 0.1 | 12 | 0.85 | 0.85 |
| BM 170 | 0.19 | 11 | 0.83 | 0.83 | CEDG 044 | 0.06 | 11 | 0.86 | 0.86 |
| DMB SSR151 | 0.08 | 14 | 0.85 | 0.85 | BM149 | 0.67 | 9 | 0.51 | 0.47 |
| CEDG 121 | 0.23 | 10 | 0.9 | 0.89 | CP 5096 | 0.27 | 11 | 0.84 | 0.83 |
| CEDG 211 | 0.11 | 14 | 0.84 | 0.84 | CP 08695 | 0.84 | 2 | 0.29 | 0.29 |
| DMB SSR001 | 0.12 | 11 | 0.82 | 0.82 | Mean | 0.25 | 11.72 | 0.80 | 0.79 |
Population genetic structure
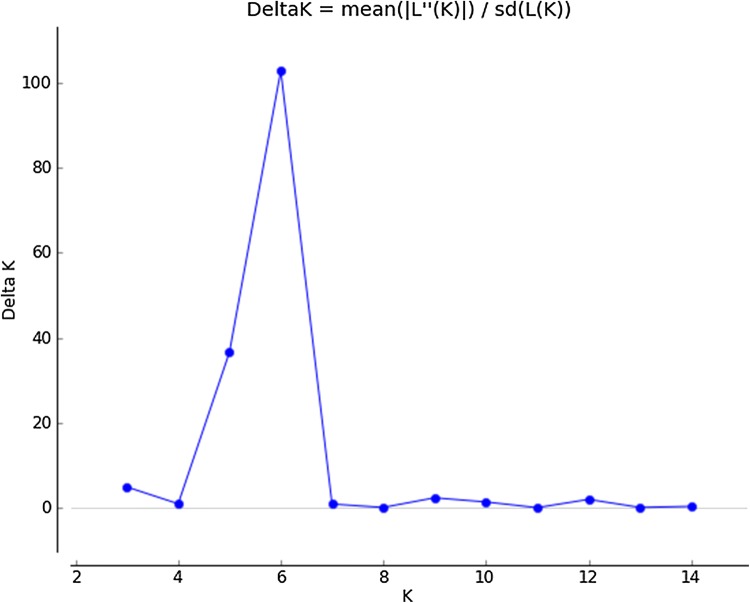
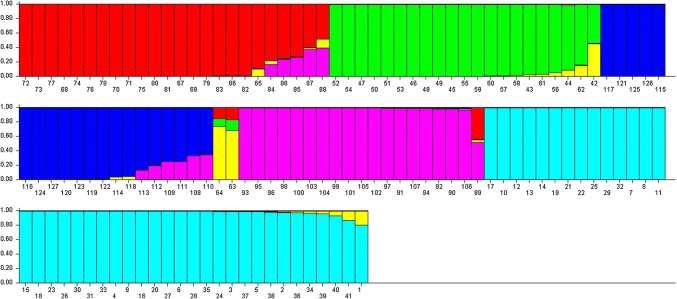
Population genetic structure of 127 mungbean genotypes was carried out and K = 6 was found suitable and showed comparable or higher likelihoods than K = 2–15 (Fig. 4). Twenty (15.74%) out of 127 mungbean genotypes (Indore Mung, EC 520024, EC 520026, EC 520029, EC 520034, EC 520034-1, ML 818, ML 512, MG 331, OBGG 52, IC 296672, IPOI 539, Kopargaon, Ganga 8, SPS 5-1, Saptari, Asha Mung, EC 550831, PDM 139 and Sona) were categorized as admixtures with varying levels of membership shared among the three groups (Fig. 5). The subpopulation-I included 24 genotypes representing 18.89% of the elite breeding lines, whereas subpopulation-II was represented by 21 genotypes (16.54%). The subpopulation-III included 20 genotypes, subpopulation-IV, 02 genotypes, subpopulation-V included 19 genotypes and subpopulation-VI included 41 genotypes sharing 15.75, 1.5, 14.96 and 32.29% of all test genotypes, respectively. The subpopulation-I consisted of most of the exotic collection and some of the varieties released by GB Pant University of Agriculture and Technology, Pantanagar and Banaras Hindu University, Varanasi. The subpopulation-II consisted of material developed by ICAR-Indian Agricultural Research Institute (IARI), New Delhi, Bhabha Atomic Research Centre (BARC), Mumbai, and Dr. Panjabrao Deshmukh Krishi Vidyapeeth (PDKV), Akola. The sobpopulation-III was characterized by advance breeding lines developed by ICAR-IIPR, Kanpur and other institutes. The subpopulation-VI was categorized as the largest subgroup which consisted majority of the released varieties and elite lines developed by ICAR-IIPR, Kanpur.
Fig. 4.
Magnitude of LnP(D) and ΔK as a function of K
Fig. 5.
Bar plot represented population genetic structure of 127 mungbean genotypes using the admixture model. Groups for each panel are represented by different colours
Marker-trait association
Association study was conducted using the MYMIV scores based on phenotypic evaluation and the allelic data of 93 genome-wide microsatellite markers. As a result, 15 marker-trait associations (MTAs) for MYMIV were observed over the environments using the most accepted maximum likelihood model (P3D mixed linear model with optimum compression) of the association analysis (Table 4). This association explained 0.41–14.45% of the total phenotypic variation through GLM whereas 0.58–13.26% of total phenotypic variation was explained by the markers through MLM. Our results showed a significant association of 15 microsatellite markers with MYMIV at P value (P < 0.05) in which five microsatellite loci viz., CEDG293, DMBSSR059, DMBSSR008, CEDG211, and CEDG121 were strongly associated with MYMIV resistance which explained about > 5.00% phenotypic variation. Among them, CEDG211, CEDG121 and DMB-SSR008 were found within the earlier reported QTL region whereas two markers viz., CEDG293 and DMB-SSR059 were found outside the region and are considered as novel identified regions.
Table 4.
Association of microsatellite markers with mungbean yellow mosaic virus resistance
| Marker | LG | R2 (E1) | P value | R2 (E2) | P value | R2 (E3) | P value | R2 (E4) | P value | R2 (E5) | P value | R2 (E6) | P value | R2 (Pooled) | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| General linear model (GLM) | |||||||||||||||
| CEDG 293 | 2 | 14.45 | 0.008 | 13.14 | 0.015 | 12.2 | 0.783 | 13.26 | 0.016 | 12.42 | 0.016 | 12.35 | 0.641 | 12.12 | 0.100 |
| CEDG 225 | 2 | 5.5 | 0.007 | 4.97 | 0.013 | 5.25 | 0.818 | 3.62 | 0.009 | 2.95 | 0.009 | 4.54 | 0.039 | 3.9 | 0.066 |
| CP00361 | 2 | 4.1 | 0.000 | 3.34 | 0.001 | 2.68 | 0.845 | 3.2 | 0.005 | 1.97 | 0.005 | 4.07 | 0.015 | 2.99 | 0.003 |
| CEDC 139 | 4 | 1.06 | 0.011 | 3.25 | 0.023 | 2.26 | 0.406 | 2.05 | 0.029 | 1.52 | 0.029 | 1.62 | 0.129 | 1.9 | 0.036 |
| DMB SSR008 | 4 | 12.24 | 0.022 | 12.41 | 0.024 | 13.04 | 0.878 | 11.34 | 0.169 | 11.84 | 0.169 | 11.91 | 0.009 | 12.87 | 0.063 |
| CEDG 121 | 6 | 3.25 | 0.010 | 1.87 | 0.072 | 2.23 | 0.960 | 1.79 | 0.187 | 2.28 | 0.187 | 1.2 | 0.200 | 1.87 | 0.193 |
| CEDG 211 | 6 | 6.05 | 0.016 | 8.7 | 0.066 | 6.52 | 0.732 | 7.85 | 0.311 | 6.15 | 0.311 | 8.52 | 0.172 | 7.48 | 0.117 |
| CEDG 191 | 6 | 4.72 | 0.004 | 4.72 | 0.028 | 8.89 | 0.850 | 7.7 | 0.095 | 6.33 | 0.095 | 5.43 | 0.146 | 5.02 | 0.068 |
| CP1038 | 6 | 3.19 | 0.005 | 2.36 | 0.008 | 3.13 | 0.902 | 1.77 | 0.157 | 1.57 | 0.157 | 2.32 | 0.091 | 1.9 | 0.137 |
| DMB-SSR 059 | 9 | 4.23 | 0.031 | 4.85 | 0.092 | 5.07 | 0.535 | 4.93 | 0.282 | 5.09 | 0.282 | 5.84 | 0.073 | 6.87 | 0.279 |
| CEDG 070 | 9 | 4.04 | 0.020 | 5.48 | 0.002 | 6.24 | 0.501 | 3.05 | 0.042 | 1.4 | 0.042 | 2.1 | 0.090 | 2.31 | 0.083 |
| CEDG 290 | 9 | 5.16 | 0.004 | 5.01 | 0.001 | 1.01 | 0.953 | 3.28 | 0.018 | 4.03 | 0.018 | 2.33 | 0.250 | 3.08 | 0.057 |
| VM 27 | 9 | 2.06 | 0.054 | 2.25 | 0.039 | 2.26 | 0.967 | 1.56 | 0.003 | 2.16 | 0.003 | 1.62 | 0.227 | 1.84 | 0.073 |
| CEDG 166 | 9 | 2.46 | 0.001 | 2.6 | 0.001 | 2.86 | 0.480 | 2.12 | 0.007 | 2.07 | 0.007 | 2.99 | 0.002 | 2.64 | 0.009 |
| Multiple linear model (MLM) | |||||||||||||||
| CEDG 293 | 2 | 13.26 | 0.008 | 12.04 | 0.015 | 11.84 | 0.7833 | 12.78 | 0.016 | 12.86 | 0.641 | 11.35 | 0.219 | 13.63 | 0.100 |
| CEDG 225 | 2 | 2.43 | 0.007 | 2.18 | 0.011 | 1.29 | 0.818 | 1.89 | 0.009 | 1.17 | 0.039 | 1.7 | 0.044 | 1.79 | 0.066 |
| CEDG 050 | 2 | 2.25 | 0.000 | 2 | 0.001 | 0.99 | 0.845 | 2.7 | 0.005 | 2.31 | 0.015 | 2.29 | 0.031 | 2.58 | 0.003 |
| CEDC 139 | 4 | 2.3 | 0.011 | 1.92 | 0.025 | 0.45 | 0.406 | 1.54 | 0.029 | 1.68 | 0.129 | 1.6 | 0.012 | 2.01 | 0.036 |
| DMB SSR008 | 4 | 11.54 | 0.010 | 11.48 | 0.075 | 12.66 | 0.960 | 11.37 | 0.187 | 11.36 | 0.200 | 11.54 | 0.192 | 11.35 | 0.193 |
| CEDG 121 | 6 | 4.76 | 0.016 | 4.7 | 0.063 | 2.84 | 0.732 | 3.23 | 0.309 | 3.26 | 0.172 | 3.26 | 0.099 | 4.53 | 0.117 |
| CEDG 211 | 6 | 5.09 | 0.004 | 5.66 | 0.023 | 5.09 | 0.850 | 7.73 | 0.093 | 4.22 | 0.146 | 6.12 | 0.025 | 5.18 | 0.068 |
| CEDG 191 | 6 | 1.3 | 0.005 | 1.35 | 0.008 | 2.29 | 0.902 | 1.21 | 0.154 | 1.14 | 0.091 | 1.43 | 0.117 | 1.39 | 0.137 |
| cp01038 | 6 | 1.63 | 0.029 | 1.42 | 0.079 | 0.83 | 0.535 | 1.44 | 0.275 | 0.58 | 0.073 | 0.84 | 0.285 | 1.03 | 0.279 |
| DMB-SSR 059 | 9 | 12.45 | 0.021 | 13.08 | 0.002 | 12.66 | 0.501 | 12.64 | 0.043 | 14.03 | 0.090 | 12.85 | 0.031 | 3.45 | 0.083 |
| CEDG 166 | 9 | 2.83 | 0.004 | 2.87 | 0.001 | 0.71 | 0.953 | 2.18 | 0.018 | 2.16 | 0.250 | 3.29 | 0.080 | 2.95 | 0.057 |
| BM 212 | 9 | 2.48 | 0.001 | 2.26 | 0.000 | 2.3 | 0.480 | 1.92 | 0.007 | 1.18 | 0.002 | 2.04 | 0.037 | 2.2 | 0.009 |
Discussion
Involvement of three different virus species in causing yellow mosaic disease in Vigna crops has been established leading to similar kind of morphological symptoms of YMD, thereby misleading us on the YMD-causing virus species. Therefore, detection of the particular virus species causing YMD is of upmost importance for its effective management. The characterization of the virus species is not possible at morphological level, as most of the molecular studies conducted earlier (Kumar et al. 2014; Singh et al. 2015; Gupta et al. 2015) also did not specified the exact species of virus leading to YMD in mungbean. Therefore, presence any of the three viruses and even a mixed infection could not be ruled out in these reports. However, now the identity of three different species of viruses leading to YMD development has been established at molecular level and the genome sequences of all these viruses are available. Therefore, the efforts should be directed towards their molecular detection which would be more precise to identify the virus specific resistance donors and develop the strategies for mungbean improvement. In our study, PCR analysis and restriction digestion were performed, which exhibited presence of MYMIV in YMD development at experimental sites.
Many of the important traits in crop plants such as yield, quality, resistance to biotic and abiotic stresses, etc. are likely to be controlled by additive genes. Nevertheless, the QTLs with minor contributions towards the trait and the sensitivity of a particular trait to the environment sometimes tend to mislead in identifying them. Molecular markers and advanced statistical methods together provide a platform to study the genetics of quantitative traits. Before the availability of mungbean genome sequence (Kang et al. 2014), work in mungbean at molecular level mainly focused on identification of transferable markers from other Vigna and related species for genetic analysis (Gupta et al. 2013; Pratap et al. 2014, 2016). Some of the transferable markers were successfully utilized in studying genetic diversity (Sangiri et al. 2007; Gwag et al. 2010; Gupta et al. 2013; Singh et al. 2014) and QTL identification for different traits by linkage mapping (Chankaew et al. 2014; Ismura et al. 2012; Alam et al. 2014). The earlier studies suggested the genetics of yellow mosaic disease resistance in mungbean and other Vigna crops to follow simple inheritance pattern and therefore it was thought to be relatively easier to manipulate in other backgrounds. Pal et al. (1991) suggested monogenic inheritance of yellow mosaic disease. However, exceptions of digenic interactions were also reported (Dwivedi and Singh 1986; Verma and Singh 1986). Even in other legumes like soybean, both single dominant monogenic (Bhattacharyya et al. 1999) and digenic recessive interaction (Singh and Mallick 1978) have been reported. Nevertheless, in most of these studies, there was an ambiguity on the species of the virus causing yellow mosaic disease. The contradictions in inheritance pattern could also be due to non-uniform distribution of whitefly population in the field, insufficient inoculum load and/or prevailing environmental conditions which might have led to inaccuracy in ascertaining yellow mosaic disease resistance, thereby leading to errors in selection of resistant genotypes. A relatively recent study suggested recessive monogenic resistance of MYMIV in blackgram (Gupta et al. 2013). CEDG180 was tagged with MYMIV resistance in blackgram which produced 136 and 163 bp amplicons in susceptible and resistance genotypes, located 12.9 cM away from the resistance gene. Likewise, linkage of SCAR-YMV1, SCAR-MYMV583, ISSR811 with yellow mosaic disease resistance in mungbean and urdbean have also been reported by Sauframanien and Gopalkrishnan (2006) and Binyamin et al. (2015). However in mungbean, no molecular evidence of monogenic control of yellow mosaic disease resistance is reported for the disease caused by MYMIV, whereas recent study on inheritance of yellow mosaic caused by MYMV clarify their monogenic inheritance pattern through SCAR markers and two SCARs namely CM9 and CM15 were found strongly associated with MYMV-resistance (Sai et al. 2017). By using the cross-transferable markers two QTLs namely qPMR1 (CEDG282-CEDG191) and qPMR2 (MBSSR238-CEDG166) with the phenotypic coverage of 20.10 and 57.81% have been identified indicating the reliability of these markers. Kitsanachandee et al. (2013) identified three QTLs for MYMIV resistance, one each on linkage group 2, 4 and 9 and were designated as qMYMIV1 (CEDG100 and cp02662), qMYMIV2 (DMB-SSR008-VR113) and qMYMIV3 (CEDG166-CEDG304), which explained 9.33%, 10.61%, 12.55% of phenotypic variation. They also reported two more, environment-specific QTLs viz., qMYMIV4 (CEDG100-cp02662) and qMYMIV5 (CEDG121-CEDG191). Chen et al. (2013) identified DMB-SSR158, a SSR marker linked to major QTL controlling MYMIV resistance on LG2 between the markers interval of CEDG100 and CEDG108 (Kajonphol et al. 2012). Alam et al. (2014) constructed a linkage map comprising 11 linkage groups and identified a new QTL, i.e., qMYMIV7 (CEDG041-VES503) for YMD resistance. Because of multiple events covered in association mapping, it has been thus used in several crop plants to identify genomic regions associated with a trait of interest which could not have been picked up by analysing a bi-parental population. In this direction, exploring QTLs by association analysis is an effective approach to trace novel QTLs controlling quantitative traits. It is based on linkage disequilibrium and diversity analysis of trait of interest. It offers the advantages of natural population and detecting multiple alleles on the same locus, targeting single genes. Because of higher frequency of spurious associations in the natural population (Rosenberg and Nordborg 2006), the evaluation of population genetic structure is a pre-requisite of association studies. The accurate population structure analysis leads to higher genetic similarity within each group. Thus, the accuracy of the associated markers depends upon whether the population structure is appropriate or not. Therefore, we used model-based structure analysis to determine the population structure of studied genotypes and this information was used in marker-trait association to reduce the chance of false positives. Structured association analysis has been reported to be successfully deployed by Nie et al. (2016) in cotton, Kumar et al. (2014) in soybean, Kumar et al. (2017a) in lentil and Noble et al. (2018) in mungbean, to reduce the false discovery rate of the associated markers.
In our study, the mungbean population of 127 genotypes was classified into 6 subpopulations, which was reasonable to eliminate the spurious association effects in the mapping of MYMIV resistance by marker-trait association analysis. Overall, the nature of population structure in the present study suggested that it was unbiased within a specific sub-population and was appropriate for association analysis. Therefore, the marker-trait association recorded in the present study should be real and the chance of obtaining false positives would be rather less (Pritchard et al. 2000; Gupta et al. 2005; Ostrowski et al. 2006; Jaiswal et al. 2012). Beside this, use of diverse breeding lines makes association mapping more cost effective. The identified QTLs for trait(s) in the background of elite breeding materials can be used directly in genomic-assisted selection (Kumar et al. 2017a, b).
In the present study 15 marker-trait associations are significantly associated with MYMIV resistance with the phenotypic coverage of 14.45 and 13.26%. However, it is not surprising to observe lower phenotypic variation in association mapping due to several reasons like including low LD and low allele frequency, less number of markers and/or accessions used in analysis (Yan et al. 2011). Based on MLM approach, three markers viz. CEDG293, CEDG225 and CEDG050 were located on LG2, which is likely to be associated with VrSSR112 loci. Likewise, CEDG139 and DMB-SSR008 (LG4); CEDG121, CEDG211, CEDG191 and cp01038 (LG6); DMB-SSR059, CEDG166 and BM212 (LG9) were found associated with their respective linkage groups and were observed to have a definite role in governing MYMIV resistance in mungbean. Three markers viz. CEDG293, DMB-SSR059 and DMB-SSR008 were found strongly associated with MYMIV in mungbean accounting to > 10.00% of the phenotypic variation. Among these the previously reported markers/QTLs involved in MYMIV resistance viz., DMB-SSR008 (Kitsanachandee et al. 2013) contributed 11.54% of phenotypic variation and rest two were detected as novel accounting to 13.26 and 12.45% of phenotypic variation, respectively. Previously, marker-trait association was conducted for a number of important traits in the legumes as well other crops (Kumar et al. 2017b; Zhang et al. 2014). These markers can be closely associated with the genes that control MYMIV resistance in mungbean and can be used in breeding program. Thus, results in the present study revealed that a number of genes/QTLs were responsible for controlling the MYMIV resistance in mungbean.
Association mapping has an ability to detect novel QTLs because it uses diverse germplasm that has more allelic diversity than bi-parental population. Therefore, marker loci associated with MYMIV resistance in the present study might be different from QTL/genes identified in earlier studies. One of the reasons for this may be that many QTLs do not segregated in the bi-parental population because only two alternative alleles are present for a trait (Jaiswal et al. 2012). In earlier studies, a marker associated significantly with a trait of interest explained low phenotypic variation (< 1%) in association mapping studies, for example, in cotton (Nie et al. 2016), and in linseed (Chandrawati et al. 2016). Kumar et al. (2014) detected two SSR markers associated with MYMIV resistance in soybean with coverage of < 1% phenotypic variation. In the present study, microsatellite markers could be associated with those genes that are involved in controlling MYMIV resistance in mungbean and two novel regions associated with MYMIV resistance on linkage group 2 and 9 were identified. The strong association and higher phenotypic coverage of CEDG293 and DMBSSR059 present on LG2 (3.3 cM) and LG9 (2.1 cM) as compared to the previously reported markers present in the QTL region, suggest their increased utility in detection of their allelic variants responsible for enhanced resistance to MYMIV. These markers can be deployed in mungbean improvement programs for introgression of MYMIV resistance.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The present research was financially supported by Science and Engineering Research Board (SERB-DST), New Delhi (YSS/2015/00484) under the startup research grant for young scientists.
Author contributions
CMS planned the work and submitted a proposal to SERB, New Delhi, India under the Start-up Research Grant Scheme for Young Scientists. CMS conducted the field experiments in different environments and genotyped the material. AP, RSB and SG helped in data analysis. CMS drafted the manuscript and SG, AP, NPS edited the manuscript. All the authors read and approved the manuscript.
Compliance with ethical standards
Conflict of interest
It is to declare that there is no conflict of interest between the co-authors.
References
- Ahmed Q. Fungicidal control of Cercospora leaf spot of mungbean (Vigna radiata L. Wilczek) Indian Phytopathol. 1985;38:418–422. [Google Scholar]
- Akaram M, Naimuddin AAK, Gupta S, Singh NP. Characterization of full genome of Dolichos yellow mosaic virus based on sequence comparison, genetic recombination and phylogenetic relationship. Ann Appl Biol. 2015;167:354–363. doi: 10.1111/aab.12231. [DOI] [Google Scholar]
- Alam AKM, Somta P, Srinives P. Identification and confirmation of quantitative trait loci controlling resistance to mungbean yellow mosaic disease in mungbean [Vigna radiata (L.) Wilczek] Mol Breed. 2014;34:1497–1506. doi: 10.1007/s11032-014-0133-0. [DOI] [Google Scholar]
- Ali MZ, Khan MAA, Kiran MM, Ahmed M, Ahmed F. Field performance of some mungbean varieties against mungbean yellow mosaic virus and Cercospora leaf spot diseases. J Exp Biosci. 2010;1:11–16. [Google Scholar]
- Bhattacharyya PK, Ram HH, Kole PC. Inheritance of resistance to yellow mosaic virus in inter-specific crosses of soybean. Euphytica. 1999;108:157–159. doi: 10.1023/A:100362071. [DOI] [Google Scholar]
- Binyamin R, Khan MA, Khan NA, Khan AI. Application of SCAR markers linked with mungbean yellow mosaic virus disease-resistance gene in Pakistan mungbean germplasm. Genet Mol Res. 2015;14:2825–2830. doi: 10.4238/2015.March.31.13. [DOI] [PubMed] [Google Scholar]
- Borah BK, Dasgupta I. Begomovirus research in India: a critical appraisal. Bio Sci. 2012;37:791–806. doi: 10.1007/s12038-012-9238-y. [DOI] [PubMed] [Google Scholar]
- Chandrawati SN, Kumar R, Kumar S, Singh PK, Yadav VK, Ranade SA, Yadav HK. Genetic diversity, population structure and association analysis in linseed. Phyiol Mol Biol Plants. 2016 doi: 10.1007/s12298-016-0408-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chankaew S, Isemura T, Isobe S, Kaga A, Tomooka N, Somta P, Hirakawa H, Shirasawa K, Vaughan DA, Srinives P. Detection of genome donor species of neglected tetraploid crop Vigna reflexo-pilosa (Creole bean) and genetic structure of diploid species based on newly developed EST-SSR markers from azukibean. PLoS ONE. 2014;9:e104990. doi: 10.1371/journal.pone.0104990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HL, Ku HS, Schafleitner R, Bains TS, Kuo GC, Liu CA, Nair RM. The major quantitative trait locus for mungbean yellow mosaic India virus resistance is tightly linked in repulsion phase to the major bruchid resistance locus in a cross between mungbean [Vigna radiata (L.) Wilczek] and its wild relative Vigna radiata ssp. sublobata. Euphytica. 2013;192:205–216. doi: 10.1007/s10681-012-0831-9. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Dwivedi S, Singh DP. Inheritance of pod pubescence and seed coat colour in blackgram. Crop Improve. 1986;13:54–57. [Google Scholar]
- Earl DA, Von-Holdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Rustgi S, Kulwal PL. Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol Biol. 2005;57:461–485. doi: 10.1007/s11103-005-0257-z. [DOI] [PubMed] [Google Scholar]
- Gupta S, Gupta DS, Anjum TK, Pratap A, Kumar J. Inheritance and molecular tagging of MYMIV resistance gene in blackgram (Vigna mungo L. Hepper) Euphytica. 2013;193:27–37. doi: 10.1007/s10681-013-0884-4. [DOI] [Google Scholar]
- Gupta SK, Souframanien J, Reddy KS. Validation of molecular markers linked to yellow mosaic virus disease resistance in diverse genetic background of black gram [Vigna mungo (L.) Hepper] Electron J Plant Breed. 2015;6:755–763. [Google Scholar]
- Gwag JG, Dixit A, Park YJ, Ma KH, Kwon SJ, Cho GT, Lee GA, Lee SY, Kang HK, Lee SH. Assessment of genetic diversity and population structure in mungbean. Genes Genom. 2010;32:299–308. doi: 10.1007/s13258-010-0014-9. [DOI] [Google Scholar]
- Isam MN, Sony SK, Borna DS. Molecular charecterization of mungbean yellow mosaic disease and coat protein gene in mungbean varieties of Bangladesh. Plant Tissue Cult Biotechnol. 2012;22(1):73–81. doi: 10.3329/ptcb.v22i1.11263. [DOI] [Google Scholar]
- Isemura T, Kaga A, Tabata S, Somta P, Srinives P, Shimizu T, Jo U, Vaughan DA, Tomooka N. Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean. PLoS ONE. 2012;7:41304. doi: 10.1371/journal.pone.0041304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal V, Mir RR, Mohan A, Balyan HS, Gupta PK. Association mapping for pre-harvest sprouting tolerance in common wheat (Triticum aestivum L.) Euphytica. 2012;188:89–102. doi: 10.1007/s10681-012-0713-1. [DOI] [Google Scholar]
- Kajonphol T, Sangsiri C, Somta P, Toojinda T, Srinives P. SSR map construction and quantitative trait loci (QTL) identification of major agronomic traits in mungbean. Sabrao J Breed Genet. 2012;44:71–86. [Google Scholar]
- Kang JY, Kim SK, Kim MY, Lestari P, Kim KH, Bo-K Ha, Jun TH, Hwang WJ, Lee T, Lee J, Shim S, Yoon MY, Jang YE, Han KS, Taeprayoon P, Yoon N, Somta P, Tanya P, Kim KS, Gwag J-G, Jung-K M, Ho-Lee Y, Park B, Bombarely A, Doyle JJ, Jackson SA, Schafleitner R, Srinive P, Varshney RK, Lee S. Genome sequence of mungbean and insights into evolution within Vigna species. Nat Commun. 2014;5:5443. doi: 10.1038/ncomms6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsanachandee R, Somta P, Chatchawankanphanich O, Akhtar KP, Shah TM, Nair RM. Detection of quantitative trait loci for mungbean yellow mosaic India virus (MYMIV) resistance in mungbean (Vigna radiata (L.) Wilczek) in India and Pakistan. Breed Sci. 2013;63:367–373. doi: 10.1270/jsbbs.63.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B, Talukdar A, Verma K, Bala I, Haris GD, Gowda S, Lal SK, Sapra RL, Singh HP. Mapping of yellow mosaic virus resistance in soybean (Glycine max L. Merr) through association mapping approach. Genetica. 2014 doi: 10.1007/s10709-014-9801-6. [DOI] [PubMed] [Google Scholar]
- Kumar J, Gupta DS, Gupta S, Dubey S, Gupta P, Kumar S. Quantitative trait loci from identification to exploitation for crop improvement. Plant Cell Rep. 2017;36:1187. doi: 10.1007/s00299-017-2127-y. [DOI] [PubMed] [Google Scholar]
- Kumar J, Gupta S, Biradar RS, Gupta P, Dubey S, Singh NP. Association of functional markers with flowering time in lentil. J Appl Genet. 2017 doi: 10.1007/s13353-017-0419-0. [DOI] [PubMed] [Google Scholar]
- Liu K, Muse SV. Power Marker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Maiti S, Basak J, Kundagrami S, Kundu A, Pal A. Molecular marker-assisted genotyping of mungbean yellow mosaic India virus resistant germplasms of mungbean and urdbean. Mol Biotechnol. 2011;47:95–104. doi: 10.1007/s12033-010-9314-1. [DOI] [PubMed] [Google Scholar]
- Malathi VG, John P. Geminiviruses infecting legumes. In: Rao GP, Lava Kumar P, Holguin-Pena RJ, editors. Characterization, diagnosis and management of plant viruses, Volume 3: vegetables and pulse crops. Texas: Stadium Press LLC; 2008. pp. 97–123. [Google Scholar]
- Naimuddin AMD, Singh NP. Yellow mosaic of mungbean and blackgram: current status and future strategies. J Food Legumes. 2016;29:77–93. [Google Scholar]
- Naimuddin K, Akram M, Gupta S. Identification of mungbean yellow mosaic India virus infecting Vigna mungo var. silvestris L. Phytopathol Mediterr. 2011;50:94–100. [Google Scholar]
- Naimuddin K, Akram M, Pratap A. First report of natural infection of Mungbean yellow mosaic India virus in two wild species of Vigna. New Dis Rep. 2011;23:21. doi: 10.5197/j.2044-0588.2011.023.021. [DOI] [Google Scholar]
- Naimuddin AM, Pratap A, Chaubey BK, Joseph KJ. PCR based identification of the virus causing yellow mosaic disease in wild Vigna accessions. J Food Legumes. 2011;24:14–17. [Google Scholar]
- Nie X, Huang C, You C, Li W, Zhao W, Shen C, Zhang B, Wang H, Yan Z, Dai B, Wang M, Zhang X, Lin Z. Genome-wide SSR-based association mapping for fiber quality in nation-wide upland cotton inbreed cultivars in China. BMC Genom. 2016;17:352. doi: 10.1186/s12864-016-2662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble TJ, Tao Y, Mace ES, Williams B, Jordan DR, Douglas CA, Mundree SG. Characterization of linkage disequilibrium and population structure in a mungbean diversity panel. Front Plant Sci. 2018;8:2102. doi: 10.3389/fpls.2017.02102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski MF, David A, Santoni S, Mckhann H, Reboud X, Corre VL, Camilleri C, Brunel D, Bouchez D, Faure B, Bataillon T. Evidence for a large-scale population structure among accessions of Arabidopsis thaliana: possible causes and consequences for the distribution of linkage disequilibrium. Mol Ecol. 2006;15:1507–1517. doi: 10.1111/j.1365-294X.2006.02865.x. [DOI] [PubMed] [Google Scholar]
- Pal SS, Dhaliwal HS, Bains SS. Inheritance of resistance to yellow mosaic virus in some Vigna spp. Plant Breed. 1991;106:168–171. doi: 10.1111/j.1439-0523.1991.tb00496.x. [DOI] [Google Scholar]
- Pant V, Gupta D, Roychoudhary N, Malathi VG, Varma A, Mukharjee SK. Molecular charecterization of Rep protein of the blackgram isolates of Indian mungbean yellow mosaic virus. J Gen Viral. 2001;82:1559–2567. doi: 10.1099/0022-1317-82-10-2559. [DOI] [PubMed] [Google Scholar]
- Pratap A, Basu PS, Gupta S, Malviya N, Rajan N, Tomar R, Madhavan L, Nadarajan N, Singh NP. Identification and characterization of sources for photo and thermo-insensitivity in Vigna species. Plant Breed. 2014;133:756–764. doi: 10.1111/pbr.12215. [DOI] [Google Scholar]
- Pratap A, Gupta S, Tomar R, Malviya N, Maurya R, Pandey VR, Mehandi S, Singh NP. Cross-genera amplification of informative microsatellite markers from common bean and scarlet runner bean for assessment of genetic diversity in mungbean (Vigna radiata) Plant Breed. 2016;135:499–505. doi: 10.1111/pbr.12376. [DOI] [Google Scholar]
- Pratap A, Chaturvedi SK, Tomar R, Rajan N, Malviya N, Thudi M, Saabale PR, Prajapati U, Varshney RK, Singh NP. Marker-assisted introgression of resistance to Fusarium wilt race 2 in Pusa 256, an elite cultivar of desi chickpea. Mol Genet Genom. 2017;292:1237–1245. doi: 10.1007/s00438-017-1343-z. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi J, Ilyas M, Mansoor S, Briddon RW. Legume yellow mosaic virus genetically isolated Begomoviruses. Mol Plant Pathol. 2007;8:343–348. doi: 10.1111/j.1364-3703.2007.00402.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg N, Nordborg M. A general population-genetic model for the production by population structure of spurious genotype-phenotype associations in discrete, admixed, or spatially distributed populations. Genetics. 2006;173:1665–1678. doi: 10.1534/genetics.105.055335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai CB, Nagrajan P, Raveendran M, Rabindran R, Kannan BJR, Senthil N. Understanding the inheritance of mungbean yellow mosaic virus resistance in mungbean. Mol Breed. 2017;37:63. doi: 10.1007/s11032-017-0650-8. [DOI] [Google Scholar]
- Sangiri C, Kaga A, Tomooka N, Vaughan D, Srinives P. Genetic diversity of the mungbean genepool on the basis of microsatellite analysis. Aust J Bot. 2007;55:837–847. doi: 10.1071/BT07105. [DOI] [Google Scholar]
- Sauframanien J, Gopalkrishnan T. ISSR and SCAR markers linked to the mungbean yellow mosaic virus (MYMV) resistance gene in blackgram. Plant Breed. 2006;125:619–622. doi: 10.1111/j.1439-0523.2006.01260.x. [DOI] [Google Scholar]
- Singh BB, Mallick AS. Inheritance of resistance to yellow mosaic in soybean. Indian J Genet. 1978;38:258–261. [Google Scholar]
- Singh A, Dikshit HK, Jain N, Yadav RN. Efficiency of SSR, ISSR and RAPD markers in molecular characterization of mungbean and other Vigna species. Indian J Biotechnol. 2014;13:81–88. [Google Scholar]
- Singh CM, Kumar R, Mishra SB, Pandey A, Arya M. Characterization of mungbean genotypes against Mungbean Yellow Mosaic Virus and cercospora leaf spot diseases under north east plain zone. Int J Agric Environ Biotechnol. 2015;8:119–125. doi: 10.5958/2230-732X.2015.00016.9. [DOI] [Google Scholar]
- Singh DP, Singh BB, Pratap A. Genetic improvement of mungbean and urdbean and their role in enhancing pulse production in India. J Genet. 2017;76:550–567. [Google Scholar]
- Singh CM, Singh P, Pratap A, Pandey R, Purwar S, Vibha DCA, Baek KH, Mishra AK. Breeding for enhancing Legumovirus resistance in mungbean: current understanding and future directions. Agronomy. 2019;9:622. doi: 10.3390/agronomy9100622. [DOI] [Google Scholar]
- Varma A, Dhar AK, Mandala B (1992) MYMV transmission and control in India. In: Green SK, Kim D (eds) Mungbean yellow mosaic disease. pp 8–27
- Varshney RK, Mohan SM, Gaur PM, Chamarthi SK, Singh VK, Srinivasan S, Swapna N, Sharma M, Pande S, Singh S, Kaur L. Marker-assisted backcrossing to introgress resistance to Fusarium wilt race 1 and Ascochyta blight in C 214, an elite cultivar of chickpea. Plant Genome. 2014;7:1–11. doi: 10.3835/plantgenome2013.10.0035. [DOI] [Google Scholar]
- Verma RPS, Singh DP. The allelic relationship of genes giving resistance to mungbean yellow mosaic virus in blackgram. Theor Appl Genet. 1986;72:737–738. doi: 10.1007/BF00266537. [DOI] [PubMed] [Google Scholar]
- Yan J, Warburton M, Crouch J. Association mapping enhancing maize (Zea mays L.) genetic improvement. Crop Sci. 2011;51:433–449. doi: 10.2135/cropsci2010.04.0233. [DOI] [Google Scholar]
- Zhang P, Liu X, Tong H, Lu Y, Li J. Association mapping for important agronomic traits in core collection of rice (Oryza sativa L.) with SSR markers. PLoS ONE. 2014;9:e111508. doi: 10.1371/journal.pone.0111508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.