The conserved core of bacterial flagellar motors reflects a shared evolutionary history that preserves the mechanisms essential for flagellar assembly, rotation, and directional switching. In this work, we describe an expanded and diversified set of core components in the Campylobacter jejuni flagellar C ring, the mechanistic core of the motor. Our work provides insight into how usually conserved core components may have diversified by gene duplication, enabling a division of labor of the ancestral protein between the two new proteins, acquisition of new roles in flagellar assembly and motility, and expansion of the function of the flagellum beyond motility, including spatial regulation of cell division and numerical control of flagellar biogenesis in C. jejuni. Our results highlight that relatively small changes, such as gene duplications, can have substantial ramifications on the cellular roles of a molecular machine.
KEYWORDS: C ring, FlhG, FliI, FliN, FliY, flagellar motor, polar flagella, type III secretion
ABSTRACT
Bacterial flagella are reversible rotary motors that rotate external filaments for bacterial propulsion. Some flagellar motors have diversified by recruiting additional components that influence torque and rotation, but little is known about the possible diversification and evolution of core motor components. The mechanistic core of flagella is the cytoplasmic C ring, which functions as a rotor, directional switch, and assembly platform for the flagellar type III secretion system (fT3SS) ATPase. The C ring is composed of a ring of FliG proteins and a helical ring of surface presentation of antigen (SPOA) domains from the switch proteins FliM and one of two usually mutually exclusive paralogs, FliN or FliY. We investigated the composition, architecture, and function of the C ring of Campylobacter jejuni, which encodes FliG, FliM, and both FliY and FliN by a variety of interrogative approaches. We discovered a diversified C. jejuni C ring containing FliG, FliM, and both FliY, which functions as a classical FliN-like protein for flagellar assembly, and FliN, which has neofunctionalized into a structural role. Specific protein interactions drive the formation of a more complex heterooligomeric C. jejuni C-ring structure. We discovered that this complex C ring has additional cellular functions in polarly localizing FlhG for numerical regulation of flagellar biogenesis and spatial regulation of division. Furthermore, mutation of the C. jejuni C ring revealed a T3SS that was less dependent on its ATPase complex for assembly than were other systems. Our results highlight considerable evolved flagellar diversity that impacts motor output, biogenesis, and cellular processes in different species.
INTRODUCTION
The bacterial flagellar motor harnesses proton flux to generate torque (1). Proton flux through a ring of stator complexes exerts torque upon a rotor (2–4); torque is transmitted through a rigid periplasmic rod and flexible surface hook to an extracellular flagellar filament that coils as a helical propeller for propulsion (1). This rotor and a switch component that controls the direction of motor rotation together form a cytoplasmic ring, the C ring (5). Torque is generated by the action of conformational changes in the stator complexes upon the C ring (2, 6–13), whereas switching of motor rotation from counterclockwise to clockwise is driven by binding of the response regulator CheY, whose activity is linked to sensing by chemosensory receptors (14). The C ring is also required for efficient flagellar biogenesis, as it is necessary for full assembly of the flagellar type III secretion system (fT3SS), which secretes extracytoplasmic axial components of the flagellar motor (15–21). Understanding how the C ring contributes to these functions is crucial to understanding how flagellar motors function and have evolved.
In the model peritrichous flagella of Escherichia coli and Salmonella species, the C ring is composed of FliG, FliM, and FliN (22–26). FliG oligomerizes into a ring that forms the upper rim of the C ring (27, 28). FliG docks the C ring to the cytoplasmic face of the transmembrane MS ring, composed of FliF, by cofolding of the C- and N-terminal domains of FliF and FliG, respectively (23, 29, 30). FliM and FliN form the switch complex beneath FliG (31, 32). The middle domain of FliM contains a CheC-like domain that forms a continuous belt in the middle of the C ring (33, 34). CheY, the response regulator of the chemotaxis signal transduction system, binds a conserved N-terminal EIDAL motif in FliM to influence clockwise or counterclockwise motor rotation for chemotaxis (16, 35–39). Structural studies suggest that homologous surface presentation of antigen (SPOA) domains in the C terminus of FliM and FliN form a continuous spiral through FliM-FliN3 protomers that compose the lower rim of the C ring (31, 40). This region of the C ring is also an assembly platform for integration of an ATPase complex into the fT3SS. Spoke-like FliH bridges anchor a FliI6-FliJ complex beneath the fT3SS inner membrane export apparatus to FliM-FliN3 (9, 41–43), where the central stalk protein FliJ can interact with the cytoplasmic ring of FlhA (44, 45). This FliI6-FliJ complex facilitates efficient secretion by the export gate by activating the fT3SS export apparatus, although its mode of action is not completely understood (46–50).
Flagellar motor C rings do not all follow this compositional blueprint. For example, Bacillus subtilis and Thermotoga maritima encode FliY in place of FliN (51–53). The domain organization of FliY is similar to that of FliM, with a phosphatase domain of the CheC/CheX/CheY family and a SPOA domain (53, 54). While FliN and FliY are mutually exclusive in most bacteria, Leptospira species and Epsilonproteobacteria, including Campylobacter and Helicobacter species, produce both FliN and FliY, although FliY in these bacteria lacks active sites for phosphatase activity (55, 56). An initial study in Helicobacter pylori indicated that FliN and FliY are required for full flagellation and motility (56, 57). Structural and interaction studies demonstrated that FliY interacts with FliM and FliN separately to assemble a three-protein complex (57). Furthermore, H. pylori FliY-FliN and FliY-FliM heterodimers interact with FliH in vitro, but how these proteins contribute to C-ring composition and architecture in H. pylori remains unknown (57). The presence of both FliY and FliN SPOA-containing proteins in Epsilonproteobacteria prompts questions regarding the function and location of these proteins, how have they diverged relative to an ancestral FliN-like protein, and what selective benefits drove retention of two FliN/FliY-like proteins.
The additional complexity of the epsilonproteobacterial C ring coincides with additional functions, although how complexity and function are related is unclear. The Campylobacter jejuni motor contains a wider C ring that generates higher torque than do many other species, facilitating motility in viscous environments by enabling interaction with a correspondingly wider ring of more stator complexes, which are positioned by large periplasmic disk structures (7–9, 58). The C. jejuni C ring also has functions unrelated to motility. We previously demonstrated that C. jejuni fliM and fliN mutants have defects in spatial regulation of septal Z ring formation to result in nonviable minicells due to cell division at poles rather than symmetrically at the midpoint (59). Many bacteria prevent FtsZ from forming Z rings at polar regions using a Min system (60). However, C. jejuni lacks a conventional Min system, instead using FlhG, a homolog of MinD (59). How the C. jejuni C ring impacts FlhG activity to prevent division at polar regions is unknown. FlhG also regulates how many flagella are assembled in polar flagellates, including C. jejuni (59, 61), and likely functions with the FlhF GTPase in many polar flagellates to control monotrichous, amphitrichous, or lophotrichous flagellation patterns (61–65). One hypothesis on how these proteins function involves FlhF toggling between ON and OFF states upon the hydrolysis of bound GTP, which is stimulated by FlhG (65). In this model, FlhF in its GTP-bound ON state initiates an undetermined early step in flagellar biogenesis (62, 64). After the production of a polar flagellum, FlhG stimulates FlhF GTPase activity to produce a GDP-bound OFF state, repressing FlhF activity and preventing assembly of more than a single polar flagellum.
Considering the potentially enhanced protein composition of flagellar C rings and diverse functions of the C ring in C. jejuni cell biology, we sought to determine the composition and architecture of the C. jejuni flagellar C ring and its contributions to flagellar biogenesis, torque generation, motility, fT3SS assembly, and cell division. To this end, we used a variety of genetic, in vivo protein interaction, functional, immunofluorescent microscopy, and in situ electron cryo-tomographic structural analyses to better characterize the C. jejuni C ring. Our results suggest that duplication (or possibly acquisition) of an ancestral FliN-like protein enabled the evolution of distinct roles in the C. jejuni C ring. This diversification of two FliN-like paralogs bestowed functional adaptations that may have coevolved with the widening of the C. jejuni C ring and facilitated the integration of regulatory mechanisms for polar flagellar assembly and the prevention of division at poles for accurate symmetrical division.
RESULTS
Evolutionary relatedness and domain structure of flagellar C-ring proteins across bacterial species.
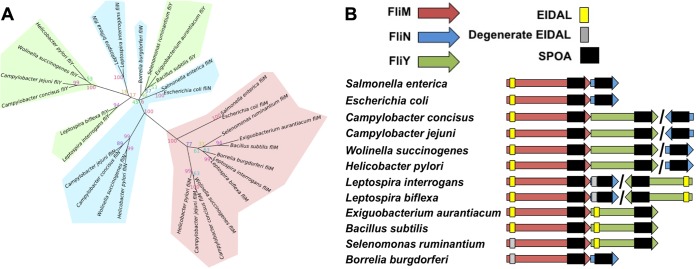
To further the understanding of the roles of the C. jejuni FliM, FliN, and FliY proteins, we first estimated their phylogenies and studied their operon neighborhoods. Our maximum likelihood phylogeny of the common SPOA domain showed segregation of FliM from FliN and FliY (Fig. 1A), together with conservation of the EIDAL motif in most FliM sequences, which is separate from their SPOA domains. Full or degenerate EIDAL motifs were also sporadically present in FliN and FliY. Unexpectedly, however, FliN and FliY did not separate into discrete clades. Instead, distinct FliN and FliY clades were found in different bacterial lineages, such as the Epsilonproteobacteria and some spirochetes, suggesting independent ancestral duplication or horizontal acquisition of a second ancestral FliN-like protein allele in these distinct lineages, leading to multiple distinct origins of contemporary FliN and FliY. Indeed, our phylogeny suggests that the spirochete FliN is more closely related to the epsilonproteobacterial FliY than to the epsilonproteobacterial FliN, and vice versa, indicating that the epsilonproteobacterial FliN and FliY do not evolutionarily (or functionally) correspond to annotated FliN and FliY proteins in other species.
FIG 1.
Phylogenetic analysis of the SPOA domain-containing family of flagellar proteins. (A) Unrooted phylogenetic tree of aligned sequences of FliM, FliN, and FliY from 12 bacteria from the spirochetes, Firmicutes, Epsilonproteobacteria, and Gammaproteobacteria. Confidence values were generated through 1,000 bootstrap repeats. Branches are shaded red for FliM, green for FliY, and blue for FliN. (B) Genomic organization and conserved encoded domains of fliM, fliN, and fliY. The SPOA domain (Pfam domain pf01052) is shown in black, and the presence of true (yellow) and potential degenerate (gray) EIDAL motifs is indicated. Adjacent genes indicate adjacent genomic locations, whereas slashes indicate distant locations.
Consistent with this, operon analysis also showed differing patterns of fliN and fliY colocation with fliM (Fig. 1B). In peritrichous flagellates such as E. coli and Salmonella species, fliM and fliN are usually organized together within an operon, but in C. jejuni, fliY is immediately downstream of fliM, while fliN is located elsewhere. Our results suggest that presence of a CheC domain does not correspond to a single distinct FliY family. Rather, diverse species have evolved multiple SPOA domain FliN homologs, with some that incorporate a CheC domain, and some that do not (see Fig. S1 in the supplemental material), with one likely being a primary structural component. In peritrichous motors, FliN is the primary SPOA structural component, whereas FliY is the primary structural component in C. jejuni, with FliN presumably functioning in another role. The function of the SPOA domain is not correlated with the presence or absence of a CheC domain. Indeed, our results are not consistent with C. jejuni FliN being any more closely related to peritrichous FliN than with C. jejuni FliY.
Protein alignments of FliM, FliN, and FliY. Multiple-sequence alignments (MSA) generated using MAFFT and filtered using the T-coffee transitive consistency score. Sourced from 12 bacterial species from the spirochetes, Firmicutes, Epsilonproteobacteria, and Gammaproteobacteria. The SPOA domains of FliM, FliY, and FliN are indicated in the red box. Download FIG S1, TIF file, 1.9 MB (2MB, tif) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Requirements of C. jejuni C-ring components for flagellar motility.
For an initial assessment of the relationship between putative C. jejuni C-ring proteins, we analyzed the production and stability of the proteins in whole-cell lysates of wild-type (WT) C. jejuni and mutants lacking the FliF MS ring protein, putative C-ring proteins (FliG, FliM, FliY, and FliN), the FliI ATPase, and the FliH spoke protein. As observed previously, FliF and FliG were dependent on each other for stability but not required for the stability of other proteins (Fig. 2A) (66). FliM, FliH, and FliI were also not required for stability of other MS ring or C-ring proteins. In contrast, FliM levels were reduced and FliN was absent in the C. jejuni ΔfliY mutant (Fig. 2A). In the ΔfliN mutant, FliY levels were reduced, and FliM levels were only slightly lower than in the WT (Fig. 2A). These findings indicate that FliY and FliN are dependent on each other for full stability, and FliM also requires FliY for stability to a degree. We were unsuccessful in generating antiserum to assess FliI production in C. jejuni.
FIG 2.
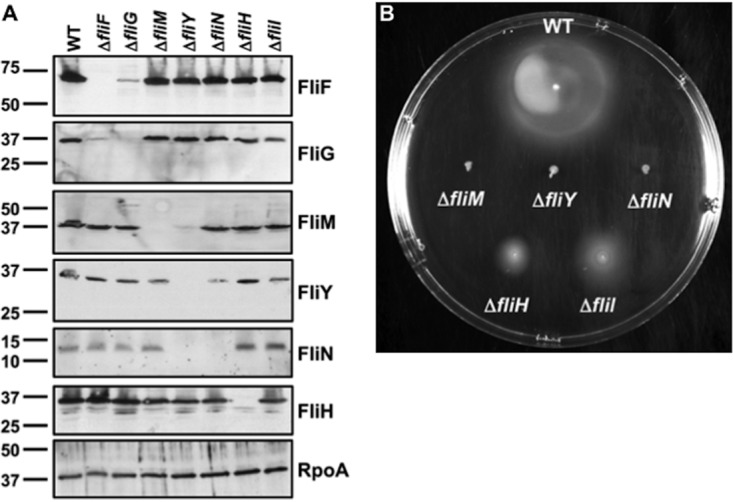
Flagellar protein stability and motility phenotypes of C. jejuni MS and C-ring mutants. (A) Immunoblot analysis of MS and C-ring protein levels in whole-cell lysates of WT C. jejuni and isogenic mutant strains. Specific antiserum to each MS and C-ring protein was used to detect each protein. The detection of RpoA served as a control to ensure equal loading of proteins across strains. (B) Motility phenotypes of WT C. jejuni and isogenic mutant strains. Motility was analyzed after 30 h of incubation at 37°C under microaerobic conditions in MH motility agar (0.4%).
We next evaluated the contribution of the C-ring proteins to flagellar motility in semisolid Mueller-Hinton (MH) motility agar and in liquid MH broth with or without 0.35% methylcellulose (MC). The addition of MC to MH (MH+MC) broth increases viscosity, which can enhance C. jejuni motility, as the bacterium characteristically swims faster in viscous environments (8, 67, 68). WT C. jejuni was motile in motility agar (Fig. 2B), and approximately 90% of WT C. jejuni cells swam normally, with frequent darting and directional changes in MH and faster swimming with turns in MH+MC broth. The C. jejuni ΔfliM and ΔfliY mutants were immobile in all media analyzed (Fig. 2B). Although the C. jejuni ΔfliN mutant did not swim in motility agar or liquid MH broth, ∼10% of the ΔfliN mutant cells showed turning motions in the MH+MC broth, indicating a partially functional motor insufficient to support propulsion. These results support divergent roles of C. jejuni FliY and FliN in flagellar motility, consistent with our phylogenetic analyses (Fig. 1A). The C. jejuni ΔfliH and ΔfliI mutants were motile in motility agar, although the motile rings were smaller than those in WT C. jejuni (Fig. 2B). Roughly 10% or less of both populations swam normally in MH and MH+MC broth, with the ΔfliH mutant displaying a slower velocity of motility than that of WT C. jejuni.
Structure and composition of the C. jejuni flagellar C ring.
We determined the structure and composition of the C. jejuni C ring by visualizing C rings in deletion mutants by electron cryo-tomography. We acquired more than 150 tomograms of motors from each of WT C. jejuni and isogenic ΔfliH, ΔfliN, ΔfliM, ΔfliY, and ΔfliM ΔfliY mutants, and we subsequently determined their structures by subtomogram averaging.
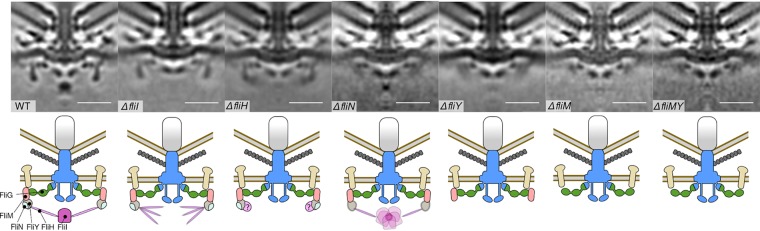
In the WT C. jejuni flagellar motor, the C ring is intact and well resolved, indicating a complete, full-occupancy structure without substantial flexibility (Fig. 3, column 1). The C ring is at the base of the flagellum, surrounding the FlhA cytoplasmic torus with the ATPase complex below (9, 43). Confirming our previous results, a higher-resolution subtomogram average of the ΔfliI mutant motor revealed a loss of FliI density but retained a wild-type C-ring structure (Fig. 3, column 2) (7).
FIG 3.
Subtomogram average flagellar motors structures of WT C. jejuni and isogenic mutants. Imaging of WT C. jejuni and ΔfliI, ΔfliH, ΔfliN, ΔfliY, ΔfliM and ΔfliM ΔfliY mutant flagellar motors revealed the distal tip of the C ring in its in situ functional state. (A) Top row, unfiltered C17 symmetrized subtomogram averages at pixel size of 8.28 Å, 25-nm scale (bar); bottom row, cartoon schematic of C-ring structures, as follows: FliG (green), FliM (pink), FliY (gray), FliN (light blue), FliH and FliI (purple), axial flagellar components (blue), stator complexes (beige), and basal disk (gray disk). Column 1, WT top (7); column 2, ΔfliI mutant composed of 205 particles; column 3, ΔfliH mutant composed of 187 particles; column 4, ΔfliN mutant composed of 281 particles; column 5, ΔfliY mutant composed of 155 particles; column 6, ΔfliM mutant composed of 187 particles; column 7, ΔfliM ΔfliY mutant composed of 212 particles. All images are 100-nm by 100-nm boxes.
To test the hypothesized role of FliH as a spoke protein that anchors the FliI6-FliJ ATPase complex to the fT3SS, we determined a subtomogram average structure of a motor after fliH deletion. Consistent with our hypothesis, the ΔfliH mutant motor lacked its ATPase complex density (Fig. 3, compare columns 1 to 3), although our resolution was insufficient to resolve FliH itself in our WT motor structure, whose structure is predominantly a narrow coiled-coil beyond the resolution of our images. The ΔfliH mutant C ring also maintained its rigidity and resembled the WT C ring, indicating that the C ring does not rely on FliH or FliI for its structural integrity. Curiously, a loss of FliH was accompanied by an additional unidentified density that appeared on the inside face of the C ring that was absent in the ΔfliI mutant (Fig. 3, columns 1 to 3). We speculated that removal of FliH disrupted secretion efficiency, leading to an accumulation of abundant chaperoned fT3SS export substrates, such as flagellin bound to its chaperone FliS, on the inner face of the C ring. To test this, we deleted fliS in the ΔfliH mutant but found that the density remained, refuting our hypothesis (Fig. S2). Next, to test whether FliI was misassembling against the C ring in the absence of FliH, we deleted fliI in a ΔfliH mutant background, but the density was again unaffected (Fig. S2). Because the unknown density is absent in the ΔfliI mutant but present in the ΔfliI ΔfliH mutant, we propose that the removal of FliH exposes a previously occluded surface that other unknown proteins can bind. These proteins might accumulate in the absence of FliH, although their identity is unclear.
Examination of the identity of the unidentified C-ring density in the ΔfliH mutant. Subtomogram average of flagellar motor structures of C. jejuni ΔfliH ΔfliS mutant (left column) and ΔfliH ΔfliI mutant (right column). The top row shows a cross-section through the center of the subtomogram average. The bottom row shows a planar section through the base of the C ring (indicated by white arrow in the side view panel). Arrowheads indicate the position of the unidentified C-ring density that appears in the C. jejuni ΔfliH mutant. Download FIG S2, TIF file, 1.9 MB (2MB, tif) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The deletion of fliN and fliY disrupted the structure of the lower rim of the C ring (Fig. 3, columns 4 and 5). The ΔfliY mutant flagellar motor showed a disordered lower C-ring rim, as well as loss of the FliI density (Fig. 3, column 5). The ΔfliN mutant motor had a similar disorder to the lower C-ring structure but unexpectedly retained a small, yet substantial, FliI density (Fig. 3, column 4). These results suggest two important features of C. jejuni C-ring composition and architecture, as follows: (i) because FliN is unstable in the absence of FliY (Fig. 2A), FliY and probably FliN are components of the C ring, and both are required to stabilize the overall C-ring structure; and (ii) FliY, but not FliN, is a component of the C ring required for anchoring FliI into the C. jejuni motor via FliH. Because of the integration of the FliI ATPase complex into the ΔfliN mutant motor but not the ΔfliY mutant motor, the C rings of these motors are different in structure and composition, which cannot be discerned by tomography.
The subtomogram average structure of the ΔfliM mutant motor showed an additional loss of density compared with ΔfliN and ΔfliY mutant flagellar motors on the cytoplasm-facing edge of the C ring (Fig. 3, column 6). This observation confirms that FliM forms a ring beneath FliG in the C. jejuni motor. The motor of the ΔfliM ΔfliY double mutant was comparable to that of the ΔfliM mutant motor, indicating that FliM directly contacts FliG to incorporate FliY into the C ring (Fig. 3, column 7). Because the FliI ATPase complex is not integrated into the motors of C. jejuni ΔfliM, ΔfliY, and ΔfliM ΔfliY mutants, either FliY alone or together with FliM is likely directly responsible for forming a platform to anchor FliI into the fT3SS via FliH spokes. The remaining densities positioned underneath and running parallel to the membrane may correspond to the globular domains of FliG as in the peritrichous motor (22).
Curiously, we saw a density corresponding to the stator complex component MotB in all mutants (7). In peritrichous motors, stator complexes associate as a function of load into wild-type motors (69–71). This observation further supports our previous speculation that MotB incorporates as a static component into the C. jejuni flagellar motor, irrespective of a functioning motor (7).
C. jejuni C-ring protein interactions.
Our subtomogram average structures revealed protein locations in the C. jejuni C ring, suggesting protein interactions that may contribute to C-ring assembly. To investigate these possible interactions, we performed in vivo coimmunoprecipitation assays with intact C. jejuni cells. To this end, C. jejuni mutants were complemented with N- or C-terminal FLAG-tagged MS ring or C-ring, FliH, or FliI proteins expressed in trans from nonnative promoters. Most FLAG-tagged proteins in the respective mutants were expressed at levels similar to those of the native proteins in WT C. jejuni and restored motility, indicating that the FLAG epitope did not hinder function (Fig. S3). The only exception was FliM-FLAG that restored motility slightly to the ΔfliM mutant compared to vector alone. Due to our inability to generate an antibody for FliI detection, we could not compare the levels of FLAG-FliI in the ΔfliI mutant with native FliI levels in WT C. jejuni.
In trans complementation of C. jejuni mutants with FLAG-tagged proteins. Plasmids expressing N- or C-terminal FLAG-tagged flagellar proteins from promoters to closely replicate expression of the native proteins from WT C. jejuni were introduced into respective C. jejuni mutants. (A) C. jejuni ΔfliF mutant expressing FLAG-FliF. (B) C. jejuni ΔfliG mutant expressing FliG-FLAG. (C) C. jejuni ΔfliM mutant expressing FliM-FLAG. (D) C. jejuni ΔfliY mutant expressing FLAG-FliY. (E) C. jejuni ΔfliN mutant expressing FLAG-FliN. (F) C. jejuni ΔfliH mutant expressing FLAG-FliH. (A to F) Each panel contains an immunoblot of the native protein from a whole-cell lysate of the wild-type strain or mutant strain containing empty vector or the mutant strain expressing the FLAG-tagged protein. An immunoblot is also shown for RpoA in whole-cell lysates to ensure equal loading of proteins across samples. All immunoblots were performed with specific antiserum to each protein. A motility assay is shown in each panel for WT C. jejuni or the mutant strain containing empty vector or the mutant strain expressing the FLAG-tagged protein. Motility was analyzed after 30 h of incubation at 37°C under microaerobic conditions. Download FIG S3, JPG file, 0.1 MB (103.1KB, jpg) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
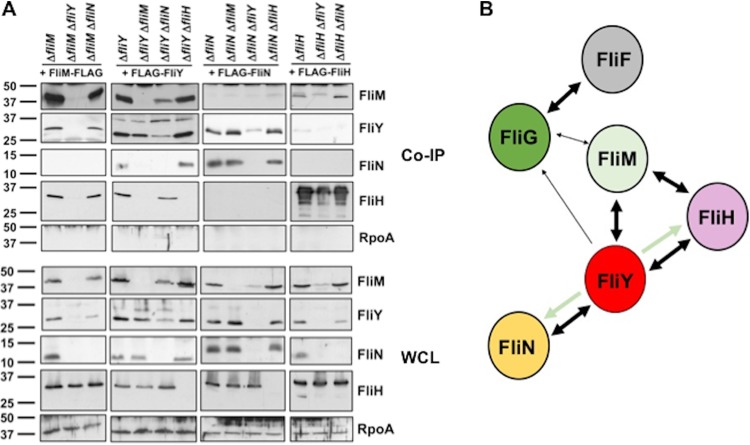
Confirming our tomography analysis that FliN is not required for assembly of the FliI ATPase via FliH (Fig. 3), the only protein we detected interacting with FliN was FliY (Fig. 4). We did not detect interactions between FLAG-FliN and FliH or FliM, which are known FliN-interacting partners in peritrichous flagella (19–21, 31, 40, 72–74). We also observed FliN in higher-order complexes (>50 kDa) after coimmunoprecipitation with either FLAG-FliY or FLAG-FliN (Fig. 4). These findings, together with our phylogenetic and cryo-tomography analyses, further support C. jejuni FliN as a C-ring component that interacts only with FliY due to an altered C-ring position and function relative to FliN in model peritrichous motors.
FIG 4.
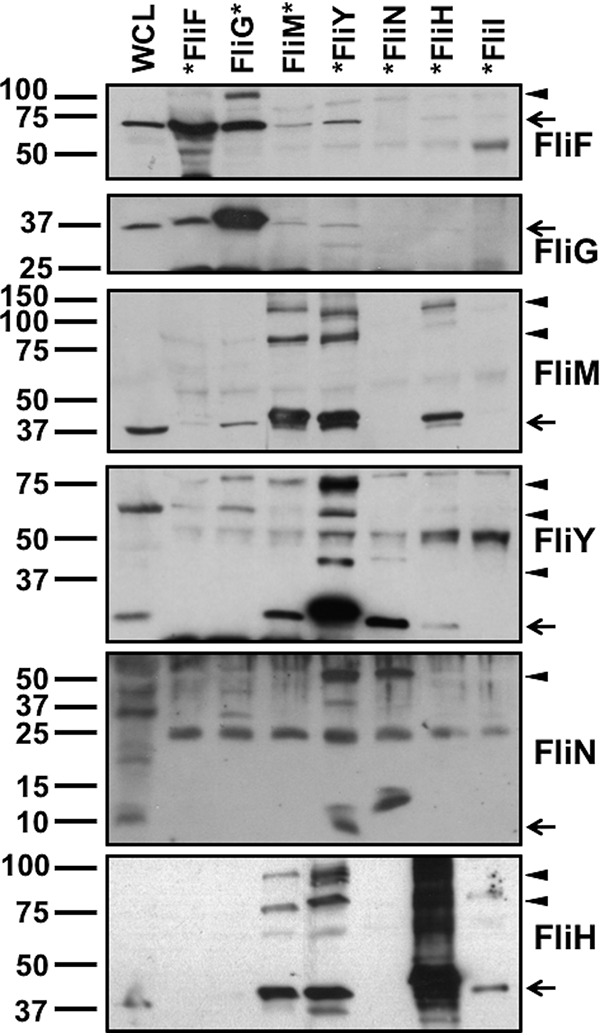
In vivo interactions between C. jejuni MS ring and C-ring proteins. N- or C-terminal FLAG-tagged proteins were expressed from plasmids in respective C. jejuni mutants and immunoprecipitated by FLAG tag antibody resin after cross-linking cells by formaldehyde. The position of the FLAG tag is indicated by the position of the asterisk (*) at the beginning or end of each protein that labels each lane at top. Each lane shows the detected proteins that coimmunoprecipitated with the FLAG-tagged protein indicated. Each protein was detected with specific antiserum. A whole-cell lysate (WCL) of WT C. jejuni was run alongside the coimmunoprecipitated proteins to indicate the size and position of the native protein. Arrows indicate the correct size of the monomer for each protein detected in the immunoblot. Arrowheads indicate bands that may represent a complex formed by the FLAG tag-immunoprecipitated protein with other proteins.
In contrast, proteins that coimmunoprecipitated with FLAG-FliY included FliM, FliN, and FliH (Fig. 4), further implicating FliY occupancy at a central hub position in the C ring enabling FliH binding to integrate the FliI ATPase into the motor and FliN incorporation into the C ring. Furthermore, the FliY-FliM interaction supports our tomography analyses that FliM contributes to stable incorporation of FliY into the side of the C ring and vice versa (Fig. 3). Reciprocal coimmunoprecipitations verified interactions of FliM-FLAG with FliY and FliH and of FLAG-FliH with FliM and FliY (Fig. 4). We also observed higher-order complexes containing FliM, FliY, and FliH after coimmunoprecipitation with any one of these proteins, suggesting that FliM, FliY, and FliH form a heteromeric complex (Fig. 4). Again, both FliM-FLAG and FLAG-FliH did not coimmunoprecipitate FliN. These findings along with the cryo-tomography analysis further suggest that both FliM and FliY contribute to the formation of a platform for FliH binding to anchor the FliI ATPase complex into the motor underneath the fT3SS export gate. Importantly, these findings strongly support C. jejuni FliY replacing the function of FliN in model peritrichous motors. Consistent with other flagellar motors (75–77), FLAG-FliI only interacted with FliH in our assays (Fig. 4), supporting the idea that C. jejuni FliH forms spokes or bridges between a FliM-FliY platform at the lower rim of the C ring and the FliI ATPase complex.
We attempted to detect interactions between the C. jejuni FliG rotor protein and other C-ring proteins. We observed weak interactions between FliG-FLAG and FliM, which is consistent with rotor-switch protein interactions observed in other flagellar systems (Fig. 4). We also observed FliM-FLAG and FLAG-FliY to coimmunoprecipitate a small amount of FliF and FliG (Fig. 4). These results imply that FliM may directly interact with FliG to tether the lower region of the C ring to the rotor, leading to the incorporation of FliY and FliN into the lower region of the C ring.
Some of the interactions inferred from these coimmunoprecipitations may have been direct, while some may require the presence or interaction of a third C-ring protein. To better understand if certain C-ring protein interactions were dependent on a third C-ring protein, we expressed FLAG-tagged proteins in trans for coimmunoprecipitation analysis in C-ring double mutants. Due to the dependence of FliM on FliY for stability, we were unable to assess whether FliM-FLAG interacted with FliH independently of FliY (Fig. 5A). Although the level of native FliY was lower in the ΔfliM ΔfliN mutant than that in the ΔfliM mutant, we clearly detected interactions between FliM-FLAG and FliY and FliH, indicating that FliM interacts directly with FliY and FliH independently of FliN (Fig. 5A and B). Thus, these data further demonstrate that FliN in C. jejuni does not contribute to FliI ATPase assembly as in other systems (19–21).
FIG 5.
Interdependencies of C-ring proteins on each other for interactions. (A) Coimmunoprecipitation (Co-IP) analysis of C-ring-protein interactions in mutants lacking another C-ring protein. C-terminal FLAG-tagged FliM or N-terminal FLAG-tagged FliY, FliN, or FliH was expressed from a plasmid in C. jejuni single or double mutants lacking specific flagellar proteins and immunoprecipitated by FLAG tag antibody resin after cross-linking cells by formaldehyde. Immunoprecipitated proteins were detected with specific antisera. The top set of immunoblots (Co-IP) shows results from coimmunoprecipitation experiments. The bottom set of immunoblots (WCL) shows levels of proteins in whole-cell lysates of mutants. RpoA is included as a control protein that should not coimmunoprecipitate with a C-ring protein and as a control to ensure equal loading of whole-cell lysates. (B) Summary model of C. jejuni C-ring-protein interactions from coimmunoprecipitation experiments. Double arrows indicate interactions verified by reciprocal coimmunoprecipitation experiments. Single arrows indicate an interaction that could only be observed by immunoprecipitation of one of two interacting partners. Thinner arrows indicate relative weaker interactions. Pale-green arrows indicate that the FliY interactions with FliH and FliN are dependent on FliM.
FLAG-FliY required FliM to form a complex with its interacting partner FliN or FliH (Fig. 5A and B). Since FliM does not interact with FliN (Fig. 4), FliM binding to FliY may alter the FliY conformation to enable interaction with FliN. Considering that our tomography analysis indicated that both FliM and FliY are required for FliI ATPase complex incorporation via FliH (Fig. 3), our coimmunoprecipitation results suggest one of the two following possibilities: FliM and FliY together form an interface to interact with FliH and integrate the FliI ATPase into the motor, or FliM binding to FliY alters FliY conformation to enable interactions with FliH, similarly to how FliM binding to FliY likely alters FliY conformation for interactions with FliN (Fig. 5B). Because we observed FLAG-FliY to interact with FliM and FliH in the ΔfliY ΔfliN mutant (Fig. 5A), FliY appears to be dependent only on FliN for full stability and not for interactions with other C-ring proteins.
Other results from our assays indicated that FLAG-FliN interactions with FliY occurred independently of FliM and FliH, supporting that FliN only binds to FliY (Fig. 5A and B). Also, FliY interacted with FliM and FliN in the absence of FliH (Fig. 5A). These findings support a model where FliM and FliY are major components of the sides of the C ring, possibly together forming an interface to interact with FliH to incorporate the FliI ATPase complex (Fig. 5B). FliN integrates into the C ring by only attaching to FliY, but FliN is important for motility (Fig. 2B and 5B). Finally, our results support the idea that a rigid, well-formed C-ring structure composed of FliM, FliY, and FliN forms in the absence of FliH (Fig. 3).
Contributions of C. jejuni C-ring proteins to flagellar biogenesis.
Due to the different composition and architecture of the C. jejuni flagellar C ring relative to those of peritrichous motors, we next investigated the contributions of C. jejuni C-ring proteins to flagellar biogenesis by analyzing flagellation levels and phenotypes of WT C. jejuni and C-ring mutants by transmission electron microscopy (TEM). As a baseline, WT C. jejuni populations contained 89% of the individual cells with what we defined as a “normal” flagellation pattern, amphitrichous flagellation (a single flagellum at both poles; 72% of the population) or a single flagellum at one pole (17% of the population; Fig. 6 and Table 1). The remaining WT cells were aflagellated (8.5%) or hyperflagellated (producing more than one flagellum at least at one pole, at 2%). Due to the lack of σ54-dependent flagellar gene expression, all C. jejuni ΔfliF and ΔfliG mutant cells were aflagellated (Table 1 and Fig. 6) (66).
FIG 6.
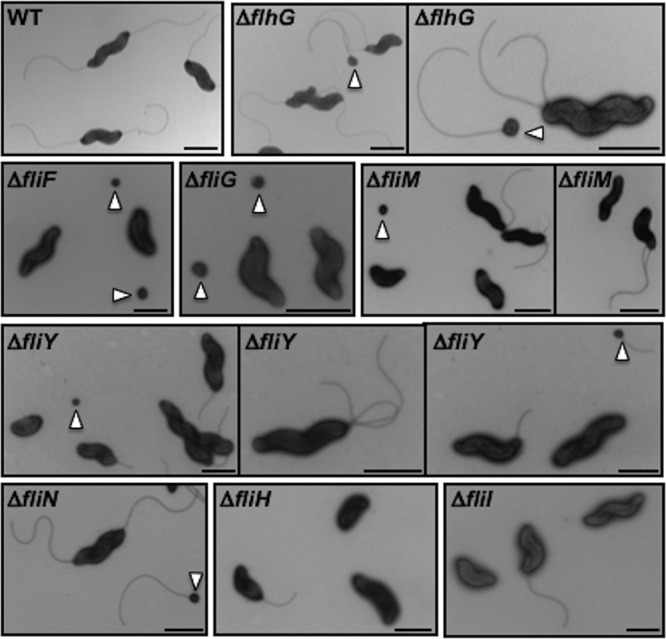
Flagellation phenotypes and minicell production of C. jejuni C-ring mutants. WT C. jejuni and isogenic mutant strains were negatively stained with uranyl acetate and examined by transmission electron microscopy. Arrowheads indicate minicells. Scale bar = 1 μm.
TABLE 1.
Quantitative analysis of flagellation phenotypes of individual WT C. jejuni and isogenic mutant cellsa
| Flagellation phenotype | % presenting phenotype (mean ± SD) in strain: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | ΔflhG | ΔfliF | ΔfliG | ΔfliM | ΔfliY | ΔfliN | ΔfliH | ΔfliI | ΔfliM ΔfliY | ΔfliM ΔfliN | ΔfliY ΔfliN | |
| All cellsb | ||||||||||||
| Normalc | 89.4 ± 0.8 | 51.8 ± 5.7 | 0 ± 0 | 0 ± 0 | 32.6 ± 0.7 | 40.1 ± 0.4 | 68.6 ± 4.4 | 12.3 ± 1.1 | 30.1 ± 4.1 | 31.0 ± 2.6 | 34.8 ± 5.3 | 29.0 ± 4.7 |
| Amphitrichous | 72.4 ± 2.5 | 8.8 ± 0.7 | 2.9 ± 0.7 | 4.7 ± 0.2 | 33.2 ± 3.2 | 0.3 ± 0.4 | 1.3 ± 1.9 | 1.3 ± 0.7 | 2.6 ± 1.1 | 0.8 ± 1.1 | ||
| One at one pole | 17.0 ± 3.3 | 43.0 ± 5.0 | 29.8 ± 1.3 | 35.4 ± 0.2 | 35.4 ± 1.2 | 11.9 ± 0.6 | 28.8 ± 6.0 | 29.7 ± 3.3 | 32.1 ± 6.4 | 28.3 ± 3.6 | ||
| Aflagellated | 8.5 ± 1.4 | 8.4 ± 2.0 | 100 ± 0 | 100 ± 0 | 62.8 ± 0.1 | 58.0 ± 0.5 | 21.7 ± 1.9 | 87.7 ± 1.1 | 69.5 ± 3.5 | 65.2 ± 1.6 | 63.8 ± 4.5 | 70.2 ± 4.7 |
| Hyperflagellatedd | 2.1 ± 0.6 | 39.8 ± 3.7 | 0 ± 0 | 0 ± 0 | 4.6 ± 0.6 | 1.9 ± 0.9 | 9.7 ± 2.5 | 0 ± 0 | 0.4 ± 0.6 | 3.8 ± 0.9 | 1.5 ± 0.9 | 0.7 ± 0 |
| Flagellated cells onlye | ||||||||||||
| Normalc | 97.7 ± 0.6 | 56.5 ± 5.0 | 87.7 ± 1.6 | 95.6 ± 2.1 | 87.5 ± 3.5 | 100 ± 0 | 98.4 ± 2.2 | 89.7 ± 3.2 | 95.7 ± 2.8 | 97.5 ± 0.3 | ||
| Hyperflagellatedd | 2.3 ± 0.6 | 43.5 ± 5.0 | 12.3 ± 1.6 | 4.4 ± 2.1 | 12.5 ± 3.5 | 0 ± 0 | 1.6 ± 2.2 | 10.9 ± 3.2 | 4.3 ± 2.8 | 2.5 ± 0.3 | ||
Two experiments were performed in which >100 individual cells were analyzed for flagellation by transmission electron microscopy.
Includes data from all cells analyzed regardless of the flagellation phenotype.
The normal flagellation phenotype includes cells that are amphitrichous (producing a single flagellum at each pole) and cells producing a single flagellum at one pole (“one at one pole”).
Cells producing 2 or more flagella at least at one pole.
Only includes the population of flagellated cells, divided into normally flagellated cells (amphitrichous and one flagellum at one pole) and hyperflagellated cells.
Although the C. jejuni ΔfliM and ΔfliY mutants were nonmotile, 32 to 40% of the cells were flagellated, with a vast majority producing a single flagellum at one pole rather than amphitrichous flagella, indicating that flagellar biogenesis was hindered (Fig. 6 and Table 1). The reduced flagellation phenotypes of these mutants are not due to reduced σ54-dependent flagellar rod and hook gene expression, as our previous analysis indicated that C. jejuni ΔfliM and ΔfliY mutants express these genes at WT levels (66). This level of flagellar biogenesis in the C. jejuni ΔfliM and ΔfliY mutants is striking, as other motile bacteria are usually aflagellated without FliM (17, 18, 78); the B. subtilis ΔfliY mutant is aflagellated, but the Bacillus cereus ΔfliY mutant produces flagella (53, 79). In contrast to the C. jejuni ΔfliM and ΔfliY mutants, more than two-thirds of ΔfliN mutant cells were flagellated, with equal proportions producing amphitrichous flagella or a single flagellum (Table 1 and Fig. 6). Again, these findings are striking, as fliN mutants in other systems are usually aflagellated (18, 78). The large differences in flagellation levels of the ΔfliN mutant compared to those of the ΔfliY (and ΔfliM) mutant supports our previous tomography and protein interaction analyses that showed that FliN has another function other than being essential for FliI ATPase integration into the motor for protein secretion and flagellation. Because the ΔfliM and ΔfliY mutants were nonmotile, and only a minority of ΔfliN mutant cells showed slight turning motions (Fig. 2B), the flagella produced by these mutants are largely nonfunctional and do not rotate, suggesting that these proteins are important for rotor function.
Of note, C. jejuni double mutants lacking any two of FliM, FliY, or FliN had flagellation levels similar to those of the ΔfliM mutant of around 30%, rather than additive effects of the mutations (Table 1 and Fig. 6 and S4A). In each double mutant, FliM, FliY, and FliN proteins were either undetectable or just above the level of detection (Fig. S4B). Thus, the C. jejuni strain producing only the FliG portion of the C ring without significant lower-C-ring structure and the FliI ATPase can produce flagella in a low but significant population of cells. These flagella, however, do not support motility.
Analysis of C. jejuni C-ring double mutants. (A) WT C. jejuni and isogenic double-mutant strains were negatively stained with uranyl acetate and examined by transmission electron microscopy. Arrowheads indicate minicells. Bar = 1 μm. (B) Immunoblots of MS and C-ring protein levels in whole-cell lysates of WT C. jejuni and isogenic double-mutant strains. A specific antiserum to each C-ring protein or FliH was used to detect each protein. The detection of RpoA served as a control to ensure equal loading of proteins across strains. Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We observed that 12% and 30% of the populations of the C. jejuni ΔfliH and ΔfliI mutants were flagellated, respectively, mostly only with a single polar flagellum produced (Fig. 6 and Table 1), confirming that FliH and FliI are required for optimal flagellar biogenesis. As with the ΔfliM and ΔfliN mutants, this level of flagellation in the ΔfliH and ΔfliI mutants is strikingly augmented compared to that in the fliH and fliI mutants in E. coli and Salmonella spp., which are rarely flagellated (47, 48, 78, 80). The flagella produced by the ΔfliH and ΔfliI mutants are functional, as they supported motility (Fig. 2B). Our findings indicate that the C. jejuni fT3SS functions with fewer parts and has less stringent requirements for its ATPase module than do other species.
C-ring proteins impact FlhG-dependent numerical control of flagellar biogenesis and spatial regulation of division.
Upon analysis of flagellation in our C. jejuni mutants, we observed noticeable hyperflagellation of certain C-ring mutants. In many polar flagellates, FlhG ATPase orthologs function as numerical regulators of flagellar biogenesis (59, 61, 81–85). As we have previously observed, the C. jejuni ΔflhG mutant displayed a hyperflagellation phenotype in 40% of all cells (and 43.5% strictly within the flagellated population; Fig. 6 and Table 1) (59, 61). Within the flagellated ΔfliM, ΔfliN, and ΔfliY mutant populations, the hyperflagellated phenotype was particularly evident. Although ΔfliM, ΔfliN, and ΔfliY mutant flagellation levels were lower than those of the WT (30 to 68% in the mutants versus ∼89% in WT C. jejuni), a greater proportion of mutant flagellated cells were hyperflagellated, with 2- to 5.5-fold higher levels of hyperflagellation relative to those of flagellated WT C. jejuni (Fig. 6 and Table 1). These findings suggest that disruption of parts of the C ring likely impacts FlhG activity for accurate numerical control of flagellar biogenesis.
FlhG also influences spatial regulation of septal Z ring formation and cell division in C. jejuni (59). In the C. jejuni ΔflhG mutant, nonviable minicells form when division occurs at polar regions rather than strictly at the midpoint for symmetrical division (59). We previously discovered that C. jejuni lacking FliF, FliM, and FliN produced an elevated minicell population (59), indicating that flagellar proteins impact FlhG activity related to spatial regulation of division, in addition to flagellar number control. Therefore, we analyzed whether C. jejuni FliY, FliH, and FliI influence FlhG-dependent spatial regulation of division. We measured the lengths of cell bodies of WT C. jejuni and mutant populations by TEM to determine the level of minicells as a proxy for symmetrical division defects. Similar to our prior analysis, about ∼90% of the WT C. jejuni cell bodies were 1 to 2 μm in length, and 7.2% were elongated (<2 μm, Fig. 7) (59). Only 1.3% of the cells were minicells (<0.5 μm in length). In comparison, over 27% of cells were minicells in the C. jejuni ΔflhG mutant population (Fig. 6 and 7). Consistent with our previous findings for the C. jejuni ΔfliF mutant, the ΔfliG, ΔfliM and ΔfliN, ΔfliY mutants produced a similar level of minicells in ∼12% of the population (Fig. 6 and 7) (59). These defects in FlhG-dependent activities for spatial regulation of division are specific for MS ring and C-ring mutants, as minicell production was not evident in the C. jejuni ΔfliH and ΔfliI mutants (Fig. 7). We only observed an enhanced elongated population in the ΔfliI mutant (Fig. 7). These findings further support the idea that C. jejuni C-ring proteins have additional functions outside motility in influencing FlhG-dependent activities for numerical regulation of flagellar biogenesis and spatial regulation of cell division.
FIG 7.
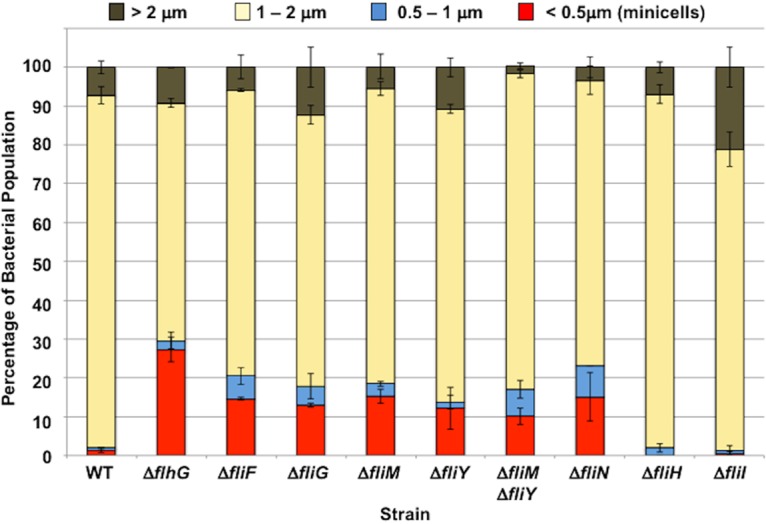
Quantitative assessment of minicell production in C. jejuni MS and C-ring mutants. The lengths of cell bodies of WT C. jejuni and isogenic mutant strains from electron micrographs were measured to determine the percentage of cell populations that are normal size cells or are minicells. Two experiments were performed in which at least 100 individual bacteria were examined per strain. The data are reported as the percentage of bacterial populations with the following cell lengths: >2 μm (brown), 1 to 2 μm (yellow), 0.5 to 1 μm (blue), and <0.5 μm, minicells (red). The data represent the average of the results from the two experiments. Bars represent standard deviations.
C. jejuni C-ring proteins impact polar localization of FlhG.
Our previous analysis indicated that FlhG with an N-terminal FLAG tag is found predominantly at the poles of C. jejuni cells (61), which is a likely requirement for FlhG to numerically regulate polar flagellar biogenesis and prevent division at the poles. We analyzed whether polar localization of native FlhG was altered in C. jejuni C-ring mutants, which might contribute to understanding how C-ring proteins influence FlhG-dependent activities. Similar to our previous analyses, we found ∼65% of WT C. jejuni cells to position FlhG exclusively at poles (Fig. 8). However, the level of cells with FlhG polarly localized dropped over 50% or more in mutants lacking the FliF MS ring protein or the FliG, FliM, FliY, and FliN C-ring proteins (Fig. 8). In contrast, the level of FlhG polar localization was at WT levels in the ΔfliH and ΔfliI mutants, which did not display any FlhG-dependent defects in numerical control of flagellar biogenesis or spatial regulation of division (Table 1 and Fig. 7). Our data support the idea that the formation of a structured C-ring structure composed of FliM, FliY, and FliN is required for efficient polar localization of FlhG so that FlhG can properly numerically regulate flagellar biogenesis and spatially regulate placement of the septal Z ring. Combined, our results implicate unique composition and architecture of the flagellar C ring in C. jejuni to impact biological activities beyond flagellar biogenesis and motility.
FIG 8.
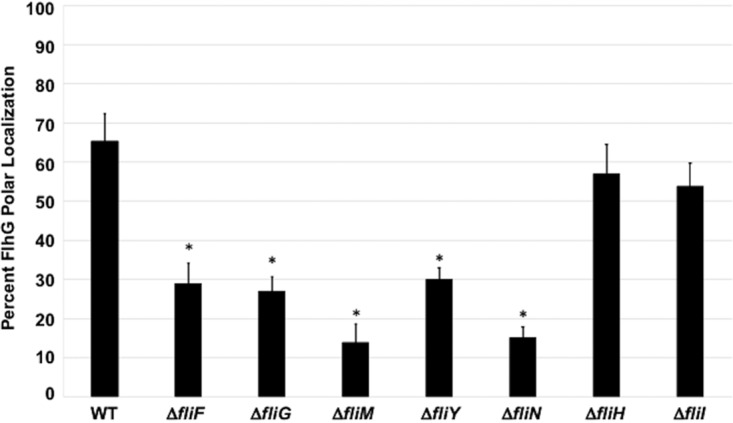
FlhG polar localization in WT C. jejuni and isogenic flagellar mutants. Strains were analyzed by immunofluorescent microscopy after staining with both FlhG and whole C. jejuni antisera. Cells in which detection of FlhG was exclusively at poles were considered positive for polar localization of FlhG. Each strain was analyzed in triplicate, and at least 100 individual cells were counted per sample. After analysis, the percentage of cells with exclusive polar localization of FlhG were averaged, and the standard deviations were determined (bars). A Student's t test was performed to determine the statistical significance of differences in polar localization of FlhG between WT and mutant strains (*, P < 0.05).
DISCUSSION
Flagellar motors across bacterial species have conserved core structures composed of proteins with conserved functions, although recent findings have shown that some motors have recruited extra proteins to form additional structures that adapt and enhance motor function (6–8, 10). Here, we show that the conserved flagellar motor core has also diversified, leading to structural alterations and enabling core components to subdivide existing roles and develop new roles. Using phylogenetic, operonic, genetic, and in situ structural analyses, we show that the C. jejuni C ring contains a paralogous duplication of an ancestral FliN-like protein to yield contemporary FliN and FliY, with FliY retaining a function more similar to that of FliN from the well-studied peritrichous E. coli and Salmonella sp. motors, and FliN diverging to a more supporting role in preserving C-ring structure for flagellar rotation. The C. jejuni C ring has also evolved to contribute to the localization of FlhG to regulate two important cellular processes in C. jejuni, numerical control of flagellar biogenesis, and spatial regulation of division. Finally, our results also reveal that the C. jejuni fT3SS can function as a secretion machine with fewer parts, being less dependent on its ATPase complex than are other T3SSs.
Our results enable speculation on how the C. jejuni C ring evolved. Our phylogenetic study suggests that an additional copy of an ancestral FliN-like protein evolved in an ancestral epsilonproteobacterium, producing two initially identical SPOA domain-containing proteins that diverged. The fact that both FliY and FliM feature CheC domains suggests that the ancestral sequence had both CheC and SPOA domains and that the CheC domain was subsequently lost by FliN proteins. Although we cannot rule out a horizontal gene transfer origin, a gene duplication is more plausible, because an exogenous homolog with a diverged binding interface is not guaranteed to be able to assemble into a preexisting structure, whereas an identical duplicate will by definition assemble. Furthermore, the spirochete FliY/FliN families show a similarly deep branching pattern. The divergence of the two ancestral FliN-like proteins gave rise to contemporary C. jejuni FliY and FliN. We propose that contemporary FliY retained the classical FliN role exemplified by FliN from peritrichous flagellar motors based on our finding that FliY is located in an operon with FliM, C. jejuni FliY has higher sequence similarity to FliN of peritrichous flagellar motors, FliY and FliM are required for the assembly of FliH to anchor the ATPase complex, and FliN is less critical for flagellar biogenesis than is FliY. We propose that FliN, meanwhile, neofunctionalized as a structural stabilizer. Sequence drift led to a loss of binding between FliN and FliM, necessitating that FliY serve as a bridge between them; this shift of FliY and FliN from homooligomer to obligate heterooligomer is seen in other heterooligomeric structures that have evolved from paralogous duplications (86, 87). Sequence drift also led FliN to lose an ability to bind FliH, as this function was fulfilled by FliY.
Whether the ancestral FliN-like protein acquired its CheC domain or whether the ancestor already had a CheC domain is unclear. It is possible that FliM represents the ancestral state that included a CheC domain and that the CheC-less FliN proteins have selectively lost their CheC domains. Indeed, the loss of a CheC domain requires evolution only of an alternative translation start site, whereas the gain of a CheC domain requires correct relative positioning of the donor CheC protein in the genome, arguing against multiple independent CheC acquisitions. Although we do not know the origins of this second allele that encodes contemporary FliN, cjj81176_0374, the gene upstream of fliN in the C. jejuni strain 81-176 genome, encodes a protein of unknown function with a predicted distantly related CheC domain echoing the typical FliY and FliM arrangement of an N-terminal CheC-like domain, followed by a C-terminal SPOA domain. This may be a relic of an ancestral state that included the CheC domain. Alternatively, it may represent an ancestral state prior to fusion of the two proteins, where the original fliY allele may have duplicated and integrated elsewhere in the genome before point mutations fused the CheC-like domain with the SPOA domain.
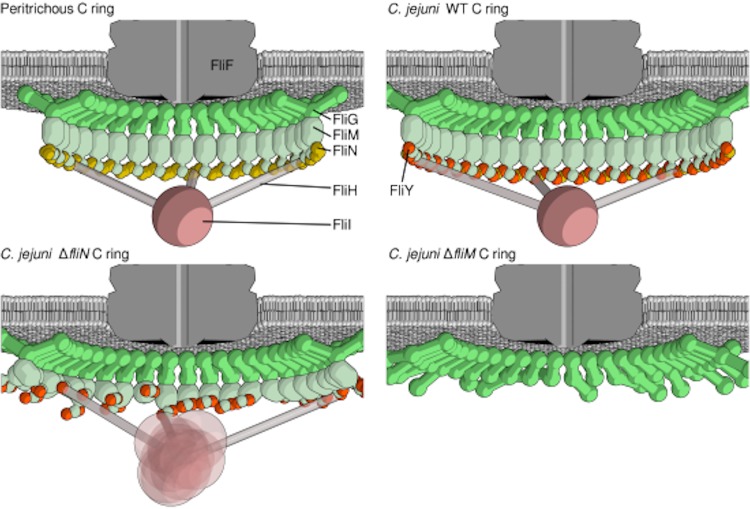
Our data enable us to propose a model for the architecture of the C. jejuni C ring whereby FliM and FliY form a structural and functional module of the C ring through their SPOA domains. FliN and FliY are often mutually exclusive, and B. subtilis FliY can restore motility to a Salmonella sp. fliN mutant (53), indicating that FliY and FliN are equivalent components of the lower rim of the C ring in different species together with FliM. Our findings show that FliN and FliY are not exchangeable in the C. jejuni C ring and possess distinct roles, whereby FliY likely forms a platform with FliM for FliH binding and anchoring the ATPase complex into the fT3SS, and FliN stabilizes a concrete C-ring architecture between the SPOA domains of FliY and FliM. Our findings suggest that FliN and FliY oligomerize with FliM differently in different species. Incorporating our results into the archetypical model from E. coli and Salmonella sp. motor of the lower rim of the C ring as a circularized spiral structure of SPOA domains formed by multimerization of a FliM-FliN3 ternary complex enables us to propose a model for the architecture of the elaborated C. jejuni C ring (Fig. 9). Our interaction data led us to propose that the SPOA domains of FliM, FliY, and FliN retain a common circularized helical architecture but with complexes forming a FliM-FliY-FliN-FliY repeat through SPOA domain interactions (Fig. 9). This arrangement would allow FliM and FliY to form an interface for FliH binding and FliI integration (even in the absence of FliN, albeit with a more disordered architecture), with FliN providing interlocking stabilizers between FliY proteins to bring adjacent complexes together (Fig. 9). While is unclear where the CheC domain from FliY fits into this model, and this model fails to incorporate previous observations of symmetric SPOA dimers for FliY, this model is the most parsimonious with our data. Our model provides a framework for future exploration to unravel how this unusual collection of proteins in a bacterium form a functional C ring that is different from that in other bacteria.
FIG 9.
Model of C-ring assembly, composition, and function in C. jejuni. Cartoon schematics show the architecture of the peritrichous C ring (top left) and the C. jejuni C ring (top right), the effect of deletion of fliN partially disrupting the C-ring and ATPase complex architecture (bottom left), and the effect of deletion of fliM further disrupting the C-ring architecture (bottom right).
Observations from the closely related epsilonproteobacterium H. pylori C-ring components have demonstrated some similar interactions that support our proposed C-ring architecture (57). Structural analysis of H. pylori FliM, FliY, FliN, and FliH suggests that an alteration of C-ring architecture and function has also occurred in this species (56, 57). Nevertheless, H. pylori FliY-FliM and FliY-FliN heterodimers can interact with FliH in vitro, which is different from what we observed in vivo in C. jejuni with only FliM and FliY contributing to FliH interactions. H. pylori also appears to have different requirements of C-ring proteins for flagellar assembly from those to C. jejuni. H. pylori fliM and fliY mutants are severely defective for flagellar biogenesis, but a fliN mutant is much less so; all of these H. pylori mutants are generally less flagellated than are the respective C. jejuni mutants (56, 57, 88). Our work combined with this analysis in H. pylori, a closely related epsilonproteobacterium, demonstrate that flagellar motors are composed of different components across bacterial species, which result in motors with different assemblies of substructures, alteration of motor output for propulsion, and roles outside flagellar motility.
Our model suggests an explanation for why, despite flagellation of the C. jejuni ΔfliM, ΔfliY, ΔfliN mutants, none of these strains are motile. These nonmotile mutants predominantly assembled a single flagellum at one pole, a flagellation pattern similar to those of the motile C. jejuni ΔfliH and ΔfliI mutants, indicating that the lack of motility in the ΔfliM, ΔfliY, and ΔfliN mutants is not due to single flagellation. The lower portion of the C ring appears to be less structured in these mutants in our tomography analysis, indicating that all three proteins are required to form a rigid C ring. We propose that disruption of this region of the C ring has consequences for FliG folding and/or assembly into the rotor for full activity. That the disruption of C ring structure has significant consequences for motility and assembly highlights an interesting apparent paradox in C-ring biology. Studies in E. coli have shown that FliM and FliN are not static components of the C ring but rather bind and dissociate dynamically. These dynamics are not detrimental to motor function in this species (89–92). However, the deletion of FliM, FliY, and FliN is highly damaging to the C. jejuni C ring. It is possible that C. jejuni C-ring components, like its stator complexes, are static components of the C ring, especially since we discovered that these proteins are important for FlhG-dependent cellular processes that can affect motility and division impacting in vivo fitness.
One remaining question is how the C. jejuni flagellar C ring impacts FlhG localization for activity to accurately control flagellar number and prevent division at the poles. All MS and C-ring mutants (but not fliH or fliI mutants) are defective in spatial regulation of division, and mutants lacking FliM, FliY, and FliN are hyperflagellated, demonstrating defects in both known functions of FlhG and implicating the C ring as contributing to FlhG biological activity. The levels of hyperflagellation and division defects are not as high, however, as those of the C. jejuni ΔflhG mutant, suggesting that other factors also influence FlhG activity. Currently, it is unknown how FlhG localizes to poles in C. jejuni. FliF and FliG like do not directly influence FlhG localization, as C. jejuni mutants lacking these proteins do not incorporate FliM, FliY, or FliN into the C ring. Thus, either complete formation of the C ring by FliM, FliY, or FliN or integration of one of these specific proteins impacts FlhG localization. As we have yet to detect a stable interaction of FlhG with any C-ring protein, the formation of a structured flagellar C ring (regardless of presence of FliH or FliI) may indirectly influence FlhG polar localization. In Vibrio cholerae, the HubP protein is involved in the polar localization of FlhG and other MinD/ParA ATPase homologs involved in chromosome segregation and localization of the chemotactic machinery (93). However, C. jejuni lacks HubP. We previously showed that FlhG has a C-terminal amphipathic helix (61); in related MinD proteins, this amphipathic helix recognizes polar curvature and inserts into the cytoplasm-inner membrane interface upon MinD binding ATP (94–97). One possibility is that C. jejuni C-ring formation influences the curvature or distortion of membrane phospholipids that is required for FlhG polar localization. Alternatively, the C ring could interact with a protein that directly interacts with FlhG for polar localization. Regardless of the mechanism, formation of the flagellar C ring likely serves as a polar determinant for polar localization of FlhG so that it can regulate flagellar number and spatial control of septal Z ring formation for accurate symmetrical division.
Our results highlight remarkable differences in the C. jejuni fT3SS to function adequately even without its ATPase complex, which is usually critical for T3SS function. Although mechanisms of T3SS are not completely clear, it is thought that ATP hydrolysis by the FliI ATPase largely functions to dissociate substrates from chaperones for delivery to the fT3SS export gate, whereas the proton motive force powers substrate secretion through the export gate (47, 48). Salmonella fliM or fliN mutants are aflagellated, presumably due to failure to assemble the FliI ATPase underneath the fT3SS export gate via C-ring-FliH interactions (17, 18, 40, 78). Furthermore, Salmonella fliH and fliI mutants rarely synthesize flagella (47, 48, 80). Disrupting FliI assembly into the C. jejuni motor, however, still allowed for a substantial proportion of flagellated cells (∼12 to 40% in the fliH, fliM, fliI, and fliY mutants). To our knowledge, the relative levels of organelle biogenesis in these C. jejuni mutants are much higher than usually reported in analogous mutants in other motile bacteria and in related injectisome T3SS mutants. These findings point to a striking minimalism for a functional fT3SS in C. jejuni. It is unclear what may be different about C. jejuni or its fT3SS to enable flagellar assembly with a minimal fT3SS machinery. Salmonella C-ring mutants can partially overcome secretion defects with elevated production of FliI or secretion substrates, with increased FliI thought to augment chaperone-free substrates or increase the activation and opening of the fT3SS export gate (1, 47, 48, 50, 98). It is possible that the C. jejuni fT3SS export gate has a naturally more open conformation, is more easily accessible by secretion substrates, or is operated by a protein motive force that is naturally higher in C. jejuni and can compensate fairly well in the absence of FliI. Alternatively, C. jejuni substrates may be produced at higher levels than in other bacteria or may be less tightly associated with chaperones for easier delivery into the fT3SS export gate. C. jejuni does use its fT3SS to also secrete Fed and Cia proteins that are not required for motility but for commensal colonization of chickens and interactions with human intestinal cells (99–104). C. jejuni may have developed an fT3SS with enhanced mechanics to secrete a specific but broader range of substrates than for other fT3SSs to be more competitive for in vivo fitness. A full understanding for how the C. jejuni fT3SS can moderately function without an ATPase complex will require a deeper understanding of how flagellar substrates are concentrated and delivered to the fT3SS export gate, whether flagellar secretion is powered differently, and the exact contribution of the FliI6-FliJ ATPase complex to secretion in C. jejuni.
An intriguing question to consider regarding C-ring composition and function is what were the evolutionary steps in developing the C-ring structure of Campylobacter species and how the C ring acquired functions to influence flagellar biogenesis, rotor function, and FlhG-dependent activities related to flagellar number control and spatial regulation of division. Because flagellar motors are widespread and likely occurred early in evolution, primordial functions of the C. jejuni C-ring proteins were for flagellar biogenesis and motility. We expect that the impact of flagellar C-ring proteins on FlhG-dependent activities likely occurred later and are specific for Campylobacter species and maybe a few other amphitrichous bacteria. We suspect that the complex composition of the C. jejuni flagellar C ring is now favorably maintained due its expanded role in the biology of C. jejuni. Our results highlight, again, that there is no one canonical flagellum; rather, the bacterial flagellum is a set of diverse evolved types and subtypes with many nuanced differences.
MATERIALS AND METHODS
Bacterial growth and media.
The C. jejuni 81-176 strain is typically grown under microaerobic conditions (85% N2, 10% CO2, 5% O2) on Mueller-Hinton (MH) agar at 37°C. For routine growth to perform most experiments, C. jejuni strains were grown from frozen stocks for 48 h under microaerobic conditions at 37°C and then restreaked onto MH agar and grown for an additional 16 h under identical conditions. As required, antibiotics were added to the MH medium at the following concentrations: 10 μg/ml trimethoprim (TMP), 15 μg/ml chloramphenicol, 50 μg/ml kanamycin, and 0.5, 1, 2, or 5 mg/ml streptomycin. All C. jejuni strains were stored at –80°C in a solution of 85% MH broth and 15% glycerol. E. coli DH5α, XL1-Blue, and BL21 strains were grown on Luria-Bertani (LB) agar or in LB broth containing the following concentrations of antibiotics when appropriate: 100 μg/ml ampicillin, 50 μg/ml kanamycin, or 15 μg/ml chloramphenicol. All E. coli strains were stored at –80°C in a solution of 80% LB broth and 20% glycerol.
Construction of mutants.
All methodologies to construct plasmids and C. jejuni mutants are described in Text S1 in the supplemental material. All bacterial strains and plasmids used in this study are listed in Tables S1 and S2.
Bacterial strains used in this study. Download Table S1, PDF file, 0.1 MB (109.1KB, pdf) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S2, PDF file, 0.1 MB (95.1KB, pdf) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetics of FliM, FliN, and FliY.
Thirty-eight protein sequences of FliM, FliN, and FliY from 12 organisms were used for this analysis. Initial multiple-sequence alignments were conducted using the package BAli-Phy. The aligned sequences were evaluated using a transitive consistency score (TCS) in the suite T-Coffee to remove residues with a value lower than 3. SeaView was used to remove gap sites in the alignment before scripts were used to trim the sequence alignment to the final 74 residues, which corresponds to the approximate common SPOA domain present in FliM, FliN, and FliY. Phylogenetic trees were generated using a maximum likelihood method in the package Garli. One thousand bootstrap repeats were generated with values for genthreshfortopoterm of 100,000 and significanttopochange of 0.00001. Operons for each organism were generated through custom scripts utilizing the python package Biography. Initial diagrams were produced showing the relative genomic location of each gene; these were manually edited and annotated with the presence of full and degenerate EIDAL motifs.
Electron microscopy sample preparation, tilt-series data collection, and tomographic data processing.
C. jejuni was grown microaerobically for 48 h on MH agar at 37°C prior to restreaking on fresh plates for 16 h. Overnight cell cultures were resuspended from plates in ∼1.5 ml of MH broth, spun down to an optical density at 600 nm (OD600) of 25, and combined with gold fiducial markers. Cell suspensions (3 μl) were applied to glow discharged Quantifoil R2/2 grids (200 mesh), blotted, and plunge frozen in a liquid ethane-propane mixture using the Vitrobot Mark 4 robot (FEI Company). The blotting conditions used were as follows: blot force, 2; blot time, 2; drain time, 1; humidity, 95%; H-value, 0.65; and no wait time. Grids were stored under liquid nitrogen until data collection.
Data were collected on a 200-kV Twin F20 microscope (FEI Company) using a Falcon II direct electron detector camera (FEI Company) with a Gatan 626 cryo-holder. The software package Leginon was used to record tilts between +54 and −54, beginning at +24 and collecting through to −54 before collecting the remainder of the tilts. The collection magnifications used were 25,000× (ΔfliY and ΔfliN mutants) for a final pixel size of 8.280 Å and 29,000× (ΔfliM and ΔfliM ΔfliY mutants) for a final pixel size of 7.002 Å. All structures were resized to a pixel size of 8.281 Å for comparison with the WT. A different defocus value was applied to each sample during collection, as follows: ΔfliY mutant, 3.5 μm; ΔfliM mutant, 2 μm; ΔfliM ΔfliY mutant, 2.5 μm; and ΔfliN mutant, 6 μm. Overnight data collection was enabled by the use of a 3-liter dewer for the cryo-trap and a nitrogen-dispensing pump controlled via Leginon to maintain cryo-holder temperatures.
Tomograms were reconstructed automatically using the RAPTOR software and the package IMOD. All data were low-pass filtered to 3.5 nm. Motors within tomograms were delineated by two points along their rotational axis. The PEET package within IMOD was then used for particle extraction, alignment, and averaging. A reference-free alignment approach with a spherical mask was used for the initial averaging, and this initial average was then used as a reference alongside a mask which excludes the inner and outer membranes from the alignment to generate the final subtomogram averages.
Motility assays.
C. jejuni strains were suspended from plates in MH broth and diluted to an OD600 of 0.8. Each bacterial strain was stabbed into semisolid MH motility medium containing 0.4% agar using an inoculation needle and then incubated for 30 h at 37°C under microaerobic conditions. When appropriate, motility agar contained chloramphenicol or kanamycin to maintain plasmids for in trans complementation of mutants. For dark-field microscopy, the cultures were further diluted 1:10 in MH broth or MH broth with 0.35% methylcellulose. Strains were immediately analyzed for motility by applying 3 μl of culture between a glass slide and glass coverslip.
In vivo immunoprecipitation of C. jejuni proteins.
Immunoprecipitation of proteins from C. jejuni for protein interaction studies were performed as previously described, with slight modifications (66, 105). C. jejuni mutant strains expressing appropriate FLAG-tagged proteins from plasmids were grown on six MH agar plates with appropriate antibiotics for 16 h at 37°C under microaerobic conditions. Bacteria were suspended from each plate in 2 ml of phosphate-buffered saline (PBS), centrifuged for 6,000 rpm for 10 min, and resuspended in 6 ml of PBS. Bacteria were cross-linked by the addition of 16.2 μl of 37% formaldehyde (final concentration, 0.1%) and incubated for 30 min at 37°C. Cross-linking was quenched with the addition 1.2 ml of 1 M glycine, followed by rocking for 10 min at 25°C. Bacteria were centrifuged at 6,000 rpm for 10 min. For osmotic lysis of bacterial cells, pellets were resuspended in 1.5 ml 200 mM Tris (pH 8.0), and then the following solutions were added in order with vortexing: 3 ml of 200 mM Tris (pH 8.0) with 1 M sucrose, 0.3 ml of 10 mM EDTA, 0.3 ml lysozyme (10 mg/ml), 9 ml H2O, and 0.9 ml 100 mM phenylmethylsulfonyl fluoride (PMSF) (105). After incubation on ice for 15 min, 15 ml of solubilization solution (50 mM Tris [pH 8.0], 10 mM MgCl2, 2% Triton X-100) was added. Samples were incubated on ice for 45 min and then centrifuged at 10,000 rpm for 20 min. The supernatant was recovered, and 45 μl of anti-FLAG M2 affinity gel resin (Sigma-Aldrich) was added to immunoprecipitate FLAG-tagged proteins along with any interacting proteins. The lysate was then rocked overnight at 4°C on a nutator. The resin was pelleted by centrifugation at 4°C at 6,000 rpm for 20 min. The resin pellet was washed three times with 3 ml of RIPA buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100), with a centrifugation step of 6,000 rpm for 15 min between each wash. The resin was then resuspended in 255 μl SDS-PAGE loading buffer and boiled for 5 min, and 30 μl was analyzed by SDS-PAGE, followed by immunoblotting with specific antisera.
Immunoblotting analysis.
The procedures for the construction of plasmids for recombinant protein expression, purification of recombinant proteins, and antiserum generation are described in the supplemental material. Preparation of C. jejuni strains for whole-cell lysates (WCLs) was performed as previously described (106). All immunoblotting analysis of WCLs or immunoprecipitated proteins was performed with equal amounts of samples from C. jejuni strains after SDS-PAGE by standard procedures. For the specific detection of proteins in WCLs, 10 μl of WCLs was analyzed to detect FliF and FliG, 12.5 μl of WCLs was analyzed to detect FliM, FliY, and RpoA, 25 μl of WCLs was analyzed to detect FliN, and 4 μl of WCLs was analyzed to detect FliH. Proteins were detected with specific murine, rabbit, or guinea pig antiserum generated previously or as described above. Primary antisera were used at the following concentrations: FliF M204, 1:2,500; (66); FliG M162, 1:2,500; (66); RpoA M251, 1:3,000; (107); FliM M186, 1:2,500; FliY M197, 1:4,000; (66), FliH GP222, 1:6,000; and FliN GP204, 1:500. All primary antisera were applied to the immunoblots for 1 to 2 h, except for the FliN antiserum, which was applied overnight. Appropriate horseradish peroxidase-conjugated goat antibodies were used as secondary antibodies to develop immunoblots.
Transmission electron microscopy.
C. jejuni strains were resuspended from MH agar into PBS, pelleted for 3 min at 13,200 rpm in a microcentrifuge, resuspended in 2% glutaraldehyde in 0.1 M cacodylate, and then incubated on ice for 1 h. Copper-coated Formvar grids were negatively glow discharged, and bacterial samples were then applied to the grids. The samples were stained with 2% uranyl acetate and visualized with an FEI Tecnai G2 Spirit BioTWIN transmission electron microscope. Flagellar numbers were determined from at least 100 individual cells and averaged from two biological replicates to determine the proportion of bacterial populations producing different flagellation phenotypes, as follows: hyperflagellated (a bacterium producing two or more flagella at one or both poles), wild type (producing a single flagellum at each pole or a flagellum at one pole with the other pole aflagellated), or aflagellated (lacking a flagellum). For analysis of cell body lengths, bacterial cell bodies from electron micrographs were visualized, measured using the ImageJ software, and placed into one of the following four categories: <0.5 μm (minicells), 0.5 to 1 μm, 1 to 2 μm, and >2 μm. After averaging, the standard deviation for each population was calculated.
Immunofluorescent microscopy.
WT C. jejuni and isogenic mutants were prepared for immunofluorescent microscopy to determine the localization of FlhG, as previously described (61). After fixation and blocking of samples on chamber slides, 70 μl of a mixture containing a 1:50 dilution of FlhG GP188 antiserum or whole C. jejuni UT527 antiserum in 3% bovine serum albumin (BSA), 1% Zwittergent, 0.1% Triton X-100, and 0.02% sodium azide in PBS was applied to each sample and incubated for 30 min at room temperature. Samples were then washed three times with blocking buffer. Seventy microliters of a mixture containing a 1:200 dilution of the appropriate secondary antibody (goat Texas Red-conjugated anti-rabbit antibody or goat isothiocyanate-conjugated anti-guinea pig antibody; Santa Cruz Biotechnology) was added to the sample for 30 min at room temperature. Samples were washed three times with blocking buffer, chamber gaskets were then removed, and two drops of Vectashield (Vector Laboratories) were added, followed by a coverslip. Slides were cured at room temperature overnight before imaging on an Applied Precision PersonalDV deconvolution microscope with an Olympus 100× lens objective and a CoolSNAP-HQ2 camera. Images were adjusted for brightness and contrast using the ImageJ software. Three separate samples were prepared for each strain, and at least 100 cells were counted for each sample. Individual cells were considered positive for FlhG polar localization if the fluorescent signal was observed only at one or both poles. After averaging, the standard deviation for each population was calculated. Student's t test was used to evaluate the statistical significance of differences in the levels of polar localization of FlhG between the WT and mutant strains.
All data and methodologies are available upon request.
Data availability.
Subtomogram averages are available in EMDB (ΔfliN, EMD-10341; ΔfliY, EMD-10342; ΔfliM, EMD-10343; ΔfliM ΔfliY, EMD-10345; ΔfliH, EMD-10454; ΔfliH ΔfliI, EMD-10455; ΔfliH ΔfliS, EMD-10456; ΔfliI, EMD-10457).
Additional Materials and Methods. Download Text S1, PDF file, 0.1 MB (151.8KB, pdf) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Paul Simpson for assistance with electron microscopy.
This work was supported by NIH grant R01AI065539 (to D.R.H.). C.J.G. was supported by NIH training grant T32 AI007520, L.D.H. was supported by a Biotechnology and Biological Sciences Research Council postgraduate training award, T.R.S.M.-P. was supported by the Francis Crick Institute (which receives its core funding from Cancer Research UK [FC001179], the UK Medical Research Council [FC001179], and the Wellcome Trust [FC001179]), and M.B. was supported by Medical Research Council grant MR/P019374/1.
Footnotes
Citation Henderson LD, Matthews-Palmer TRS, Gulbronson CJ, Ribardo DA, Beeby M, Hendrixson DR. 2020. Diversification of Campylobacter jejuni flagellar C-ring composition impacts its structure and function in motility, flagellar assembly, and cellular processes. mBio 11:e02286-19. https://doi.org/10.1128/mBio.02286-19.
REFERENCES
- 1.Erhardt M, Namba K, Hughes KT. 2010. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol 2:a000299. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou J, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci U S A 95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol 180:2729–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blair DF, Berg HC. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi S, Aizawa S, Kihara M, Isomura M, Jones CJ, Macnab RM. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella Typhimurium. J Bacteriol 168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moon KH, Zhao X, Manne A, Wang J, Yu Z, Liu J, Motaleb MA. 2016. Spirochetes flagellar collar protein FlbB has astounding effects in orientation of periplasmic flagella, bacterial shape, motility, and assembly of motors in Borrelia burgdorferi. Mol Microbiol 102:336–348. doi: 10.1111/mmi.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beeby M, Ribardo DA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR. 2016. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci U S A 113:E1917–E1926. doi: 10.1073/pnas.1518952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaban B, Coleman I, Beeby M. 2018. Evolution of higher torque in Campylobacter-type bacterial flagellar motors. Sci Rep 8:97. doi: 10.1038/s41598-017-18115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossmann FM, Beeby M. 2018. Insights into the evolution of bacterial flagellar motors from high-throughput in situ electron cryotomography and subtomogram averaging. Acta Crystallogr D Struct Biol 74:585–594. doi: 10.1107/S2059798318007945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojima S, Blair DF. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Blair DF. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol 273:428–439. doi: 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd SA, Blair DF. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol 266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 14.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 15.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán JE, Aizawa SI. 1998. Supramolecular structure of the Salmonella Typhimurium type III protein secretion system. Science 280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 16.Sockett H, Yamaguchi S, Kihara M, Irikura VM, Macnab RM. 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella Typhimurium. J Bacteriol 174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi S, Fujita H, Ishihara A, Aizawa S, Macnab RM. 1986. Subdivision of flagellar genes of Salmonella Typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol 166:187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irikura VM, Kihara M, Yamaguchi S, Sockett H, Macnab RM. 1993. Salmonella Typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol 175:802–810. doi: 10.1128/jb.175.3.802-810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González-Pedrajo B, Minamino T, Kihara M, Namba K. 2006. Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol Microbiol 60:984–998. doi: 10.1111/j.1365-2958.2006.05149.x. [DOI] [PubMed] [Google Scholar]
- 20.McMurry JL, Murphy JW, Gonzalez-Pedrajo B. 2006. The FliN-FliH interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry 45:11790–11798. doi: 10.1021/bi0605890. [DOI] [PubMed] [Google Scholar]
- 21.Minamino T, Yoshimura SD, Morimoto YV, Gonzalez-Pedrajo B, Kami-Ike N, Namba K. 2009. Roles of the extreme N-terminal region of FliH for efficient localization of the FliH-FliI complex to the bacterial flagellar type III export apparatus. Mol Microbiol 74:1471–1483. doi: 10.1111/j.1365-2958.2009.06946.x. [DOI] [PubMed] [Google Scholar]
- 22.Stock D, Namba K, Lee LK. 2012. Nanorotors and self-assembling macromolecular machines: the torque ring of the bacterial flagellar motor. Curr Opin Biotechnol 23:545–554. doi: 10.1016/j.copbio.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Francis NR, Irikura VM, Yamaguchi S, DeRosier DJ, Macnab RM. 1992. Localization of the Salmonella Typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci U S A 89:6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol 235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 25.Zhao R, Amsler CD, Matsumura P, Khan S. 1996. FliG and FliM distribution in the Salmonella Typhimurium cell and flagellar basal bodies. J Bacteriol 178:258–265. doi: 10.1128/jb.178.1.258-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao R, Pathak N, Jaffe H, Reese TS, Khan S. 1996. FliN is a major structural protein of the C-ring in the Salmonella Typhimurium flagellar basal body. J Mol Biol 261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 27.Baker MA, Hynson RM, Ganuelas LA, Mohammadi NS, Liew CW, Rey AA, Duff AP, Whitten AE, Jeffries CM, Delalez NJ, Morimoto YV, Stock D, Armitage JP, Turberfield AJ, Namba K, Berry RM, Lee LK. 2016. Domain-swap polymerization drives the self-assembly of the bacterial flagellar motor. Nat Struct Mol Biol 23:197–203. doi: 10.1038/nsmb.3172. [DOI] [PubMed] [Google Scholar]
- 28.Kim EA, Panushka J, Meyer T, Carlisle R, Baker S, Ide N, Lynch M, Crane BR, Blair DF. 2017. Architecture of the flagellar switch complex of Escherichia coli: conformational plasticity of FliG and implications for adaptive remodeling. J Mol Biol 429:1305–1320. doi: 10.1016/j.jmb.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch MJ, Levenson R, Kim EA, Sircar R, Blair DF, Dahlquist FW, Crane BR. 2017. Co-Folding of a FliF-FliG split domain forms the basis of the MS:C ring interface within the bacterial flagellar motor. Structure 25:317–328. doi: 10.1016/j.str.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kihara M, Miller GU, Macnab RM. 2000. Deletion analysis of the flagellar switch protein FliG of Salmonella. J Bacteriol 182:3022–3028. doi: 10.1128/jb.182.11.3022-3028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDowell MA, Marcoux J, McVicker G, Johnson S, Fong YH, Stevens R, Bowman LA, Degiacomi MT, Yan J, Wise A, Friede ME, Benesch JL, Deane JE, Tang CM, Robinson CV, Lea SM. 2016. Characterisation of Shigella Spa33 and Thermotoga FliM/N reveals a new model for C-ring assembly in T3SS. Mol Microbiol 99:749–766. doi: 10.1111/mmi.13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul K, Gonzalez-Bonet G, Bilwes AM, Crane BR, Blair D. 2011. Architecture of the flagellar rotor. EMBO J 30:2962–2971. doi: 10.1038/emboj.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park SY, Lowder B, Bilwes AM, Blair DF, Crane BR. 2006. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc Natl Acad Sci U S A 103:11886–11891. doi: 10.1073/pnas.0602811103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sircar R, Borbat PP, Lynch MJ, Bhatnagar J, Beyersdorf MS, Halkides CJ, Freed JH, Crane BR. 2015. Assembly states of FliM and FliG within the flagellar switch complex. J Mol Biol 427:867–886. doi: 10.1016/j.jmb.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathews MA, Tang HL, Blair DF. 1998. Domain analysis of the FliM protein of Escherichia coli. J Bacteriol 180:5580–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toker AS, Macnab RM. 1997. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J Mol Biol 273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 37.Bren A, Eisenbach M. 1998. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol 278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 38.Ahn DR, Song H, Kim J, Lee S, Park S. 2013. The crystal structure of an activated Thermotoga maritima CheY with N-terminal region of FliM. Int J Biol Macromol 54:76–83. doi: 10.1016/j.ijbiomac.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Szurmant H, Bunn MW, Cannistraro VJ, Ordal GW. 2003. Bacillus subtilis hydrolyzes CheY-P at the location of its action, the flagellar switch. J Biol Chem 278:48611–48616. doi: 10.1074/jbc.M306180200. [DOI] [PubMed] [Google Scholar]
- 40.Notti RQ, Bhattacharya S, Lilic M, Stebbins CE. 2015. A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat Commun 6:7125. doi: 10.1038/ncomms8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin Z, Lin WT, Zhu S, Franco AT, Liu J. 2017. Imaging the motility and chemotaxis machineries in Helicobacter pylori by cryo-electron tomography. J Bacteriol 199:e00695. doi: 10.1128/JB.00695-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ibuki T, Imada K, Minamino T, Kato T, Miyata T, Namba K. 2011. Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nat Struct Mol Biol 18:277–282. doi: 10.1038/nsmb.1977. [DOI] [PubMed] [Google Scholar]
- 43.Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM. 2013. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bange G, Kummerer N, Engel C, Bozkurt G, Wild K, Sinning I. 2010. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc Natl Acad Sci U S A 107:11295–11300. doi: 10.1073/pnas.1001383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ibuki T, Uchida Y, Hironaka Y, Namba K, Imada K, Minamino T. 2013. Interaction between FliJ and FlhA, components of the bacterial flagellar type III export apparatus. J Bacteriol 195:466–473. doi: 10.1128/JB.01711-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minamino T, Morimoto YV, Kinoshita M, Aldridge PD, Namba K. 2014. The bacterial flagellar protein export apparatus processively transports flagellar proteins even with extremely infrequent ATP hydrolysis. Sci Rep 4:7579. doi: 10.1038/srep07579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minamino T, Namba K. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- 48.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. 2008. Energy source of flagellar type III secretion. Nature 451:489–492. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- 49.Terashima H, Kawamoto A, Tatsumi C, Namba K, Minamino T, Imada K. 2018. In vitro reconstitution of functional type III protein export and insights into flagellar assembly. mBio 9:e00988-18. doi: 10.1128/mBio.00988-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erhardt M, Mertens ME, Fabiani FD, Hughes KT. 2014. ATPase-independent type-III protein secretion in Salmonella enterica. PLoS Genet 10:e1004800. doi: 10.1371/journal.pgen.1004800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward E, Kim EA, Panushka J, Botelho T, Meyer T, Kearns DB, Ordal G, Blair DF. 2018. Organization of the flagellar switch complex of Bacillus subtilis. J Bacteriol 201:e00626-18. doi: 10.1128/JB.00626-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sircar R, Greenswag AR, Bilwes AM, Gonzalez-Bonet G, Crane BR. 2013. Structure and activity of the flagellar rotor protein FliY: a member of the CheC phosphatase family. J Biol Chem 288:13493–13502. doi: 10.1074/jbc.M112.445171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bischoff DS, Ordal GW. 1992. Identification and characterization of FliY, a novel component of the Bacillus subtilis flagellar switch complex. Mol Microbiol 6:2715–2723. doi: 10.1111/j.1365-2958.1992.tb01448.x. [DOI] [PubMed] [Google Scholar]
- 54.Szurmant H, Muff TJ, Ordal GW. 2004. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J Biol Chem 279:21787–21792. doi: 10.1074/jbc.M311497200. [DOI] [PubMed] [Google Scholar]
- 55.Liao S, Sun A, Ojcius DM, Wu S, Zhao J, Yan J. 2009. Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol 9:253. doi: 10.1186/1471-2180-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lowenthal AC, Hill M, Sycuro LK, Mehmood K, Salama NR, Ottemann KM. 2009. Functional analysis of the Helicobacter pylori flagellar switch proteins. J Bacteriol 191:7147–7156. doi: 10.1128/JB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lam KH, Xue C, Sun K, Zhang H, Lam WWL, Zhu Z, Ng JTY, Sause WE, Lertsethtakarn P, Lau KF, Ottemann KM, Au S. 2018. Three SpoA-domain proteins interact in the creation of the flagellar type III secretion system in Helicobacter pylori. J Biol Chem 293:13961–13973. doi: 10.1074/jbc.RA118.002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beeby M. 2015. Motility in the epsilon-proteobacteria. Curr Opin Microbiol 28:115–121. doi: 10.1016/j.mib.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Balaban M, Hendrixson DR. 2011. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog 7:e1002420. doi: 10.1371/journal.ppat.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lutkenhaus J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem 76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- 61.Gulbronson CJ, Ribardo DA, Balaban M, Knauer C, Bange G, Hendrixson DR. 2016. FlhG employs diverse intrinsic domains and influences FlhF GTPase activity to numerically regulate polar flagellar biogenesis in Campylobacter jejuni. Mol Microbiol 99:291–306. doi: 10.1111/mmi.13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kazmierczak BI, Hendrixson DR. 2013. Spatial and numerical regulation of flagellar biosynthesis in polarly flagellated bacteria. Mol Microbiol 88:655–663. doi: 10.1111/mmi.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuhmacher JS, Rossmann F, Dempwolff F, Knauer C, Altegoer F, Steinchen W, Dorrich AK, Klingl A, Stephan M, Linne U, Thormann KM, Bange G. 2015. MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc Natl Acad Sci U S A 112:3092–3097. doi: 10.1073/pnas.1419388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuhmacher JS, Thormann KM, Bange G. 2015. How bacteria maintain location and number of flagella? FEMS Microbiol Rev 39:812–822. doi: 10.1093/femsre/fuv034. [DOI] [PubMed] [Google Scholar]
- 65.Bange G, Kummerer N, Grudnik P, Lindner R, Petzold G, Kressler D, Hurt E, Wild K, Sinning I. 2011. Structural basis for the molecular evolution of SRP-GTPase activation by protein. Nat Struct Mol Biol 18:1376–1380. doi: 10.1038/nsmb.2141. [DOI] [PubMed] [Google Scholar]
- 66.Boll JM, Hendrixson DR. 2013. A regulatory checkpoint during flagellar biogenesis in Campylobacter jejuni initiates signal transduction to activate transcription of flagellar genes. mBio 4:e00432-13. doi: 10.1128/mBio.00432-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berg HC, Turner L. 1979. Movement of microorganisms in viscous environments. Nature 278:349–351. doi: 10.1038/278349a0. [DOI] [PubMed] [Google Scholar]
- 68.Ferrero RL, Lee A. 1988. Motility of Campylobacter jejuni in a viscous environment: comparison with conventional rod-shaped bacteria. J Gen Microbiol 134:53–59. doi: 10.1099/00221287-134-1-53. [DOI] [PubMed] [Google Scholar]
- 69.Blair DF, Berg HC. 1988. Restoration of torque in defective flagellar motors. Science 242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 70.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A 103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. 2013. Load-dependent assembly of the bacterial flagellar motor. mBio 4:e00551-13. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown PN, Mathews MA, Joss LA, Hill CP, Blair DF. 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol 187:2890–2902. doi: 10.1128/JB.187.8.2890-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paul K, Blair DF. 2006. Organization of FliN subunits in the flagellar motor of Escherichia coli. J Bacteriol 188:2502–2511. doi: 10.1128/JB.188.7.2502-2511.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sarkar MK, Paul K, Blair DF. 2010. Subunit organization and reversal-associated movements in the flagellar switch of Escherichia coli. J Biol Chem 285:675–684. doi: 10.1074/jbc.M109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Minamino T, MacNab RM. 2000. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol 35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- 76.Minamino T, MacNab RM. 2000. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol Microbiol 37:1494–1503. doi: 10.1046/j.1365-2958.2000.02106.x. [DOI] [PubMed] [Google Scholar]
- 77.Auvray F, Ozin AJ, Claret L, Hughes C. 2002. Intrinsic membrane targeting of the flagellar export ATPase FliI: interaction with acidic phospholipids and FliH. J Mol Biol 318:941–950. doi: 10.1016/S0022-2836(02)00172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S. 1992. Morphological pathway of flagellar assembly in Salmonella Typhimurium. J Mol Biol 226:433–446. doi: 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- 79.Senesi S, Celandroni F, Salvetti S, Beecher DJ, Wong AC, Ghelardi E. 2002. Swarming motility in Bacillus cereus and characterization of a fliY mutant impaired in swarm cell differentiation. Microbiology 148:1785–1794. doi: 10.1099/00221287-148-6-1785. [DOI] [PubMed] [Google Scholar]
- 80.González-Pedrajo B, Fraser GM, Minamino T, Macnab RM. 2002. Molecular dissection of Salmonella FliH, a regulator of the ATPase FliI and the type III flagellar protein export pathway. Mol Microbiol 45:967–982. doi: 10.1046/j.1365-2958.2002.03047.x. [DOI] [PubMed] [Google Scholar]
- 81.Correa NE, Peng F, Klose KE. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription heirarchy. J Bacteriol 187:6324–6332. doi: 10.1128/JB.187.18.6324-6332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dasgupta N, Arora SK, Ramphal R. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 182:357–364. doi: 10.1128/jb.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dasgupta N, Ramphal R. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 183:6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ono H, Takashima A, Hirata H, Homma M, Kojima S. 2015. The MinD homolog FlhG regulates the synthesis of the single polar flagellum of Vibrio alginolyticus. Mol Microbiol 98:130–141. doi: 10.1111/mmi.13109. [DOI] [PubMed] [Google Scholar]
- 85.Kondo S, Imura Y, Mizuno A, Homma M, Kojima S. 2018. Biochemical analysis of GTPase FlhF which controls the number and position of flagellar formation in marine Vibrio. Sci Rep 8:12115. doi: 10.1038/s41598-018-30531-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Finnigan GC, Hanson-Smith V, Stevens TH, Thornton JW. 2012. Evolution of increased complexity in a molecular machine. Nature 481:360–364. doi: 10.1038/nature10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wollenberg K, Swaffield JC. 2001. Evolution of proteasomal ATPases. Mol Biol Evol 18:962–974. doi: 10.1093/oxfordjournals.molbev.a003897. [DOI] [PubMed] [Google Scholar]
- 88.Tsang J, Hoover TR. 2015. Basal body structures differentially affect transcription of RpoN- and FliA-dependent flagellar genes in Helicobacter pylori. J Bacteriol 197:1921–1930. doi: 10.1128/JB.02533-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan J, Branch RW, Hosu BG, Berg HC. 2012. Adaptation at the output of the chemotaxis signalling pathway. Nature 484:233–236. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lele PP, Branch RW, Nathan VS, Berg HC. 2012. Mechanism for adaptive remodeling of the bacterial flagellar switch. Proc Natl Acad Sci U S A 109:20018–20022. doi: 10.1073/pnas.1212327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delalez NJ, Berry RM, Armitage JP. 2014. Stoichiometry and turnover of the bacterial flagellar switch protein FliN. mBio 5:e01216-14. doi: 10.1128/mBio.01216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Branch RW, Sayegh MN, Shen C, Nathan VSJ, Berg HC. 2014. Adaptive remodelling by FliN in the bacterial rotary motor. J Mol Biol 426:3314–3324. doi: 10.1016/j.jmb.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamaichi Y, Bruckner R, Ringgaard S, Moll A, Cameron DE, Briegel A, Jensen GJ, Davis BM, Waldor MK. 2012. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev 26:2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Szeto TH, Rowland SL, Habrukowich CL, King GF. 2003. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J Biol Chem 278:40050–40056. doi: 10.1074/jbc.M306876200. [DOI] [PubMed] [Google Scholar]
- 95.Szeto TH, Rowland SL, Rothfield LI, King GF. 2002. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci U S A 99:15693–15698. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu Z, Lutkenhaus J. 2003. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol Microbiol 47:345–355. doi: 10.1046/j.1365-2958.2003.03321.x. [DOI] [PubMed] [Google Scholar]
- 97.Zhou H, Lutkenhaus J. 2003. Membrane binding by MinD involves insertion of hydrophobic residues within the C-terminal amphipathic helix into the bilayer. J Bacteriol 185:4326–4335. doi: 10.1128/jb.185.15.4326-4335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Konishi M, Kanbe M, McMurry JL, Aizawa S. 2009. Flagellar formation in C-ring-defective mutants by overproduction of FliI, the ATPase specific for flagellar type III secretion. J Bacteriol 191:6186–6191. doi: 10.1128/JB.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barrero-Tobon AM, Hendrixson DR. 2012. Identification and analysis of flagellar coexpressed determinants (Feds) of Campylobacter jejuni involved in colonization. Mol Microbiol 84:352–369. doi: 10.1111/j.1365-2958.2012.08027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barrero-Tobon AM, Hendrixson DR. 2014. Flagellar biosynthesis exerts temporal regulation of secretion of specific Campylobacter jejuni colonization and virulence determinants. Mol Microbiol 93:957–974. doi: 10.1111/mmi.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Buelow DR, Christensen JE, Neal-McKinney JM, Konkel ME. 2011. Campylobacter jejuni survival within human epithelial cells is enhanced by the secreted protein CiaI. Mol Microbiol 80:1296–1312. doi: 10.1111/j.1365-2958.2011.07645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Christensen JE, Pacheco SA, Konkel ME. 2009. Identification of a Campylobacter jejuni-secreted protein required for maximal invasion of host cells. Mol Microbiol 73:650–662. doi: 10.1111/j.1365-2958.2009.06797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. 1999. Bacterial secreted proteins are required for the internaliztion of Campylobacter jejuni into cultured mammalian cells. Mol Microbiol 32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- 104.Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, Mickelson J. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol 186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Johnson TL, Scott ME, Sandkvist M. 2007. Mapping critical interactive sites within the periplasmic domain of the Vibrio cholerae type II secretion protein EpsM. J Bacteriol 189:9082–9089. doi: 10.1128/JB.01256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sommerlad SM, Hendrixson DR. 2007. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J Bacteriol 189:179–186. doi: 10.1128/JB.01199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Joslin SN, Hendrixson DR. 2009. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J Bacteriol 191:2656–2667. doi: 10.1128/JB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein alignments of FliM, FliN, and FliY. Multiple-sequence alignments (MSA) generated using MAFFT and filtered using the T-coffee transitive consistency score. Sourced from 12 bacterial species from the spirochetes, Firmicutes, Epsilonproteobacteria, and Gammaproteobacteria. The SPOA domains of FliM, FliY, and FliN are indicated in the red box. Download FIG S1, TIF file, 1.9 MB (2MB, tif) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Examination of the identity of the unidentified C-ring density in the ΔfliH mutant. Subtomogram average of flagellar motor structures of C. jejuni ΔfliH ΔfliS mutant (left column) and ΔfliH ΔfliI mutant (right column). The top row shows a cross-section through the center of the subtomogram average. The bottom row shows a planar section through the base of the C ring (indicated by white arrow in the side view panel). Arrowheads indicate the position of the unidentified C-ring density that appears in the C. jejuni ΔfliH mutant. Download FIG S2, TIF file, 1.9 MB (2MB, tif) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In trans complementation of C. jejuni mutants with FLAG-tagged proteins. Plasmids expressing N- or C-terminal FLAG-tagged flagellar proteins from promoters to closely replicate expression of the native proteins from WT C. jejuni were introduced into respective C. jejuni mutants. (A) C. jejuni ΔfliF mutant expressing FLAG-FliF. (B) C. jejuni ΔfliG mutant expressing FliG-FLAG. (C) C. jejuni ΔfliM mutant expressing FliM-FLAG. (D) C. jejuni ΔfliY mutant expressing FLAG-FliY. (E) C. jejuni ΔfliN mutant expressing FLAG-FliN. (F) C. jejuni ΔfliH mutant expressing FLAG-FliH. (A to F) Each panel contains an immunoblot of the native protein from a whole-cell lysate of the wild-type strain or mutant strain containing empty vector or the mutant strain expressing the FLAG-tagged protein. An immunoblot is also shown for RpoA in whole-cell lysates to ensure equal loading of proteins across samples. All immunoblots were performed with specific antiserum to each protein. A motility assay is shown in each panel for WT C. jejuni or the mutant strain containing empty vector or the mutant strain expressing the FLAG-tagged protein. Motility was analyzed after 30 h of incubation at 37°C under microaerobic conditions. Download FIG S3, JPG file, 0.1 MB (103.1KB, jpg) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of C. jejuni C-ring double mutants. (A) WT C. jejuni and isogenic double-mutant strains were negatively stained with uranyl acetate and examined by transmission electron microscopy. Arrowheads indicate minicells. Bar = 1 μm. (B) Immunoblots of MS and C-ring protein levels in whole-cell lysates of WT C. jejuni and isogenic double-mutant strains. A specific antiserum to each C-ring protein or FliH was used to detect each protein. The detection of RpoA served as a control to ensure equal loading of proteins across strains. Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains used in this study. Download Table S1, PDF file, 0.1 MB (109.1KB, pdf) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids used in this study. Download Table S2, PDF file, 0.1 MB (95.1KB, pdf) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additional Materials and Methods. Download Text S1, PDF file, 0.1 MB (151.8KB, pdf) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Subtomogram averages are available in EMDB (ΔfliN, EMD-10341; ΔfliY, EMD-10342; ΔfliM, EMD-10343; ΔfliM ΔfliY, EMD-10345; ΔfliH, EMD-10454; ΔfliH ΔfliI, EMD-10455; ΔfliH ΔfliS, EMD-10456; ΔfliI, EMD-10457).
Additional Materials and Methods. Download Text S1, PDF file, 0.1 MB (151.8KB, pdf) .
Copyright © 2019 Henderson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.