Abstract
Recent studies have shown that global translational reprogramming is an early activation event in pattern-triggered immunity, when plants recognize microbe-associated molecular patterns. However, it is not fully known whether translational regulation also occurs in subsequent immune responses, such as effector-triggered immunity (ETI). In this study, we performed genome-wide ribosome profiling in Arabidopsis upon RPS2-mediated ETI activation and discovered that specific groups of genes were translationally regulated, mostly in coordination with transcription. These genes encode enzymes involved in aromatic amino acid, phenylpropanoid, camalexin, and sphingolipid metabolism. The functional significance of these components in ETI was confirmed by genetic and biochemical analyses. Our findings provide new insights into the diverse translational regulation in the plant immune responses and demonstrate that translational coordination of metabolic gene expression is an important strategy for ETI.
Keywords: Translational regulation, Ribosome profiling, Effector-triggered immunity, ETI, phenylalanine, phenylpropanoids, helper receptors
Short Summary:
This article presents a genome-wide ribosome profiling analysis of RPS2-mediated effector-triggered immunity (ETI). During ETI, specific groups of genes were found to be translationally regulated in coordination with transcription. One main group of these genes were involved in aromatic amino acid biosynthesis. Substantial genetic and biochemical data demonstrate that translational regulation of metabolic pathways is an important strategy for ETI.
INTRODUCTION
With the recent application of high-throughput sequencing to ribosome footprinting (Ribo-seq), translational reprogramming has been found to play a significant role in plant responses to various environmental stimuli including pathogens, light, hypoxia, drought, ethylene, and heat stress (Juntawong et al., 2014; Lei et al., 2015; Liu et al., 2013; Merchante et al., 2015; Meteignier et al., 2017; Xu et al., 2017a; Yángüez et al., 2013). Ribo-seq analysis of pattern-triggered immunity (PTI), the first layer of active immune response to microbe-associated molecular patterns (MAMPs), showed that translational reprogramming occurs early in this immune response, most likely before the major transcriptional event (Xu et al., 2017a). However, it is not fully understood how translational regulation is involved in subsequent immune responses, including effector-triggered immunity (ETI), which is induced through recognition of pathogen effectors by the host nucleotide binding site-leucine rich repeat (NBS-LRR) domain containing immune receptors; for example, AvrRpm1 and AvrRpt2 effectors are recognized by RPM1 and RPS2, respectively, to confer resistance (Grant et al., 1995; Mindrinos et al., 1994). Recently, a translational response to the pathogen effector, AvrRpm1, expressed in Arabidopsis as a transgene was reported (Meteignier et al., 2017). This study showed that recognition of AvrRpm1 through RPM1 could influence the translational status of thousands of transcripts, including the stress-responsive translational regulator Target of Rapamycin (TOR) kinase. However, fundamental questions still remain to be answered. Since ETI is the ultimate immune response because it often leads to the programmed cell death (PCD), is it possible that ETI-mediated PCD involves a global translation shutdown? Is global translational regulation distinct between PTI and ETI? Are the targets of translational regulation during ETI specific or similar for different immune receptors?
To answer these fundamental questions, we performed a global translatome analysis of the response to the bacterial pathogen Pseudomonas syringae pv. maculicola (Psm) ES4326 carrying the effector gene AvrRpt2 (Psm ES4326/AvrRpt2) using the Ribo-seq strategy (Ingolia et al., 2009; Xu et al., 2017a). Here, we discovered a targeted translational response in coordination with transcriptional regulation in a subset of genes involved in effector perception/signaling and several metabolic pathways. We further demonstrated genetically that these metabolic genes are specifically important for immune response during ETI. Together, our study provides new insights into translational regulation during ETI, which link metabolic dynamics to the immune response in plants.
RESULTS
ETI-induced TBF1 translation occurs later than the PTI-induced translation
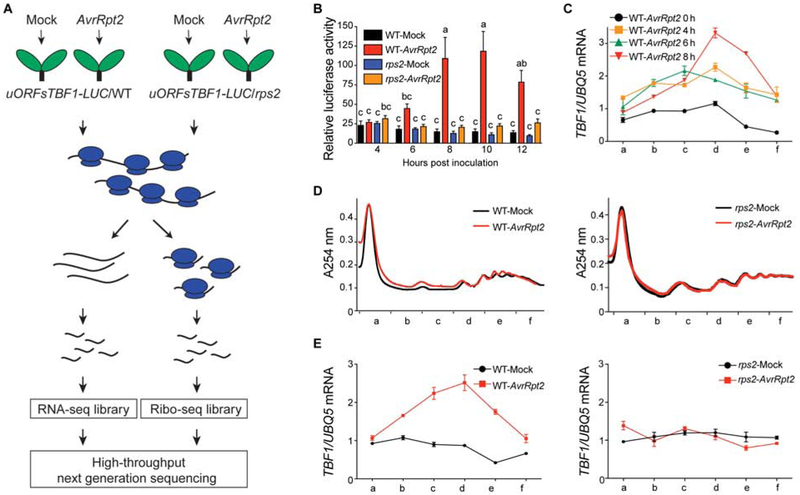
To determine the optimal condition for the global translatome analysis, we used the constitutively transcribed 35S:uORFsTBF1-LUC reporter in wild type (WT) and as a control, the NBS-LRR receptor mutant rps2, which fails to recognize the AvrRpt2 effector (Mindrinos et al., 1994) (Figure 1A). Translation of the luciferase reporter is controlled by the upstream open reading frames (uORFs) of the immune transcription factor gene TBF1 (uORFsTBF1), which has been shown to be rapidly induced by multiple pathogenic signals (Xu et al., 2017b). We found a dramatic induction of the luciferase activity by Psm ES4326/AvrRpt2 between 6 and 8 hours post infiltration (hpi) in WT, while no significant changes were detected in the rps2 mutant (Figure 1B), indicating that the observed translational change in the reporter was dependent on the RPS2 receptor. Consistent with the induction of the luciferase reporter, the endogenous TBF1 mRNA was enriched in polysome-associated fractions at 8 hpi in response to Psm ES4326/AvrRpt2 (Figure 1C). In contrast to WT, the rps2 mutant failed to increase translation of the endogenous TBF1 mRNA (Figures 1D and 1E).
Figure 1. Translational activities during ETI.
(A) Schematic of RNA-seq (RS) and Ribo-seq (RF) library construction using 35S:uORFsTBF1-LUC/WT and 35S:uORFsTBF1-LUC/rps2 plants. (B) Translation of the 35S:uORFsTBF1-LUC reporter in wild type (WT) and the rps2 mutant after Mock (10 mM MgCl2) or Psm ES4326/AvrRpt2 (AvrRpt2) treatment (OD600nm = 0.02). Relative LUC activity was normalized to the level at 1 hour post infiltration (hpi). Data are mean ± SEM; n = 6 biological replicates. Error bars with different letters represent statistically significant differences based on Tukey’s test (P < 0.05; one-way ANOVA). (C) TBF1 mRNA associated with polysomal fractions at different time points after Psm ES4326/AvrRpt2 treatment (OD600nm = 0.02) (mean ± SD; n = 4 technical replicates from one representative experiment). TBF1 mRNA abundance was normalized to the level of UBQ5 mRNA in all fractions. Lower-case letters in the x-axis indicate polysomal fractions in the polysome profile obtained by sucrose density gradient fractionation. (D and E) Polysome profiling of global translational activity at 8 hpi (D) and TBF1 mRNA translational activity (E) calculated as ratios of polysomal/total mRNA in WT (left panel) and rps2 (right panel) after Psm ES4326/AvrRpt2 treatment (OD600nm = 0.02). Transcript levels of TBF1 were normalized against UBQ5 levels determined by qRT-PCR and ratios of polysomal fractions over the total mRNA are presented. Data are mean ± SD; n = 4 technical replicates.
See also Figures S1 and S2.
Notably, ETI-induced TBF1 translation occurred later than the PTI-induced translation induction (Xu et al., 2017a). Moreover, we did not detect a significant difference between the overall polysome profiles of mock-treated or Psm ES4326/AvrRpt2-inoculated samples collected at 8 hpi (Figure 1D), suggesting that, instead of a global shutdown of translation, ETI may involve targeted changes in active translation, including induction of TBF1 translation (Figure 1E).
Global translatome analysis reveals distinct translational regulation during ETI in coordination with transcriptional regulation
To identify genes that undergo targeted changes in active translation during ETI, we collected Arabidopsis leaves at 8 hours after either mock or Psm ES4326/AvrRpt2 infection (OD600nm = 0.02) and generated libraries for RNA-seq (RS) and Ribo-seq (RF) analyses. The library construction and quality control are detailed in Methods (Figures S1 and S2). We compared the Psm ES4326/AvrRpt2-inoculated samples versus mock-inoculated samples and found 983 and 203 genes transcriptionally up-regulated (RSup) and down-regulated (RSdn), respectively, in WT plants (Data S1). Differential analysis of RF fold change (RFfc) detected 926 genes with increased ribosome occupancy (RFup) and 156 genes with decreased ribosome occupancy (RFdn) (Table S1). In the rps2 mutant, only a small number of genes responded differentially in RS (20 RSup and 21 RSdn) and RF (15 RFup and 6 RFdn) (Data S2; Figure S2H). Considering that similar observations were made in a previous microarray experiment with the same Psm ES4326/AvrRpt2 (Gu et al., 2016) and a previous RNA-seq experiment with Pseudomonas syringae pv. tomato carrying the effector gene AvrRpt2 (Mine et al., 2018), the small RS and RF changes in the rps2 mutant were attributable to the specific sampling time used in this analysis.
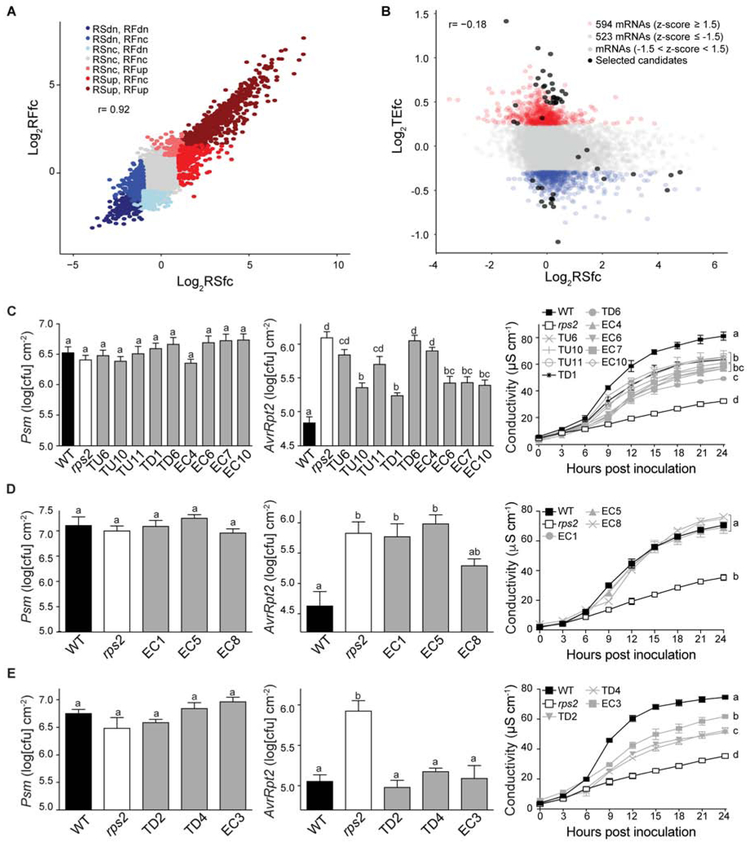
Interestingly, in contrast to the poor correlation between transcriptional and translational changes during PTI (r = 0.41) (Xu et al., 2017a), changes in transcription and translation during ETI showed a relatively strong correlation (r = 0.92; Figure 2A), indicating that the majority of translational changes during ETI were coordinated with changes in transcription. Nevertheless, in combination with transcriptional induction, significant translational induction is detected for the TBF1 transcript in WT (Figure S3A), implying that translational regulation indeed occurs during ETI. To identify translationally regulated genes during ETI, we calculated translational efficiency (TE) according to the previously reported formula (Ingolia et al., 2009; Xu et al., 2017a) and identified 594 mRNAs showing increased TE fold change (TEfc) and 523 mRNAs displaying decreased TEfc (|z| ≥ 1.5) (Figures 2B and S3B; Data S1). We performed dispersion analysis to filter out genes with small changes in RS and RF and found 157 mRNAs showing increased translational efficiency and 109 mRNAs showing decreased translational efficiency (Figure S1H).
Figure 2. Identification of new immune regulators based on global analysis of translational changes during RPS2-mediated ETI.
(A and B) Relationships between RSfc and RFfc (A), and between RSfc and TEfc (B) in WT. dn, down; nc, no change. Black dots, candidates selected for ETI phenotype testing. (C–E) Growth of Psm ES4326 (left) or Psm ES4326/AvrRpt2 (middle) (OD600nm = 0.001), and ion leakage analysis (right) caused by Psm ES4326/AvrRpt2 (OD600nm = 0.01) in phenotypic group A (C), group B (D), and group C (E) of new immune regulators. WT and rps2 were used as controls. TU, TEup; TD, TEdn; EC, ETI candidate. Different letters indicate values that are significantly different based on Tukey’s test (P < 0.01; one-way ANOVA). For ion leakage analysis, the last time point was used for statistical analysis. Data are mean ± SEM; n = 8 biological replicates for bacterial growth and n = 3 biological replicates for ion leakage analysis.
Identification of translationally responsive regulators of ETI
For genetic analysis of potential novel regulators of ETI, we included the top TEup (TU) or TEdn (TD) genes based on our TE (Figure 2B) and dispersion analyses (Figure S1H). We further extended our candidates (EC) from TE changed genes to include those with RF-up changes such as genes encoding Toll interleukin-1 receptor nucleotide-binding site domain (TIR-NBS), leucine-rich repeat domain (LRR), and TIR-NBS-LRR domain-containing proteins and proteins involved in sphingolipid metabolism, which were implicated in ETI in prior studies (Aboul-Soud et al., 2009; Berkey et al., 2012; Nandety et al., 2013; Padmanabhan et al., 2009; Sánchez-Rangel et al., 2015; Swiderski et al., 2009; Zhang et al., 2010). Using these selection criteria, we identified 50 candidate genes (27 TU, 11 TD, 12 EC; Data S3; Figures 2B and S1H).
We obtained available T-DNA insertional mutants for 26 of these genes (11 TU, 6 TD, 9 EC) and tested them for Psm ES4326/AvrRpt2-mediated ETI resistance by measuring both in planta bacterial growth as well as ion leakage resulting from PCD (Data S3). Through these measurements, we found 15 mutants compromised in at least one of these two phenotypes (Figures 2C-2E; Table S1). It is worth noting that the defects detected in these mutants were ETI-specific, because the mutants displayed similar growth of the isogenic Psm ES4326 lacking the AvrRpt2 effector as WT (Figures 2C-2E, left panel). Based on the patterns of Psm ES4326/AvrRpt2 growth and ion leakage analysis, the ETI mutants can be further categorized into three distinct groups. Group A showed reduced PCD, accompanied with increased Psm ES4326/AvrRpt2 bacterial growth compared to WT (Figure 2C). This is a typical mutant phenotype for a positive regulator of ETI. The nine genes in this group include inositol phosphorylceramide (a sphingolipid) synthase 2, the abscisic acid receptor PYL1, TIR-NBS-LRR, TIR-NBS and LRR proteins, amino acid permease 3, and a chaperone DnaJ-domain protein. Group B mutants showed more Psm ES4326/AvrRpt2 bacterial growth, but WT levels of ion leakage (Figure 2D). The three mutants in Group B represent genes encoding two transporters (one for transporting histidine and lysine, and the other for a golgi nucleotide sugar transporter important for sphingolipid metabolism), and one TIR-NBS-LRR immune receptor. Group C mutants showed reduced ion leakage upon Psm ES4326/AvrRpt2 treatment, but without a detectable increase in Psm ES4326/AvrRpt2 pathogen growth compared to WT (Figure 2E). Genes represented by Group C mutants may have a more significant role in ETI-associated PCD than restricting pathogen growth (Coll et al., 2010). They included a phosphate transporter, one enzyme in photorespiration, and a D111/G-patch domain-containing protein which may be involved in RNA binding and processing based on functional studies of its homologs (Aravind and Koonin, 1999).
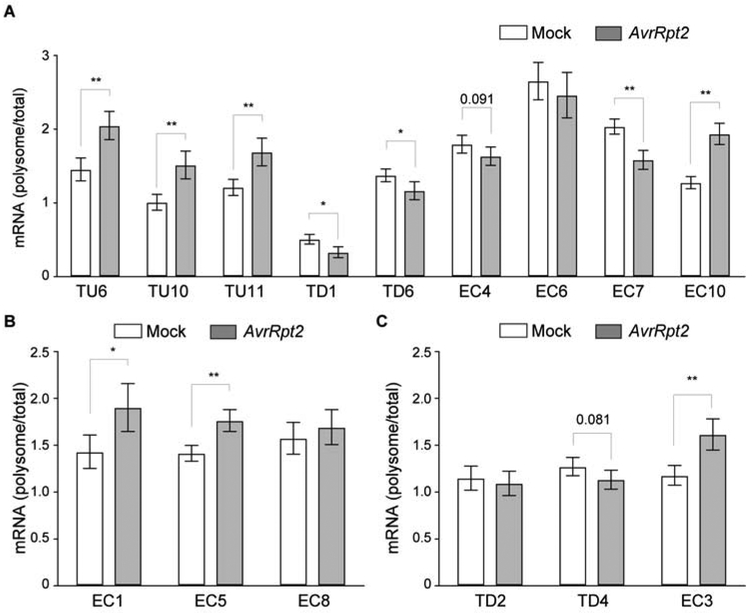
To further confirm that these ETI-regulated genes are indeed translationally regulated, we performed ribosome pelleting experiments. Consistent with our Ribo-seq result, we found that most of these candidate mRNAs displayed a consistent change in their association with the ribosome upon ETI induction, suggesting a contribution of translational regulation to their expression (Figure 3). Only three mRNAs, EC6, EC8, and TD2, did not recapitulate the Ribo-seq dynamics, probably due to differences in the methodology. These data indicate that even though translational induction is highly coordinated with transcriptional activation, translational regulation plays a significant role in regulating ETI. To dissect how much the transcriptional and translational components contribute to these individual candidates in defense, future study using mutants separating these processes will be required.
Figure 3. mRNA association with ribosomes upon ETI induction.
(A–C) Ribosomal associations of ETI-gene mRNAs were calculated as polysomal/total mRNA fractions with mock or ETI (AvrRpt2) induction. Expression levels were normalized against UBQ5. (A), (B), and (C) correspond to Figure 2C, 2D, and 2E, respectively. Data are mean ± SD; n = 3 three biological replicates. Data were combined using linear mixed effect model (lme4) with experiment as random effects; * P < 0.05 and ** P < 0.01 as determined by student's t-test.
See also Table S1.
Phenotypes of these ETI-deficient mutants confirmed the involvement of sphingolipids in ETI (Berkey et al., 2012; Sánchez-Rangel et al., 2015) and suggested possible roles for ABA perception and amino acid transporters in the immune response beyond basal resistance (Lim and Lee, 2015; Yang et al., 2014). Another striking result of the mutant analysis is the involvement of additional receptors including TIR-NBS-LRRs, a TIR-NBS, and a LRR in conferring AvrRpt2-induced resistance, which is known to be mediated by the coiled coil (CC)-NBS-LRR, RPS2. This result suggests that these additional proteins are likely “helper receptors” aiding in completion of RPS2 activation. Alternatively, these additional receptors may function downstream of RPS2 activation to establish full resistance (Bonardi et al., 2011). In contrast to the rps2 mutant, single knockout mutants of these receptors only partially compromised RPS2-mediated ETI (EC6, EC7, EC8, and EC10; Figures 2C and 2D). How these helper receptors contribute to RPS2-mediated ETI, and whether they interact with RPS2 or its cellular target RIN4 (Belkhadir et al., 2004; Day et al., 2005) will require further investigation.
Translational dynamics of metabolic pathway genes during ETI
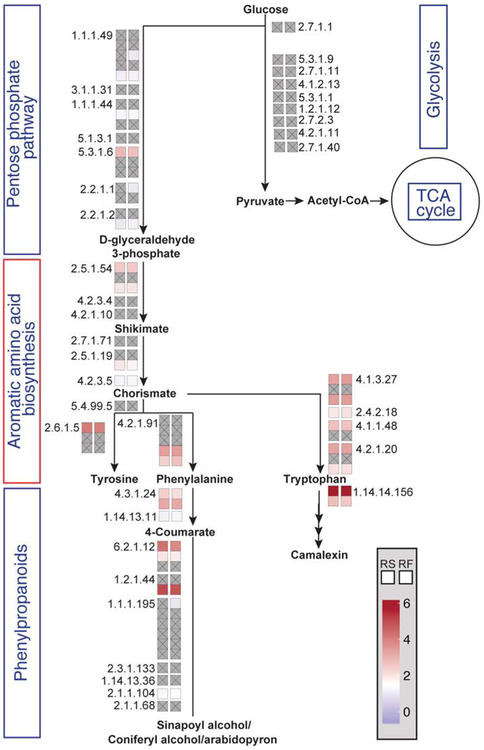
To broaden our investigation on the transcriptional and translational changes that occur during ETI, we performed Gene Ontology (GO) enrichment analysis on the genes differentially regulated in either RS or RF (Data S4) to highlight and compare the physiological pathways that were affected transcriptionally and/or translationally during ETI. In addition to the expected biotic stress response GO terms enriched in both RS and RF samples, we found GO terms associate with several metabolic pathways including “cellular aromatic compound metabolic process” and “indole-containing compound metabolism”. Enrichment of these terms suggests that metabolic pathways involved in aromatic amino acid biosynthesis may be regulated during ETI. To further study ETI-associated metabolic dynamics, we focused on RS and RF changes for the genes involved in metabolic pathways linked to the aromatic amino acid biosynthesis, including glycolysis, the pentose phosphate pathway, as well as the phenylpropanoid and camalexin biosynthesis pathways (Figure 4; Table S2). Interestingly, in our previous PTI analysis (Xu et al., 2017a), fewer genes in those pathways showed changes in RS and RF (Figure S4). To determine whether these changes are specific to aromatic amino acids or general to other amino acids as well, we analyzed RF changes for all the amino acid biosynthesis pathways. We found that although some fluxes were linked to certain amino acids including alanine, methionine, and glutamine, translational induction was mostly associated with enzymes involved in aromatic amino acid biosynthetic pathways (Figure S5; Table S3). These results suggest that induction of ETI may involve dynamic changes in plants’ primary metabolism. We next examined the “cellular aromatic compound metabolic process” GO-term enriched in RSup and RFup genes to identify possible differences in ETI-induced transcriptional and translational activities. We found that “phenylpropanoid metabolic process” is uniquely enriched in the RFup genes, but absent from the RSup genes (Figure S6), suggesting that translational up-regulation of this secondary metabolic pathway branched from the aromatic amino acid, phenylalanine, may be important for the induction of ETI.
Figure 4. Transcriptional and translational dynamics of specific metabolic pathways during ETI.
The schematic representation of metabolic pathways with each enzymatic step (EC number) was generated using MetaCyc Metabolic Pathway Database (Caspi et al., 2018). The fold changes are shown with colors for transcription (left box) and translation (right box). Gray box with inscribed x indicates no significant change detected.
See also Figures S4, S5, Data S4, Tables S2-S3.
Dynamics of amino acid metabolism during ETI
To test the hypothesis that plants actively control amino acid biosynthesis as a defense mechanism during ETI, we first performed amino acid profiling at 8 hpi and found that several amino acids had either an increase or a decrease in their levels during ETI (Figure 5A). Among the three aromatic amino acids, phenylalanine level was significantly increased while tyrosine and tryptophan levels were decreased. Interestingly, the reduction in tryptophan is contrary to the up-regulation of genes within the tryptophan biosynthetic pathway (Figure 4). The observed decrease in the tryptophan level may be due to a potential increase in the biosynthesis of the tryptophan-derived antimicrobial phytoalexin, camalexin (Lemarié et al., 2015), through the dramatic increases in both RS and RF (66 fold) of the key camalexin biosynthetic enzyme, tryptophan N-monooxygenase, CYP79B2 (EC 1.14.14.156; Figures 4 and S7A). Moreover, the CYP79B2 mRNA had a slight increase of ribosomal association upon ETI induction in the ribosome pelleting experiment, suggesting a potential contribution of translational regulation (Figure S7B). Indeed, the cyp79B2 mutant showed reduced PCD, indicating the importance of camalexin biosynthesis in this RPS2-mediated response (Figure S7C). The increase in phenylalanine is consistent with the observed increase in both RS and RF of its biosynthesis genes (Figure 4). It is also consistent with the translational up-regulation of genes within the phenylpropanoid pathway since phenylalanine is a precursor of phenylpropanoid biosynthesis, which has been associated with plant stress responses (Dixon and Paiva, 1995). The decreased tyrosine level during ETI might be associated with the phenylalanine increase, because tyrosine shares the upstream biosynthetic pathway with phenylalanine and is known to be the best amino donor for cytosolic phenylalanine biosynthesis (Yoo et al., 2013).
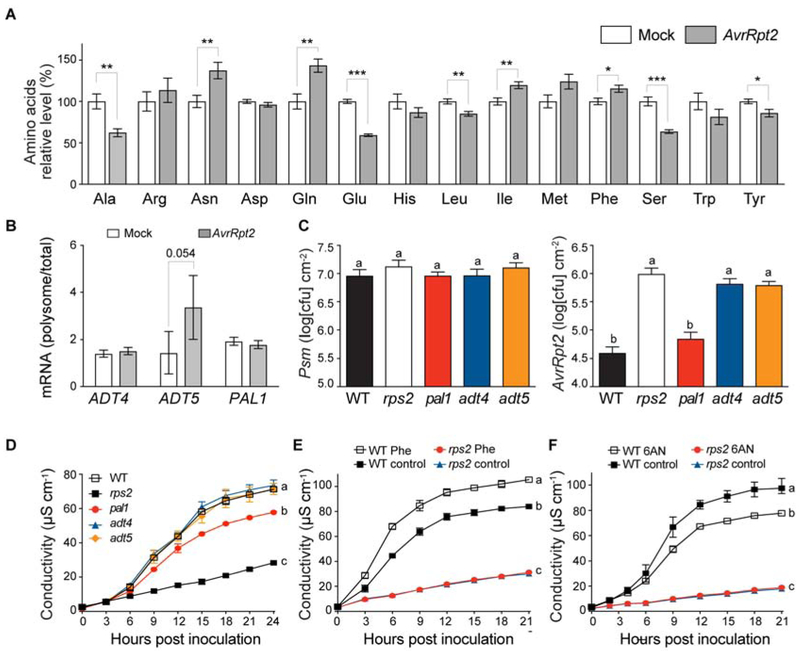
Figure 5. Phenylalanine and its derivatives are important for ETI response.
(A) Levels of amino acids during ETI. Data are mean ± SEM; n = 8 biological replicates from 2 experiments. * P < 0.05, ** P < 0.01, *** P < 0.001 as determined by student's t-test (Met P = 0.063). (B) Ribosomal associations of ADT4, ADT5, and PAL1 mRNAs were calculated as polysomal/total RNA fractions with mock or ETI (AvrRpt2) induction. Expression levels were normalized against UBQ5. Data are mean ± SD; n = 3 biological replicates. Data were combined using linear mixed effect model (lme4) with experiment as random effects and student's t-test was performed. (C and D) Growth of Psm ES4326 (left), and Psm ES4326/AvrRpt2 (right) (C), and ion leakage caused by Psm ES4326/AvrRpt2 (D) (OD600nm = 0.001 for bacterial growth and 0.01 for ion leakage). Data are mean ± SEM; n = 8 biological replicates for bacterial growth and n = 3 biological replicates for ion leakage analysis. (E) Ion leakage analysis with exogeneous phenylalanine (Phe) treatment (5 mM) in WT and rps2 plants. Data are mean ± SEM; n = 4 biological replicates. (F) Ion leakage analysis with exogeneous 6-aminonicotinamide (6AN) treatment (1.25 mM) in WT and rps2 plants. Data are mean ± SEM; n = 3 biological replicates. (C-F) Different letters indicate values that are significantly different based on Tukey’s test (P < 0.01; one-way ANOVA). For ion leakage analysis, the last time point was used for statistical analysis.
See also Figures S6 and S7.
Contribution of phenylalanine and phenylalanine-derived phenylpropanoid pathways to PCD and ETI
We next tested whether phenylalanine and its secondary metabolites in the phenylpropanoid pathway are important to confer ETI in plants. We examined genes in these metabolic pathways and found those encoding arogenate dehydratase 4 and 5 (ADT4 and ADT5, respectively) for phenylalanine synthesis (EC 4.2.1.91), and phenylalanine ammonia lyase 1 (PAL1) for conversion of phenylalanine to phenylpropanoid compounds were among the RF up-regulated genes (EC 4.3.1.24; Figure 4; Table S2). The ribosome pelleting experiment confirmed the pattern of ETI-induced association with the ribosome for the ADT5 mRNA (Figure 5B). The importance of PAL1 for plant immunity has been shown previously (Huang et al., 2010; Kim and Hwang, 2014), but its involvement in RPS2-mediated ETI is unknown.
To determine their roles in ETI, we obtained adt4, adt5, and pal1 mutants and compared bacterial growth and PCD in these mutants with WT. We observed more bacterial growth specific to Psm ES4326/AvrRpt2 that carries the effector in adt4 and adt5 than WT (Figure 5C), but with no significant difference in PCD (Figure 5D). In contrast, we observed a significant reduction in ETI-associated PCD in pal1 without a clear increase in Psm ES4326/AvrRpt2 growth (Figures 5C and 5D). These results support a positive role for phenylalanine and its secondary metabolites in ETI, both at the level of PCD regulation and at the level of immunity.
To eliminate possible direct effects of Phe or Phe derivatives on pathogen growth, we made use of lines carrying a dexamethasone (DEX)-inducible AvrRpt2 transgene in WT and rps2 backgrounds (Axtell et al., 2001). Induction of the bacterial effector AvrRpt2 expression in the plants by DEX-treatment can lead to PCD in an RPS2-dependent manner. We tested the effect of phenylalanine treatment 3 hours prior to DEX induction and observed an increase in the level of ion leakage in the phenylalanine-treated plants compared to the control (Figure 5E). To complement increasing the level of phenylalanine through exogenous application, we also looked for a way to reduce the level of phenylalanine. Since it has been previously suggested that the pentose phosphate pathway contributes to the biosynthesis of aromatic amino acids (e.g. phenylalanine) by producing the aromatic amino acid precursor erythrose 4-phosphate (E4P) (Khan, 2017), we hypothesized that inhibition of pentose phosphate pathway will lead to a decrease in phenylalanine synthesis, and thus reduced PCD. To test this hypothesis, we treated plants with 6-aminonicotinamide (6AN), an inhibitor of glucose-6-phosphate dehydrogenase (G6PD), the first enzyme of the pentose phosphate pathway, and found a significant reduction in AvrRpt2-mediated PCD (Figure 5F). Together, these results suggest that during ETI, both transcription and translation of metabolic enzymes contribute to increased levels of phenylalanine and its derivatives, which directly affect effector-triggered PCD and immunity.
DISCUSSION
Our study on global translational regulation during ETI establishes a link between immune response and metabolic dynamics. Global translational regulation during ETI is distinct from that during PTI, suggesting that plants operate complicated and dynamic translational mechanisms in response to specific immune stimuli. Unlike our PTI study (Xu et al., 2017a), a search for potential consensus in the 5′ leader sequences and 3′ UTRs did not yield any cis element in the mRNAs with ETI-mediated RF changes. This suggests that the coordinated translational change upon ETI may be accomplished through modifications of the translational machinery instead of specific sequences in the mRNAs. Alternatively, such mRNA cis-elements might be difficult to capture due to the small number of genes with large TE changes.
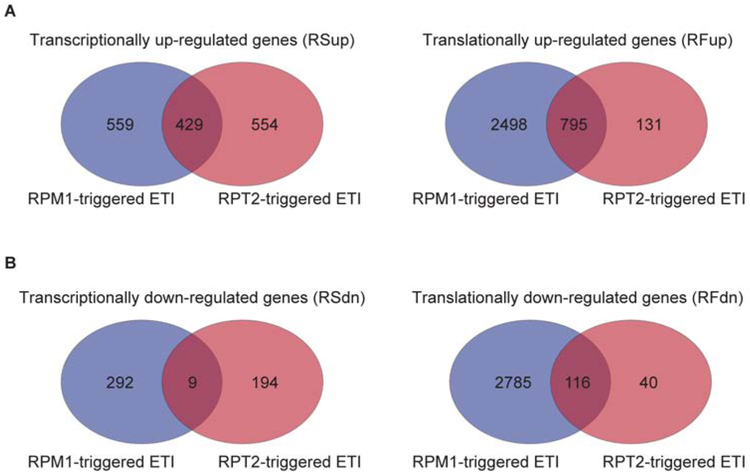
We compared the AvrRpt2 response to previously reported translational regulation in response to in planta expression of a different effector molecule, AvrRpm1 (Meteignier et al., 2017) to determine whether the translational effect observed during ETI is specific for the individual effector molecule. We found that 50% of up- and 5% of down-regulated genes from the RPS2-mediated transcriptional response overlapped with the RPM1-mediated transcriptional response, while 80% up and 75% down regulated genes overlapped in the translational response (Figure 6; Data S5). This result suggests that although AvrRpt2 and AvrRpm1 are recognized by different NBS-LRRs, there is not only a significant overlap in the downstream transcriptional events as previously reported (Mine et al., 2018), but translational activities as well. Recognition of AvrRpm1 by RPM1 also induced genes in metabolic pathways including “indole-containing compound metabolic process” and “aromatic amino acid family metabolic process” (Meteignier et al., 2017), suggesting that translational changes in metabolic pathways during ETI are likely a general pattern. Furthermore, eight of the eleven TIR-NBS domain-containing genes and two of the three LRR genes in the RPS2-mediated response also showed significant changes in association with ribosomes after DEX-induced AvrRpm1 expression (Meteignier et al., 2017) (Data S5). This result suggests that beyond initial recognition of AvrRpm1 and AvrRpt2 by their cognate receptor RPM1 and RPS2, respectively, other helper receptors likely contribute to core ETI signaling. Future experiments are needed to test whether any of the commonly identified translationally regulated genes are also required for ETI mediated by RPM1.
Figure 6. Venn diagrams of transcriptional and translational responses from the AvrRpm1 and AvrRpt2 datasets.
(A) Venn diagrams showing numbers of overlapping and non-overlapping transcriptionally up-regulated genes (RSup) and translationally up-regulated genes (RFup) between the AvrRpm1 (blue) and AvrRpt2 (red) datasets. (B) Venn diagrams showing numbers of overlapping and non-overlapping transcriptionally down-regulated genes (RSdn) and translationally down-regulated genes (RFdn) between the AvrRpm1 and AvrRpt2 datasets. RS, RNA-seq; RF, ribosomal footprinting; fc, fold change; up, up-regulated; dn, down-regulated.
See also Data S5.
Accumulating evidence indicates that amino acid metabolism plays an active role in plant immunity. Exogenous application of proline induced hypersensitive response-like cell death in Arabidopsis (Deuschle et al., 2004), whereas threonine has been shown to directly inhibit the growth of Hyaloperonospora arabidopsidis (Stuttmann et al., 2011). Also, lysine-derived pipecolic acid has been reported as a key regulator of systemic acquired resistance (Hartmann et al., 2018; Návarová et al., 2012). Our study not only discovered that phenylalanine is important for ETI, but also establishes that translational regulation is a key regulatory mechanism of phenylalanine biosynthetic pathways during ETI. Furthermore, through our global translatome analysis, we discovered novel regulators and metabolic pathways involved in RPS2-mediated ETI. While these metabolic pathways have been suggested to play a role in plant defense by many studies based on cDNA array, RNA-seq, and/or metabolic profiling (Aliferis et al., 2014; Misra et al., 2016; Scheideler et al., 2002; Schwachtje et al., 2018; Ward et al., 2010), our discovery provides a strong piece of evidence that plants actively control biosynthetic pathways for phenylalanine and phenylalanine-derived compounds at both transcriptional and translational levels to activate the ETI defense program.
METHODS
Plant materials and growth conditions
Plants were grown on soil (Metro Mix 360) at 22 °C under a 12/12-h light/dark cycle with 55% humidity. The Arabidopsis transgenic lines carrying the 35S:uORFsTBF1-LUC construct in WT background was previously described (Xu et al., 2017a) and in rps2 background was generated by genetic crossing. adt4 (SALK_065483), adt5 (SALK_088171), pal1 (SALK_000357) and cyp79B2 (SALK_113348) and other T-DNA insertion mutants used in this study were obtained from the Arabidopsis Biological Resource Center (Data S3 and Data S6).
LUC activity measurement
To record the 35S:uORFsTBF1-LUC reporter activity, 4-week-old transgenic plants were sprayed with 1 mM luciferin (Gold Biotechnology) 12 h before infiltration with either Psm ES4326/AvrRpt2 (OD600nm = 0.02) or 10 mM MgCl2. Luciferase activity was recorded in a CCD camera-equipped box (Nightshade Company) with each exposure time of 20 min.
Ribosome pelleting and Ribo-seq library construction
Ribo-seq libraries were constructed as previously described (Ingolia et al, 2012; Xu et al., 2017a). Briefly, leaves (0.3 g) from 4-week-old transgenic plants were collected, ground in liquid nitrogen, and resuspended in 2 ml of pre-chilled polysome extraction buffer (PEB). Ribosomes were purified using a sucrose cushion after ultra-centrifugation at 4 °C (70,000 rpm for 4 h). For the ribosome pelleting experiment, the samples were washed twice with cold water and resuspended in TRIzol (Ambion) to extract ribosome-associated mRNA. For Ribo-seq library construction, the pellet was washed twice with cold water and resuspended in 300 μl of RNase I digestion buffer. The supernatant was then transferred to new tubes and incubated with 10 μl of RNase I (100 U μl−1) at 25 °C for 60 min. Then, 15 μl of SUPERase-In (20 U μl−1) was added to halt the reaction. Subsequent steps of library construction were performed as previously reported (Figure 1A; Ingolia et al, 2012; Xu et al., 2017a).
RNA-seq library construction
RNA-seq library was constructed as previously described (Xu et al., 2017a). Briefly, 0.75 ml TRIzol® LS (Ambion) was added to the 0.25 ml lysate saved from the Ribo-seq library construction. Total RNA was extracted, quantified and qualified using Nanodrop (Thermo Fisher Scientific Inc). 50-75 μg of total RNA was used for mRNA purification with Dynabeads® Oligo (dT)25 (Invitrogen). 20 μl of the purified poly (A) mRNA was mixed with 20 μl of 2× fragmentation buffer (2 mM EDTA, 10 mM Na2CO3, 90 mM NaHCO3) and incubated at 95 °C for 40 min before cooling on ice. 500 μl of cold water, 1.5 μl of GlycoBlue, and 60 μl of cold 3 M sodium acetate were then added to the samples and mixed. Subsequently, 600 μl of isopropanol was added and incubated at −80 °C for at least 30 min to precipitate RNA. Samples were then centrifuged at 4°C (12,000 rpm for 30 min) to remove all liquid and air-dried for 10 min before resuspension in 5 μl of 10 mM Tris (pH 8.0). Remaining steps were performed equivalently to the Ribo-seq library preparation.
Quality check of Ribo-seq and RNA-seq
Library quality was validated with bioanalyzer profile, which detected the expected peak at ~176 bp. In the RF samples, we detected a peak length distribution at 30 nt, consistent with the expected length protected by a ribosome, but no specific peak in the RS data (Figures S1A and S2A). RF coverage was the highest at the translation initiation codon and showed a preference for reads in the first frame (Figures S1B, S1C, S2B, and S2C). Detected coverage of ribosome footprints in 5’ leader sequence and 3’UTR is affected by digestion buffer conditions (Hsu et al., 2016). Reads in 5’ leader sequence and 3’UTR could occur under buffer conditions allowing the capture of ribosomes in different conformations, in mature plants, and in response stress. Moreover, there is a possibility that other RNA-binding proteins interacting with 5’ leader sequences and 3’UTRs under certain circumstance as described previously (Dunn et al., 2013; Miettinen and Björklund, 2015). Hierarchical clustering and Pearson correlation of expression showed strong reproducibility among replicates with the same treatment for each genotype (Figures S1D, S1E, S2D, and S2E), and the range of RFfc and RSfc appeared to be similar (Figures S1F and S2F). Strong correlations between mRNA abundance (RS) and ribosome association (RF) were detected within the same treatment (i.e., Mock or AvrRpt2) for WT (Figure S1G) and rps2 plants (Figure S2G).
Polysome profiling
Polysome profiling was performed as previously reported (Xu et al., 2017a). Briefly, 0.5 g of 4-week-old leaves were ground in liquid nitrogen and resuspended in 2 ml of pre-chilled PEB buffer. 1 ml of crude lysate was loaded to a sucrose gradient (15-60%) and centrifuged at 4 °C (35,000 rpm for 10 h). Polysome-associated mRNA was isolated through fractionation of the sucrose gradient.
Psm ES4326/AvrRpt2 bacterial growth and ion leakage measurement
For bacterial growth assays, 4-week-old plants were infiltrated with Psm ES4326/AvrRpt2 or Psm ES4326 (OD600nm = 0.001). Infected and mock-infiltrated leaves were collected 3 days post infiltration. Leaf discs were ground in solution containing 10 mM MgCl2 and plated in dilution series onto plates containing King’s B medium (KB) supplemented with 100 μg ml−1 streptomycin and 10 μg ml−1 tetracycline for Psm ES4326/AvrRpt2 or KB with 100 μg ml−1 streptomycin for Psm ES4326. Bacterial growth was scored 3 days after plating. For ion leakage measurement, plants were infiltrated with Psm ES4326/AvrRpt2 (OD600nm = 0.01). After 1 h, leaf discs were collected from infiltrated leaves and ion leakage was measured every 3 h for 24 h using the conductivity meter (Thermo Scientific).
Chemical applications
1.25 mM 6-aminonicotinamide (6AN) (Sigma-Aldrich) dissolved in 1.25% dimethyl sulfoxide (DMSO) or mock (1.25 % DMSO only) was sprayed one day prior to 50 μM DEX induction. 5 mM L-phenylalanine (Sigma-Aldrich) or mock (water only) was sprayed 3 h prior to DEX spray. Leaf discs were collected from infiltrated leaves 1 h after DEX induction and ion leakage was measured every 3 h for 21-24 h.
Quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from approximately 50 mg of leaf tissue based on manufacturer's instruction using TRIzol (Ambion). After DNase I (Ambion) treatment, cDNA synthesis was performed by the instruction of Superscript® III Reverse Transcriptase (Invitrogen) using oligo (dT). qRT-PCR was performed with FastStart Universal SYBR Green Master (Roche). For polysome profiling, mRNA was extracted from each fraction while total mRNA was extracted from the crude lysate. Expression level in each fraction was normalized to total mRNA abundance.
Amino acid profiling and high performance liquid chromatography (HPLC) analysis
Approximately 0.2 g of leaves from 4-week-old plants were collected, ground in liquid nitrogen, and resuspended in 0.8 ml of extraction buffer containing 2-amino-2-methyl-1-propanol hydrochloride (Sigma) in 75% ethanol (pH was adjusted to pH 10 by NaOH). As an internal standard, 60 nmol aminoadipic acid (Sigma-Aldrich) was added into the extraction buffer. The extract was centrifuged at 4°C (12,000 rpm for 30 min). Supernatant was collected and filtered through a 10 kDa MWCO Amicon column (EMD-Millipore) by centrifugation at 4°C (3,500 rpm for 90 min). The filtered solution was dried with SpeedVac (45°C for 3 h; Eppendorf), and the pellet was stored at −20°C for 30 min and dissolved in 50 μl of water. 10 μl of the final reaction mixture was derivatized with o-phthalaldehyde (Agilent) and analyzed on the Agilent 1100 HPLC system using the ZORBAX Eclipse AAA column (3.5 μm, 3.0 × 140 mm; Agilent) at a flow rate of 0.45 ml min−1 with a 40 min linear gradient of 0 to 30 % methanol and acetone in 15 mM ammonium acetate buffer, pH 7.8.
Bioinformatic and statistical analyses
Bioinformatic and statistical analyses were performed as previously described (Xu et al., 2017a). Briefly, Bowtie2 was used to align reads to the Arabidopsis TAIR10 genome (Langmead and Salzberg, 2012). Read assignment to genes was achieved using HT-seq (Anders et al., 2015) for transcriptome and translatome libraries by exon and CDS, respectively. Transcriptome and translatome fold changes (i.e., RSfc and RFfc) were calculated using DESeq2 (Love et al., 2014). TE was calculated by combining reads for all genes that passed RPKM ≥ 1 in CDS threshold in all biological replicates and normalizing Ribo-seq RPKM to RNA-seq RPKM as reported. Density plot was presented using IGB (Nicol et al., 2009). Statistical analysis was done by one-way ANOVA with Tukey’s Honest Significant Difference test for Figure 1B, Figures 2C-2E, Figure 5C-5F, and Figure S7C. Statistical analysis was done by student's t-test for Figure 5A. GraphPad Prism 6 was used for all the statistical analyses. For qPCR data (Figures 1B and 1D), n indicates technical repeats, which were verified by two or three independent experiments. All other replicates indicate biological replicates.
The RS and RF data presented in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO: GSE124115). During review process, sequencing data can be assessed through this link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124115
Supplementary Material
Data S1. RSfc, RFfc, and TEfc by AvrRpt2 treatment in WT.
Data S2. RSfc and RFfc by AvrRpt2 treatment in rps2.
Data S3. List of candidate genes analyzed for ETI phenotypes.
Data S4. Gene Ontology (GO) terms for RS and RF-altered genes by AvrRpt2 treatment.
Data S5. List of RSfc and RFfc genes from the AvrRpm1 and AvrRpt2 datasets.
Data S6. ABRC mutant stocks and primers used in this study.
Figure S1. Related Figure 1. Quality of the WT RNA-seq and Ribo-seq libraries. (A) Length distribution of total reads from RS and RF libraries in WT plants. (B) Nucleotide resolution of the read density surrounding translation initiation and translation termination sites using the 15th nucleotide of 30 nt reads of RF. Genes with RPKM ≥ 1 and the length of 5’ LS ≥ 1 nt, CDS ≥ 30 nt, and 3’ UTR ≥ 1 nt were analyzed. 5’ LS, 5’ leader sequence; CDS, coding sequence; 3’ UTR, 3’ untranslated region. (C) Ribosome occupancy in frame of coding regions from RF libraries. (D) Hierarchical clustering showing the reproducibility between RS (left) and RF (right) within three replicates. Darker color means greater correlation. (E) Correlations between triplicates (Rep1/2/3) of RS and RF samples. Data are shown as the correlations of log2(RPKM) in CDS for expressed genes with RPKM in CDS ≥ 1. (F) Histogram of transcriptional (log2(RSfc)) and translational (log2(RFfc)) responses to Psm ES4326/AvrRpt2 in WT. RS, RNA-seq; RF, ribosomal footprinting; fc, fold change; μ, mean; δ, standard deviation. (G) Correlation between RS and RF in the same sample. r, Pearson correlation coefficient. (H) Dispersion analysis between RS and RF fold changes. Relationships between RSfc and RFfc, which pass RPKM > 1 with highlighted genes above 90th percentile dispersion between RSfc and RFfc. Selected candidates are highlighted in black.
Figure S2. Related Figure 1. Quality of the rps2 RNA-seq and Ribo-seq libraries. (A) Length distribution of total reads from RS and RF libraries in rps2 mutant plants. (B) Nucleotide resolution of the read density surrounding translation initiation and translation termination sites using the 15th nucleotide of 30 nt reads of RF. Genes with RPKM ≥ 1 and the length of 5’ LS ≥ 1 nt, CDS ≥ 30 nt, and 3’ UTR ≥ 1 nt were analyzed. 5’ LS, 5’ leader sequence; CDS, coding sequence; 3’ UTR, 3’ untranslated region. (C) Ribosome occupancy in frame of coding regions from RF libraries. (D) Hierarchical clustering showing the reproducibility between RS (left) and RF (right) within two replicates. Darker color means greater correlation. (E) Correlations between duplicates (Rep1/2) of RS and RF samples. Data are shown as the correlations of log2(RPKM) in CDS for expressed genes with RPKM in CDS ≥ 1. r, Pearson correlation coefficient. (F) Histogram of transcriptional (log2(RSfc)) and translational (log2(RFfc)) responses to Psm ES4326/AvrRpt2 in rps2. RS, RNA-seq; RF, ribosomal footprinting; fc, fold change; μ, mean; δ, standard deviation. (G) Correlation between RS and RF in the same sample. r, Pearson correlation coefficient. (H) Relationship between RSfc and RFfc in rps2. dn, down; nc, no change.
Figure S3. Related Figure 2. Normalized distribution of RNA-seq and Ribo-seq reads for TBF1, and representative TEup and TEdn candidates. (A and B) Read maps from the RNA-seq (RS) and Ribo-seq (RF) data for the endogenous TBF1 mRNA in WT and rps2 (A), TEup (AT1G77380) and TEdn (AT5G20380) genes in WT (B). Read coverage was normalized to uniquely mapped reads with the Integrated Genome Browser (IGB) (Nicol et al., 2009).
Figure S4. Related to Figure 4. Transcriptional and translational dynamics of specific metabolic pathways during PTI and ETI. Expression dynamics observed in the pentose phosphate pathway, phenylalanine, and phenylpropanoid biosynthesis during PTI and ETI. Pie charts depict overall dynamics of all genes in selected KEGG pathways. Genes showing significant difference during PTI were selected and compared with those during ETI. For AT4G39950, Log2 Fold Change is presented in the figure.
Figure S5. Related Figure 4. Dynamic RF changes in amino acid biosynthetic pathways. Enzymes with RFup are highlighted with red arrows and described in numbered order in Table S3.
Figure S6. Related Figure 5. GO-term enrichment analysis for transcriptionally or translationally up-regulated and down-regulated genes. (A and B) Virtual Plant program was used to identify GO-term enrichments of transcriptionally up-regulated (A) and translationally up-regulated (B) genes in response to Psm ES4326/AvrRpt2. (C and D) Selected GO-term enrichments (highlighted in rectangles in A and B) for RSup (C) and RFup (D) genes.
Figure S7. Related Figure 5. Dynamic transcriptional and translational changes in the camalexin biosynthesis pathway during ETI. (A) Schematic representation of the camalexin biosynthetic pathway showing each enzymatic step. The fold changes are shown with colors for transcription (left box) and translation (right box). In these biosynthetic pathways, genes encoding enzymes responsible for first three steps were shown as up-regulated genes from the RS or RF dataset. (B) Ribosomal association of the CYP79B2 mRNA was calculated as the polysomal/total RNA fraction in response to mock or ETI induction. Expression levels were normalized against UBQ5. Data were combined using linear mixed effect model (lme4) with experiment as random effects; Student's t-test was performed. (C) Growth of Psm ES4326 (left) or Psm ES4326/AvrRpt2 (middle), and ion leakage analysis (right) caused by Psm ES4326/AvrRpt2 (OD600nm = 0.001 for bacterial growth and 0.01 for ion leakage) in WT, rps2, and cyp79B2 plants. Different letters indicate values that are significantly different based on Tukey’s test (P < 0.01; one-way ANOVA). For ion leakage analysis, the last time point was used for statistical analysis. Data are mean ± SEM; n = 8 biological replicates for bacterial growth and n = 3 biological replicates for ion leakage analysis.
Table S1. List of candidate genes showing RPS2-mediated ETI phenotypes.
Table S2. List of genes in metabolic pathways in Figure 4.
Table S3. List of RFup genes in amino acid metabolic pathways.
ACKNOWLEDGMENTS
We thank George Dubay for assisting amino acid analysis, Jenny Shen for helping with MetaCyc analysis, Musoki Mwimba for linear modeling of ribosome pelleting experiment, and Paul J. Zwack, Sophia G. Zebell, and Dong lab members for critical comments on this project. This study was supported by grants from NIH R35GM118036-02, NSF IOS 1645589 and HHMI-GBMF (through Grant GBMF3032) to X. Dong and Hargitt fellowship to H.Y. No conflict of interest is declared.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aboul-Soud MA, Chen X, Kang JG, Yun BW, Raja MU, Malik SI, and Loake GJ (2009). Activation tagging of ADR2 conveys a spreading lesion phenotype and resistance to biotrophic pathogens. New Phytol. 183, 1163–1175. [DOI] [PubMed] [Google Scholar]
- Aliferis KA, Faubert D, and Jabaji S (2014). A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS One 9, e111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, and Koonin EV (1999). G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci 24, 342–344. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, McNellis TW, Mudgett MB, Hsu CS, and Staskawicz BJ (2001). Mutational analysis of the Arabidopsis RPS2 disease resistance gene and the corresponding Pseudomonas syringae avrRpt2 avirulence gene. Mol. Plant Microbe. Interact 14, 181–188. [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, and Dangl JL (2004). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16, 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkey R, Bendigeri D, and Xiao S (2012). Sphingolipids and plant defense/disease: the "death" connection and beyond. Front. Plant Sci 3, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, and Dangl JL (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. U.S.A 108, 16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Billington R, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Midford PE, Ong Q, Ong WK, et al. (2018). The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 46, D633–D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, and Epple P (2010). Arabidopsis type I metacaspases control cell death. Science 330, 1393–1397. [DOI] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Huang J, Chisholm ST, Li D, and Staskawicz BJ (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17, 1292–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle K, Funck D, Forlani G, Stransky H, Biehl A, Leister D, van der Graaff E, Kunze R, and Frommer WB (2004). The role of Δ1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell 16, 3413–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, and Paiva NL (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, and Weissman JS (2013). Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife 2, e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, and Dangl JL (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Gu Y, Zebell SG, Liang Z, Wang S, Kang BH, and Dong X (2016). Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell 166, 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M, Zeier T, Bernsdorff F, Reichel-Deland V, Kim D, Hohmann M, Scholten N, Schuck S, Bräutigam A, Hölzel T, et al. (2018). Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173, 456–469. [DOI] [PubMed] [Google Scholar]
- Hsu PY, Calviello L, Wu HL, Li FW, Rothfels CJ, Ohler U, and Benfey PN (2016). Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 113, E7126–E7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, and Chen Z (2010). Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153, 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, and Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, and Bailey-Serres J (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 111, E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, et al. (2010). VirtualPlant: a software platform to support systems biology research. Plant Physiol. 152, 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AS (2017). Flowering plants: Structure and industrial products. (New Jersey: John Willey & Sons; ) [Google Scholar]
- Kim DS, and Hwang BK (2014). An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot 65, 2295–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Shi J, Chen J, Zhang M, Sun S, Xie S, Li X, Zeng B, Peng L, Hauck A, et al. (2015). Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 84, 1206–1218. [DOI] [PubMed] [Google Scholar]
- Lemarié S, Robert-Seilaniantz A, Lariagon C, Lemoine J, Marnet N, Levrel A, Jubault M, Manzanares-Dauleux MJ, and Gravot A (2015). Camalexin contributes to the partial resistance of Arabidopsis thaliana to the biotrophic soilborne protist Plasmodiophora brassicae. Front. Plant Sci 6, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CW, and Lee SC (2015). Arabidopsis abscisic acid receptors play an important role in disease resistance. Plant Mol. Biol 88, 313–324. [DOI] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Wu JF, Lin WD, Wu YC, Tsai TY, Tsai HL, and Wu SH (2013). Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25, 3699–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Brumos J, Yun J, Hu Q, Spencer KR, Enríquez P, Binder BM, Heber S, Stepanova AN, and Alonso JM (2015). Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163, 684–697. [DOI] [PubMed] [Google Scholar]
- Meteignier LV, El Oirdi M, Cohen M, Barff T, Matteau D, Lucier JF, Rodrigue S, Jacques PE, Yoshioka K, and Moffett P (2017). Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in Arabidopsis. J. Exp. Bot 68, 2333–2344. [DOI] [PubMed] [Google Scholar]
- Miettinen TP, and Björklund M (2015). Modified ribosome profiling reveals high abundance of ribosome protected mRNA fragments derived from 3' untranslated regions. Nucleic Acids Res. 43, 1019–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, and Ausubel FM (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Mine A, Seyfferth C, Kracher B, Berens ML, Becker D, and Tsuda K (2018). The defense phytohormone signaling network enables rapid, high-amplitude transcriptional reprogramming during effector-triggered immunity. Plant Cell 30, 1199–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra BB, de Armas E, and Chen S (2016). Differential metabolomic responses of PAMP-triggered immunity and effector-triggered immunity in Arabidopsis suspension cells. Metabolomics 12, 61. [Google Scholar]
- Návarová H, Bernsdorff F, Döring AC, and Zeier J (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24, 5123–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandety RS, Caplan JL, Cavanaugh K, Perroud B, Wroblewski T, Michelmore RW, and Meyers BC (2013). The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 162, 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG, Raja A, and Loraine AE (2009). The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25, 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan M, Cournoyer P, and Dinesh-Kumar SP (2009). The leucine-rich repeat domain in plant innate immunity: a wealth of possibilities. Cell Microbiol. 11, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rangel D, Rivas-San Vicente M, de la Torre-Hernández ME, Nájera-Martínez M, and Plasencia J (2015). Deciphering the link between salicylic acid signaling and sphingolipid metabolism. Front. Plant Sci 6, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheideler M, Schlaich NL, Fellenberg K, Beissbarth T, Hauser NC, Vingron M, Slusarenko AJ, and Hoheisel JD (2002). Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J. Biol. Chem 277, 10555–10561. [DOI] [PubMed] [Google Scholar]
- Schwachtje J, Fischer A, Erban A, and Kopka J (2018). Primed primary metabolism in systemic leaves: a functional systems analysis. Sci. Rep 8, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuttmann J, Hubberten HM, Rietz S, Kaur J, Muskett P, Guerois R, Bednarek P, Hoefgen R and Parker JE (2011). Perturbation of Arabidopsis amino acid metabolism causes incompatibility with the adapted biotrophic pathogen Hyaloperonospora arabidopsidis. Plant Cell 23, 2788–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski MR, Birker D, and Jones JD (2009). The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant Microbe. Interact 22, 157–165. [DOI] [PubMed] [Google Scholar]
- Ward JL, Forcat S, Beckmann M, Bennet M, Miller SJ, Baker JM, Hawkins ND, Vermeer CP, Lu C, Lin W, et al. (2010). The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 63, 443–457. [DOI] [PubMed] [Google Scholar]
- Xu G, Greene GH, Yoo H, Liu L, Marqués J, Motley J, and Dong X (2017a). Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 25, 487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Yuan M, Ai C, Liu L, Zhuang E, Karapetyan S, Wang S, and Dong X (2017b). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yángüez E, Castro-Sanz AB, Fernández-Bautista N, Oliveros JC, and Castellano MM (2013). Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. PLoS One 8, e71425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Postel S, Kemmerling B, and Ludewig U (2014). Altered growth and improved resistance of Arabidopsis against Pseudomonas syringae by overexpression of the basic amino acid transporter AtCAT1. Plant Cell Environ. 37, 1404–1414. [DOI] [PubMed] [Google Scholar]
- Yoo H, Widhalm JR, Qian Y, Maeda H, Cooper BR, Jannasch AS, Gonda I, Lewinsohn E, Rhodes D, and Dudareva N (2013). An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine: Phenylpyruvate aminotransferase. Nat. Commun 4, 2833. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang Y, Fang B, Gannon P, Ding P, Li X, and Zhang Y (2010). Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRKY70. Plant Cell 22, 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. RSfc, RFfc, and TEfc by AvrRpt2 treatment in WT.
Data S2. RSfc and RFfc by AvrRpt2 treatment in rps2.
Data S3. List of candidate genes analyzed for ETI phenotypes.
Data S4. Gene Ontology (GO) terms for RS and RF-altered genes by AvrRpt2 treatment.
Data S5. List of RSfc and RFfc genes from the AvrRpm1 and AvrRpt2 datasets.
Data S6. ABRC mutant stocks and primers used in this study.
Figure S1. Related Figure 1. Quality of the WT RNA-seq and Ribo-seq libraries. (A) Length distribution of total reads from RS and RF libraries in WT plants. (B) Nucleotide resolution of the read density surrounding translation initiation and translation termination sites using the 15th nucleotide of 30 nt reads of RF. Genes with RPKM ≥ 1 and the length of 5’ LS ≥ 1 nt, CDS ≥ 30 nt, and 3’ UTR ≥ 1 nt were analyzed. 5’ LS, 5’ leader sequence; CDS, coding sequence; 3’ UTR, 3’ untranslated region. (C) Ribosome occupancy in frame of coding regions from RF libraries. (D) Hierarchical clustering showing the reproducibility between RS (left) and RF (right) within three replicates. Darker color means greater correlation. (E) Correlations between triplicates (Rep1/2/3) of RS and RF samples. Data are shown as the correlations of log2(RPKM) in CDS for expressed genes with RPKM in CDS ≥ 1. (F) Histogram of transcriptional (log2(RSfc)) and translational (log2(RFfc)) responses to Psm ES4326/AvrRpt2 in WT. RS, RNA-seq; RF, ribosomal footprinting; fc, fold change; μ, mean; δ, standard deviation. (G) Correlation between RS and RF in the same sample. r, Pearson correlation coefficient. (H) Dispersion analysis between RS and RF fold changes. Relationships between RSfc and RFfc, which pass RPKM > 1 with highlighted genes above 90th percentile dispersion between RSfc and RFfc. Selected candidates are highlighted in black.
Figure S2. Related Figure 1. Quality of the rps2 RNA-seq and Ribo-seq libraries. (A) Length distribution of total reads from RS and RF libraries in rps2 mutant plants. (B) Nucleotide resolution of the read density surrounding translation initiation and translation termination sites using the 15th nucleotide of 30 nt reads of RF. Genes with RPKM ≥ 1 and the length of 5’ LS ≥ 1 nt, CDS ≥ 30 nt, and 3’ UTR ≥ 1 nt were analyzed. 5’ LS, 5’ leader sequence; CDS, coding sequence; 3’ UTR, 3’ untranslated region. (C) Ribosome occupancy in frame of coding regions from RF libraries. (D) Hierarchical clustering showing the reproducibility between RS (left) and RF (right) within two replicates. Darker color means greater correlation. (E) Correlations between duplicates (Rep1/2) of RS and RF samples. Data are shown as the correlations of log2(RPKM) in CDS for expressed genes with RPKM in CDS ≥ 1. r, Pearson correlation coefficient. (F) Histogram of transcriptional (log2(RSfc)) and translational (log2(RFfc)) responses to Psm ES4326/AvrRpt2 in rps2. RS, RNA-seq; RF, ribosomal footprinting; fc, fold change; μ, mean; δ, standard deviation. (G) Correlation between RS and RF in the same sample. r, Pearson correlation coefficient. (H) Relationship between RSfc and RFfc in rps2. dn, down; nc, no change.
Figure S3. Related Figure 2. Normalized distribution of RNA-seq and Ribo-seq reads for TBF1, and representative TEup and TEdn candidates. (A and B) Read maps from the RNA-seq (RS) and Ribo-seq (RF) data for the endogenous TBF1 mRNA in WT and rps2 (A), TEup (AT1G77380) and TEdn (AT5G20380) genes in WT (B). Read coverage was normalized to uniquely mapped reads with the Integrated Genome Browser (IGB) (Nicol et al., 2009).
Figure S4. Related to Figure 4. Transcriptional and translational dynamics of specific metabolic pathways during PTI and ETI. Expression dynamics observed in the pentose phosphate pathway, phenylalanine, and phenylpropanoid biosynthesis during PTI and ETI. Pie charts depict overall dynamics of all genes in selected KEGG pathways. Genes showing significant difference during PTI were selected and compared with those during ETI. For AT4G39950, Log2 Fold Change is presented in the figure.
Figure S5. Related Figure 4. Dynamic RF changes in amino acid biosynthetic pathways. Enzymes with RFup are highlighted with red arrows and described in numbered order in Table S3.
Figure S6. Related Figure 5. GO-term enrichment analysis for transcriptionally or translationally up-regulated and down-regulated genes. (A and B) Virtual Plant program was used to identify GO-term enrichments of transcriptionally up-regulated (A) and translationally up-regulated (B) genes in response to Psm ES4326/AvrRpt2. (C and D) Selected GO-term enrichments (highlighted in rectangles in A and B) for RSup (C) and RFup (D) genes.
Figure S7. Related Figure 5. Dynamic transcriptional and translational changes in the camalexin biosynthesis pathway during ETI. (A) Schematic representation of the camalexin biosynthetic pathway showing each enzymatic step. The fold changes are shown with colors for transcription (left box) and translation (right box). In these biosynthetic pathways, genes encoding enzymes responsible for first three steps were shown as up-regulated genes from the RS or RF dataset. (B) Ribosomal association of the CYP79B2 mRNA was calculated as the polysomal/total RNA fraction in response to mock or ETI induction. Expression levels were normalized against UBQ5. Data were combined using linear mixed effect model (lme4) with experiment as random effects; Student's t-test was performed. (C) Growth of Psm ES4326 (left) or Psm ES4326/AvrRpt2 (middle), and ion leakage analysis (right) caused by Psm ES4326/AvrRpt2 (OD600nm = 0.001 for bacterial growth and 0.01 for ion leakage) in WT, rps2, and cyp79B2 plants. Different letters indicate values that are significantly different based on Tukey’s test (P < 0.01; one-way ANOVA). For ion leakage analysis, the last time point was used for statistical analysis. Data are mean ± SEM; n = 8 biological replicates for bacterial growth and n = 3 biological replicates for ion leakage analysis.
Table S1. List of candidate genes showing RPS2-mediated ETI phenotypes.
Table S2. List of genes in metabolic pathways in Figure 4.
Table S3. List of RFup genes in amino acid metabolic pathways.