Abstract
Circulating rare cells in the blood are of great significance for both materials research and clinical applications. For example, circulating tumor cells (CTCs) have been demonstrated as useful biomarkers for “liquid biopsy” of the tumor. Circulating fetal nucleated cells (CFNCs) have shown potential in noninvasive prenatal diagnostics. However, it is technically challenging to detect and isolate circulating rare cells due to their extremely low abundance compared to hematologic cells. Nanostructured substrates offer a unique solution to address these challenges by providing local topographic interactions to strengthen cell adhesion and large surface areas for grafting capture agents, resulting in improved cell capture efficiency, purity, sensitivity, and reproducibility. In addition, rare-cell retrieval strategies, including stimulus-responsiveness and additive reagent-triggered release on different nanostructured substrates, allow for on-demand retrieval of the captured CTCs/CFNCs with high cell viability and molecular integrity. Several nanostructured substrate-enabled CTC/CFNC assays are observed maturing from enumeration and subclassification to molecular analyses. These can one day become powerful tools in disease diagnosis, prognostic prediction, and dynamic monitoring of therapeutic response—paving the way for personalized medical care.
Keywords: circulating fetal nucleated cells, circulating tumor cells, liquid biopsy, microfluidics, nanostructured substrates
1. Introduction
The blood circulatory system is a complex network responsible for the flow of blood cells, nutrients, oxygen, and other biochemical components in the human body. While erythrocytes (red blood cells, RBCs), leukocytes (white blood cells, WBCs), and platelets constitute the vast majority of cellular components in the blood, studies have found that detecting circulating rare cells in peripheral blood can be of great significance for both materials research and clinical applications.[1] Examples include, but are not limited to, circulating tumor cells (CTCs),[2] circulating fetal nucleated cells (CFNCs),[3] circulating stem cells,[4] endothelial cells,[5] plasma cells,[6] and mesenchymal cells.[7] Some of these have better-defined roles in diagnostic applications (such as CTCs, CFNCs, and certain subcategories of circulating stem cells), though others remain underexplored. In this review article, we pay close attention to CTCs and CFNCs for that nanomaterial-embedded platforms have largely grown in parallel with the research on these entities. The difficulties in utilizing CTCs or CFNCs in clinical settings also represent the challenges in developing nanotechnology-based circulating rare-cell assays.
The current gold standard for diagnosis of solid tumors in the majority of cancers is the characterization of tumor tissues acquired via invasive procedures, e.g., surgical excision or needle biopsy.[8] CTCs are tumor cells that either passively shed into or actively enter the circulation[9] (Figure 1). Over the past few decades, “liquid biopsy”[10] approaches based on the detection and characterization of CTCs have gradually evolved from the initial CTC enrichment and enumeration for prognosis to understanding disease biology and monitoring tumor progression.[2,11] The number of CTCs in the blood varies with tumor types and disease stage but generally falls in a range of a few to hundreds (per milliliter) among a large number (109 mL−1) of hematologic cells.[2,12] As numerous studies have shown across different cancers, CTCs have substantial similarities to the primary tumor tissue specimens, including their genomic alterations,[13,14] gene expression,[15,16] protein expression,[17–19] and cellular function.[20,21] Once appropriately harvested, CTCs can be subjected to morphologic,[22–24] phenotypic,[19,25,26] genomic,[27,28] transcriptomic,[29–32] and even functional[33,34] characterizations, which could provide insightful information for understanding underlying tumor biology. Detection and enumeration of CTCs have demonstrated their significance in prognosis of breast,[35] prostate,[36,37] and colorectal cancers.[38] It is of great hope that CTCs can serve as a surrogate tumor tissue source for conducting noninvasive diagnostics, particularly in settings where tumor biopsy is challenging or not accessible.[39,40] Even in cases where tumor biopsies are possible, serial blood draws are far more feasible and favorable in clinical practice than serial tumor biopsies. Because CTCs can be detected over the course of the disease, they provide an opportunity for real-time and dynamic monitoring of therapeutic interventions as well as the evolving malignant process.[41] Currently, CTCs share many proposed clinical utilities with other circulating entities, particularly circulating tumor DNA (ctDNA).[42] However, unlike ctDNA (which is highly fragmented and confounded by substantial background), CTCs’ membranes provide a natural barrier, which protects the fragile biomolecular contents (e.g., genomic DNA, RNA, and proteins) from degradation, guaranteeing their intactness for downstream molecular analysis. Ultimately, CTCs are expected to reveal more biological insights by allowing the integration of multilayer information (e.g., phenotype, genomic, and functional), which adds even more values to the characterization of underlying tumor biology.
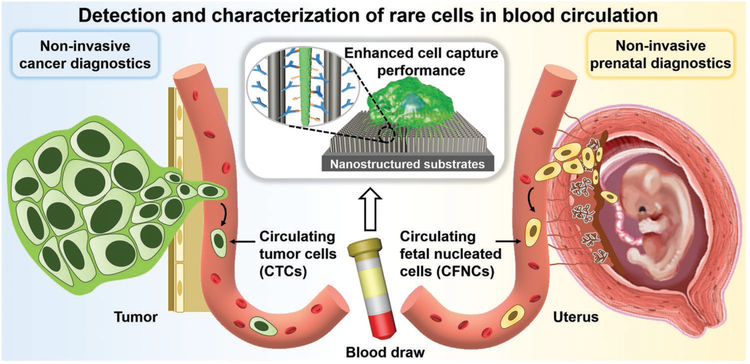
Figure 1.
Conceptual illustration depicting how nanostructured substrates can be utilized for achieving detection and characterization of CTCs and CFNCs with enhanced cell-capture performance. CTCs break away from either a primary tumor mass or metastatic sites to intravasate and circulate in the peripheral blood, and are used as tumor liquid biopsy for conducting noninvasive cancer diagnostics. Much like CTCs’ role in cancer diagnosis, CFNCs in the maternal circulation open up the opportunity for implementing noninvasive prenatal diagnostics.
Much like cancer diagnosis, prenatal diagnosis also relies heavily upon invasive tissue sampling and can potentially benefit from circulating rare-cell analyses. Although noninvasive prenatal screening technologies based on cell-free fetal DNA (cffDNA) are widely implemented in obstetric clinics for detecting fetal aneuploidy (e.g., trisomies 21, 18, and 13),[43] the confirmative diagnosis of fetal chromosomal aberrations, genetic disorders, and many other abnormalities still requires invasive procedures,[44] namely, amniocentesis (performed at gestational age (GA) = 15–18 weeks) or chorionic villus sampling (performed at GA = 10–13 weeks). Through these procedures, fetal cells are collected for karyotyping and other molecular analyses.[45] However, it is noted that amniocentesis and chorionic villus sampling are accompanied by a significant risk of miscarriage (0.6–2%)[46] and other complications. Over the past half-century,[47] tremendous research endeavors[3,48] in the search for noninvasive prenatal diagnostics (NIPD) have focused on circulating fetal nucleated cells. In contrast to fragmented cffDNA (typically hundreds of base pairs) which is confounded by a large amount of maternal DNA and is limited to screening use, CFNCs in maternal circulation house pure and intact fetal genomic DNA, and are an ideal candidate for conducting NIPD. In addition, intact CFNCs may provide opportunities for additional analysis such as multiomic and functional assessments, which will reveal further insights into placental and fetal biology. The presence of CFNCs (Figure 1) has long been documented[49,50] at a level of <6 cells mL−1 of maternal blood in normal pregnancy[51] among a background of 109 maternal hematologic cells mL−1. Among CFNCs that have been identified in maternal circulation, circulating trophoblasts (cTBs)[48] and circulating fetal nucleated red blood cells (fNRBCs)[52] have been studied most extensively. cTBs are detached from the placenta and feature distinctively large cell sizes, as well as representative information of fetal karyotype and genotype in most cases.[53] fNRBCs are directly derived from embryonic tissue and have intact fetal genetic information. Both cTBs and fNRBCs have short lifespans (of only a few days), which make it nearly impossible to isolate CFNCs from previous pregnancies.
The low abundance of CTCs and CFNCs among a high number of background hematologic cells represents the primary technical challenge[54] for the realization of their detection and characterization. It has been a major focus of the field to develop methods with ultra-high sensitivity and specificity. Among the most commonly used cell-sorting technologies, label-free approaches such as gradient centrifugation[55] and filter-based technologies[56,57] isolate CTCs/CFNCs from other hematopoietic components based on their differences in density and size, respectively. These methods offer simple and scalable enrichment solutions for rare-cell isolation, though with suboptimal sensitivity and a high probability of target cell loss. Dielectrophoresis[58] is another example of label-free cell sorting that enriches CTCs and CFNCs from peripheral blood mononuclear cells by their different dielectric properties. Approaches based on specific labeling of CTCs/CFNCs have also been developed, such as fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting. FACS is often performed on flow cytometry and is a powerful cell-sorting technology for analyzing and isolating subpopulations of immunofluorescently labeled CTCs/CFNCs.[59–62] Immunomagnetic separation has also become particularly popular over the past two decades,[63–65] with CellSearch (Menarini Silicon Biosystems Inc., Huntington Valley, PA) assay[66] being cleared by the U.S. Food and Drug Administration (FDA) for detection of CTCs in metastatic colorectal,[38] breast,[35,67] and prostate[68,69] cancers. With appropriate cell labeling, immunomagnetic cell separation may enrich CTCs and CFNCs with certain cellular markers (e.g., antiepithelial cell adhesion molecule (EpCAM)[70] for CTCs of epithelial origin), or deplete the background cells with certain cellular markers (e.g., WBCs using anti-CD45[71]) and collect the remaining CTCs/CFNCs. Newly developed immunomagnetic isolation methods, e.g., MagSweeper,[72] AdnaTest,[73] magnetic sifters[74,75] and nanoparticle-mediated magnetic ranking,[19,76,77] have further improved the efficiency and speed of cell detection and isolation. It is worth noting that tremendous research efforts have also been devoted to the development of microfluidics-based circulating rare-cell assays,[23,78–84] such as CTC-chip,[85] herringbone-chip (HB-Chip),[86] micro-Hall detector (μHD) chip,[87] geometrically enhanced differential immunocapture microdevices,[88] and deterministic lateral displacement (DLD) patterned microchips.[89,90] These devices further improved CTC/CFNC capture efficiency. Several review articles[50,91–95] have summarized these aforementioned technologies for detection and characterization of CTCs or CFNCs, providing coverage and scope different from this review article.
In recent decades, medical research in the fields of nanomaterials and nanotechnology has made great progress in improving the efficiency of CTC/CFNC enrichment, while at the same time reducing the costs of CTC/CFNC characterization.[112] It has been documented that nanoscale components in the tissue microenvironment (including cell-surface structures, e.g., microvilli[96] and filopodia,[97,98] and extracellular matrix[99]) provide structural and biochemical support that regulate cell behaviors,[100,101] fates[102–104] and functions[105,106] (e.g., morphology,[107] adhesion,[108,109] viability,[110,111] migration,[112,113] and differentiation[114–116]). Nanostructured substrates[117,118] mimic the nanoscale features found in the tissue microenvironment, offering a simple but effective solution to enhance the performance of CTC/CFNC enrichment.[119,120] The rationale of using nanostructured substrates for CTC/CFNC detection and characterization lies in the increased surface area available for contact between nanofeatures on the substrates and nanoscale cell-surface components, allowing for more binding sites to achieve highly efficient affinity capture.
This review article summarizes the recent development of nanostructured substrates capable of detecting and characterizing CTCs and CFNCs. The article starts from a systemic review of different categories of nanostructured substrates for capturing circulating rare cells, followed by discussing their extended utilities for cell retrieval. Figure 2 lays out the representative nanostructured substrates and controlled cell retrieval strategies in chronological order of publications. Technologies for the detection and isolation of CTCs have been the most widely developed. On the other hand, CFNCs are relatively difficult to study due to the technical and regulatory barriers in obtaining and analyzing maternal blood and other pregnancy-related biospecimens. It is therefore not surprising to see that the majority of circulating rare-cell assays (including many examples in this review) were initially developed for CTCs, with only a few being later adopted for detecting and/or isolating CFNCs. We also present examples of successful coupling of nanostructured substrate-embedded devices with CTC enumeration, CTC morphological analysis, and downstream CTC/CFNC molecular analysis (e.g., gene sequencing or protein analysis). The resulting CTC/CFNC analysis data support the clinical application in the field of noninvasive cancer and prenatal diagnostics. At the end of the review, we discuss the remaining challenges and future perspectives for improving the performance of nanostructured substrate-based circulating rare-cell assays and promoting their roles in the era of precision medicine.
Figure 2.
Classification of nanostructured substrates developed for detection and characterization of circulating rare cells (i.e., CTCs and CFNCs). i) High aspect ratios: nanowires/rods/tubes. Reproduced with permission.[121] Copyright 2009, John Wiley and Sons. Nanofibers. Reproduced with permission.[122] Copyright 2012, John Wiley and Sons. ii) Low aspect ratios: nanodots. Reproduced with permission.[123] Copyright 2011, John Wiley and Sons. Dendrimers. Reproduced with permission.[124] Copyright 2011, John Wiley and Sons. Nanoparticles/spheres. Reproduced with permission.[125] Copyright 2012, John Wiley and Sons. Nanosheets/films. Reproduced with permission.[126] Copyright 2013, Springer Nature. iii) Hierarchical nanostructures: fractal structures. Reproduced with permission.[127] Copyright 2013, John Wiley and Sons. Nano–micro structures. Reproduced with permission.[128] Copyright 2013, John Wiley and Sons. Controlled CTC/CFNC retrieval strategies include: i) Laser microdissection. Reproduced with permission.[129] Copyright 2013, John Wiley and Sons. ii) Stimulus-responsiveness: thermoresponsiveness. Reproduced with permission.[130] Copyright 2013, John Wiley and Sons. Electrical stimulation. Reproduced with permission.[131] Copyright 2014, John Wiley and Sons. Mechanoresponsiveness. Reproduced with permission.[132] Copyright 2015, John Wiley and Sons. Photoresponsiveness. Reproduced with permission.[133] Copyright 2016, American Chemical Society. iii) Additive reagent-triggered release: chemical reaction. Reproduced with permission.[134] Copyright 2013, American Chemical Society. Enzymatic digestion. Reproduced with permission.[135] Copyright 2013, John Wiley and Sons. DNA hybridization. Reproduced with permission.[136] Copyright 2016, John Wiley and Sons.
2. Nanostructured Substrates for Circulating Rare-Cell Capture
Nanostructured substrates with embedded characteristic features ranging between 1 and 1000 nm, provide a unique interface which can facilitate the interactions with cells. In the presence of affinity capture agents, nanostructured substrates exhibited enhanced affinity for targeted circulating rare cells (i.e., CTCs and CFNCs). Unlike the flat substrates, the enhanced CTC/CFNC affinity observed for nanostructured substrates can be attributed to the increased surface area available for contact between nanofeatures on the substrates and nanoscale cell-surface components, allowing for more binding sites to achieve highly efficient affinity capture.[137] Various fabrication methods—including etching,[138] template-assisted synthesis,[139] wet-chemical approaches,[140] lithography techniques,[141] electrospinning,[142] and chemical vapor deposition,[143] etc.—have been adopted to introduce different nanoscale characteristic features onto substrates made of a wide spectrum of materials.[118] Existing nanostructured substrates used for CTC/CFNC capture are classified in Figure 2 according to their morphological characteristics, including aspect ratios (the ratio of height to diameter), shapes, orientations, and compositions: i) high aspect ratios (≥2:1), including nanowires[121]/rods[144]/tubes,[145] nanofibers,[122] etc.; ii) low aspect ratios (<2:1), including nanodots,[123] dendrimers,[124] nanoparticles/spheres,[125] and nanosheets/films,[126] etc.; iii) hierarchical nanostructures, including fractal nanostructures[127,146] and nano–micro structures.[128] All the morphological features affect the cell-substrate interactions, including 1) substrate surface contact area, 2) capture agent loading capacity, and 3) other physical interactions of the cellular surface components (i.e., filopodia or lamellipodia), which significantly contribute to the cell capture performances. Other unique features, e.g., materials, capture agents, and integration of microfluidic components, as well as the working mechanisms, also contribute to the cell isolation performance. These different morphological and unique features, working mechanisms, cell isolation performances, and advantages/limitations of representative nanostructured substrates were listed in Table 1.
Table 1.
Summary of nanostructured substrates for circulating rare-cell isolation.
| Category | Substrate | Materials | Capture agents | Microfluidics | Cell line isolation study | Advantage/limitation | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|
| Yield | Purity | Viability | |||||||
| High aspect ratio | Nanowires, nanorods, and nanotubes | SiNWs | EpCAM antibody | − | C: 45–65% | N/A | 84–91% | Efficient and reproducible capture of CTCs | [121] |
| EpCAM antibody | + | C: 95% | N/A | N/A | High efficiency achieved with microfluidics | [158] | |||
| EpCAM antibody | + |
C: 94% R: 80% |
≈200 WBCs | N/A | Click reaction/disulfide cleavage-mediated CTC capture/ release with high sensitivity and specificity | [225] | |||
| A549cell aptamer | + |
C: 80% R: 85% |
95% | 78–83% | Nuclease digestion of DNA aptamers to release CTCs | [135] | |||
| Aptamer cocktail | + | C: >50% | N/A | N/A | Limited to the relatively low affinity of the aptamer | [160] | |||
| PNIPAAm-SiNWs | EpCAM antibody | − | C: >70% R: 90% | N/A | 90% | Thermoresponsive capture and release | [130] | ||
| EpCAM antibody | + | R: >70% | 88–98% | >90% | High purity via two rounds of thermoresponsive isolation | [219] | |||
| EpCAM antibody | − | R: 99% | 97% | ≈95% | Hydrophobic interaction-mediated capture/release | [220] | |||
| PolyAAPBA-SiNW | EpCAM antibody | − |
C: 60% R: 98% |
N/A | 95% | pH and glucose dual-responsive CTC release | [134] | ||
| Au NC-SiNWs | EpCAM antibody | − | C: 88% | N/A | 95% | Enhanced surface area for antibody binding | [150] | ||
| QNWs | EpCAM antibody | − | C: ≈65% | N/A | N/A | Efficiency and rapid analysis of CTCs | [151] | ||
| Lipid-QNWs | EpCAM antibody | − | C: ≈92% | ≈71% | N/A | Lipid preventing nonspecific cell adhesion | [154] | ||
| SS-biotin-Ppy NWs | EpCAM antibody | − | C: >93% R: ≈98% | N/A | >90% | Dual electrical- and GSH-responsive CTC release | [152] | ||
| AuNWs | Aptamer-sgc8c | − |
C: 83% R: 96% |
N/A | 90% | CTC release by reductive cleavage of Au—S bonds | [153] | ||
| PLGA nanopillar | EpCAM antibody | + | C: 70% | N/A | N/A | Integrating with LCM to isolate single cTBs | [159] | ||
| PEDOT nanorods | EpCAM antibody | − |
C: 70% R: 90% |
N/A | 97% | Bioelectronic interfaces, capable of releasing CTCs | [155,156] | ||
| EpCAM antibody | + |
C: ≈75% R: 95% |
46% | >95% | Glycan stimulation enabled CTC efficient release | [222] | |||
| BSA-TiO2 nanorods | EpCAM aptamer | − | C: 85–95% | ≈96% | N/A | Using BSAto inhibit nonspecific cell adhesion | [144] | ||
| Polystyrene nanotubes | EpCAM antibody | − | C: 60–80% | N/A | ≈98% | High cell viability achieved by the soft nature of PS | [157] | ||
| Nanofibers | TiO2 | EpCAM antibody | − | C: 40–70% | N/A | N/A | Enhanced cell affinity | [122] | |
| MnO2 | EpCAM antibody | − |
C: 80% R: 88% |
N/A | ≈90% | CTC release by oxalic acid dissolution of MnO2 | [161] | ||
| PLGA | EpCAM antibody | + | C: ≈80% | N/A | >80% | Single CTC isolation by LCM | [163] | ||
| CD146 antibody | + | C: 87% | N/A | N/A | Single CMC isolation by LMD technique | [129] | |||
| HA | + | C: 80% | N/A | N/A | Only capture of CD44-positive tumor cell | [164] | |||
| Chitosan | EpCAM aptamer | − |
C: ≈96% R: ≈98% |
N/A | ≈90% | DNA hybridization with aptamer to release CTCs | [136] | ||
| Polystyrene | EpCAM antibody | C: =89% | N/A | =99% | Fibrous network with a “trap effect” | [167] | |||
| PEO/PEDOT:PSS | EpCAM antibody | + |
C: 90% R: 87% |
N/A | >95% | CTC capture/release via electrochemical doping/ dedoping of PEDOT:PSS | [174] | ||
| PEI/PVA | RGD peptides | + |
C: ≈92% R: ≈93% |
33% | ≈89% | Only capture and release of αvβ3-overexpressing CTCs | [224] | ||
| Other | Horizontal HNTs | EpCAM antibody | − | C: 92% | 100 WBCs | N/A | Easy self-assembly process and high capture efficiency | [179] | |
| Porous polyAPBA | PBA groups | − |
C: 78% R: 94% |
N/A | 90% | Fructose-mediated release of CTCs | [180] | ||
| GNRs- gelatin | EpCAM antibody | − |
C: 92% R: 92% |
N/A | 90% | Photoresponsive site-specific CTC release | [133] | ||
| Low aspect ratio |
Nanodots | PEDOT-COOH | EpCAM antibody | − | N/A | N/A | N/A | Higher capture efficiency than smooth films | [123] |
| Biotin-Ppy | EpCAM antibody | − |
C: 90% R: 90% |
N/A | ≈90% | Electric stimulation for rapidly releasing CTCs | [184] | ||
| Dendrimers | PAMAM | EpCAM or EGFR antibody | − | C: ≈70% | N/A | N/A | Local multivalent binding with enhanced stability | [124,185] | |
| EpCAM, HER-2, and PSA antibody | C: 82% | 90% | N/A | Enhance sensitivity and specificity by combing cell rolling and multivalent binding | [186,187] | ||||
| Nano-particles (NPs) and nano-spheres | Fe3O4 | Tf | C: 84% | N/A | N/A | Only targeting TfRs-positive cell lines | [125] | ||
| MnO2 | EpCAM antibody | − |
C: ≈81% R: 92% |
98% | 90% | CTC release via reduction of MnO2 by oxalic acid | [189] | ||
| Au | EpCAM aptamer | + | C: >90% | 70% | N/A | High-affinity binding effect using AuNP-aptamers | [191] | ||
| EpCAM antibody | + |
C: ≈98% R: >91% |
N/A | >80% | GSH-triggered AuNP-thiol exchange to release CTCs | [192] | |||
| Chitosan | EpCAM aptamer | − | C: 90% | 97% | N/A | Decreased nonspecific cell adhesion achieved by PEG | [193] | ||
| Gelatin-polystyrene | EpCAM antibody | + |
C: 75–96% R: 93% |
N/A | ≈90% | Thermo or mechanoresponsive release of CTCs | [132] | ||
| Biotin-Ppy | CD147 antibody | + | R: ≈95% | N/A | N/A | Rapidly electrical release of fNRBCs | [195] | ||
| Gelatin | CD147 antibody | + | C: >80% R:89% | 85% | 90% | Enzymatic dissolution of GNPs to release fNRBCs | [196] | ||
| Nanosheets and nanofilms | GO nanosheets | EpCAM antibody | + | C: 73% | N/A | N/A | High sensitivity | [126,197] | |
| PAPDEA-GO composites | EpCAM antibody | + |
C: 85–95% R: ≈95% |
N/A | 92% | Thermoresponsive release of CTCs | [198] | ||
| rGO nanofilms | EpCAM antibody | C: 67–93% | N/A | N/A | High efficiency achieved by the rough and low-stiffness nature of rGO | [199] | |||
| ALG/PAH nanofilms | EpCAM antibody | + |
C: 80% R: 95% |
53% | 90% | Enzymatic degradation of nanofilms to release CTCs | [230] | ||
| Other | Nanorough glass | - | + | C: 88–95% | N/A | N/A | Label-free CTC capture; problems of specificity | [205] | |
| Hierarchical nanostructure | Fractal nanostructures | FAuNSs | EpCAM antibody | − |
C: 62% R: 98% |
N/A | 95% | Electrochemical release of CTCs via Au–S cleavage |
[127] |
| Fractal ITO nanowire | EpCAM antibody | − | C: 89% | N/A | 96% | High CTC capture efficiency and viability | [146] | ||
| TiO2 nanosisal | EpCAM antibody | − | C: ≈58% | N/A | 97% | Trypsinization-based cell release lacking specificity | [212] | ||
| Nano-micro hierarchical substrates | LIPs | EpCAM antibody | C: 62% | N/A | N/A | Topographic interactions at both micro- and nanoscale | [208] | ||
| Cell replica surfaces | EpCAM antibody | − | C: 53–62% | N/A | N/A | Topographic interactions at both micro- and nanoscale | [209] | ||
| Flowerlike HZnPNS | EpCAM antibody | − |
C: 90% R: 88% |
63% | 92% | CTC release via sodium citrate-induced dissolution | [211] | ||
| EpCAM aptamer | + |
C: 90% R: 86% |
N/A | 81% | DNA hybridization with aptamer to release CTCs | [231] | |||
| rGO/ZnO foam | EpCAM antibody | − | C: 80% | N/A | N/A | Increased cell-substrate contact frequency | [128] | ||
| ZnO nanograss | EpCAM antibody | + |
C: 80% R: 90% |
N/A | 90–95% | Precise control of acid condition and release time | [223] | ||
| TiO2 nanorods-Si micropillars | EpCAM antibody | + | C: 77% | N/A | N/A | A sensitive nano-bio interface | [213] | ||
| GO-MNPson nickel micropillars | EpCAM antibody | + |
C: 70% R: 93% |
N/A | 78% | Magnetic capture and release of CTCs | [214] | ||
| Fe2O3 NPs on nickel squares | EpCAM antibody | + | C:94% | N/A | 93% | Magnetic capture and release of CTCs | [215] | ||
| AuNPson DLD-based micropillars | EpCAM aptamer | + |
C: 89% R: 80% |
N/A | 96% | CTC release via thiol exchange reaction | [216] | ||
(−) Without microfluidics; (+) With microfluidics; C: capture yield; R: release yield; N/A: not available.
2.1. Nanostructured Substrates with High Aspect Ratios
2.1.1. Nanowires, Nanotubes, and Nanorods
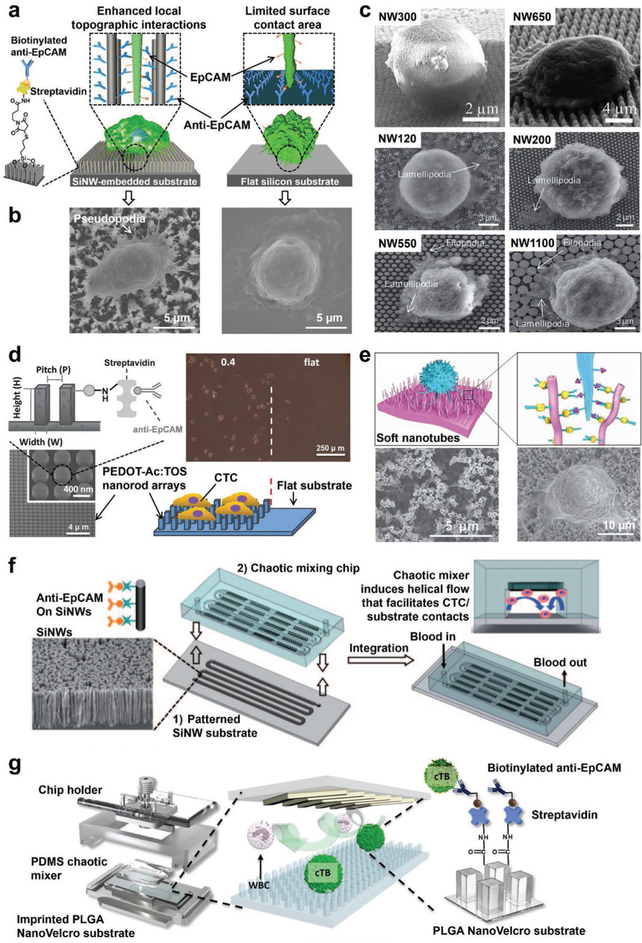
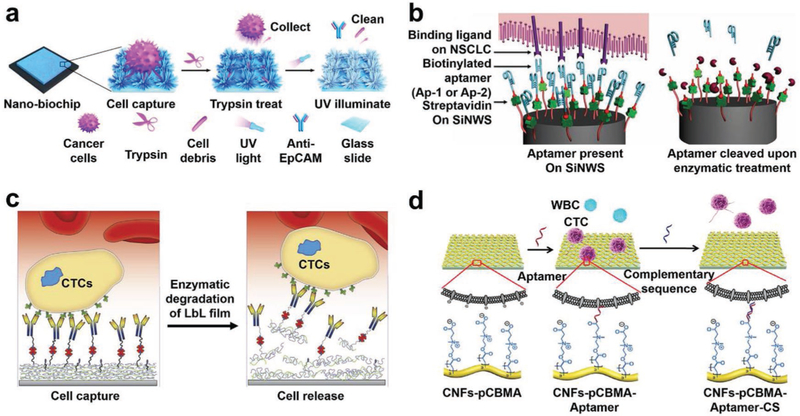
Silicon nanowire (SiNW)-embedded substrates were first explored for rare-cell capture in 2009.[121] Ag nanoparticle-templated wet etching[138] was employed to create high aspect ratios of SiNWs (diameters = 100–200 nm, lengths = 1–20 μm) on silicon wafers. N-Hydroxysuccinimide/maleimide chemistry was used to covalently conjugate streptavidin onto the surfaces of SiNW-embedded substrates. Biotinylated antibodies were then grafted onto SiNW-embedded substrates to confer the specificity to capture the circulating rare cells of interest. In studies testing such an SiNW-based cell capturing system on EpCAM-positive MCF-7 breast cancer cells and PC3 prostate cancer cells, the resulting anti-EpCAM-grafted substrates exhibited dramatically improved cell capture performance (45–65%) compared to that observed for a flat silicon substrate (Figure 3a).[121] Much like the tight attachment of two fabric strips of a Velcro fastener, the nanoscale topographic interactions between SiNWs and cell surface features combined with antibody-mediated biorecognition tightly binds CTCs to the SiNWs. Because of these Velcro-like interactions, the researchers named this substrate the “NanoVelcro” substrate.[119,120,147] Under scanning electron microscopy (SEM), cells captured on SiNW substrates formed semielliptical shapes with numerous pseudopodia (e.g., lamellipodia or filopodia), while cells captured on flat silicon substrates were observed to have spherical morphologies with few nanoscale cellular protrusions (Figure 3b). With the local topographic interactions leading to the formation of pseudopodia, SiNW substrates experience firmer cell adhesion,[148,149] improving the cell capture efficiency. The aspect ratios of SiNWs, with diameters (120–1100 nm) and interwire spacings (35–800 nm), were found to influence the cell morphology and pseudopodia formation (Figure 3c).[96] The effective contact area of nanowire-embedded substrates was linearly related to the capture yield. In other words, more compact SiNW arrays and smaller diameters of nanowires demonstrated higher capture yield. Spacings between SiNWs of less than 100 nm showed significantly reduced capture yield, and spacings of more than 800 nm significantly reduced specificity. Interestingly, the increase of SiNW lengths (1–20 μm) improved the capture yield for EpCAM-positive cells but had a negligible effect on the capture of EpCAM-negative cells.[121] The increase of nanowire length likely has a positive effect on capture efficiency by increasing the surface area available to bind more antibodies for immunoaffinity-based specific cell recognition.
Figure 3.
Nanowire, nanorod, and nanotube-embedded substrates developed for CTC or CFNC capture. a) An anti-EpCAM-functionalized SiNW-embedded substrate (i.e., NanoVelcro cell-affinity substrate) achieved highly efficient capture of CTCs by employing enhanced local topographic interactions in contrast to an anti-EpCAM functionalized flat silicon substrate with limited surface contact area. Biotinylated anti-EpCAM was grafted onto these substrates to endow CTC recognition with specificity. b) SEM images of CTCs captured on an SiNW-embedded substrate (left) and a flat silicon substrate (right), respectively. Reproduced with permission.[121] Copyright 2009, John Wiley and Sons. c) SEM images showing cell morphologies and behaviors on SiNWs with differing interwire spacings (ranging from 38 to 790 nm) and diameters (ranging from 120 to 1100 nm). Reproduced with permission.[96] Copyright 2014, Royal Society of Chemistry. d) A tosylate (TOS)-doped PEDOT nanorod array-embedded substrate with anti-EpCAM conjugation achieved higher capture performance than a flat substrate with anti-EpCAM. Reproduced with permission.[155] Copyright 2014, John Wiley and Sons. e) An anti-EpCAM functionalized soft PS nanotube-embedded substrate provides a large contact area for CTC capture. SEM images showed the structures of PS nanotubes and the captured cells. Reproduced with permission.[157] Copyright 2013, Springer Nature. f) Schematic diagram of the configuration and operational mechanism of a NanoVelcro Chip for capturing CTCs, which consists of a patterned SiNW-embedded substrate with anti-EpCAM-coating, and an overlaid PDMS chaotic micromixer. Reproduced with permission.[158] Copyright 2011, John Wiley and Sons. g) An imprinted PLGA NanoVelcro Chip developed for cTB capture. Reproduced with permission.[159] Copyright 2017, American Chemical Society.
SiNWs can be fabricated not only by Ag nanoparticle-templated wet etching but also through a rapid thermal chemical vapor deposition. Gold nanocluster (Au NC)-coated SiNWs were fabricated with Au NCs (thicknesses = 3–5 nm, density = 3 × 1012 cm−2) evenly covering the entire sidewalls of the SiNWs (diameters = 50–160 nm, length = 5–10 μm), providing large surface areas for anti-EpCAM conjugation.[150] Compared to the previous anti-EpCAM-modified SiNWs, these anti-EpCAM-modified and Au NC-coated SiNWs had a significantly increased CTC capture yield of 88%.
In addition to SiNWs, research efforts were devoted to exploring the use of different nanomaterials such as quartz,[151] polymers[152] and gold[153] to fabricate nanowire-embedded substrates. Transparent quartz nanowire arrays (QNWs, diameters = 80–100 nm, lengths = 250–350 nm)[151] were introduced on a quartz wafer using a combination of polystyrene nanoparticle (PS NP)-templated colloidal lithography and chemical etching. Anti-EpCAM-functionalized QNWs showed a capture efficiency of 65%. Other researchers have attempted to coat lipids on the QNWs to serve as a functionalized bilayer for facilitating antibody-based cell capture and preventing nonspecific cell adhesion.[154] As for polymer nanowires, one example is the disulfide-biotin-doped polypyrrole nanowires (SS-biotin-Ppy NWs, diameter = 200 nm, length = 2 μm)[152] which were prepared by anodic alumina oxide (AAO) nanopore-templated electropolymerization. Anti-EpCAM-conjugated Ppy NWs had a capture yield of 93% for cancer cell lines. Recently, gold nanowires (AuNWs, diameter = 208 nm, length = 5.7 μm, spacings = 110–130 nm)[153] were prepared by using AAO-templated electrochemical deposition. DNA aptamers against tyrosine kinase 7 (a cell membrane protein of human leukemic lymphoblasts, e.g., CCRF-CEM cells) were modified on the AuNWs via Au–S chemistry and realized a capture yield of 83% and 90% cell viability.
Additionally, vertically grown nanorod and nanotube arrays, with structures similar to nanowires, have been developed as sensitive biosensors for CTC/CFNC detection. Both conducting polymers and metallic oxides can serve as candidate materials for fabricating nanorods. Electrically conducting polymers like poly(3,4-ethylenedioxythiophene) (PEDOT), have advantages including few structural defects, good biocompatibility, manufacturing flexibility, excellent electrical transport, and electrochemical charge-discharge capabilities. For this reason, researchers utilized PEDOT to fabricate large-scale nanorod arrays as 3D bioelectronic interfaces for CTC analyses (Figure 3d).[155,156] The PEDOT nanorods were synthesized on the indium tin oxide (ITO) glass substrate by a combination of chemical oxidative polymerization and poly(dimethylsiloxane) (PDMS) transfer printing methods, using Si nanorod arrays (diameter = 0.4 μm, heights = 0.4–1.2 μm) as masters. Compared with an anti-EpCAM-modified flat substrate, the anti-EpCAM-modified PEDOT nanorods captured more CTCs with a capture yield of 70% and cell viability of 97%.[155] Titanium dioxide (TiO2) nanorods (diameters = 160–300 nm) composed of nanoparticles with diameters of 30–50 nm, were synthesized on the F-doped SnO2 (FTO) substrate by hydrothermal reaction.[144] TiO2 nanorods functionalized with BSA and DNA aptamer (against EpCAM) had a cell capture yield of 85–95%. Nanotube-embedded substrates can be made of several materials, such as carbon[145] and PS.[157] One such nanotube is the soft PS nanotube (PS NT, diameter = 200 nm; Figure 3e),[157] which was fabricated by an AAO-templated replication method. The physical absorption of biotinylated bovine serum albumin (biotin-BSA) to materials through hydrophobic interactions was employed to immobilize SA and biotinylated anti-EpCAM onto the PS NT-embedded substrate, providing an inert surface to prevent nonspecific molecular/cell adhesion. Cells captured on the PS NT-embedded substrate protruded filopodia to contact the surface structures. Capture yields of 60–80% were obtained using this PS NT-embedded substrate.
To further improve CTC-capture performance, nanostructure-embedded substrates were integrated with microfluidic components. An SiNW-embedded microfluidic platform, a.k.a., NanoVelcro Chip (Figure 3f) was developed by combining the SiNW-embedded substrate and an overlaid PDMS chaotic micromixer, featuring an 88 cm long serpentine chaotic mixing channel.[158] The chaotic micromixer induces a vertical flow of the cell suspension and increases the frequency of CTC-substrate contact. A cell capture yield of 95% was achieved with artificial blood samples using an optimal flow rate of 1 mL h−1. NanoVelcro Chips were able to capture more CTCs than Cell-Search Assay.[66] Additionally, an imprinted poly(lactic-co-glycolic acid) (PLGA) NanoVelcro Chip was prepared for cTB enrichment from maternal blood by integrating an anti-EpCAM-coated PLGA nanopillar-embedded substrate with the PDMS chaotic micromixer (Figure 3g).[159] Via chlorobenzene-assisted nanoimprinting, a set of poly(methyl methacrylate) nanopillar features (diameter = 200 nm, length = 1.5 μm, spacing = 800 nm) was transferred from PDMS replicates onto the PLGA film that was spin-coated on a laser microdissection (LMD) slide. The resulting imprinted PLGA NanoVelcro substrate had the advantage of optical transparency, allowing it to be used with laser capture microdissection (LCM) to isolate individual cTBs. By combining this with a PDMS chaotic mixer and using anti-EpCAM as the capture agent, >70% capture efficiency was achieved for cTBs at an optimal flow rate of 1 mL h−1. Aside from antibodies, aptamer cocktails against nonsmall cell lung cancer (NSCLC) cell line subtypes were also exploited using SiNW-embedded NanoVelcro Chips.[160] The combination of multiple aptamers exhibited synergistic effects and achieved high-affinity differential capture for NSCLC CTC subtypes, providing a possible way for characterizing the heterogeneity of CTC population.
2.1.2. Nanofibers
Unlike the nanowires, nanotubes, and nanorods mentioned above, which are vertically aligned on the substrates, nanofiber-embedded substrates feature horizontally deposited nanofibers with extremely high aspect ratios. Electrospinning deposition[142] offered a versatile and straightforward fabrication approach for the preparation of ultralong polymer nanofibers with controllable diameters (10 nm to 10 μm), from a diversity of soluble and fusible polymer precursors. Additionally, inorganic additives (e.g., TiO2[122]) can be blended into the polymer precursors to generate composite nanofibers. After calcination at high temperature to thermally decompose polymeric components, inorganic nanofibers can be obtained with intact morphologies. Substrates with electrospun polymeric and inorganic nanofibers are classified as a subcategory of nanostructured substrates and exhibit enhanced capture performance for CTCs and CFNCs. The diameters, lengths, and density of the embedded nanofibers affect their CTC/CFNC-capture affinity and can be optimized through controlling electrospinning parameters, e.g., applied voltage, concentration of precursory solutions, the distance between the injection nozzle and substrates, and deposition time/density. Additionally, the physical and chemical surface properties of nanofibers also contribute to the CTC/CFNC capture sensitivities and specificities.
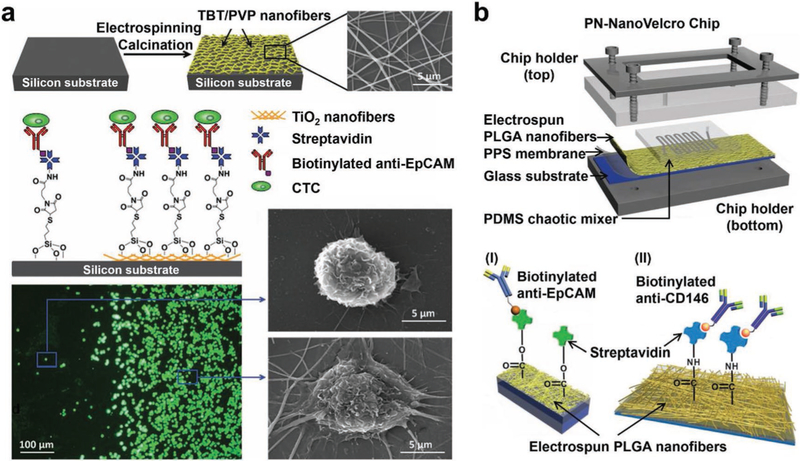
TiO2 nanofiber (TiNF)-embedded substrates (Figure 4a) were first prepared by conducting electrospinning deposition of precursory solution (containing titanium n-butoxide and polyvinyl pyrrolidone) onto Si wafers, followed by calcination at high temperature.[122] The horizontally packed ultralong TiNFs had diameters of 100–300 nm. After grafting CTC capture agent, i.e., anti-EpCAM, onto TiNFs via biotin–streptavidin conjugation, TiNF-embedded substrates were subjected to cell capture study in the presence of EpCAM-positive colorectal cancer cells, affording capture yields of 40–70%. A substrate with both flat topography and TiNF coating was prepared for comparing their differential cell-capture affinity. As shown in the fluorescence micrograph image (Figure 4a), the TiNF-coated area captured much more cells than the flat Si surface. SEM imaging study revealed that the cells captured on the TiNF-coated area presented fully outspread pseudopodia structures in contrast to those on the flat Si surface. This phenomenon is consistent with the results observed for vertically aligned nanowires (Figure 3b), strongly supporting the general applicability of the nanostructured substrates for achieving enhanced cell-capture performance. Similarly, calcinated manganese dioxide (MnO2) nanofibers[161] with a much smaller diameter (20 nm) were also deposited onto glass substrates, allowing for CTC affinity capture with 80% of efficiency.
Figure 4.
Conjugation of capture antibodies onto nanofiber-embedded substrates enables highly efficient CTC enrichment. a) TiNF-embedded substrates were prepared via electrospinning deposition of horizontally oriented precursory nanofibers, followed by calcination and anti-EpCAM conjugation. Fluorescent micrographs (bottom left) and SEM images (bottom right) suggest that the resulting TiNF-embedded substrates demonstrated a higher CTC capture efficiency and affinity than that observed for flat silicon substrates, respectively. Reproduced with permission.[122] Copyright 2012, John Wiley and Sons. b) Schematic representation of a PN-NanoVelcro Chip consisting of an overlaid PDMS chaotic micromixer and a transparent PN-NanoVelcro substrate. i) Biotinylated anti-EpCAM (a prostate cancer-specific capture agent), reproduced with permission.[163] Copyright 2013, John Wiley and Sons or ii) biotinylated anti-CD146 (a melanoma-specific capture agent) were covalently conjugated on the PN-NanoVelcro substrates, Reproduced with permission.[129] Copyright 2013, John Wiley and Sons.
A wide spectrum of polymer materials—including PLGA,[129,162–164] chitosan,[136,165,166] PS,[167] and cellulose acetate,[168] polyvinyl alcohol/polyethyleneimine (PVA/PEI),[169–171] nylon-6/poly(sulfobetaine methacrylate)/poly(acrylic acid) (nylon-6/PSBMA/PAA),[172] polystyrene/poly(styrene-co-maleic anhydride),[173] poly(ethylene oxide)/poly(3,4-ethylenedioxyt hiophene):polystyrene sulfonate (PEO/PEDOT:PSS),[174] and poly(N-isopropylacrylamide)/poly(benzophenone) (PNIPAAm/PBP)[175])—were used in the preparation of polymer nanofiber-embedded substrates for conducting CTC/CFNC capture. Among different polymer nanofiber-embedded substrates, PLGA nanofibers (diameter = 130 nm) were first deposited onto glass substrates for capturing CTCs. By integrating the PLGA nanofiber-embedded substrate with a PDMS chaotic micromixer, a PLGA nanofiber microchip, a.k.a., PN-NanoVelcro Chip[129,163] (Figure 4b) was created for both CTC enrichment and single-CTC isolation in conjunction with the use of downstream LMD technique. In the presence of anti-EpCAM capture agent, PN-NanoVelcro Chips demonstrated superior performance for capturing CTCs from blood samples collected from prostate cancer patients.[163] When a melanoma-specific capture agent (anti-CD146) was used, the devices were capable of capturing circulating melanoma cells (CMCs)[129] from melanoma patients. PN-NanoVelcro Chips exhibited CTC/CMC capture performance of 75–90% when artificial CTC/CMC samples were used in calibration studies.
A natural polymer, chitosan, was also adopted for the preparation of polymer nanofiber-embedded substrates for cell capture.[136,165,166] Chitosan nanofibers (diameter = 190 nm) electrospun onto glass substrates were modified with Zwitterionic poly(carboxybetaine methacrylate) (PCBMA) brushes.[136] These PCBMA brushes provided polyvalent carboxyl groups to immobilize DNA aptamers against EpCAM to capture gastric cancer cells with a yield of 96% in the culture medium 54–66% in 1 mL blood. Electrospun PS nanofibers are another type of polymer nanofibers which were integrated into a 3D micro/nano-scale fibrous network with a “trap effect” for CTC capture.[167] The 3D PS network possessed micrometer-sized pores to provide cell fitting traps, nanoscale fibers to increase surface area, anti-EpCAM for specific biorecognition, and microbeads to enhance cell adhesion. The capture performance of breast cancer cells was up to 89% in culture medium and 52–63% in whole blood. Besides antibodies and aptamers, hyaluronic acid (HA),[176] actobionic acid (LA),[177] and folic acid (FA)[178] (which exhibit good affinity with different types of cancer cells) can also be used to capture CTCs in conjunction with nanofiber-embedded substrates. By conjugating HA onto PLGA nanofiber-embedded substrates, the integrated microchip demonstrated a capture efficiency of 80% for CD44-positive tumor cells.[164] Similarly, PVA/PEI nanofiber-embedded substrates with HA conjugation displayed an 85% capture efficiency for cervical cancer cells,[169] and those substrates with LA conjugation showed comparable performance in capturing liver cancer cells.[170]
2.1.3. Miscellaneous Nanostructured Substrates with High Aspect Ratios
Unlike the nanostructured substrates with vertically aligned nanotubes, horizontally oriented nanotubes were deposited onto substrates for enhanced CTC capture. For example, halloysite nanotubes (HNTs, Al2Si2O5(OH)4) were self-assembled into horizontally aligned strips (widths = 50–120 μm) on a glass substrate.[179] These HNT had diameters of 30–70 nm, lengths of 200–1500 nm, and aspect ratios of 2.8–50. The anti-EpCAM-functionalized HNT-coating substrate had a CTC capture yield of 92%. Nanoporous substrates, e.g., porous poly(aminophenylboronic acid) (polyAPBA) nanostructured substrates,[180] nanoporous anodic aluminum oxide-embedded substrates,[181] quartz nanohole arrays,[182] and bionic TiO2 inverse opal photonic crystal (IOPC) structure,[183] have also been used to capture CTCs with enhanced performance. For instance, porous polyAPBA nanostructured substrates were prepared on solid substrates by altering the nucleation and growth rates for polymerization of 3-aminophenylboronic acid (3-APBA) monomer.[180] Based on the affinity of phenylboronic acid groups to the glycans or sialic acids expressed on CTC surfaces, porous polyAPBA nanostructured substrates exhibited a CTC capture yield of 78%.
2.2. Nanostructured Substrates with Low Aspect Ratios
2.2.1. Nanodots
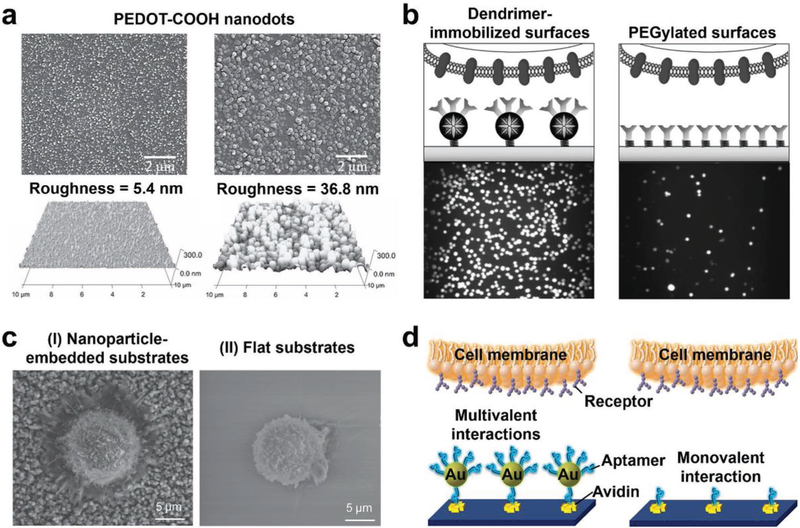
Among different low-aspect-ratio nanostructures, conducting polymer nanodots were first employed to achieve enhanced CTC capture. An electropolymerization approach was employed to deposit carboxylic acid group functionalized poly(3,4-ethylenedioxy)thiophenes (PEDOT-COOH) onto ITO glass in dichloromethane solutions containing the monomer precursor and electrolytes. The sizes and densities of the resulting PEDOT-COOH nanodots on the ITO substrates (Figure 5a) were altered by the applied electrochemical potential (1.0–1.4 V), affecting their CTC capture performance. The carboxylic acid groups on the PEDOT-COOH nanodots allowed for covalent attachment via N-hydroxysuccinimide-mediated bioconjugation. These PEDOT-COOH nanodot-embedded substrates showed 4–5 times higher capture efficiency than smooth PEDOT-COOH films.[123] This enhancement was due to the synergistic effect of nanostructure matching and ligand-receptor interaction between the nanodots and CTCs. Similarly, the biotin-Ppy mentioned above was also used to fabricate nanodots in the microchannel surfaces via electrochemical deposition.[184] By conjugating with anti-EpCAM, the nanodot-embedded microfluidic device had capture efficiencies of 90% for CTCs in the culture medium and 65% for CTCs in whole blood.
Figure 5.
Nanodot, dendrimer, nanoparticle, and nanosphere-embedded substrates developed for enhanced CTC capture. a) SEM (top) and AFM (bottom) images of PEDOT-COOH nanodots with different diameters and roughness. Reproduced with permission.[123] Copyright 2011, John Wiley and Sons. b) Schematic (top) and fluorescence (bottom) images of cancer cells captured on multiple antibody-functionalized PAMAM dendrimer-immobilized surfaces (left) and linear poly(ethylene glycol)-immobilized (PEGylated) surfaces (right), respectively. Reproduced with permission.[124] Copyright 2011, John Wiley and Sons. c) SEM images showing cell morphologies and behaviors on anti-EpCAM modified I) nanoparticle-embedded substrates and II) flat substrates, respectively. Reproduced with permission.[190] Copyright 2013, John Wiley and Sons. d) Schematic representation of the multivalent interactions between AuNP–aptamer-embedded substrates and cell membrane, and the monovalent interactions between aptamers only and receptors. Reproduced with permission.[191] Copyright 2013, American Chemical Society.
2.2.2. Dendrimers
Nanoscale polymer dendrimers can be conjugated with preorienting ligands, enabling the multivalent ligand-receptor binding to improve the affinity for capturing CTCs. Poly(amidoamine) (PAMAM) dendrimers (diameters = 8–10 nm) were coated on an epoxy-functionalized glass slide, followed by conjugation with multiple capture antibodies, e.g., anti-EpCAM, epidermal growth factor receptor antibody (anti-EGFR), and epidermal growth factor receptor-2 antibody (anti-HER2)).[124,185] The 3D structure of dendrimers (Figure 5b) could organize ligands into a small spatial area, reduce the deformation energy (entropy) of ligand-receptor binding, and promote local multivalent binding with enhanced stability. These PAMAM dendrimer-embedded substrates carried an average of 2.8 or 4.9 antibody molecules per dendrimer and had an average CTC capture yield of 70%. Furthermore, by combining the dendrimer-embedded substrate with the E-selectin-induced cell rolling, a CTC device named “CapioCyte” was prepared for differential capturing CTCs with an enhanced capture yield of 82% and a purity of 90%.[186,187]
2.2.3. Nanoparticles and Nanospheres
Nanoparticles and nanospheres can have a variety of physical and chemical properties, as well as different compositions of interfacial layers (i.e., inorganic or organic molecules). Nanostructured substrates fabricated by assembling nanoparticles or nanospheres onto substrates exhibit increased surface area, which enhances rare-cell capture. The first example is PEG-crosslinked Fe3O4 nanoparticles (diameter = 25 nm), which were covalently attached to silane-functionalized glass substrates.[125] By covalently grafting transferrin (Tf) onto PEG-Fe3O4 nanoparticle-embedded substrates, the substrates showed enhanced capture performance (84%) for colon cancer cells with overexpressed transferrin receptors (TfRs). Similarly, TiO2 nanoparticles (diameter = 400 nm),[188] MnO2 nanoparticles (diameter = 200 nm),[189] and candle soot nanoparticles (diameters = 19–43 nm)[190] can be deposited on flat glass substrates and functionalized with anti-EpCAM to give a variety of nanoparticle-embedded substrates which all showed improved CTC capture performance (yields ≈80%). As shown in SEM images (Figure 5c), cells captured on nanoparticle-embedded substrates extended more pseudopodia than those captured on flat substrates. Gold nanoparticles (AuNPs, diameter = 13.6 nm) can serve as efficient multivalent ligand scaffolds for grafting DNA aptamers (Figure 5d).[191,192] By integrating AuNP–aptamers (hydrodynamic diameter = 61.8 nm) into herringbone microfluidic devices, AuNP–aptamer-embedded substrates were fabricated for high-affinity CTC capture.[191] The multivalent aptamers on AuNPs (≈95 aptamers per AuNP) greatly enhanced the affinity of aptamers and CTCs, resulting in a capture yield of >90% and purity of 70%.
Polymer nanoparticles such as chitosan and PS nanoparticles have also been coated on substrates to enhance CTC and CFNC capture. Chitosan nanoparticles (diameter = 250 nm) were coated on substrates via electrospray and functionalized with aptamers to capture CTCs with yields of 90% in culture medium and 45–60% in whole blood.[193] PS nanoparticles (diameter = 166 nm) were assembled in a herringbone microfluidic device and used to capture EpCAM-positive CTCs with yields of 75–96%.[132] Furthermore, HAp/chitosan,[194] biotin-Ppy,[195] and gelatin[196] nanoparticles have also been assembled in microchips and functionalized with anti-CD147 (glycosylated protein expressing on erythroid precursors) for capturing fNRBCs from maternal blood samples. These polymer nanoparticle-embedded microchips had an average capture yield of 80%, purity of 85%, and viability of 90% for fNRBCs.
2.2.4. Nanosheets and Nanofilms
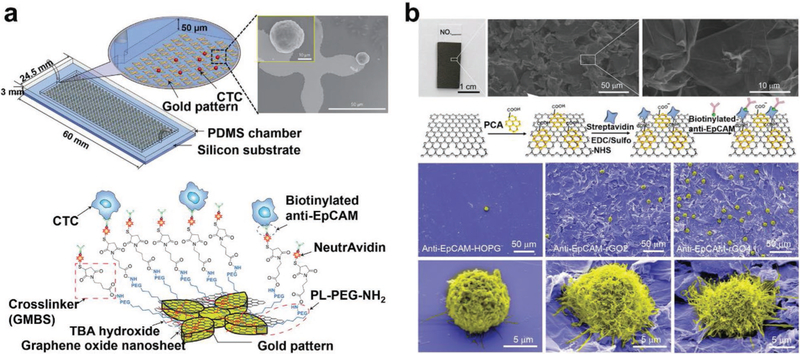
2D nanomaterials (i.e., nanosheets or nanofilms) offer the advantages of large surface areas and rough textures, facilitating sufficient cell-substrate contact and enhancing CTC capture affinity. For instance, GO nanosheet-embedded devices (a.k.a., GO chips) were fabricated by depositing GO nanosheets onto gold-patterned silicon substrates, followed by the conjugation of anti-EpCAM capture agent and integration with a PDMS microfluidic channel (Figure 6a).[126,197] These GO chips exhibited a CTC capture yield of 73%, which was five times higher than that of the control silicon devices without GO nanosheets. Furthermore, thermal-sensitive polymer–GO Chips were prepared via the drop-cast deposition of polymer–GO nanocomposite films (thicknesses = 3–4 μm) onto patterned glass substrates.[198] After grafting anti-EpCAM, these polymer-GO Chips achieved an enhanced CTC capture yield of 85–95%. Similarly, reduced GO (rGO) film with a petal-like wrinkled architecture was also used to capture CTCs (Figure 6b).[199] Prepared by vacuum filtration and thermal reduction, rGO films were conjugated with anti-EpCAM and had CTC capture yields of 67–93%. Other nanofilms like HAp/chitosan nanofilms[200,201] and gold nanofilms[202] have also been deposited on substrates and applied for capturing CTCs with enhanced performance.
Figure 6.
Nanosheet and nanofilm-embedded substrates for CTC isolation. a) Schematic view of a GO nanosheet-embedded microfluidic chip and SEM images of CTCs captured on the anti-EpCAM functionalized GO nanosheets that were absorbed on gold patterns. Reproduced with permission.[126] Copyright 2013, Springer Nature. b) rGO nanofilms with a petal-like wrinkled architecture were functionalized with anti-EpCAM for CTC capture. SEM images showing CTC capture performance and CTC morphologies on anti-EpCAM-modified smooth highly ordered pyrolytic graphite (HOPG) film, rGO film-embedded substrates with Ra of 0.83 and 19.42 μm, respectively. Reproduced with permission.[199] Copyright 2015, John Wiley and Sons.
2.2.5. Miscellaneous Nanostructured Substrates with Low Aspect Ratios
The advent of DNA origami technology[203] allows for the creation of a verity of nanoscale DNA 3D structures. Nanoscale DNA tetrahedrons (sizes = 6–10 nm) were prepared via self-assembly of DNA building blocks and immobilized onto gold electrodes to give DNA tetrahedron-embedded substrates. Subsequently, aptamers that specifically recognize EpCAM were conjugated on CTCs using a unique multibranched hybridization chain reaction amplification. These aptamer-grafted CTCs were captured on the DNA tetrahedron-embedded substrates. Horseradish peroxidases were tagged onto the immobilized CTCs, enabling the devices to electrochemically detect CTCs with enhanced sensitivity.[204] By combining photolithography and reactive ion etching, nanorough islands (Rq = 1–150 nm) were generated on glass wafers for CTC capture.[205] In the absence of capture agents, these nanorough glass substrates exhibited a differential affinity to cancer cells rather than WBCs. Although these substrates were potentially applicable for CTC capture, concerns have been raised about the lack of both capture specificity and understanding of the molecular and cellular mechanisms behind this differential affinity.
2.3. Hierarchical Nanostructures
Hierarchical nanostructures represent a class of integrated architectures which are composed of either nanoscale building blocks in multiple dimensions or multiscale components.[206] By mimicking natural hierarchical structures, such as trees with trunks and branches,[127,146] flowers with multiple layers of petals,[207] or even cell surfaces with microvilli,[208,209] in the nanoscale configuration, hierarchical nanostructure-embedded substrates have their unique advantages of high surface contact areas and synergistic interactions (at both nanoscale and microscale) for improving the affinity of capturing CTCs.
2.3.1. Fractal Nanostructures
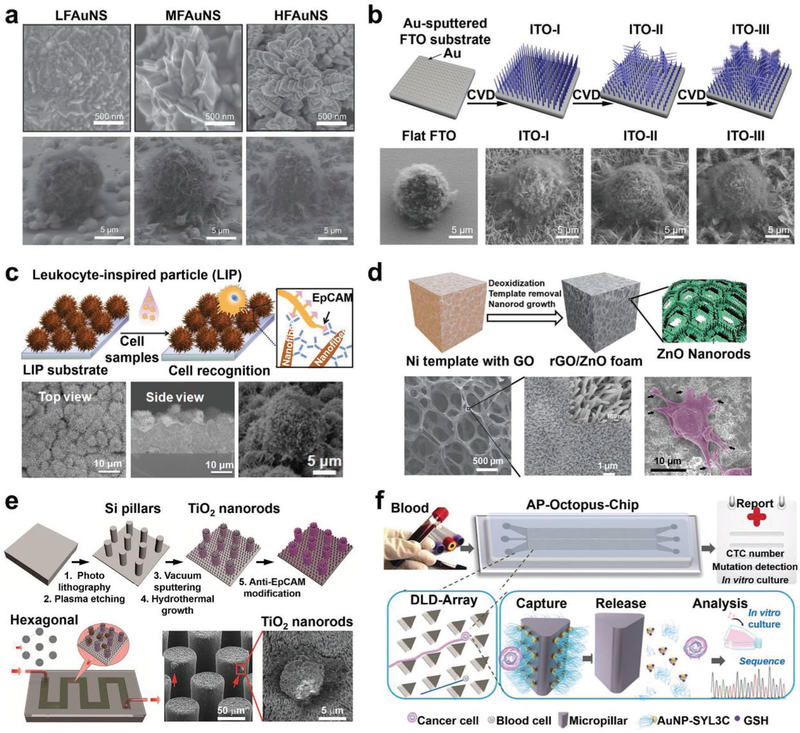
A representative hierarchical nanostructure is the fractal nanostructures which have branching patterns and can be split into several self-similar parts at nanoscale.[210] For example, fractal gold nanostructures (FAuNSs), which were electrochemically deposited on ITO substrates exhibited enhanced CTC capture performance.[127] By altering the electrochemical potential and supporting electrolyte, the fractal dimensions of FAuNS can be modulated. Anti-EpCAM was grafted onto the FAuNS-embedded substrates via the biotin–streptavidin-mediated conjugation. As shown in the SEM images (Figure 7a), with the fractal dimensions of the FAuNS increased, the cells captured on the FAuNS-embedded substrates possessed more well-expanded filopodia structures. The increase of fractal dimensions also led to improved cell capture performance. The substrates with the highest dimension of FAuNSs exhibited a CTC capture efficiency of 62%. Similarly, a fractal ITO nanowire-embedded substrate with both vertical and horizontal nanowire branches was fabricated by CVD, providing a new type of nanostructured substrates for efficient CTC capture (Figure 7b).[146] After conjugating with anti-EpCAM, fractal ITO nanowire-embedded substrates had an improved CTC capture yield of 89% while the ITO nanowire-embedded substrate without branches had a CTC capture yield of 67%.
Figure 7.
Hierarchical nanostructure-embedded substrates developed for CTC capture. a) SEM images showing cell morphologies and behaviors on FAuNSs with different fractal dimensions including low FAuNS (LFAuNS, fractal dimension ≈ 2.40), moderate FAuNS (MFAuNS, fractal dimension ≈ 2.54) and high FAuNS (HFAuNS, fractal dimension ≈ 2.70). Reproduced with permission.[127] Copyright 2013, John Wiley and Sons. b) A fractal ITO nanowire-embedded substrate with both vertical and horizontal nanowire branches was fabricated by CVD. SEM images (bottom) reveal cell morphologies and behaviors on a flat FTO substrate and different ITO nanowire-embedded substrates with/without branches, respectively. Reproduced with permission.[146] Copyright 2016, American Chemical Society. c) Schematic (top) and SEM images (bottom) of an anti-EpCAM functionalized LIP-embedded substrate for capturing CTCs. Reproduced with permission.[208] Copyright 2014, John Wiley and Sons. d) Freestanding reduced GO composite foams with ZnO nanorods (rGO/ZnO foam) were prepared and functionalized with anti-EpCAM for CTC capture. SEM images (bottom) show the structure of rGO/ZnO foams and cell morphologies on rGO/ZnO/anti-EpCAM foams. Reproduced with permission.[128] Copyright 2013, John Wiley and Sons. e) A nano–micro hierarchical microfluidic device was fabricated by growing TiO2 nanorod array on hexagonally patterned Si micropillars. Reproduced with permission.[213] Copyright 2016, Springer Nature. f) A schematic drawing of the AP-Octopus-Chip developed for CTC isolation. Reproduced with permission.[216] Copyright 2019, John Wiley and Sons.
2.3.2. Nano–Micro Hierarchical Substrates
Bioinspired nano–micro hierarchical substrates including leukocyte-inspired particles (LIPs),[208] cell replica surfaces,[209] flowerlike substrates,[207,211] and TiO2 nanosisal-like substrate,[212] have been prepared for improving CTC capture performance. Representatively, LIPs with vertically burgeoned nanofibers (Figure 7c) were prepared by the combination of thermal oxidation and CVD.[208] After grafting anti-EpCAM, LIP-embedded substrates had a capture yield of 62% for CTCs in the culture medium, and 50–58% for CTCs in whole blood. Hierarchically topographic interactions occurred between LIPs and CTCs at both microscale and nanoscale, and the increased surface area helps increase its affinity to capture cells. Cell replica surfaces were fabricated by the silica bioreplication to faithfully replicate the surface structural features of MCF7, PC3, or T24 cells.[209] With anti-EpCAM conjugation, these cell replica-embedded substrates captured CTCs with efficiencies ranging from 53% to 62%. Flowerlike zinc phosphate-based hierarchical nanostructured substrate (HZnPNS) was transformed from ZnO nanowires by a low-temperature hydrothermal method.[211] Anti-EpCAM-modified HZnPNS had a capture efficiency of 90%.
Another type of nano–micro hierarchical nanostructures is fabricated by growing nanostructures on microstructures embedded on substrates, producing dual-scale cell capture effects. Yin et al. prepared a hierarchical nanostructured graphene platform[128] by growing ZnO nanorod arrays (diameters = 40–60 nm, lengths = 1–2 μm) on a freestanding rGO foam (rGO/ZnO foam) in an aqueous solution (Figure 7d). The microporosity of rGO foam allowed small RBCs to pass through. Meanwhile, the high density of ZnO nanorods increased the surface area for grafting anti-EpCAM. The rGO/ZnO/anti-EpCAM foam had a CTC capture yield of 80%. Microposts or micropillars have been utilized to fabricate “CTC-chips,”[85] and DLD patterned microchips,[89,90] making significant progress in CTC capture and isolation. To further improve the capture performance, nanostructures are superposed on microposts or micropillars to generate new nano–micro hierarchical substrates. For example, TiO2 nanorods were grown on hexagonally patterned Si micropillars (Figure 7e).[213] By conjugating with anti-EpCAM and further integrating with microfluidics, this nano–micro hierarchical substrate showed a CTC capture yield of 77%. Additionally, anti-EpCAM-functionalized nanoparticles or nanospheres such as GO-coated Fe3O4 magnetic nanoparticles (GO–MNPs, diameter = 250 nm)[214] and SiO2-coated Fe2O3 nanospheres (diameter ≈ 357 nm)[215] can be immobilized on nickel micropillars or squares which were aligned in microfluidic chips for capturing CTCs under an external magnetic field. The staggered microscale pillars or squares increased cell contact frequency, while the nanoparticles or nanospheres offered a high density of anti-EpCAM, resulting in enhanced CTC capture yields of 70–94%. In another example, by mimicking the multivalent tentacles of the octopus, an aptamer-tailed octopus-chip (AP-Octopus-Chip; Figure 7f) was engineered by immobilizing multivalent aptamer-functionalized nanospheres (apt-AuNPs, 250 aptamers per AuNP) on DLD-based rotated triangular micropillars that were embedded in a microfluidic chip.[216] CTCs with diameters larger than the critical diameter (Dc = 13 μm) had frequent collisions with micropillars, while WBCs with smaller diameters flowed through with much less chance of collision, resulting in a size-selective contact enhancement. The rotated triangular micropillars generated a smooth, hydrodynamic gradient and decreased the flow velocity, increasing the time available for cells to contact the micropillars. Additionally, multivalent aptamers on AuNPs had a stronger binding affinity with CTCs than individual aptamers. As a result, the AP-Octopus-Chip achieved capture yields of 89% for CTCs in culture medium and 74–84% for CTCs in whole blood.
3. Strategies for Rare-Cell Retrieval from Nanostructured Substrates
The aforementioned nanostructured substrates offer powerful technologies for highly efficient CTC/CFNC capture. However, as the demand for postcapture molecular and functional analyses increases, many nanostructured substrates encountered the challenge of retrieving captured CTCs/CFNCs from nanostructured substrates while maintaining cell integrity and viability, and minimum contamination of surrounding cells. To retrieve the captured cells, one has to overcome the adhesive forces that were used to increase cell capture efficiency, i.e., capture agent–cell affinity, and focal adhesion.[217] It is reported that flowing fluid shear stress as large as 180 dyne cm−2 can release approximately 50% of cells captured by antibody in a microchannel.[188] However, this approach leaves many of the released cells damaged or killed. It is reasonable to expect an even larger force would be required if this approach is to be used in nanostructured substrate-based CTC/CFNC capture devices, given their enhanced adhesive forces. Counteracting the adhesive forces without destroying cells has become the primary focus of ongoing technologic development. In addition, as many nanostructured substrate-based CTC/CFNC capture devices do allow nonspecific adhesion of background hematologic cells, a cell retrieval approach more selective to CTCs or CFNCs is desired for the ease of downstream analysis. The CTC/CFNC retrieval strategies listed in Figure 2 have been developed to retrieve the captured cells selectively or nonselectively, with or without physical or chemical destruction of the nanostructured substrates. Here we categorize these cell retrieval approaches on nanostructured substrates into i) laser microdissection;[129] ii) stimulus-responsiveness, e.g., photoresponsiveness,[218] thermoresponsiveness,[130] electrical stimulation,[131] mechanoresponsiveness[132] and magnetic field switch;[214] iii) additive reagent-triggered release, e.g., chemical reaction,[134] enzymatic digestion,[135] and DNA hybridization.[136] Some of the approaches can be performed in a focal or selective manner to harvest individual target cells with the assistance of fluorescence microscopy. Release efficiency (percentage of retrieved cells in captured cells), operating time, and viability and purity of the retrieved cells are the major parameters for evaluating different rare-cell retrieval strategies.
3.1. Laser Microdissection
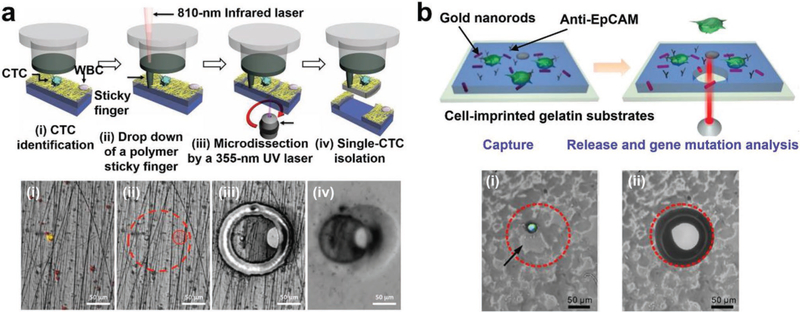
Laser microdissection represents the most straightforward approach to isolate individual CTCs/CFNCs that are immobilized on transparent nanostructured substrates,[129,163] allowing for downstream molecular analysis of the recovered CTCs/CFNCs.[159] The PN-NanoVelcro substrates[129] were first prepared by depositing electrospun PLGA nanofibers onto commercial LMD glass slides with predeposited PPS membranes, followed by conjugation of CD-146 antibodies. After capturing CMCs from melanoma patients’ blood samples, an LMD microscope equipped with a 355 nm laser beam was used to cut out the PLGA nanofibers together with the PPS membranes to isolate CMCs. The individually isolated CMCs were then subjected to mutation analysis by Sanger sequencing.[129] To avoid cell loss caused by the static charge, the process was further improved[163] by using an infrared LCM cap to drop down an IR-laser “sticky finger” to facilitate the isolation and characterization of CTCs (Figure 8a) in prostate cancer patients’ blood samples. In addition to CTC and CMC analyses, imprinted PLGA NanoVelcro Chips coated with anti-EpCAM have been utilized to enrich, isolate, and characterize individual cTBs in maternal blood for NIPD applications.[159] After being captured on the substrates, single cTBs were isolated with LCM techniques, followed by downstream genetic analysis. However, LMD/LCM is costly, labor-intensive, and time-consuming. The released cells are often nonviable due to the accompanied staining procedure or prolonged operating time. Further, the exposure of cells to UV light may damage their genetic materials and denature cellular proteins, undermining the downstream molecular characterization of these isolated cells.
Figure 8.
LCM and photoresponsive approaches used for CTC retrieval from nanostructured substrates. a) Schematic and micrograph images showing the process of single-CTC isolation from PN-NanoVelcro Chips with the use of LCM (a.k.a., PN-NanoVelcro/LCM). Reproduced with permission.[163] Copyright 2013, John Wiley and Sons. b) Schematic and micrograph images of the NIR-responsive approach for cell retrieval, enabling the capture and site-specific release of CTCs from the GNR-embedded gelatin substrates. Reproduced with permission.[133] Copyright 2016, American Chemical Society.
3.2. Stimulus-Responsiveness
The use of stimulus-responsive molecules or surfaces on nanostructured substrates offers an opportunity for retrieving CTCs/CFNCs in response to a specific physical stimulus such as changes in light, temperature, electrical potential, mechanical force, or magnetic field. These stimulus-responsive components usually have modulable properties such as wettability, solubility, adsorption ability, and cell affinity, that lead to a dramatic transformation or dissociation of their nanostructures and a disruption of cell adhesive properties.
3.2.1. Photoresponsiveness
The photoresponsive approach for cell retrieval enables a simple and site-specific recovery of CTCs immobilized on nanostructure-embedded substrates. Near-infrared (NIR) laser-responsive substrates were developed for conducting efficient CTC capture and site-specific release by the combined use of the photothermal transfer effect of gold nanorods (GNRs) and the thermoresponsive dissolution of gelatin hydrogel (Figure 8b).[133] Prepared by imprinting target cancer cells on GNR-embedded gelatin hydrogels and functionalized with anti-EpCAM, the substrates were capable of capturing CTCs with enhanced performance (92%). By locally irradiating a specific area of the substrate (where CTCs were immobilized) with a NIR laser beam, the embedded GNRs absorbed the laser beam, leading to the increased local temperature of gelatin hydrogel. At this temperature, gelatin hydrogels underwent a transition from gel to sol, resulting in the site-specific release of individual CTCs. This photoresponsive approach gave a CTC release efficiency of 92%, and the viability of released CTCs was as high as 90%.
3.2.2. Thermoresponsiveness
Thermoresponsive polymer brushes (i.e., PNIPAAm) which can undergo temperature-dependent conformational changes show an important application in the controllable and gentle bulk release of captured CTCs from nanostructured substrates. Created by coating the polymer brushes onto SiNW-embedded substrates and functionalizing anti-EpCAM onto these polymer brushes, thermoresponsive NanoVelcro substrates were able to capture and release CTC efficiently at 37 and 4 °C, respectively (Figure 9a).[130] The conformational changes of polymer brushes in response to temperature changes could effectively alter the accessibility of the anti-EpCAM, allowing for rapid CTC retrieval while maintaining CTC viability (90%) and molecular integrity. Furthermore, this thermoresponsive NanoVelcro substrate was integrated into a microfluidic device with a Peltier cooling/heating pad to make a CTC purification system with improved CTC purity (88–98%) after two rounds of capture/release.[219] In another similar study, anti-EpCAM was conjugated to the hydrophobic anchor biotin-BSA which could be adsorbed on the PNIPAAm-coated SiNWs at 37 °C and desorbed from the hydrophilic surface at 20 °C (Figure 9b).[220] This thermoresponsive mechanism resulted in 99% of captured MCF-7 cells being retrieved from PNIPAAm-coated SiNW substrate.
Figure 9.
Thermoresponsive CTC retrieval strategies applied on nanostructured substrates. a) Thermoresponsive NanoVelcro substrates for efficient CTC capture/release at 37 and 4 °C caused by the exposure/internalization of anti-EpCAM grafted on polymer brushes of NanoVelcro substrates. Reproduced with permission.[130] Copyright 2013, John Wiley and Sons. b) PNIPAAm-coated SiNW substrates for reversible cell capture/release due to the change of hydrophobic and hydrophilic surface properties between 37 and 20 °C. Reproduced with permission.[220] Copyright 2012, John Wiley and Sons. c) A schematic view of the fabrication and working mechanisms of thermal-sensitive PAPDEA–GO composite-embedded microfluidic devices for CTC capture/release. Reproduced with permission.[198] Copyright 2016, John Wiley and Sons. d) Thermoresponsive CTC capture/release mechanisms using gelatin-coated PDMS substrates. Reproduced with permission.[132] Copyright 2015, John Wiley and Sons.
In addition to polymer brushes, the thermoresponsive change of solubility of copolymers and gelatin was also employed for modulating CTC capture/release. Copolymer poly(N-acryloyl piperidine-co-N,N-diethyl acrylamide) (PAPDEA) is insoluble above 13 °C but becomes soluble below this temperature. Thermoresponsive PAPDEA–GO composite-embedded microfluidic devices[198] (Figure 9c) were first fabricated by drop-casting the PAPDEA–GO on the substrates and functionalized with anti-EpCAM for enhanced CTC capture (efficiencies = 85–95%). After performing CTC capture in the devices, CTCs were released with 92–95% efficiencies and 92% viability by flowing PBS solution at 5 °C. This low temperature induced the dissolution of PAPDEA–GO composite, allowing for a mild CTC retrieval condition. Gelatin undergoes a gel-to-sol transition when the temperature exceeds 32 °C. Gelatin-coated PDMS substrates[132] were prepared by introducing biotinylated gelatin onto streptavidin-grafted PDMS substrates, followed by deposition of streptavidin-grafted polystyrene nanoparticles. After anti-EpCAM conjugation, the nanoparticle-embedded substrates were employed for CTC capture (efficiencies of 75–96%) at room temperature. Upon raising the experimental temperature to 37 °C (Figure 9d), gelatin polymer within the device underwent a gel-to-sol transition, leading to the bulk release of CTCs captured in the devices with an average efficiency of 93%.
3.2.3. Electrical Stimulation
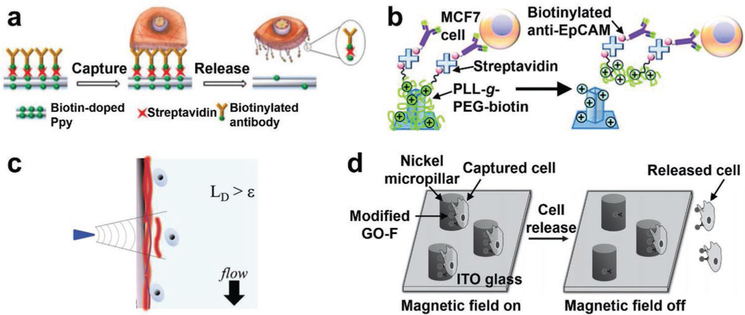
By incorporating electrically responsive functional groups into the cell affinity substrates, CTCs/CFNCs immobilized on the substrates can be triggered for cell retrieval by electrical stimulation. These electrical stimulation-mediated CTC/CFNC retrieval approaches exhibited advantages of short time-consuming (varying from a few seconds to minutes), high efficiency, and high cell viability. For example, by applying a negative electrochemical potential of −1.2 V, the CTCs captured on the aforementioned FAuNS-embedded substrates[127] (Figure 5a) could be released as a result of reductive cleavage of the Au–S bonds. The general CTC recovery performance was 98% and the recovered CTCs had a viability of 95%. Similarly, AuNW-embedded substrates[153] modified with thiol-terminal aptamers are also capable of capturing and releasing CTCs via the formation and reductive cleavage of Au–S bonds.
Electrical stimulation was employed to achieve on-demand drug release from conducting polymer materials such as Ppy[184] and PEDOT.[156,174] The same strategy was adopted to incorporate biotin into Ppy nanostructure-embedded microfluidic devices[184,221] to enable CTC capture and electrically stimulated CTC release (Figure 10a). The devices exhibited a CTC capture efficiency >90%, and a CTC release efficiency >90% upon the application of a potential of −0.8 V. The negative electrochemical potential led to the biotin release from the Ppy nanostructure-embedded substrates as a result of the shrinkage of conductive polymer backbones. This electrically stimulated CTC release happened fairly quickly (2–15 s). fNRBCs could also be electrically retrieved by using biotin-Ppy nanoparticle-embedded microchips, where anti-CD147 was used as the capture agent.[195] PEDOT nanorod-embedded bioelectronic substrates[156] have also been utilized for efficient CTC capture/release. Here, poly-(l)-lysine-graft-poly-ethylene-glycol (PLL-g-PEG-biotin) was introduced onto the substrates via charge interactions, allowing for conjugation of anti-EpCAM capture agent (Figure 10b). After CTC capture, enhanced CTC release performance was achieved by carrying out cyclic voltammetry between −0.8 and 0.5 V. This strategy was also employed on PEO/PEDOT:PSS nanofiber-embedded indium tin oxide electrodes for programming capture/release of CTCs with a capture efficiency of 90% and a release efficiency of 87%.[174]
Figure 10.
Electrically stimulated, mechanoresponsive, and magnetic field-mediated CTC retrieval on nanostructured substrates. a) CTC capture and electrically stimulated release mechanisms of biotin-Ppy nanostructure-embedded substrates. Reproduced with permission.[184] Copyright 2014, Ivyspring International Publisher. b) Electrically stimulated CTC release via the desorption of PLL-g-PEG-biotin from PEDOT nanorod-embedded bioelectronic substrates. Reproduced with permission.[156] Copyright 2015, Royal Society of Chemistry. c) The mechanoresponsive cell retrieval strategy to recover individual CTCs from gelatin-based PDMS substrates. Reproduced with permission.[132] Copyright 2015, John Wiley and Sons. d) The magnetic field-mediated CTC capture/release mechanisms of the GO–Fe3O4 nanoparticle-decorated micropillar devices. Reproduced with permission.[214] Copyright 2013, John Wiley and Sons.
3.2.4. Mechanoresponsiveness
Mechanical cell retrieval methods were previously considered unfavorable due to the risk of damaging cells. It is worth noting that recent advances in material science have led to several mechanoresponsive nanostructured substrates that can effectively release cells without causing significant damage. Shear-responsive dissolution of thixotropic hydrogels (such as gelatin that was coated on the aforementioned PDMS substrates[132]) is a unique mechanoresponsive strategy for individually releasing captured CTCs (Figure 10c). After capturing CTCs with anti-EpCAM on gelatin-coated PDMS substrates, a frequency-controlled microtip was used to produce a tunable vibration at the surface of substrates, generating a normal inertial force to cause the flow (i.e., shear-thinning) of gelatin nanocoating and the selective release of single CTCs. The retrieved CTCs possessed 92% viability and were suitable for downstream single-cell functional and molecular analysis.
3.2.5. Magnetic Field
The use of magnetic nanoparticles in the microfluidic devices offers an opportunity for capture/release of CTCs by simply switching on/off external magnetic fields without applying any complicated external stimuli.[214,215,221] In a new category of micropillar-embedded microfluidic devices,[214] anti-EpCAM-conjugated GO–Fe3O4 nanoparticles were densely packed onto the surfaces of micropillars to enhance CTC capture (efficiency > 70%) in the presence of magnetic fields. After removing magnetic fields, the captured CTCs could be released as a result of the detachment of magnetic nanoparticles from micropillars (Figure 10d). A similar mechanism was implemented in the Fe2O3 nanosphere-embedded microfluidic system.[215] Moreover, magnetic up-conversion nanoparticles could be pretagged onto CTCs to facilitate CTC capture/release in SiNW-embedded microfluidic chips by applying magnetic fields.[221]
3.3. Additive Reagent-Triggered Release
3.3.1. Chemical Reaction
A competitive binding approach relies on the binding of ligand molecules to receptors, antibodies, or other macromolecules. In the presence of high-affinity ligands, thermodynamic principles drive the ligand exchange for releasing the ligands with low binding affinity. Such a competitive binding approach has been adopted for on-demand CTC retrieval in conjunction with the use of nanostructured substrates. At a physiologically compatible pH value (pH 7–9), boronic acids can reversibly bind with 1, 2- or 1, 3-diols in carbohydrate molecules, leading to the formation of cyclic ester complexes. Polymerization of boronic acid-containing 3-acrylamidophenylboronicacid (3-AAPBA) onto SiNW arrays led to the production of polyAAPBA-SiNW-embedded substrates.[134] The polyAAPBA-SiNW-embedded substrates exhibited a dual-responsive CTC capture/release mechanism via precise control of both pH and glucose concentration. Under a weakly acidic condition (pH 6.8), the boronic acid groups on SiNW arrays bind with sialic acid on CTC membranes, enabling CTC capture. Under an alkaline condition (pH 7.8), the exposure of 70 × 10−3 m glucose led to the release of immobilized CTCs as a result of competitive binding between glucose and boronic acid group (Figure 11a). Similarly, the aforementioned 3D porous polyAPBA nanostructured substrates also employed the competitive binding between fructose and boronic acid for CTC capture/release.[180] Recently, boronic acid-grafted PEDOT NanoVelcro chips were developed to purify prostate cancer CTCs.[222] Anti-EpCAM was conjugated onto NanoVelcro substrates via boronic acid–oligosaccharide bonding, conferring CTC capture specificity and sensitivity to the devices. In this case, sorbitol was introduced into the devices, and the competitive binding between sorbitol and boronic acid resulted in CTC release with an efficiency of 95% and viability of 96% (Figure 11b). CTCs purified by this chip provided well-preserved RNA transcripts for downstream analysis.
Figure 11.
Chemical reaction-mediated CTC retrieval strategies on nanostructured substrates. a) pH and glucose dual-responsive SiNW-embedded substrates for capturing and releasing CTCs. Reproduced with permission.[134] Copyright 2013, American Chemical Society. b) PBA-grafted PEDOT Nano-Velcro chips with anti-EpCAM conjugation for purifying CTCs. Reproduced with permission.[222] Copyright 2017, John Wiley and Sons. c) A schematic representation showing the fabrication process and CTC capture/release mechanisms of NP-HBCTC-Chips. Reproduced with permission.[192] Copyright 2017, American Chemical Society. d) Oxalic acid induced the reduction and dissolution of MnO2 nanofibers to release captured CTCs. Reproduced with permission.[161] Copyright 2015, AIP Publishing. e) A schematic illustration of “Click Chips” leveraging bioorthogonal ligation-mediated CTC capture and disulfide cleavage-driven CTC release in an SiNW-embedded microfluidic system.
The Au–S bonds utilized to graft CTC capture agents onto AuNP-embedded substrates can be cleaved through the competitive binding of other thiol molecules like glutathione (GSH), enabling CTC retrieval from AuNP-embedded substrates. AuNP-HBCTC-Chips were prepared by assembling thiol-modified AuNPs in HBCTC-Chips and conjugating anti-EpCAM onto these AuNPs (Figure 11c).[192] CTCs were captured by the anti-EpCAM-conjugated AuNPs in the device with efficiencies ranging from 68% to 96%. Upon the exposure to GSH for 30 min, CTCs were released from the AuNP-embedded substrates as a result of the competitive binding of GSH and AuNPs which led to the cleavage of Au–S bonds sandwiched between AuNPs and anti-EpCAM. This approach achieved the release of CTCs with an efficiency of 90% and the viability of 78%. Similarly, the aforementioned AP-Octopus-Chip (Figure 7d) also employed the GSH-mediated CTC retrieval strategy to achieve CTC release from the aptamer-modified AuNPs with an efficiency of 80% and the viability of 96%.[216]
Acidic treatment of nanostructured substrates could facilitate the recovery of CTCs by breaking cell-substrate interactions as a result of the dissolution of the embedded nanosubstrates or cleavage of chemical linkers that graft capture agents. MnO2 could be transformed into water-soluble Mn2+ ions after oxalic acid reduction. MnO2 nanospheres[189] and nanofibers[161] that were functionalized with anti-EpCAM for capturing CTCs could be dissolved by oxalic acid, resulting in the release of captured CTCs (efficiencies = 88–92%; Figure 11d). The aforementioned anti-EpCAM-modified HZnPNS[211] could also be rapidly dissolved by sodium citrate into the form of Zn2+, releasing 88% of captured CTCs with a viability of 92%. Similarly, ZnO nanograss-embedded substrates with anti-EpCAM conjugation were reported to capture CTCs and then release them by treating with 0.1 M hydrochloric acid (HCl) solution (pH 5.6).[223] Alternatively, benzoic imine presented an acid-sensitive linker and was utilized to graft arginine–glycine–aspartic acid (RGD) peptides on PVA/PEI nanofiber-embedded microfluidic devices.[224] The RGD peptides captured αvβ3-overexpressing cancer cells in the devices. Under weakly acidic conditions (pH 6.8), these immobilized cancer cells were released from nanofiber-embedded microfluidic devices as a result of the acid-sensitive cleavage of benzoic imine. It is worth noting that acid concentrations and treatment time must be carefully controlled to avoid negatively impacting cell viability.
Redox-sensitive disulfide bonds can serve as linkers for grafting capture agents and can be destroyed by reductants such as GSH, tris(2-carboxyethyl)phosphine (TCEP) and dithiothreitol (DTT) to release CTCs from the nanostructured substrates. GSH (50 × 10−3 m)-mediated disulfide cleavage has been utilized to release CTCs that were captured on anti-EpCAM-modified SS-biotin-Ppy nanowire-embedded substrates,[152] anti-EpCAM-conjugated flowerlike substrates,[207] and HA-functionalized chitosan nanofiber-embedded substrates.[166] The efficiencies were all above 85%, and the viabilities ranged from 80% to 98%. However, the incubation time of 40–60 min for GSH treatment is relatively long. Dong et al. created “Click Chips”[225] that leverage bioorthogonal ligation-mediated CTC capture and disulfide cleavage-driven CTC release in an SiNW-embedded microfluidic system (Figure 11e). CTCs were captured by inverse-electron-demand Diels-Alder reactions between trans-cyclooctene (TCO) and tetrazine (Tz) with enhanced capture efficiency (94% with TCO-anti-EpCAM as low as 0.1 ng) and specificity (≈2000 WBCs). The incorporation of disulfide bonds into the surface linkers tethering Tz motifs allowed for the disulfide cleavage-driven CTC release from Click Chips upon exposure of 50 × 10−3 m DTT for 12 min with an efficiency of 80% and significantly enhanced specificity (≈200 WBCs). This rapid CTC purification platform is ideal for performing CTC-mRNA assays. Similarly, TCEP was also used to mediate the disulfide cleavage-driven CTC release in FA-modified PEI/PVA nanofiber-embedded devices with efficiencies of 69–98%.[171]
3.3.2. Enzymatic Digestion
Enzymes can directly digest the proteins or nucleic acids that mediate CTC capture on nanostructured substrates, allowing for the release of captured CTCs. The most commonly used approach has been to use proteolytic enzymes, like trypsin (0.05–0.25%), to resuspend cells via digestion of extracellular adhesive proteins at 37 °C for a few minutes. Trypsin was used to recover MCF7 cancer cells that were captured on TiO2 nanosisal-like substrates (Figure 12a)[212] and BT20 cancer cells that were captured by a quartz nanohole-embedded microfluidic system with a release efficiency of 90% and cell viability of 75%.[182] However, trypsinization is a nonspecific cell retrieval strategy that often results in low purity of CTCs in the final cell suspension. The duration of trypsinization should be carefully controlled since prolonged trypsinization leads to cell membrane damage and decreased viability. Nucleases, including exonucleases and endonucleases, are another enzymatic tool that can be used in the development of biosensors.[226–228] Nucleases can offer a specific method for releasing captured CTCs from aptamer-immobilized substrates through the cleavage of the phosphodiester bonds of DNA aptamers.[229] NSCLC CTCs captured by aptamer cocktails on NanoVelcro chips could be recovered by treatment with benzonase nuclease at 37 °C for 15 min (Figure 12b). Through the digestion of DNA aptamers, the nuclease-mediated CTC retrieval approach achieved a release efficiency of 85% and cell viability of 78–83%.[135]
Figure 12.
Enzymatic digestion and DNA hybridization-mediated CTC retrieval from nanostructured substrates. a) TiO2 nanosisal-like substrates captured CTCs with anti-EpCAM and released CTCs upon trypsin treatment. Reproduced with permission.[212] Copyright 2015, American Chemical Society. b) Benzonase nuclease-mediated digestion of DNA aptamers to release captured NSCLC CTCs from NanoVelcro chips. Reproduced with permission.[135] Copyright 2013, John Wiley and Sons. c) Enzymatically degradable ALG/PAH nanofilm-embedded substrates for CTC capture/release. Reproduced with permission.[230] Copyright 2015, Elsevier. d) Aptamer recognition and DNA hybridization-mediated CTC capture and release on pCBMA-chitosan nanofiber-embedded substrates. Reproduced with permission.[136] Copyright 2016, John Wiley and Sons.
Enzyme-degradable nanostructured substrates provide an alternative means for recovering CTCs/CFNCs. Anionic polymers such as alginate (ALG) can be degraded by bacterial enzymes like alginate lyase through the cleavage of the polymeric backbones. By assembling anionic polymers together with the oppositely charged cationic polymers like poly(allylamine hydrochloride) (PAH), layer-by-layer ALG/PAH nanofilms were generated on the internal walls of HBCTC-Chip for conducting CTC capture and release.[230] After using anti-EpCAM to capture CTCs, ALG/PAH nanofilms were degraded by introducing alginate lyase into the devices, resulting in CTC release with 95% efficiency and 90% viability (Figure 12c). Similarly, the aforementioned gelatin nanoparticle-embedded microchips achieved the capture/release of fNRBCs.[196] After capturing fNRBCs with anti-CD147, the gelatin nanoparticles were degraded by matrix metalloproteinase-9, resulting in the release of captured fNRBCs with an efficiency of 89% and viability of 90%.
3.3.3. DNA Hybridization
The hybridization of aptamers with the complementary oligonucleotides has emerged as a unique approach for CTC retrieval from aptamer-grafted nanostructured substrates. There are two different mechanisms for DNA hybridization-mediated CTC retrieval. The first mechanism is the hybridization of complementary oligonucleotides with the functional regions of aptamers to cause a conformational change of aptamers and destruction of aptamer-cell affinity. The aforementioned chitosan nanofiber-embedded substrates with aptamer conjugation have achieved efficient CTC capture and release (Figure 12d).[136,165] After CTC capture, complementary oligonucleotides were hybridized with aptamers and opened their secondary structures, resulting in CTC release with an efficiency of 98%. The second mechanism is the hybridization of complementary oligonucleotides with the nonfunctional regions of aptamers to induce strand displacement of immobilized DNA linkers and the release of aptamers along with captured CTCs. By employing this mechanism, aptamer-functionalized HZnPNS-embedded microfluidic chips could release captured CTCs with an efficiency of 86% and viability of 81%.[231]
4. Clinical Applications of Nanostructured Substrate-Based CTC Assays
CTC represents an emerging biomarker for predicting prognosis. The goal of performing “liquid biopsy” using CTCs is to assess tumor characteristics in a noninvasive manner with low cost and high feasibility, paving the way toward personalized oncology. CellSearch, as the only FDA cleared CTC assay, has demonstrated CTC count as an independent indicator of survival in many metastatic cancers.[66] However, as the demand for in-depth analysis of CTCs continues to grow, there has been a constant call for next-generation CTC assays that consume less blood (compared to 7.5 mL per assay for CellSearch) and have higher sensitivity and better compatibility with downstream molecular and functional characterization. Nanostructured substrate-based CTC isolation/capture technologies have attracted attention due to higher cell capture efficiencies and sensitivities that could greatly reduce blood consumption. The cell retrieval strategies further facilitate downstream molecular and functional analyses of CTCs with improved accuracy and potentially a significantly reduced cost. In this section, we highlight several applications of nanostructured substrate-based CTC assays (Figure 13) covering i) enumeration, morphologic analysis, and subclassification,[20,21] ii) genetic and genomic analysis, iii) transcriptomic profiling, and iv) protein analysis, with emphasis on their potential clinical value.
Figure 13.
Clinical applications of nanostructured substrate-enabled CTC assays. a) Enumeration. Reproduced with permission.[232] Copyright 2018, John Wiley and Sons. Morphologic analysis. Reproduced with permission.[120] Copyright 2018, Elsevier. Subclassification. Reproduced with permission.[24] Copyright 2015, John Wiley and Sons. b) Genetic and genomic analysis (e.g., fluorescence in situ hybridization (FISH). Reproduced with permission.[198] Copyright 2016, John Wiley and Sons. Whole genome amplification (WGA). Reproduced with permission.[120] Copyright 2018, Elsevier. Sanger sequencing and single-CTC whole genome sequencing (WGS). Reproduced under the terms of the CC-BY license.[233] Copyright 2015, Jiang et al., published by Oncotarget). c) Transcriptomic profiling (e.g., next-generation RNA sequencing. Reproduced with permission.[192] Copyright 2017, American Chemical Society. Single-cell reverse transcription-polymerase chain reaction (RT-PCR) analysis and NanoString nCounter). d) Protein analysis (e.g., multiple protein expression. Reproduced with permission.[87] Copyright 2012, American Association for the Advancement of Science). Reproduced with permission.[236] Copyright 2018, American Chemical Society.
4.1. CTC Enumeration, Morphologic Analysis, and Subclassification
CTC enumeration has shown its value in detecting the early development of metastasis, assessing therapeutic response, and evaluating overall survival (OS) in clinical trials.[10,41,237] It has been well recognized that CTCs present a heterogeneous population,[54] with some subsets possibly providing more clinically insightful information than others. Many nanostructured substrate-embedded CTC assays capture CTCs over large surface areas with minimal vertical variation, serving as ideal settings for imaging. With the aid of high-resolution imaging technologies, morphologic variations and distinct phenotypic subpopulations can be identified, many of which are closely associated with disease progression and adverse outcomes.
4.1.1. Prostate Cancer CTCs
As an early proof-of-concept, prostate cancer CTCs were used as the model to optimize and test the capture performance of the original NanoVelcro Chip (with anti-EpCAM conjugation).[158] A three-color immunocytochemistry (ICC) protocol was adopted by using anti-CK for identification of epithelial-derived CTCs, anti-CD45 for identification of WBCs and DAPI for nuclear staining (Figure 14a). CTCs (CK+/CD45−/DAPI+, 10 μm < sizes < 40 μm) were distinguished from the background WBCs (CK−/CD45+/DAPI+, sizes < 15 μm) and cellular debris. The NanoVelcro Chip could detect CTCs using down to 1 mL blood and exhibited consistency in CTC capture performance for patient blood samples.[238] A side-by-side comparison of NanoVelcro Chips and CellSearch assay for 100 prostate cancer patients showed concordant CTC enumeration results (Figure 14b),[120,147] including a significant reduction in CTC count after the initiation of anticancer therapy and elevation at tumor progression. It was noted that on occasion WBCs appeared CK-positive and might be falsely counted as CTCs in CellSearch, resulting in exceptionally high CTC counts in a few cases. This problem was minimized with the use of higher resolution fluorescence microscopy and routine cytopathologic review of NanoVelcro assays. These approaches not only improved the accuracy of CTC identification but also inspired researchers to perform more detailed morphologic analysis for cellular and subcellular features of CTCs.
Figure 14.
Nanostructured substrate-enabled enumeration and subclassification of prostate cancer CTCs. a) A three-color ICC protocol (left) and representative fluorescent images (right) for distinguishing CTCs from nonspecifically trapped WBCs. Cytokeratin was shown in green, CD45 in orange and nuclear staining in blue (scale bars, 10 μm). Reproduced with permission.[120] Copyright 2018, Elsevier. b) Comparison of CTC enumeration by NanoVelcro Chip and CellSearch Assay for mCRPC patients. c) Three subpopulations of CTCs captured by NanoVelcro Chips, i.e., vsnCTCs, snCTCs, and lnCTCs, were identified by ICC protocol and nuclear size measurements (scale bars, 10 μm). (Left) Reproduced with permission.[24] Copyright 2015, John Wiley and Sons. Prostate cancer patients were placed into three cohorts according to their metastatic status. Right reproduced with permission.[120] Copyright 2018, Elsevier. d) Examples of CTC clusters captured by the graphene oxide chip (left). CK 7/8 was shown in red (scale bar, 10 μm). CTC clusters had a heterogeneous size ranging from two to eight cells within one cluster (right). Reproduced under the terms of the CC-BY License.[197] Copyright 2018, Wiley-VCH.
In a subsequent study, a detailed morphological analysis focusing on subcellular and nuclear features of CTCs identified a morphologic subset of CTCs associated with lethal visceral progression in prostate cancer. CTC enumeration was performed with NanoVelcro CTC assay using serial blood specimens from a wide spectrum of prostate cancer patients with stages ranging from localized to advanced metastatic disease.[239] The mathematical modeling and unsupervised clustering on the CTC nuclear size distribution identified three distinct subpopulations of CTCs (Figure 14c) that were named very-small-nuclear CTCs (vsnCTCs, nuclear size < 8.5 μm), small-nuclear CTCs (snCTCs, nuclear size between 8.5 and 15.0 μm), and large-nuclear CTCs (lnCTCs, nuclear size > 15.0 μm). VsnCTCs and snCTCs were more frequently seen in metastatic prostate cancer patients’ blood. Most importantly, the presence of vsnCTCs was highly associated with visceral metastases (VM). Further investigations on the correlation of vsnCTCs and VM showed that vsnCTCs might serve as a putative biomarker for VM in metastatic castration-resistant prostate cancer (mCRPC).[24] These discoveries were attributed to the addition of morphologic categorization (i.e., nuclear size) to the enumeration of CTCs. Similar findings involving CTCs with smaller nuclei and their association with more aggressive variants of mCRPC were reported by other groups using non-nanostructured substrate-based CTC detection methodologies.[25,240]
Multicellular aggregates of CTCs (more than two or three CTCs in a group) termed CTC clusters are more competent than individual CTCs in forming metastasis. CTC clusters have been shown to be potentially useful markers for metastasis in prostate cancer patients.[197] The aforementioned GO Chips[126] have been used to isolate both individual CTCs and CTC clusters (Figure 14d) from whole blood samples from 41 patients with mCRPC.[197] These CTC clusters consisted of 2–8 CTCs per cluster with the majority containing less than 4 CTCs. The fraction of captured CTCs that were part of CTC clusters varied widely in prostate patients from 0% to 54.8%.
4.1.2. Breast Cancer CTCs
As one of the most extensively studied cancers in the liquid biopsy field, breast cancer has been used to verify the feasibility of many nanostructured substrate-based CTC assays. The microfluidic device equipped with a gelatin-coated nanostructured substrate was tested using blood samples collected from breast cancer patients.[132] CTCs were detected in 7 out of 8 metastatic breast cancer patients (0–159 CTCs/3.5 mL, median = 13 CTCs/3.5 mL, mean = 29 ± 18.7 CTCs/3.5 mL). The NP-HBCTC-Chip (a herringbone microfluidic chip coated with gold nanoparticles) was designed to release captured CTCs upon GSH treatment and tested on blood samples from four metastatic breast cancer patients.[192] The study reported CTC counts ranging from 6 to 12 CTCs mL−1 (median = 7.4 CTCs mL−1, mean = 8.2 ± 2.7 CTCs mL−1). Clusters of CTCs were observed in both studies. Despite success in demonstrating the feasibility of the devices, these two studies were not able to provide correlation to clinical outcomes due to the small patient cohort. More recently, the GO chip[126] was validated in a cohort of 47 metastatic breast cancer patients.[241] The device detected CTC counts ranging from 0 to 27 CTCs mL−1 with an average of 2.33, 1.52, and 5.83 cells mL−1 in ER/PR+, HER2+, and triple-negative breast cancer (TNBC), respectively. The study also found the association of shorter PFS and OS in patients with >1 CTC mL−1. CTCs with HER2 expression were further correlated with the HER2 status of the primary tumor. 22 of 33 cases showed a discrepancy in HER2 status between primary tumor and expression on the corresponding CTCs. Further stratification of breast cancer CTCs with EMT markers (vimentin, or N-cadherin) showed significantly more epithelial CTCs compared to HER2+ or TNBCs, and more CTCs in EMT state (CK+/EMT+) in HER2+ or TNBC patients.
4.1.3. Colorectal Cancer CTCs
Colorectal cancer is another one of the most commonly tested cancers for testing the feasibility of nanostructured substrate-based CTC technologies.[122,133,201] One study utilized nylon-6/PSBMA/PAA to generate triple-blend fibrous mats via electrospinning for CTC capture.[172] The assay was tested on 19 patients who underwent a concurrent colonoscopy and successfully detected CTCs from 3 stage II, 4 stage III, and one stage IV colorectal cancer patients. The assay detected CTCs in only 2 of 4 (50%) stage I patients and had 2 false positive events in 7 patients without identifiable tumors via colonoscopy. It is worth noting that pathological analysis of biopsies from colonoscopies took about one week, while CTC detection from a peripheral blood specimen required only 6 hours in this study, which highlights the clinical benefit of liquid biopsy.
4.1.4. Pancreatic Cancer CTCs
CTCs also show potential to serve as an adjuvant biomarker for initial diagnosis and staging of pancreatic ductal adenocarcinoma (PDAC), a highly lethal disease difficult to obtain adequate tissue for diagnosis. CTC detection in PDAC patients has been challenging due to the low numbers of CTCs in peripheral blood. To address this issue, the highly sensitive NanoVelcro CTC assay was tested on blood samples from 100 patients (28 nonadenocarcinoma and 72 PDAC patients.[242] CTCs (CK+ and/or CEA+/CD45−/DAPI+, size ≥ 6 μm; Figure 15a) were detected in 54/72 patients with pathologically confirmed primary PDAC (sensitivity = 75.0%, specificity = 96.4%). A cutoff of ≥3 CTCs per 4 mL blood sample distinguished metastatic PDAC patients from the local/regional PDAC patients with an 85.2% sensitivity, 86.7% specificity, 79.3% positive predictive value, 90.7% negative predictive value, and an area under receiver operating characteristic (AUROC) curve of 0.885. A subsequent investigation of 126 PDAC patients showed an association between the CTC counts and clinicopathologic variables as well as clinical outcomes.[243] There was a correlation between CTC counts and cancer stages, and CTC count was found to be an independent predictor of OS in multivariate analysis (Figure 15b) and a univariate predictor of recurrence-free survival after surgery. Further, the patients with occult metastatic disease had survival similar to those with radiologically visible metastatic lesions, and significantly different from those with early-stage or locally advanced diseases.
Figure 15.
Clinical applications of CTC enumeration by NanoVelcro Chips. a) Identification of pancreatic cancer CTC via a four-color ICC (CK+ and/or CEA+/CD45−/DAPI+, left), and correlation of CTC counts with PDAC patients’ diagnostic and staging information (right). Reproduced with permission.[242] Copyright 2016, Springer Nature. b) OS for PDAC patients with early stage (n = 40, blue), locally advanced stage (n = 12, yellow), metastasis stage (n = 25, green), and occult metastasis stage (n = 12, orange). Reproduced with permission.[243] Copyright 2018, Springer Nature. c) Serial CTC enumeration by four different aptamer cocktails for a patient with NSCLC who was receiving chemotherapy. The results correlated with radiographic changes of the tumor and showed a shifting of CTC subsets during treatment. PR, partial response. PD, progressive disease. Reproduced with permission.[160] Copyright 2016, John Wiley and Sons. d) Summary of CTC enumeration of total CTCs and vimentin-positive CTCs for 8 healthy donors and 61 patients at different stages of HCC (top). Survival analyses showed a correlation between vimentin-positive CTC counts and OS as well as time to recurrence. Reproduced with permission.[232] Copyright 2018, John Wiley and Sons.
4.1.5. Lung Cancer CTCs
The exploit of aptamer cocktails in NanoVelcro Chips has enabled differential capture of CTCs in blood samples collected from 11 NSCLC patients.[160] Four different aptamer cocktails provided different CTC counts which constituted a distinct profile for each patient. This result suggested potentially differential enrichment of CTC subsets by different aptamer cocktails. To investigate the clinical value of CTC subpopulations in treatment monitoring, CTCs were enumerated with four different aptamer cocktails in parallel from four NSCLC patients before and after Pemetrexed disodium and Cisplatin treatment. The change of CTC counts after treatment showed concordance with the change of tumor status in patients’ computed tomography (CT) images (Figure 15c). The occasional discrepancies observed in CTC counts by different aptamer cocktails were in line with the heterogeneity of CTC population, which supported the idea that some CTC subsets may reflect the disease status better in certain clinical context. Overall, the parallel enumeration of CTC subpopulations with multiple aptamer cocktails may provide more comprehensive information than enumeration with a single aptamer and may serve as a valuable tool to monitor treatment response.
4.1.6. Kidney Cancer CTCs
Renal cell carcinoma (RCC) constitutes the vast majority of kidney cancer cases, with clear cell RCC being the most common subtype.[244] Conventional anti-EpCAM-based assays are inefficient in capturing RCC CTCs likely due to variable EpCAM expression in RCC CTCs.[245,246] A new combination of anticarboanhydrase 9 and anti-CD147 was tested in conjunction with NanoVelcro CTC assay to achieve effective RCC CTC capture and detection. Blood samples from 72/76 clear cell RCC patients were detected with CTCs (sensitivity = 94.7%).[247] The total CTC counts were much higher in clear cell RCC patients than those with benign kidney tumors, and even higher in advanced stages (III and IV > I and II). The number of CTC subsets that expressed vimentin stratified RCC patients and highly correlated with their clinical stages. The findings again highlight the improved sensitivity in detecting CTCs and the value of subclassifying them, both facilitated by the nanostructured substrate-based CTC capture approaches.
4.1.7. Liver Cancer CTCs
Liver cancer became the sixth and fourth leading cause of new cancer diagnosis and cancer-related death worldwide in 2018, respectively.[248] As the most common histologic type of liver cancer, hepatocellular carcinoma (HCC) shows a steady increase in both incidence rate and mortality rate in the United States.[249] It has a high recurrence rate and poor prognosis after recurrence despite curative treatment (such as surgical resection or liver transplantation).[250,251] The NanoVelcro CTC assay has been developed for prognostication and treatment selection for HCC patients.[232] An antibody cocktail targeting asialoglycoprotein receptor, glypican-3, and EpCAM, was utilized to capture HCC CTCs in peripheral blood samples collected from 80 HCC patients. CTCs were detected in 59/61 HCC patients (sensitivity = 97%) with higher CTC count correlating with advanced stages of HCC (median 3 CTCs/4 mL blood (range 0–15) in early stage, 9 CTCs/4 mL blood (range 0–14) for locally advanced disease, and 12 CTCs/4 mL blood (range 2–23) for metastatic HCC) and portal vein invasions. A cutoff of >2 CTCs per 4 mL of blood discriminated patients with HCC from those without HCC, with a sensitivity of 84.2%, specificity of 88.5%, and an AUROC of 0.92. The number of vimentin-positive CTCs correlated with tumor stages, recurrence, and OS (Figure 15d), with >1 vimentin-positive CTCs per 4 mL blood being associated with worse OS, and earlier recurrence after the treatment. These results suggest a potential role for vimentin-positive CTC subsets in indicating aggressiveness and occult metastases of the underlying disease and providing guidance for treatment selection.
4.1.8. Other Cancer CTCs
In addition to studying the common cancer types, researchers also explored the use of nanostructured substrate-based technologies for capture and analysis of CTCs from a few solid tumors with lower incidences. The aforementioned “CapioCyte” device, which utilized multivalent binding, E-selectin-medicated biomimetic cell rolling, and multiple capture antibodies (anti-EpCAM, anti-HER2, and anti-EGFR), was tested on cancer patients receiving radiotherapy.[187] CTCs were detected from the preradiotherapy blood samples collected from the 20 patients with head and neck squamous cell carcinoma. A reduction of CTC count was observed in 12/14 patients undergoing serial CTC enumeration, among which 11 patients had a complete response to radiotherapy. An elevation of CTC count was observed in 2/3 patients who had residual disease. The NanoVelcro system was also tested on patients with gestational choriocarcinoma in a multicenter study.[252] The CTCs were captured with anti-EpCAM and anti-CD147 and later identified by the expression of β-HCG. A cutoff of ≥6 CTCs was established as the prognostic threshold for PFS and OS in a 90-patient training cohort and validated in a separate 90-patient validation cohort. Among the 180 patients participating in the project, 96% of the GC patients had ≥2 CTCs per 7.5 mL of blood. The CTC counts in patients with distant metastases were much higher than those without. Using post-treatment CTC enumeration data from 106 patients receiving chemotherapy, univariate analyses identified the CTC count after the first cycle of chemotherapy a strong predictor of OS.
4.2. Genomic Analysis of CTCs Captured by Nanostructured Substrates
Genomic analysis of CTCs may provide real-time insights into the biological mechanisms of tumorigenesis, progression, metastasis, recurrence and treatment resistance. The rarity of CTCs in the peripheral blood has been the greatest challenge in obtaining qualified genomic information from CTCs. Nanostructured substrate-based CTC assays, as discussed in previous sections, help isolate CTCs with higher sensitivity and better integrity. With the aid of WGA, it is now possible to analyze CTC-derived genomic DNA for alterations such as single-nucleotide variations (SNVs), copy number variation, and structural variation (SVs).
4.2.1. Targeted Mutational Analysis
Years of research in cancer biology have identified numerous mutations in cancer cells that are associated with prognosis, susceptibility, and/or resistance to therapies. Therefore, it has become one of the major arms of precision oncology to search for these mutations to guide clinical decision making.[253] Several nanostructured substrate-based platforms, particularly those coupled with cell retrieval strategies, have successfully obtained high-quality CTCs for targeted mutation analysis. In general, the retrieved CTCs were lysed to extract genomic materials for amplification by WGA and/or target-specific PCR, and further subjected to sequencing or allele-specific detection (e.g., probe hybridization). Using the thermoresponsive Nano-Velcro Chip as an example, CTCs were purified through multiple cycles of thermocontrolled capture and release.[219] The resulting CTC suspension was subjected to EGFR mutation analysis (Figure 16a). Both L858R and T790M point mutations could be detected in CTCs acquired from 7 NSCLC patients’ blood, and the results were consistent with those observed in matching tumor tissues. In an index patient who received first-generation EGFR inhibitor (i.e., gefitinib), loss of L858R mutation and emergence of T790M mutation was observed by serial CTC analysis. The finding coincided with the development of clinical resistance to the inhibitor with radiographic progression of the lung tumor (Figure 16b). Other researchers utilized thermoresponsive polymer–GO Chips in conjunction with FISH technique to detect HER2 gene amplification in the isolated CTCs.[198]
Figure 16.
DNA mutational analysis of CTCs isolated by NanoVelcro Chips. a) A workflow summarizing EGFR mutation detection starting from NSCLC CTC purification via thermoresponsive NanoVelcro system, followed by WGA, PCR and quality control, to Sanger sequencing. b) Monitoring evolution of hotspot mutations in EGFR gene through serial CTC analysis with concurrent CT images for an index NSCLC patient at timepoints I) before gefitinib (first-generation EGFR inhibitor) treatment, II) 3 months post-treatment, and III) tumor relapse because of resistance to gefitinib. Before gefitinib treatment, L858R mutation was detected in both CTCs and tumor tissues. After the initiation of gefitinib, tumor regression was observed. T790M mutation, but not L858R mutation, was detected at the time of radiographic progression. a and b) Reproduced with permission.[120] Copyright 2018, Elsevier. c) WGS on single prostate cancer CTCs isolated by PN-NanoVelcro/LCM assay revealed that SNVs and gene rearrangements in tumor suppressor genes including RB1, BRCA2, and PTEN were shared by CTCs and tumor tissues, but not by WBCs. Reproduced under the terms of the CC-BY License.[233] Copyright 2015, Jiang et al. Published by Oncotarget.
As the researchers began to recognize the heterogeneity of CTCs, concerns were raised that bulk analysis of isolated CTCs runs the risk of missing some important genomic information of minor CTC subsets. Single-CTC analyses were successfully performed in some studies to assess specific mutations in individual CTCs. In one study, the researchers were able to detect oncogenic driver mutations (e.g., hotspot mutations in BRAF,[129] KRAS,[254] or EGFR[234]) in single CTCs using the PN-NanoVelcro assay coupled with LMD technology and subsequent Sanger sequencing. With the same platform but a different capture antibody (anti-CD146), single CMCs were isolated from the peripheral blood collected from melanoma patients (stage IV) with known BRAFV600E mutation by standard sequencing of tissue biopsy and identified by their Mart1+/HMW-MAA+/CD45− characteristics under fluorescence microscopy.[129] Following WGA, allele-specific PCR, and Sanger sequencing, BRAFV600E mutation was detected in CMC with a strong signal-to-noise ratio. The detection of BRAFV600E mutation in CMCs suggests a potential role for CTC-based mutational analysis before initiating BRAF inhibitor (e.g., vemurafenib) therapy to assess the sensitivity to treatment. To further assess the sensitivity of single-CTC sequencing, 5 pancreatic cancer patients bearing KRAS point mutations were involved in a study using PN-NanoVelcro/LCM assay.[254] Pancreatic CTCs were captured with anti-EpCAM, identified by their CEA+/CD45− characteristics, dissected using LCM, and then subjected to Sanger sequencing to detect codon-12 activating KRAS mutations. Of all the dissected CTCs from 5 patients, 21/44 (47.7%) had KRAS mutations. One patient had KRASG12V (35G to T) and 4 patients had KRASG12D (35G to A) mutations, which was consistent with the results of tumor tissue sequencing. Recently, the PN-NanoVelcro/LCM assay was utilized to detect EGFR mutations in individual CTCs harvested from 72 NSCLC patients.[234] Seven EGFR alterations could be detected by targeted sequencing, including 19-Del, L858R, T790M, 20-Ins, G719X, S768I, and L861Q. These results showed high concordance to primary tumor tissues and the small discrepancy highlighted the heterogeneity in CTC population. Again, the results of T790M mutations in CTCs correlated with acquired resistance to EGFR inhibitor (i.e., gefitinib). In another example, researchers used photothermal GNR-embedded gelatin nanostructured substrates with NIR-mediated targeted cell retrieval to harvest individual CTCs and detect KRAS and P53 gene mutations.[133] Individual CTCs were also isolated from a variety of cancers by mechanoresponsive gelatin-PDMS substrates with success in detecting oncogenic driver mutations such as PIK3CA and EGFR hotspot mutations.[132]
4.2.2. Whole Genome Sequencing
The development of single-cell technologies facilitates genome-wide analysis of individual CTCs. Zhao et al. used the PN-NanoVelcro/LCM assay to isolate single prostate cancer CTCs for whole exome analysis (WES),[163] and reached 25–80% whole exome coverage and a mean depth of 29 to 48×. This was the first case to utilize single-CTC WES to compare CTCs with WBCs. In a subsequent study, WGS was successfully performed on individual prostate cancer CTCs with 95% sequencing coverage, an average 30× depth, and overall quality comparable with tumor tissue sequencing.[233] The study found that 29% of somatic SNVs (shared by primary or metastatic tumors) coexist in CTCs (Figure 16c); 86% of CTC clonal mutations could be traced back to primary or metastatic tumors. SVs in important tumor suppressor genes such as BRCA2, PTEN, and RB1 shared between CTCs and the matching tumor tissues were also identified through single CTC WGS. This study not only highlighted the improvement of CTC isolation achieved by nanostructured substrates but also demonstrated the feasibility of characterizing tumor heterogeneity and biological evolution of individual CTCs.
4.3. Transcriptomic Profiling
In addition to genomic sequencing, transcriptomic profiling has been widely used in cancer research and helped gain tremendous knowledge in tumor biology and classification.[255] It is expected that transcriptome analysis in CTCs could provide valuable information about cancer biology. In one study, breast cancer CTCs, purified by ligand exchange on NP-HBCTC-Chip, were subjected to RNA sequencing.[192] The researchers successfully identified gene signatures associated with disease progression, patient survival, risk of metastasis, and epithelial to mesenchymal transition in CTCs. By using GO chips, genes related to EMT, cell proliferation, metastasis, apoptosis, inflammation, and stromal components were found to be differentially expressed in CTCs as well.[241] Patients with a high level of ERBB2, VIM, and CD44 showed a significantly higher probability of systematic progression. Measurement of RNA biomarkers (such as new noncoding RNAs) in CTCs was also made possible, as demonstrated in a study purifying prostate cancer CTCs using sorbitol stimulation on the PBA-grafted PEDOT NanoVelcro chip.[222] The harvested CTCs retained well-preserved RNA transcripts for downstream RT-quantitative PCR (RT-qPCR) analysis. Several prostate cancer-specific genes (e.g., AR-FL, AR-V7, KLK3, FOLH1), as well as long noncoding RNA (SChLAP1), were detected in 94% (16/17) of patients, with levels being significantly higher in patients with meta-static disease (Figure 17a). GO Chip-based prostate cancer CTC analysis also demonstrated that RNA expression of the isolated CTCs had significant relationships with OS (Figure 17b).[197] Recently, efforts were made to transform tumor tissue-based RNA classification into a CTC-based RNA panel for assessment of tumor biology and response to therapy. In one example, the thermoresponsive NanoVelcro system was used to isolate prostate cancer CTCs from mCRPC patients, which were then subjected to RNA quantification in the NanoString nCounter platform.[235] The researchers focused on a gene panel derived from one of the three subtypes defined in Prostate cancer Classification System (PCS)[256] and tested 31 blood samples from 23 patients receiving androgen receptor signaling inhibitor (ARSI) (Figure 17c). Compared with ARSI-sensitive samples (ARSI-S, n = 17), ARSI-resistant samples (ARSI-R, n = 14) had significantly higher PCS1 Z scores. The changes in the PCS1 Z score were consistent with the changes of eight patients from initial ARSI-S to later ARSI-R. This approach demonstrated the potential for a CTC-based noninvasive method to detect gene expression associated with therapeutic resistance, which may facilitate early detection of drug resistance and better patient-specific therapy selection.
Figure 17.
Transcriptomic Profiling of CTCs isolated by different nanostructured substrates. a) RNA signatures of prostate cancer CTCs purified by sorbitol stimulation on the PBA-grafted PEDOT NanoVelcro Chips from 17 prostate cancer patients, 7 healthy donors, positive control (PTC) and negative control (NTC). Reproduced with permission.[222] Copyright 2017, John Wiley and Sons. b) Kaplan–Meier curves obtained by GO Chip-based prostate cancer CTC analysis demonstrating RNA expression had significant relationships with OS. Reproduced under the terms of the CC-BY License.[197] Copyright 2018, Wiley-VCH. c) RNA expression of target genes in CTC-PCS1 panel (top) obtained from blood samples of 31 mCRPC patients by combing the thermoresponsive NanoVelcro system with the NanoString nCounter platform. The resulting PCS1 Z scores were correlated with the clinical drug sensitivity status of mCRPC patients, among which patients with abiraterone acetate and enzalutamide sensitivity (ARSI-S) were labeled in blue, while patients with abiraterone acetate and enzalutamide resistance were labeled in red. Receiving operating characteristics curve analysis of PCS1 Z score (bottom middle) shows an area under curve (AUC) of 0.75 in distinguishing ARSI-R from ARSI-S patients. An increase in PCS1 Z score was observed in serial CTC samples from individual mCRPC patients who progressed from ARSI-S to ARSI-R states (bottom right).
4.4. Protein Analysis
Protein analysis is also regarded as an important aspect in comprehensively understanding CTCs. Most CTC assays required ICC protocol targeting specific cytoplasmic and membranous proteins to identify CTCs from background WBCs. In addition to more routine staining, disease or cancer-related proteins (such as HER2 and EGFR) were also commonly stained. For example, Issadore et al. reported simultaneous analysis of multiple proteins including EpCAM, HER2, and EGFR on individual CTCs by using a panel of magnetic nanoparticles in a microfluidic chip-combined micro-hall detector.[87] Wu et al. proposed a multifunctional nanosphere-based micropillar chip for efficient phenotype analysis of individual CTCs.[236] By using red fluorescent magnetic nanospheres targeting EpCAM and green fluorescent nanospheres targeting HER2, CTCs labelled with magnetic tags were trapped on the size-selective micropillar chip and analyzed via fluorescence signals. Seven breast cancer patients were detected with HER2-expressing CTC subpopulations. Despite the success in assessing a small number of proteins in and on CTCs, more comprehensive protein characterization with higher throughput methodologies has yet to be explored on nanostructured substrate-based CTC assays.
5. Nanostructured Substrate-Embedded Assays for Noninvasive Prenatal Diagnostic Applications
5.1. Circulating Trophoblasts
As a subcategory of CFNCs, cTBs are one of the ideal targets for detecting the expression of unique markers, fetal karyotypes, and genotypes (except in rare scenarios involving placenta mosaicism). Using the imprinted PLGA NanoVelcro Chips (Figure 18a),[159] cTBs from maternal blood were effectively enriched using an anti-EpCAM capture agent, followed by ICC treatment. The devices were scanned by a fluorescence microscope, cTBs (Hoechst+/CK7+/HLA-G+/CD45−, 12 μm < sizes < 20 μm) were identified from nonspecifically captured WBCs (Hoechst+/CK7-/HLAG−/CD45+, 8 μm < sizes < 15 μm; Figure 18b). On average, 3–15 cTBs per 2 mL were detected in maternal blood collected from pregnant women (GA = 8–23 weeks). Individual cTBs were isolated by LCM for WGA and downstream genetic testing, e.g., short tandem repeats (STR) and array comparative genomic hybridization (aCGH). STR fingerprints obtained from cTBs, maternal blood, and the matching umbilical cord tissues could confirm the feto-parental relationship (Figure 18c). Fetal genders and chromosomal abnormalities (such as trisomy 21) were accurately detected in isolated cTBs by aCGH (Figure 18d).
Figure 18.
A cTB-based NIPD approach enabled by imprinted PLGA NanoVelcro Chips. a) A general workflow composed of i) three-step cTB enrichment/isolation, including RBC depletion, affinity capture, and LCM isolation, and ii) downstream genetic characterization, including WGA, aCGH, and STR. b) Fluorescence images of an isolated cTB and a WBC. c) STR genomic fingerprinting confirmed the fetal-parental relationship between cTBs and maternal cells, and the result was consistent with that of cTBs and the matching umbilical tissue. d) aCGH data from three cTBs showed fetal trisomy 21. Reproduced with permission.[159] Copyright 2017, American Chemical Society.
5.2. Circulating Fetal Nucleated Red Blood Cells
In addition to placenta-originated cTBs, fNRBCs directly derived from embryonic tissue are an alternative CFNC source for implementing NIPD. Researchers have introduced anti-CD147-functionalized nanoparticles, e.g., HAp/chitosan NPs,[194] biotin-Ppy NPs,[195] and gelatin NPs[196] into microchips to enrich fNRBCs from maternal peripheral blood collected from pregnant women (GA = 10–30 weeks). The target fNRBCs were identified using an ICC protocol (e.g., ε-globin+/CD71+/DAPI+/CD45−). A higher number of fNRBCs were observed in the blood samples from those pregnant women who conceived a fetus with chromosomal aneuploidy than those with normal pregnancies, showing that the high counts of fNRBCs may indicate abnormal gravidity.[195] Furthermore, fetal chromosomal aneuploidies, e.g., trisomy 18/21/XXX syndrome, were detected in fNRBCs using the FISH technique, the findings of which were in accordance with the results of amniocentesis and karyotyping. The comparison of cTB and fNRBC-based NIPD using nanostructured substrates are summarized in Table 2.
Table 2.
Comparison of cTB and fNRBC-based NIPD using nanostructured substrates.
| CFNC-based NIPD | cTB | fNRBC |
|---|---|---|
| Origin | Fetal | Fetal |
| Physical character | 12 μm < size < 20 μm | 10 μm < size < 18 μm |
| Marker expression | EpCAM, CK7, and HLA-G | CD147, CD71.CD36, GPA, and ε-HbF |
| Physiologic window for detection | GA > 6 Weeks | GA > 6 Weeks |
| Association with abnormal conditions | cTB enumeration correlates with abnormal fetal or placental development | High counts of fNRBCs indicate abnormal gravidity |
| Utilities in NIPD | Detection of genome-wide abnormalities (covering chromosomal abnormalities, insertion or deletion, and genetic mutations) and fetal gender | |
| Analysis technique | ICC, FISH, aCGH, STR | |
| Major challenge for assay development | Rarity, phenotypic viability of cTBs, reproducibility, and scalability of tests | Rarity, potential maternal NRBC contamination, reproducibility, and scalability of tests |
| Ref. | [159,257] | [80,194–196] |
6. Conclusion and Future Perspectives
Many advances have been made in detection, isolation, and characterization of circulating rare cells—especially CTCs and CFNCs. These have mostly been driven by interdisciplinary cooperative research efforts spanning the fields of materials science, chemistry, nanotechnology, bioengineering, cancer biology, oncology, maternal-fetal medicine, and other related specialties. Recent advances in the field of nanomaterials have led to the development of nanostructured substrate-embedded assays, which enables a diversity of in-depth detection and characterization of CTCs/CFNCs with drastically enhanced performance and reduced costs. Initially, such efforts were focused upon CTC enrichment and enumeration using the nanostructured substrates, allowing for morphologic analysis and subclassification of CTCs. Later, the addition of rare-cell retrieval strategies to nanostructured substrates allowed CTCs/CFNCs to be purified with high molecular integrity and cell viability, thereby enabling downstream molecular and functional analyses (such as genomic/transcriptomic profiling and ex vivo culture). Information from such CTC-derived characterizations demonstrated potential utility in cancer diagnosis, monitoring, and prognosis, bringing the field of oncology closer toward the ultimate goal of personalized care. Similarly, the CFNC-derived genetic testing and whole-genome profiling data suggested the feasibility of nanostructured substrate-embedded assays for noninvasive prenatal diagnostics.
Despite these advances, there is still space to increase the purity of CTC/CFNCs. The heterogeneity of CTCs also has an effect on the accuracy and reliability of the results. Looking toward the future, research endeavors in developing nanostructured substrate-embedded circulating rare-cell assays will be devoted to i) acquiring a fundamental understanding of how the underlying chemical/physical properties of nanostructured substrates affect the capture performance, viability, and molecular integrity of circulating rare cells; ii) exploring new nanomaterials, chemistry, and capture/release mechanisms on nanostructured substrates in order to reduce the hematologic cell contamination and improve the viability and molecular integrity of the isolated/purified circulating rare cells; iii) combining different capture agents (e.g., antibody, aptamer cocktails) and new markers to further increase the specificity of cell capture and address the heterogeneity in circulating rare cell populations; iv) exploiting a wider variety of multiomic analytical technologies with single-cell resolution to better characterize the heterogeneous cells of interest; v) studying other types of circulating rare cells (e.g., circulating stem cells, endothelial cells, plasma cells, mesenchymal cells) and noncellular components (e.g., extracellular vesicles[95,258,259]) that also carry molecular information about the disease microenvironment. Last but not least, the interactions between circulating rare cells and hematological cells deserve additional attention. Studies using microfluidic devices[23,260] and the CellSearch assay[261] have demonstrated that CTCs or CTC clusters can interact with other circulating entities like neutrophils and platelets. A more recent study pointed out that the WBC–CTC clusters were relatively rare events (less than 3.5%), played an important role in the formation of metastases, and correlated with a poor prognosis in cancer patients.[262] However, the roles of these cell complexes in the diagnostic or theranostic processes are mostly unknown. There is currently a lack of research efforts in using nanostructured substrates to study the interactions between circulating rare cells and other hematologic entities. We anticipate the use of nanostructured substrates, with the advantages of their variability and unique features, can lead to more discoveries about these cell complexes, improving our understanding of the immune system, and potentially making a great impact in the field of immune oncology.
Following the development of nanostructured substrate-embedded assays, challenges remain in the translation of these methods to fulfill unmet needs in the areas of cancer biology, clinical oncology, and maternal-fetal medicine. The lack of standardization and normalization of manufacturing nanostructured substrates, processing samples, and analyzing data, raises serious concerns about reliability and reproducibility when it comes to dissemination of the technologies in clinical trial settings. Therefore, more research efforts will be driven by the needs of i) pursuing more interdisciplinary collaboration for clinical validation of nanostructured substrate-embedded rare-cell assays and establishing standardization for the transition of these platforms to clinical applications; ii) differentiating CTC subpopulations based on distinct phenotypes (e.g., morphological characteristics, behaviors, and molecular fingerprints) to address heterogeneity in tumors and CTCs; iii) integrating and interpreting multiomic characterizations of CTCs alongside concurrent disease tissues to establish the relationship between CTCs and the overarching diseases; iv) performing analyses on serial blood samples in order to monitor the dynamic changes in CTC populations and their molecular signatures to better understand disease evolution; v) producing and using CTC-derived cell lines to gain a more comprehensive and profound understanding of oncogenic/resistance mechanisms, and to assess which therapeutic options may be beneficial to individual patients.
While the feasibility of performing CFNC-derived genetic testing and whole-genome profiling has been demonstrated by several nanostructured substrate-embedded assays, clinical adoption for NIPD applications requires further improvements in reproducibility and scalability. Advances in genomic analyses may enable the detection of genomic copy number variations within the megabase size range, as well as the detection of de novo point mutations. Additionally, enumeration and whole-genome profiling of CFNCs could potentially be used synergistically for screening abnormal placental dysplasia. Such advancements would be transformative for the fields of prenatal care.
Overall, we anticipate more powerful nanostructured substrate-based circulating rare-cell assays to be introduced and more integral clinical validation to be completed. Ultimately, future regulatory and commercial efforts will be needed to bring these tests to market.
Acknowledgements
J.D. and J.-F.C. contributed equally to this work. This work was supported by the National Institutes of Health (R21 CA151159, R33 CA157396, R33 CA174562, R44 CA180482, U01 CA198900, R01 CA218356, R21 CA235340, and U01 EB026421). Jiantong Dong gratefully acknowledges financial support from the China Scholarship Council (201706010064).
Biographies

Jiantong Dong received her B.S. degree in chemistry from Shandong University in 2014. She then joined the Molecular Recognition and Biochemical Analysis Laboratory to pursue her Ph.D. degree under the supervision of professor Meiping Zhao in the College of Chemistry and Molecular Engineering at Peking University. She is currently a joint Ph.D. student in professor Hsian-Rong Tseng’s research group in the Department of Molecular and Medical Pharmacology in David Geffen School of Medicine at UCLA. Her research interest concerns the development of nanomaterial-enabled technologies for analysis of circulating biomarkers.

Yazhen Zhu is currently an assistant researcher in the Department of Molecular and Medical Pharmacology in David Geffen School of Medicine at UCLA. She received her M.D. degree in clinical medicine at Wuhan University in 2005 and her Ph.D. degree in pathology at Fudan University in 2010. She served as an attending physician before joining professor Hsian-Rong Tseng’s research team at UCLA Medical School in 2017. Her research interests focus on the technical development and clinical translation of novel noninvasive diagnostic solutions for cancer patients and pregnant women.

Hsian-Rong Tseng is a professor of Molecular and Medical Pharmacology in Crump Institute for Molecular Imaging and California NanoSystems Institute in David Geffen School of Medicine at UCLA. He received his Ph.D. degree in organic chemistry from National Taiwan University in 1998. He then worked in the Department of Chemistry and Biochemistry at UCLA as a postdoctoral researcher (2000–2003) under the supervision of professor Sir J. Fraser Stoddart. After joining UCLA’s faculty in 2003, his research team has been working on the development of biomimetic nanomaterials for clinical applications in the field of circulating tumor biomarkers.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/adma.201903663.
Contributor Information
Jiantong Dong, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, Los Angeles, CA 90095, USA; Beijing National Laboratory for Molecular Sciences, MOE Key Laboratory of Bioorganic Chemistry and Molecular Engineering, College of Chemistry and Molecular Engineering, Peking University, Beijing 100871, P. R. China.
Jie-Fu Chen, Department of Pathology and Immunology, School of Medicine, Washington University in St. Louis, St. Louis, MO 63110, USA.
Matthew Smalley, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Meiping Zhao, Beijing National Laboratory for Molecular Sciences, MOE Key Laboratory of Bioorganic Chemistry and Molecular Engineering, College of Chemistry and Molecular Engineering, Peking University, Beijing 100871, P. R. China.
Zunfu Ke, Department of Pathology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong 510080, P. R. China.
Yazhen Zhu, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, Los Angeles, CA 90095, USA.
Hsian-Rong Tseng, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles, Los Angeles, CA 90095, USA.
References
- [1].Schreier S, Borwornpinyo S, Udomsangpetch R, Triampo W, Ann. Transl. Med 2018, 6, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].van de Stolpe A, Pantel K, Sleijfer S, Terstappen LW, den Toonder JM, Cancer Res. 2011, 71, 5955. [DOI] [PubMed] [Google Scholar]
- [3].Béroud C, Karliova M, Bonnefont JP, Benachi A, Munnich A, Dumez Y, Lacour B, Paterlini-Bréchot P, Lancet 2003, 361, 1013. [DOI] [PubMed] [Google Scholar]
- [4].Pang R, Law WL, Chu AC, Poon JT, Lam CS, Chow AK, Ng L, Cheung LW, Lan XR, Lan HY, Tan VP, Yau TC, Poon RT, Wong BC, Cell Stem Cell 2010, 6, 603. [DOI] [PubMed] [Google Scholar]
- [5].Goon PK, Lip GY, Boos CJ, Stonelake PS, Blann AD, Neoplasia 2006, 8, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bianchi G, Kyle RA, Larson DR, Witzig TE, Kumar S, Dispenzieri A, Morice WG, Rajkumar SV, Leukemia 2013, 27, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xu L, Li G, J. Orthop. Transl 2014, 2, 1. [Google Scholar]
- [8].Kasraeian S, Allison DC, Ahlmann ER, Fedenko AN, Menendez LR, Clin. Orthop. Rel. Res 2010, 468, 2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chaffer CL, Weinberg RA, Science 2011, 331, 1559. [DOI] [PubMed] [Google Scholar]
- [10].Alix-Panabieres C, Pantel K, Cancer Discovery 2016, 6, 479. [DOI] [PubMed] [Google Scholar]
- [11].Kaiser J, Science 2010, 327, 1072. [DOI] [PubMed] [Google Scholar]
- [12].Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB, Lerner RA, Bruce RH, Proc. Natl. Acad. Sci. USA 2004, 101, 10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, Zong C, Bai H, Chapman AR, Zhao J, Xu L, An T, Ma Q, Wang Y, Wu M, Sun Y, Wang S, Li Z, Yang X, Yong J, Su XD, Lu Y, Bai F, Xie XS, Wang J, Proc. Natl. Acad. Sci. USA 2013, 110, 21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, Trombetta JJ, Lu D, Tallapragada N, Tahirova N, Kim S, Blumenstiel B, Sougnez C, Lowe A, Wong B, Auclair D, Van Allen EM, Nakabayashi M, Lis RT, Lee GS, Li T, Chabot MS, Ly A, Taplin ME, Clancy TE, Loda M, Regev A, Meyerson M, Hahn WC, Kantoff PW, Golub TR, Getz G, Boehm JS, Love JC, Nat. Biotechnol 2014, 32, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, Ulman MJ, Xega K, Contino G, Alagesan B, Brannigan BW, Milos PM, Ryan DP, Sequist LV, Bardeesy N, Ramaswamy S, Toner M, Maheswaran S, Haber DA, Nature 2012, 487, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kalinich M, Bhan I, Kwan TT, Miyamoto DT, Javaid S, LiCausi JA, Milner JD, Hong X, Goyal L, Sil S, Choz M, Ho U, Kapur R, Muzikansky A, Zhang H, Weitz DA, Sequist LV, Ryan DP, Chung RT, Zhu AX, Isselbacher KJ, Ting DT, Toner M, Maheswaran S, Haber DA, Proc. Natl. Acad. Sci. USA 2017, 114, 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kalinsky K, Mayer JA, Xu X, Pham T, Wong KL, Villarin E, Pircher TJ, Brown M, Maurer MA, Bischoff FZ, Clin. Transl. Oncol 2015, 17, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wallwiener M, Hartkopf AD, Riethdorf S, Nees J, Sprick MR, Schonfisch B, Taran FA, Heil J, Sohn C, Pantel K, Trumpp A, Schneeweiss A, BMC Cancer 2015, 15, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Poudineh M, Aldridge PM, Ahmed S, Green BJ, Kermanshah L, Nguyen V, Tu C, Mohamadi RM, Nam RK, Hansen A, Sridhar SS, Finelli A, Fleshner NE, Joshua AM, Sargent EH, Kelley SO, Nat. Nanotechnol 2017, 12, 274. [DOI] [PubMed] [Google Scholar]
- [20].Alix-Panabieres C, Bartkowiak K, Pantel K, Mol. Oncol 2016, 10, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pantel K, Alix-Panabieres C, Clin. Chem 2016, 62, 328. [DOI] [PubMed] [Google Scholar]
- [22].Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, Brannigan BW, Kapur R, Stott SL, Shioda T, Ramaswamy S, Ting DT, Lin CP, Toner M, Haber DA, Maheswaran S, Cell 2014, 158, 1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B, Engstrom A, Zhu H, Sundaresan TK, Miyamoto DT, Luo X, Bardia A, Wittner BS, Ramaswamy S, Shioda T, Ting DT, Stott SL, Kapur R, Maheswaran S, Haber DA, Toner M, Nat. Methods 2015, 12, 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen JF, Ho H, Lichterman J, Lu YT, Zhang Y, Garcia MA, Chen SF, Liang AJ, Hodara E, Zhau HE, Hou S, Ahmed RS, Luthringer DJ, Huang J, Li KC, Chung LW, Ke Z, Tseng HR, Posadas EM, Cancer 2015, 121, 3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Beltran H, Jendrisak A, Landers M, Mosquera JM, Kossai M, Louw J, Krupa R, Graf RP, Schreiber NA, Nanus DM, Tagawa ST, Marrinucci D, Dittamore R, Scher HI, Clin. Cancer Res 2016, 22, 1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Scher HI, Graf RP, Schreiber NA, McLaughlin B, Jendrisak A, Wang Y, Lee J, Greene S, Krupa R, Lu D, Bamford P, Louw JE, Dugan L, Vargas HA, Fleisher M, Landers M, Heller G, Dittamore R, Cancer Res 2017, 77, 5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gulbahce N, Magbanua MJM, Chin R, Agarwal MR, Luo X, Liu J, Hayden DM, Mao Q, Ciotlos S, Li Z, Chen Y, Chen X, Li Y, Zhang RY, Lee K, Tearle R, Park E, Drmanac S, Rugo HS, Park JW, Drmanac R, Peters BA, Cancer Res 2017, 77, 4530. [DOI] [PubMed] [Google Scholar]
- [28].Carter L, Rothwell DG, Mesquita B, Smowton C, Leong HS, Fernandez-Gutierrez F, Li Y, Burt DJ, Antonello J, Morrow CJ, Hodgkinson CL, Morris K, Priest L, Carter M, Miller C, Hughes A, Blackhall F, Dive C, Brady G, Nat. Med 2017, 23, 114. [DOI] [PubMed] [Google Scholar]
- [29].Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, Arora KS, Desai N, Dahl DM, Sequist LV, Smith MR, Kapur R, Wu CL, Shioda T, Ramaswamy S, Ting DT, Toner M, Maheswaran S, Haber DA, Science 2015, 349, 1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kwan TT, Bardia A, Spring LM, Giobbie-Hurder A, Kalinich M, Dubash T, Sundaresan T, Hong X, LiCausi JA, Ho U, Silva EJ, Wittner BS, Sequist LV, Kapur R, Miyamoto DT, Toner M, Haber DA, Maheswaran S, Cancer Discovery 2018, 8, 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Miyamoto DT, Lee RJ, Kalinich M, LiCausi JA, Zheng Y, Chen T, Milner JD, Emmons E, Ho U, Broderick K, Silva E, Javaid S, Kwan TT, Hong X, Dahl DM, McGovern FJ, Efstathiou JA, Smith MR, Sequist LV, Kapur R, Wu CL, Stott SL, Ting DT, Giobbie-Hurder A, Toner M, Maheswaran S, Haber DA, Cancer Discovery 2018, 8, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jack RM, Grafton MM, Rodrigues D, Giraldez MD, Griffith C, Cieslak R, Zeinali M, Kumar Sinha C, Azizi E, Wicha M, Tewari M, Simeone DM, Nagrath S, Adv. Sci 2016, 3, 1600063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, Polanski R, Burt DJ, Simpson KL, Morris K, Pepper SD, Nonaka D, Greystoke A, Kelly P, Bola B, Krebs MG, Antonello J, Ayub M, Faulkner S, Priest L, Carter L, Tate C, Miller CJ, Blackhall F, Brady G, Dive C, Nat. Med 2014, 20, 897. [DOI] [PubMed] [Google Scholar]
- [34].Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, Wittner BS, Stojanov P, Brachtel E, Sgroi D, Kapur R, Shioda T, Ting DT, Ramaswamy S, Getz G, Iafrate AJ, Benes C, Toner M, Maheswaran S, Haber DA, Science 2014, 345, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen L, Hayes DF, Engl N. J. Med 2004, 351, 781. [DOI] [PubMed] [Google Scholar]
- [36].Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M, Scher HI, Clin. Cancer Res 2007, 13, 7053. [DOI] [PubMed] [Google Scholar]
- [37].Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B, Anand A, Smith K, Maslak P, Doyle GV, Terstappen LW, Lilja H, Heller G, Fleisher M, Scher HI, Clin. Cancer Res 2007, 13, 2023. [DOI] [PubMed] [Google Scholar]
- [38].Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LWMM, Meropol NJ, J. Clin. Oncol 2008, 26, 3213. [DOI] [PubMed] [Google Scholar]
- [39].Chen JF, Lu YT, Cheng S, Tseng HR, Figlin RA, Posadas EM, J. Hematol. Oncol 2017, 10, 63. [PubMed] [Google Scholar]
- [40].Lin E, Cao T, Nagrath S, King MR, Annu. Rev. Biomed. Eng 2018, 20, 329. [DOI] [PubMed] [Google Scholar]
- [41].Moon DH, Lindsay DP, Hong S, Wang AZ, Adv. Drug Delivery Rev 2018, 125, 143. [DOI] [PubMed] [Google Scholar]
- [42].Alix-Panabières C, Pantel K, Cancer Discovery 2016, 6, 479. [DOI] [PubMed] [Google Scholar]
- [43].Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, Tomlinson MW, Pereira L, Spitz JL, Hollemon D, Cuckle H, Musci TJ, Wapner RJ, Engl N. J. Med 2015, 372, 1589. [DOI] [PubMed] [Google Scholar]
- [44].Wapner RJ, Semin. Perinatol 2005, 29, 401. [DOI] [PubMed] [Google Scholar]
- [45].Dugoff L, Norton ME, Kuller JA, Am. J. Obstet. Gynecol 2016, 215, B2. [DOI] [PubMed] [Google Scholar]
- [46].Mujezinovic F, Alfirevic Z, Obstet. Gynecol 2007, 110, 687. [DOI] [PubMed] [Google Scholar]
- [47].Walknowska J, Conte FA, Grumbach MM, Lancet 1969, 293, 1119. [DOI] [PubMed] [Google Scholar]
- [48].Breman AM, Chow JC, U’Ren L, Normand EA, Qdaisat S, Zhao L, Henke DM, Chen R, Shaw CA, Jackson L, Yang Y, Vossaert L, Needham RH, Chang EJ, Campton D, Werbin JL, Seubert RC, Van den Veyver IB, Stilwell JL, Kaldjian EP, Beaudet AL, Prenatal Diagn. 2016, 36, 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bianchi DW, J. Pediatr. Surg 2007, 42, 12. [DOI] [PubMed] [Google Scholar]
- [50].Beaudet AL, Am. J. Med. Genet., Part C 2016, 172, 123. [DOI] [PubMed] [Google Scholar]
- [51].Emad A, Bouchard EF, Lamoureux J, Ouellet A, Dutta A, Klingbeil U, Drouin R, Prenatal Diagn. 2014, 34, 538. [DOI] [PubMed] [Google Scholar]
- [52].Ganshirt D, Smeets FW, Dohr A, Walde C, Steen I, Lapucci C, Falcinelli C, Sant R, Velasco M, Garritsen HS, Holzgreve W, Fetal Diagn. Ther 1998, 13, 276. [DOI] [PubMed] [Google Scholar]
- [53].Jain CV, Kadam L, van Dijk M, Kohan-Ghadr HR, Kilburn BA, Hartman C, Mazzorana V, Visser A, Hertz M, Bolnick AD, Fritz R, Armant DR, Drewlo S, Sci. Transl. Med 2016, 8, 363re4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Alix-Panabières C, Pantel K, Nat. Rev. Cancer 2014, 14, 623. [DOI] [PubMed] [Google Scholar]
- [55].Kwon KH, Jeon YJ, Hwang HS, Lee KA, Kim YJ, Chung HW, Pang MG, Prenatal Diagn. 2007, 27, 1245. [DOI] [PubMed] [Google Scholar]
- [56].Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Brechot C, Paterlini-Brechot P, Am. J. Pathol 2000, 156, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mouawia H, Saker A, Jais JP, Benachi A, Bussieres L, Lacour B, Bonnefont JP, Frydman R, Simpson JL, Paterlini-Brechot P, Reprod. BioMed. Online 2012, 25, 508. [DOI] [PubMed] [Google Scholar]
- [58].Hu X, Bessette PH, Qian J, Meinhart CD, Daugherty PS, Soh HT, Proc. Natl. Acad. Sci. USA 2005, 102, 15757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].He W, Wang H, Hartmann LC, Cheng JX, Low PS, Proc. Natl. Acad. Sci. USA 2007, 104, 11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lewis DE, Schober W, Murrell S, Nguyen D, Scott J, Boinoff J, Simpson JL, Bischoff FZ, Elias S, Cytometry 1996, 23, 218. [DOI] [PubMed] [Google Scholar]
- [61].Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bäuerle T, Wallwiener M, Holland-Letz T, Höfner T, Sprick M, Scharpff M, Marmé F, Sinn HP, Pantel K, Weichert W, Trumpp A, Nat. Biotechnol 2013, 31, 539. [DOI] [PubMed] [Google Scholar]
- [62].Herzenberg LA, Bianchi DW, Schroder J, Cann HM, Iverson GM, Proc. Natl. Acad. Sci. USA 1979, 76, 1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Xiong K, Wei W, Jin Y, Wang S, Zhao D, Wang S, Gao X, Qiao C, Yue H, Ma G, Xie HY, Adv. Mater 2016, 28, 7929. [DOI] [PubMed] [Google Scholar]
- [64].Tibbe AG, de Grooth BG, Greve J, Liberti PA, Dolan GJ, Terstappen LW, Nat. Biotechnol 1999, 17, 1210. [DOI] [PubMed] [Google Scholar]
- [65].Busch J, Huber P, Pfluger E, Miltenyi S, Holtz J, Radbruch A, Prenatal Diagn. 1994, 14, 1129. [DOI] [PubMed] [Google Scholar]
- [66].Riethdorf S, O’Flaherty L, Hille C, Pantel K, Adv. Drug Delivery Rev 2018, 125, 102. [DOI] [PubMed] [Google Scholar]
- [67].Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K, Clin. Cancer Res 2007, 13, 920. [DOI] [PubMed] [Google Scholar]
- [68].de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D, Clin. Cancer Res 2008, 14, 6302. [DOI] [PubMed] [Google Scholar]
- [69].Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, Heller G, Lancet Oncol. 2009, 10, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S, Hum. Pathol 2004, 35, 122. [DOI] [PubMed] [Google Scholar]
- [71].Yang L, Lang JC, Balasubramanian P, Jatana KR, Schuller D, Agrawal A, Zborowski M, Chalmers JJ, Biotechnol. Bioeng 2009, 102, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, Jeffrey SS, Davis RW, Proc. Natl. Acad. Sci. USA 2009, 106, 3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, Lotan TL, Zheng Q, De Marzo AM, Isaacs JT, Isaacs WB, Nadal R, Paller CJ, Denmeade SR, Carducci MA, Eisenberger MA, Luo J, Engl N. J. Med 2014, 371, 1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Earhart CM, Hughes CE, Gaster RS, Ooi CC, Wilson RJ, Zhou LY, Humke EW, Xu L, Wong DJ, Willingham SB, Schwartz EJ, Weissman IL, Jeffrey SS, Neal JW, Rohatgi R, Wakelee HA, Wang SX, Lab Chip 2014, 14, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Park SM, Wong DJ, Ooi CC, Kurtz DM, Vermesh O, Aalipour A, Suh S, Pian KL, Chabon JJ, Lee SH, Jamali M, Say C, Carter JN, Lee LP, Kuschner WG, Schwartz EJ, Shrager JB, Neal JW, Wakelee HA, Diehn M, Nair VS, Wang SX, Gambhir SS, Proc. Natl. Acad. Sci. USA 2016, 113, E8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mohamadi RM, Besant JD, Mepham A, Green B, Mahmoudian L, Gibbs T, Ivanov I, Malvea A, Stojcic J, Allan AL, Lowes LE, Sargent EH, Nam RK, Kelley SO, Angew. Chem., Int. Ed 2015, 54, 139. [DOI] [PubMed] [Google Scholar]
- [77].Labib M, Green B, Mohamadi RM, Mepham A, Ahmed SU, Mahmoudian L, Chang IH, Sargent EH, Kelley SO, J. Am. Chem. Soc 2016, 138, 2476. [DOI] [PubMed] [Google Scholar]
- [78].Huang R, Barber TA, Schmidt MA, Tompkins RG, Toner M, Bianchi DW, Kapur R, Flejter WL, Prenatal Diagn. 2008, 28, 892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lee D, Sukumar P, Mahyuddin A, Choolani M, Xu G, J. Chromatogr. A 2010, 1217, 1862. [DOI] [PubMed] [Google Scholar]
- [80].Kantak C, Chang CP, Wong CC, Mahyuddin A, Choolani M, Rahman A, Lab Chip 2014, 14, 841. [DOI] [PubMed] [Google Scholar]
- [81].Jackson JM, Witek MA, Kamande JW, Soper SA, Chem. Soc. Rev 2017, 46, 4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hajba L, Guttman A, TrAC, Trends Anal. Chem 2014, 59, 9. [Google Scholar]
- [83].Hyun KA, Jung HI, Lab Chip 2014, 14, 45. [DOI] [PubMed] [Google Scholar]
- [84].Myung JH, Hong S, Lab Chip 2015, 15, 4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M, Nature 2007, 450, 1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP Jr., Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M, Proc. Natl. Acad. Sci. USA 2010, 107, 18392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Issadore D, Chung J, Shao H, Liong M, Ghazani AA, Castro CM, Weissleder R, Lee H, Sci. Transl. Med 2012, 4, 141ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ, Lab Chip 2010, 10, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Huang LR, Cox EC, Austin RH, Sturm JC, Science 2004, 304, 987. [DOI] [PubMed] [Google Scholar]
- [90].Liu Z, Zhang W, Huang F, Feng H, Shu W, Xu X, Chen Y, Biosens. Bioelectron 2013, 47, 113. [DOI] [PubMed] [Google Scholar]
- [91].Li Y-Q, Chandran BK, Lim CT, Chen X, Adv. Sci 2015, 2, 1500118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Dharmasiri U, Witek MA, Adams AA, Soper SA, Annu. Rev. Anal. Chem 2010, 3, 409. [DOI] [PubMed] [Google Scholar]
- [93].Green BJ, Saberi Safaei T, Mepham A, Labib M, Mohamadi RM, Kelley SO, Angew. Chem., Int. Ed 2016, 55, 1252. [DOI] [PubMed] [Google Scholar]
- [94].Myung JH, Park SJ, Wang AZ, Hong S, Adv. Drug Delivery Rev. 2018, 125, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Li W, Wang H, Zhao Z, Gao H, Liu C, Zhu L, Wang C, Yang Y, Adv. Mater 2018, 10.1002/adma.201805344. [DOI] [PubMed] [Google Scholar]
- [96].Wang S, Wan Y, Liu Y, Nanoscale 2014, 6, 12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Woo KM, Chen VJ, Ma PX, J. Biomed. Mater. Res 2003, 67A, 531. [DOI] [PubMed] [Google Scholar]
- [98].Wan Y, Wang Y, Liu Z, Qu X, Han B, Bei J, Wang S, Biomaterials 2005, 26, 4453. [DOI] [PubMed] [Google Scholar]
- [99].Martinez E, Engel E, Lopez-Iglesias C, Mills CA, Planell JA, Samitier J, Micron 2008, 39, 111. [DOI] [PubMed] [Google Scholar]
- [100].Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF, Biomaterials 1999, 20, 573. [DOI] [PubMed] [Google Scholar]
- [101].Stevens MM, George JH, Science 2005, 310, 1135. [DOI] [PubMed] [Google Scholar]
- [102].Park J, Bauer S, von der Mark K, Schmuki P, Nano Lett. 2007, 7, 1686. [DOI] [PubMed] [Google Scholar]
- [103].Mosqueira D, Pagliari S, Uto K, Ebara M, Romanazzo S, Escobedo-Lucea C, Nakanishi J, Taniguchi A, Franzese O, Di Nardo P, Goumans MJ, Traversa E, Pinto-do-Ó P, Aoyagi T, Forte G, ACS Nano 2014, 8, 2033. [DOI] [PubMed] [Google Scholar]
- [104].Liu X, Wang S, Chem. Soc. Rev 2014, 43, 2385. [DOI] [PubMed] [Google Scholar]
- [105].Bettinger CJ, Langer R, Borenstein JT, Angew. Chem., Int. Ed 2009, 48, 5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Saracino GA, Cigognini D, Silva D, Caprini A, Gelain F, Chem. Soc. Rev 2013, 42, 225. [DOI] [PubMed] [Google Scholar]
- [107].Kandere-Grzybowska K, Campbell C, Komarova Y, Grzybowski BA, Borisy GG, Nat. Methods 2005, 2, 739. [DOI] [PubMed] [Google Scholar]
- [108].Chang WC, Lee LP, Liepmann D, Lab Chip 2005, 5, 64. [DOI] [PubMed] [Google Scholar]
- [109].Karuri NW, Liliensiek S, Teixeira AI, Abrams G, Campbell S, Nealey PF, Murphy CJ, J. Cell Sci 2004, 117, 3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE, Science 1997, 276, 1425. [DOI] [PubMed] [Google Scholar]
- [111].Park SY, Park SY, Namgung S, Kim B, Im J, Kim JY, Sun K, Lee KB, Nam JM, Park Y, Hong S, Adv. Mater 2007, 19, 2530. [Google Scholar]
- [112].Yim EK, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW, Biomaterials 2005, 26, 5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Yousaf MN, Houseman BT, Mrksich M, Angew. Chem., Int. Ed 2001, 40, 1093. [PubMed] [Google Scholar]
- [114].Dang JM, Leong KW, Adv. Mater 2007, 19, 2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CD, Oreffo RO, Nat. Mater 2007, 6, 997. [DOI] [PubMed] [Google Scholar]
- [116].Jan E, Kotov NA, Nano Lett. 2007, 7, 1123. [DOI] [PubMed] [Google Scholar]
- [117].Yoon HJ, Kozminsky M, Nagrath S, ACS Nano 2014, 8, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Wang L, Asghar W, Demirci U, Wan Y, Nano Today 2013, 8, 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Lin M, Chen J-F, Lu Y-T, Zhang Y, Song J, Hou S, Ke Z, Tseng H-R, Acc. Chem. Res 2014, 47, 2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Jan YJ, Chen JF, Zhu Y, Lu YT, Chen SH, Chung H, Smalley M, Huang YW, Dong J, Chen LC, Yu HH, Tomlinson JS, Hou S, Agopian VG, Posadas EM, Tseng HR, Adv. Drug Delivery Rev 2018, 125, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR, Angew. Chem., Int. Ed 2009, 48, 8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q, He R, Hong L, Liu W, Guo S, Liu K, Tseng HR, Xiong B, Zhao XZ, Adv. Mater 2012, 24, 2756. [DOI] [PubMed] [Google Scholar]
- [123].Sekine J, Luo SC, Wang S, Zhu B, Tseng HR, Yu HH, Adv. Mater 2011, 23, 4788. [DOI] [PubMed] [Google Scholar]
- [124].Myung JH, Gajjar KA, Saric J, Eddington DT, Hong S, Angew. Chem., Int. Ed 2011, 50, 11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Banerjee SS, Paul D, Bhansali SG, Aher ND, Jalota-Badhwar A, Khandare J, Small 2012, 8, 1657. [DOI] [PubMed] [Google Scholar]
- [126].Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, Simeone DM, Nagrath S, Nat. Nanotechnol 2013, 8, 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Zhang P, Chen L, Xu T, Liu H, Liu X, Meng J, Yang G, Jiang L, Wang S, Adv. Mater 2013, 25, 3566. [DOI] [PubMed] [Google Scholar]
- [128].Yin S, Wu Y-L, Hu B, Wang Y, Cai P, Tan CK, Qi D, Zheng L, Leow WR, Tan NS, Wang S, Chen X, Adv. Mater. Interfaces 2014, 1, 1300043. [Google Scholar]
- [129].Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang W-H, He R, Zhao X-Z, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng H-R, Angew. Chem., Int. Ed 2013, 52, 3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY, Nakao A, Garcia MA, Song M, Lee T, Xiong B, Luo S-C, Tseng H-R, Yu H.-h., Adv. Mater 2013, 25, 1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Jeon S, Moon JM, Lee ES, Kim YH, Cho Y, Angew. Chem., Int. Ed 2014, 53, 4597. [DOI] [PubMed] [Google Scholar]
- [132].Reategui E, Aceto N, Lim EJ, Sullivan JP, Jensen AE, Zeinali M, Martel JM, Aranyosi AJ, Li W, Castleberry S, Bardia A, Sequist LV, Haber DA, Maheswaran S, Hammond PT, Toner M, Stott SL, Adv. Mater 2015, 27, 1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Lv S-W, Liu Y, Xie M, Wang J, Yan X-W, Li Z, Dong W-G, Huang W-H, ACS Nano 2016, 10, 6201. [DOI] [PubMed] [Google Scholar]
- [134].Liu H, Li Y, Sun K, Fan J, Zhang P, Meng J, Wang S, Jiang L, J. Am. Chem. Soc 2013, 135, 7603. [DOI] [PubMed] [Google Scholar]
- [135].Shen Q, Xu L, Zhao L, Wu D, Fan Y, Zhou Y, Ouyang WH, Xu X, Zhang Z, Song M, Lee T, Garcia MA, Xiong B, Hou S, Tseng HR, Fang X, Adv. Mater 2013, 25, 2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Sun N, Liu M, Wang J, Wang Z, Li X, Jiang B, Pei R, Small 2016, 12, 5090. [DOI] [PubMed] [Google Scholar]
- [137].Albuschies J, Vogel V, Sci. Rep 2013, 3, 1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Peng KQ, Yan YJ, Gao SP, Zhu J, Adv. Mater 2002, 14, 1164. [Google Scholar]
- [139].Das B, McGinnis SP, Appl. Phys. A: Mater. Sci. Process 2000, 71, 681. [Google Scholar]
- [140].Tan C, Zhang H, Nat. Commun 2015, 6, 7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Gurkan UA, Fan Y, Xu F, Erkmen B, Urkac ES, Parlakgul G, Bernstein J, Xing W, Boyden ES, Demirci U, Adv. Mater 2013, 25, 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Sun B, Long YZ, Zhang HD, Li MM, Duvail JL, Jiang XY, Yin HL, Prog. Polym. Sci 2014, 39, 862. [Google Scholar]
- [143].MacDonald RA, Laurenzi BF, Viswanathan G, Ajayan PM, Stegemann JP, J. Biomed. Mater. Res., Part A 2005, 74A, 489. [DOI] [PubMed] [Google Scholar]
- [144].Sun N, Li X, Wang Z, Zhang R, Wang J, Wang K, Pei R, ACS Appl. Mater. Interfaces 2016, 8, 12638. [DOI] [PubMed] [Google Scholar]
- [145].Abdolahad M, Taghinejad M, Taghinejad H, Janmaleki M, Mohajerzadeh S, Lab Chip 2012, 12, 1183. [DOI] [PubMed] [Google Scholar]
- [146].Zhang F, Jiang Y, Liu X, Meng J, Zhang P, Liu H, Yang G, Li G, Jiang L, Wan LJ, Hu JS, Wang S, Nano Lett. 2016, 16, 766. [DOI] [PubMed] [Google Scholar]
- [147].Chen JF, Zhu Y, Lu YT, Hodara E, Hou S, Agopian VG, Tomlinson JS, Posadas EM, Tseng HR, Theranostics 2016, 6, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Hanson L, Lin ZC, Xie C, Cui Y, Cui B, Nano Lett. 2012, 12, 5815. [DOI] [PubMed] [Google Scholar]
- [149].Kim DJ, Seol JK, Lee G, Kim GS, Lee SK, Nanotechnology 2012, 23, 395102. [DOI] [PubMed] [Google Scholar]
- [150].Park G-S, Kwon H, Kwak DW, Park SY, Kim M, Lee J-H, Han H, Heo S, Li XS, Lee JH, Kim YH, Lee J-G, Yang W, Cho HY, Kim SK, Kim K, Nano Lett. 2012, 12, 1638. [DOI] [PubMed] [Google Scholar]
- [151].Lee SK, Kim GS, Wu Y, Kim DJ, Lu Y, Kwak M, Han L, Hyung JH, Seol JK, Sander C, Gonzalez A, Li J, Fan R, Nano Lett. 2012, 12, 2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Hong WY, Jeon SH, Lee ES, Cho Y, Biomaterials 2014, 35, 9573. [DOI] [PubMed] [Google Scholar]
- [153].Zhai TT, Ye D, Zhang QW, Wu ZQ, Xia XH, ACS Appl. Mater. Interfaces 2017, 9, 34706. [DOI] [PubMed] [Google Scholar]
- [154].Lou HY, Zhao W, Hanson L, Zeng C, Cui Y, Cui B, Langmuir 2017, 33, 1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Hsiao YS, Luo SC, Hou S, Zhu B, Sekine J, Kuo CW, Chueh DY, Yu HH, Tseng HR, Chen P, Small 2014, 10, 3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Hsiao Y-S, Ho B-C, Yan H-X, Kuo C-W, Chueh D-Y, Yu H.-h., Chen P, J. Mater. Chem. B 2015, 3, 5103. [DOI] [PubMed] [Google Scholar]
- [157].Liu X, Chen L, Liu H, Yang G, Zhang P, Han D, Wang S, Jiang L, NPG Asia Mater. 2013, 5, e63. [Google Scholar]
- [158].Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR, Angew. Chem., Int. Ed 2011, 50, 3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Hou S, Chen JF, Song M, Zhu Y, Jan YJ, Chen SH, Weng TH, Ling DA, Chen SF, Ro T, Liang AJ, Lee T, Jin H, Li M, Liu L, Hsiao YS, Chen P, Yu HH, Tsai MS, Pisarska MD, Chen A, Chen LC, Tseng HR, ACS Nano 2017, 11, 8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Zhao L, Tang C, Xu L, Zhang Z, Li X, Hu H, Cheng S, Zhou W, Huang M, Fong A, Liu B, Tseng H-R, Gao H, Liu Y, Fang X, Small 2016, 12, 1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Liu HQ, Yu XL, Cai B, You SJ, He ZB, Huang QQ, Rao L, Li SS, Liu C, Sun WW, Liu W, Guo SS, Zhao XZ, Appl. Phys. Lett 2015, 106, 093703. [Google Scholar]
- [162].Ma K, Chan CK, Liao S, Hwang WY, Feng Q, Ramakrishna S, Biomaterials 2008, 29, 2096. [DOI] [PubMed] [Google Scholar]
- [163].Zhao L, Lu YT, Li F, Wu K, Hou S, Yu J, Shen Q, Wu D, Song M, OuYang WH, Luo Z, Lee T, Fang X, Shao C, Xu X, Garcia MA, Chung LW, Rettig M, Tseng HR, Posadas EM, Adv. Mater 2013, 25, 2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Xu G, Tan Y, Xu T, Yin D, Wang M, Shen M, Chen X, Shi X, Zhu X, Biomater. Sci 2017, 5, 752. [DOI] [PubMed] [Google Scholar]
- [165].Wang Z, Sun N, Liu M, Cao Y, Wang K, Wang J, Pei R, ACS Sens. 2017, 2, 547. [DOI] [PubMed] [Google Scholar]
- [166].Wang M, Xiao Y, Lin L, Zhu X, Du L, Shi X, Bioconjugate Chem. 2018, 29, 1081. [DOI] [PubMed] [Google Scholar]
- [167].Ma L, Yang G, Wang N, Zhang P, Guo F, Meng J, Zhang F, Hu Z, Wang S, Zhao Y, Adv. Healthcare Mater 2015, 4, 838. [DOI] [PubMed] [Google Scholar]
- [168].Zhao Y, Zhu X, Liu H, Luo Y, Wang S, Shen M, Zhu M, Shi X, J. Mater. Chem. B 2014, 2, 7384. [DOI] [PubMed] [Google Scholar]
- [169].Zhao Y, Fan Z, Shen M, Shi X, Adv. Mater. Interfaces 2015, 2, 1500256. [Google Scholar]
- [170].Zhao Y, Fan Z, Shen M, Shi X, RSC Adv. 2015, 5, 70439. [Google Scholar]
- [171].Xiao YC, Wang MY, Lin LZ, Du LF, Shen MW, Shi XY, Mater. Chem. Front 2018, 2, 891. [Google Scholar]
- [172].Tseng H-C, Lee A-W, Wei P-L, Chang Y-J, Chen J-K, J. Mater. Chem. B 2016, 4, 6565. [DOI] [PubMed] [Google Scholar]
- [173].Yoon J, Yoon HS, Shin Y, Kim S, Ju Y, Kim J, Chung S, Nanomedicine 2017, 13, 1617. [DOI] [PubMed] [Google Scholar]
- [174].Yu CC, Ho BC, Juang RS, Hsiao YS, Naidu RVR, Kuo CW, You YW, Shyue JJ, Fang JT, Chen P, ACS Appl. Mater. Interfaces 2017, 9, 30329. [DOI] [PubMed] [Google Scholar]
- [175].Kim YJ, Ebara M, Aoyagi T, Angew. Chem., Int. Ed 2012, 51, 10537. [DOI] [PubMed] [Google Scholar]
- [176].Zhong Y, Zhang J, Cheng R, Deng C, Meng F, Xie F, Zhong Z, J. Controlled Release 2015, 205, 144. [DOI] [PubMed] [Google Scholar]
- [177].Selim KK, Ha Y-S, Kim S-J, Chang Y, Kim T-J, Lee GH, Kang I-K, Biomaterials 2007, 28, 710. [DOI] [PubMed] [Google Scholar]
- [178].Nie L, Li F, Huang X, Aguilar ZP, Wang YA, Xiong Y, Fu F, Xu H, ACS Appl. Mater. Interfaces 2018, 10, 14055. [DOI] [PubMed] [Google Scholar]
- [179].He R, Liu MX, Shen Y, Long ZR, Zhou CR, J. Mater. Chem. B 2017, 5, 1712. [DOI] [PubMed] [Google Scholar]
- [180].Ouyang J, Chen M, Bao W-J, Zhang Q-W, Wang K, Xia X-H, Adv. Funct. Mater 2015, 25, 6122. [Google Scholar]
- [181].Kumeria T, Kurkuri MD, Diener KR, Parkinson L, Losic D, Biosens. Bioelectron 2012, 35, 167. [DOI] [PubMed] [Google Scholar]
- [182].Lim JT, Yoon YS, Lee WY, Jeong JT, Kim GS, Kim TG, Lee SK, Nanoscale 2017, 9, 17224. [DOI] [PubMed] [Google Scholar]
- [183].Xu H, Dong B, Xiao Q, Sun X, Zhang X, Lyu J, Yang Y, Xu L, Bai X, Zhang S, Song H, ACS Appl. Mater. Interfaces 2017, 9, 30510. [DOI] [PubMed] [Google Scholar]
- [184].Jeon S, Hong W, Lee ES, Cho Y, Theranostics 2014, 4, 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Myung JH, Roengvoraphoj M, Tam KA, Ma T, Memoli VA, Dmitrovsky E, Freemantle SJ, Hong S, Anal. Chem 2015, 87, 10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Myung JH, Gajjar KA, Chen J, Molokie RE, Hong S, Anal. Chem 2014, 86, 6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Myung JH, Eblan MJ, Caster JM, Park SJ, Poellmann MJ, Wang K, Tam KA, Miller SM, Shen C, Chen RC, Zhang T, Tepper JE, Chera BS, Wang AZ, Hong S, Clin. Cancer Res 2018, 24, 2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].He R, Zhao L, Liu Y, Zhang N, Cheng B, He Z, Cai B, Li S, Liu W, Guo S, Chen Y, Xiong B, Zhao XZ, Biomed. Microdevices 2013, 15, 617. [DOI] [PubMed] [Google Scholar]
- [189].Huang Q, Chen B, He R, He Z, Cai B, Xu J, Qian W, Chan HL, Liu W, Guo S, Zhao XZ, Yuan J, Adv. Healthcare Mater 2014, 3, 1420. [DOI] [PubMed] [Google Scholar]
- [190].Yang G, Liu H, Liu X, Zhang P, Huang C, Xu T, Jiang L, Wang S, Adv. Healthcare Mater 2014, 3, 332. [DOI] [PubMed] [Google Scholar]
- [191].Sheng W, Chen T, Tan W, Fan ZH, ACS Nano 2013, 7, 7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Park MH, Reategui E, Li W, Tessier SN, Wong KH, Jensen AE, Thapar V, Ting D, Toner M, Stott SL, Hammond PT, J. Am. Chem. Soc 2017, 139, 2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Sun N, Wang J, Ji L, Hong S, Dong J, Guo Y, Zhang K, Pei R, Small 2015, 11, 5444. [DOI] [PubMed] [Google Scholar]
- [194].He ZB, Guo F, Feng C, Cai B, Lata JP, He RX, Huang QQ, Yu XL, Rao L, Liu HQ, Guo SS, Liu W, Zhang YZ, Huang TJ, Zhao XZ, J. Mater. Chem. B 2017, 5, 226. [DOI] [PubMed] [Google Scholar]
- [195].Feng C, He Z, Cai B, Peng J, Song J, Yu X, Sun Y, Yuan J, Zhao X, Zhang Y, Theranostics 2018, 8, 1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Wei XY, Cai B, Chen KK, Cheng L, Zhu YZ, Wang ZX, Sun Y, Liu W, Guo SS, Zhang YZ, Zhao XZ, Sens. Actuators, B 2019, 281, 131. [Google Scholar]
- [197].Kozminsky M, Fouladdel S, Chung JS, Wang Y, Smith DC, Alva A, Azizi E, Morgan T, Nagrath S, Adv. Sci 2019, 6, 1801254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Yoon HJ, Shanker A, Wang Y, Kozminsky M, Jin Q, Palanisamy N, Burness ML, Azizi E, Simeone DM, Wicha MS, Kim J, Nagrath S, Adv. Mater 2016, 28, 4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Li Y, Lu Q, Liu H, Wang J, Zhang P, Liang H, Jiang L, Wang S, Adv. Mater 2015, 27, 6848. [DOI] [PubMed] [Google Scholar]
- [200].Cheng B, He Z, Zhao L, Fang Y, Chen Y, He R, Chen F, Song H, Deng Y, Zhao X, Xiong B, Int. J. Nanomed 2014, 9, 2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Chen YY, Cheng BR, He ZB, Wang SY, Wang ZM, Sun M, Song HB, Fang Y, Chen FF, Xiong B, J. Cancer 2016, 7, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Chiu WJ, Ling TK, Chiang HP, Lin HJ, Huang CC, ACS Appl. Mater. Interfaces 2015, 7, 8622. [DOI] [PubMed] [Google Scholar]
- [203].Rothemund PWK, Nature 2006, 440, 297. [DOI] [PubMed] [Google Scholar]
- [204].Zhou G, Lin M, Song P, Chen X, Chao J, Wang L, Huang Q, Huang W, Fan C, Zuo X, Anal. Chem 2014, 86, 7843. [DOI] [PubMed] [Google Scholar]
- [205].Chen WQ, Weng SN, Zhang F, Allen S, Li X, Bao LW, Lam RHW, Macoska JA, Merajver SD, Fu JP, ACS Nano 2013, 7, 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Ren Z, Guo Y, Liu C-H, Gao P-X, Front. Chem 2013, 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Dou X, Li P, Jiang S, Bayat H, Schonherr H, ACS Appl. Mater. Interfaces 2017, 9, 8508. [DOI] [PubMed] [Google Scholar]
- [208].Meng J, Liu H, Liu X, Yang G, Zhang P, Wang S, Jiang L, Small 2014, 10, 3735. [DOI] [PubMed] [Google Scholar]
- [209].Wang W, Cui H, Zhang P, Meng J, Zhang F, Wang S, ACS Appl. Mater. Interfaces 2017, 9, 10537. [DOI] [PubMed] [Google Scholar]
- [210].Zhang P, Wang S, ChemPhysChem 2014, 15, 1550. [DOI] [PubMed] [Google Scholar]
- [211].Guo S, Xu J, Xie M, Huang W, Yuan E, Liu Y, Fan L, Cheng S, Liu S, Wang F, Yuan B, Dong W, Zhang X, Huang W, Zhou X, ACS Appl. Mater. Interfaces 2016, 8, 15917. [DOI] [PubMed] [Google Scholar]
- [212].Meng J, Zhang P, Zhang F, Liu H, Fan J, Liu X, Yang G, Jiang L, Wang S, ACS Nano 2015, 9, 9284. [DOI] [PubMed] [Google Scholar]
- [213].Qiu J, Zhao K, Li L, Yu X, Guo W, Wang S, Zhang X, Pan C, Wang ZL, Liu H, Nano Res. 2017, 10, 776. [Google Scholar]
- [214].Yu X, He R, Li S, Cai B, Zhao L, Liao L, Liu W, Zeng Q, Wang H, Guo SS, Zhao XZ, Small 2013, 9, 3895. [DOI] [PubMed] [Google Scholar]
- [215].Tang M, Wen CY, Wu LL, Hong SL, Hu J, Xu CM, Pang DW, Zhang ZL, Lab Chip 2016, 16, 1214. [DOI] [PubMed] [Google Scholar]
- [216].Song Y, Shi Y, Huang M, Wang W, Wang Y, Cheng J, Lei Z, Zhu Z, Yang C, Angew. Chem., Int. Ed 2019, 58, 2236. [DOI] [PubMed] [Google Scholar]
- [217].Zheng Q, Iqbal SM, Wan Y, Biotechnol. Adv 2013, 31, 1664. [DOI] [PubMed] [Google Scholar]
- [218].Sada T, Fujigaya T, Niidome Y, Nakazawa K, Nakashima N, ACS Nano 2011, 5, 4414. [DOI] [PubMed] [Google Scholar]
- [219].Ke Z, Lin M, Chen JF, Choi JS, Zhang Y, Fong A, Liang AJ, Chen SF, Li Q, Fang W, Zhang P, Garcia MA, Lee T, Song M, Lin HA, Zhao H, Luo SC, Hou S, Yu HH, Tseng HR, ACS Nano 2015, 9, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Liu H, Liu X, Meng J, Zhang P, Yang G, Su B, Sun K, Chen L, Han D, Wang S, Jiang L, Adv. Mater 2013, 25, 922. [DOI] [PubMed] [Google Scholar]
- [221].Wang C, Ye M, Cheng L, Li R, Zhu W, Shi Z, Fan C, He J, Liu J, Liu Z, Biomaterials 2015, 54, 55. [DOI] [PubMed] [Google Scholar]
- [222].Shen MY, Chen JF, Luo CH, Lee SJ, Li CH, Yang YL, Tsai YH, Ho BC, Bao LR, Lee TJ, Jan YJ, Zhu YZ, Cheng S, Feng FY, Chen P, Hou S, Agopian V, Hsiao YS, Tseng HR, Posadas EM, Yu H. h., Adv. Healthcare Mater 2018, 7, 1700701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Hui L, Su Y, Ye T, Liu Z, Tian Q, He C, Zhao Y, Chen P, Wang X, Han W, Luo Y, Wang B, ACS Appl. Mater. Interfaces 2018, 10, 207. [DOI] [PubMed] [Google Scholar]
- [224].Xiao Y, Wang M, Lin L, Du L, Shen M, Shi X, Nanomedicine 2019, 14, 183. [DOI] [PubMed] [Google Scholar]
- [225].Dong J, Jan YJ, Cheng J, Zhang RY, Meng M, Smalley M, Chen P-J, Tang X, Tseng P, Bao L, Huang T-Y, Zhou D, Liu Y, Chai X, Zhang H, Zhou A, Agopian VG, Posadas EM, Shyue J-J, Jonas SJ, Weiss PS, Li M, Zheng G, Yu H.-h., Zhao M, Tseng H-R, Zhu Y, Sci. Adv 2019, 5, eaav9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Dong J, Wu T, Xiao Y, Xu L, Fang S, Zhao M, Chem. Commun 2016, 52, 11923. [DOI] [PubMed] [Google Scholar]
- [227].Dong J, Wu T, Xiao Y, Chen L, Xu L, Li M, Zhao M, Biosens. Bioelectron 2018, 100, 333. [DOI] [PubMed] [Google Scholar]
- [228].Dong J, Zhao M, TrAC, Trends Anal. Chem 2016, 80, 190. [Google Scholar]
- [229].Chen L, Liu X, Su B, Li J, Jiang L, Han D, Wang S, Adv. Mater 2011, 23, 4376. [DOI] [PubMed] [Google Scholar]
- [230].Li W, Reategui E, Park MH, Castleberry S, Deng JZ, Hsu B, Mayner S, Jensen AE, Sequist LV, Maheswaran S, Haber DA, Toner M, Stott SL, Hammond PT, Biomaterials 2015, 65, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Guo S, Huang HY, Deng XJ, Chen YQ, Jiang ZR, Xie M, Liu SM, Huang WH, Zhou X, Nano Res. 2018, 11, 2592. [Google Scholar]
- [232].Court CM, Hou S, Winograd P, Segel NH, Li QW, Zhu Y, Sadeghi S, Finn RS, Ganapathy E, Song M, French SW, Naini BV, Sho S, Kaldas FM, Busuttil RW, Tomlinson JS, Tseng HR, Agopian VG, Liver Transplant. 2018, 24, 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Jiang R, Lu YT, Ho H, Li B, Chen JF, Lin M, Li F, Wu K, Wu H, Lichterman J, Wan H, Lu CL, OuYang W, Ni M, Wang L, Li G, Lee T, Zhang X, Yang J, Rettig M, Chung LW, Yang H, Li KC, Hou Y, Tseng HR, Hou S, Xu X, Wang J, Posadas EM, Oncotarget 2015, 6, 44781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Jiang W, Wang H, Cui Y, Lei Y, Wang Y, Xu D, Jiang N, Chen Y, Sun Y, Zhang Y, Cao J, Ke Z, Int. J. Nanomed 2018, 13, 1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Jan YJ, Yoon J, Chen J-F, Teng P-C, Yao N, Cheng S, Lozano A, Chu GCY, Chung H, Lu Y-T, Chen P-J, Wang JJ, Lee Y-T, Kim M, Zhu Y, Knudsen BS, Feng FY, Garraway IP, Gao AC, Chung LWK, Freeman MR, You S, Tseng H-R, Posadas EM, Theranostics 2019, 9, 2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Wu LL, Tang M, Zhang ZL, Qi CB, Hu J, Ma XY, Pang DW, Anal. Chem 2018, 90, 10518. [DOI] [PubMed] [Google Scholar]
- [237].Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS, Clin. Cancer Res 2014, 20, 2553. [DOI] [PubMed] [Google Scholar]
- [238].Lu YT, Zhao L, Shen Q, Garcia MA, Wu D, Hou S, Song M, Xu X, Ouyang WH, Ouyang WW, Lichterman J, Luo Z, Xuan X, Huang J, Chung LW, Rettig M, Tseng HR, Shao C, Posadas EM, Methods 2013, 64, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Chen JF, Ho H, Lichterman J, Lu YT, Zhang Y, Garcia MA, Chen SF, Liang AJ, Hodara E, Zhau HE, Hou S, Ahmed RS, Luthringer DJ, Huang J, Li KC, Chung LW, Ke Z, Tseng HR, Posadas EM, Cancer 2015, 121, 3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].McDaniel AS, Ferraldeschi R, Krupa R, Landers M, Graf R, Louw J, Jendrisak A, Bales N, Marrinucci D, Zafeiriou Z, Flohr P, Sideris S, Crespo M, Figueiredo I, Mateo J, de Bono JS, Dittamore R, Tomlins SA, Attard G, BJU Int. 2017, 120, E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Kim TH, Yoon HJ, Fouladdel S, Wang Y, Kozminsky M, Burness ML, Paoletti C, Zhao L, Azizi E, Wicha MS, Nagrath S, Adv. Biosyst 2019, 3, 1800278. [DOI] [PubMed] [Google Scholar]
- [242].Ankeny JS, Court CM, Hou S, Li Q, Song M, Wu D, Chen JF, Lee T, Lin M, Sho S, Rochefort MM, Girgis MD, Yao J, Wainberg ZA, Muthusamy VR, Watson RR, Donahue TR, Hines OJ, Reber HA, Graeber TG, Tseng HR, Tomlinson JS, Br. J. Cancer 2016, 114, 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Court CM, Ankeny JS, Sho S, Winograd P, Hou S, Song M, Wainberg ZA, Girgis MD, Graeber TG, Agopian VG, Tseng HR, Tomlinson JS, Ann. Surg. Oncol 2018, 25, 1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM, Eur. Urol 2016, 70, 93. [DOI] [PubMed] [Google Scholar]
- [245].Went P, Dirnhofer S, Salvisberg T, Amin MB, Lim SD, Diener PA, Moch H, Am. J. Surg. Pathol 2005, 29, 83. [DOI] [PubMed] [Google Scholar]
- [246].Rossi E, Fassan M, Aieta M, Zilio F, Celadin R, Borin M, Grassi A, Troiani L, Basso U, Barile C, Sava T, Lanza C, Miatello L, Jirillo A, Rugge M, Indraccolo S, Cristofanilli M, Amadori A, Zamarchi R, Br. J. Cancer 2012, 107, 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Liu S, Tian Z, Zhang L, Hou S, Hu S, Wu J, Jing Y, Sun H, Yu F, Zhao L, Wang R, Tseng H-R, Zhau HE, Chung LWK, Wu K, Wang H, Wu JB, Nie Y, Shao C, Oncotarget 2016, 7, 59877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, Ca-Cancer J. Clin 2018, 68, 394. [DOI] [PubMed] [Google Scholar]
- [249].El-Serag HB, Davila JA, Petersen NJ, McGlynn KA, Ann. Intern. Med 2003, 139, 817. [DOI] [PubMed] [Google Scholar]
- [250].Zimmerman MA, Ghobrial RM, Tong MJ, Hiatt JR, Cameron AM, Hong J, Busuttil RW, Arch. Surg 2008, 143, 182. [DOI] [PubMed] [Google Scholar]
- [251].Shah SA, Cleary SP, Wei AC, Yang I, Taylor BR, Hemming AW, Langer B, Grant DR, Greig PD, Gallinger S, Surgery 2007, 141, 330. [DOI] [PubMed] [Google Scholar]
- [252].Hou M, Zheng Y, Ding Z, He S, Xu M, Chen X, Zhang H, Zeng C, Sun C, Jiang W, Wang H, Shen H, Zhang Y, Liu J, Sun S, Jiang N, Cui Y, Sun Y, Chen Y, Cao J, Wang C, Li M, Zhang Y, Wang J, Lin M, Ke Z, Biomater. Sci 2019, 7, 1200. [DOI] [PubMed] [Google Scholar]
- [253].Chae YK, Pan AP, Davis AA, Patel SP, Carneiro BA, Kurzrock R, Giles FJ, Mol. Cancer Ther 2017, 16, 2645. [DOI] [PubMed] [Google Scholar]
- [254].Court CM, Ankeny JS, Sho S, Hou S, Li Q, Hsieh C, Song M, Liao X, Rochefort MM, Wainberg ZA, Graeber TG, Tseng HR, Tomlinson JS, J. Mol. Diagn 2016, 18, 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Rhodes DR, Chinnaiyan AM, Nat. Genet 2005, 37, S31. [DOI] [PubMed] [Google Scholar]
- [256].You S, Knudsen BS, Erho N, Alshalalfa M, Takhar M, Al-Deen Ashab H, Davicioni E, Karnes RJ, Klein EA, Den RB, Ross AE, Schaeffer EM, Garraway IP, Kim J, Freeman MR, Cancer Res. 2016, 76, 4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Chen P-J, Teng P-C, Zhu Y, Jan YJ, Smalley M, Afshar Y, Chen L-C, Pisarska MD, Tseng H-R, Curr. Obstet. Gynecol. Rep 2019, 8, 1. [PMC free article] [PubMed] [Google Scholar]
- [258].Yang B, Chen Y, Shi J, Adv. Mater 2019, 31, 1802896. [DOI] [PubMed] [Google Scholar]
- [259].Dong J, Zhang RY, Sun N, Smalley M, Wu Z, Zhou A, Chou S-J, Jan YJ, Yang P, Bao L, Qi D, Tang X, Tseng P, Hua Y, Xu D, Kao R, Meng M, Zheng X, Liu Y, Vagner T, Chai X, Zhou D, Li M, Chiou S-H, Zheng G, Di Vizio D, Agopian VG, Posadas E, Jonas SJ, Ju S-P, Weiss PS, Zhao M, Tseng H-R, Zhu Y, ACS Appl. Mater. Interfaces 2019, 11, 13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Jiang X, Wong KHK, Khankhel AH, Zeinali M, Reategui E, Phillips MJ, Luo X, Aceto N, Fachin F, Hoang AN, Kim W, Jensen AE, Sequist LV, Maheswaran S, Haber DA, Stott SL, Toner M, Lab Chip 2017, 17, 3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Jansson S, Bendahl P-O, Larsson A-M, Aaltonen KE, Rydén L, BMC Cancer 2016, 16, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, Scheidmann MC, Donato C, Scherrer R, Singer J, Beisel C, Kurzeder C, Heinzelmann-Schwarz V, Rochlitz C, Weber WP, Beerenwinkel N, Aceto N, Nature 2019, 566, 553. [DOI] [PubMed] [Google Scholar]