Summary
Taste systems detect a vast diversity of toxins, which are perceived as bitter. When a species adapts to a new environment, its taste system must adapt to detect new death threats. We deleted each of six commonly expressed bitter Gustatory receptors (Grs) from Drosophila melanogaster. Systematic analysis revealed that requirements for these Grs differed for the same tastant in different neurons, and for different tastants in the same neuron. Responses to some tastants in some neurons required four Grs, including Gr39a.a. Deletions also produced increased or novel responses, supporting a model of Gr-Gr inhibitory interactions. Co-expression of four Grs conferred several bitter responses to a sugar neuron. We then examined bitter coding in three other Drosophila species. We found major evolutionary shifts. One shift depended on the concerted activity of seven Grs. This work shows how the complex logic of bitter coding provides the capacity to detect innumerable hazards and the flexibility to adapt to new ones.
Keywords: Taste; Drosophila; taste receptor, bitter coding; evolution; D. simulans; D. sechellia; D. erecta
eTOC Blurb
Dweck and Carlson show that responses of some neurons to bitter tastants require four Gustatory receptors (Grs), and that coexpression of four Grs confers several bitter responses to sugar neurons. Loss of some Grs produces increased or novel responses, suggesting Gr-Gr inhibition. Two classes of bitter neurons arise in D. melanogaster via 7 Grs.
Introduction
The natural world abounds in toxins. Plants, which cannot flee their predators, synthesize toxic chemicals that deter them [1]. Animals, in turn, have evolved sensitive systems to detect toxins [2–4]. Taste systems allow recognition of many toxic chemicals as bitter. Such toxins are detected and encoded by bitter-sensing taste cells through mechanisms that are poorly understood. As a species evolves to inhabit a new niche, its taste system must evolve to detect the chemical threats in its new environment. Little is known about the logic of bitter coding, and even less is known about how this coding evolves.
Drosophila detects and avoids a wide variety of compounds that taste bitter to humans [5]. The main taste organ of the Drosophila head is the labellum, which contains 31 taste sensilla (Figure 1A) [6]. These sensilla fall into three morphological classes: Large (L), Intermediate (I) and Small (S) [7, 8]. Each sensillum contains a pore at the distal end. When the sensillum contacts a potential food source, compounds enter via the pore and may activate taste neurons within.
Figure 1. Labellar sensilla and sample phenotypes.
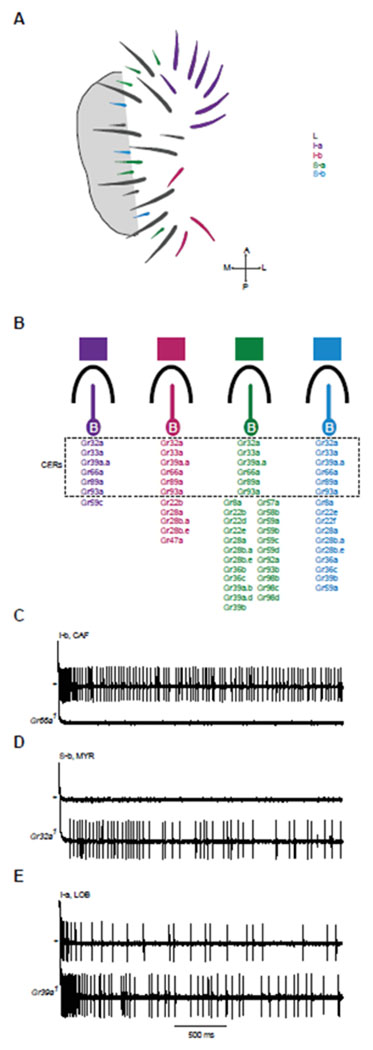
(A) Map of taste sensilla on the labellum. (B) The four classes of sensilla that contain a bitter-sensitive neuron, indicated by “B”. These sensilla all contain other neurons that are not shown. The bitter-sensitive neuron of each sensillum class expresses a different subset of Gr-GAL4 drivers, as indicated. Five of these drivers are expressed ubiquitously in all four classes and identify receptors that are called Commonly Expressed Receptors (CERs). Other drivers are expressed in subsets of sensillum types. Although one of these other drivers, Gr93a-GAL4, is expressed in a subset of sensilla, immunohistochemical analysis indicates that the Gr93a receptor is expressed in all bitter neurons; it is therefore classified as a CER and indicated as expressed in each sensillum type [10]. (C) Example of a physiological response that is lost in a CER mutant. CAF elicits a strong response from I-b in control flies, but not in Gr66a flies. (D) A novel response. MYR elicits no response from S-b in control flies, but elicits a response from Gr32a flies. (E) An increased response. LOB elicits a response from control flies, and a stronger response from Gr39a flies. Panels A and B are adapted from [19]. See also Table S1.
An electrophysiological analysis of the 31 sensilla with a diverse panel of bitter tastants identified four functional classes of bitter-sensitive sensilla, I-a, I-b, S-a, and S-b, each with a distinguishable response profile [5]. These I and S sensilla each have a neuron that responds to bitter compounds and that expresses members of the Gustatory receptor (Gr) family of taste receptors [9].
A collection of Gr-GAL4 drivers representing the entire family of 68 Gr receptors was used to map Grs to these bitter-sensing neurons [5]. The molecular map identified four classes of neurons, in good agreement with the physiological map (Figure 1B). Five Gr-GAL4 drivers were expressed in all bitter-sensing classes of the labellum, and 24 other drivers were expressed in some but not all classes. The five ubiquitous Gr-GAL4 drivers identified “Commonly Expressed Receptors” (CERs): Gr32a, Gr33a, Gr39a.a, Gr66a, and Gr89a (Figure 1B). A sixth receptor, Gr93a, was detected in all bitter-sensing classes by immunohistochemistry [10]. We therefore now classify it as a CER.
A variety of studies have shown that some individual receptors are required for the response to particular bitter compounds in certain sensilla [10–16]. Most of these studies have examined one or a small number of tastants in one or a small number of sensilla. These studies have demonstrated that three of the CERs, Gr32a, Gr33a, and Gr66a, are required for responses to various bitter compounds.
We have analyzed the requirements for all six CERs. We generated new deletions of five CER genes using CRISPR-Cas9 genome editing. These five deletions, and an existing Gr93a deletion [10], were each crossed into a standard genetic background and tested against a panel of 21 diverse bitter compounds in all classes of sensilla.
The results reveal that within an individual sensillum, different CERs underlie the response to different tastants. The response to an individual tastant requires different CERs in different sensilla. Responses to some tastants in some sensilla require four CERs, including Gr39a, which had not previously been implicated in bitter taste. The removal of each CER produces not only decreased responses, but also increased or novel responses to various tastants in various sensilla, supporting a model of extensive Gr-Gr inhibitory interactions. Analysis of bitter coding in three other Drosophila species revealed major evolutionary shifts in neuronal response specificities and in functional organization. One unique evolutionary feature of D. melanogaster arises through the concerted activities of seven Gr genes.
Results
We used CRISPR-Cas9 genome editing to create mutant alleles of Gr32a, Gr33a, Gr39a.a, Gr66a, and Gr89a. Each of these new mutant alleles, and an existing deletion of Gr93a [10], were backcrossed to our control genetic background for five generations to minimize genetic background effects.
We then tested the six mutants with a panel of 21 bitter compounds [5, 17, 18] in all four sensillum types (I-a, I-b, S-a, and S-b). Several kinds of phenotypes were observed. In 24% of the 504 mutation-tastant-sensillum combinations, the response was reduced compared to the response observed in our control genetic background (Figure 1C). The reduction was partial in approximately half of these cases and complete in the others.
By contrast, in some cases the response was greater in the mutant than in the control. We distinguished between a phenotype in which the control genotype shows no response but the mutant exhibits a response (“novel response”, Figure 1D, 4% of combinations), and a phenotype in which the control genotype shows a response but the mutant shows a greater response (“increased response”, Figure 1E, 4%).
Requirements for CERs differ among tastants and sensilla
Control I-a sensilla responded strongly to quinine (QUI), sparteine (SPS), escin (ESC), denatonium (DEN), berberine (BER), and lobeline (LOB), in agreement with previous findings (Figure 2 and Table S2; for ease of visualization, the tastants are listed in a different order for each sensillum type) [5, 19, 20]. The responses of I-a to all six of these tastants depend on three receptors, Gr32a, Gr33a, and Gr66a; mutations of any of these receptors result in a complete or near-complete loss of response. Responses to these six tastants were robust in mutants of the other three CERs. The reduced responses to these six tastants, represented in brown letters, are indicated in Figure 3.
Figure 2. Responses to tastants of bitter-sensing sensilla in CER mutants.
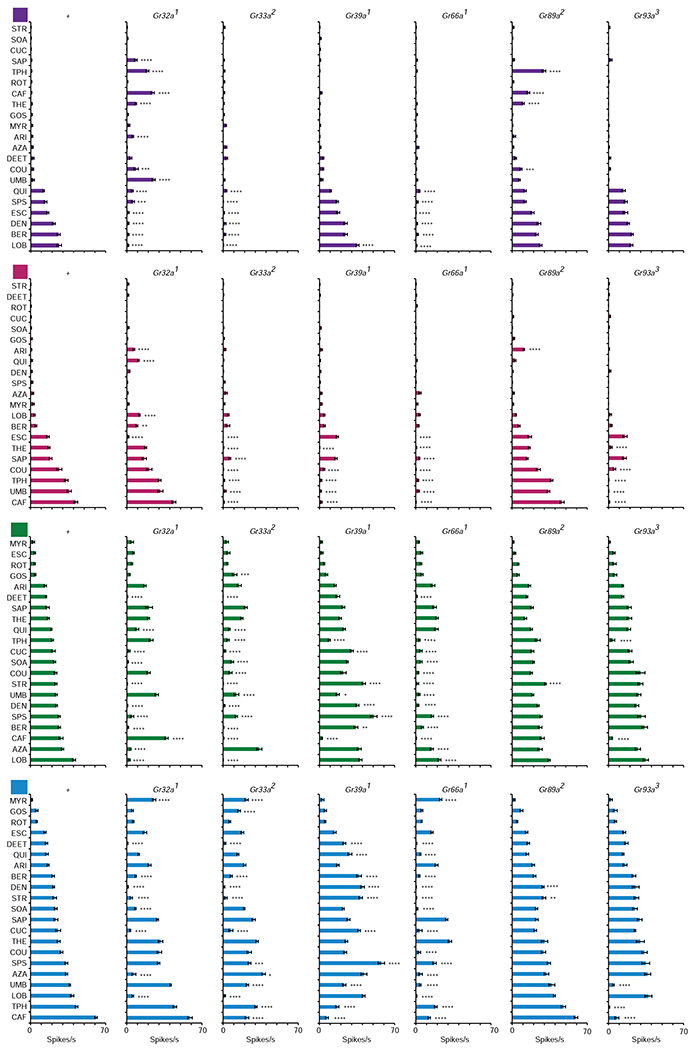
Each class of sensilla is tested with each of the 21 indicated tastants in each of the indicated 7 genotypes. For ease of visualization, the order of tastants is listed differently for each sensillum class. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; One-way ANOVA followed by Dunnett’s multiple comparison test, n≥10 for 98% of the 587 genotype-tastant-sensillum class combinations; 7≤n<10 for the remaining 2%. See also Figure S1 and Table S2.
Figure 3. Summary of phenotypes produced by mutations of different CERs in different sensilla.
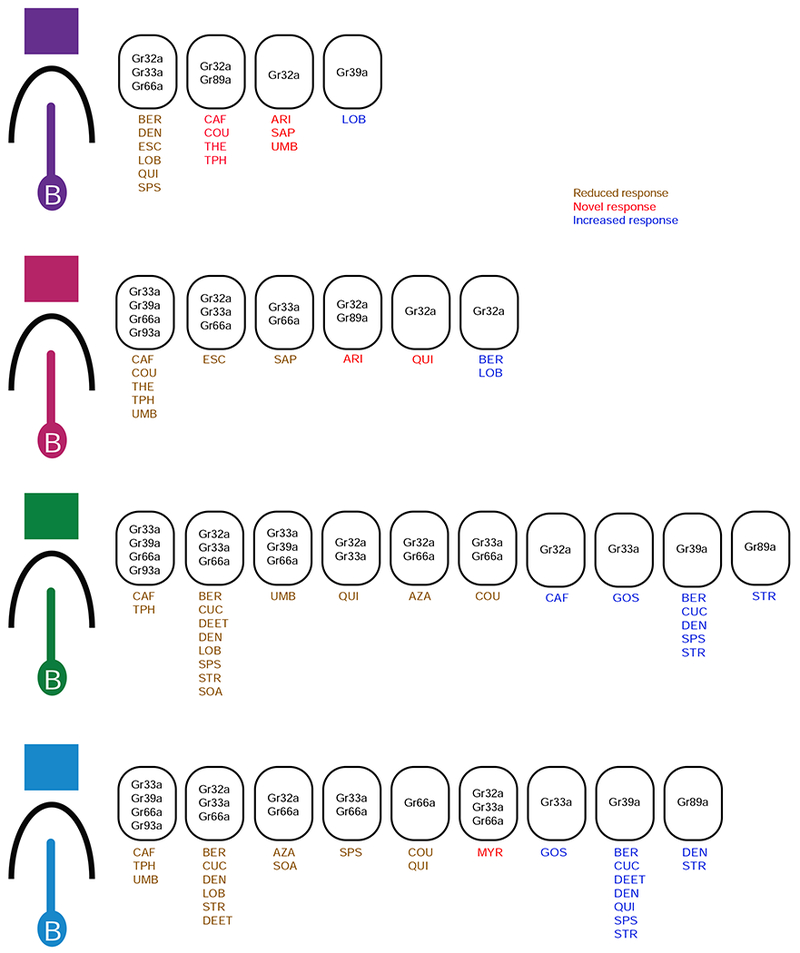
In each sensillum type, mutations of the receptors within the same oval all produce the same kind of phenotype, i.e. reduced, novel, or increased responses, to the indicated tastants. Thus in I-a sensilla, deletions of either Gr32a, Gr33a, or Gr66a produce a reduced response to BER, DEN, ESC, LOB, QUI, and SPS; loss of Gr32a or Gr89a produce a novel response to CAF, COU, THE, and TPH; loss of Gr32a but not Gr89a produces novel responses to ARI, SAP, and UMB. See also Figure S2.
I-b sensilla respond robustly to seven tastants of the panel: escin (ESC), theobromine (THE), saponin (SAP), coumarin (COU), theophylline (TPH), umbelliferone (UMB) and caffeine (CAF). Unlike I-a, in I-b the responses to different tastants required different subsets of CERs (Figures 2 and 3). Responses to five tastants (THE, COU, TPH, UMB, and CAF) required four receptors, Gr33a, Gr39a, Gr66a, and Gr93a. Response to ESC required three receptors: Gr32a, Gr33a, and Gr66a. Response to SAP required only two receptors, Gr33a and Gr66a.
S-a sensilla respond to a broad spectrum of tastants (Figure 2), which may reflect the broad array of receptors that they express (Figure 1B). As in I-b, responses to different tastants depended on different subsets of CERs. Responses to CAF and TPH were again reduced by removal of any of four CERs, but most responses were reduced by removal of any of three, either Gr32a, Gr33a, or Gr66a in most cases, or Gr33a, Gr39a, or Gr66a in one case. Each of three other tastants required two CERs, but a different pair in each case.
S-b sensilla also express many Grs and responded broadly to many tastants. Some responses required four CERs, some three, some two, and some only one.
In summary, there is no CER that is required for response to all tastants in all sensilla. Rather, the requirements for CERs differ among tastants and among sensilla. Within an individual sensillum, responses to different tastants depend on different CERs, and the response to an individual tastant depends on different CERs in various sensilla. Deletion of one CER gene, Gr89a, did not reduce the response to any tastant in any sensillum. Responses to one tastant, aristolochic acid (ARI), were not reduced by removal of any CERs, a finding consistent with the discovery that responses to ARI depend on TRPA1 [21].
Deletion of CERs produced novel responses
Deletion of CERs produced 10 novel responses, in I-a, I-b or S-b (Figures 2 and 3; red in Figure 3). The novel responses were to a variety of compounds: seven compounds in I-a, two in I-b, and one in S-b. No novel responses were observed in S-a. Novel responses were observed following removal of some, but not all CERs.
In I-a, the deletion of either Gr32a or Gr89a produced novel responses to four compounds. A novel response to three other tastants, including ARI, could be produced by deletion of Gr32a but not by deletion of Gr89a. By contrast, in I-b a novel response to ARI could be produced by either Gr32a or Gr89a. All 10 of the novel responses were observed at lower concentrations (Figure S2).
Deletion of CERs produced increased responses
In each of the four sensillum types we found examples in which deletion of a CER led to an increase in the magnitude of a response observed in control sensilla (Figures 2 and 3). Most of these increases were observed in mutants of Gr39a, but deletions of Gr32a, Gr33a, and Gr89a also produced increased responses.
Removal of some receptors had dual effects. For example, while the Gr39a deletion increased responses to seven tastants in S-b, it decreased responses to three other tastants in S-a.
Gr39a.a is required for normal feeding and oviposition behavior
The Gr39a locus had previously been found to be required for normal sexual behavior [22]. Given our finding that it is required for physiological response to the bitter compounds CAF, COU, THE, TPH, and UMB in I-b, and for responses to some of these compounds in S-a and S-b (Figures 2 and 3), we wondered if it played a role in feeding and oviposition preferences.
We tested the Gr39a deletion mutant in the FLIC (Fly Liquid-Food Interaction Counter) assay (Figure 4A) [23, 24]. We gave the fly a choice between two wells, one containing sucrose alone and the other containing sucrose and varying concentrations of a bitter compound. Whereas the control showed strong avoidance of the food containing bitter compounds, this avoidance was abolished or severely reduced by the Gr39a deletion in the case of all five bitter compounds (Figure 4B). As a control, we also tested DEN: the strong physiological responses to DEN were not reduced by deletion of Gr39a (Figure 2), and behavioral responses were not reduced either (Figure 4B).
Figure 4. Gr39a.a is required for normal feeding and oviposition behavior.
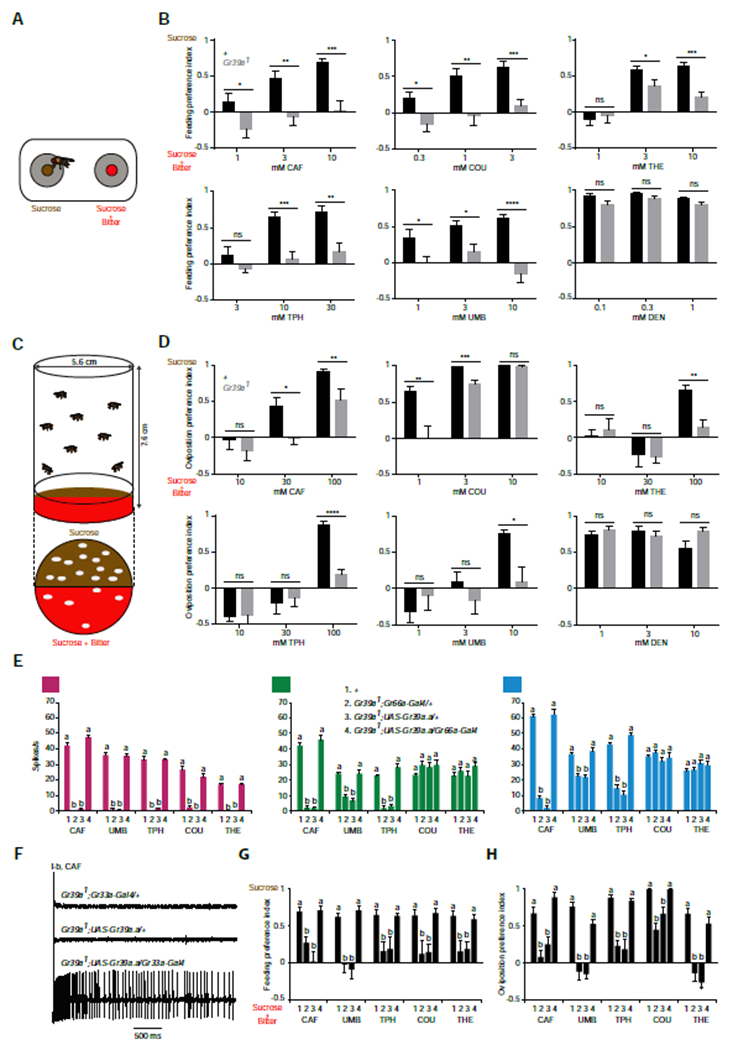
(A) Measuring feeding preference with the FLIC assay. Flies are given a choice between a solution containing sucrose alone or sucrose mixed with a bitter compound. (B) Gr39a mutants have a reduced preference for all bitter compounds tested except DEN. Mann-Whitney test, n=9-12. (C) Oviposition preference paradigm. The oviposition preference is based on the number of eggs laid on each half of the plate. (D) Gr39a mutants have a reduced preference for all bitter compounds tested except DEN. Mann-Whitney test, n=9-10. (E) The physiological defect of Gr39a can be fully rescued by expression of the Gr39a.a splice form. One-way ANOVA followed by Tukey’s multiple comparison test, n=6-12. (F) Sample traces from the rescue experiment shown in E. (G) The feeding preference is rescued fully by a Gr39a.a transgene. Each tastant is tested at 10 mM except that COU was tested at 3 mM. One-way ANOVA followed by Tukey’s multiple comparison test, n=10-18 (H) The oviposition preference is rescued fully by a Gr39a.a transgene. CAF, THE, and TPH were tested at 100 mM; UMB and COU were tested at 10 mM and 3 mM respectively. One-way ANOVA followed by Tukey’s multiple comparison test, n = 10. In (E-H), values indicated with different letters are significantly different. The key shown for (E) applies to (G,H). See also Figure S3.
In an oviposition paradigm, female flies were given a choice between a substrate containing sucrose alone or a mixture of sucrose and a bitter compound (Figure 4C). Consistent with previous results, control flies found low concentrations of some bitter compounds attractive, whereas high concentrations of all bitter compounds were aversive [25–27]. Deletion of Gr39a reduced the magnitude of preferences at one or more concentrations of all five bitter compounds (Figure 4D). As a control, oviposition preference was unchanged when DEN was tested.
Gr39a is one of a small number of Gr genes that is alternatively spliced [9, 28]. Our Gr39a deletion removes not only Gr39a.a, but also three other Gr39a splice forms. We asked whether the phenotypes could be rescued by the Gr39a.a splice form, which was the only splice form expressed in all bitter-sensitive sensilla in a Gr-GAL4 analysis [5]. Expression of Gr39a.a completely restored the loss of electrophysiological response to I-b, S-a, and S-b (Figures 4E and 4F). The feeding and oviposition phenotypes with all five tastants were also completely rescued by expression of Gr39a.a (Figures 4G and 4H).
We constructed a 32-dimensional physicochemical space in which each dimension represents a molecular descriptor, e.g. carbon chain length [29]. The five bitter compounds that depend on Gr39a.a, CAF, COU, THE, TPH, and UMB, cluster together in this space (Figure S3A). The mean Euclidean distance among these five tastants is shorter than the mean distance among all 21 tastants (0.4 ± 0.04 vs. 2.4 ± 0.06; p<0.0001, Mann-Whitney). The five clustered tastants are composed of exactly two fused heterocyclic rings (Figure S3B). In summary, the compounds that depend on a particular CER have a particular structure.
Gr39a.a operates in conjunction with three other Grs
Gr39a.a is one of several receptors that are required for response to CAF, COU, THE, TPH, and UMB. Besides Gr39a.a, Gr33a, Gr66a, and Gr93a are also required for responses to all five of these compounds in I-b sensilla (Figures 2 and 3). One possible interpretation of this surprisingly complex requirement is that these four Grs interact cooperatively to detect these tastants. An earlier study had misexpressed Gr33a, Gr66a, and Gr93a in a sweet-sensing neuron and had not detected a response to caffeine [14]. We asked whether Gr39a.a together with this trio would confer a response to bitter compounds.
We coexpressed all four Grs in a sugar-sensing neuron, using a Gr5a-GAL4 driver. Although parental control strains responded only to sucrose, expression of Gr33a, Gr39a.a, Gr66a, and Gr93a conferred response to all five bitter compounds (Figures 5A and 5B). We confirmed these results by testing responses at lower concentrations of all five tastants (Figure 5C). We also found that Gr93a, Gr33a, and Gr66a were essential components (Figure S4).
Figure 5. Coexpression of four Grs confers bitter response to sugar neurons.
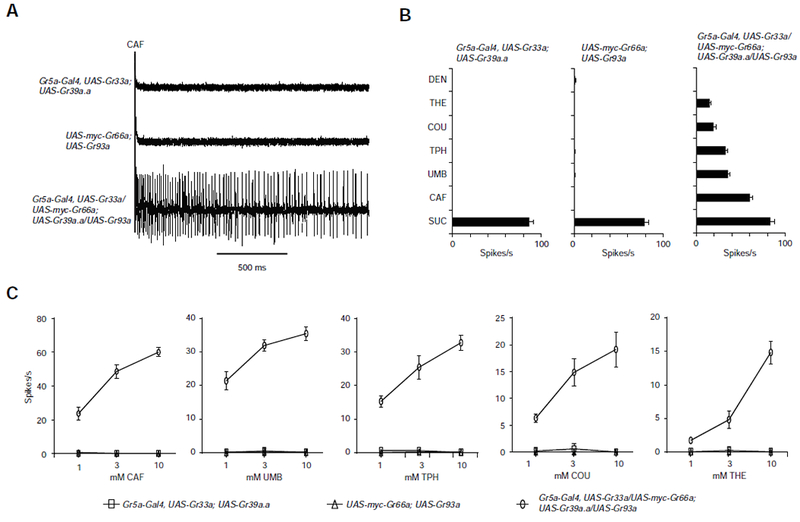
(A) Coexpression of four Grs in a sugar-sensitive neuron confers response to CAF. (B) Coexpression of four Grs in a sugar-sensitive neuron confers response to five bitter compounds. N=65-10. (C) Coexpression confers response across several concentrations of five bitter compounds. Recordings were from L sensilla. n=10. See also Figure S4.
Evolution of taste coding by the Drosophila labellum
The two classes of I sensilla are morphologically similar, but functionally distinct. CAF, COU, THE, TPH, and UMB elicit responses from I-b but not I-a; LOB, BER, and DEN elicit responses from I-a but not I-b. We wondered how such striking differences in response profiles evolve.
The sensitivities of taste sensilla presumably evolve to meet the needs of the organism to detect toxins in its environment. As species adapt to new environments, new threats must be detected. Little is known, however, about the mechanisms underlying the evolution of taste coding.
We extended our analysis to include the complete repertoire of taste sensilla on the labella of D. simulans, D. sechellia, and D. erecta, which diverged from D. melanogaster 2-3 million years ago (MYA), 3-5 MYA, and 10-12 MYA, respectively [30–33]. The numbers and positions of most sensilla are similar among the four species, suggesting that the developmental logic of patterning is broadly conserved (Figure S5A). In all species, none of the L sensilla respond to any of the bitter tastants (Figure S5B and Table S3). All species contain two S sensilla that showed no bitter responses. Hierarchical cluster analysis revealed that in each species, the sensilla divide into several functional classes (Figure 6). In each species, all L sensilla cluster together with two S sensilla to form a class that responds to none of the tested bitter compounds. All other functional classes are composed uniformly of sensilla of the same morphology, i.e. either small or intermediate. In each species, two functional classes consist exclusively of S sensilla, which we call S-a and S-b.
Figure 6. Clustering of labellar sensilla into functional classes.
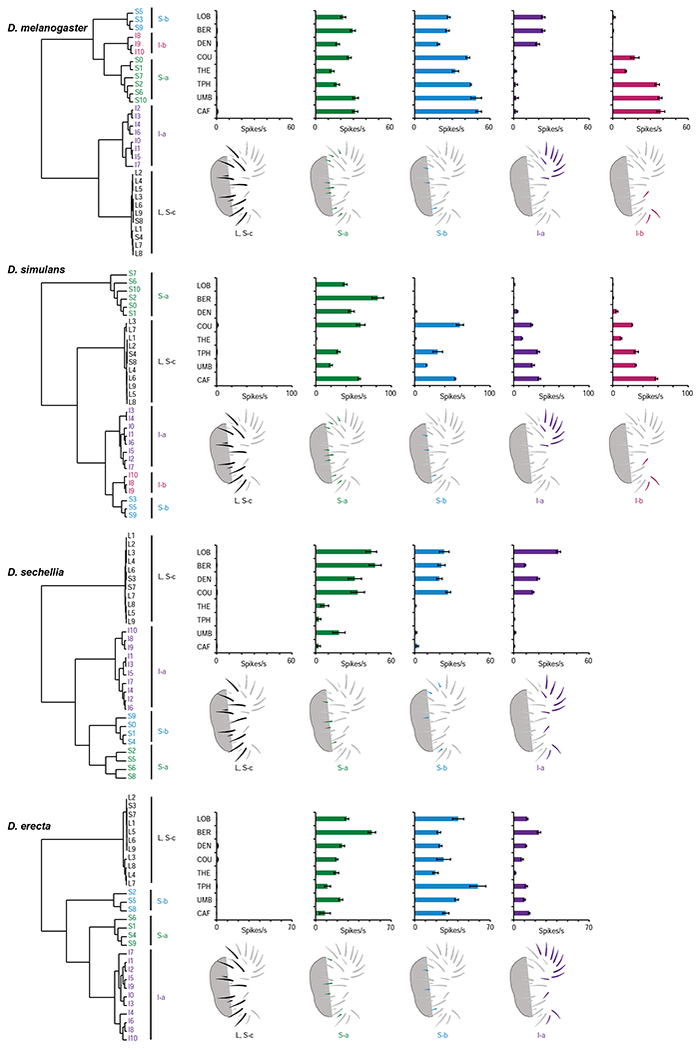
The graphs show the mean response to each tastant for all sensilla of a functional class. The maps show the distribution of the sensilla of each class. Data are from Figure 6. n=3-10. See also Figure S5 and Table S3.
I sensilla have undergone major evolutionary shifts in their functional organization (Figure 6). Only in D. melanogaster do the I sensilla fall into two functional classes whose response profiles are starkly different, i.e. complementary with respect to the test panel. In D. simulans, there are again two classes, but their profiles are strikingly similar; they are distinguishable primarily by their different magnitudes of response to a single tested tastant, CAF (p<0.0001 for differences in CAF response, Mann-Whitney; p < 0.01 for differences in entire profiles, R = 0.75; ANOSIM based on Bray-Curtis similarity; n=10). In D. sechellia and D. erecta, all I sensilla fall into a single class.
Other species show interesting similarities between the profiles of certain S and I classes. For example, in D. simulans, the profile of S-b is reminiscent of both I-a and Ib profiles (Figure 6). In D. sechellia, the profiles of S-a, S-b, and I-a are similar.
Major evolutionary shifts in sensitivity to particular compounds have occurred. For example, all bitter-sensing labellar sensilla in D. simulans respond to TPH and CAF; none do in D. sechellia (Figure S5B). THE elicits robust responses from D. melanogaster, but little or no response from sensilla in D. simulans or D. sechellia. D. simulans has evolved exceptionally strong responses to BER, stronger than any responses in D. melanogaster (Figure S5B). These differences between species may reflect differences in the prevalence of toxins that threaten these species in their natural environments. Alternatively, an individual species might be more or less sensitive to the lethality of a specific toxin, thereby increasing or decreasing the need to detect it with high sensitivity and reliability.
Molecular basis of the dichotomy in I sensilla of D. melanogaster
In the other three species all I sensilla were identical or very similar in their response profiles. How, then, did the two distinct response profiles of I sensilla evolve in D. melanogaster? Why do I-b, but not I-a sensilla, respond to CAF, COU, THE, TPH, and UMB?
We earlier carried out experiments with a Gr, Gr59c, which is not a CER, but rather is expressed in I-a but not I-b [19]. We found that misexpression of Gr59c in I-b induced a change in its response profile to one that was similar to I-a (Figure 7A). This finding suggested that in I-a, Gr59c acts in the suppression of responses to CAF and the other tastants that elicit responses from I-b. Consistent with this hypothesis, in a 17kb deletion mutant lacking Gr59c and several other genes, I-a sensilla underwent a phenotypic switch and adopted a profile like that of I-b, responding to CAF, COU, THE, TPH, and UMB (Figure 7A).
Figure 7. Regulation of I-a identity by seven Grs.
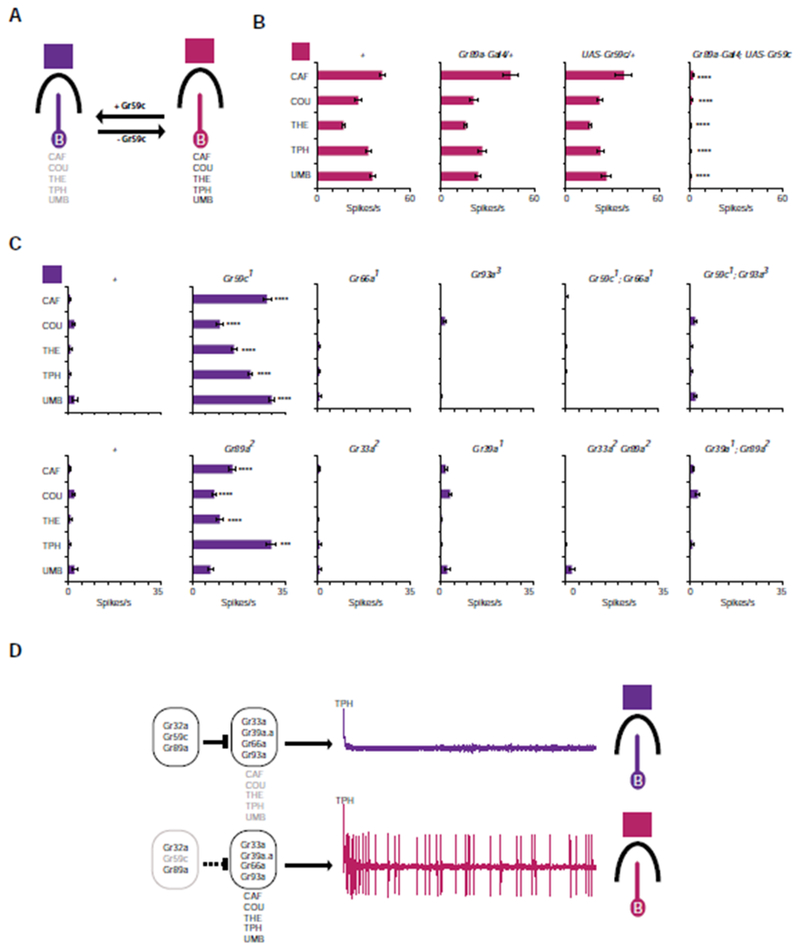
(A) Summary of previous data showing that misexpression of Gr59c in I-b induced a change in response profile to one resembling I-a, and that loss of Gr59c from I-a induced a change in profile to one resembling I-b [19]. (B) Confirmation that misexpression of Gr59c in I-b suppresses response to bitter compounds. The values for the control genotype “+” were from Figure 2. One-way ANOVA followed by Dunnett’s multiple comparison test, n=6-10. (C) New mutations confirm that loss of Gr59c or Gr89a from I-a leads to novel responses to TPH, CAF, THE, and COU; these responses depend on Gr66a, Gr93a, Gr33a, and Gr39a as shown. One-way ANOVA followed by Dunnett’s multiple comparison test, n=10-23. (D) Model for regulation of bitter response in I sensilla. Expression of four receptors, Gr33a, Gr39a.a, Gr66a, and Gr93a, together confer response to CAF, COU, THE, TPH, and UMB. In I-a, Gr32a, Gr59c, and Gr89a are all required to block the four receptors from conferring responses to these tastants. See also Figure S6.
To investigate the molecular determinants of the I-a and I-b taste profiles, we first confirmed the finding that ectopic expression of Gr59c in I-b sensilla suppressed response to these tastants (Figure 7B); it also suppressed responses to most of these tastants in S sensilla (Figure S6A). We then used CRISPR-Cas9 genome editing to construct a Gr59c deletion whose endpoints lie within Gr59c. This more precise mutation also induced a phenotypic switch from I-a to I-b (Figure 7C, Figure S6B). These results support a model in which Gr59c acts in I-a to suppress the response to CAF, COU, THE, TPH, and UMB. Our analysis of CER deletions showed that Gr32a and Gr89a also suppress responses to CAF, TPH, THE, and COU in I-a (Figures 2 and 3). These results suggested that Gr59c, Gr32a, and Gr89a cooperate to suppress these responses in I-a; they cannot cooperate in I-b, which lacks Gr59c. In support of this notion of cooperation, we found that expression of Gr59c in I-b did not suppress response to the tested bitter compounds in Gr32a or Gr89a mutants (Figure S6C).
Next we asked whether the phenotypic switch from I-a to I-b depended on any other CERs. Since we had found that four Grs, Gr66a, Gr93a, Gr33a, and Gr39a, are necessary for the response of CAF, COU, THE, TPH, and UMB in I-b sensilla (Figure 2), and together they confer response to these tastants in sugar-sensing neurons (Figure 5), we asked whether they are required for the phenotypic switch from I-a to I-b that is observed in mutants of Gr59c and Gr89a.
To address this question we constructed a series of double mutants containing each of the four Gr mutations, Gr66a, Gr93a, Gr33a, and Gr39a, together with either the Gr59c or Gr89a deletion. All four genes, Gr66a, Gr93a, Gr33a, and Gr39a, are required for the phenotypic switch from I-a to I-b; in none of the double mutants did I-a respond to CAF, COU, THE, TPH, or UMB (Figure 7C).
Thus the generation of the two distinct I sensillum types in D. melanogaster require the deployment and activity of seven Gr genes. Taken together, the results support a model in which Gr39a.a, Gr33a, Gr66a, and Gr93a cooperate to induce the response to CAF, COU, THE, TPH, or UMB in I-b, but that Gr32a, Gr59c, and Gr89a together suppress this induction in I-a (Figure 7D).
Discussion
The logic of bitter coding in D. melanogaster
One basic principle illustrated by our study is that the logic of bitter signaling is much more complex than that of olfactory signaling. Most olfactory receptor neurons express a single Odor receptor (Or) gene, together with a single co-receptor gene, Orco [34, 35]. The tuning profile of the neuron depends on the Or [36, 37]; trafficking of the Or to the membrane and its signaling depend on Orco [35, 38]. By contrast, requirements for signaling within bitter neurons are highly variable, depending on both the tastant and the neuron. For most bitter compounds several CERs are required to ensure a normal response, in addition to whichever Grs may be required from among those that are expressed in subsets of bitter-sensing neurons.
Within an individual neuron type, the responses to different tastants rely on different CERs. For example, in S-b sensilla, CAF depended on Gr33a, Gr39a, Gr66a, and Gr93a, whereas AZA depended on Gr32a and Gr66a. For an individual tastant, responses rely on different CERs in different neuron types. For example, SPS depended on Gr32a in S-a but not in S-b.
The number of CERs that are required for response was highly variable (Figures 2 and 3). The responses to several tastants in I-b sensilla depend on four CERs, Gr39a.a, Gr33a, Gr66a, and Gr93a. At the other extreme, SAP and THE depend on none of the CERs in S-a and S-b, and ARI depends on none of the tested CERs in any sensillum.
Not only does the response of I-b sensilla to several bitter compounds (CAF, COU, THE, TPH, and UMB) require Gr39a.a, Gr33a, Gr66a, and Gr93a, but the requirement is absolute: mutation of any of these four Grs virtually eliminates response. This finding argues against a model in which the four Grs act in two parallel, independent mechanisms that each confers a partial response within an I-b neuron. Rather, the finding supports a model in which the four Grs are obligate components of a single mechanism.
We know of no previous case in which four Grs, or four distinct members of any insect chemoreceptor family, have been shown to be required for a chemosensory response. Expression of the four Grs in a sugar-sensing cell also conferred response to these tastants. The four Grs could form a single heteromeric receptor, or perhaps two heteromeric complexes of which one acts as a receptor and the other a co-receptor that functions in its trafficking or signalling. A recent cryo-EM study revealed a tetrameric structure for Orco, which is a member of the superfamily that includes Grs [39]. It will be interesting to determine whether Grs also form tetramers.
Our finding that some neurons require different subsets of Grs for response to different tastants supports a model in which multiple heteromeric receptors form within the neuron. For example, in S-b, different heteromeric receptors might mediate responses to CAF and AZA. According to this model, the coexpression of many Grs within a neuron would allow the formation of diverse multimeric receptors. This kind of intraneuronal combinatorial assembly of receptors could contribute to the ability of the taste system to detect a vast diversity of bitter compounds.
Consistent with this concept, mutations of CERs in some cases reduced but did not abolish response. Modest reductions in response occurred primarily in S sensilla, which express the greatest number of Grs [5]. Perhaps the proliferation of Grs in these sensilla provides multiple receptors through which a tastant can signal.
The simplest model of Gr function would predict that loss of any Gr would produce either a decrease in response, or no change in response if the Gr had a redundant function. However, we observed many cases in which the deletion of a Gr gene led to an increase in an existing response or a novel response.
This diversity of phenotypes has not been reported in the olfactory system. It supports a model of extensive Gr-Gr interactions within bitter neurons. A Gr may inhibit another Gr by binding to it, activate another Gr by binding to it, or compete with another Gr for binding to a required signaling factor. This model was elaborated largely to explain the results of a gain-of-function study in which a series of Grs were systematically misexpressed in bitter neurons [19].
The present loss-of-function study supports the model in two ways. First, the increased responses we observed when a CER was mutated may be interpreted in terms of inhibition. Such a Gr may partially inhibit the activity of another Gr by binding to it: when the former Gr is eliminated, the latter Gr then signals at a higher level. Second, the novel responses we observed when a CER was mutated may also be explained by inhibition. Such a Gr may completely inhibit the activity of another Gr by binding to it. Thus when the former Gr is mutated, the latter Gr provides a signal that is not observed in wild type.
Deletion of most CERs produced at least some decreases in response in at least some neurons. Loss of Gr89a, however, did not reduce the response to any tastant in any bitter neuron. Gr89a may be required for response to a bitter compound that we did not test, or its role may be largely redundant with another Gr, at least in the labellum. Loss of Gr89a was, however, accompanied by a novel response to TPH and some other tastants in I-a sensilla, suggesting that it interacts with and inhibits other Grs in these sensilla.
When we began this study we were attracted by a model in which CERs were largely distinct from other Grs in function, analogous to the distinction between Orco and Ors. Our finding that expression of four CERs in sugar neurons confers a response to bitter compounds does not support this model in its simplest form.
The evolution of bitter coding in Drosophila
We analyzed the coding of bitter tastants on the labella of three additional Drosophila species. Several conclusions emerged.
The functional organization of the sensilla was conserved in some respects. The numbers and positions of sensilla, although not identical, were largely conserved across species. L sensilla did not respond to bitter compounds in any species; S and I sensilla responded in all species. In each species bitter-sensitive sensilla fell into several functional classes, each class composed solely of either S or I sensilla.
Other features showed evolutionary plasticity. Each class of S and I sensilla was distinct from all other classes in all other species (p < 0.0001; R = 0.95-0.96 for S-a, S-b, and I sensilla; ANOSIM based on Bray-Curtis similarity). Whereas in D. melanogaster the S sensilla are broadly tuned and the I sensilla are more narrowly tuned, in D. simulans and D. sechellia S-b sensilla are more narrowly tuned and have response profiles similar to those of their I sensilla counterparts. Thus tuning breadth can expand or contract over evolutionary time.
Sensitivity to individual tastants has also waxed or waned during the evolution of these species, e.g. the conspicuous absence or near absence of responses to CAF, TPH and THE in D. sechellia. D. sechellia is highly specialized in that it reproduces on the fruit of Morinda citrifolia, a tree of the coffee family [40]. Perhaps the loss of CAF (caffeine) response in D. sechellia represents an evolutionary mechanism for preventing high-level activation of aversive taste circuits by compounds in this fruit.
The evolutionary plasticity of bitter responses across Drosophila species is in contrast to the relatively conserved olfactory response profiles among antennal sensilla [41]. Most of the odor stimuli to which flies respond have a positive valence and signify the presence of a food source [42, 43]. The bitter stimuli tested here have a negative valence and signify the presence of a potentially lethal toxin. Perhaps a system that detects death threats is under particularly strong selective pressure to adapt rapidly to new environments.
The ability of the bitter detection system to adapt rapidly may be especially important if their host plants are evolving new toxins rapidly. Many plants are in an evolutionary arms race with insect herbivores and are under selective pressure to produce new toxins to defend themselves [44–46]. The rapid evolution of bitter coding in Drosophila may reflect the need to recognize toxins that plants evolve to defend themselves against any of a variety of other herbivores.
Gr39a has evolved quickly and has been implicated in sexual behavior [22]. We found that Gr39a is essential for the physiological response to bitter tastants and for feeding and oviposition behaviors. Gr39a is like Gr32a in being required for both sexual behavior and labellar bitter response [11, 14, 47]. However, unlike Gr32a, Gr39a is alternatively spliced, and a large exon that has been especially dynamic over evolutionary time encodes most of the Gr39a.a splice form [9, 22, 28, 48]. This splice form completely rescued the taste phenotypes. Perhaps another Gr39a splice form rescues the sexual behavior phenotype. We note that a Gr39a.a-GAL4 driver is expressed not only in the labellum, but also in the legs [5, 49, 50].
Strikingly, most of the increased responses that we observed in our genetic analysis were observed in the Gr39a mutant (Figure 3). Gr39a.a could play a central role in the regulation of taste responses by inhibiting other Grs. Changes in the structure of Gr39a.a could translate into changes in the levels or activity of several Gr complexes, thereby providing a flexible and efficient regulatory mechanism useful in adapting to new environments.
In a larger sense, interactions among Grs could provide a general mechanistic explanation for the relatively rapid evolution of bitter coding. The model of a network of Grs capable of both positive and negative interactions provides a means of adaptation that is not available to olfactory neurons that express only a single Or and Orco [34, 35].
We identified a mechanism through which two classes of I sensilla with different response profiles originate in D. melanogaster via the concerted action of seven Gr genes. No comparable mechanism has been identified in the olfactory system of Drosophila species. Rather, evolution of the olfactory system has been shown to proceed via changes in receptor sequences [51–53], expansion of the number of ORNs expressing a particular receptor [41, 54, 55], or changes in central circuitry [56].
Taken together our analysis supports a model of taste coding that contains many degrees of freedom. There are multiple bitter-sensing neurons with distinct response profiles, multiple Grs in each bitter-sensing neuron, and multiple splice forms for some Grs. At least seven Grs are required to specify the identities of the two I sensillum types in D. melanogaster, and our results support a model in which interactions among Grs underlie bitter coding. Together these degrees of freedom endow the taste system with a rich capacity to evolve and to facilitate adaptation to new chemical environments.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, John R. Carlson (john.carlson@yale.edu). All unique and stable reagents generated in this study are available from the Lead Contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila stocks
Flies were reared on corn syrup and soy flour culture medium (Archon Scientific) at 25°C and 60% relative humidity in a 12: 12-hour light–dark cycle. Gr93a3 (LB27592) was obtained from the Bloomington Drosophila Stock Center. UAS-myc-Gr66a was obtained from Dr. Craig Montell. D. simulans (14021-0251.001), D. sechellia (14021-0248.27) and D. erecta (14021-0224.01) were obtained from the Drosophila Species Stock Center.
METHOD DETAILS
Transgenic flies
Gr-GAL4 drivers (Gr5a-Gal4, Gr66a-Gal4 and Gr89a-Gal4) are described in Weiss et al. (2011). UAS-Gr lines were generated by amplification of Gr coding regions from Canton-S cDNA. UAS-Gr33a, UAS-Gr59c and UAS-Gr93a were cloned into the pUAST expression vector and inserted into the genome using random transposon integration. UAS-Gr39a.a was cloned from gDNA into the pBI-UASCG expression vector and inserted into the genome with the phiC31 site-specific integration system (site #8622).
Gr Deletions
Gr deletions were generated using CRISPR/Cas9 homologous recombination. Guide RNAs (gRNAs) were designed using the flyCRISPR Optimal Target Finder. The gRNA plasmid was generated using pCFD4 (Addgene 49411), following the protocol described in (Port et al., 2014). Gibson Assembly was performed using Gibson Assembly Master Mix (New England BioLabs, E2611S). gRNA plasmid and a donor plasmid (pDsRed-attP, Addgene 51019) with homologous arms and a DsRed marker were injected into embryos by Bestgene, Inc. (Chino Hills, CA). DsRed positive alleles were then backcrossed to our control w1118 Canton-S line for five generations. Oligonucleotides used to generate gRNA and donor vectors, and to confirm Gr deletions, are shown in Table S1.
Bitter Tastants
Bitter tastants were obtained at the highest available purity from Sigma-Aldrich. The compounds are structurally diverse, including naturally occurring alkaloids, terpenoids, and phenolic compounds, and some synthetic compounds such as the insect repellent DEET (N, N-Diethyl-meta-toluamide). All tastants were tested at high concentrations, near the limit of their solubility. All tastants were dissolved in 30 mM tricholine citrate (TCC), an electrolyte that inhibits the water neuron [57]. All tastants were prepared fresh and used for no more than one day. For each set of experiments, all fly lines were tested in the same day by a single tastant. For electrophysiological recordings, tastants were tested at the following concentrations unless otherwise indicated: aristolochic acid (ARI), 1 mM; azadirachtin (AZA), 1 mM; berberine chloride (BER), 1 mM; caffeine (CAF), 10 mM; coumarin (COU), 10 mM; cucurbitacin I hydrate (CUC), 1 mM; N, N-Diethyl-m-toluamide (DEET), 10 mM; denatonium benzoate (DEN), 10 mM; escin (ESC), 1 mM; gossypol from cotton seeds (GOS), 1 mM; (−)-lobeline hydrochloride (LOB), 1 mM; myricetin(MYR), 1 mM; quinine (QUI), 1 mM; rotenone (ROT), 1 mM; saponin from quillaja bark (SAP), 0.1%; D-(+)-sucrose octaacetate (SOA), 1 mM; sparteine sulfate salt (SPS), 10 mM; strychnine nitrate salt (STR), 10 mM; theobromine (THE), 10 mM; theophylline (TPH), 10 mM; umbelliferone (UMB), 10 mM. All compounds were stirred for 24 hours. THE and UMB were additionally heated to increase their solubility, then cooled and tested while in solution.
Electrophysiology
Electrophysiological recordings were performed with the tip-recording method [58], with some modifications. 5-7 days old mated female flies were used. Flies were immobilized in pipette tips, and the labellum was placed in a stable position on a glass coverslip. A reference tungsten electrode was inserted into the eye of the fly. The recording electrode consists of a fine glass pipette (10–15μm tip diameter) and connects to an amplifier with a silver wire. This pipette performs the dual function of recording electrode and container for the stimulus. Recording started the moment the glass capillary electrode was brought into contact with the tip of the sensillum. Signals were amplified (10x; Syntech Universal AC/DC Probe; www.syntech.nl), sampled (10,667 samples/s), and filtered (100–3000 Hz with 50/60-Hz suppression) via a USB-IDAC connection to a computer (Syntech). Action potentials were extracted using Syntech Auto Spike 32 software. Responses were quantified by counting the number of spikes generated over a 500 ms period after contact. Response to the TCC diluent was not subtracted in any case; TCC responses were small and are shown in Table S2. When recording from sensilla of a particular class, e.g. I-a, all sensilla of that class, i.e. I0-I6, were tested. Sensilla on both halves of the labellum were tested.
Two-Choice Feeding Assay
Two-choice feeding experiments were performed using the FLIC (Fly Liquid-Food Interaction Counter) Drosophila Behavior System. In this paradigm, liquid food is contained in a well that is surrounded by a metal ring. To reach the food, a fly needs to stand on the ring; when the fly feeds, it closes an electrical circuit between the metal ring, the liquid food, and another metal sheet at the bottom of the well. An electrical signal is generated when the fly feeds, and such electrical signals serve as a good measure of ingestion [23]. This system was from Sable Systems International (North Las Vegas, NV) and was described by Ro et al. (2014). The FLIC Monitor Software (version 2.1, downloaded from wikiflic.com) was used to collect raw data from the Drosophila Feeding Monitor (DFM). Before experiments, 5–7 days old mated flies were transferred to a vial containing moistened Kimwipes and starved at room temperature for 18–21 hours. Each of the two channels of the DFM was loaded with either 100 mM sucrose alone or or a mixture of 100 mM sucrose and the indicated concentration of bitter compound. An individual fly was introduced in an arena through a hole in an acrylic ceiling using an aspirator and allowed to choose between sucrose alone and the mixture of sucrose and bitter compound. The two-choice feeding assay was performed for 3 hours. To automate the analysis of raw FLIC data, we wrote custom software in the R programming language (Tam, 2017). Source code for the software can be obtained at https://github.com/edrictam/FLIC-analysis (with a copy archived at https://github.com/elifesciences-publications/FLIC-Analysis. Feeding Preference indices were calculated as (Total number of sips from sucrose alone-Total number of sips from sucrose and bitter compound)/Total number of sips. For each set of experiments, all fly lines were tested at the same time. Tastants were prepared fresh in water on the same day of the experiment.
Two-Choice Oviposition Assay
The two-choice oviposition assay was modified from Joseph et al. (2009), except that corn meal food was replaced with 1% agar containing 100 mM sucrose. Oviposition plates consisted of plastic Petri dishes (60 X 15 mm, Falcon), which were divided into two halves; each half contained either sugar or sugar mixed with a bitter compound. Twenty-five newly eclosed flies (five males, twenty females) were transferred to fly food vials and kept at 25°C and 60% relative humidity in a 12: 12-hour light-dark cycle. Flies, when 5-7d old, were placed into an oviposition cage (Genesee Scientific) without anesthesia through a small funnel that fits in the lid of the cage, and left for 24 hours in the dark. Eggs were counted from each substrate. An oviposition preference index was calculated as follows: (number of eggs on sucrose substrate – number of eggs on sucrose-bitter substrate)/(total number of eggs on both substrates).
QUANTIFICATION AND STATISTICAL ANALYSIS
Hierarchical cluster analyses were performed using Ward’s method with PAST (paleontological statistics software package for education and data analysis; Hammer et al., 2001). This technique organizes the data into clusters based on the response profiles of each sensillum to the panel of tastants. Euclidean distances were calculated according to Ward’s classification method for the hierarchical cluster analysis. Other statistical tests were performed in GraphPad Prism (version 6.01). All error bars are SEM. Molecular descriptors were calculated by Dragon (http://www.talete.mi.it). Descriptors were z-scores normalized for principal component analysis. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
DATA AND CODE AVAILABILITY
This study did not generate any datasets or new code. All raw data is available upon request.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Aristolochic acid (ARI) | Sigma-Aldrich | Cat# A5512 |
| Azadirachtin (AZA) | Sigma-Aldrich | Cat# A7430 |
| Berberine chloride (BER) | Sigma-Aldrich | Cat# Y0001149 |
| Caffeine (CAF) | Sigma-Aldrich | Cat# C1778 |
| Coumarin (COU) | Sigma-Aldrich | Cat# C4261 |
| Cucurbitacin I hydrate (CUC) | Sigma-Aldrich | Cat# C4493 |
| N,N-Diethyl-m-toluamide (DEET) | Sigma-Aldrich | Cat# 36542 |
| Denatonium benzoate (DEN) | Sigma-Aldrich | Cat# D5765 |
| Escin (ESC) | Sigma-Aldrich | Cat# E1378 |
| (±)-Gossypol from cotton seeds (GOS) | Sigma-Aldrich | Cat# G8761 |
| (−)-Lobeline hydrochloride (LOB) | Sigma-Aldrich | Cat# 141879 |
| Myricetin (MYR) | Sigma-Aldrich | Cat# 72576 |
| Quinine (QUI) | Sigma-Aldrich | Cat# 22620 |
| Rotenone (ROT) | Sigma-Aldrich | Cat# 45656 |
| Saponin (SAP) | Sigma-Aldrich | Cat# 47036 |
| D-(+)-sucrose octaacetate (SOA) | Sigma-Aldrich | Cat# W303801 |
| Sparteine sulfate salt (SPS) | Sigma-Aldrich | Cat# 234664 |
| Strychnine nitrate salt (STR) | Sigma-Aldrich | Cat# S2880 |
| Theobromine (THE) | Sigma-Aldrich | Cat# T4500 |
| Theophylline (TPH) | Sigma-Aldrich | Cat# T1633 |
| Umbelliferone (UMB) | Sigma-Aldrich | Cat# H24003 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: wCS; Gr32a1 | Figure 2 | N/A |
| D. melanogaster: wCS; Gr33a2 | Figure 2 | N/A |
| D. melanogaster: wCS; Gr39a1 | Figure 2 | N/A |
| D. melanogaster: wCS; Gr66a1 | Figure 2 | N/A |
| D. melanogaster: wCS; Gr89a2 | Figure 2 | N/A |
| D. melanogaster: wCS; Gr93a3 | Bloomington Drosophila Stock Center | Stock# 27592 |
| D. melanogaster: wCS; Gr66a-Gal4 | Dr. John Carlson’s lab | [5] |
| D. melanogaster: wCS; UAS-Gr39a.a | Dr. John Carlson’s lab | N/A |
| D. melanogaster: w; UAS-Gr33a | Dr. John Carlson’s lab | N/A |
| D. melanogaster: w; UAS-Gr39a.a | Dr. John Carlson’s lab | N/A |
| D. melanogaster: w; UAS-Gr66a | Dr.Craig Montell’s Lab | [13] |
| D. melanogaster: w; UAS-Gr93a | Dr. John Carlson’s lab | N/A |
| D. melanogaster: w; Gr5a-Gal4 | Dr. John Carlson’s lab | [5] |
| D. melanogaster: wCS; Gr59c1 | Figure 7 | N/A |
| D. melanogaster: wCS; Gr89a-Gal4 | Dr. John Carlson’s lab | [5] |
| D. melanogaster: wCS; UAS-Gr59c | Dr. John Carlson’s lab | [5] |
| D. simulans | Drosophila Species Stock Center | Stock# 14021-0251.001 |
| D. sechellia | Drosophila Species Stock Center | Stock# 14021-0248.27 |
| D. erecta | Drosophila Species Stock Center | Stock# 14021-0224.01 |
Highlights.
Responses of some neurons to some bitter tastants require 4 Grs, including Gr39a.a
Co-expression of 4 Grs confers several bitter responses to a sugar neuron
Loss of some Grs produces larger or new responses, suggesting Gr-Gr inhibition
Evolutionary shifts in taste sensilla have arisen in D. melanogaster via 7 Grs
ACKNOWLEDGMENTS
We thank Zina Berman for support and the other members of the Carlson laboratory for discussion. Supported by a Merck fellowship from the Life Sciences Research Foundation to H.K.M.D and NIH R01 DC11697, NIH R01 DC02147, and NIH R01 DC04729 to J.R.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Ibanez S, Gallet C, and Despres L (2012). Plant insecticidal toxins in ecological networks. Toxins (Basel) 4, 228–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liman ER, Zhang YV, and Montell C (2014). Peripheral coding of taste. Neuron 81, 984–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soldano A, Alpizar YA, Boonen B, Franco L, Lopez-Requena A, Liu G, Mora N, Yaksi E, Voets T, Vennekens R, et al. (2016). Gustatory-mediated avoidance of bacterial lipopolysaccharides via TRPA1 activation in Drosophila. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wada-Katsumata A, Silverman J, and Schal C (2013). Changes in taste neurons support the emergence of an adaptive behavior in cockroaches. Science 340, 972–975. [DOI] [PubMed] [Google Scholar]
- 5.Weiss LA, Dahanukar A, Kwon JY, Banerjee D, and Carlson JR (2011). The molecular and cellular basis of bitter taste in Drosophila. Neuron 69, 258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stocker RF (1994). The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res 275, 3–26. [DOI] [PubMed] [Google Scholar]
- 7.Hiroi M, Marion-Poll F, and Tanimura T (2002). Differentiated response to sugars among labellar chemosensilla in Drosophila. Zoolog Sci 19, 1009–1018. [DOI] [PubMed] [Google Scholar]
- 8.Shanbhag SR, Park SK, Pikielny CW, and Steinbrecht RA (2001). Gustatory organs of Drosophila melanogaster: fine structure and expression of the putative odorant-binding protein PBPRP2. Cell Tissue Res 304, 423–437. [DOI] [PubMed] [Google Scholar]
- 9.Clyne PJ, Warr CG, and Carlson JR (2000). Candidate taste receptors in Drosophila. Science 287, 1830–1834. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Moon SJ, and Montell C (2009). Multiple gustatory receptors required for the caffeine response in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 106, 4495–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y, Kim SH, and Montell C (2010). Avoiding DEET through insect gustatory receptors. Neuron 67, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y, Moon SJ, Wang Y, and Montell C (2015). A Drosophila Gustatory Receptor Required for Strychnine Sensation. Chem Senses 40, 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon SJ, Kottgen M, Jiao Y, Xu H, and Montell C (2006). A taste receptor required for the caffeine response in vivo. Current biology : CB 16, 1812–1817. [DOI] [PubMed] [Google Scholar]
- 14.Moon SJ, Lee Y, Jiao Y, and Montell C (2009). A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr Biol 19, 1623–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poudel S, Kim Y, Kim YT, and Lee Y (2015). Gustatory receptors required for sensing umbelliferone in Drosophila melanogaster. Insect Biochem Mol Biol 66, 110–118. [DOI] [PubMed] [Google Scholar]
- 16.Poudel S, and Lee Y (2016). Gustatory Receptors Required for Avoiding the Toxic Compound Coumarin in Drosophila melanogaster. Mol Cells 39, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoonhoven LM, Van Loon JJA, and Dicke M (2005). Insect-Plant Biology (Oxford: Oxford University Press; ). [Google Scholar]
- 18.Wiener A, Shudler M, Levit A, and Niv MY (2012). BitterDB: a database of bitter compounds. Nucleic Acids Res 40, D413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delventhal R, and Carlson JR (2016). Bitter taste receptors confer diverse functions to neurons. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung HY, Jeong YT, Lim JY, Kim H, Oh SM, Hwang SW, Kwon JY, and Moon SJ (2017). Heterogeneity in the Drosophila gustatory receptor complexes that detect aversive compounds. Nat Commun 8, 1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SH, Lee Y, Akitake B, Woodward OM, Guggino WB, and Montell C (2010). Drosophila TRPA1 channel mediates chemical avoidance in gustatory receptor neurons. Proc Natl Acad Sci U S A 107, 8440–8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe K, Toba G, Koganezawa M, and Yamamoto D (2011). Gr39a, a highly diversified gustatory receptor in Drosophila, has a role in sexual behavior. Behavior genetics 41, 746–753. [DOI] [PubMed] [Google Scholar]
- 23.Itskov PM, Moreira JM, Vinnik E, Lopes G, Safarik S, Dickinson MH, and Ribeiro C (2014). Automated monitoring and quantitative analysis of feeding behaviour in Drosophila. Nature communications 5, 4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ro J, Harvanek ZM, and Pletcher SD (2014). FLIC: high-throughput, continuous analysis of feeding behaviors in Drosophila. PLoS One 9, e101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph RM, Devineni AV, King IF, and Heberlein U (2009). Oviposition preference for and positional avoidance of acetic acid provide a model for competing behavioral drives in Drosophila. Proc Natl Acad Sci U S A 106, 11352–11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph RM, and Heberlein U (2012). Tissue-specific activation of a single gustatory receptor produces opposing behavioral responses in Drosophila. Genetics 192, 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang CH, Belawat P, Hafen E, Jan LY, and Jan YN (2008). Drosophila egg-laying site selection as a system to study simple decision-making processes. Science 319, 1679–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson HM, Warr CG, and Carlson JR (2003). Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A 100 Suppl 2, 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haddad R, Khan R, Takahashi YK, Mori K, Harel D, and Sobel N (2008). A metric for odorant comparison. Nat Methods 5, 425–429. [DOI] [PubMed] [Google Scholar]
- 30.Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, and Presgraves DC (2012). Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res 22, 1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hey J, and Kliman RM (1993). Population genetics and phylogenetics of DNA sequence variation at multiple loci within the Drosophila melanogaster species complex. Mol Biol Evol 10, 804–822. [DOI] [PubMed] [Google Scholar]
- 32.Lachaise D, and Silvain JF (2004). How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica 120, 17–39. [DOI] [PubMed] [Google Scholar]
- 33.Russo CA, Takezaki N, and Nei M (1995). Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol 12, 391–404. [DOI] [PubMed] [Google Scholar]
- 34.Couto A, Alenius M, and Dickson BJ (2005). Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol 15, 1535–1547. [DOI] [PubMed] [Google Scholar]
- 35.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, and Vosshall LB (2004). Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714. [DOI] [PubMed] [Google Scholar]
- 36.Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, and Carlson JR. (2003). Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841. [DOI] [PubMed] [Google Scholar]
- 37.Hallem EA, Ho MG, and Carlson JR (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979. [DOI] [PubMed] [Google Scholar]
- 38.Benton R, Sachse S, Michnick SW, and Vosshall LB (2006). Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol 4, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butterwick JA, Del Marmol J, Kim KH, Kahlson MA, Rogow JA, Walz T, and Ruta V (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones CD (2005). The genetics of adaptation in Drosophila sechellia. Genetica 123, 137–145. [DOI] [PubMed] [Google Scholar]
- 41.Stensmyr MC, Dekker T, and Hansson BS (2003). Evolution of the olfactory code in the Drosophila melanogaster subgroup. Proc Biol Sci 270, 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dweck HKM, Ebrahim SAM, Retzke T, Grabe V, Weissflog J, Svatos A, Hansson BS, and Knaden M (2018). The Olfactory Logic behind Fruit Odor Preferences in Larval and Adult Drosophila. Cell reports 23, 2524–2531. [DOI] [PubMed] [Google Scholar]
- 43.Kreher SA, Mathew D, Kim J, and Carlson JR (2008). Translation of sensory input into behavioral output via an olfactory system. Neuron 59, 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson JP, Gleason CA, Foley RC, Thrall PH, Burdon JB, and Singh KB (2010). Plants versus pathogens: an evolutionary arms race. Funct Plant Biol 37, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornell HV, and Hawkins BA (2003). Herbivore responses to plant secondary compounds: a test of phytochemical coevolution theory. Am Nat 161, 507–522. [DOI] [PubMed] [Google Scholar]
- 46.Kareiva P (1999). Coevolutionary arms races: is victory possible? Proc Natl Acad Sci U S A 96, 8–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan P, Manoli DS, Ahmed OM, Chen Y, Agarwal N, Kwong S, Cai AG, Neitz J, Renslo A, Baker BS, et al. (2013). Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell 154, 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gardiner A, Barker D, Butlin RK, Jordan WC, and Ritchie MG (2008). Evolution of a complex locus: exon gain, loss and divergence at the Gr39a locus in Drosophila. PLoS One 3, e1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon JY, Dahanukar A, Weiss LA, and Carlson JR (2014). A map of taste neuron projections in the Drosophila CNS. J Biosci 39, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ling F, Dahanukar A, Weiss LA, Kwon JY, and Carlson JR (2014). The molecular and cellular basis of taste coding in the legs of Drosophila. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 7148–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mansourian S, Enjin A, Jirle EV, Ramesh V, Rehermann G, Becher PG, Pool JE, and Stensmyr MC (2018). Wild African Drosophila melanogaster Are Seasonal Specialists on Marula Fruit. Curr Biol 28, 3960–3968 e3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prieto-Godino LL, Rytz R, Cruchet S, Bargeton B, Abuin L, Silbering AF, Ruta V, Dal Peraro M, and Benton R (2017). Evolution of Acid-Sensing Olfactory Circuits in Drosophilids. Neuron 93, 661–676 e666. [DOI] [PubMed] [Google Scholar]
- 53.Shaw KH, Johnson TK, Anderson A, de Bruyne M, and Warr CG (2019). Molecular and Functional Evolution at the Odorant Receptor Or22 Locus in Drosophila melanogaster. Mol Biol Evol 36, 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dekker T, Ibba I, Siju KP, Stensmyr MC, and Hansson BS (2006). Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Current biology : CB 16, 101–109. [DOI] [PubMed] [Google Scholar]
- 55.Linz J, Baschwitz A, Strutz A, Dweck HK, Sachse S, Hansson BS, and Stensmyr MC (2013). Host plant-driven sensory specialization in Drosophila erecta. Proc Biol Sci 280, 20130626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seeholzer LF, Seppo M, Stern DL, and Ruta V (2018). Evolution of a central neural circuit underlies Drosophila mate preferences. Nature 559, 564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wieczorek H, and Wolff G (1989). The labellar sugar receptor of Drosophila. J. Comp. Physiol. A 164, 825–834. [Google Scholar]
- 58.Hodgson ES, Lettvin JY, and Roeder KD (1955). Physiology of a primary chemoreceptor unit. Science 122, 417–418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any datasets or new code. All raw data is available upon request.


