Abstract
Mutations in genes encoding isocitrate dehydrogenases (IDHs) 1 and 2 are common cancer-related genetic abnormalities. Malignancies with mutated IDHs exhibit similar pathogenesis, metabolic pattern, and resistance signature. However, an effective therapy against IDH1-mutated solid tumor remains unavailable. In the present study, we showed that acquisition of IDH1 mutation results in the disruption of NADP+/NADPH balance and an increased demand for glutathione metabolism. Moreover, the nuclear factor erythroid 2–related factor 2 (Nrf2) plays a key protective role in IDH1-mutated cells by prompting glutathione synthesis and ROS scavenging. Pharmacological inhibition of the Nrf2/glutathione pathway via brusatol administration exhibited a potent tumor suppressive effect on IDH1-mutated cancer in vitro and in vivo. Our findings highlight a possible therapeutic strategy that could be valuable for IDH1-mutated cancer treatment.
Keywords: IDH1-mutated glioma, Nrf2, glutathione, antioxidant
Introduction
Isocitrate dehydrogenases (IDHs) are a family of enzymes that mediate the oxidative decarboxylation of isocitrate to α-ketoglutarate. These enzymes depend on NAD+/NADP+, as they reduce NAD+/NADP+ for NADH/NADPH production (1). While IDH1 and IDH2 isozymes are homodimers that use NADP+ as a cofactor, IDH3 is a heterotetramer that uses NAD+ as a cofactor (2). Genetic abnormalities in IDH1/2 are common in multiple types of human tumors. For example, mutations in IDH1/2 have been found in over 80% of World Health Organization grade II/III gliomas, including astrocytoma and oligodendroglioma (3). These mutations are found in 73% of secondary glioblastomas, which are derived from lower-grade gliomas, but are less frequent in primary glioblastoma multiforme (GBM) (4). Furthermore, the IDH1/2 mutations are commonly identified in acute myeloid leukemia (AML), central chondrosarcoma, central/periosteal chondromas, and cholangiocarcinoma (5,6). However, despite the widespread prevalence of these mutations, effective therapies for IDH-mutated solid tumors remain unavailable.
The majority of cancer-associated IDH mutations are amino acid substitutions of an arginine residue in its catalytic center. For IDH1, the 132 arginine (R) residue is frequently altered to histidine (H) or cysteine (C). The R132H (73.67%) and R132C (13.35%) variant comprise over 87% of all IDH1 mutations in human. A seminal study by Dang et al.(7) revealed that these amino acid substitutions in IDH1 lead to a neomorphic activity of the enzyme, which is the NADPH-dependent consumption of α-ketoglutarate for 2-hydroxyglutarate (2-HG) production. The accumulation of 2-HG has been reported to associate with glioma oncogenesis by inhibiting α-ketoglutarate dioxygenases (8-10). Several pioneer studies also suggest that IDH mutants are associated with depletion of NADPH and GSH, accompanied with elevated reactive oxygen species (ROS) levels (11,12). The neomorphic catalytic function, 2-HG accumulation and occurrence of oxidative stress suggest a distinctive oncogenesis mechanism, which could be exploited as a therapeutic vulnerability in IDH1-mutated malignancies.
In the present study, we investigated the association between mutant IDH1 enzyme and ROS levels. Furthermore, we analyzed the role of nuclear factor erythroid 2–related factor 2 (Nrf2) in the regulation of glutathione (GSH) metabolism that maintains cellular redox homeostasis and survival. Moreover, we investigated the efficacy of Nrf2/GSH metabolism blockade as a therapeutic approach in IDH1-mutated malignancies.
Material and Methods
Cell culture
The U251 cell was obtained from Sigma in 2015. Cells were cultured in DMEM medium supplemented with 10% heat inactivated FBS (Thermo Fisher, Waltham, MA) and antibiotics (Thermo Fisher) at 37°C in a humidified air with 5% CO2. Brain tumor initiating cell (BTIC) TS603 (IDH1 R132H) was obtained from Dr. Timothy Chan in 2017 (13). BTIC GSC827 (IDH1 wild-type) and GSC923 (IDH1 wild-type) were previously established in our laboratory, which are derived from patient sample following the approval of National Cancer Institute Institutional Review Board (14). All BTIC lines were cultured in NBE media as previously described (15). All cell lines are PCR tested negative for mycoplasma. For inducing gene expression, cells were treated with 100 ng/mL doxycycline (Gold Biotechnology) for at least 24 hr.
Reagents and treatment condition
Brusatol was purchased from Sigma and dissolved in DMSO. The final concentration used is 40 nM in vitro. Cells were treated with brusatol for 24-72 hr among different experiments in vitro. N-acetylcysteine (NAC, Sigma) was dissolved in PBS. The final concentration used is 2.5 mM. Catalase (Sigma) was dissolved in PBS, the final concentration used is 500 U/mL. Manntol (Sigma) was dissolved in PBS, the final concentration used is 50 mM. MnTBAP (Millipore) was dissolved in DMSO, the final concentration used is 100 μM. AGI-5198 was purchased from Cellagen Technology and dissolved in DMSO. The final concentration used is 1 μM in vitro.
Plasmid, lentivirus and stable cell line generation
The coding sequence of IDH1 gene was inserted into pLVX-TetOne-Puro (Clontech) using restriction sites EcoRI/BamHI. The R132C/H variants were introduced by QuikChange Lightning site-directed mutagenesis kit (Agilent) and verified by Sanger sequencing. The lentivirus was packaged in HEK293T cells using pMD2.G (Addgene #12259) and psPAX2 (Addgene #12260) system. Virus was directly added into cell culture medium. Stable expression cell lines were selected by puromycin (1-2µg/mL).
NADP+/NADPH quantification
NADP+/NADPH was quantified by NADP+/NADPH-Glo assay (Promega). Cells were seeded in 96-well plate at 4,000 cells per well. Cells were lysed in 50 µL 0.2N NaOH with 1% dodecyltrimethylammonium bromide. The lysate was separated into two vials for NADP+ quantification or NADPH quantification. The luminescence signal was recorded by a Polarstar Optima plate reader (BMG LABTECH). NADP+/NADPH ratio was calculated through luminescence signal.
ROS quantification
The quantity of ROS was measured by ROS-Glo H2O2 assay (Promega). Cells were seeded in 96-well plate at 4,000 cells per well. Cells were incubated with H2O2 substrate for 4 hr and lysed in ROS-Glo detection solution. The ROS level was measured by luminescence signal that recorded by a Polarstar Optima plate reader (BMG LABTECH).
GSH/GSSG quantification
Cellular level of glutathione was quantified using GSH/GSSG-Glo assay (Promega). Cells were seeded in 96-well plate at 4,000 cells per well. Cells were lysed in total glutathione lysis reagent or oxidized glutathione lysis reagent. The lysate was then treated with luciferin generation reagent and the luminescence signal was recorded by a Polarstar Optima plate reader (BMG LABTECH). GSH/GSSG ratio was calculated through luminescence signal.
Live cell imaging
To quantify the level of oxidative damage, cells were seeded in 8-well chamber slides (Ibidi) at 10,000 cells per well. After treatment, cells were probed with Image-iT Lipid Peroxidation reagent (10 μM, 30 min, Thermo Fisher) or MitoSOX (5 μM, 10 min, Thermo Fisher). Fluorescent signal was recorded using a Lionheart FX automated microscope (BioTek). Oxidative stress was evaluated by measuring fluorescent intensity with ImageJ software.
Western blot
Western blot was performed as previously described (16). Briefly, Cells were seeded in 6-well plate at 3-5×105 per well. After treatment, cells were lysed in RIPA buffer supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Fisher). The lysate was separated by NuPAGE Bis Tris gel (Thermo Fisher) and transferred to PVDF membrane (Millipore). The membranes were blocked in Superblock blocking buffer (Thermo Fisher) followed by primary antibody. The quantity of target protein was revealed by HRP-conjugated secondary antibody and chemiluminescence assay (Bio-Rad). The primary antibodies used in this study are listed as follow: Nrf2 (Abcam ab62352, 1:1,000), GCLC (Abcam ab41463, 1:2,000), GCLM (Proteintech 14241-1-AP, 1:2,000), SLC7A11 (Proteintech, ab37185, 1:2,000), EGFP (Thermo Fisher, A-11122, 1:2,000), β-actin (CST, 4967, 1:5,000), and HA-tag (Origene, 1:2,000).
Real-time PCR
Quantitative real-time PCR was performed as previously described (17). Total RNA was extracted form cultured cells by PureLink RNA mini kit (Thermo Fisher), and reverse transcript to cDNA using Superscript IV VILO Master Mix (Thermo Fisher). Genes related to oxidative stress were analyzed by Power SYBR Green Master Mix. Primers used in the present study include NQO1 (QT00050281), HMOX1 (QT00092645), NFE2L2 (QT00027384) and ACTB (QT00095431).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed as previously described (18). In brief, cells were seeded in 150 mm plate at 1.2×107 cells per plate. After treatment, cells were fixed with 1% formaldehyde for 10 min. Cells were collected in ice cold PBS and extract for chromatin complex. The chromatin was sheared by sonication and precipitated using Nrf2 antibody (Active Motif). The copy number of antioxidant genes were measured by quantitative real-time PCR. The primers used in the present study are listed as follow: GCLC.F: 5’-CGC AGT TGT TGT GAT ACA GCC-3’; GCLC.R: 5’-GGA CTG AGA CTT TGC CCT AAG AA-3’; GCLM.F: 5’-ATT CCA AAC TGA GGG AGC TGT TT-3’; GCLM.R: 5’-ATG AGT AAC GGT TAC GAA GCA CT-3’; NQO1.F: 5’-GTG TGA CAG AGG CCT CAA AA-3’; NQO1.R: 5’-TGA TCC CTG GAC TCT CTT GG-3’; SLC7A11.F: 5’-AGC TTC CCA CAA AGT CGA AG-3’; SLC7A11.R: 5’-ACA TTC CTG CTT GTC TTG GT-3’.
Small interference RNA
Small interference RNA was designed and synthesized by Integrated DNA Technologies. Cells were seeded in 6-well plate at 5×105 cells per well. Fifty picomole of siRNA was transfected into cells by using Lipofectamine RNAiMAX (Thermo Fisher) based on manufacturer’s protocol. The suppression of gene expression was validated by quantitative real-time PCR. Small interference RNAs used in the present study are listed as follow: siGCLC.1.F: 5’-ACA AUU GGA CAG AUA GUA GCC AAC UGA-3’; siGCLC.1.R: 5’-AGU UGG CUA CUA UCU GUC CAA UUG T-3’; siGCLC.2.F: 5’-UAA AUA UUG GUA CAU UGA UGA CAA CCU-3’; siGCLC.2.R: 5’-GUU GUC AUC AAU GUA CCA AUA UUT A-3’; siGCLM.1.F: 5’-AAG GUU UUU UGG AUA CAA UCA UGA AGC-3’; siGCLM.1R: 5’-UUC AUG AUU GUA UCC AAA AAA CCT T-3’; siGCLM.2.F: 5’-CCU UCU UUU AGC UUG UAA AAU GUA GCC-3’; siGCLM.2.R: 5’-CUA CAU UUU ACA AGC UAA AAG AAG G-3’. AllStar Negative control siRNA (QIAGEN) was used as control.
Annexin V/Propidium iodide (PI) apoptosis analysis
Cell apoptosis level was analyzed by Annexin V/PI apoptosis kit (Thermo Fisher) according to manufacturer’s protocol. Cells were seeded in 6-well plate at 3-5×105 cells per well. After treatment, cells were harvested and incubated with FITC-conjugated Annexin V and PI for 20 min on ice. Cell samples were analyzed by FACS Canto II (BD Biosciences) flow cytometer.
Luciferase reporter assay
The Nrf2-associated transcriptional activity of was determined using reporter plasmid pGL4.37[luc2P/ARE/Hygro] (Promega) containing antioxidant response element (ARE). Nine hundred nanogram of reporter plasmid and 0.1 mg of pRL-CMV were transfected into 105 cells in 12-well plate using Lipofectamine 3000. Luminescence was measured by Dual-luciferase reporter assay system (Promega) according to manufacturer’s protocol.
Immunoprecipitation
Nrf2 ubiquitination was quantified by immunoprecipitation assay as previously described (19). Cells were seeded in 6-well plate at 3-5×105 cells per well and transfected with EGFP-Nrf2 (Addgene 21549) and ubiquitin (Addgene 18712) plasmids using Lipofectamine 3000. Cells were incubated with MG-132 (MedChem Express) to suppress proteasome activity. Cells were lysed in RIPA buffer supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Fisher) and 1% SDS. EGFP-conjugated Nrf2 were precipitated using EGFP antibody and Dynabeads Protein G immunoprecipitation kit (Thermo Fisher). Protein ubiquitination was measured by western blot.
Cycloheximide (CHX) pulse chase assay
CHX pulse chase assay was performed as previously described (20). Cells were seeded in 6-well plate at 3-5×105 cells per well. Cells were transfected with EGFP-Nrf2 plasmid and exposed with 50 μg/mL CHX (Sigma). Total protein was extracted and Nrf2 residue was analyzed through western blot and EGFP immunoblot.
Xenograft
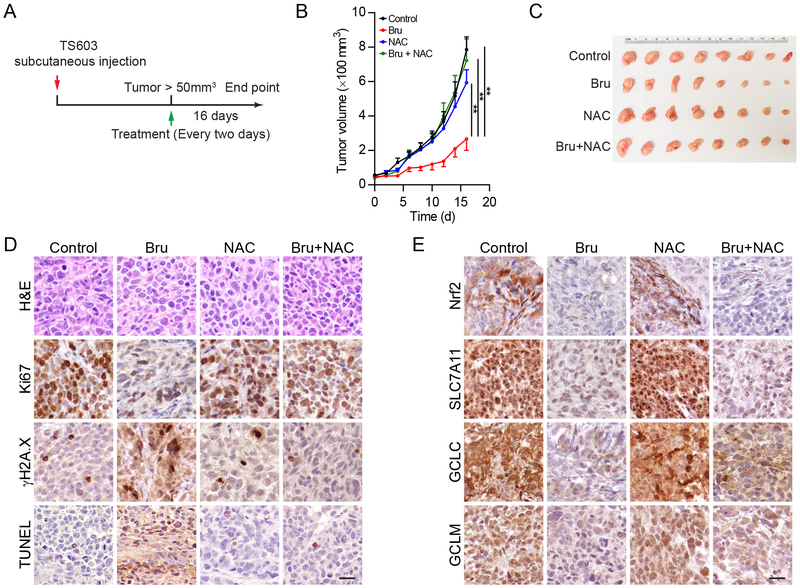
Six-to-eight-week-old NSG mice (The Jackson Laboratory) were subcutaneous injected with 5×106 TS603 cells in 100 μl PBS. Once the tumors reached over 50 mm3, mice were randomly allocated into four groups and treated i.p. with DMSO (8 μl DMSO in 100 μl PBS), brusatol (Bru, 2 mg/kg, 8 μl DMSO in 100 μl PBS)(21,22), NAC (50 mg/mL, dissolved in 100 μl PBS containing 8 μl DMSO), or Bru + NAC every other day for a total of five times. Tumor size was measured using Vernier calipers. Sixteen days after brusatol treatment, mice were sacrificed, and tumors were harvested for analysis. All animal studies were conducted in accordance with the principles and procedures outlined in the NIH Guide for the Care and Use of Animals and approved by the Animal Care and Use Committee of the National Institute of Health.
Statistical analysis
Statistical analysis was performed with Student’s t-test between two data groups. Differences among groups were analyzed using one-way ANOVA test followed by Student’s t-test as the post statistical analysis. All tests were two-sided, the results were presented as mean ± SEM. A *p value < 0.05 was considered as statistically significant. All of the analysis was conducted using GraphPad Prism 7.01 (GraphPad Software).
Results
Neomorphic activity in cancer-associated mutant IDH1 triggers oxidative stress
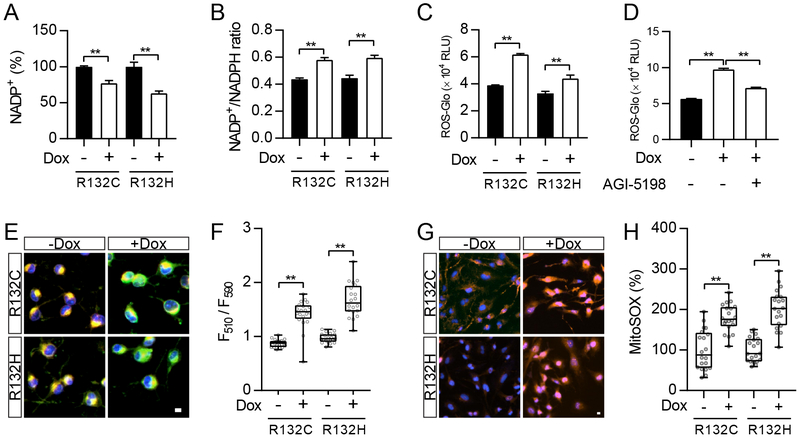
To better understand the effect of IDH1 mutants on redox homeostasis, we established a doxycycline-induced IDH1 mutant U251 cell line (Supplementary Figure 1A). We noticed that upon the expression of mutant IDH1 enzymes, the overall quantity of NADP decreased, in both of its oxidized (NADP+) and reduced (NADPH) forms (Figure 1A). Moreover, the NADP+/NADPH ratio significantly increased (Figure 1B), suggesting that the neomorphic enzyme activity of mutant IDH1 exhausted the cellular pool of NADPH. The balance between NADP+ and NADPH is a critical factor to maintain cellular redox homeostasis, as NADPH is a general cofactor in reductive biosynthetic reactions, and to provide electrons for metabolic pathways, such as the reduction of glutathione disulfide (GSSG) back to GSH and ROS neutralization (23). Further, by quantification of H2O2, we showed that mutant IDH1 enzymes led to severe oxidative stress in both U251 cells and BTIC TS603 (Figure 1C and Supplementary Figure 2A). Such an increase in ROS levels depended on the presence of mutant enzymes. The treatment with AGI-5198, a specific inhibitor of mutant IDH1 (13), reduced ROS accumulation in cells expressing the IDH1 R132H variant (Figure 1D). Further, the elevated ROS levels led to oxidative stress to macromolecules and subcellular organelles, evidenced by increased lipid peroxidation (Figure 1E and F) and mitochondrial ROS level (Figure 1G and H). The ROS scavenger catalase and MnTBAP, but not mannitol, abrogated the accumulation of ROS in IDH1-mutated cells, indicating the majority form of oxidative stress is derived from hydrogen peroxide and superoxide anion (Supplementary Figure 1B).
Figure 1.
Cancer-associated IDH1 mutants trigger oxidative stress
A. NADP+ level was measured in U251 cells with doxycycline (Dox)-induced expression of IDH1 mutant enzymes (R132C and R132H). **p<0.01.
B. Measurement of NADP+/NADPH ratio in U251 cells with expression of IDH1 mutant enzymes. **p<0.01.
C. ROS-Glo measurement in U251 cells with expression of IDH1 mutant enzymes. **p<0.01.
D. ROS-Glo measurement in IDH1-R132H U251 cells with AGI-5198 treatment (1 μM, 24 hr). **p<0.01.
E. Lipid peroxidation staining measures membrane oxidative damage in U251 cells with expression of IDH1 mutant enzymes. Bar = 10 µm.
F. Quantification of lipid peroxidation in E. **p<0.01.
G. MitoSOX staining measures mitochondrial ROS in IDH1-mutated U251 cells. Bar = 10 µm.
H. Quantification of MitoSOX signal in G. **p<0.01.
Glutathione metabolism supports redox balance and survival in IDH1-mutated cells
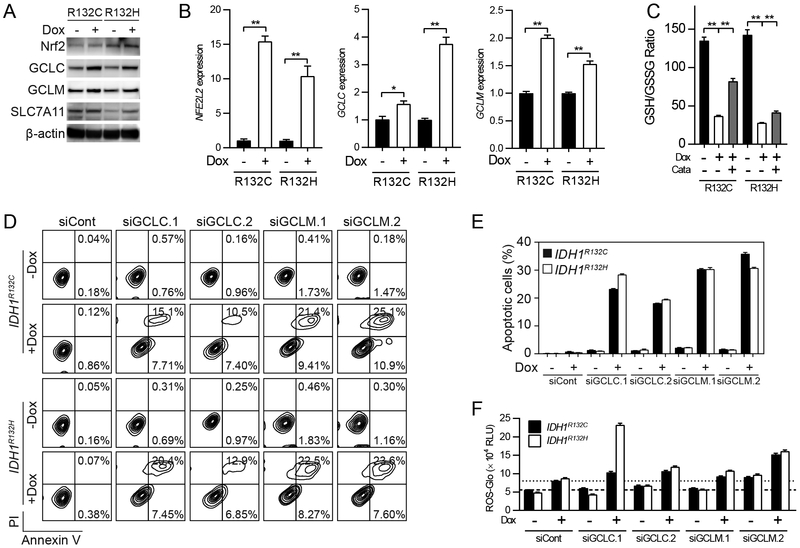
Considering the remarkable ROS accumulation in IDH1-mutated cells, we speculate that antioxidant pathways, such as GSH-dependent ROS scavenging systems, may be triggered to maintain redox homeostasis. To test this hypothesis, we firstly measured the protein levels of GSH synthesis enzymes by western blot. We found that upon introduction of pathogenic mutant IDH1 enzymes, the levels of key enzymes in GSH biosynthesis, such as glutamate-cysteine ligase (GCLC, catalytic subunit; GCLM, modifier subunit), and cystine/glutamate transporter (SLC7A11, xCT transporter), increased (Figure 2A). We also noticed that the levels of Nrf2 protein (NFE2L2), the major transcriptional factor responsible for ROS sensing, increased in IDH1-mutated cells. The enhancement of Nrf2 and GSH-dependent ROS scavenging pathways in IDH1-mutated U251 cells, as well as IDH1-mutated BTIC TS603, were also confirmed by quantitative PCR (Figure 2B and Supplementary Figure 2B). To further understand the role of GSH in IDH1-mutated cells, we quantified GSH/GSSG levels in U251 cells expressing R132C/H IDH1 mutants. We recorded a substantial decrease in the GSH/GSSG ratio compared to that in cells expressing wild-type IDH1, suggesting that there is an elevated demand from GSH-dependent ROS scavenging (Figure 2C). We also measured GSH/GSSG levels in BTICs, the result consistently showed that IDH1-mutated BTIC TS603 has lower GSH/GSSG ratio compared with IDH1 wild-type BTIC GSC827 and GSC 923 (Supplementary Figure 2C). Moreover, the addition of an exogenous antioxidant enzyme, catalase, partially restored the GSH/GSSG ratio, indicating that the GSH/GSSG imbalance could be a result of GSH oxidation by hydrogen peroxide decomposition pathways (e.g., glutathione peroxidases). Importantly, glutathione biosynthesis exhibited a critical protective role in cells with mutant IDH1 enzyme. A loss-of-function experiment showed that 72 hr after genetic silencing of GCLC/GCLM resulted in remarkable apoptotic changes. The annexin V/PI apoptosis assay showed that apoptotic cell population increased remarkably upon small interference RNA treatment (R132H, siCont = 0.45% vs siGCLM.2 = 31.2%, Figure 2D and E). Moreover, western blot confirmed that cleaved caspase-3 was increased in U251 IDH1 R132H cells after genetic silencing of GCLC and GCLM (Supplementary Figure 1C). ROS scavenger catalase restored caspase-3 cleavage in the presence of GCLC and GCLM RNA interference (Supplementary Figure 1D). The increase of apoptosis was not observed when mutant IDH1 enzyme is absent, suggesting that the blockade of GSH metabolism is selectively toxic to IDH1-mutated cells. Furthermore, we recorded a great increase in cytoplasmic ROS levels after 48 hr of small interference RNA treatment, suggesting that apoptosis could be caused by ROS-derived cellular damage (Figure 2F).
Figure 2.
GSH de novo synthesis support cellular physiology in IDH1-mutated cells
A. Western blot measures the expression of GSH synthesis enzymes in U251 cells with IDH1 mutant expression. β-actin was used as internal control.
B. Quantitative real-time PCR analysis measures mRNA level of GSH synthesis enzymes. *p<0.05, **p<0.01.
C. GSH and GSSG level was measured in IDH1-mutated U251 cells. Catalase (Cata) was used as exogenous ROS scavenger (500U/mL, 24 hr). **p<0.01.
D. Annexin V/PI apoptotic analysis in IDH1-mutated U251 cells with genetic silencing of GCLC and GCLM.
E. Quantification of apoptotic cells in D.
F. ROS-Glo assay in IDH1-mutated U251 cells with genetic silencing of GCLC and GCLM.
Activation of Nrf2 antioxidant pathway in IDH1-mutated cells
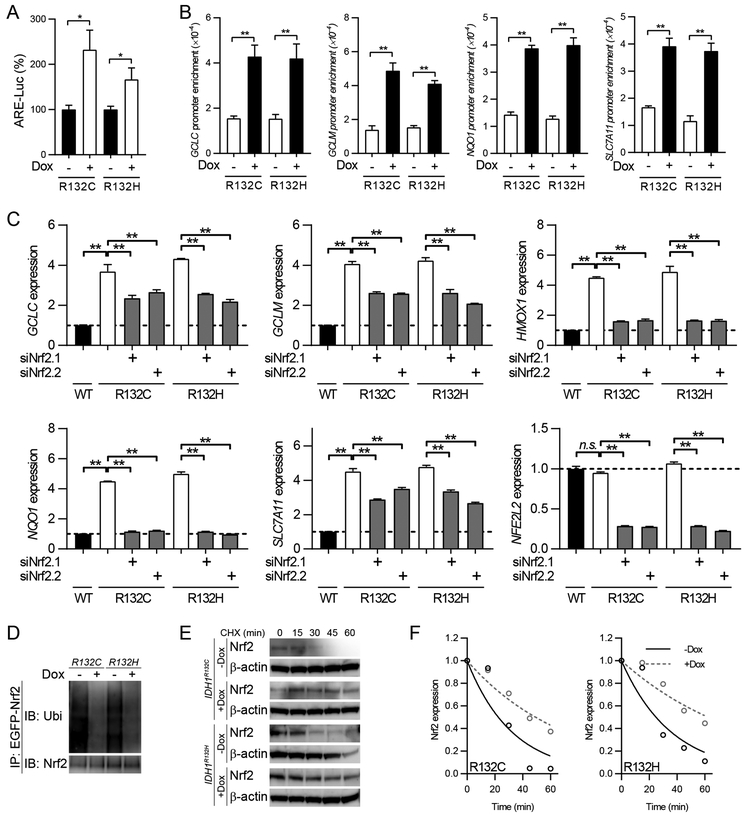
The transcription factor Nrf2 is a basic leucine zipper (bZIP) protein that regulates the cellular responses to oxidative stress by activating the expression of antioxidant genes (24). Under physiological conditions, Nrf2 is tightly controlled by interaction with Kelch-like ECH-associated protein 1 (Keap1), which is a protein adaptor for E3 ubiquitin ligases, and proteasomal degradation. When cells are challenged by oxidative stress, Keap1-Nrf2 interaction is disrupted and the dissociated Nrf2 translocate into the nucleus for transcriptional activation (25). In IDH1-mutated cells, the increased ROS levels may trigger Nrf2 stabilization and gene transcription, which could be relevant to prompt GSH synthesis. To test this hypothesis, we firstly evaluated Nrf2-associated gene transcription via antioxidant response element (ARE)-luciferase reporter assay. We found that mutant IDH1 expression was associated with enhanced Nrf2-dependent transcriptional activation (Figure 3A). Furthermore, through chromatin immunoprecipitation assay (ChIP), we demonstrated that the affinity of Nrf2 to antioxidant gene promoters was strongly enhanced after mutant IDH1 introduction, indicating that Nrf2 transactivation plays a central role in ROS homeostasis in IDH1-mutated cells (Figure 3B). Consistent with these findings, genetic silencing of Nrf2 resulted in downregulation of antioxidant genes, such as GCLC, GCLM, HMOX1, NQO1, and SLC7A11, in IDH1-mutated cells (Figure 3C and Supplementary Figure 1E). The induction of Nrf2 transcription activity was accompanied by its prolonged protein stability. Immunoprecipitation assay showed that Nrf2 ubiquitination is compromised in the presence of mutant IDH1 (Figure 3D). Furthermore, the protein stability of Nrf2 was elevated when mutant IDH1 enzymes were expressed (Figure 3E and F). The protein half-lives of Nrf2 were prolonged from 22.4 min to 49.2 min (R132C) or 62.3 min (R132H).
Figure 3.
Nrf2 regulates GSH metabolism in IDH1-mutated cells
A. ARE-luciferase reporter assay was performed in U251 cells with IDH1 mutant expression. *p<0.05.
B. ChIP PCR assay showed antioxidant genes promoter affinity of Nrf2 in IDH1-mutated U251 cells. **p<0.01.
C. Real-time PCR assay showed mRNA level of antioxidant genes after 48 hr genetic silencing of Nrf2 in U251 cells with IDH1 mutant expression. **p<0.01.
D. Immunoprecipitation assay measures Nrf2 ubiquitination in U251 cells with IDH1 mutant expression.
E. CHX pulse chase assay measures Nrf2 protein stability in IDH1-mutated U251 cells.
F. Quantification of Nrf2 half lives from results in E.
Suppressing Nrf2/glutathione axis results in oxidative damage in IDH1-mutated cells
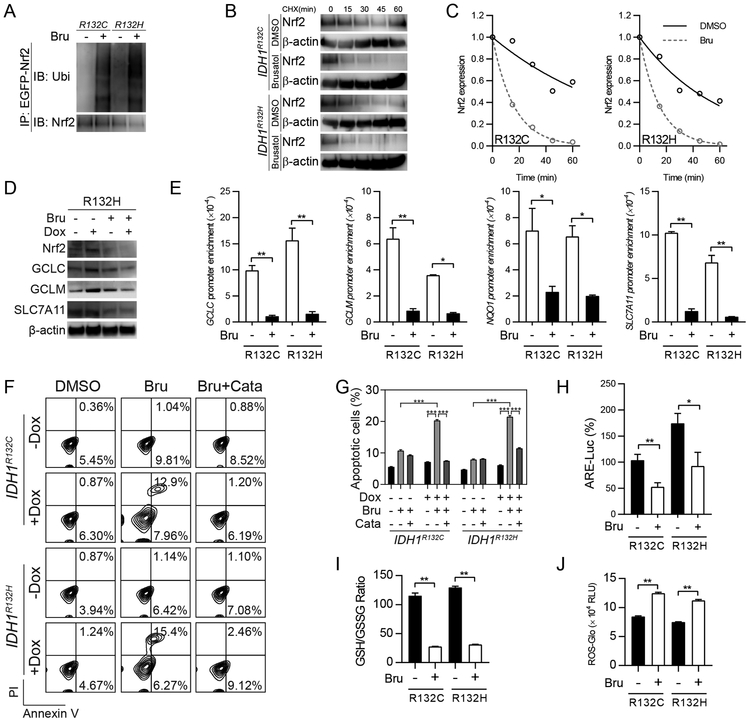
Considering the central role of Nrf2 in the physiology of IDH1-mutated cells, blockade of Nrf2/GSH axis may be an effective therapeutic approach for tumors with IDH1 R132 variants. To investigate this, we tested a Nrf2 inhibitor, brusatol, in IDH1-mutated cells. Brusatol has been shown to strongly reduce Nrf2 transcriptional activity and enhance chemosensitivity in transformed cells (21,26). Here, we confirmed that brusatol promoted Nrf2 degradation in IDH1-mutated cells, as evidenced by increased Nrf2 protein ubiquitination (Figure 4A). Moreover, cycloheximide (CHX) pulse chase assay confirmed that Nrf2 protein stability is compromised upon brusatol treatment (Figure 4B). The protein half-lives decreased for both IDH1 R132C (67.1 min vs 11.1 min) and R132H (41.6 min vs 10.34 min) variants (Figure 4C). Further, western blot analysis showed that Nrf2 protein levels drastically decreased after brusatol treatment (Figure 4D). Accordingly, ChIP-PCR assay showed that the Nrf2 affinity for DNA sharply decreased in the presence of brusatol (Figure 4E).
Figure 4.
Suppressing Nrf2/GSH axis results in oxidative damage in IDH1-mutated cells
A. Immunoprecipitation assay measures Nrf2 ubiquitination in U251 cells with IDH1 mutant expression after brusatol treatment (40nM, 12 hr).
B. CHX pulse chase assay measures Nrf2 protein stability in IDH1-mutated U251 cells after brusatol treatment.
C. Quantification of Nrf2 half lives from results in B.
D. Western blot measures the expression of GSH synthesis enzymes with brusatol treatment (40nM, 24 hr) in IDH1-mutated U251 cells.
E. ChIP PCR assay showed antioxidant genes promoter affinity of Nrf2 with brusatol (40nM, 24 hr) in IDH1-mutated U251 cells. *p<0.05, **p<0.01.
F. Annexin V/PI apoptosis assay showed apoptotic changes in IDH1-mutated U251 cells with brusatol treatment (40nM, 72 hr). Exogenous antioxidant Catalase (Cata) was used as exogenous ROS scavenger.
G. Quantification of apoptotic cells in F.***p<0.001.
H. ARE-luciferase reporter assay showed Nrf2-associated gene transcription with brusatol (40nM, 24 hr) in IDH1-mutated U251 cells. *p<0.05, **p<0.01.
I. GSH/GSSG measurement in IDH1-mutated U251 cells with brusatol treatment (40nM, 24 hr). **p<0.01.
J. ROS-Glo measurement in IDH1-mutated U251 cells with brusatol treatment (40nM, 24 hr). **p<0.01.
Importantly, the suppression of Nrf2 activity resulted into exacerbated oxidative damage and cell death in IDH1-mutated cells. Annexin V/PI flowcytometry assay showed that brusatol increased apoptotic rates by 1.9-fold and 2.7-fold in IDH1 R132C and R132H U251 cells, respectively (Figure 4F and G). Consistently, brusatol also resulted in cell apoptosis in IDH1-mutated BTIC TS603, but the trend was much less in IDH1 wild-type BTICs (Supplementary Figure 2D and E). We noticed that brusatol treatment resulted in reduced ARE-luciferase activity, suggesting that Nrf2 activity is suppressed in these cells (Figure 4H). Quantification of GSH revealed that brusatol further reduced GSH availability in IDH1-mutated cells. Brusatol decreased the GSH/GSSG ratio by 75.8% and 75.9% in IDH1 R132C and R132H cells, respectively (Figure 4I). Accordingly, the cytoplasmic levels of ROS were significantly elevated by brusatol treatment (Figure 4J).
Targeting Nrf2/glutathione axis suppresses IDH1-mutated xenografts
The aforementioned in vitro experiments strongly indicate that the blockade of GSH metabolism could be a valuable approach to suppress malignancies with mutant IDH1 enzymes. To better test this hypothesis, we established a xenograft mice model based on a patient-derived IDH1-mutated cell line TS603 (Figure 5A). Patient-derived TS603 glioma cells with intrinsic mutant IDH1 enzyme were injected into NSG immunocompromised mice to establish xenograft tumor. When the tumor mass approaches 50 mm3, mice were treated with either brusatol and/or the exogenous antioxidant acetylcysteine (NAC). Tumor growth curve showed that brusatol significantly reduced the expansion of tumor mass (Figure 5B and C). Notably, NAC abolished the suppressive effect of brusatol, suggesting ROS played a critical role in brusatol effects on tumor growth. No significant loss of body weight was observed during the treatment (Supplementary Figure 1F). Histological analysis revealed that brusatol treatment reduced the expression of antioxidant genes such as Nrf2, SLC7A11, GCLC and GCLM (Figure 5D). NAC slightly restored antioxidant gene expression in the xenografts. On the other hand, brusatol led to reduced expression of Ki67, but elevated levels of DNA damage markers, γH2A.X and TUNEL (Figure 5E). Similarly, NAC treatment minimized cytotoxicity in IDH1-mutated xenografts.
Figure 5.
Targeting Nrf2/GSH axis suppresses IDH1-mutated xenografts
A. Schematic illustration for the xenograft and treatment schedule.
B. Tumor growth curve of TS603 xenografts. n=10 for each group. **p<0.01.
C. Gross anatomy of TS603 xenografts.
D. Immunohistochemistry assay showed the expression of Ki67, γH2A.X and TUNEL assay in tumor sections. Bar = 50 μm.
E. Immunohistochemistry assay showed the expression of Nrf2, SLC7A11, GCLC and GCLM in tumor sections. Bar = 50 μm.
Discussion
IDH1-mutated malignancies and therapeutic approaches
Mutations of IDH1/2 genes are widespread genetic abnormalities detected in several types of human malignancies, including lower grade glioma, leukemia, chondroma, chondrosarcoma, and cholangiocarcinoma. Cancer-associated IDH mutations cause amino acid substitution of an arginine residue in the IDH enzyme catalytic center. Biochemical studies showed that IDH1 R132 mutants have elevated affinity for both NADPH (Km=0.44 µM) and α-ketoglutarate (Km=965 µM), indicating that the mutant enzyme prefers NADPH and α-ketoglutarate, whereas wild-type IDH enzyme prefers NADP+ and isocitrate for its catalytic function (7,27).
Several pioneered studies showed that direct targeting mutant IDH1 enzyme is an effective treatment for IDH1-mutated hematopoietic malignancies, such as relapsed or refractory AML (28). Regarding IDH1-mutated solid tumors, Rohle et al. (13) showed that the inhibition of mutant IDH1 delayed IDH1-mutated xenograft expansion in vivo. However, preliminary data from several early phase clinical trials showed modest impact on objective response rate and delay of progression of IDH1-mutated solid tumors. More effective therapies, such as developing refined inhibitors of mutant IDH1, or targeting IDH-related pathways, have been urged to improve disease outcome of IDH1-mutated malignancies. Besides direct targeting the mutant enzyme, several lines of evidence show that metabolic reprogramming in IDH1-mutated cells could be targeted to synergize with conventional chemo/radiotherapies. We and other colleagues demonstrated that NAD+ depletion, as well as 2-HG-mediated deficiency in homologous DNA recombination, establish vulnerability to poly ADP ribose polymerase (PARP) inhibitors in IDH1-mutated cells (29-32). The glutaminase inhibitor CB-839 has also been proposed to be useful for the treatment of IDH1-mutated cancers (33,34). In the present study, we extended the investigation of effective therapy for IDH1-mutated cancer and discovered that Nrf2/GSH metabolism could be another therapeutic vulnerability in malignancies that harbor IDH1 mutation.
IDH-mutated cells develop dependency on glutathione ROS scavenging
The production of ROS is involved in several aspects of cancer biology, such as genomic instability, loss of growth control, cellular motility, and tumor invasiveness (35,36). On the other hand, excessive ROS is harmful to biological molecules, resulting in oxidative damage to DNA, lipid and proteins (37). Maintaining appropriate ROS levels is key to cancer cells during oncogenesis and therapeutic resistance. Glutathione is an endogenous antioxidant tripeptide that participates in the elimination of reactive molecules, such as free radicals, peroxides, lipid peroxides, and metals. The thiol group in reduced glutathione is responsible for its reducing activity, which alleviates oxidative stress through direct reduction of disulfide bonds in the cysteine residues of cytoplasmic proteins and eliminates ROS through the glutathione-ascorbate cycle (38). For IDH1-mutated malignancies, several pioneered studies suggested the correlation with glutathione depletion and ROS accumulation (39). Although failed control of intracellular ROS has been indicated with tumorigenesis process (40), there is still lack of a direct evidence showing IDH1-mutant derived ROS promote tumor development.
In the present study, we found that IDH1-mutated malignancies exhibit a tendency to suffer oxidative stress, as the introduction of mutant IDH1 is closely associated with elevated ROS levels in cytoplasm and mitochondria (Figure 1C to H). Similarly, recent research suggests that tumoral glutathione levels negatively correlate with 2-HG, suggesting that IDH-mutated cells have elevated demands for glutathione (41). Our findings showed that, in IDH1-mutated cells, the key regulatory enzymes of glutathione biosynthesis were upregulated to meet the increased demands of endogenous antioxidant systems (Figure 2A to C). Loss-of-function experiments demonstrated that blocking GSH synthesis led to remarkably elevated ROS levels, oxidative stress and apoptotic changes (Figure 2D and E). Overall, our findings suggest that upregulation of glutathione-based ROS scavenging pathways play a central role to maintain cellular homeostasis in IDH1-mutated cancers.
Nrf2-regulated glutathione metabolism
The multifunctional transcriptional factor, Nrf2, governs the cellular response to oxidative stress by triggering antioxidant gene transcription. It regulates a variety of genes for electrophile and oxidant metabolism, as well as genes that support cellular survival under stress conditions (42). In the present study, we recorded enhanced Nrf2 transcriptional activity that was associated with the presence of mutant IDH1 enzyme (Figure 3A and B), suggesting that Nrf2-driven antioxidant response is a compensatory response to IDH1-associated oxidative stress. As a further validation, it was shown that several known Nrf2 transcription targets, such as GCLC, GCLM, HMOX1, NQO1 and SLC7A11, were upregulated in IDH1-mutated cells (Figure 3C). Importantly, these genes play central role in cysteine uptake and de novo glutathione biosynthesis, indicating that the protective effect of Nrf2 is a result of increased intracellular GSH pool. Protein stability tests showed that Nrf2 was less ubiquitinated and degraded in IDH1-mutated cells, which would lead to the activation of antioxidant expression (Figure 3D-F). Moreover, the activation of Nrf2/antioxidant pathway not only relieves the metabolic stress for IDH1-mutated cells but may also support cellular viability and promote growth advantage during oncogenesis. Our findings highlighted the role of Nrf2-dependent GSH metabolism in IDH1-mutated cells, indicating a selective vulnerability of IDH1-mutated malignancies.
Targeting Nrf2/glutathione metabolism as a new strategy for IDH1-mutated malignancies
Considering the critical role of Nrf2/GSH metabolism in cancer biology and therapeutic resistance, several attempts have been made to achieve specific targeting of this pathway using small molecular compounds. For example, by high-throughput screening assay, Singh et al. (43) reported that the small molecule compound ML-385 exhibits inhibitory effect on Nrf2 transcriptional activity. However, the effective dosage of ML-385 is too high for further preclinical studies in animals. For another example, Ren et al. (21) reported that brusatol, a quassinoid compound, exhibits potent inhibitory effect on Nrf2 transcriptional activity. In our study, we confirmed that brusatol is able to block Nrf2 activity in IDH1-mutated cells, which strongly suppressed antioxidant pathways, such as de novo glutathione biosynthesis (Figure 4). Interestingly, brusatol has been used as a sensitizer for conventional chemotherapy (21,26,44). In contrast, our study showed that at a similar dosage, brusatol monotherapy is sufficient to cause apoptotic changes in IDH1-mutated cells (45). We speculate that the brusatol potent efficacy is due to the induction of ROS levels, which leads to cell death in concert with the inhibition of endogenous antioxidants expression. The xenograft experiments confirmed this hypothesis. The introduction of an exogenous ROS scavenger, NAC, compromised the tumor-suppressing effect of brusatol, suggesting that ROS play a key role in brusatol-induced cytotoxicity (Figure 5).
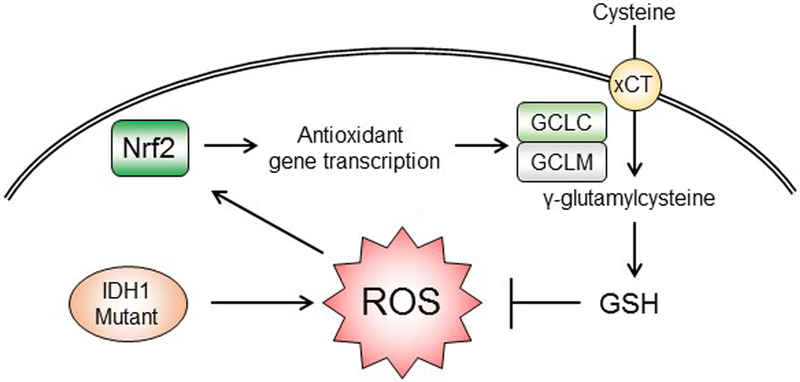
Overall, our study showed that neomorphic activity of mutant IDH1 enzyme results in mitochondrial and cytoplasmic ROS accumulation by disrupting NADP+/NADPH balance. The major regulator of antioxidant responses, Nrf2, controls the glutathione de novo synthesis and plays a central role in the cellular physiology of IDH1-mutated cells. Blockade of Nrf2/antioxidant pathway exhibited selective cytotoxicity in cells with IDH1 mutation (Figure 6). Our findings highlight the importance of GSH metabolism in IDH1-mutated cells and indicates a novel therapeutic approach for malignancies with IDH1 mutation.
Figure 6.
Targeting Nrf2/GSH axis for IDH1-mutated cancers
Cancer-associated IDH1 mutants cause metabolic deficiency and accumulation of oxidative stress. Nrf2 plays a key protective role in IDH1-mutated cells, by transcribing GSH synthesis enzymes. The enhanced de novo GSH synthesis neutralizes excessive ROS, and therefore avoids oxidative damages to macromolecules. Targeting Nrf2/GSH could be a novel therapeutic strategy in this type of human malignancies.
Supplementary Material
Acknowledgement
This work was supported by the Intramural Research Program of the NIH, NCI, CCR.
Footnotes
The authors declare no potential conflicts of interest
Reference
- 1.Alp PR, Newsholme EA, Zammit VA. Activities of citrate synthase and NAD+-linked and NADP+-linked isocitrate dehydrogenase in muscle from vertebrates and invertebrates. Biochem J 1976;154(3):689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol 2016;27(4):599–608 doi 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 3.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360(8):765–73 doi 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res 2009;15(19):6002–7 doi 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 5.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 2011;224(3):334–43 doi 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 6.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 2012;17(1):72–9 doi 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462(7274):739–44 doi 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011;19(1):17–30 doi 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 2012;483(7390):484–8 doi 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012;483(7390):479–83 doi 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolker S, Pawlak V, Ahlemeyer B, Okun JG, Horster F, Mayatepek E, et al. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d-2-hydroxyglutaric aciduria. Eur J Neurosci 2002;16(1):21–8. [DOI] [PubMed] [Google Scholar]
- 12.Latini A, Scussiato K, Rosa RB, Llesuy S, Bello-Klein A, Dutra-Filho CS, et al. D-2-hydroxyglutaric acid induces oxidative stress in cerebral cortex of young rats. Eur J Neurosci 2003;17(10):2017–22. [DOI] [PubMed] [Google Scholar]
- 13.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 2013;340(6132):626–30 doi 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ene CI, Edwards L, Riddick G, Baysan M, Woolard K, Kotliarova S, et al. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PLoS One 2012;7(12):e51407 doi 10.1371/journal.pone.0051407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang C, Iyer RR, Yu AC, Yong RL, Park DM, Weil RJ, et al. beta-Catenin signaling initiates the activation of astrocytes and its dysregulation contributes to the pathogenesis of astrocytomas. Proc Natl Acad Sci U S A 2012;109(18):6963–8 doi 10.1073/pnas.1118754109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Asthagiri AR, Iyer RR, Lu J, Xu DS, Ksendzovsky A, et al. Missense mutations in the NF2 gene result in the quantitative loss of merlin protein and minimally affect protein intrinsic function. Proc Natl Acad Sci U S A 2011;108(12):4980–5 doi 10.1073/pnas.1102198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang Y, Lu Y, Caisova V, Liu Y, Bullova P, Huynh TT, et al. Targeting NAD+/PARP DNA repair pathway as a novel therapeutic approach to SDHB-mutated cluster I pheochromocytoma and paraganglioma. Clin Cancer Res 2018. doi 10.1158/1078-0432.CCR-17-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang C, Sun MG, Matro J, Huynh TT, Rahimpour S, Prchal JT, et al. Novel HIF2A mutations disrupt oxygen sensing, leading to polycythemia, paragangliomas, and somatostatinomas. Blood 2013;121(13):2563–6 doi 10.1182/blood-2012-10-460972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C, Rahimpour S, Lu J, Pacak K, Ikejiri B, Brady RO, et al. Histone deacetylase inhibitors increase glucocerebrosidase activity in Gaucher disease by modulation of molecular chaperones. Proc Natl Acad Sci U S A 2013;110(3):966–71 doi 10.1073/pnas.1221046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C, Wang H, Zhu D, Hong CS, Dmitriev P, Zhang C, et al. Mutant glucocerebrosidase in Gaucher disease recruits Hsp27 to the Hsp90 chaperone complex for proteasomal degradation. Proc Natl Acad Sci U S A 2015;112(4):1137–42 doi 10.1073/pnas.1424288112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, et al. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci U S A 2011;108(4):1433–8 doi 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Lu Y, Celiku O, Li A, Wu Q, Zhou Y, et al. Targeting IDH1-Mutated Malignancies with NRF2 Blockade. J Natl Cancer Inst 2019. doi 10.1093/jnci/djy230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med 2001;30(11):1191–212. [DOI] [PubMed] [Google Scholar]
- 24.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 1997;236(2):313–22. [DOI] [PubMed] [Google Scholar]
- 25.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 1999;13(1):76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olayanju A, Copple IM, Bryan HK, Edge GT, Sison RL, Wong MW, et al. Brusatol provokes a rapid and transient inhibition of Nrf2 signaling and sensitizes mammalian cells to chemical toxicity-implications for therapeutic targeting of Nrf2. Free Radic Biol Med 2015;78:202–12 doi 10.1016/j.freeradbiomed.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Losman JA, Kaelin WG Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev 2013;27(8):836–52 doi 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, et al. Durable Remissions with Ivosidenib in IDH1-Mutated Relapsed or Refractory AML. N Engl J Med 2018;378(25):2386–98 doi 10.1056/NEJMoa1716984. [DOI] [PubMed] [Google Scholar]
- 29.Lu Y, Kwintkiewicz J, Liu Y, Tech K, Frady LN, Su YT, et al. Chemosensitivity of IDH1-Mutated Gliomas Due to an Impairment in PARP1-Mediated DNA Repair. Cancer Res 2017;77(7):1709–18 doi 10.1158/0008-5472.CAN-16-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tateishi K, Higuchi F, Miller JJ, Koerner MVA, Lelic N, Shankar GM, et al. The Alkylating Chemotherapeutic Temozolomide Induces Metabolic Stress in IDH1-Mutant Cancers and Potentiates NAD(+) Depletion-Mediated Cytotoxicity. Cancer Res 2017;77(15):4102–15 doi 10.1158/0008-5472.CAN-16-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, Bai H, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med 2017;9(375) doi 10.1126/scitranslmed.aal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y, Liu Y, Pang Y, Pacak K, Yang C. Double-barreled gun: Combination of PARP inhibitor with conventional chemotherapy. Pharmacol Ther 2018;188:168–75 doi 10.1016/j.pharmthera.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 2010;70(22):8981–7 doi 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emadi A, Jun SA, Tsukamoto T, Fathi AT, Minden MD, Dang CV. Inhibition of glutaminase selectively suppresses the growth of primary acute myeloid leukemia cells with IDH mutations. Exp Hematol 2014;42(4):247–51 doi 10.1016/j.exphem.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Behrend L, Henderson G, Zwacka RM. Reactive oxygen species in oncogenic transformation. Biochem Soc Trans 2003;31(Pt 6):1441–4 doi 10.1042/. [DOI] [PubMed] [Google Scholar]
- 36.Reczek CR, Chandel NS. The Two Faces of Reactive Oxygen Species in Cancer. Annual Review of Cancer Biology 2017;1(1):79–98 doi 10.1146/annurev-cancerbio-041916-065808. [DOI] [Google Scholar]
- 37.Dixon SJ, Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat Chem Biol 2014;10(1):9–17 doi 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 38.Meister A Glutathione, ascorbate, and cellular protection. Cancer Res 1994;54(7 Suppl):1969s–75s. [PubMed] [Google Scholar]
- 39.Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio II, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev 2012;26(18):2038–49 doi 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature 2000;408(6809):239–47 doi 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 41.Andronesi OC, Arrillaga-Romany IC, Ly KI, Bogner W, Ratai EM, Reitz K, et al. Pharmacodynamics of mutant-IDH1 inhibitors in glioma patients probed by in vivo 3D MRS imaging of 2-hydroxyglutarate. Nat Commun 2018;9(1):1474 doi 10.1038/s41467-018-03905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 2007;47:89–116 doi 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 43.Singh A, Venkannagari S, Oh KH, Zhang YQ, Rohde JM, Liu L, et al. Small Molecule Inhibitor of NRF2 Selectively Intervenes Therapeutic Resistance in KEAP1-Deficient NSCLC Tumors. ACS Chem Biol 2016;11(11):3214–25 doi 10.1021/acschembio.6b00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun X, Wang Q, Wang Y, Du L, Xu C, Liu Q. Brusatol Enhances the Radiosensitivity of A549 Cells by Promoting ROS Production and Enhancing DNA Damage. Int J Mol Sci 2016;17(7) doi 10.3390/ijms17070997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai SJ, Liu Y, Han S, Yang C. Brusatol, an NRF2 inhibitor for future cancer therapeutic. Cell Biosci 2019;9:45 doi 10.1186/s13578-019-0309-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.