Abstract
The tumor microenvironment (TME) is a key determinant of metastatic efficiency. We performed a quantitative high throughput screen (qHTS) of diverse medicinal-chemistry tractable scaffolds (44,420 compounds) and pharmacologically active small molecules (386 compounds) using a layered organotypic, robust assay representing the ovarian cancer (OvCa) metastatic TME. This 3D model contains primary human mesothelial cells, fibroblasts and extracellular matrix, to which fluorescently-labeled OvCa cells are added. Initially, 100 compounds inhibiting OvCa adhesion/invasion to the 3D model in a dose-dependent manner were identified. Of those, eight compounds were confirmed active in five high-grade serous OvCa cell lines, and were further validated in secondary in vitro and in vivo biological assays. Two tyrosine kinase inhibitors, PP-121 and Milciclib, and a previously unreported compound, NCGC00117362, were selected because they had potency at 1μ in vitro. Specifically, Milciclib and NCGC00117362 inhibited OvCa adhesion, invasion and proliferation, while PP-121 inhibited OvCa invasion and proliferation. Using in situ kinase profiling and cellular thermal shift assays, we found that Milciclib targeted Cdk2 and Cdk6, and PP-121 targeted mTOR. In vivo, all three compounds prevented OvCa adhesion/invasion and metastasis, prolonged survival and reduced omental tumor growth in an intervention study. To evaluate the clinical potential of NCGC00117362, structure-activity-relationship studies were performed. Four close analogs of NCGC00117362 efficiently inhibited cancer aggressiveness in vitro and metastasis in vivo. Collectively, these data show that a complex 3D culture of the TME is effective in qHTS. The three compounds identified have promise as therapeutics for prevention and treatment of OvCa metastasis.
Introduction
Approximately 70% of patients with OvCa present with metastatic disease, for which we currently have no available targeted treatments. Indeed, OvCa is the leading gynecologic cancer, with an estimated 17,000 newly diagnosed cases in the United States in 2018 (1). The overall survival rate of patients with advanced high-grade serous OvCa is extremely low despite aggressive treatment with surgery and chemotherapy (2). This poor prognosis is due to widespread metastasis at the time of diagnosis and therapeutic resistance. Therefore, new therapies targeting OvCa metastasis are essential if we are to significantly improve the prognosis for OvCa patients.
The cellular and extracellular components of the tumor microenvironment (TME) play a critical role in cancer metastasis and drug resistance (3,4). Metastasis of high-grade serous OvCa mainly involves the detachment of OvCa cells from the primary tumor and their subsequent transcoelomic dissemination in the peritoneal fluid to mesothelium-lined metastatic sites within the confines of the abdominal and pleural cavities. The OvCa tumor cells and the cells in the surrounding peritoneal tumor microenvironment interact closely and play a critical role in the progression and metastasis of OvCa (reviewed in (5–7)). For the past several years, we and others showed that adipocytes (8), fibroblasts (9), macrophages (10) and mesothelial cells (11–15) change their phenotype to cancer-associated stromal cells that promote OvCa cell growth through bi-directional communication.
The first organotypic model that recapitulated the mesothelium surface histology of the omentum and peritoneum was based on a detailed analysis of the omentum and peritoneum of patients without OvCa (16). Recently, this model was further developed to perform high throughput drug screening (HTS) in 384- and 1536-well format to screen 2,000 compounds (17,18). Specifically, primary human omental mesothelial cells and fibroblasts are mixed with extracellular matrix (ECM) to re-construct, in vitro, the superficial tissue layer of the mesothelium, the principal site of OvCa metastasis (19–21).
The goal of this study was to identify inhibitors of OvCa metastasis by performing a fully robotic qHTS cell based assay using five high-grade serous OvCa cell lines, primary human stromal cells, and extracellular matrices (ECM). Over 44,000 new, structurally-diverse small molecule compounds and clinical drugs were screened using the complex 3D qHTS assay. Through a series of confirmatory and secondary in vitro and in vivo assays, three compounds that inhibit OvCa metastasis in vivo were identified.
Materials and Methods
Reagents
Two collections of compounds assembled at the NCATS were screened A collection of 44,420 diverse small molecule drugs; and the Mechanism Interrogation PlatE 4.0 collection of 386 pharmacologically active small molecules (22). The active compounds were re-purchased for repeat and secondary assays. Milciclib (catalog #PHA-848125), MK-0752 (#HY-10974) and PP-121 (#HY-10372) were purchased from MedChem Express. Ouabain (#1076) and AMG-51 (#SYN-111) were purchased from Fisher Scientific. Vardenafil (#Y0001647) and Oridonin (#O9639) were purchased from Sigma-Aldrich. Peruvoside (# P227570) was purchased from Toronto Research Chemicals. NCGC00117362 (#D233–0871), NCGC00111761 (#C686–0165), NCGC00117328 (#D233–0834), NCGC00117505 (#D244–0327), NCGC00117477 (#D244–0252), NCGC00117166 (#D233–0497), NCGC00115018 (#D053–0260), NGC00117330 (#D233–0835) and NCGC00117364 (#D233–0885) were purchased from ChemDiv. Collagen type I (rat-tail), and fibronectin (human) were purchased from BD Biosciences. Rabbit anti-phospho m-TOR against Ser2448 (#2971), rabbit anti-mTOR (#2972), rabbit anti-phospho Rb against Ser807/811 (#8516, clone D20B12), mouse anti-Rb (#9309), mouse anti-Cdk6 (#3136), lysis buffer (#9803), and 3x Blue Loading Buffer (#56036) were purchased from Cell Signaling Technology. Mouse anti-Cdk1/Cdk2 (#sc-53219) was purchased from Santa Cruz Biotechnology Inc.
Cell lines
The Ovcar4 cells were purchased from the DCTD Tumor/Cell Line Repository (#0507673). The COV318 cells were obtained from Gottfried Koneczny at UCLA. Dr. Gordon B. Mills (originally at M.D. Anderson Cancer Center) provided the SKOV3ip1 and HeyA8 cells. The CaOV3 cells were purchased from the American Type Culture Collection (#HTB-75). The Ovcar5 cells were obtained from UCSF (23). The Tyk-nu cells were obtained from UCLA (24). The Kuramochi cells were purchased from the JCRB Cell Bank (#JCRB0098). The Snu-119 cells were purchased from the Korean Cell Line Bank (#00119). All of these OvCa cell lines were cultured in DMEM with 10% FBEssence, 1% L-glutamine, and 1% penicillin-streptomycin (absent in CETSA assays). The 59M cells were obtained from the European Collection of Cell Cultures, and cultured in DMEM with 2mM glutamine, 1mM sodium pyruvate,10μg/ml insulin and 10% FBEssence (23). The GFP/luciferase-labeled OvCa cells were constructed utilizing a lentivirus vector expressing copepod GFP (# CD511B-1) and the lentivirus packing kit (# LV500A-1) from System Biosciences (9). The ID8p53−/− were provided by Ian McNeish (25) and are cultured in DMEM with 4 % fetal-bovine serum, 1x insulin-transferrin-selenium, 1% L-glutamine and 1% penicillin-streptomycin. All cell lines were passaged 3–8 times after thawing before used in described experiments. The cell lines were banked in liquid nitrogen and one vial of each passage were confirmed Mycoplasma negative using the STAT-Myco kit and validated using short tandem repeat DNA fingerprinting with the AmpFℓSTR Identifier kit and compared with known fingerprints by IDEXX BioAnalytics Laboratories (Columbia, MO).
Primary HTS assay
Specimens of fresh human omentum were obtained from patients undergoing surgery for benign conditions who gave written informed consent before surgery. The protocol was approved by the University of Chicago Institutional Review Board. Primary human mesothelial cells and fibroblasts were isolated from normal human omentum, purity were verified by vimentin, calretinin and cytokeratin immunohistochemistry and used within two passages of isolation (12,16).
For the 1536-well format, 40 primary human omental fibroblasts and 400 mesothelial cells were seeded with 0.02 μg fibronectin and 0.02 μg collagen type I in 4μl of growth media (2.3mm2, Fig. S1a). After a 48-hour incubation at 37°C, 1,200 SKOV3ip1-GFP were seeded in 3μl of serum-free media (growth media minus fetal bovine serum) on top of primary human omental cells. The compounds were screened in four doses (0.36–46 μM), and the plates contained the positive control (Tomatine) in eight doses (0.035–75 μM) and DMSO (equal volume controls). The compounds or controls were added to each well immediately after addition of the cancer cells. The plates were incubated at room temperature (RT) for 2 hours and then at 37°C for 16 hours. Following incubation, the media was aspirated and each well washed with PBS (5μl) and then fixed with 4% paraformaldehyde (PFA; 5μl). After 15 minutes the PFA solution was aspirated, PBS (5μl) was added, and the number of GFP-labeled cells analyzed using a fluorescent cytometer (TTP LabTech Acumen eX3; Hertfordshire, UK).
Confirmatory and counter assays
The 3D HTS assay was plated in a 384-well format. Primary human omental fibroblasts (400) and mesothelial cells (4000) were seeded with collagen type I in 40μl of growth media (0.06 cm2). After 48 hours incubation at 37°C/ 5% CO2/ 95% relative humidity, SKOV3ip1-GFP, HeyA8-GFP, Kuramochi-GFP, COV318-GFP, SNU119-GFP, Tyk-nu-GFP or Ovcar4-GFP OvCa cells (12,000) were seeded in 40μl of serum-free media (growth media minus fetal bovine serum) on top of primary human omental cells (final volume of 80ul total). The library compounds dissolved in DMSO at 46 μM final concentration, positive control (Tomatine) at 10 μM final concentration, or DMSO (0.5% final concentration) were added to each well immediately after the addition of cancer cells. The plates were incubated at RT for 2 hours before being placed in an incubator at 37°C. After a 16 hour incubation, each well was washed with phosphate-buffered saline (PBS, 40 μl) followed by fixation with 4% paraformaldehyde. The number of fluorescent cells in the assay was analyzed using a fluorescent cytometer (TTP Labtech Acumen eX3).
For the confirmatory assay, the dose response of the compounds was tested using 12 concentrations (10 nM to 100 μM). The compounds dissolved in DMSO or DMSO (equal volume control) were added to each well immediately after the addition of cancer cells. The plates were incubated, treated and fixed as described above and the number of fluorescent cells analyzed using the fluorescent cytometer.
Counter assay
OvCa cells (4,000 SKOV3ip1-GFP or 2,000 HeyA8-GFP) were seeded in 40 μl of growth media and incubated for 24 hours. Compounds dissolved in DMSO at 12 concentrations (10 nM to 100 μM) or DMSO (equal volume control) were added. The plates were incubated at RT for 2 hours before being placed in an incubator at 37°C. After a 16 hour incubation at 37°C, the CellTiter-Glo® cell viability assay (Promega, Madison, WI) was performed and analyzed using a luminescent plate reader (BioTek Synergy NEO2).
The compound’s area under the curve (AUC) for activity outcomes from the confirmatory and counter screens was calculated based on the data analysis and dose-response curve fittings. The compounds were clustered hierarchically using TIBCO Spotfire 6.0.0 (Spotfire Inc.).
Secondary biological in vitro assays
The secondary biological assays were miniaturized for high-throughput analysis. The 3D culture was assembled on black-walled 384-well plates for adhesion and growth assays using the iPipette from Apricot Designs. For invasion assays, the 3D culture was assembled on pre-coated (7μg of collagen type I) 96-well transwell inserts (BD Biosciences) (12,16). The compounds were re-purchased as described above. Compounds dissolved in DMSO were tested at 1, 2, 5, and 10 μM concentrations, and DMSO (equal volumes) was the control.
Adhesion
8,000 fluorescently-labeled Tyk-nu-GFP, Ovcar5-GFP or Kuramochi-GFP OvCa cells were mixed with the compounds and seeded in 40 μl of serum-free media on top of the 3D culture (0.33cm2, n=5–15). After 1-hour incubation at 37°C, the wells were washed with PBS, fixed with PFA and cell number computed using a Spectramax i3 MiniMax 300 imaging cytometer (Molecular Devices; San Jose, CA).
Invasion
8,000 Tyk-nu-GFP, Ovcar5-GFP or Kuramochi-GFP cells were seeded in 40μl of serum-free media in the upper chamber of a 96-well transwell plate (0.134cm2, n=5–10) pre-coated with the 3D culture (12). The compounds were added to the cancer cells in the upper chamber, growth media (100 μl) was placed in the bottom chamber, and the plates were incubated at 37°C for 24–48 hours. All cells were removed from the top chamber and the invaded OvCa cells were quantified using the Spectrmax i3 MiniMax 300 imaging cytometer.
Proliferation
2,000 Tyk-nu-GFP, 1,000 Ovcar5-GFP or 4,000 Kuramochi-GFP cells were seeded in 40 μl of serum-free media on top of the 3D culture (0.33cm2, n=5–15). The compounds were added after 30 minutes, plates were incubated for 72 hours at 37°C, and total number of cells was counted using the Spetramax i3 MiniMax 300 imaging cytometer.
Animal experiments
Female C57BL/6NCrl (C57BL/6; #027) mice and female HSD:Athymic Nude-Foxn1nu (athymic nude; #069(nu)/070(nu/+) mice at age 5–6 weeks and approximately 20 grams were purchased from Charles River and Envigo respectively. All procedures involving animal care were approved by the Committee on Animal Care at the University of Chicago.
In vivo adhesion/invasion assay
Mice were randomized into groups (n=5) and injected intra-peritoneally (i.p.) with 4 million ID8p53−/− luciferase/GFP cells mixed with compounds (5 μM) or DMSO (equal volume). Sixteen hours (the same time span as the primary screen) after cancer cell injection the mice were sacrificed (5 mice/ group). The omentum was removed and omental lysates were prepared, and total luciferase activity was quantified as described previously (26).
In vivo prevention metastasis assay
Mice were randomized into groups (n=5) and injected i.p. with ID8 cells (4×106) mixed with compounds (5 μM) or DMSO (equal volume) on day 1 (5 mice/group). Forty-five days after injection the mice were sacrificed (17). The tumor colonies were counted, collected, and weighed.
In vivo prevention survival assay
Mice were randomized into groups (n=5) injected i.p. with Ovcar5 (5×106) mixed with compounds (5 μM) or DMSO (equal volume) on day 1. Mice were treated twice after inoculation (48 and 96 hours) with i.p. injection of compounds (1 mg/kg/day) in PBS (200 μl). The mice were sacrificed at the first signs of distress (17).
In vivo intervention assay
Mice were injected i.p. with 4 million ID8p53−/− luciferase/GFP cells. Twenty-one days after cancer cell injection the mice were randomized (n=5) into groups and treated with i.p. injection of compounds (5mg/kg/day) or DMSO (equal volume) in PBS (200μl) daily for 10 days. The mice were sacrificed 1 day after last treatment. The omental tumors were weighed. The concentration of Milciclib tested is lower than the doses reported in clinical trials.
Kinase selectivity assays
Tyk-nu cells were grown in Corning hyperflasks, washed with PBS then treated with Milciclib or PP-121 for 16 hours (100, 10, 1, and 0.1 μM). The cells were washed with PBS, trypsinized, neutralized and pelleted. The pellets were snap-frozen in liquid nitrogen and sent to ActiveX Biosciences Inc. (La Jolla, CA) to perform in situ kinase profiling (KiNativ) (27).
Immunoblots
RIPA buffer was used to lyse cells for cellular p-mTOR, mTOR, pRb and Rb analysis. Cell Signaling Lysis Buffer was used to lyse cells for pRb and Rb detection. Equal amounts of protein were added to each well of an SDS-PAGE gel and resolved. Proteins were transferred to a nitrocellulose membrane. The membranes were blocked in 5% milk tris-buffered saline with 0.1% tween. The membranes were incubated in primary antibodies 1:1000 dilution in 5% bovine-serum albumin tris-buffered saline overnight at 4°C. The next day, the blot was incubated with horseradish peroxidase-conjugated IgG secondary antibody at RT for one hour at a 1:5000 dilution, and visualized using chemiluminescence reagents.
Cellular thermal shift assay (CETSA)
Tyk-nu cells (3×105) were suspended in 100 μL of PBS containing complete Mini EDTA-free protease inhibitor cocktail. In order to evaluate Cdk2 or Cdk6 aggregation, temperature cell samples in thin-wall PCR stripe were incubated for 3 min between 45 and 65°C using Bio-Rad T100 Thermal Cycler. The negative control sample was incubated at RT. Cdk2 or Cdk6 thermal stability upon inhibitor binding was evaluated by incubation of the cell suspensions containing DMSO solutions of inhibitor in 100 μM to 1 nM concentrations and inhibitor solvent control sample at 52°C. Afterwards samples were centrifuged at 3000 x g for 40 min at 4°C. Supernatants containing non-aggregated fraction of protein were separated by SDS-PAGE (28). Protein was visualized by immunoblotting using mouse anti-Cdk1/Cdk2 and mouse anti-Cdk6 diluted 1:1000. Intensity of the bands on the membranes was quantified in ImageJ software. The charts of the relative amount of Cdk2 or Cdk6 (normalized to RT control) stabilized in the presence of Milciclib or PP-121 in range of concentrations were prepared in GraphPad Prism version 7.04.
Statistical analysis
Confirmatory (n=8), adhesion (n=5–15), invasion (n=5–8) and growth (n=8–15) assays were conducted in at least three independent experiments. The mean and standard deviation or standard error of the mean are reported. All statistical analyses were performed using GraphPad Prism (GraphPad). For experiments comparing two groups, data was analyzed using a two-tailed Mann Whitney U test to account for non-normal distribution of the data. For experiments with more than two groups, one-way ANOVA followed by Dunnett’s multiple comparisons test (DMSO vs. each of the other groups) was used. Differences were considered significant if p< 0.05. Kaplan-Meier survival estimates were calculated to assess group differences in the survival study. For the survival assay, a log-rank test was performed to compare the four treatment groups and individual comparisons of DMSO vs. treatment were reported (Bonferroni-adjusted).
Results
Use of a 3D organotypic assay for qHTS
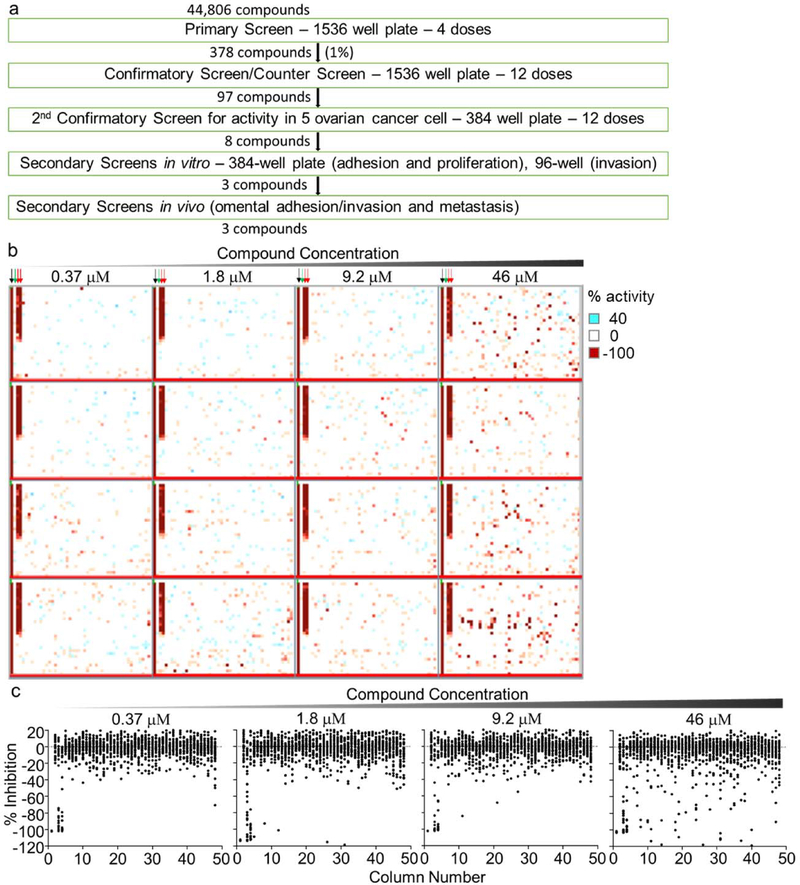
Our screening approach to evaluate the effect of small molecule compounds on the early steps of OvCa metastasis is shown in Fig. 1a. A 3D organotypic assay was implemented onto a fully automated robotic platform and 44,806 compounds screened. In this assay, primary human fibroblasts and mesothelial cells were embedded in fibronectin and collagen and allowed to grow for 48 hours in 1536-well plates. The GFP-labeled cancer cells were then added, followed by compound treatment, and a short incubation at RT of 2 hours (Fig. S1a, b, methods). After an additional 16 hours incubation at 37 °C, the adhered/ invaded cancer cells were quantified using a fluorescent cytometer, after the non-adhered cells were washed. Each compound was tested at 4 doses, 0.37, 1.8, 9.2, and 46 μM (Fig. 1b, c). The expansion of the HTS platform for the screening of 44,806 compounds did not compromise the quality of the HTS assay, which was still robust and reproducible with signal-to-background ratios of 22–63-fold and Z’-factor values of 0.4–0.84 (Fig. S1c).
Figure 1. A 3D organotypic model used in qHTS to identify inhibitors of OvCa metastasis.
a. An outline of the screening strategy and number of active compounds after each assay. b-c. 1,536-well HTS primary screening assay (4-dose response). Adhesion and invasion of fluorescently labelled (GFP) SKOV3ip1 cells to the 3D organotypic model (16h) was measured using a fluorescence cytometer. b. Representative images of the four compound plates are shown. Inhibition of adhesion/invasion depicted in red; no activity in white. The controls, no cancer cells added (−100% inhibition, black arrow) dimethyl sulfoxide (0 % inhibition, green arrow) and Tomatine (pharmacological control, red arrows) are shown c. Scatter % inhibition plots of the assay plates for the four compound doses tested. Active compounds were selected using a combination of parameters including curve response, class scoring, maximum response, efficacy and compound concentrations that give half-maximal response values (EC50).
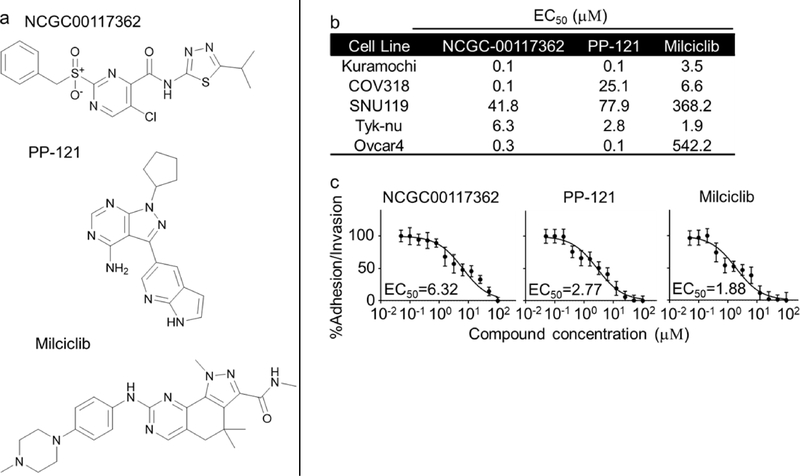
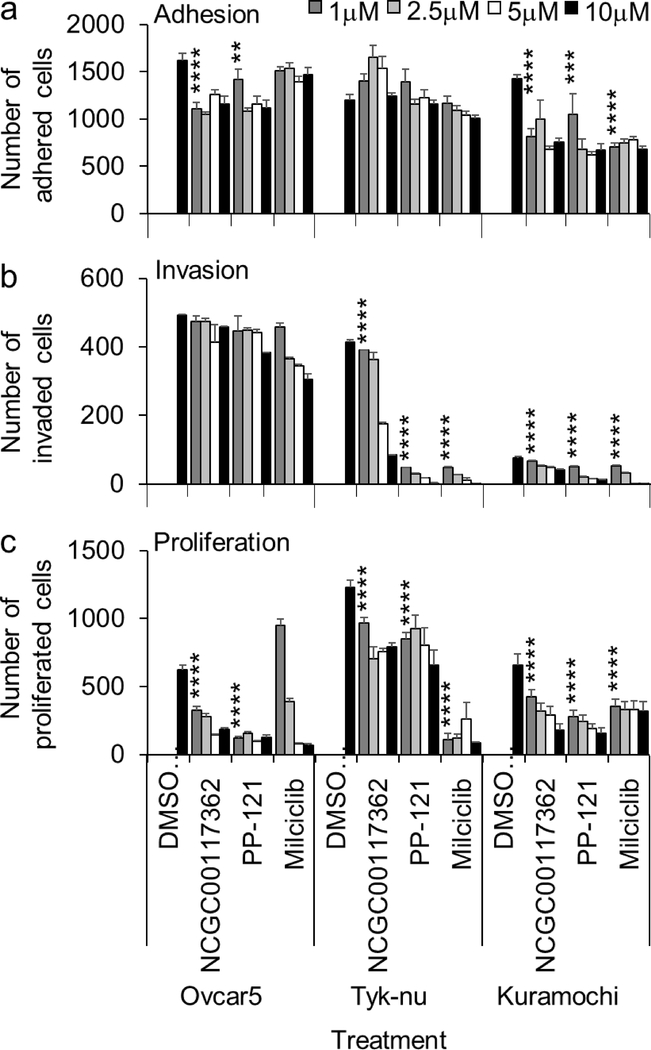
From the primary screen, we identified 378 compounds that inhibited the adhesion/ invasion of SKOV3ip1 cells in a dose-dependent manner. In a confirmatory assay, the compounds were re-tested in the 3D adhesion/ invasion HTS assay using a 12-point response in SKOV3ip1 and HeyA8 cells (Fig. S2). A counter screen was performed to identify and eliminate compounds that affected OvCa cell viability at a similar doses and time of the assay (Fig. S2). Ninety-seven out of 378 compounds were confirmed to have activity with both the HeyA8 and SKOV3ip1 cell lines, at a dose that was not cytotoxic. The 97 compounds were further tested in a multi-dose adhesion/ invasion assay with five additional serous OvCa cell lines (29) (CaOV3, Ovcar4, Kuramochi, Snu-119 and Tyk-nu). In this testing, eight out of the initial 97 compounds were identified to be active in all five serous OvCa cell lines (Fig. 2 and S3). The structure of the compounds, NCGC00117362, PP-121 and Milciclib, is shown in Fig. 2a, and the structure of the compounds, AMG-51, MK-0752, NCGC00161703, Oridonin and Vardenafil, is shown in Fig. S3a. The eight compounds were further tested at 4 doses in three additional functional screens using 3 serous OvCa cell lines, Ovcar5, Tyk-nu, and Kuramochi (Fig. 3 and S4a–c). Compounds were considered active in the functional assays if they inhibited adhesion, invasion or proliferation in two of the three cell lines, at a concentration of 1 μM. Three compounds (NCGC00117362, PP-121, and AMG-51) inhibited adhesion to the 3D organotypic culture after 1 hour (Fig. 3a and S4a). Four compounds (NCGC00117362, PP-121, Milciclib and NCGC00161703) inhibited OvCa cell invasion through the 3D organotypic culture after 24 hours (Fig. 3b and S4b). Finally, three compounds (NCGC00117362, PP-121 and Milciclib) decreased OvCa cell growth after 72 hours when co-cultured with the 3D organotypic culture (Fig. 3c and S4c).
Figure 2. Activity of hit compounds in secondary confirmatory assay.
a. Chemical structure of NCGC001177362, Milciclib and PP-121. b-c. The three compounds were tested at 12-doses in five ovarian cancer (OvCa) cell lines for their inhibitory effect on OvCa cell adhesion/ invasion to the 3D organotypic model (16h). b. The EC50 for each compound in each cell line in adhesion/ invasion assay to 3D organotypic model (16h). c. Representative dose-response curves of Tyk-nu OvCa cell adhesion/ invasion to the 3D organotypic model (16h, mean+/− standard deviation, n=8).
Figure 3. Activity of hit compounds in secondary in vitro biological assays.
Secondary in vitro screens (adhesion, invasion and proliferation) were performed. The effect of compounds at 4-doses were tested in three ovarian cancer (OvCa) cell lines. OvCa cell adhesion (2h; a) and proliferation (96h; c) were tested in 384-well plates on the 3D organotypic culture. OvCa cancer invasion (24–48h; b) invasion was tested using a 96-well Boyden chamber lined with collagen type I as ECM and the 3D organotypic culture. Mean+/− standard deviation. **, p<0.01. ***, p<0.001. ****, p<0.0001, n=5–8. DMSO, dimethyl sulfoxide.
Efficacy of the compounds in in vivo metastasis assays
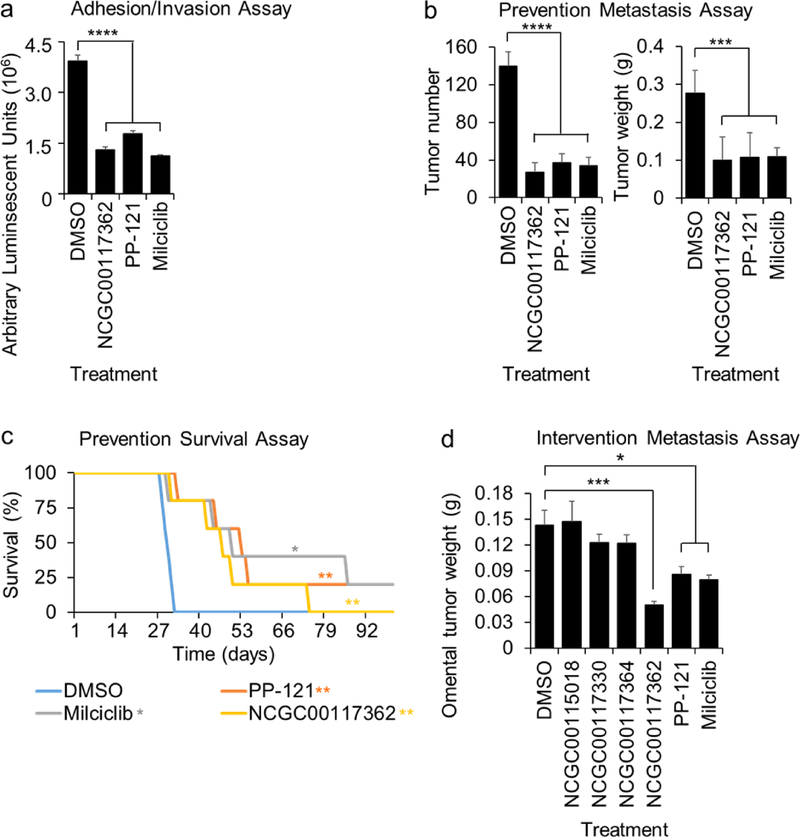
The effect of early treatment (prevention study) was tested using three in vivo assays. First, we performed a short-term adhesion/invasion assay in the mouse peritoneal cavity (8,11,17), using the same 16 hour time-point used in the in vitro studies. Luciferase-labeled ID8p53−/− OvCa cells were mixed with each of the eight most efficient compounds (NCGC00117362, PP-121, Milciclib, AMG-51, MK-0752, NCGC00161703, Oridonin and Vardenafil), injected i.p. into mice, and cancer cell adhesion/invasion measured. Consistent with the in vitro studies, all eight compounds inhibited adhesion/invasion to the omentum in vivo (Fig. 4a and S4d). Because three compounds (NCGC11007362, PP-121, and Milciclib) significantly inhibited at least two key in vitro OvCa functions in early metastasis at a low dose (1 μM) and inhibited in vivo OvCa cell adhesion/invasion at 5 μM, we evaluated the efficacy of these compounds further in vivo.
Figure 4. Activity of hit compounds in in vivo biological assays.
a-c. in vivo screens (adhesion/invasion and prevention metastasis assay). The compounds were tested at a 5μM dose. a. In Vivo Adhesion/Invasion Assay. Luciferase-labeled ID8p53−/− cells (5 million) were mixed with the indicated compound and injected into C57BL/6 mice. The mice were sacrificed at 16h and the luciferase signal in the omentum was measured. b-c. Five million Ovcar5 ovarian cancer cells were injected i.p. with NCGC1177362, PP-121 or Milciclib (5 μM), and i.p. treatment repeated 48 and 96 h post-cancer cell injection (3 total treatments: 10 mg/kg or equal volume of DMSO, n=5). b. In Vivo Prevention Metastasis Assay. Forty-five days post cancer cell injection the weight and number of tumors was determined (Mean+/− standard deviation). c. In Vivo Prevention Survival Study. Mice were sacrificed once they showed signs of distress, and Kaplan–Meier curves were calculated. d. In Vivo Intervention Metastasis Assay. 21 days post cancer cell injection the mice were i.p. treated daily with NCGC00117362, PP-121 or Milciclib (5mg/kg/day) and sacrificed 10 days later. The omental tumor weight was determined (mean+/− standard deviation). *, p<0.05. **, p<0.01. ***, p<0.001. ****, p<0.0001, n=5. DMSO, dimethyl sulfoxide.
We next tested these three compounds in a metastasis prevention assay using the ID8p53−/− OvCa cells syngeneic model (30,31). The OvCa cells were mixed with each of the three compounds (NCGC00117362, PP-121 or Milciclib) and injected i.p. After 2 more treatments (day 2, 4), which were followed by 41 days of observation, NCGC00117362, PP-121 and Milciclib were found to reduce tumor number by at least 70% and tumor weight by 64% (Fig. 4b). Next, the three compounds were tested in a survival assay (12,17) in which mice were injected i.p. with ID8p53−/− cells using the same treatment schedule as described for the previous in vivo assay. The treatment of mice with NCGC00117362, PP-121 and Milciclib increased overall survival significantly (median survival: 47, 53 and 50 days, respectively; Fig. 4c) when compared to vehicle treated control mice (median survival: 30 days). There was no significant difference in survival comparing the three treatment groups.
Last, the three compounds were tested in an intervention treatment assay in which mice were injected i.p. with ID8p53−/− cells and 21 days post cancer cell injection the mice were treated i.p. with compounds (5mg/kg/day) for 10 consecutive days. The treatment of mice with NCGC00117362, PP-121 and Milciclib decreased omental tumor weight significantly (Fig. 4d). NCGC00117362 treatment inhibited omental tumor growth significantly more than PP-121 (p=0.007) or Milciclib (p=0.002) treatment.
Activity of the two kinase inhibitors in ovarian cancer cells
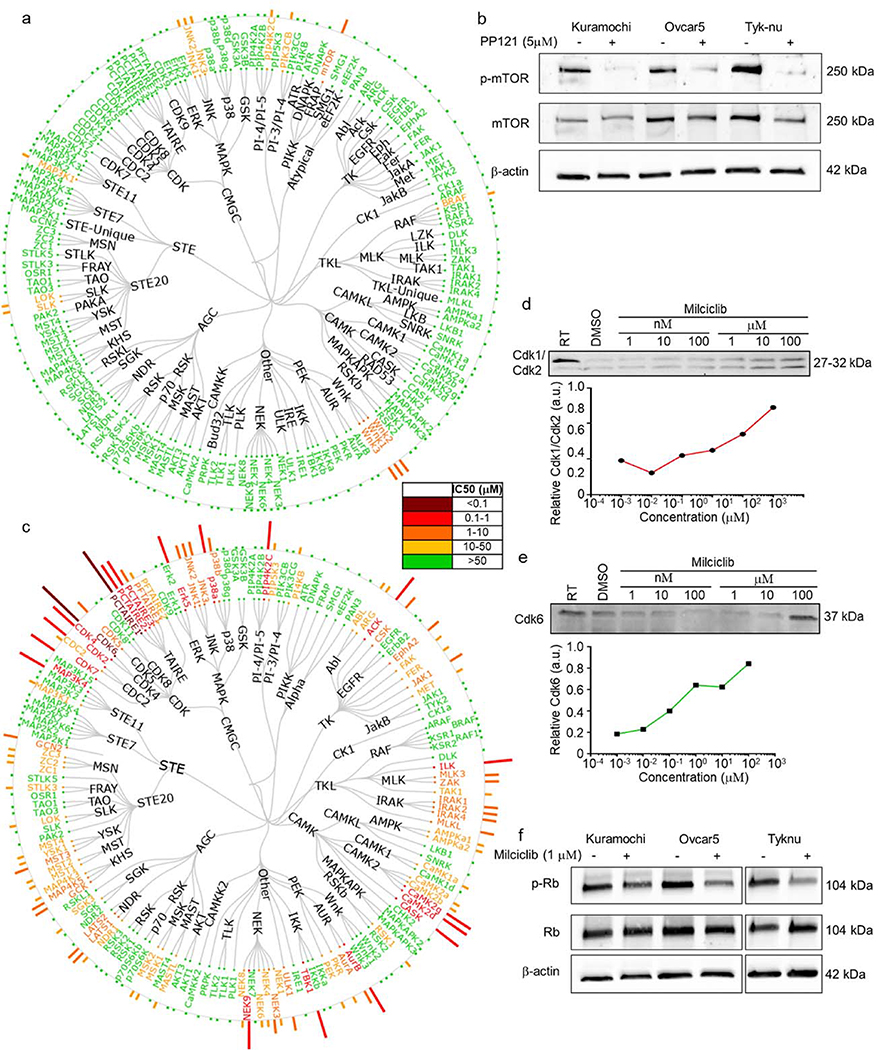
Because PP-121 and Milciclib are both kinase inhibitors, we investigated their kinase selectivity profile. Using in situ kinase profiling of 156 kinases (Fig. 5a), PP-121 in Tyk-nu cells inhibited mTOR and WNK1–3.Using immunoblotting we confirmed that PP-121 inhibits phosphorylation of mTOR in all three OvCa cell lines (Ovcar5, Kuramochi and Tyk-nu) (Fig 5b). Because multiple mTOR inhibitors are currently being used clinically, we wanted to determine if other mTOR inhibitors inhibit metastasis. However, early i.p. treatment with Everolimus or Temsirolimus, two mTOR inhibitors, had no effect on OvCa adhesion/invasion to the omentum in vivo (Fig. S5).
Figure 5. Kinase selectivity profile of PP-121 and Milciclib in ovarian cancer cells.
a. Tyk-nu cells were treated with increasing concentrations of PP-121 for 16h, and in situ kinase profiling performed. Dendrogram representation of kinase inhibitor activity. Potency depicted by length of the bar for each kinase, and color code for concentration active. b. Immunoblot analysis of phosphorylated and total mTOR after PP121 (5 μM) or DMSO control treatment for 24 hours in Kuramochi, Ovcar5 and Tyk-nu cells. DMSO, dimethyl sulfoxide. c. Tyk-nu cells were treated with increasing concentrations of Milciclib for 16h, and in situ kinase profiling performed. Dendrogram representation of kinase inhibitor activity. Potency depicted by length of the bar for each kinase, and color code for concentration active. d-e. Cellular thermal shift assays of Cdk1/Cdk2 (d) and Cdk6 (e) with increasing concentrations of Milciclib (1 nM to 100 μM). f. Immunoblot analysis of phosphorylated and total retinoblastoma (Rb) after Milciclib (1 μM) or dimethyl sulfoxide (DMSO) control treatment for 24 hours in Kuramochi, Ovcar5 and Tyk-nu cells. RT, room temperature.
Using the same in situ kinase profiling described above, we found that Milciclib inhibited a plethora of kinases (Fig. 5c), including cyclin-dependent kinases (CDK)2/4/6. Using CETSA in Tyk-nu cells, we determined that Milciclib interacts directly with Cdk1/Cdk2 and Cdk6 (Fig. 5d–e). The downstream effector of CDK is retinoblastoma (Rb) protein, which upon phosphorylation allows the cell cycle to proceed through G1 (32). In three OvCa cell lines (Ovcar5, Tyk-nu and Kuramochi; Fig. 5f), Milciclib inhibited phosphorylation of Rb, which indicates that one of its mechanisms of action is cell cycle arrest. This inhibition was independent of cyclin E1 levels, since Milciclib similarly inhibited the invasion and proliferation of OvCa cells expressing low or high levels of cyclin E1 inhibitor (Fig. S6).
Next, we investigated whether Milciclib and NCGC00117362 could regulate the phosphorylation of mTOR, or whether PP-121 and NCGC00117362 could regulate the phosphorylation of Rb. Milciclib and NCGC00117362 did not regulate the phosphorylation of mTOR, while PP-121 and NCGC00117362 did not control the phosphorylation of Rb (Fig. S7).
Structure-activity relationship of NCGC00117362
The third lead compound, NCGC00117362, has not been previously reported to affect tumor growth or metastasis. Gene expression profiling of OvCa cells after 24 hours of treatment with NCGC00117362 revealed significant changes in the expression of genes associated with NF-kB survival, MAPK signaling and GPCR ligand binding. (Table S1). Consistent with the pathway analysis, NCGC00117362, but not PP-121 or Milciclib downregulated the expression of Rac2, IL1β, and MMP-14 (Fig. S8).
To evaluate the relationship between the chemical structure of this compound and its biological activity, structure activity relationship (SAR) studies were performed. Four close SAR analogues of the NCGC00117362 compound (Fig. 6a) and three inert control compounds (Fig. 6b) from the same chemical series were identified, purchased and tested using the screening paradigm described in Fig. 1a. The four SAR analogs significantly inhibited adhesion/invasion, while the three inert control compounds had no effect on the adhesion/invasion of five serous OvCa cell lines to the 3D HTS assay (Fig. S9). Interestingly, NCGC00117362 and one other compound, NCGC0017328, inhibited OvCa adhesion (Fig. 6c), while all SAR analogues inhibited OvCa cell invasion (Fig. 6d). Only NCGC00117362 inhibited OvCa proliferation (Fig. 6e). The inert control compounds had no effect on OvCa adhesion, invasion or proliferation (Fig. 6c–e). Consistent with the in vitro studies, only the four SAR analogues inhibited OvCa adhesion/invasion to the omentum in vivo (Fig. 6f and S5). Furthermore, i.p. treatment of mice with the SAR analogs prior to OvCa cell implantation on the intra-abdominal surface, reduced tumor weight and tumor number by at least 30% when compared to control mice (Fig. 6g). Conversely, inert control compounds had not effect on omental tumor growth when treatment was given 21 days post OvCa cell injection in a syngeneic mouse model (Fig. 4d).
Figure 6. In vitro and in vivo testing of close analogs of the novel active compound, NCGC00117362.
a. Chemical structure of NCGC00117362 and 4 analogs. b. Chemical structure of 3 inert control compounds. c-e. Secondary in vitro screens were performed. The effect of compounds at 4-doses (1, 2.5, 5 and 10 μM) was tested in three ovarian cancer (OvCa) cell lines. c, e. OvCa cell adhesion (2h; c) and proliferation (96h; e) were tested in 384-well plates on the 3D organotypic culture. d. OvCa invasion (24–48h) was tested using a 96-well Boyden chamber lined with collagen type I as ECM and the 3D organotypic culture. f-g. Secondary in vivo screens were performed. f. Adhesion/Invasion Assay. The analog compounds were tested at a 5μM dose. Luciferase-labeled ID8p53−/− cells (5 million) were mixed with the indicated compound and injected into C57BL/6 mice. The mice were sacrificed at 16h and the luciferase signal in the omentum was measured using a luminometer. g. Prevention Metastasis Assay. Five million Ovcar5 cells were injected i.p. with individual compounds (5 μM), and i.p. treatment continued at 48 and 96 h post-cancer cell injection (10 mg/kg or equal volume of dimethyl sulfoxide (DMSO), n=5). Forty-five days post cancer cell injection the weight and number of tumors determined. Mean+/− standard deviation. *, p<0.05. **, p<0.01. ***, p<0.001. ****, p<0.0001, n=4–8.
Discussion
A previously developed complex organotypic assay that reconstructs important attributes of the OvCa metastatic microenvironment (17,18) was used to screen 44,802 compounds in qHTS. The compounds screened included new, structurally-diverse compounds and existing approved or investigational drugs that could potentially be re-purposed for OvCa therapy. This 1536-well primary assay, simulating the setting of OvCa metastasis and the systematic follow-up approach after the primary screen (Fig. 1A), proved to be successful strategies that were able to identify inhibitors of OvCa metastasis in vivo. The activity of compounds, which blocked atleast two OvCa functions (adhesion, invasion and/or proliferation) in multiple OvCa cell lines in vitro, was confirmed in three in vivo assays using xenograft and syngeneic mouse models with prevention and intervention treatment regimens.
Three compounds with both in vitro and in vivo activity were identified using our screening strategy, the two kinase inhibitors, PP-121 and Milciclib, and a new compound, NCGC00117362. Using an unbiased kinase screening platform, we found that PP-121 specifically bound to mTOR and inhibited mTOR phosphorylation at low doses. PP-121 has been reported as a strong multi-targeted kinase inhibitor, with activity against PDGFR, Hck, mTOR, VEGFR2, Src and Abl. It is also known to block pro-tumorigenic signaling pathways, including Akt-mTOR and NF-κB (33). In glioblastoma cell lines, it has been shown that PP-121 blocks proliferation (33) and in thyroid cancer cell lines it inhibits thyroid cancer cell migration, invasion, cell viability and tumor growth (34). Therefore, PP-121 is a dual inhibitor of tyrosine and phosphoinositide kinases that targets major pro-tumorigenic signaling pathways.
Milciclib bound cyclin-dependent kinases at low doses in OvCa cells. Specifically, Milciclib interacted with CDK6 and PCTAIRE1 (CDK16). Moreover, Milciclib inhibited Rb phosphorylation, which is downstream of CDK activity as previously reported in melanoma (35), glioma (36) and the A2680 OvCa (37) cell lines. The effect of Milciclib on cancer cells is not limited to OvCa, it has also shown efficacy in non-small cell lung, melanoma, colon, pancreatic and prostate cancer subcutaneous tumor growth in vivo (37), and is currently in Phase I and II clinical trials (, , ) in patients with unresectable/metastatic hepatocellular carcinoma, thymic carcinoma, and thymoma. These reports, and the data presented here, support the further clinical development of Milciclib towards clinical testing in epithelial OvCa.
The third compound, NCGC00117362, is in a group of structurally similar small molecules, which reliably inhibited in vitro and in vivo OvCa activity in SAR studies. NCGC00117362 downregulated multiple signaling pathways, including NF-kB and MAPK, important signaling pathways in high-grade serous OvCa (38,39). The efficiency of NCGC00117362 and its four SAR analogues in in vitro and in vivo studies supports our view that the structure of this compound may provide a promising lead for the further development of a candidate that is clinically effective against metastatic disease.
That the NCGC00117362 compound was identified using our systematic 3D approach, and the fact that all analogues also proved efficient against the mechanisms tested with the HTS assay and subsequent in vitro and in vivo biological studies, shows that our approach can very effectively identify drug candidates for further clinical development. In this study, we proved that a 3D model could be used with high reproducibility in a HTS setting with tens of thousands of compounds. Of note, we established the assay at the University of Chicago and transferred the technology to NCATS, showing that the method may be adapted for various HTS settings in different laboratories. Although the in vivo microenvironment is not fully recreated in this model (it lacks endothelial and immune cells), the model successfully incorporates primary human cells in a dependable, consistent, and reproducible 1536-well assay format. The modular concept of the 3D model can be used to screen for inhibitors of other cancers by customizing it to simulate the microenvironments specific to those cancers. For example, the effect of different drugs on the specific metastatic sites of castrate-resistant prostate cancer could be investigated by recreating the specific TME of lymph nodes, bone, or liver (40).
In the future, the 3D model could be further developed and individualized by testing drugs using syngeneic primary patient-derived cancer and host cells at the time of the initial surgery and diagnosis, establishing a truly individualized therapy. In summary, our complex physiologically-relevant model was successfully used in qHTS to identify potential therapeutics for the prevention of OvCa metastasis. If these agents reach clinical use, they could be beneficial for patients after optimal tumor reduction to reduce the re-seeding of cancer cells in the abdominal cavity.
Supplementary Material
Acknowledgements
We thank Gail Isenberg for editing the manuscript. This work was supported by the Ovarian Cancer Research Fund Alliance (OCRFA)- Liz Tilberis Early Career Award 545674 (H Kenny), an HTS award from the Chicago Biomedical Consortium with funding from the Searle Funds at The Chicago Community Trust (E Lengyel and H Kenny), Bears Care - the charitable beneficiary of the Chicago Bears Football Club (EL and HAK), R01CA111882 (E Lengyel), and by the Intramural Research Program (Division of Preclinical Innovation, National Center for Advancing Translational Sciences). The Cellular Screening Center and Human Tissue Resource Center Cores at the University of Chicago are funded by the Cancer Center Support Grant (P30CA014599).
Abbreviations list
- OvCa
ovarian cancer
- TME
tumor microenvironment
- HTS
high throughput screening
- ECM
extracellular matrix
- CETSA
cellular thermal shift assay
- NCATS
National Center for Advancing Translational Sciences
- DMSO
dimethyl sulfoxide
- GFP
green-fluorescent protein
- RT
room temperature
- AUC
area under the curve
- PBS
phosphate-buffered saline
- SAR
structure activity relationship
- i.p.
intaperitoneal
Footnotes
Conflict of Interest
The authors declare no potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68(1):7–30 doi 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Vaughan S, Coward JI, Bast RC, Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: Recommendations for improving outcomes. Nature Reviews 2011;11:719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19(11):1423–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hui L, Chen Y. Tumor microenvironment: Sanctuary of the devil. Cancer Lett 2015;368(1):7–13 doi 10.1016/j.canlet.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 5.Hansen JM, Coleman RL, Sood AK. Targeting the tumour microenvironment in ovarian cancer. Eur J Cancer 2016;56:131–43 doi 10.1016/j.ejca.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitra AK. Ovarian cancer metastasis: A unique mechanism of dissemination In: Xu K, editor. Tumor Metastasis: InTech; 2016. [Google Scholar]
- 7.Yeung TL, Leung CS, Yip KP, Au Yeung CL, Wong ST, Mok SC. Cellular and molecular processes in ovarian cancer metastasis. A Review in the Theme: Cell and Molecular Processes in Cancer Metastasis. Am J Physiol Cell Physiol 2015;309(7):C444–56 doi 10.1152/ajpcell.00188.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med 2011;17(11):1498–503 doi 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitra AK, Zillhardt M, Hua YJ, Tiwari P, Murmann AE, Peter ME, et al. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov 2012;2(12):1100–8 doi 10.1038/onc.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin M, Li X, Tan S, Zhou HJ, Ji W, Bellone S, et al. Tumor-associated macrophages drive spheroid formation during early transcoelomic metastasis of ovarian cancer. J Clin Invest 2016;126(11):4157–73 doi 10.1172/jci87252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenny HA, Chiang CY, White EA, Schryver EM, Habis M, Romero IL, et al. Mesothelial cells promote early ovarian cancer metastasis through fibronectin secretion. J Clin Invest 2014;124(10):4614–28 doi 10.1172/JCI74778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenny HA, Kaur S, Coussens LM, Lengyel E. The initial steps of ovarian cancer cell metastasis are mediated by MMP-2 cleavage of vitronectin and fibronectin. J Clin Invest 2008;118(4):1367–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rynne-Vidal A, Jimenez-Heffernan JA, Fernandez-Chacon C, Lopez-Cabrera M, Sandoval P. The mesothelial origin of carcinoma associated-fibroblasts in peritoneal metastasis. Cancers (Basel) 2015;7(4):1994–2011 doi 10.3390/cancers7040872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandoval P, Jiménez-Heffernan JA, Rynne-Vidal A, Pérez-Lozano ML, Gilsanz A, Ruiz-Carpio V, et al. Carcinoma-associated fibroblasts derive from mesothelial cells via mesothelial to mesenchymal transition in peritoneal metastasis. J Pathol 2013;231(4):517–31. [DOI] [PubMed] [Google Scholar]
- 15.Rynne-Vidal A, Au-Yeung CL, Jimenez-Heffernan JA, Perez-Lozano ML, Cremades-Jimeno L, Barcena C, et al. Mesothelial-to-mesenchymal transition as a possible therapeutic target in peritoneal metastasis of ovarian cancer. J Pathol 2017. doi 10.1002/path.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenny HA, Krausz T, Yamada SD, Lengyel E. Use of a novel 3D culture model to elucidate the role of mesothelial cells, fibroblasts and extra-cellular matrices on adhesion and invasion of ovarian cancer cells to the omentum. Int J Cancer 2007;121(7):1463–72 doi 10.1002/ijc.22874. [DOI] [PubMed] [Google Scholar]
- 17.Kenny HA, Lal-Nag M, White EA, Shen M, Chiang CY, Mitra AK, et al. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat Commun 2015;6:6220 doi 10.1038/ncomms7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lal-Nag M, McGee L, Guha R, Lengyel E, Kenny HA, Ferrer M. A High-Throughput Screening model of the tumor microenvironment for ovarian cancer cell growth. SLAS Discov 2017;22(5):494–506 doi 10.1177/2472555216687082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lengyel E Ovarian cancer development and metastasis. Am J Pathol 2010;177(3):1053–64 doi 10.2353/ajpath.2010.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auersperg N, Wong AST, Choi KC, Kang SK, Leung PCK. Ovarian surface epithelium: Biology, endocrinology and pathology. Endocr Rev 2001;22(2):255–88. [DOI] [PubMed] [Google Scholar]
- 21.Levanon K, Crum CP, Drapkin R. New insights into the pathogenesis of serous ovarian cancer and its clinical impact. J Clin Oncol 2008;26(32):5284–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathews Griner LA, Guha R, Shinn P, Young RM, Keller JM, Liu D, et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A 2014;111(6):2349–54 doi 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louie KG, Hamilton TC, Winker MA, Behrens BC, Tsuruo T, Klecker RW Jr., et al. Adriamycin accumulation and metabolism in adriamycin-sensitive and -resistant human ovarian cancer cell lines. Biochem Pharmacol 1986;35(3):467–72. [DOI] [PubMed] [Google Scholar]
- 24.Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proceedings of the National Academy of Sciences USA 2011;108(30):12372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walton J, Blagih J, Ennis D, Leung E, Dowson S, Farquharson M, et al. CRISPR/Cas9-Mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high-grade serous carcinoma. Cancer Res 2016;76(20):6118–29 doi 10.1158/0008-5472.CAN-16-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenny HA, Lengyel E. MMP-2 functions as an early response protein in ovarian cancer metastasis. Cell cycle 2009;8(5):683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem Biol 2011;18(6):699–710 doi 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckert MA, Coscia F, Chryplewicz A, Chang JW, Hernandez KM, Pan S, et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019;569(7758):723–8 doi 10.1038/s41586-019-1173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coscia F, Watters KM, Curtis M, Eckert MA, Chiang CY, Tyanova S, et al. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat Commun 2016;7:12645 doi 10.1038/ncomms12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaur S, Kenny HA, Jagadeeswaran S, Zillhardt M, Montag AG, Kistner E, et al. ß3-integrin expression on tumor cells inhibits tumor progression, reduces metastasis, and is associated with a favorable prognosis in patients with ovarian cancer. Am J Pathol 2009;175(5):2184–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenny HA, Leonhardt P, Ladanyi A, Yamada SD, Montag A, Im HK, et al. Targeting the urokinase plasminogen activator receptor inhibits ovarian cancer metastasis. Clin Cancer Res 2011;17(3):459–71 doi 10.1158/1078-0432.CCR-10-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. eLife 2014;3 doi 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apsel B, Blair JA, Gonzalez B, Nazif TM, Feldman ME, Aizenstein B, et al. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat Chem Biol 2008;4(11):691–9 doi 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Che HY, Guo HY, Si XW, You QY, Lou WY. PP121, a dual inhibitor of tyrosine and phosphoinositide kinases, inhibits anaplastic thyroid carcinoma cell proliferation and migration. Tumour Biol 2014;35(9):8659–64 doi 10.1007/s13277-014-2118-3. [DOI] [PubMed] [Google Scholar]
- 35.Caporali S, Alvino E, Starace G, Ciomei M, Brasca MG, Levati L, et al. The cyclin-dependent kinase inhibitor PHA-848125 suppresses the in vitro growth of human melanomas sensitive or resistant to temozolomide, and shows synergistic effects in combination with this triazene compound. Pharmacol Res 2010;61(5):437–48 doi 10.1016/j.phrs.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 36.Albanese C, Alzani R, Amboldi N, Degrassi A, Festuccia C, Fiorentini F, et al. Anti-tumour efficacy on glioma models of PHA-848125, a multi-kinase inhibitor able to cross the blood-brain barrier. Br J Pharmacol 2013;169(1):156–66 doi 10.1111/bph.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albanese C, Alzani R, Amboldi N, Avanzi N, Ballinari D, Brasca MG, et al. Dual targeting of CDK and tropomyosin receptor kinase families by the oral inhibitor PHA-848125, an agent with broad-spectrum antitumor efficacy. Mol Cancer Ther 2010;9(8):2243–54 doi 10.1158/1535-7163.Mct-10-0190. [DOI] [PubMed] [Google Scholar]
- 38.Pohl G, Ho CL, Kurman RJ, Bristow RE, Wang TL, Shih L. Inactivation of the mitogen-activated protein kinase pathway as a potential target-based therapy in ovarian serous tumors with kras or braf mutations. Cancer Res 2005;65(5):1994–2000. [DOI] [PubMed] [Google Scholar]
- 39.Leizer AL, Alvero AB, Fu HH, Holmberg JC, Cheng YC, Silasi DA, et al. Regulation of inflammation by the NF-kappaB pathway in ovarian cancer stem cells. Am J Reprod Immunol 2011;65(4):438–47 doi 10.1111/j.1600-0897.2010.00914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halabi S, Kelly WK, Ma H, Zhou H, Solomon NC, Fizazi K, et al. Meta-Analysis Evaluating the Impact of Site of Metastasis on Overall Survival in Men With Castration-Resistant Prostate Cancer. J Clin Oncol 2016;34(14):1652–9 doi 10.1200/jco.2015.65.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.