Abstract
The cargo-specific removal of organelles via selective autophagy is important to maintain neuronal homeostasis. Genetic studies indicate that deficits in these pathways are implicated in neurodegenerative diseases, including Parkinson’s and Amyotrophic lateral sclerosis. Here, we review our current understanding of the pathways that regulate mitochondrial quality control, and compare these mechanisms to those regulating ER and the clearance of protein aggregates. Research suggests that there are multiple mechanisms regulating the degradation of specific cargos, such as dysfunctional organelles and protein aggregates. These mechanisms are critical for neuronal health, as neurons are uniquely vulnerable to impairment in organelle quality control pathways due to their morphology, size, polarity and postmitotic nature. We highlight the consequences of dysregulation of selective autophagy in neurons, and discuss current challenges in correlating noncongruent findings from in vitro and in vivo systems.
Keywords: selective autophagy, mitophagy, neurodegeneration, mitochondrial quality control, ER-phagy
1. INTRODUCTION: BULK VS. SELECTIVE AUTOPHAGY IN NEURONS
Neurons are highly polarized, terminally differentiated cells that transmit information through the propagation of electrical signals. Due to these unique properties, neurons rely heavily on the targeted delivery and regulated removal of proteins and organelles along the axon. Defects in the removal of aggregated proteins and damaged or aged organelles can lead to cellular stress and eventually neurodegeneration or death. Furthermore, there is strong genetic evidence linking mutations in intracellular trafficking proteins to a range of neurodegenerative diseases [1].
In the neuron, multiple maintenance mechanisms are in place to degrade cellular debris. Macroautophagy (hereafter referred to as autophagy) is the major clearance pathway to nonselectively remove dysfunctional proteins and organelles in the cell [2]. This evolutionarily conserved pathway involves the engulfment of cytoplasmic components into a double-membraned vesicle or autophagosome which fuses with late endosomes or lysosomes to form autolysosomes. Engulfed cargos are degraded by lysosomal enzymes which allow for recycling of cellular components [3–5]. The protein machinery driving this pathway, autophagy related proteins (ATGs), has been well investigated in yeast and mammals (for reviews, see [6–8]).
In neurons, autophagy is critical for maintaining cellular homeostasis, as neuron-specific knockouts (KO) of key ATG proteins is sufficient to induce cellular degeneration [3, 4]. A constitutively active pathway has been described that functions under basal conditions [9, 10]. In vitro and in vivo, autophagosomes are formed in the distal axon and mature through fusion with lysosomes as the organelle is retrogradely trafficked to the soma [11–16]. Observations to date suggest that this constitutive pathway for autophagy takes up cargo nonselectively, engulfing both protein aggregates and mitochondrial fragments at the axon terminal [13, 17].
In contrast to the nonselective bulk removal of cytoplasmic components, selective autophagy involves the degradation of specific cargos through the autophagic pathway [18, 19]. Autophagosome formation is mediated by specific autophagy receptors. These receptors are cargo recognition proteins, and contain unique domains that allow them to simultaneously bind, directly or indirectly, to the cargo targeted for degradation (usually through ubiquitination) and to the phagophore or growing autophagosome via interaction with membrane-associated mATG8 proteins. The sequestration of each organelle or aggregate that is targeted and cleared by this pathway may require specific cues, although all pathways converge downstream in their use of the core components of the nonselective pathway to generate autophagosomes that engulf the cargo [18]. Specific types of selective autophagy may use multiple autophagy receptors or single receptors may mediate the removal of several types of organelles, suggesting some redundancy in the system [19]. Since it is crucial to remove damaged or aged proteins and organelles from the neuron, the promiscuity of the receptors may act as a failsafe mechanism.
Several types of selective autophagy have been reported to date, including the removal of mitochondria by mitophagy, endoplasmic reticulum (ER) by ER-phagy, aggregated proteins by aggrephagy, ribosomes via ribophagy, peroxisomes by pexophagy, etc. Much of this work was performed in non-neuronal systems. But given the key role for autophagy in the maintenance of neuronal health, and the accumulating genetic data implicating autophagy deficits in neurodegeneration, it is essential to develop a mechanistic understanding of the neuron-specific aspects of autophagy. Here, we summarize progress examining selective autophagy in neurons, highlighting the three best understood mechanisms: mitophagy, ER-phagy, and aggrephagy. We discuss how damaged organelles signal for degradation and the molecular players required for cargo recognition and sequestration. Neurons are highly polarized cells with distinct somal, dendritic and axonal compartments; as both dendritic and axon terminals may be located far from the soma, these cells face unique challenges. We emphasize the diverse mechanisms that mediate quality control in neurons, and focus on how dynamics, long-distance trafficking, and compartmentalization contribute to the overall effectiveness of the pathway. Finally, we discuss the pathophysiological roles of these pathways and implications to disease states.
2. SELECTIVE AUTOPHAGY
Many of the proteins identified as autophagy receptors have a microtubule-associated protein 1A/1B-light chain 3 (LC3)-interacting region (LIR) that mediates binding to the growing autophagosome, and a ubiquitin binding domain (UBAN, UBA, or UBZ) which allows for binding to the damaged protein or organelle [19]. It should be noted that ubiquitin-independent selective autophagy has also been reported [20]. Additionally, most autophagy receptors are cytosolic proteins that translocate to the cargo upon activation of the pathway. One exception is Nip-like protein X (NIX), a mitophagy receptor with a transmembrane domain that is found on the outer mitochondrial membrane [21, 22]. Selectivity can be further enhanced by post-translational modifications of autophagy receptors or by receptor oligomerization. For example, oligomerization of the autophagy receptor p62 enhances and stabilizes LC3 binding [23]. Similarly, phosphorylation or ubiquitination of receptors can stabilize cargo binding [18, 24, 25].
Although the types of selective autophagy differ by the organelles that are sequestered, commonalities among the parallel pathways do exist [18]. For instance, all of the pathways are thought to initiate through some type of stress, perturbation, or age-dependent turnover of the organelle or protein aggregate. The damaged organelle or aggregate cues receptor-mediated specificity for degradation, and all use the components of the autophagic pathway for elimination. Selective autophagy pathways appear to be important for neuronal homeostasis, as they are implicated in neurodegenerative diseases. Mutations in the mitophagy components OPTN and TBK1 cause familial forms of amyotrophic lateral sclerosis (ALS; [26, 27]) and mutations in PINK1 and Parkin cause familial forms of Parkinson’s disease (PD; [28–30]). Similarly, mutations in the autophagy receptor p62 cause ALS, frontotemporal dementia (FTD), and Paget’s disease of bone [31], and mutations in the ER-phagy receptor family with sequence similarity 134 member B (FAM134B) cause sensory and autonomic neuropathy [32].
3. MITOPHAGY
Mitophagy is the selective sequestration and elimination of damaged mitochondria [33–35]. Neurons, in particular, rely heavily on the mitochondrial network to support local energy demands at synaptic sites. Thus, the prompt removal of dysfunctional mitochondria is critical for neuronal health and maintenance. In vivo, damage to mitochondria occurs through aging, localized reactive oxygen species (ROS) production, exposure to toxins, hypoxia, or developmental programming. Experimentally, mitochondrial damage and mitophagy can be initiated via multiple triggers, including oxidative stress through antioxidant removal, chemical uncouplers (i.e. CCCP, carbonyl cyanide m-chlorophenyl hydrazone; Antimycin A; Oligomycin; rotenone), hypoxia, iron chelation, and the local generation of ROS [36–38], although not all of these approaches are likely to effectively model physiological changes in mitochondrial health.
Initial investigation of the mitophagy pathway started with the discovery of two genes that were linked to familial PD, park6 encoding PINK1 (PTEN-induced kinase 1) and park2 encoding Parkin [28–30]. In immortalized cells lines these proteins were found to be critical for the clearance of damaged mitochondria. Since these early discoveries, multiple mitophagy pathways have been reported (Figure 1; [21, 39, 40]). Some of these pathways do not involve either PINK1 or Parkin, sparking questions as to which pathways are most relevant in vivo and under which circumstances. In general, it is thought that the multiple mitophagy mechanisms, identified to date, work in parallel to maintain mitochondrial health. However, whether there is a predominant pathway whose dysregulation leads to the progressive functional and structural degeneration of the neuron is highly debated.
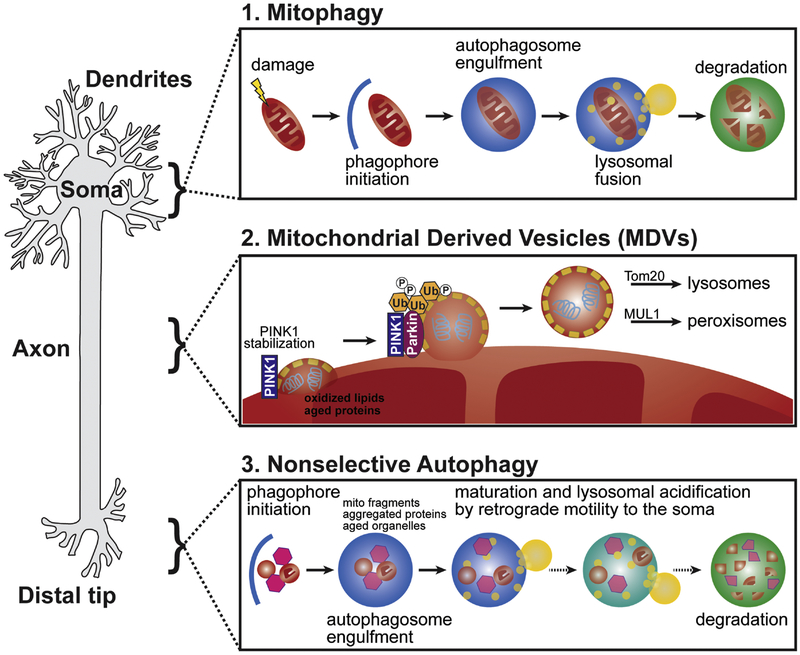
Figure 1: The mechanisms of neuronal mitochondrial quality control.
1. Damaged mitochondria are removed from the cell via a process termed mitophagy. Organelles are marked for degradation, sequestered by an autophagosome, and degraded by lysosomal fusion. Additional mechanisms exist to remove organelle components, including mitochondrial derived vesicles (MDVs) and the nonselective removal of mitochondrial fragments by autophagy. 2. MDVs are budded vesicles that are thought to contain oxidized lipids and damaged/aged proteins. These small vesicles utilize PINK1/Parkin phospho-ubiquitination signaling and undergo cargo-based targeting to peroxisomes and lysosomes. Additional budded vesicles have been reported, such as SNPH-positive vesicles that are Parkin-independent. 3. In the distal axon, mitochondrial fragments, protein aggregates, and other cytosolic components are nonselectively engulfed by the constitutive formation autophagosomes. Cargos are degraded through lysosomal fusion and acidification as autophagosomes mature through retrograde trafficking. Each of these molecularly distinct signaling pathways provides a different aspect of mitochondrial regulation.
3.1. PINK1/Parkin-dependent mitophagy
PINK1/Parkin-dependent mitophagy is the best described pathway for mitochondrial quality control and initially was characterized in immortalized cell lines. This is a highly regulated process that involves the translocation of several proteins to the damaged organelle. Under basal conditions, the serine-threonine kinase PINK1 is proteolytically processed at the outer mitochondrial membrane by a mitochondrial intermembrane protease presenilin-associated rhomboid-like protein (PARL), which keeps the relative PINK1 concentration low [41, 42]. Under cellular or environmental stress, mitochondrial depolarization leads to a failure in PINK1 processing and its subsequent accumulation on the outer mitochondrial membrane (OMM) [43, 44]. Autophosphorylation of the kinase enhances activity which leads to the recruitment, phosphorylation, and activation of Parkin, an E3 ubiquitin ligase [43, 45, 46]. Additionally, PINK1 phosphorylates ubiquitin [47, 48]. Together, these proteins create a feedforward amplification mechanism whereby ubiquitin binding and PINK1-phosphorylation of Parkin induce full activation of the protein and thus conjugation of ubiquitin chains to OMM substrates (i.e. voltage-dependent anion channel [VDAC] and Mitofusin [Mfn] 2); in turn, this leads to new sites for PINK1 phosphorylation and the resulting amplification of the initial response [47, 49–51]. As a result, phospho-ubiquitination of OMM proteins by PINK1 and Parkin generates a specific signal and a platform for the recruitment of autophagy receptors for organelle degradation.
3.1.1. PINK1/Parkin-dependent mitophagy in nonneuronal cell lines
In non-neuronal cell lines overexpressing Parkin, multiple autophagy receptors translocate to mitochondria after mitophagy induction via CCCP or local ROS generation, including optineurin (OPTN), nuclear dot protein 52 kDa or calcium-binding and coiled-coil domain-containing protein 2 (NDP52/CALCOCO2), tax1 binding protein 1 (TAX1BP1), neighbor of BRCA1 gene 1 protein (NBR1), and p62/sequestosome 1 (SQSTM1) [24, 38, 52–54]. Translocation and mitochondrial interactions are mediated by the ubiquitin domain found in all of the proteins. Initial studies suggested p62 might mediate PINK1/Parkin-dependent mitophagy [53], but additional work showed that p62 can cluster damaged mitochondria but does not facilitate mitophagy [38, 52, 54]. To investigate the functional contribution of each receptor in mitophagy, one study generated a cell line lacking all five receptors (OPTN, NDP52, TAX1BP1, NBR1, and p62). In this penta-KO background, cells failed to remove damaged mitochondria, but overexpression of OPTN, NDP52, or, to a lesser extent, TABX1BP1 rescued mitophagy [52]; similar results were reported using transient knock-down (KD) in a wild-type (WT) background [24, 38]. While defects were seen in the penta-KO cells, individual depletion of either OPTN or NDP52 was not reported to cause a long-term defect in mitochondrial clearance [52]. In contrast, transient depletion of OPTN was shown to cause a significant delay in mitophagy, indicating a kinetic impairment upon OPTN loss [24, 38].
Autophagy receptor specificity is enhanced by posttranslational modifications, such as the phosphorylation of autophagy receptors by TANK-binding kinase 1 (TBK1). TBK1 has been shown to phosphorylate multiple receptors at sites that are relevant to selective autophagy, for example OPTN, NDP52, TAX1BP1, and p62 [25, 55–57]. However, interactions with OPTN appear to be the most functionally relevant. TBK1 binds and phosphorylates OPTN at sites in or near both the UBAN and LIR motifs which activates the protein [24, 25, 52, 56]. For instance, TBK1-dependent phosphorylation of OPTN S177 is necessary for efficient LC3 recruitment [24, 52, 56]. TBK1 is co-translocated with OPTN to the surface of damaged organelles, where phosphorylation helps to stabilize OPTN on the OMM [24, 25]. In cells in which TBK1 was either depleted or pharmacologically inhibited, autophagosome engulfment of damaged mitochondria was impaired [24]. Additionally, expression of disease-associated mutants in either OPTN or TBK1 disrupted the pathway leading to accumulation of damaged mitochondria [24, 25, 38, 52]. More recently, TBK1 was shown to cooperate with NDP52 to recruit induction complex components of the autophagic machinery, UNC-51-like autophagy acting kinase 1 (ULK1) through interactions with FAK family kinase-interacting protein of 200 KDa (FIP200) [58]. Surprisingly, NDP52 and TBK1 could target ULK1 to cargo and mediate autophagosome biogenesis in the absence of mATG8s, indicating that selective autophagy receptors might participate more upstream in the signaling pathway than was initially thought.
A novel family of mitochondrial matrix proteins have been implicated in mitophagy signaling, illustrating further redundancy in the PINK1/Parkin-dependent mitophagy pathway. In nonneuronal cell lines, 4-nitrophenylphosphatase domain and non-neuronal SNAP25-like protein homolog 1 (NIPSNAP1) and NIPSNAP2 accumulated on the OMM of depolarized mitochondria [59]. NIPSNAP1 is highly expressed in dopaminergic neurons of mice [60] and the homologs appear to be redundant [59]. NIPSNAP1 immunoprecipitated with Parkin and interacted with LC3B and autophagy receptors, p62 and NDP52, to target the sequestration of damaged organelles. When NIPSNAP1 and NIPSNAP2 were depleted from cells, Parkin was efficiently recruited to depolarized mitochondria, but autophagy receptor recruitment was impaired, suggesting NIPSNAPs are required for autophagy receptor recruitment and mitophagy. Consistent with a role of mitophagy in PD, zebrafish models show loss of NIPSNAP1 leads to increased mitochondrial dysfunction, fewer dopaminergic neurons, and impaired motor neuron function [59].
3.1.2. PINK1/Parkin-dependent mitophagy in primary neurons
Work in nonneuronal cell lines has established a critical role of PINK1 and Parkin in mitophagy, but studies translating these findings to mitophagy in primary neurons or in vivo have yielded some surprising results. Further, the unique characteristics of neurons have raised additional questions, such as whether all neuronal mitochondria are subject to the same quality control mechanisms and whether mitophagy is spatially regulated, a point we return to below.
Thus far, work in neurons has confirmed the molecular players involved, suggesting that the PINK1-Parkin mitophagy pathway is conserved across cell types (Figure 2). In cultured neurons, Parkin is selectively recruited to damaged mitochondria after induction of mitophagy with multiple damaging agents, including CCCP, Antimycin A, and rotenone [37, 48, 61, 62]. Interestingly, Parkin translocation was shown to be differentially regulated depending on the damaging agent and by the presence or absence of antioxidants in the culture media [62]. Further, Parkin translocation has been reported to occur over a long time scale, ~18 hours after damage [61], much slower than the temporal translocation dynamics observed in HeLa cells where LC3 engulfment can be observed within an hour of insult [24, 38]. In contrast to the observations of Cai et al. (2012), local damage to axonal mitochondria using the mitochondrially-targeted photosensitizer mitoKillerRed was reported to occur within an hour [37].
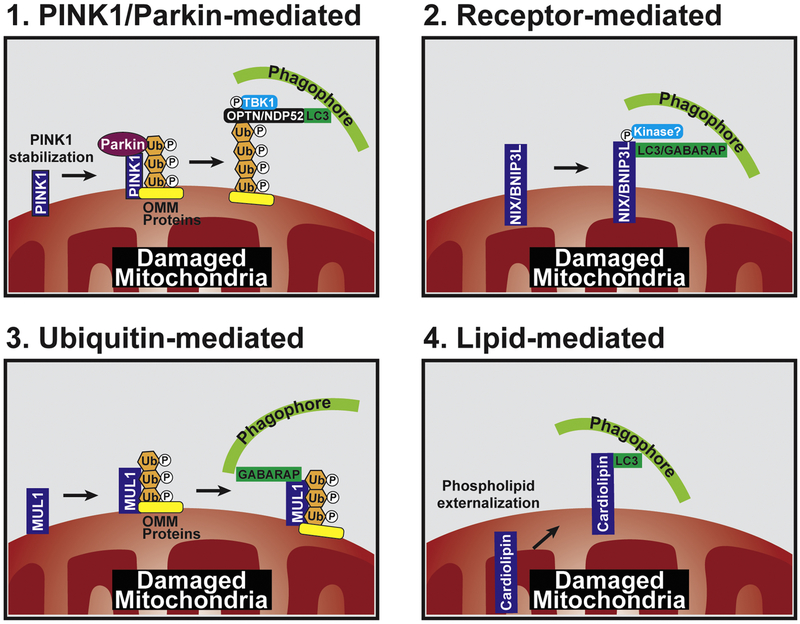
Figure 2: The diverse pathways to induce mitophagy.
Within the neuron, multiple mechanisms exist to signal cargo engulfment, including PINK1/Parkin, OMM receptors, ubiquitin cascades, and lipids. 1. PINK1/Parkin-mediated mitophagy initiates with PINK1 stabilization on the OMM, which recruits Parkin. Phospho-ubiquitination of OMM substrates (VDAC, Mfn2, etc.) by PINK1/Parkin recruits autophagy receptors, OPTN and NDP52. Phosphorylation of OPTN by TBK1 helps to stabilize it on the mitochondrial surface. These autophagy receptors link the damaged organelle to the autophagosome through interactions with ubiquitin and LC3 on the mitochondrial membrane and growing phagophore, respectively. 2. Several autophagy receptors are anchored within the OMM, including BNIP3 and NIX/BNIP3L. Highlighted is the role of NIX/BNIP3L, which is phosphorylated by an unknown kinase, leading to activation and binding of NIX/BNIP3L to LC3 or GABARAP on the phagophore. 3. Additional E3 ligases have been reported to regulate mitophagy independent of Parkin. MUL1 has similar substrates to Parkin and can directly bind to GABARAP, suggesting that it can function independently to mediate engulfment. 4. Externalization of cardiolipin, normally found on the IMM, to the OMM is a unique mechanism for lipid-mediated mitophagy. Cardiolipin can bind directly to LC3 to induce mitophagy.
A ‘double’ hit strategy of antioxidant removal coupled to treatment with a mitochondrial damaging agent has been used to induce mitophagy in neurons but is not needed in nonneuronal cells [37, 48, 62]. Moreover, only a subset of mitochondria appears to enter the mitophagic pathway after global treatment of neurons. In contrast, the integrity of the mitochondrial network in nonneuronal cells was lost and the majority of organelles were sequestered following similar treatments. It may be that post-mitotic neurons have additional quality control mechanisms in place to preserve mitochondrial health in order to prevent the turnover of the entire network; these mechanisms may be key in terminally differentiated and post-mitotic cells.
3.1.3. PINK1/Parkin-dependent mitophagy in vivo
PD is a neurodegenerative disease that leads to the progressive degeneration of dopaminergic neurons in the substantia nigra. Since loss-of-function mutations in park2 and park6 genes are linked to familial PD [28–30], transgenic models were generated to recapitulate aspects of the disease in vivo. Thus far, most of the work has utilized mouse and fly model systems. Interestingly, loss of PINK1 or Parkin in mouse models does not cause a robust PD phenotype, such as dopaminergic neuron loss. However, these phenotypes are observed in Drosophila mutant flies. These thought-provoking findings have pushed the field to generate new mitochondrial probes to better understand mitophagy in vivo and question our understanding of this mechanism.
Rodent models:
KO of neither PINK1 nor Parkin in mice was reported to be lethal. An initial Parkin KO mouse line unexpectedly displayed only minor phenotypes, with no loss of the dopaminergic neurons, a hallmark of PD in humans [63, 64]. As a result, additional Parkin KO mice were generated through the deletion of different exons compared to the first two lines generated. Still, these mice failed to exhibit any substantial motor or behavioral phenotype, or degeneration of dopaminergic neurons [65, 66]. In two different lines of PINK1 KO mice dopaminergic loss was not observed [67, 68]. In aged mice, dopamine levels were lower than in control mice but dopamine turnover was upregulated, without significant changes in the number of dopaminergic neurons [67]. However, striatal slices exhibited decreased evoked dopaminergic release [68].
Additionally, triple KO mice lacking PINK1, Parkin, and DJ1 (a protein encoded by park7, another gene linked to familial PD; [69]) showed no loss of dopaminergic neurons after twenty-four months [70]. Interestingly, KO rat models of PINK1 and DJ1 showed significant dopaminergic neuron loss at eight months, with motor defects starting at four months; similar to mice, Parkin KO rats had no phenotype [71]. Collectively, these data point to a non-essential role of PINK1 and Parkin in neuronal mitophagy, suggesting other mitochondrial quality control pathways play a more significant role than was initially proposed. It is possible that PINK1 and Parkin are crucial in humans but the shorter lifespan of mice fails to allow for full manifestation of the disease, as the average onset of PD in humans is in the early sixties.
Drosophila models:
In sharp contrast to the mouse models, which only displayed small changes in dopaminergic levels, null and mutant flies of either PINK1 or Parkin showed clear PD phenotypes. Multiple studies reported a subset of dopaminergic neurons had morphology defects or abnormal shrinkage [72, 73]. In addition, locomotive defects, reduced lifespan, and sterility were all observed [74, 75]. Consistent with the Parkin phenotype in flies, PINK1 null or mutant flies exhibited defects in dopaminergic neuronal degeneration with flight muscle and locomotive defects [76–78]. These results seem contradictory to the mouse models’ findings that suggest a minimal role for PINK1 and Parkin in neuronal mitophagy. Furthermore, work in nonneuronal cell lines and primary neuronal cultures corroborates with the Drosophila results, suggesting PINK1 and Parkin are important in facilitating mitophagy. However, overexpression of these mitophagy-associated proteins in culture may upregulate PINK1/Parkin-dependent mitophagy, making it appear to be more significant than it truly is in vivo. It is possible that other mitophagy pathways play a more significant role than were initially proposed, such as partially redundant mechanisms that utilize alternative E3 ligases or PINK1/Parkin-independent mitophagy pathways.
Mitochondrial form and function:
In addition to investigating the onset of PD, the PINK1 and Parkin KO model systems have been used to study the role of these proteins in vivo. No morphological changes were observed in striatal mitochondria from mice depleted of Parkin [79]. However, one study using PINK1 deficient mice reported no gross changes to the ultrastructure, but an increase in large swollen mitochondria in the striatum of old mice compared to young mice [80]. Consistent with the later finding, transmission electron microscopy (TEM) of flight muscles in Parkin or PINK1 null or mutant flies illustrated muscle degeneration with enlarged swollen mitochondria and disrupted cristae [74, 75, 77]. Mitochondrial fragmentation was also observed in primary mouse embryonic fibroblasts (MEFs) from PINK1 KO mice [81]. In Drosophila motor neurons, somal mitochondria appeared abnormal but axonal mitochondria were unaffected [82, 83], suggesting tissue- and compartment-specific phenotypes. Thus, loss of PINK1 and Parkin in Drosophila and mice, to a lesser extent, alters mitochondrial morphology.
The muscle degeneration and morphological phenotypes of PINK1 mutant flies can be rescued by overexpression of human PINK1, illustrating functional conservation between species [77, 78]. These phenotypes could also be rescued by overexpression of Parkin, but not DJ1 [76–78]. It is important to note that PINK1 expression could not rescue Parkin phenotypes and double mutants phenocopied individual PINK1 and Parkin mutants [76, 78]. These findings provide strong in vivo evidence that 1) PINK1 and Parkin function in a common pathway and 2) Parkin is downstream from PINK1 [76–78].
Functionally, loss of PINK1 in mice and flies altered mitochondrial membrane potential, increased ROS production, decreased mitochondrial DNA, impaired ATP synthesis, and decreased ATP levels [76, 81, 82]. Mitochondria isolated from PINK1 KO mice had significantly lower Ca2+ load capacity [67] and striatum ATP levels were reduced in vivo [81]. In motor axons of PINK1 null Drosophila, mitochondrial flux was altered with no change in organelle density or length [82]; however, Parkin nulls had a decreased number of axonal mitochondria [83]. Purified dopaminergic neurons from Parkin Drosophila mutants accumulate enlarged depolarized mitochondria [84]. Despite significant gross morphological changes, null PINK1 and Parkin mice exhibit decreased expression of proteins involved in oxidative stress, resulting in impaired mitochondrial respiration and increased sensitivity to ROS [79]. These findings are consistent with reports from Parkin loss-of-function mutants in Drosophila [75, 85]. In Drosophila, PINK1 and Parkin have a role in the maintenance of mitochondrial integrity in vivo and protein depletion leads to PD phenotypes. However, in mice there is sparse evidence implicating PINK1 and Parkin in mitochondrial maintenance and no reports of PD phenotypes in KO neurons. It is possible that mice have developed compensatory mechanisms or tolerate a higher threshold for mitochondrial damage than flies or humans. Mitochondrial dysregulation and impairment are early cellular manifestations that ultimately lead to tissue degeneration and this is apparent in Drosophila models. But more work is needed to fully understand the disparate findings in these model systems.
Mitochondrially-targeted fluorescent probes:
Due to the difficulty of studying mitophagy in vivo, transgenic mice with mitochondrially-targeted fluorescent probes were generated, including mito-Timer, mt-Keima, and mito-QC. Mito-Timer is a variant of DsRed fluorescent protein that localizes to the inner mitochondrial membrane via a targeting sequence and transitions from green to red over time (~48hrs) allowing for temporal monitoring of mitochondria [86]. The mito-Timer construct is routinely used in neuronal cultures, but the reporter mice have primarily been used to study cardiac mitochondria [87, 88]. There is potential to for the reporter mice to be used in other tissues, such as the brain, as the slow oxidation rate of mito-Timer make it an ideal probe to study relative age-related mitochondrial dynamics.
Generation of mt-Keima reporter mice took advantage of the Keima protein, a coral-derived pH-dependent fluorescent probe that is resistant to lysosomal degradation; the Keima moiety is targeted to the mitochondrial matrix through the mitochondrial targeting sequence of cytochrome c oxidase subunit-VIII (COX-VIII) [89–91]. Mitophagic flux can be monitored through ratiometric analysis. At physiological pH the shorter wavelength excitation dominates, but fluorescence shifts to longer wavelength excitation in acidic environments. In vivo, basal mitophagy levels were lower in the cortex, substantia nigra, and striatum compared to the dentate gyrus and lateral ventricle, illustrating tissue-specific differences in mice [89]. Additionally, mitophagy was reduced in the dentate gyrus of aged mice compared to young mice [89]. In contrast, basal mitophagy was shown to increase with age in dopaminergic neurons of genetically engineered flies expressing the mt-Keima reporter [92]. This increase in frequency was suppressed by depletion of Parkin, demonstrating a critical role of the PINK1/Parkin mitophagy pathway in age-dependent basal mitochondrial turnover. Aging has been correlated with mitochondrial dysfunction, accumulation of mitochondrial DNA mutations, and mitochondrial morphological defects [93–97]. It is possible that mitochondrial impairment is due to a decrease in mitophagy with aging. On the other hand, mitophagy could be upregulated with age as a result of increased mitochondrial dysfunction to maintain organelle homeostasis. As with autophagy, mitophagy modulation impacts neuronal longevity and lifespan [74, 97, 98], but more work is needed to clarify the role of mitophagy in aging.
The mito-QC reporter mouse line has a constitutive knock-in of mCherry-GFP-FIS1 [99, 100]. Mitophagy is monitored by the loss of the pH-sensitive GFP signal, while the fluorescence of the fused mCherry moiety is retained in the acidic environment produced by fusion of the autophagosome with lysosomes. As a result, the dually labeled mitochondrial network appears yellow and damaged organelles undergoing mitophagy are red. Tissue-wide analysis illustrated that constitutive mitophagy occurs in multiple organs, providing further evidence that mitophagy is a homeostatic pathway. Immunolabeling of brain sections showed that Purkinje neurons undergo pronounced mitophagy, and suggested high turnover in the soma but minimal turnover in the axonal projections and dendritic arbors [99]. To further investigate the role of PINK1 in mitophagy in vivo, a cross between PINK1 KO and mito-QC reporter mice was generated [101]. Surprisingly, in tyrosine hydroxylase (TH)-positive dopaminergic neurons of the midbrain, basal mitophagy was indistinguishable between WT and PINK1 KO conditions. Similar findings, using the same reporter, were observed in Drosophila dopaminergic neurons of PINK1 or Parkin mutant flies [102]. This controversial data suggests the role of PINK1 in basal mitophagy is minimal and that other quality control pathways are simultaneously functioning to maintain mitochondrial health. Nonetheless, genetic and clinical data clearly demonstrate the significance of PINK1 and Parkin in mitophagy [28, 29], so more work is needed to understand the functional role of these proteins as compared to other cellular pathways for mitochondrial quality control in neurons.
3.2. Alternative mitophagy pathways independent of PINK1 and Parkin
While PINK1/Parkin-dependent mitophagy is the best understood pathway to date, several findings summarized above have raised questions as to whether this pathway is functionally dominant in vivo. The mild phenotypes displayed by the PINK1 and Parkin KO mice [63, 65, 67, 68] suggest that other compensatory quality control mechanisms are in place. In addition, basal mitophagy in Drosophila was suggested to be a rare event [83]. Recently, more focus has been placed on understanding alternative mitophagy pathways and how these mechanisms collectively cooperate to remove damaged organelles (Figure 2).
3.2.1. Receptor mediated mitophagy
Instead of the requirement of Parkin translocation to damaged mitochondria, receptor mediated mitophagy utilizes autophagic receptors that contain transmembrane domains, directing integration into the OMM. These receptors contain LIR motifs, allowing them to independently recruit and bind the autophagic machinery to mediate the engulfment of damaged mitochondria [21, 103]. Thus, these receptors directly induce mitophagy independent of PINK1/Parkin. Several receptors of this type have been reported, including NIX (also known as BCL2/adenovirus E1B 19kDa interacting protein 3-like, BNIP3L). Structurally, NIX/BNIP3L is anchored to the OMM via a C-terminal transmembrane domain and interacts with multiple mATG8s, such as LC3 and GABA type A receptor-associated protein (GABARAP), through a N-terminus LIR domain [22, 104]. Phosphorylation of NIX/BNIP3L enhances GABARAP binding [104], but the specific kinases and phosphatases that regulate this process are yet to be identified. The role of NIX/BNIP3L in mitophagy was initially described in erythroblasts where genetic ablation revealed NIX/BNIP3L plays an important role in programmed mitochondrial clearance and maturation [105, 106]. Similar findings for programmed mitophagy were reported in retinal ganglion cells (RGCs) during development and differentiation [107].
Interestingly, in human fibroblasts derived from an asymptomatic carrier of a Parkin mutation, mitochondrial turnover still occurred despite dysfunctional Parkin, suggesting the involvement of an alternative pathway [108]. Mitophagy was shown to be mediated by NIX/BNIP3L; this pathway was unaffected by the additional KD of PINK1. The independent role of NIX/BNIP3L in mitophagy was further confirmed in PINK1 and Parkin PD patient cell lines where overexpression or pharmacological activation of NIX/BNIP3L restored mitophagy [108]. In response to hypoxia, NIX/BNIP3L is transcriptionally upregulated by hypoxia-inducible factor (HIF) or forkhead box O3 (FOXO3) to induce mitophagy [109–111]. The functions of NIX/BNIP3L and its homolog BCL2/adenovirus E1B 19kDa interacting protein 3 (BNIP3) are now starting to be investigated in injury and disease models. Current work suggests that NIX/BNIP3L has a neuroprotective role in ischemic stroke [112], despite its homolog having a detrimental role in ischemic-hypoxic brain and spinal cord injury [113, 114]. Future work to elucidate these roles could help to provide potential therapeutic targets.
3.2.2. Lipid mediated mitophagy
Lipids can serve as an elimination signal to initiate autophagosome engulfment of damaged mitochondria. Cardiolipin is a mitochondrial phospholipid that is primarily found in the inner mitochondrial membrane [115–117]. The unique conical shape of cardiolipin allows it to interact with proteins located inside and outside of the mitochondria [116, 117]. As a result, cardiolipin has been reported to participate in the maintenance of mitochondrial membrane fluidity/stability and electron transport chain efficiency, fusion/fission, mitochondrial dynamics, cell death, and more recently mitophagy [116, 118–122]. In damaged neuronal mitochondria, cardiolipin is externalized to the OMM where it binds LC3 to drive the engulfment of the damaged mitochondria by the autophagosome [122]. In neurons, the redistribution of cardiolipin was facilitated by phospholipid scramblase-3 (PLS3) [122], but other proteins have been reported to mediate cardiolipin externalization, including NDPK-D/NM23-H4 in SH-SY5Y cells [123, 124] and Tafazzin (TAZ) in primary MEFs [125]. This interaction is facilitated by the negatively charged head group of cardiolipin and basic residues in LC3. Downregulation of cardiolipin or mutagenesis of LC3 at sites predicted to interact with the phospholipid decreased mitochondrial engulfment [122]. In primary neurons treated with rotenone to induce mitophagy, the fold change of externalized cardiolipin was greater than that seen in HeLa cells, suggesting cell types have variable responses or thresholds to noxious stimuli and differentially regulate their mitophagy pathways.
3.2.3. Ubiquitin mediated mitophagy
Additional E3 ligases have been identified that can substitute for Parkin in the removal of damaged mitochondria [21]. Mitochondrial ubiquitin ligase 1 (MUL1; otherwise known as MAPL, GIDE, and MULAN) is an E3 ligase that is localized to the OMM and interacts with substrates through a cytosolic C-terminal RING finger [126–128]. In nonneuronal cells, MUL1 was shown to play a role in mitochondrial dynamics, as over-expression caused mitochondria to cluster and fragment [126, 127]. Regulation of MUL1 is mediated through interactions with the mitochondrial fission protein dynamin 1 like protein (Drp1) [129] and the mitochondrial fusion protein Mfn [130]; it should be noted that these proteins are also substrates of Parkin. In Drosophila and mouse neurons, MUL1 can suppress PINK1 or Parkin mutant phenotypes by negatively regulating Mfn through ubiquitin-mediated degradation [130]. Additionally, loss of MUL1 has no effect on Parkin-mediated mitophagy and double mutants of MUL1 with either PINK1 or Parkin had more severe phenotypes than individual nulls. These data suggest MUL1 functions in a parallel pathway and both MUL1 and Parkin regulate Mitofusin activity to maintain mitochondrial health. Furthermore, a yeast-two hybrid screen determined MUL1 interacts with GABARAP, and further analysis identified an LIR motif in the MUL1 RING domain [131]. Thus, MUL1 can regulate mitophagy through multiple mechanisms, providing further evidence that the lack of phenotype in PINK1 and Parkin KO mouse models could be due to compensation by other mitophagy regulators or pathways.
3.3. Additional aspects of mitochondrial quality control in neurons
3.3.1. Nonselective mitochondrial clearance
Besides the selective removal of damaged or aged organelles, mitochondrial fragments are engulfed by autophagosomes in a nonselective manner in the axon terminals of neurons (Figure 1). This distinct autophagy pathway is a constitutive mechanism that does not appear to involve the selective receptors discussed above [12, 13, 132]. In cultured neurons, LC3-positive organelles form constitutively in the distal axon and are retrogradely transported to the soma [11–13, 133, 134]. Similar findings have been reported in other model systems, including Caenorhabditis elegans and Drosophila [14, 15, 135]. Autophagosomes mature during trafficking to the soma by fusion with lysosomes, leading to efficient degradation of cargo. The cargos observed in this nonselective uptake include mitochondrial fragments and both aggregated and cytosolic proteins [13, 17, 35]. In primary dorsal root ganglion neurons (DRGs), the mitochondria contained in autophagosomes appear fragmented and are morphologically distinct from the rest of the axonal mitochondrial population [13], suggesting that mitochondrial fragments are generated in the distal axon, and are then taken up nonspecifically into constitutively forming autophagosomes. The subsequent breakdown of engulfed mitochondrial fragments was disrupted when the retrograde transport of axonal autophagosomes to the cell soma was blocked [17]. Thus, autophagosome motility appears to be required for effective fusion with lysosomes and cargo degradation. Potentially, this pathway maintains the health of axonal or synaptic mitochondria localized at considerable distances from the soma, but more work is required to clarify the mechanisms involved in the generation and engulfment of mitochondrial fragments within the distal axon.
3.3.2. Mitochondrial derived vesicles
Recent work identified mitochondrial derived vesicle (MDV) formation as a novel aspect of mitochondrial cell biology [39]. MDVs are small vesicles that bud off of an organelle and undergo cargo-based targeting to peroxisomes [127] and lysosomes [136] (Figure 1). The SNARE syntaxin 17 mediates MDV-endolysosomal fusion through interactions with homotypic fusion and vacuole protein sorting (HOPS) tethering complex [137]. MDVs are produced in a constitutive pathway in which vesicles form independent of mitochondrial depolarization, but the mechanism can be further induced by oxidative stress [136]. MDV biogenesis was disrupted by depletion of PINK1 or overexpression of PD-associated Parkin mutants, indicating this quality control pathway is also regulated by these proteins [138]. However, it is a distinct pathway from mitophagy because the autophagic machinery is not required [136].
In response to mild perturbations in neurons, syntaphilin (SNPH; a mitochondrial anchoring protein [139]) is released from stressed axonal mitochondria, triggering motor-driven motility and the formation of SNPH-positive vesicles derived from mitochondria [140]. These vesicles form independent of Parkin, Drp1, and the autophagy machinery, so they are apparently distinct from the MDVs previously described [127, 136–138]. This novel pathway is also upregulated in neurons from early symptomatic stages, but not later stages, of familial ALS-linked and Alzheimer’s Disease (AD)-mutant mice, suggesting it is a quality control mechanism that responds quickly to pathological stresses [140]. The generation of SNPH-positive vesicles appear to be an independent pathway that is activated prior to PINK1/Parkin-dependent mitophagy. As a result, damaged proteins and lipids can be removed to preserve the health of the parent organelle before complete removal of the mitochondrion is required [35, 39]. This additional quality control mechanism may be critical in the maintenance of axonal mitochondria.
3.3.3. Mitophagy compartmentalization in neurons
While multiple studies have reported mitophagy can be induced by mitochondrial damage or stress in neurons, there are discrepancies in regard to the localization of neuronal mitophagy. Since the prompt removal of damaged mitochondria is critical for maintaining cellular health, it has been proposed that local mitophagy occurs throughout the cell. Yet, primary culture and in vivo based studies have suggested otherwise. In cortical neurons, Parkin-positive mitochondria were found to accumulate in the somatodendritic compartment where they underwent LC3 sequestration and lysosomal degradation [61]. Neurons treated with CCCP displayed an increase in retrograde motility of mitochondria, suggesting damaged mitochondria are trafficked back to the soma for degradation [61]. Conversely, another report described local mitophagy in the axon following treatment of neurons with Antimycin A or upon selective local damage induced by mitoKillerRed [37]. This axonal mitophagy pathway was shown to require PINK1 and Parkin, with clearance mediated by local autophagosome-lysosome fusion.
The spatial aspects of mitophagy have also been examined in vivo; these studies suggest that neuronal mitophagy is a compartmentally restricted mechanism. In PINK1 and Parkin mutant flies, axonal mitochondria displayed normal morphology with a decrease in mitochondrial flux, but no mitochondrial accumulation was observed in the axons of motor neurons [82, 83]. However, depletion of either protein resulted in altered somatic mitochondrial morphology, with mitochondria appearing more tubular and reticular than in control flies [82, 83]. Additional evidence from examination of Purkinje cells from the mito-QC mouse suggests somatodendritic compartmentalization of neuronal mitophagy [99]. It is possible that differences between induced mitophagy in culture and basal mitophagy in vivo contribute to the discrepancies reported.
3.3.4. Axonal mitochondrial transport
Since mitochondrial biogenesis occurs primarily in the soma, newly synthesized organelles need to be trafficked along the axon to replenish mitochondria at sites located distally along the axon [141, 142]. Conversely, retrograde transport may play important roles in clearance and repair [61], although there is some debate on the relative contribution of this mechanism. Axonal mitochondria move bidirectionally, with long-distance transport mediated by kinesin and dynein motors. The motility of axonal mitochondria decreases during neuronal maturation, with most organelles generally immobile and localized to presynaptic sites in mature neurons [143]. Mitochondrial motility is regulated by mitochondrial Rho (Miro), a small GTPase localized to the OMM that binds kinesin adaptors, trafficking kinesin-binding protein (TRAK) 1 and 2 (otherwise known as Milton in Drosophila) [144–147].
In the axon, there appears to be a relationship between mitochondrial motility and mitophagy, where motility disruptions isolate organelles, facilitating their removal [141, 148]. Upon mitochondrial damage, Miro is removed from the OMM, arresting mitochondrial motility. Miro has been linked to PINK1, Parkin, and Leucine-rich repeat kinase 2 (LRRK2; another PD-associated protein encoded by the gene park8 [149–151]); these proteins promote Miro degradation to halt mitochondria and remove organelles via mitophagy [148, 152, 153]. These observations are consistent with the report that local mitophagy can occur in the axon following mitochondrial damage [37]. Interestingly, mutations in LRRK2 are the most common cause of PD [154]. In LRRK2 mutant neurons, Miro remains on damaged mitochondria, delaying efficient degradation [152]. Furthermore, LRRK2 was still recruited to damaged mitochondria in PD fibroblast lines expressing mutant PINK1 and Parkin, suggesting LRRK2 functions in a parallel pathway to PINK1/Parkin, which converges on Miro [152]. In addition to regulating Miro, LRRK2 has been implicated in alterations to mitochondrial morphology and function, RAB10 phosphorylation, and modulating Parkin/Drp1 interactions, all of which could potentially play a role in the regulation of mitophagy by LRRK2 [155–159]; more work is clearly needed to clarify the mechanisms involved.
Another axonal quality control mechanism facilitated by transport is fusion/fission events. Through these events, dysfunctional mitochondria can fuse with healthy organelles and subsequently undergo fission to evenly distribute components [160]. Fusion/fission events can also help to isolate damaged mitochondria to facilitate sequestration and elimination.
3.4.5. Interplay of neuronal mitophagy pathways and mitochondrial quality control
The mild phenotypes displayed in PINK1 and Parkin KO mice in vivo, suggest other mitochondrial mechanisms may be at play [63, 65, 67, 68]. This concept is further supported by the findings that loss of PINK1 in mice and loss of PINK1 or Parkin in flies had no effect on basal mitophagy [101, 102]. The recent focus on exploring additional mitophagy pathways independent of PINK1/Parkin has increased our understanding of receptor-, lipid- and ubiquitin-mediated mitophagy [21]. Currently, it is unclear to what extent these mechanisms cooperate to contribute to mitochondrial turnover. It may be that the various mitophagy pathways are developmentally- or tissue-specifically regulated or that there is a common dominant pathway and the others are failsafe mechanisms. Additionally, it is not understood whether all mitochondria are subject to the same quality control mechanisms. Since neurons are highly polarized, it would be practical to utilize multiple quality control mechanisms to maintain health. For example, in axons, SNPH-positive vesicles formed independent of and prior to PINK/Parkin-dependent mitophagy [140]. This finding supports a model where vesicle budding is an axonal pathway that occurs under low levels of stress to preserve mitochondrial health, where the mitophagy pathway is activated under more severe stress or to remove terminal unrecoverable mitochondria. Further work is needed to understand the interplay among the various mitophagy pathways and related mitochondrial quality control mechanisms.
3.4. Pathophysiological role of mitophagy
3.4.1. Neurodegenerative diseases
Mitochondrial dynamics and regulation are important to homeostasis, as disruption of mitophagy is implicated in multiple neurodegenerative diseases, including PD, AD, and Huntington’s disease (HD) [161]. PD is characterized by the loss of dopaminergic neurons in the substantia nigra and proteins that regulate mitophagy have been linked to the disease. As previously mentioned, cell-based experiments directly link PINK1 and Parkin to mitophagy. PD patients also have elevated mitochondrial DNA deletions in the substantia nigra, further associating defects in mitochondrial quality control with PD [95, 162]. KO mice do not display PD-phenotypes [63, 65, 67, 68], but Drosophila mutant flies have locomotive defects, reduced lifespan, and dopaminergic degeneration [72–74, 76, 78]. Also, PINK1 KO rats showed dopaminergic loss and motor defects [71]. Both model systems, however, exhibit mitochondrial dysfunction. In mutant flies, swollen mitochondria with disrupted cristae were observed in flight muscle cells which correlated with muscle degeneration [74, 75, 77, 92]. In neurons, mitochondrial impairments and mitophagy defects are upstream of neurodegeneration. Therefore, the lack of phenotype in mice points to compensatory quality control pathways that are sufficient to maintain neuronal health and prevent neurons from crossing a critical threshold that induces degeneration.
Selective autophagy receptors OPTN and p62, as well as their kinase TBK1, are implicated in ALS [26, 27, 163]; mutations in OPTN and TBK1 are additionally associated with glaucoma [164, 165]. It is interesting that some of the disease-associated mutants of PD and ALS converge on a single mitochondrial quality control pathway, yet the disease etiology and symptoms induced by these mutations are diverse. ALS is characterized by the degeneration and loss of motor neurons, whereas PD primarily affects dopaminergic neurons. Since TBK1 phosphorylates OPTN and p62, impaired mitochondrial turnover and protein aggregation may both contribute to neurodegeneration in ALS. Future work to elucidate the role of these proteins in vivo will be important in deciphering the molecular signaling necessary for each neurodegenerative disease.
AD is characterized by neuronal degeneration leading to cognitive dysfunction and memory loss [166, 167]. The pathophysiology of AD involves formation of intracellular neurofibrillary tangles from tau accumulation and extracellular plaques containing amyloid-β (Aβ) peptides derived from the amyloid precursor protein (APP). This age-related disease is complex, as multiple pathways may contribute to degeneration, such as oxidative stress, mitochondrial impairment, and dysregulation of autophagic and endolysosomal clearing mechanisms [168]. In both familial and sporadic AD, mitochondrial accumulation is observed [167, 169–171]. It is thought that reduced mitochondrial clearance contributes to tau and Aβ pathologies that are hallmarks of the disease, but the underlying molecular mechanisms are not well understood [167, 171]. Both Aβ fragments and tau have been shown to interact with mitochondrial proteins resulting in functional impairment [172–175]. Additionally, mitochondrial proteostasis induction delayed Aβ toxicity and reduced amyloid aggregation in multiple AD model systems [169]. Recent work reported that mitophagy stimulation reduced Aβ pathology and reversed cognitive decline through the phagocytic activity of microglia in AD mouse models; complementary results were observed in C. elegans AD models [171]. The authors proposed a cooperative relationship among damaged mitochondria, mitophagy, tau neurofibrillary tangles, and Aβ plaques whereby defective signaling cascades exacerbate one another, giving rise to AD pathology and neurodegeneration [171].
HD is caused by the abnormal expansion of a trinucleotide (CAG) repeat in the gene that encodes the protein Huntingtin (Htt). Expansion beyond thirty-six repeats is detrimental, with disease severity correlating with expansion length [176]. HD is characterized by psychiatric disturbances, cognitive decline, and motor dysfunction [168, 176, 177]. The polyglutamine repeat in the N-terminus of the protein is prone to misfolding and aggregation [178, 179]. Due to its unique size and number of interacting partners, Htt is implicated in multiple cellular pathways, many of which have been suggested to contribute to the pathogenesis of HD, such as mitochondrial and autophagosome impairment. In patients and HD model systems, mitochondrial respiration and Ca2+ buffering are altered, and abnormal mitochondrial dynamics have been observed [180–185]. Morphological alterations have also been observed, including cristae disruption and increased mitochondrial fission through Drp1 interaction and activation [184]. Mitochondrial dysregulation is prevalent in multiple neurodegenerative diseases, but whether dysfunction is causative or correlative is still poorly understood. Further work is needed to elucidate the exact mechanisms that contribute to disease states, which could provide new avenues for therapy.
3.4.2. Innate immunity
The inflammatory signaling pathway can be hyperactivated by impaired removal of damaged mitochondria, eventually leading to chronic inflammation and inflammatory diseases [186]. A microarray to characterize transcriptional alterations in Parkin mutants in Drosophila identified activation of genes in the innate immune response [85]. Similar changes were seen in striatal gene expression in PINK KO mice [67]. These observations are consistent with the finding that Type 1 interferons (IFNs), cytokines that regulate the innate immune response, are upregulated in post-mortem brains of AD and PD patients and have been implicated in aging. Also, mitochondrial stress induced by rotenone or 1-methyl-4-phenyl-1,2,3,6 tetrahydropyridine (MPTP) treatment upregulates a proinflammatory response in cultured neurons and glia, suggesting mitophagy may alleviate inflammation [187, 188]. Specifically, rotenone induces type 1 IFN signaling, which results in a proinflammatory response in glia, resulting in downstream glia-dependent neurotoxicity [188]. Recent work has identified an inflammatory phenotype in PINK1 and Parkin KO mice after acute and chronic mitochondrial stress by excessive exercise and mitochondrial DNA mutations, respectively [189]. This phenotype could be rescued by depletion of stimulator of interferon genes (STING), a regulator of the type 1 IFN signaling response. Together these data suggest mitophagy, via PINK1 and Parkin, may alleviate inflammation and neurotoxicity by efficiently clearing damaged mitochondria.
4. OTHER FORMS OF SELECTIVE AUTOPHAGY: ER-PHAGY AND AGGREPHAGY IN NEURONS
While mitophagy is the best-characterized pathway for selective autophagy in neurons, there are a growing number of cellular cargos known to be selectively targeted for degradation by autophagy, including the ER, peroxisomes, lipid droplets, ribosomes, and protein aggregates. All of these degradative pathways likely contribute to neuronal health by regulating organelle turnover or proteostasis, both critical in terminally differentiated cells that must last the lifetime of the organism. Research on the impact of some of these pathways on neuronal homeostasis is at an early stage, but recently there has been significant progress in our understanding of ER-phagy (also referred to as reticulophagy) as an essential quality control mechanism to maintain the endoplasmic reticulum (ER) [190]. In contrast, the clearance of aggregated proteins via aggrephagy has long been considered to be critical to neuronal health, due to the pronounced accumulation of protein aggregates in major neurodegenerative diseases such as HD and ALS.
4.1. ER-phagy in neurons
Focused ion-beam scanning electron microscopy (FIB-SEM) of mouse brain tissue has produced 3D reconstructions of the ER in neurons. Images emphasize the extension of ER tubules throughout all neuronal compartments and highlight the numerous contacts made by the ER with organelles, such as mitochondria, and the plasma membrane [191]. Given this extensive ER structure, it is unsurprising that active turnover via autophagy is required to remodel and renew this organelle.
A key pathway for this remodeling involves FAM134B or reticulophagy regulator 1 (RETREG1), an ER-anchored member of the reticulon family of proteins, which binds to both LC3 and GABARAP via a LIR motif and can remodel proteins through its reticulon homology domain (RHD; [192]). Mutations in FAM134B cause sensory and autonomic neuropathy [32], indicating a key role for this protein in maintaining neuronal health. Consistent with a role in selective autophagy, overexpression of FAM134B causes fragmentation of the ER, followed by autophagosome sequestration and lysosomal degradation [192]. Conversely, depletion of FAM134B or expression of disease-associated mutants causes ER expansion. In mice, disruption of the gene encoding FAM134B induced neurodegeneration of peripheral sensory neurons, similar to the phenotypes seen in human patients [32].
Recent insight into the mechanisms driving ER-phagy came from recent molecular dynamics simulations of the RHD of FAM134B [193]. The RHD domain can induce membrane curvature in a lipid bilayer and clustering of the FAM134B-RHD amplifies membrane deformation. Thus, FAM134B and other reticulon-like ER-phagy receptors facilitate the membrane remodeling necessary for the autophagy of ER sheets [193].
Additional ER-associated receptors involved in the selective removal of ER have been identified, including cell cycle progression 1 (CCPG1; [194]), reticulon 3 (RTN3; [195]), SEC62 [196], atlastin 3 (ATL3; [197]), and testis expressed 265 (TEX264; [198, 199]); these proteins contain LIR motifs that link the ER to the growing phagophore. ATL3 is of particular interest, as it is a dynamin-like GTPase from the atlastin family that binds specifically to GABARAP rather than LC3 [197]. ATL3 is mutated in hereditary sensory and autonomic neuropathy type 1 (HSAN1), a neurological disorder characterized by nerve abnormalities in the legs and feet [200, 201]. Disease-associated mutations impair binding to GABARAP and disrupt starvation-induced ER-phagy in cell lines [197], although the effects on ER-phagy in neurons have not yet been tested. Further, ATL3 has been shown to affect ER-mitochondrial contacts, while disease-linked mutations affect mitochondrial distribution [202] and axon outgrowth [203] in primary cortical neurons.
TEX264 is a newly identified ER-phagy receptor, a single-pass transmembrane protein localized to the ER with a LIR motif that can bind to LC3B and GABARAP proteins [198, 199]. TEX264 has a major role in remodeling the ER proteome in response to nutrient deprivation (An et al., 2019), but the role of this protein in neuronal ER-phagy remains to be assessed. Structural analysis of TEX264 identified a required element, an intrinsically disordered region (IDR) near the LIR motif, that is predicted to extend 25 nm [198]. Similar IDRs were identified in other ER-phagy receptors, including FAM134B, CCPG1, RTN3 and SEC62, leading to the hypothesis that the IDR is required to bridge the gap between the ribosome-studded ER membrane and the developing autophagosome [198].
The growing number of ER-phagy receptors raises interesting questions about functional specificity versus redundancy. One possibility is that these receptors are selective for the turnover of specific types of ER. For example, FAM134B facilitates the turnover of curved ER sheets [192], while RTN3 and ATL3 regulate the turnover of ER tubules [195, 197]. An alternative idea is that the receptors mediate the turnover of different ER-associated proteins [199]. Further, these receptors may be differentially induced by cellular perturbations such as ER stress or nutrient deprivation [197]. Finally, ER-phagy receptors may be differentially expressed within tissues or during development. Thus, some cell types may be uniquely susceptible to mutations in specific components, like the vulnerability of sensory neurons to mutations in either FAM134B or ATL3 [32, 200, 201]. Further work is clearly required on this emerging area of neuronal cell biology.
4.2. Aggrephagy in neurons
One consistent hallmark of major neurodegenerative diseases, for instance ALS, AD, and HD, is the accumulation of protein aggregates. These observations suggest that neurons are uniquely vulnerable to the accumulation of damaged or misfolded proteins, potentially because of their long-lived nature and highly-polarized morphologies. Most cytosolic proteins are cleared by the ubiquitin proteasome system [204], but larger protein aggregates are cleared by autophagy. Some of this clearance is nonselective, as aggregates of either mutant superoxide dismutase (SOD) 1 or mutant Htt are readily engulfed by axonal autophagosomes [13, 17]. However, there is strong evidence for the selective recognition and removal of protein aggregates or aggrephagy, which is suggested to be mediated by several receptors, like p62 and autophagy-linked FYVE protein (Alfy).
p62, also known as SQSTM1, is implicated in the selective removal of aggregated proteins. Missense mutations in p62 lead to a range of disorders, including ALS, FTD, and Paget’s disease of bone [31]. However, KO of p62 in the mouse does not result in an obvious phenotype of neurodegeneration [5]. Although, alterations in kinase activity were apparent in KO mouse brain, along with the accumulation of phosphorylated tau [205]. Behavioral assays indicate increased anxiety and depression in p62 KO mice [205].
In aggrephagy, p62 oligomerizes, mediates phase separation of ubiquitinated/misfolded proteins, and interacts with both LC3B and linear ubiquitin, leading to the bending of membrane around cargo and the effective sequestration of protein aggregates [23, 206]. Recently, p62 was shown to directly interact with FIP200, a scaffolding protein that is part of the autophagic machinery and acts upstream of mATG8 proteins; binding is enhanced by p62 phosphorylation [207]. In reconstituted systems, LC3B can outcompete FIP200 for binding to p62, suggesting that p62 acts upstream during initiation and at later stages during sequestration of ubiquitin condensates in aggrephagy [207]. p62 can also work in concert with other components of the aggrephagy pathway, such as Alfy [208]. In addition to its functional role in clearing protein aggregates, p62 accumulation is frequently used as a proxy for defective autophagy in cells or tissues.
Alfy, otherwise known as WDFY3, is a molecular scaffold that mediates interactions between cargo and the core members of the autophagy machinery, for example mATG8 proteins [209]. In stable cell lines, Alfy acts as an adaptor for aggrephagy [208, 210, 211], but its involvement in removing cellular debris in neurons is less clear. Alfy is highly expressed throughout the central nervous system of mice, and deletion of the protein is perinatal lethal; pups are born but exhibit abnormal brain morphology and striking neuronal development defects [209, 212]. At the cellular level, there are no widespread autophagy deficits in Alfy KO mice, but there is a deficit in the selective clearance of ubiquitinated proteins aggregates and polyglutamine inclusions in KO MEFs [209].
4.3. Selective autophagy: similar problems, distinct solutions?
There are clear parallels among the three selective autophagy pathways reviewed here. Mitophagy, ER-phagy, and aggrephagy all involve the recruitment of specific receptors that also bind members of the ATG8 family, such as LC3B to facilitate autophagosome formation around selected cargos. In the case of mitophagy and ER-phagy, multiple receptors have been identified that may be partially redundant. For example, mutations in the mitophagy receptor OPTN are sufficient to cause either ALS or glaucoma, but cellular studies suggest that NDP52 can fully compensate for the loss of OPTN when expressed at a high enough level [52]; however, this has not yet been demonstrated in neurons. Similarly, KO of the specific ER-phagy receptor FAM134B has a surprisingly limited effect in the mouse, but does result in the degeneration of peripheral sensory neurons, consistent with sensory neuropathy seen in human patients with FAM134B mutations [32]. Careful studies will be required to determine whether the multiple receptors mediating either mitophagy or ER-phagy, identified to date, are functionally redundant within neurons, where phenotypic differences may result from tissue-specific expression patterns. Alternatively, activation of these receptors may be governed by distinct regulatory control mechanisms, allowing the neuron to react more specifically to metabolic or environmental perturbations. Thus far, neuronal aggrephagy appears distinct from this paradigm, as p62 has a conserved function in the induction of protein aggregates within neurons that does not appear to be redundant with any other receptor.
Several studies have addressed the possible crossover of autophagy receptors between mitophagy and aggrephagy. For example, OPTN was reported to clear protein aggregates in a ubiquitin-independent manner [213]. Similarly, the aggrephagy receptor p62 was initially implicated in mitophagy, but multiple studies have since shown that while p62 can cluster damaged mitochondria, it does not facilitate their engulfment by the autophagosome [38, 52]. The aggrephagy receptor Alfy has also been implicated in mitochondrial quality control in neurons, although the mechanism involved requires further investigation [214]. In contrast, no crossover of ER-phagy receptors to other selective clearance pathways has been reported. The picture for now remains one in which receptors bind relatively specifically to cellular cargos targeted for degradation, although this may change in future as the clearance of other organelles, such as peroxisomes and lysosomes are explored in more detail. Importantly, there is also substantial literature implicating known mitophagy or aggrephagy receptors such as OPTN or p62 in other cellular pathways, particularly those involved in interferon signaling or inflammation. Exploring the links between autophagy and inflammatory signaling will be an exciting area for future research.
Downstream of receptor recruitment, the pathways for selective autophagy all converge. Binding to LC3B facilitates the formation of an autophagosome around the cargo to be degraded. Some receptors bind equally well or better to other members of the ATG8 family, such as GABARAP; the implications of this selectivity for autophagosome formation deserve further study. Once autophagosomes form, they must fuse with lysosomes to become degradative organelles and this aspect of the pathway does not seem to differ significantly. One interesting question is whether different cellular stressors effectively upregulate factors leading to more efficient cargo degradation, such as induction of lysosomal protein synthesis via transcription factor EB (TFEB). Again, more work is required to see how this may function in neurons, both in vitro and in vivo.
5. CONCLUSIONS
We have highlighted our current understanding of the selective removal of mitochondria, ER, and aggregated proteins in neurons. Although the signaling cascades are diverse, the pathways discussed here converge on the use of autophagic and endolysosomal machinery to remove cargo, emphasizing the importance of autophagy in the maintenance of neuronal health. Genetic findings continue to identify novel proteins and disease-associated mutations, providing more molecular pieces to the puzzle that is neurodegeneration. For example, our understanding of mitophagy has benefited from the consideration of both PINK1/Parkin-mediated mitochondrial turnover and other complementary pathways that contribute to regulating neuronal mitochondria. Additionally, more nuanced approaches are needed to understand the molecular cues and basis for defects. Moving towards in vivo studies, it is important to appreciate the molecular overlap that exists amongst pathways, as mitochondrial dysfunction is implicated in multiple neurodegenerative diseases. It will be important to understand the molecular signaling and how that contributes to each disease-state. Mitophagy, ER-phagy, and aggrephagy are the best characterized selective autophagy pathways in neurons, but other mechanisms like pexophagy, nucleophagy, and lysophagy have been studied in yeast and nonneuronal cell lines. Going forward, it will be of interest to see whether these mechanisms are conserved and how they contribute to neuronal homeostasis.
HIGHLIGHTS.
Selective autophagy is a degradative pathway that removes specific cargos through the autophagic machinery
In neurons, multiple quality control pathways work together to maintain mitochondrial health
Parallel pathways for selective autophagy regulate ER quality control and clear protein aggregates
Deciphering the signaling mechanisms of selective autophagy in vivo could provide novel therapeutic targets for neurodegenerative diseases
ACKNOWLEDGMENTS
The authors thank members of the Holzbaur lab for thoughtful insights and comments on the manuscript. This work was supported by the HHMI Hanna H. Gray Fellowship to C.S.E and the National Institutes of Health R37 NS060698 to E.L.F.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATIONS OF INTEREST
None
REFERENCES
- [1].Millecamps S, Julien JP. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci. 2013;14:161–76. [DOI] [PubMed] [Google Scholar]
- [2].Ohsumi Y Historical landmarks of autophagy research. Cell Res. 2014;24:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. [DOI] [PubMed] [Google Scholar]
- [4].Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. [DOI] [PubMed] [Google Scholar]
- [5].Komatsu M, Wang QJ, Holstein GR, Friedrich VL Jr., Iwata J, Kominami E, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104:14489–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Microb Cell. 2016;3:588–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Klionsky DJ, Codogno P, Cuervo AM, Deretic V, Elazar Z, Fueyo-Margareto J, et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. 2010;6:438–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Maday S, Holzbaur EL. Compartment-Specific Regulation of Autophagy in Primary Neurons. J Neurosci. 2016;36:5933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hollenbeck PJ. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J Cell Biol. 1993;121:305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Maday S, Holzbaur EL. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell. 2014;30:71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stavoe AK, Hill SE, Hall DH, Colon-Ramos DA. KIF1A/UNC-104 Transports ATG-9 to Regulate Neurodevelopment and Autophagy at Synapses. Dev Cell. 2016;38:171–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Neisch AL, Neufeld TP, Hays TS. A STRIPAK complex mediates axonal transport of autophagosomes and dense core vesicles through PP2A regulation. J Cell Biol. 2017;216:441–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jin EJ, Kiral FR, Ozel MN, Burchardt LS, Osterland M, Epstein D, et al. Live Observation of Two Parallel Membrane Degradation Pathways at Axon Terminals. Curr Biol. 2018;28:1027–38 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wong YC, Holzbaur EL. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci. 2014;34:1293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Anding AL, Baehrecke EH. Cleaning House: Selective Autophagy of Organelles. Dev Cell. 2017;41:10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–78. [DOI] [PubMed] [Google Scholar]
- [20].Khaminets A, Behl C, Dikic I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016;26:6–16. [DOI] [PubMed] [Google Scholar]
- [21].Villa E, Marchetti S, Ricci JE. No Parkin Zone: Mitophagy without Parkin. Trends Cell Biol. 2018;28:882–95. [DOI] [PubMed] [Google Scholar]
- [22].Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wurzer B, Zaffagnini G, Fracchiolla D, Turco E, Abert C, Romanov J, et al. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. Elife. 2015;4:e08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Moore AS, Holzbaur EL. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci U S A. 2016;113:E3349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A. 2016;113:4039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–6. [DOI] [PubMed] [Google Scholar]
- [27].Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Muller K, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–6. [DOI] [PubMed] [Google Scholar]
- [28].Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. [DOI] [PubMed] [Google Scholar]
- [29].Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. [DOI] [PubMed] [Google Scholar]
- [30].Valente EM, Brancati F, Caputo V, Graham EA, Davis MB, Ferraris A, et al. PARK6 is a common cause of familial parkinsonism. Neurol Sci. 2002;23 Suppl 2:S117–8. [DOI] [PubMed] [Google Scholar]
- [31].Sanchez-Martin P, Komatsu M. p62/SQSTM1 - steering the cell through health and disease. J Cell Sci. 2018;131. [DOI] [PubMed] [Google Scholar]
- [32].Kurth I, Pamminger T, Hennings JC, Soehendra D, Huebner AK, Rotthier A, et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet. 2009;41:1179–81. [DOI] [PubMed] [Google Scholar]
- [33].Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem. 2007;282:5617–24. [DOI] [PubMed] [Google Scholar]
- [34].Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Misgeld T, Schwarz TL. Mitostasis in Neurons: Maintaining Mitochondria in an Extended Cellular Architecture. Neuron. 2017;96:651–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ashrafi G, Schlehe JS, LaVoie MJ, Schwarz TL. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol. 2014;206:655–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:E4439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33:2142–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Whitworth AJ, Pallanck LJ. PINK1/Parkin mitophagy and neurodegeneration-what do we really know in vivo? Curr Opin Genet Dev. 2017;44:47–53. [DOI] [PubMed] [Google Scholar]
- [41].Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Takatori S, Ito G, Iwatsubo T. Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci Lett. 2008;430:13–7. [DOI] [PubMed] [Google Scholar]
- [43].Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–6. [DOI] [PubMed] [Google Scholar]
- [49].Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56:360–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].McLelland GL, Goiran T, Yi W, Dorval G, Chen CX, Lauinger ND, et al. Mfn2 ubiquitination by PINK1/parkin gates the p97-dependent release of ER from mitochondria to drive mitophagy. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sun Y, Vashisht AA, Tchieu J, Wohlschlegel JA, Dreier L. Voltage-dependent anion channels (VDACs) recruit Parkin to defective mitochondria to promote mitochondrial autophagy. J Biol Chem. 2012;287:40652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. [DOI] [PubMed] [Google Scholar]
- [54].Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Matsumoto G, Shimogori T, Hattori N, Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24:4429–42. [DOI] [PubMed] [Google Scholar]
- [56].Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell. 2015;60:7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, et al. Spatiotemporal Control of ULK1 Activation by NDP52 and TBK1 during Selective Autophagy. Mol Cell. 2019;74:347–62 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Princely Abudu Y, Pankiv S, Mathai BJ, Hakon Lystad A, Bindesboll C, Brenne HB, et al. NIPSNAP1 and NIPSNAP2 Act as “Eat Me” Signals for Mitophagy. Dev Cell. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nautiyal M, Sweatt AJ, MacKenzie JA, Mark Payne R, Szucs S, Matalon R, et al. Neuronal localization of the mitochondrial protein NIPSNAP1 in rat nervous system. Eur J Neurosci. 2010;32:560–9. [DOI] [PubMed] [Google Scholar]
- [61].Cai Q, Zakaria HM, Simone A, Sheng ZH. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol. 2012;22:545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Joselin AP, Hewitt SJ, Callaghan SM, Kim RH, Chung YH, Mak TW, et al. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum Mol Genet. 2012;21:4888–903. [DOI] [PubMed] [Google Scholar]
- [63].Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–35. [DOI] [PubMed] [Google Scholar]
- [64].Itier JM, Ibanez P, Mena MA, Abbas N, Cohen-Salmon C, Bohme GA, et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet. 2003;12:2277–91. [DOI] [PubMed] [Google Scholar]
- [65].Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci U S A. 2005;102:2174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Von Coelln R, Thomas B, Savitt JM, Lim KL, Sasaki M, Hess EJ, et al. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci U S A. 2004;101:10744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Akundi RS, Huang Z, Eason J, Pandya JD, Zhi L, Cass WA, et al. Increased mitochondrial calcium sensitivity and abnormal expression of innate immunity genes precede dopaminergic defects in Pink1-deficient mice. PLoS One. 2011;6:e16038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kitada T, Pisani A, Porter DR, Yamaguchi H, Tscherter A, Martella G, et al. Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proc Natl Acad Sci U S A. 2007;104:11441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–9. [DOI] [PubMed] [Google Scholar]
- [70].Kitada T, Tong Y, Gautier CA, Shen J. Absence of nigral degeneration in aged parkin/DJ-1/PINK1 triple knockout mice. J Neurochem. 2009;111:696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Dave KD, De Silva S, Sheth NP, Ramboz S, Beck MJ, Quang C, et al. Phenotypic characterization of recessive gene knockout rat models of Parkinson’s disease. Neurobiol Dis. 2014;70:190–203. [DOI] [PubMed] [Google Scholar]
- [72].Whitworth AJ, Theodore DA, Greene JC, Benes H, Wes PD, Pallanck LJ. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc Natl Acad Sci U S A. 2005;102:8024–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Cha GH, Kim S, Park J, Lee E, Kim M, Lee SB, et al. Parkin negatively regulates JNK pathway in the dopaminergic neurons of Drosophila. Proc Natl Acad Sci U S A. 2005;102:10345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Pesah Y, Pham T, Burgess H, Middlebrooks B, Verstreken P, Zhou Y, et al. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development. 2004;131:2183–94. [DOI] [PubMed] [Google Scholar]
- [76].Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. [DOI] [PubMed] [Google Scholar]
- [77].Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. [DOI] [PubMed] [Google Scholar]
- [79].Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, et al. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–22. [DOI] [PubMed] [Google Scholar]
- [80].Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Heeman B, Van den Haute C, Aelvoet SA, Valsecchi F, Rodenburg RJ, Reumers V, et al. Depletion of PINK1 affects mitochondrial metabolism, calcium homeostasis and energy maintenance. J Cell Sci. 2011;124:1115–25. [DOI] [PubMed] [Google Scholar]
- [82].Devireddy S, Liu A, Lampe T, Hollenbeck PJ. The Organization of Mitochondrial Quality Control and Life Cycle in the Nervous System In Vivo in the Absence of PINK1. J Neurosci. 2015;35:9391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Sung H, Tandarich LC, Nguyen K, Hollenbeck PJ. Compartmentalized Regulation of Parkin-Mediated Mitochondrial Quality Control in the Drosophila Nervous System In Vivo. J Neurosci. 2016;36:7375–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Burman JL, Yu S, Poole AC, Decal RB, Pallanck L. Analysis of neural subtypes reveals selective mitochondrial dysfunction in dopaminergic neurons from parkin mutants. Proc Natl Acad Sci U S A. 2012;109:10438–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Greene JC, Whitworth AJ, Andrews LA, Parker TJ, Pallanck LJ. Genetic and genomic studies of Drosophila parkin mutants implicate oxidative stress and innate immune responses in pathogenesis. Hum Mol Genet. 2005;14:799–811. [DOI] [PubMed] [Google Scholar]
- [86].Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, et al. “Fluorescent timer”: protein that changes color with time. Science. 2000;290:1585–8. [DOI] [PubMed] [Google Scholar]
- [87].Stotland A, Gottlieb RA. alpha-MHC MitoTimer mouse: In vivo mitochondrial turnover model reveals remarkable mitochondrial heterogeneity in the heart. J Mol Cell Cardiol. 2016;90:53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Williams JA, Zhao K, Jin S, Ding WX. New methods for monitoring mitochondrial biogenesis and mitophagy in vitro and in vivo. Exp Biol Med (Maywood). 2017;242:781–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sun N, Yun J, Liu J, Malide D, Liu C, Rosvira II, et al. Measuring In Vivo Mitophagy. Mol Cell. 2015;60:685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Sun N, Malide D, Liu J, Rovira II, Combs CA, Finkel T. A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat Protoc. 2017;12:1576–87. [DOI] [PubMed] [Google Scholar]
- [91].Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18:1042–52. [DOI] [PubMed] [Google Scholar]
- [92].Cornelissen T, Vilain S, Vints K, Gounko N, Verstreken P, Vandenberghe W. Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Bernhardt D, Muller M, Reichert AS, Osiewacz HD. Simultaneous impairment of mitochondrial fission and fusion reduces mitophagy and shortens replicative lifespan. Sci Rep. 2015;5:7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hebert SL, Marquet-de Rouge P, Lanza IR, McCrady-Spitzer SK, Levine JA, Middha S, et al. Mitochondrial Aging and Physical Decline: Insights From Three Generations of Women. J Gerontol A Biol Sci Med Sci. 2015;70:1409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–7. [DOI] [PubMed] [Google Scholar]
- [96].Simonetti S, Chen X, DiMauro S, Schon EA. Accumulation of deletions in human mitochondrial DNA during normal aging: analysis by quantitative PCR. Biochim Biophys Acta. 1992;1180:113–22. [DOI] [PubMed] [Google Scholar]
- [97].Sun N, Youle RJ, Finkel T. The Mitochondrial Basis of Aging. Mol Cell. 2016;61:654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci U S A. 2013;110:8638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].McWilliams TG, Prescott AR, Allen GF, Tamjar J, Munson MJ, Thomson C, et al. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. 2016;214:333–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].McWilliams TG, Ganley IG. Investigating Mitophagy and Mitochondrial Morphology In Vivo Using mito-QC: A Comprehensive Guide. Methods Mol Biol. 2019;1880:621–42. [DOI] [PubMed] [Google Scholar]
- [101].McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, Barini E, et al. Basal Mitophagy Occurs Independently of PINK1 in Mouse Tissues of High Metabolic Demand. Cell Metab. 2018;27:439–49 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Lee JJ, Sanchez-Martinez A, Zarate AM, Beninca C, Mayor U, Clague MJ, et al. Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. J Cell Biol. 2018;217:1613–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Liu L, Sakakibara K, Chen Q, Okamoto K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014;24:787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Rogov VV, Suzuki H, Marinkovic M, Lang V, Kato R, Kawasaki M, et al. Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Sci Rep. 2017;7:1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Esteban-Martinez L, Sierra-Filardi E, McGreal RS, Salazar-Roa M, Marino G, Seco E, et al. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J. 2017;36:1688–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Koentjoro B, Park JS, Sue CM. Nix restores mitophagy and mitochondrial function to protect against PINK1/Parkin-related Parkinson’s disease. Sci Rep. 2017;7:44373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Real PJ, Benito A, Cuevas J, Berciano MT, de Juan A, Coffer P, et al. Blockade of epidermal growth factor receptors chemosensitizes breast cancer cells through up-regulation of Bnip3L. Cancer Res. 2005;65:8151–7. [DOI] [PubMed] [Google Scholar]
- [111].Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–73. [PubMed] [Google Scholar]
- [112].Yuan Y, Zheng Y, Zhang X, Chen Y, Wu X, Wu J, et al. BNIP3L/NIX-mediated mitophagy protects against ischemic brain injury independent of PARK2. Autophagy. 2017;13:1754–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Shi RY, Zhu SH, Li V, Gibson SB, Xu XS, Kong JM. BNIP3 interacting with LC3 triggers excessive mitophagy in delayed neuronal death in stroke. CNS Neurosci Ther. 2014;20:1045–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Yu D, Li M, Nie P, Ni B, Zhang Z, Zhou Y. Bcl-2/E1B-19KD-Interacting Protein 3/Light Chain 3 Interaction Induces Mitophagy in Spinal Cord Injury in Rats Both In Vivo and In Vitro. J Neurotrauma. 2018;35:2183–94. [DOI] [PubMed] [Google Scholar]
- [115].Gebert N, Joshi AS, Kutik S, Becker T, McKenzie M, Guan XL, et al. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr Biol. 2009;19:2133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Schlame M Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res. 2008;49:1607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Pointer CB, Klegeris A. Cardiolipin in Central Nervous System Physiology and Pathology. Cell Mol Neurobiol. 2017;37:1161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Sathappa M, Alder NN. The ionization properties of cardiolipin and its variants in model bilayers. Biochim Biophys Acta. 2016;1858:1362–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Mancuso DJ, Kotzbauer P, Wozniak DF, Sims HF, Jenkins CM, Guan S, et al. Genetic ablation of calcium-independent phospholipase A2{gamma} leads to alterations in hippocampal cardiolipin content and molecular species distribution, mitochondrial degeneration, autophagy, and cognitive dysfunction. J Biol Chem. 2009;284:35632–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Petrosillo G, Matera M, Casanova G, Ruggiero FM, Paradies G. Mitochondrial dysfunction in rat brain with aging Involvement of complex I, reactive oxygen species and cardiolipin. Neurochem Int. 2008;53:126–31. [DOI] [PubMed] [Google Scholar]
- [121].Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12:1154–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Kagan VE, Jiang J, Huang Z, Tyurina YY, Desbourdes C, Cottet-Rousselle C, et al. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 2016;23:1140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Schlattner U, Tokarska-Schlattner M, Ramirez S, Tyurina YY, Amoscato AA, Mohammadyani D, et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. J Biol Chem. 2013;288:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Hsu P, Liu X, Zhang J, Wang HG, Ye JM, Shi Y. Cardiolipin remodeling by TAZ/tafazzin is selectively required for the initiation of mitophagy. Autophagy. 2015;11:643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 2008;3:e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, Rachubinski RA, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–8. [DOI] [PubMed] [Google Scholar]
- [128].Zhang B, Huang J, Li HL, Liu T, Wang YY, Waterman P, et al. GIDE is a mitochondrial E3 ubiquitin ligase that induces apoptosis and slows growth. Cell Res. 2008;18:900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Yun J, Puri R, Yang H, Lizzio MA, Wu C, Sheng ZH, et al. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife. 2014;3:e01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Ambivero CT, Cilenti L, Main S, Zervos AS. Mulan E3 ubiquitin ligase interacts with multiple E2 conjugating enzymes and participates in mitophagy by recruiting GABARAP. Cell Signal. 2014;26:2921–9. [DOI] [PubMed] [Google Scholar]
- [132].Evans CS, Holzbaur ELF. Autophagy and mitophagy in ALS. Neurobiol Dis. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Cheng XT, Zhou B, Lin MY, Cai Q, Sheng ZH. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J Cell Biol. 2015;209:377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Lee S, Sato Y, Nixon RA. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J Neurosci. 2011;31:7817–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Soukup SF, Kuenen S, Vanhauwaert R, Manetsberger J, Hernandez-Diaz S, Swerts J, et al. A LRRK2-Dependent EndophilinA Phosphoswitch Is Critical for Macroautophagy at Presynaptic Terminals. Neuron. 2016;92:829–44. [DOI] [PubMed] [Google Scholar]
- [136].Soubannier V, McLelland GL, Zunino R, Braschi E, Rippstein P, Fon EA, et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22:135–41. [DOI] [PubMed] [Google Scholar]
- [137].McLelland GL, Lee SA, McBride HM, Fon EA. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J Cell Biol. 2016;214:275–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Kang JS, Tian JH, Pan PY, Zald P, Li C, Deng C, et al. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 2008;132:137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Lin MY, Cheng XT, Tammineni P, Xie Y, Zhou B, Cai Q, et al. Releasing Syntaphilin Removes Stressed Mitochondria from Axons Independent of Mitophagy under Pathophysiological Conditions. Neuron. 2017;94:595–610 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Zheng YR, Zhang XN, Chen Z. Mitochondrial transport serves as a mitochondrial quality control strategy in axons: Implications for central nervous system disorders. CNS Neurosci Ther. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Sheng ZH. Mitochondrial trafficking and anchoring in neurons: New insight and implications. J Cell Biol. 2014;204:1087–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Lewis TL Jr., Turi GF, Kwon SK, Losonczy A, Polleux F. Progressive Decrease of Mitochondrial Motility during Maturation of Cortical Axons In Vitro and In Vivo. Curr Biol. 2016;26:2602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Fransson A, Ruusala A, Aspenstrom P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J Biol Chem. 2003;278:6495–502. [DOI] [PubMed] [Google Scholar]
- [145].Fransson S, Ruusala A, Aspenstrom P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem Biophys Res Commun. 2006;344:500–10. [DOI] [PubMed] [Google Scholar]
- [146].van Spronsen M, Mikhaylova M, Lipka J, Schlager MA, van den Heuvel DJ, Kuijpers M, et al. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron. 2013;77:485–502. [DOI] [PubMed] [Google Scholar]
- [147].Wang X, Schwarz TL. The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136:163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann Neurol. 2002;51:296–301. [DOI] [PubMed] [Google Scholar]
- [150].Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. [DOI] [PubMed] [Google Scholar]
- [151].Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–7. [DOI] [PubMed] [Google Scholar]
- [152].Hsieh CH, Shaltouki A, Gonzalez AE, Bettencourt da Cruz A, Burbulla LF, St Lawrence E, et al. Functional Impairment in Miro Degradation and Mitophagy Is a Shared Feature in Familial and Sporadic Parkinson’s Disease. Cell Stem Cell. 2016;19:709–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, et al. Parkinson’s disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 2012;8:e1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson’s disease. Physiol Rev. 2011;91:1161–218. [DOI] [PubMed] [Google Scholar]
- [155].Bonello F, Hassoun SM, Mouton-Liger F, Shin YS, Muscat A, Tesson C, et al. LRRK2 impairs PINK1/Parkin-dependent mitophagy via its kinase activity: pathologic insights into Parkinson’s disease. Hum Mol Genet. 2019. [DOI] [PubMed] [Google Scholar]
- [156].Mortiboys H, Johansen KK, Aasly JO, Bandmann O. Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology. 2010;75:2017–20. [DOI] [PubMed] [Google Scholar]
- [157].Wauters F, Cornelissen T, Imberechts D, Martin S, Koentjoro B, Sue C, et al. LRRK2 mutations impair depolarization-induced mitophagy through inhibition of mitochondrial accumulation of RAB10. Autophagy. 2019:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Wang X, Yan MH, Fujioka H, Liu J, Wilson-Delfosse A, Chen SG, et al. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum Mol Genet. 2012;21:1931–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Papkovskaia TD, Chau KY, Inesta-Vaquera F, Papkovsky DB, Healy DG, Nishio K, et al. G2019S leucine-rich repeat kinase 2 causes uncoupling protein-mediated mitochondrial depolarization. Hum Mol Genet. 2012;21:4201–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Palikaras K, Tavernarakis N. Mitophagy in neurodegeneration and aging. Front Genet. 2012;3:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–20. [DOI] [PubMed] [Google Scholar]
- [163].Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–6. [DOI] [PubMed] [Google Scholar]
- [164].Rezaie T, Child A, Hitchings R, Brice G, Miller L, Coca-Prados M, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–9. [DOI] [PubMed] [Google Scholar]
- [165].Fingert JH, Robin AL, Stone JL, Roos BR, Davis LK, Scheetz TE, et al. Copy number variations on chromosome 12q14 in patients with normal tension glaucoma. Hum Mol Genet. 2011;20:2482–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Citron M Alzheimer’s disease: strategies for disease modification. Nat Rev Drug Discov. 2010;9:387–98. [DOI] [PubMed] [Google Scholar]
- [167].Kerr JS, Adriaanse BA, Greig NH, Mattson MP, Cader MZ, Bohr VA, et al. Mitophagy and Alzheimer’s Disease: Cellular and Molecular Mechanisms. Trends Neurosci. 2017;40:151–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Martinez-Vicente M Neuronal Mitophagy in Neurodegenerative Diseases. Front Mol Neurosci. 2017;10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Sorrentino V, Romani M, Mouchiroud L, Beck JS, Zhang H, D’Amico D, et al. Enhancing mitochondrial proteostasis reduces amyloid-beta proteotoxicity. Nature. 2017;552:187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Baloyannis SJ. Mitochondrial alterations in Alzheimer’s disease. J Alzheimers Dis. 2006;9:119–26. [DOI] [PubMed] [Google Scholar]
- [171].Fang EF, Hou Y, Palikaras K, Adriaanse BA, Kerr JS, Yang B, et al. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat Neurosci. 2019;22:401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Casley CS, Land JM, Sharpe MA, Clark JB, Duchen MR, Canevari L. Beta-amyloid fragment 25–35 causes mitochondrial dysfunction in primary cortical neurons. Neurobiol Dis. 2002;10:258–67. [DOI] [PubMed] [Google Scholar]
- [173].Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science. 2004;304:448–52. [DOI] [PubMed] [Google Scholar]
- [174].Amadoro G, Corsetti V, Florenzano F, Atlante A, Ciotti MT, Mongiardi MP, et al. AD-linked, toxic NH2 human tau affects the quality control of mitochondria in neurons. Neurobiol Dis. 2014;62:489–507. [DOI] [PubMed] [Google Scholar]
- [175].Corsetti V, Florenzano F, Atlante A, Bobba A, Ciotti MT, Natale F, et al. NH2-truncated human tau induces deregulated mitophagy in neurons by aberrant recruitment of Parkin and UCHL-1: implications in Alzheimer’s disease. Hum Mol Genet. 2015;24:3058–81. [DOI] [PubMed] [Google Scholar]
- [176].Costa V, Scorrano L. Shaping the role of mitochondria in the pathogenesis of Huntington’s disease. EMBO J. 2012;31:1853–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Purdon SE, Mohr E, Ilivitsky V, Jones BD. Huntington’s disease: pathogenesis, diagnosis and treatment. J Psychiatry Neurosci. 1994;19:359–67. [PMC free article] [PubMed] [Google Scholar]
- [178].Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–58. [DOI] [PubMed] [Google Scholar]
- [179].DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–3. [DOI] [PubMed] [Google Scholar]
- [180].Costa V, Giacomello M, Hudec R, Lopreiato R, Ermak G, Lim D, et al. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol Med. 2010;2:490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Mitchell IJ, Cooper AJ, Griffiths MR. The selective vulnerability of striatopallidal neurons. Prog Neurobiol. 1999;59:691–719. [DOI] [PubMed] [Google Scholar]
- [182].Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–6. [DOI] [PubMed] [Google Scholar]
- [183].Pickrell AM, Fukui H, Wang X, Pinto M, Moraes CT. The striatum is highly susceptible to mitochondrial oxidative phosphorylation dysfunctions. J Neurosci. 2011;31:9895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Trushina E, Dyer RB, Badger JD 2nd, Ure D, Eide L, Tran DD, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Gkikas I, Palikaras K, Tavernarakis N. The Role of Mitophagy in Innate Immunity. Front Immunol. 2018;9:1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Main BS, Zhang M, Brody KM, Ayton S, Frugier T, Steer D, et al. Type-1 interferons contribute to the neuroinflammatory response and disease progression of the MPTP mouse model of Parkinson’s disease. Glia. 2016;64:1590–604. [DOI] [PubMed] [Google Scholar]
- [188].Main BS, Zhang M, Brody KM, Kirby FJ, Crack PJ, Taylor JM. Type-I interferons mediate the neuroinflammatory response and neurotoxicity induced by rotenone. J Neurochem. 2017;141:75–85. [DOI] [PubMed] [Google Scholar]
- [189].Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561:258–62. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [190].Grumati P, Dikic I, Stolz A. ER-phagy at a glance. J Cell Sci. 2018;131. [DOI] [PubMed] [Google Scholar]
- [191].Wu Y, Whiteus C, Xu CS, Hayworth KJ, Weinberg RJ, Hess HF, et al. Contacts between the endoplasmic reticulum and other membranes in neurons. Proc Natl Acad Sci U S A. 2017;114:E4859–E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–8. [DOI] [PubMed] [Google Scholar]
- [193].Bhaskara RM, Grumati P, Garcia-Pardo J, Kalayil S, Covarrubias-Pinto A, Chen W, et al. Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat Commun. 2019;10:2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, et al. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell. 2018;44:217–32 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Grumati P, Morozzi G, Holper S, Mari M, Harwardt MI, Yan R, et al. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016;18:1173–84. [DOI] [PubMed] [Google Scholar]
- [197].Chen Q, Xiao Y, Chai P, Zheng P, Teng J, Chen J. ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Curr Biol. 2019;29:846–55 e6. [DOI] [PubMed] [Google Scholar]
- [198].Chino H, Hatta T, Natsume T, Mizushima N. Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol Cell. 2019. [DOI] [PubMed] [Google Scholar]
- [199].An H, Ordureau A, Paulo JA, Shoemaker CJ, Denic V, Harper JW. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol Cell. 2019;74:891–908 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Fischer D, Schabhuttl M, Wieland T, Windhager R, Strom TM, Auer-Grumbach M. A novel missense mutation confirms ATL3 as a gene for hereditary sensory neuropathy type 1. Brain. 2014;137:e286. [DOI] [PubMed] [Google Scholar]
- [201].Kornak U, Mademan I, Schinke M, Voigt M, Krawitz P, Hecht J, et al. Sensory neuropathy with bone destruction due to a mutation in the membrane-shaping atlastin GTPase 3. Brain. 2014;137:683–92. [DOI] [PubMed] [Google Scholar]
- [202].Krols M, Asselbergh B, De Rycke R, De Winter V, Seyer A, Muller FJ, et al. Sensory neuropathy-causing mutations in ATL3 affect ER-mitochondria contact sites and impair axonal mitochondrial distribution. Hum Mol Genet. 2019;28:615–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Behrendt L, Kurth I, Kaether C. A disease causing ATLASTIN 3 mutation affects multiple endoplasmic reticulum-related pathways. Cell Mol Life Sci. 2019;76:1433–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Kleiger G, Mayor T. Perilous journey: a tour of the ubiquitin-proteasome system. Trends Cell Biol. 2014;24:352–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom AL, Kemppainen R, Cox N, et al. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106:107–20. [DOI] [PubMed] [Google Scholar]
- [206].Sun D, Wu R, Zheng J, Li P, Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28:405–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Turco E, Witt M, Abert C, Bock-Bierbaum T, Su MY, Trapannone R, et al. FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol Cell. 2019;74:330–46 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Lystad AH, Ichimura Y, Takagi K, Yang Y, Pankiv S, Kanegae Y, et al. Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep. 2014;15:557–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Dragich JM, Kuwajima T, Hirose-Ikeda M, Yoon MS, Eenjes E, Bosco JR, et al. Autophagy linked FYVE (Alfy/WDFY3) is required for establishing neuronal connectivity in the mammalian brain. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–44. [DOI] [PubMed] [Google Scholar]
- [211].Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Orosco LA, Ross AP, Cates SL, Scott SE, Wu D, Sohn J, et al. Loss of Wdfy3 in mice alters cerebral cortical neurogenesis reflecting aspects of the autism pathology. Nat Commun. 2014;5:4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, et al. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci. 2013;126:580–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Napoli E, Song G, Panoutsopoulos A, Riyadh MA, Kaushik G, Halmai J, et al. Beyond autophagy: a novel role for autism-linked Wdfy3 in brain mitophagy. Sci Rep. 2018;8:11348. [DOI] [PMC free article] [PubMed] [Google Scholar]