ABSTRACT
Background
Transcription factor 7–like 2 (TCF7L2) genetic variants that predispose individuals to type 2 diabetes (T2D) show inconsistent associations with anthropometric traits. Interaction between TCF7L2 genotypes and dietary factors may help explain these observations.
Objective
We aimed to examine the potential modulation of TCF7L2-rs7903146 and rs12255372 on anthropometric markers by a Mediterranean diet (MedDiet).
Methods
Cross-sectional analysis was conducted in 1120 participants (aged 45–75 y) of the Boston Puerto Rican Health Study. Anthropometric variables were measured, and polymorphisms were genotyped using standardized protocols. Diet was assessed using a validated FFQ. The MedDiet was defined based on adherence to 9 food and nutrient components using sex-specific population-based median cut-offs; high adherence was defined as meeting ≥4 components. Haplotypes were tested for association with obesity traits, independently and via interaction with the MedDiet.
Results
TCF7L2-rs7903146 showed significant interaction with the MedDiet influencing BMI, weight, and waist circumference. The T risk-allele carriers (CT + TT) with a high MedDiet score had lower weight (77.3 ± 1.0 compared with CC 80.9 ± 1.0 kg; P = 0.013) and waist circumference (99.2 ± 0.9 compared with CC 102.2 ± 0.9 cm; P = 0.021), when compared with CC participants. A low MedDiet score resulted in no significant differences between genotypes. For TCF7L2-rs12255372, we found significant interactions with the MedDiet for weight (P-interaction = 0.034) and BMI (P-interaction = 0.036). The T allele carriers with a higher MedDiet score showed a trend of lower but no significant differences when compared with CC participants for BMI (P = 0.19), weight (P = 0.09), and waist circumference (P = 0.11). We found significant interactions between the 2 risk-carrying haplotypes and the MedDiet compared with the common haplotype (GC), with lower BMI (β ± SE, TT: −1.53 ± 0.68; P-interaction = 0.024), weight (TT: −4.16 ± 1.77; P-interaction = 0.019), and waist circumference (GT: −5.07 ± 2.50; P-interaction = 0.042) at a high MedDiet score.
Conclusion
Puerto Ricans with the TCF7L2-rs7903146 and rs12255372 T2D risk genotypes, although still high, had better anthropometric profiles when adhering to a MedDiet, suggesting that this diet may offset unfavorable genetic predisposition.
Keywords: TCF7L2 polymorphism, Mediterranean diet, anthropometric markers, gene–diet interaction, Puerto Rican, Hispanics/Latinos
Introduction
Accumulated evidence from genome-wide association studies (GWASs) has identified gene variants associated with type 2 diabetes (T2D) (1, 2). However, few GWASs or single nucleotide polymorphism (SNP) replication studies of those identified gene variants have been conducted among US Hispanics/Latinos (3) who show a disproportionally high prevalence of T2D (4). Recently, Qi et al. conducted a GWAS in a cross-sectional study of Hispanic/Latino adults residing in the United States and found that a common genetic variant (rs7903146) within the transcription factor 7–like 2 (TCF7L2), locus, was a top SNP, explaining 0.28% of the variance in T2D (5). Mechanistically, TCF7L2 plays a role in the transcription of the proglucagon gene and consequently glucagon-like peptide 1 (GLP-1) synthesis (6). Some studies have found that GLP-1 concentrations are higher after the ingestion of fat but not carbohydrate (7–9), suggesting that functional variants of TCF7L2 may modulate the response to weight loss depending on diet composition. Previous studies have consistently shown strong associations between TCF7L2-rs7903146 and TCF7L2-rs12255372 and T2D as well as other related traits (10–12), especially among nonobese individuals (5, 13). The literature has been contradictory with regards to other anthropometric and cardiometabolic traits. Some studies, but not all (14, 15), have found associations with weight (14, 16), body composition (14), metabolic syndrome (17, 18), and cardiometabolic risk profiles (19).
The study of gene–diet interactions aims to explain how diet may contribute to the modulation of gene variant effects. Epidemiological studies have shown that carbohydrate quality (20, 21), dietary fat (22), and animal protein (15) interact with TCF7L2 SNPs and diabetes risk and glucose measures. Data from randomized controlled trials, such as the Diabetes Prevention Program and the Diabetes Prevention Study, showed that lifestyle or environmental factors could modulate the genetic effects of TCF7L2 polymorphisms (10, 23). Although numerous studies have looked at TCF7L2 polymorphism–nutrient interactions and T2D (10, 20–23), few studies have focused on anthropometric outcomes (14, 16, 24) and even fewer have focused on diverse populations such as Hispanics/Latinos. Results from the POUNDS LOST trial (16) showed that individuals with the TCF7L2-rs12255372 risk genotype had lower BMI, total fat mass, and trunk fat mass (P-interaction < 0.05) when consuming a diet lower in total fat compared with those without the risk genotype, suggesting that those dietary changes may help achieve better adiposity parameters in individuals with the risk genotype.
Over recent years, nutrition research aiming to identify dietary effects has shifted from studying isolated nutrients or foods to a more holistic approach that investigates overall dietary quality (25). The Mediterranean diet (MedDiet) has emerged as a scientifically sound healthy dietary pattern (26). Despite this, few studies have assessed interactions between the MedDiet and TCF7L2 on anthropometric measures. One study showed a borderline significant interaction between the MedDiet and TCF7L2-rs7903146 on weight gain, where participants with 1 or 2 risk alleles experienced less weight gain when adherence to the MedDiet was high (27). However, independent replication of studies in different populations is important to support stronger genetic-based personalized recommendations. Therefore, our objective was to examine the potential modulation of TCF7L2-rs7903146 (C > T) and rs12255372 (G > T) on anthropometric markers using the MedDiet score among Puerto Ricans living in Boston, a population with a high prevalence of obesity and T2D (28, 29).
Methods
Study population
The design and procedures of the Boston Puerto Rican Health Study (BPRHS), an ongoing longitudinal cohort study, have been described in detail previously (29). Briefly, eligible participants included subjects of self-reported Puerto Rican descent, aged 45–75 y, living in the Boston, MA, area, and able to answer interview questions in English or Spanish. Participants were recruited during 2004–2009 using door-to-door enumeration supplemented with community recruitment approaches. One participant per qualified household was randomly invited to participate. In addition, participants were identified by random approach during community festivals/fairs, referrals, or calls in response to flyers, radio, or television. The baseline data include 1500 participants, of which 1194 had complete data for the MedDiet, and 1385 had available genotype data. In total, the present analysis includes 1120 participants with available data for both SNPs and the MedDiet. The study protocol was approved by the Institutional Review Boards at Tufts Medical Center, Tufts University, and Northeastern University. Written informed consent was obtained from all participants.
Dietary assessment and Mediterranean score
Dietary intake was assessed using a semiquantitative FFQ specifically developed and validated for this population that asked about usual dietary intake consumed over the past 12 mo (30). Nutrition Data System for Research (NDS_R) software (Nutrition Coordinating Center) was used to calculate nutrient intake. Reported serving equivalents of individual foods were used to create food groups. For mixed dishes, we disaggregated the intake into individual food items that were then added to the appropriate food group. Participants with energy intakes ≤600 or ≥4800 kcal/d (≤2510 or ≥20,083 kJ/d) or with ≥10 questions left blank in the FFQ were excluded.
The MedDiet was previously defined in this population (31) and was slightly modified from the original score described by Trichopoulou et al. (32). Specifically, we replaced the total grains group with whole grains because the high intake of refined grains in this population (33) would have confounded the results. The 9 components were scored using the sex-specific population median adjusted for total energy using the residual method. A score of 0 was assigned to a participant consuming below the median for healthful components (or above the median for unfavorable components), and 1 point was assigned for the opposite. The added points for all components equaled a range of 0–9; higher values indicate greater observance of a Mediterranean pattern. The MedDiet was categorized as low (meeting <4 components) and high adherence (meeting ≥4 components), based on the median of the population sample.
Genotyping TCF7L2 gene variants
DNA was isolated from buffy coats of peripheral blood using QIAamp DNA Blood Mini kits (Qiagen) according to the vendor's recommended protocol. The SNPs rs7903146 and rs12255372 were genotyped using the Applied Biosystems TaqMan SNP genotyping system (34). For all genotyping, blinded no-template controls and replicates of DNA samples were incorporated in each DNA sample plate, which were routinely checked by laboratory personnel. Based on internal quality control as well as that estimated independently by external laboratories, the genotyping error rate was <1%. For the rs7903146 SNP, recessive (24), dominant (14, 20, 27), and codominant (13, 16) models have been described in the literature. In the present study, a dominant model was used as it is more commonly reported. The effects of the rs12255372 SNP are presented as a codominant model based on the literature (16). Since both SNPs have been described to be in moderately strong linkage disequilibrium (LD) (r2 = 0.77, HapMap data), instead of calculating genetic score based on 2 SNPs, we conducted haplotype analysis based on these 2 SNPs using the haplo.stats R package.
Anthropometric outcomes
Standing height, weight, and waist and hip circumference were measured in duplicate using standard protocols (35, 36); the average value was used. BMI was calculated as weight (kg) divided by height (m) squared (kg/m2).
Covariates
Age, sex, educational attainment, marital status, household income, medical history, and use of medication were self-reported by the participants. Acculturation was measured with a 10-item psychological acculturation scale that assesses the degree of psychological attachment to either mainland USA or Puerto Rican culture (37). Information about social support and network was collected using the Norbeck Social Support Questionnaire (38). Social activities were assessed using the Social and Community Support and Assistance Questionnaire (38). A validated questionnaire was used to assess perceptions of one's life as stressful (Perceived Stress Scale) (39, 40).
Smoking information was collected with a comprehensive questionnaire asking about type and frequency of tobacco product use. Smoking status was defined as never, former, or current. Physical activity was assessed by a modified Paffenbarger questionnaire (41) from the Harvard Alumni Activity Survey; a score was defined by multiplying the self-reported hours spent in heavy, moderate, light, or sedentary activities in 24 h by weighing factors that parallel the rate of oxygen consumption of each activity. T2D was defined as fasting glucose ≥126 mg/dL or medication use.
Statistical analysis
Statistical tests were conducted using SAS version 9.4 (SAS Institute) and haplo.stats (version 1.7.9) R package. Differences in demographic, socioeconomic, and lifestyle characteristics of the participants by TCF7L2 SNPs genotype were calculated using ANOVA, t-test, and chi-square test. General linear regression models were run to determine associations between continuous outcomes across genotypes adjusted for age, sex, smoking, physical activity, education, income, acculturation, social support, and perceived stress. Population admixture was estimated using principle components analysis based on 100 informative ancestry markers (42). All analyses were adjusted for the estimated population admixture using the first major principal component with linear regression models (42). Interactions between SNPs or haplotypes and the MedDiet were conducted by assuming dominant and codominant genetic models. Means and P values between each category of the genotype by strata of MedDiet were adjusted for multiple comparisons using Tukey's tests. Statistical power of our analysis was calculated using Quanto software. For SNPs with a minor allele frequency of 0.3 and a sample size of n = 1120, we had 92% power to detect gene*environment interactions that contribute R2 = 1% to BMI (mean = 32, SD = 6.7), or waist circumference (mean = 102, SD = 15), or weight (mean = 80, SD = 17). Statistical significance was set at P < 0.05.
Results
The minor allele frequency was 0.30 for the rs7903146 risk allele (T) and 0.29 for the rs12255372 risk allele (T). Both variants were in Hardy–Weinberg equilibrium (P = 0.630 for rs7903146 and P = 0.668 for rs12255372). There were no significant differences in baseline characteristics by TCF7L2 genotype except for TCF7L2-rs7903146, for which those carrying the T allele had a higher psychological acculturation score but lower BMI and waist circumference than those with the major CC genotype (Table 1). In sensitivity analysis we tested the association between the 2 SNPs and the anthropometric measures based on T2D, obesity status, and metabolic syndrome (Supplemental Tables 1 and 2). We found a gene–diabetes interaction for TCF7L2-rs7903146 on BMI and weight and in both SNPs for waist circumference, such that participants with T2D and the risk allele showed lower anthropometric values (Supplemental Table 1). No significant interactions were found for SNPs and adherence to the MedDiet on anthropometric traits except for TCF7L2-rs12255372 and weight. However, for TCF7L2-rs7903146 participants with metabolic syndrome with higher genetic predisposition had significantly lower BMI, weight, and waist circumference only when the adherence to the MedDiet was high (Supplemental Table 2). No significant interactions were found with age or sex (data not shown).
TABLE 1.
Baseline characteristics of the Boston Puerto Rican Health Study participants by TCF7L2-rs7903146 and TCF7L2-rs12255372 variants1
| TCF7L2-rs7903146 | TCF7L2-rs12255372 | ||||||
|---|---|---|---|---|---|---|---|
| Baseline | CC | CT + TT | P value | GG | GT | TT | P value |
| n, % | 539 (48) | 581(52) | 553 (49) | 476 (43) | 91 (8) | ||
| Age, y | 57.0 ± 7.4 | 57.2 ± 7.7 | 0.645 | 56.7 ± 7.5 | 57.3 ± 7.7 | 57.7 ± 7.1 | 0.252 |
| Sex, % female | 72.4 | 67.9 | 0.069 | 69.5 | 71.9 | 65 | 0.287 |
| Educational attainment higher than 8th grade, % | 52.4 | 53.5 | 0.880 | 52.4 | 53.1 | 57.3 | 0.822 |
| Total household income, $/y | 17,507 ± 18,116 | 18,266 ± 20,454 | 0.479 | 17,320 ± 17,770 | 18,762 ± 21,661 | 17,355 ± 16,508 | 0.418 |
| Has health insurance, % | 94.7 | 95.7 | 0.399 | 94.8 | 96.0 | 94.0 | 0.478 |
| Current smoker, % | 24.4 | 24.8 | 0.334 | 25.8 | 24.0 | 21.6 | 0.228 |
| Physical activity score2 | 31.3 ± 4.6 | 31.7 ± 4.9 | 0.094 | 31.5 ± 4.6 | 31.7 ± 5.0 | 31.1 ± 4.7 | 0.399 |
| Psychological acculturation score3 | 18.0 ± 6.1 | 18.8 ± 7.1 | 0.025 | 18.2 ± 6.6 | 18.7 ± 7.2 | 18.1 ± 6.8 | 0.278 |
| Perceived stress score4 | 23.8 ± 9.6 | 23.5 ± 9.4 | 0.678 | 24. 1 ± 9.4 | 23.0 ± 9.6 | 24.4 ± 8.8 | 0.080 |
| Social support5 | 0.4 ± 0.9 | 0.4 ± 0.9 | 0.879 | 0.4 ± 1.7 | 0.5 ± 0.9 | 0.4 ± 0.8 | 0.821 |
| Mediterranean diet score6 | 4.3 ± 1.7 | 4.5 ± 1.7 | 0.332 | 4.4 ± 1.7 | 4.5 ± 1.6 | 4.4 ± 1.6 | 0.588 |
| Total energy intake, kcal/d | 2265 ± 1247 | 2253 ± 1147 | 0.849 | 2277 ± 1242 | 2228 ± 1106 | 2293 ± 1346 | 0.744 |
| Fat, % energy | 31.7 ± 5.5 | 31.9 ± 5.6 | 0.595 | 31.8 ± 5.4 | 31.9 ± 5.6 | 31.7 ± 6.1 | 0.926 |
| Carbohydrates, % energy | 47.7 ± 7.2 | 46.9 ± 7.2 | 0.071 | 47.5 ± .3 | 47.1 ± 7.2 | 46.7 ± 7.5 | 0.460 |
| Protein, % energy | 16.7 ± 3.1 | 17.0 ± 3.2 | 0.122 | 16.7 ± 3.2 | 17.0 ± 3.2 | 17.1 ± 3.2 | 0.164 |
| BMI, kg/m2 | 32.3 ± 6.7 | 31.4 ± 6.5 | 0.009 | 31.9 ± 6.7 | 31.7 ± 6.51 | 32.2 ± 7.0 | 0.737 |
| Weight, kg | 81.2 ± 17.4 | 79.3 ± 17.0 | 0.068 | 80.5 ± 17.7 | 79.4 ± 16.7 | 81.5 ± 17.4 | 0.892 |
| Waist circumference, cm | 102.6 ± 15.1 | 101.1 ± 14.6 | 0.041 | 101.9 ± 15.1 | 101.6 ± 14.7 | 102.2 ± 14.0 | 0.344 |
| Type 2 diabetes, % | 37.1 | 41.8 | 0.073 | 38.1 | 40.2 | 45.3 | 0.311 |
1Continuous variables are expressed as mean ± SD; categorical variables as percentages.
Assessed by a modified Paffenbarger questionnaire from the Harvard Alumni Activity Survey; a score was defined by multiplying the self-reported hours spent in heavy, moderate, light, or sedentary activities in 24 h by weighing factors that parallel the rate of oxygen consumption of each activity. Higher scores are indicative of greater physical activity.
Measured with a 10-item psychological acculturation scale that assesses the degree of psychological attachment to either USA or Puerto Rican culture. Higher scores are indicative of greater USA orientation.
Perceived Stress Scale score ranges from 0–56, with a higher score indicative of higher stress.
Social support participants were asked to identify important persons in their life and to indicate their perception of how these important persons could support them emotionally or assist them in time of need.
Mediterranean diet score assessed with a modified Trichopoulou et al. definition (32) for adherence to 9 foods or nutrients using sex-specific population-based median cut-offs; range 0–9 points; higher score indicative of better adherence to the Mediterranean diet.
First, we tested individual SNP interaction with MedDiet on obesity traits. Significant interactions with the MedDiet score were found for TCF7L2-rs7903146 on weight (P-interaction = 0.024) and waist circumference (P-interaction = 0.026) (Table 2). Individuals with the T risk allele showed significantly lower anthropometric measures at high adherence to the MedDiet than CC individuals (weight: CT + TT: 77.3 ± 1.0 kg compared with CC: 80.9 ± 1.0 kg, P = 0.013; waist circumference: CT + TT: 99.2 ± 0.9 cm compared with CC: 102.2 ± 0.9 cm, P = 0.021) after multivariable adjustment. At low MedDiet adherence, weight and waist circumference were similar by TCF7L2-rs7903146 genotype. For this SNP, the interaction with MedDiet was marginally nonsignificant for BMI (P-interaction = 0.06), although those with the rs7903146 T allele had significantly lower BMI (31.0 ± 0.4) than those with the CC genotype (32.2 ± 0.4) at high MedDiet adherence (P = 0.033).
TABLE 2.
Interactions between TCF7L2-rs7903146 and TCF7L2-rs12255372 variants and Mediterranean diet score for anthropometric measures in the Boston Puerto Rican Health Study1
| BMI, kg/m2 | Weight, kg | Waist circumference, cm | |||||
|---|---|---|---|---|---|---|---|
| n | Low MedDiet | High MedDiet | Low MedDiet | High MedDiet | Low MedDiet | High MedDiet | |
| TCF7L2-rs7903146 | |||||||
| CC | 539 | 32.7 ± 0.4 | 32.2 ± 0.4 | 81.9 ± 1.1 | 80.9 ± 1.0 | 103.6 ± 0.9 | 102.2 ± 0.9 |
| CT + TT | 581 | 32.8 ± 0.4 | 31.0 ± 0.4 | 82.2 ± 1.0 | 77.3 ± 1.0 | 103.9 ± 0.9 | 99.2 ± 0.9 |
| P value | 0.94 | 0.033 | 0.838 | 0.013 | 0.871 | 0.021 | |
| P-interaction2 | 0.06 | 0.024 | 0.026 | ||||
| TCF7L2-rs12255372 | |||||||
| GG | 553 | 32.3 ± 0.4 | 32.0 ± 0.4 | 81.2 ± 1.0 | 80.3 ± 1.0 | 103.2 ± 0.9 | 101.5 ± 0.9 |
| GT | 476 | 33.0 ± 0.4 | 31.4 ± 0.4 | 82.3 ± 1.2 | 78.4 ± 1.1 | 104.1 ± 1.0 | 100.3 ± 0.9 |
| TT | 91 | 33.7 ± 1.0 | 30.6 ± 1.0 | 84.8 ± 2.5 | 75.7 ± 2.6 | 104.6 ± 2.1 | 97.6 ± 2.3 |
| P value | 0.175 | 0.193 | 0.18 | 0.09 | 0.56 | 0.11 | |
| P-interaction | 0.036 | 0.034 | 0.132 | ||||
1Values are means (± SE) adjusted for age, sex, household income, health insurance, educational attainment, physical activity, smoking status, psychological acculturation, social support, perceived stress, type 2 diabetes, and population structure.
P value for interaction terms were obtained from the corresponding multivariate-adjusted models by including an interaction term as independent predictors of each outcome.
Mediterranean diet (MedDiet) score assessed with a modified Trichopoulou et al. definition (32) for adherence to 9 foods or nutrients using sex-specific population-based median cut-offs; range 0–9 points; higher score indicative of better adherence to the MedDiet. Low MedDiet defined as <4 points, and high MedDiet defined as ≥4 points, based on the median of the sample.
Similarly, the MedDiet score interacted significantly with TCF7L2-rs12255372 on BMI (P-interaction = 0.036) and weight (P-interaction = 0.034); individuals with the TT risk genotype had lower anthropometric measures at high MedDiet adherence, whereas those with the GG genotype had lower anthropometric measures at low MedDiet adherence. However, these individual differences by genotype within the MedDiet adherence category were not statistically significant. The T allele carriers with a higher MedDiet score showed a trend of lower but no significant differences when compared with CC participants for BMI (P = 0.19), weight (P = 0.09), and waist circumference (P = 0.11). No significant interaction was found for waist circumference (Table 2).
As the 2 SNPs are in LD (r2 = 0.77) and not independent, we constructed haplotypes based on the 2 SNPs to examine the combined effect of both genetic variants. The most common haplotype was GC with a frequency of 63.2% (Table 3). The lowest frequency was found for the TC haplotype (6.3%). We found significant interactions between haplotype TT (frequency = 0.23) and MedDiet on BMI (P-interaction = 0.024), weight (P-interaction = 0.019), and between haplotype GT and MedDiet on waist circumference (P-interaction = 0.042) and weight (P-interaction = 0.048) with reference to the common haplotype GC (Table 4). In addition, the overall P value for interaction was significant for BMI (P = 0.039) and weight (P = 0.014). After stratifying the population into low (<4 points) and high (≥4 points) MedDiet score, participants with the GT haplotype and a high MedDiet score had significantly lower BMI (P = 0.026), waist circumference (P = 0.015), and body weight (P = 0.009) compared with those with the GC haplotype (Figure 1). On the other hand, when adherence to the MedDiet was low, there was no significant difference between participants with or without the GT haplotype. Furthermore, participants with the TT haplotype and a high MedDiet score had lower body weight when compared with those with the GC haplotype, whereas participants with the TT haplotype and a low MedDiet score showed no significant difference in body weight when compared to those with the GC haplotype (Figure 1).
TABLE 3.
Estimated haplotype frequency (for the TCF7L2-rs7903146 and TCF7L2-rs12255372 variants) in the Boston Puerto Rican Health Study1
| Haplotype no. | Haplotype name | rs12255372 allele | rs7903146 allele | Haplotype frequency |
|---|---|---|---|---|
| 1 | GC | G | C | 0.632 |
| 2 | GT | G | T | 0.074 |
| 3 | TC | T | C | 0.063 |
| 4 | TT | T | T | 0.231 |
1Haplotype frequencies were estimated based on the genotypes of 2 SNPs TCF7L2-rs7903146 and TCF7L2-rs12255372 of 1120 participants using haplo.stats (version 1.7.9) R package.
TABLE 4.
Interactions between haplotypes and Mediterranean diet score for anthropometric measures1
| β-Coefficient ± SE (unadjusted model) | P-interaction (unadjusted model) | β-Coefficient ± SE (multivariable model)2 | P-interaction (multivariable model)2 | Overall P value for interaction (multivariable model)2 | |
|---|---|---|---|---|---|
| BMI, kg/m2 | 0.039 | ||||
| Haplotype GC | Ref | Ref | |||
| GT | −4.36 ± 2.50 | 0.182 | −1.67 ± 1.09 | 0.124 | |
| TC | −3.09 ± 2.66 | 0.060 | −1.97 ± 1.15 | 0.085 | |
| TT | −2.67 ± 1.56 | 0.041 | −1.53 ± 0.68 | 0.024 | |
| Waist circumference, cm | |||||
| Haplotype GC | Ref | Ref | 0.05 | ||
| GT | −4.36 ± 2.50 | 0.083 | −5.07 ± 2.50 | 0.042 | |
| TC | −3.09 ± 2.66 | 0.246 | −3.38 ± 2.63 | 0.198 | |
| TT | −2.67 ± 1.56 | 0.088 | −2.97 ± 1.55 | 0.056 | |
| Weight, kg | |||||
| Haplotype GC | Ref | Ref | 0.014 | ||
| GT | −4.17 ± 2.90 | 0.152 | −5.62 ± 2.83 | 0.048 | |
| TC | −5.35 ± 3.08 | 0.083 | −5.83 ± 2.99 | 0.052 | |
| TT | −3.62 ± 1.81 | 0.046 | −4.16 ± 1.77 | 0.019 | |
Mediterranean diet (MedDiet) score assessed with a modified Trichopoulou et al. definition (32) for adherence to 9 foods or nutrients using sex-specific population-based median cut-offs; range 0–9 points; higher score indicative of better adherence to the MedDiet. Low MedDiet defined as <4 points, and high MedDiet defined as ≥4 points, based on the median of the sample. Haplotype frequencies were estimated based on the genotypes of 2 SNPs TCF7L2-rs7903146 and TCF7L2-rs12255372 of 1120 participants using haplo.stats (version 1.7.9) R package.
2Values are β-coefficients ± SE adjusted for age, sex, household income, health insurance, educational attainment, physical activity, smoking status, psychological acculturation, social support, perceived stress, type 2 diabetes, and population structure. P value for interaction terms were obtained from the corresponding multivariate-adjusted models by including an interaction term as independent predictors of each outcome.
FIGURE 1.
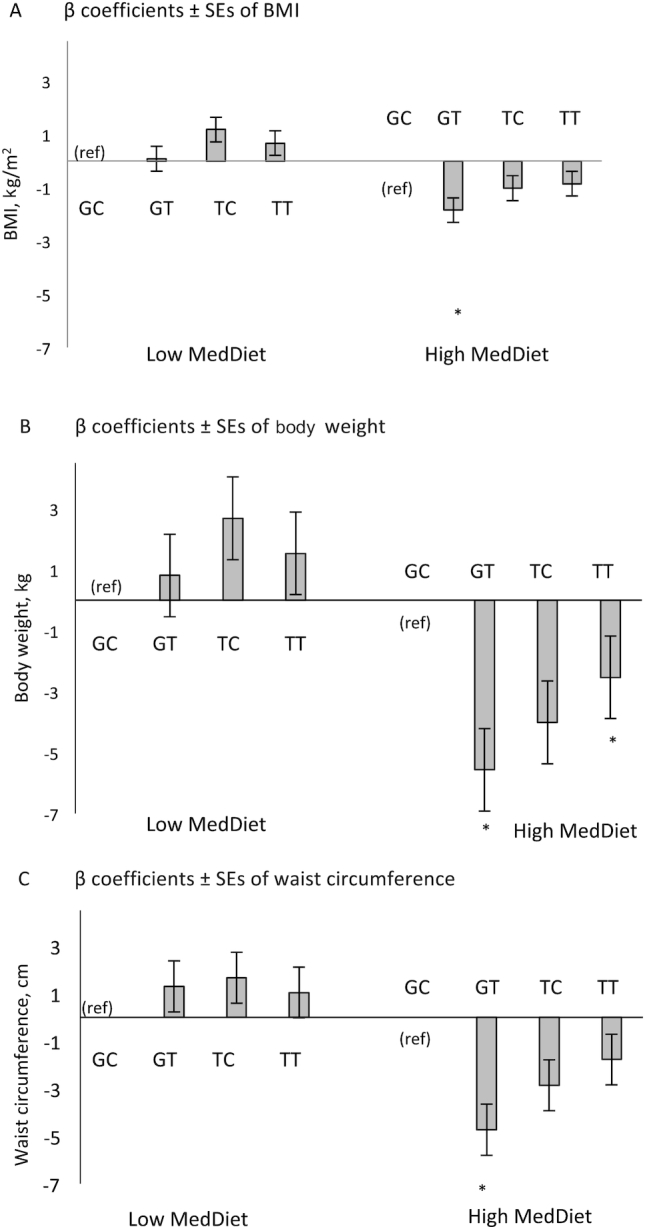
β coefficients (SE) of BMI (A), body w eight (B), and waist circumference (C), by haplotype (GC [ref], GT, TC, TT) and Mediterranean diet (MedDiet) adherence in the Boston Puerto Rican Health Study. The MedDiet score was assessed with a modified Trichopoulou et al. definition ( 32) for adherence to 9 foods or nutrients using sex-specific population-based median cut-offs; range 0–9 points; higher score indicative of better adherence to the MedDiet. Low MedDiet was defined as <4 points (n = 582), and high MedDiet was defined as ≥4 points (n = 538), based on the median of the sample. Values were obtained from generalized linear models adjusted by age, sex, household income, health insurance, educational attainment, physical activity, smoking status, psychological acculturation, social support, perceived stress, ancestral population admixture, type 2 diabetes. *P < 0.05 .
Discussion
We found that Puerto Ricans living in Boston with the TCF7L2-rs7903146 and rs12255372 T2D risk genotypes, although still high, had better anthropometric profiles when adherence to the MedDiet was high, suggesting that unfavorable genetic predisposition may be offset by a healthy diet. Analysis of haplotypes based on 2 risk alleles combined magnified the associations for those with higher genetic risk, who had lower BMI, weight, and waist circumference only when they adhered to the MedDiet.
Previous research supports our results of different cardiometabolic outcomes in response to diet and genetic risk associated with the risk alleles of rs7903146 and rs12255372. A randomized controlled trial with obese participants following a 10-wk intervention with hypoenergetic diets varying in fat content (low fat 20–25% from energy or high fat 40–45% from energy) showed that obese individuals with the TCF7L2-rs7903146 risk allele had better responses in weight loss and adiposity outcomes, although not BMI. (24) In addition, Roswall et al. (27) found a borderline significant interaction between the MedDiet and TCF7L2-rs7903146 on weight gain (P = 0.05), with a better response for those carrying 1 or 2 risk alleles. We also found that participants carrying the rs7903146 T risk allele had significantly lower weight and waist circumference when adherence to the MedDiet was high, but no significant interaction was found for BMI. Mattei et al. reported interactions between fat intake and TCF7L2-rs12255372 in the POUNDS LOST trial, a 2-y trial with 4 energy-reduced diets, whereby BMI and other body adiposity measures decreased more in participants with the TT risk genotype when consuming a diet low in total fat (20%) in comparison to a diet with 40% of energy from fat (16). However, they did not find any interaction with rs7903146. In our study, similar and consistent results were found for TCF7L2-rs12255372, where a significant interaction was found with the MedDiet on BMI and weight but not waist circumference. When adherence to the MedDiet was low, no significant differences were found for any anthropometric outcomes by genotype. Previous studies have suggested that the association of the TCF7L2 gene and T2D might be modulated by obesity with a higher effect in lean compared with obese individuals (13). As the prevalence of obesity is high in our population, we were not able to observe any difference between groups. TCF7L2 gene–BMI interaction or gene–waist circumference interaction and elevated blood glucose have been described recently in Chinese (43) and other populations (13), however, we did not find any significant interaction. We found an interaction with T2D on anthropometric measures. Previous literature has shown that the TCF7L2-T2D risk allele was associated with lower BMI in T2D cases but not in controls (44). Similar results were found in a meta-analysis (45) and other European populations (46), however, identifying the specific mechanisms behind these relations will require further research.
In our study, the presence of risk alleles from both TCF7L2 SNPs magnified the difference in 3 anthropometric measures studied in the context of low, compared with high, MedDiet adherence and suggests that participants who carry the GT haplotype could have better weight-related outcomes when their adherence to the MedDiet is high. However, not all studies have reported a better response to diet for those with the risk alleles. In a 9-mo lifestyle intervention of participants with T2D (n = 309), Haupt et al. found that contrary to other studies, the risk allele carriers for rs7903146 showed a lower reduction in BMI (P ≤ 0.011) and total body fat (P-dominant = 0.016; P-additive = 0.07) (14). However, in the Finnish Diabetes Prevention Study and in the Diabetes Prevention Program, the higher risk of T2D associated with the SNPs was mitigated by a lifestyle intervention (10, 23). Another study showed that high dietary saturated fat intake (≥15.5% of energy) accentuated the deleterious effects of rs7903146 on metabolic syndrome risk, which includes waist circumference (47). The fact that the expression of TCF7L2 mRNA in cell lines has been found to be 1.5- and 3-fold higher for individuals homozygous for both rs7903146 and rs12255372 risk alleles compared with individuals homozygous for the common alleles (48), may support our findings of a stronger difference in anthropometric variables when these 2 genetic risk markers were evaluated based on haplotypes. However, these 2 SNPs are also in LD with other variants; further studies elucidating how they act together are warranted.
We did not evaluate single macronutrients but rather the whole quality of the diet. As described in previous research, small changes in diet quality alone may have a strong effect on cardiometabolic outcomes (26, 49). Our previous research studying diet quality and cardiometabolic outcomes in Puerto Rican adults showed that, among various dietary patterns, the MedDiet was the most strongly associated with cardiometabolic markers, including adiposity, insulin, and inflammation (50). Puerto Ricans with a high adherence to the MedDiet consumed foods that are more akin to traditional Puerto Rican cuisine: homemade soups and stews, root crops, fruit juice, green bananas, oatmeal, light bread, beans and legumes, fish and shellfish, canned tuna, milk and cheese, corn and olive oils, and beer. We conclude that population-based cut-offs (as used by the MedDiet score) may be appropriate in capturing the distribution of food intake in diverse populations (50). One study evaluated the effect of a TCF7L2-rs7903146–MedDiet interaction on metabolic markers using baseline data (cross-sectional analysis), and on stroke incidence using longitudinal data from a randomized controlled trial (19). They showed that higher adherence to a MedDiet was associated with lower fasting glucose, total cholesterol, LDL-cholesterol, and triglycerides in TT individuals at baseline and that these same individuals had a lower risk of stroke when randomized to a MedDiet supplemented with extra virgin olive oil and nuts, compared with CC individuals. Based on the components of the MedDiet, especially olive oil and nuts, this diet may have a high percentage of dietary fat, which might contradict the previous literature reporting the benefits of a low-fat diet (16, 24). However, the type of fat (47) and quality of the diet (10, 19, 23) might be more important than quantity when advising those predisposed to disease through genetic risk.
A previous study hypothesized that carriers of the rs7903146 risk allele variant were selectively adapted to maintain weight stability under low-protein conditions and that they may have evolved with a plant-based diet (similar to the MedDiet). Therefore, this selection for lean mass preservation could positively benefit those carrying that variant under such dietary conditions (15). Other pathways have also been explored to elucidate how TCF7L2 could regulate body weight; for example, through hunger-satiety hormones that influence weight loss (44); however, in our study, energy intake was not different by genotype or strata of MedDiet. TCF7L2 is also essential for transcription of the proglucagon gene and thus for GLP-1, which has been found to reduce insulinotropic effects in T allele carriers, and may regulate body weight through appetite, adipose tissue metabolism, and insulin signaling (6). More research is warranted to determine the exact pathways and the combination of all macronutrients interacting together in the context of a denominated high-quality diet such as the MedDiet, to modulate adiposity measures in carriers of the risk allele.
Our results emphasize the importance of adhering to a healthy diet, especially in a population that has a high risk of cardiometabolic conditions (28, 29) and poor dietary behaviors, including a low intake of fruit and vegetables and whole grains, and a high intake of unhealthy fats, added sugars, and sodium (51). Thus, identifying cardiometabolic genetic risk predisposition in a high-risk population and nutrition precision approaches such as dietary patterns (i.e. MedDiet) that may offset this predisposition, could offer a strategy to tailor prevention and treatment of cardiometabolic conditions based on individual genetic characteristics.
Some limitations should be acknowledged. First, the nature of self-reported dietary data may lead to measurement error; however, we used an FFQ specifically developed for this population and validated against biomarkers of intake (30). Second, the cross-sectional study design does not allow us to establish causality, so the results should be interpreted with caution. Third, the generalizability of the results should be limited because Puerto Ricans living in Boston may have specific characteristics not shared by Puerto Ricans or Hispanics/Latinos in other locations; nonetheless, our carefully adjusted models should account for the main sociodemographic differences. Residual confounding is always a concern; however, we were able to adjust for documented potential confounders, especially for population admixture, which is critical in genetic studies of admixed populations (42).
In conclusion, our results suggest that in a high-risk population of Boston Puerto Rican adults, individuals with TCF7L2 risk variants might have a better anthropometric profile with high adherence to a MedDiet, suggesting that an unfavorable predisposition may be counterbalanced by a healthy diet. Additional studies should clarify the role of dietary patterns and risk alleles on anthropometric markers on populations at-risk of T2D as this may help with precision medicine strategies.
Supplementary Material
Acknowledgments
The authors’ responsibilities were as follows—MS-P: designed the analysis, conducted statistical analysis, interpreted the results, and wrote the manuscript; JM: contributed to study design, analyzed data, and interpreted the results; JMO and KLT: contributed to study design and interpreted the results; JMO, CES, and C-QL: supervised the genotyping analysis and quality control; and all authors contributed meaningfully to the manuscript and read and approved the final manuscript. MS-P and JM had primary responsibility for the final content.
Notes
Supported by NIH grants P50HL105185, P01AG023394, R01AG055948, and K01-HL120951 and by the USDA, under agreement no. 8050-51000-098-00D. CES is supported by K08 HL112845.
Author disclosures: MS-P, CES, C-QL, KLT, JMO, and JM, no conflicts of interest.
Supplemental Tables 1 and 2 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: GLP-1, glucagon-like peptide 1; GWAS, genome-wide association study; LD, linkage disequilibrium; MedDiet, Mediterranean diet; SNP, single-nucleotide polymorphism; TCF7L2, transcription factor 7–like 2; T2D, type 2 diabetes.
References
- 1. Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A et al.. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G et al.. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL et al.. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) Consortium Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qi Q, Stilp AM, Sofer T, Moon JY, Hidalgo B, Szpiro AA, Wang T, Ng MCY, Guo X, Chen Y et al.. Genetics of type 2 diabetes in U.S. Hispanic/Latino individuals: results from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Diabetes. 2017;66:1419–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin T, Liu L.. The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol Endocrinol. 2008;22:2383–92. [DOI] [PubMed] [Google Scholar]
- 7. Paniagua JA, de la Sacristana AG, Sanchez E, Romero I, Vidal-Puig A, Berral FJ, Escribano A, Moyano MJ, Perez-Martinez P, López-Miranda J et al.. A MUFA-rich diet improves posprandial glucose, lipid and GLP-1 responses in insulin-resistant subjects. J Am Coll Nutr. 2007;26:434–44. [DOI] [PubMed] [Google Scholar]
- 8. Eller LK, Ainslie PN, Poulin MJ, Reimer RA. Differential responses of circulating amylin to high-fat vs. high-carbohydrate meal in healthy men. Clin Endocrinol (Oxf). 2008;68:890–7. [DOI] [PubMed] [Google Scholar]
- 9. Potts JE, Gray LJ, Brady EM, Khunti K, Davies MJ, Bodicoat DH. The effect of glucagon-like peptide 1 receptor agonists on weight loss in type 2 diabetes: a systematic review and mixed treatment comparison meta-analysis. PLoS One. 2015;10:e0126769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D; Diabetes Prevention Program Research G. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med. 2006;355:241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang C, Qi L, Hunter DJ, Meigs JB, Manson JE, van Dam RM, Hu FB. Variant of transcription factor 7-like 2 (TCF7L2) gene and the risk of type 2 diabetes in large cohorts of U.S. women and men. Diabetes. 2006;55:2645–8. [DOI] [PubMed] [Google Scholar]
- 12. Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A et al.. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–3. [DOI] [PubMed] [Google Scholar]
- 13. Corella D, Coltell O, Sorli JV, Estruch R, Quiles L, Martinez-Gonzalez MA, Salas-Salvado J, Castaner O, Aros F, Ortega-Calvo M et al.. Polymorphism of the transcription factor 7-like 2 gene (TCF7L2) interacts with obesity on type-2 diabetes in the PREDIMED Study emphasizing the heterogeneity of genetic variants in type-2 diabetes risk prediction: time for obesity-specific genetic risk scores. Nutrients. 2016;8(12):793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haupt A, Thamer C, Heni M, Ketterer C, Machann J, Schick F, Machicao F, Stefan N, Claussen CD, Häring HU et al.. Gene variants of TCF7L2 influence weight loss and body composition during lifestyle intervention in a population at risk for type 2 diabetes. Diabetes. 2010;59:747–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fisher E, Meidtner K, Angquist L, Holst C, Hansen RD, Halkjaer J, Masala G, Ostergaard JN, Overvad K, Palli D et al.. Influence of dietary protein intake and glycemic index on the association between TCF7L2 HapA and weight gain. Am J Clin Nutr. 2012;95:1468–76. [DOI] [PubMed] [Google Scholar]
- 16. Mattei J, Qi Q, Hu FB, Sacks FM, Qi L. TCF7L2 genetic variants modulate the effect of dietary fat intake on changes in body composition during a weight-loss intervention. Am J Clin Nutr. 2012;96:1129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan Y, North KE, Heiss G, Klein R, Girman CJ, Lange EM, Pankow JS, Brancati FL, Boerwinkle E. Transcription factor 7-like 2 (TCF7L2) polymorphism and context-specific risk of impaired fasting glucose in African American and Caucasian adults: the atherosclerosis risk in communities (ARIC) study. Diabetes Metab Res Rev. 2010;26:371–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Delgado-Lista J, Perez-Martinez P, Garcia-Rios A, Phillips CM, Williams CM, Gulseth HL, Helal O, Blaak EE, Kiec-Wilk B, Basu S et al.. Pleiotropic effects of TCF7L2 gene variants and its modulation in the metabolic syndrome: from the LIPGENE study. Atherosclerosis. 2011;214:110–6. [DOI] [PubMed] [Google Scholar]
- 19. Corella D, Carrasco P, Sorli JV, Estruch R, Rico-Sanz J, Martinez-Gonzalez MA, Salas-Salvado J, Covas MI, Coltell O, Arós F et al.. Mediterranean diet reduces the adverse effect of the TCF7L2-rs7903146 polymorphism on cardiovascular risk factors and stroke incidence: a randomized controlled trial in a high-cardiovascular-risk population. Diabetes Care. 2013;36:3803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fisher E, Boeing H, Fritsche A, Doering F, Joost HG, Schulze MB. Whole-grain consumption and transcription factor-7-like 2 (TCF7L2) rs7903146: gene-diet interaction in modulating type 2 diabetes risk. Br J Nutr. 2009;101:478–81. [DOI] [PubMed] [Google Scholar]
- 21. Cornelis MC, Qi L, Kraft P, Hu FB. TCF7L2, dietary carbohydrate, and risk of type 2 diabetes in US women. Am J Clin Nutr. 2009;89:1256–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruchat SM, Elks CE, Loos RJ, Vohl MC, Weisnagel SJ, Rankinen T, Bouchard C, Perusse L. Evidence of interaction between type 2 diabetes susceptibility genes and dietary fat intake for adiposity and glucose homeostasis-related phenotypes. J Nutrigenet Nutrigenomics. 2009;2:225–34. [DOI] [PubMed] [Google Scholar]
- 23. Wang J, Kuusisto J, Vanttinen M, Kuulasmaa T, Lindstrom J, Tuomilehto J, Uusitupa M, Laakso M. Variants of transcription factor 7-like 2 (TCF7L2) gene predict conversion to type 2 diabetes in the Finnish Diabetes Prevention Study and are associated with impaired glucose regulation and impaired insulin secretion. Diabetologia. 2007;50:1192–200. [DOI] [PubMed] [Google Scholar]
- 24. Grau K, Cauchi S, Holst C, Astrup A, Martinez JA, Saris WH, Blaak EE, Oppert JM, Arner P, Rössner S et al.. TCF7L2 rs7903146-macronutrient interaction in obese individuals' responses to a 10-wk randomized hypoenergetic diet. Am J Clin Nutr. 2010;91:472–9. [DOI] [PubMed] [Google Scholar]
- 25. Cespedes EM, Hu FB. Dietary patterns: from nutritional epidemiologic analysis to national guidelines. Am J Clin Nutr. 2015;101:899–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dinu M, Pagliai G, Casini A, Sofi F. Mediterranean diet and multiple health outcomes: an umbrella review of meta-analyses of observational studies and randomised trials. Eur J Clin Nutr. 2018;72(1):30–43. [DOI] [PubMed] [Google Scholar]
- 27. Roswall N, Angquist L, Ahluwalia TS, Romaguera D, Larsen SC, Ostergaard JN, Halkjaer J, Vimaleswaran KS, Wareham NJ, Bendinelli B et al.. Association between Mediterranean and Nordic diet scores and changes in weight and waist circumference: influence of FTO and TCF7L2 loci. Am J Clin Nutr. 2014;100:1188–97. [DOI] [PubMed] [Google Scholar]
- 28. Heiss G, Snyder ML, Teng Y, Schneiderman N, Llabre MM, Cowie C, Carnethon M, Kaplan R, Giachello A, Gallo L et al.. Prevalence of metabolic syndrome among Hispanics/Latinos of diverse background: the Hispanic Community Health Study/Study of Latinos. Diabetes Care. 2014;37:2391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tucker KL, Mattei J, Noel SE, Collado BM, Mendez J, Nelson J, Griffith J, Ordovas JM, Falcon LM. The Boston Puerto Rican Health Study, a longitudinal cohort study on health disparities in Puerto Rican adults: challenges and opportunities. BMC Public Health. 2010;10:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tucker KL, Bianchi LA, Maras J, Bermudez OI. Adaptation of a food frequency questionnaire to assess diets of Puerto Rican and non-Hispanic adults. Am J Epidemiol. 1998;148:507–18. [DOI] [PubMed] [Google Scholar]
- 31. Ye X, Scott T, Gao X, Maras JE, Bakun PJ, Tucker KL. Mediterranean diet, healthy eating index 2005, and cognitive function in middle-aged and older Puerto Rican adults. J Acad Nutr Diet. 2013;113:276–81. e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trichopoulou A, Costacou T, Bamia C, Trichopoulos D. Adherence to a Mediterranean diet and survival in a Greek population. N Engl J Med. 2003;348:2599–608. [DOI] [PubMed] [Google Scholar]
- 33. Van Rompay MI, McKeown NM, Castaneda-Sceppa C, Ordovas JM, Tucker KL. Carbohydrate nutrition differs by diabetes status and is associated with dyslipidemia in Boston Puerto Rican adults without diabetes. J Nutr. 2013;143:182–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lai CQ, Tucker KL, Parnell LD, Adiconis X, Garcia-Bailo B, Griffith J, Meydani M, Ordovas JM. PPARGC1A variation associated with DNA damage, diabetes, and cardiovascular diseases: the Boston Puerto Rican Health Study. Diabetes. 2008;57:809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Najjar MF, Kuczmarski RJ.. Anthropometric data and prevalence of overweight for Hispanics: 1982–84. Vital Health Stat 11. 1989;239:1–106. [PubMed] [Google Scholar]
- 36. Chumlea WC, Guo SS, Wholihan K, Cockram D, Kuczmarski RJ, Johnson CL. Stature prediction equations for elderly non-Hispanic white, non-Hispanic black, and Mexican-American persons developed from NHANES III data. J Am Diet Assoc. 1998;98:137–42. [DOI] [PubMed] [Google Scholar]
- 37. Tropp LR, Erkut S, Coll CG, Alarcon O, Vazquez Garcia HA. Psychological acculturation: development of a new measure for Puerto Ricans on the U.S. mainland. Educ Psychol Meas. 1999;59:351–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Norbeck JS. Modification of life event questionnaires for use with female respondents. Res Nurs Health. 1984;7:61–71. [DOI] [PubMed] [Google Scholar]
- 39. Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. J Health Soc Behav. 1983;24:385–96. [PubMed] [Google Scholar]
- 40. Ramirez MT, Hernandez RL.. Factor structure of the Perceived Stress Scale (PSS) in a sample from Mexico. Span J Psychol. 2007;10:199–206. [DOI] [PubMed] [Google Scholar]
- 41. Paffenbarger RS Jr., Hyde RT, Wing AL, Lee IM, Jung DL, Kampert JB. The association of changes in physical-activity level and other lifestyle characteristics with mortality among men. N Engl J Med. 1993;328:538–45. [DOI] [PubMed] [Google Scholar]
- 42. Lai CQ, Tucker KL, Choudhry S, Parnell LD, Mattei J, Garcia-Bailo B, Beckman K, Burchard EG, Ordovas JM. Population admixture associated with disease prevalence in the Boston Puerto Rican health study. Hum Genet. 2009;125:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li L, Wang J, Ping Z, Li Y, Wang C, Shi Y, Zhou W, Zhang L. Interaction analysis of gene variants of TCF7L2 and body mass index and waist circumference on type 2 diabetes. Clinical nutrition (Edinburgh, Scotland). 2019;pii: S0261-5614:(19):30032–9. [DOI] [PubMed] [Google Scholar]
- 44. Helgason A, Palsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen Y, Chen G, Reynisdottir I et al.. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39:218–25. [DOI] [PubMed] [Google Scholar]
- 45. Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J et al.. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cauchi S, Choquet H, Gutierrez-Aguilar R, Capel F, Grau K, Proenca C, Dina C, Duval A, Balkau B, Marre M et al.. Effects of TCF7L2 polymorphisms on obesity in European populations. Obesity (Silver Spring). 2008;16:476–82. [DOI] [PubMed] [Google Scholar]
- 47. Phillips CM, Goumidi L, Bertrais S, Field MR, McManus R, Hercberg S, Lairon D, Planells R, Roche HM. Dietary saturated fat, gender and genetic variation at the TCF7L2 locus predict the development of metabolic syndrome. J Nutr Biochem. 2012;23:239–44. [DOI] [PubMed] [Google Scholar]
- 48. Pang DX, Smith AJ, Humphries SE. Functional analysis of TCF7L2 genetic variants associated with type 2 diabetes. Nutr Metab Cardiovasc Dis. 2013;23:550–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sotos-Prieto M, Bhupathiraju SN, Mattei J, Fung TT, Li Y, Pan A, Willett WC, Rimm EB, Hu FB. Association of changes in diet quality with total and cause-specific mortality. N Engl J Med. 2017;377:143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mattei J, Sotos-Prieto M, Bigornia SJ, Noel SE, Tucker KL. The Mediterranean diet score is more strongly associated with favorable cardiometabolic risk factors over 2 years than other diet quality indexes in Puerto Rican adults. J Nutr. 2017;147:661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mattei J, Bhupathiraju S, Tucker KL. Higher adherence to a diet score based on American Heart Association recommendations is associated with lower odds of allostatic load and metabolic syndrome in Puerto Rican adults. J Nutr. 2013;143:1753–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


