Abstract
Objective: Autophagy plays important roles in tumor occurrence and development. The present study aimed to investigate the association between autophagy and apoptosis in clear cell renal carcinoma cells (ccRCCs). Methods: Atg7-overexpressing and -knockdown RCC 786-O cells (pLenti6.3-ATG7 and sh-ATG7-2 lv) were established using lentiviral transfection and interference shRNA. pLenti6.3-GFP and sh-scramb-con lv were used as controls. Cells were cultured in medium with or without the apoptosis inhibitor Z-VAD-FMK. Cell apoptosis were detected by flow cytometry. Cell proliferation was determined by MTT assay. Expression of apoptotic pathway proteins was measured by Western blot. Results: The apoptosis rate of Z-VAD-FMK-treated cells was significantly decreased compared with untreated cells (P < 0.05). However, no significant difference in the apoptosis rate was detected among cell groups with different autophage level. The Z-VAD-FMK treatment induced significant changes in apoptosis rate in all cell groups, but only slightly changed the cell proliferation. When cell apoptosis were inhibited by Z-VAD-FMK, the cell viability in pLenti6.3-ATG7 group was significantly reduced compared with 786-O control (P < 0.05), whereas the cell viability in sh-ATG7-2 lv was significantly enhanced (P < 0.05) indicating that cell proliferation was closely associated with the level of autophage. The expression of caspase proteins in pLenti6.3-ATG7 was significantly higher compared with sh-ATG7-2 lv group (P < 0.05). Conclusion: autophagy and apoptosis are independent processes of PCD in human ccRCC 786-O cells. Autophagy is the main type of PCD and may be closely associated with apoptosis through the classical death receptor, mitochondria and endoplasmic reticulum apoptosis pathway.
Keywords: Renal carcinoma cell (RCC), autophagy, apoptosis, ATG 7, shRNA
Introduction
Autophagy is a self-degradation process during which unnecessary or dysfunctional cellular components are digested by the lysosomal machinery. Autophagy frequently occurs as an adaptive response to stresses such as nutrition deprivation, hypoxic stress, etc., which maintains cellular energy and thereby promotes cellular survival, whereas it may promote cell death through excessive degradation of cellular, and has been known as type II programmed cell death (PCD) [1].
Studies have suggested that autophage is related to the occurrence and progression of several diseases including cancer. In recent years, autophage has therefore become a research hotspot in cancer biology [2,3]. Autophagy plays a double-edged role in cancer: both tumor suppression and tumor cell survival [4,5]. The process of autophagy is strictly controlled by over 30 autophagy-related (Atg) genes [6]. As one of the essential autophagy-related proteins, ATG7 mediates the formation of ATG5-ATG12-ATG16 complex and promotes the formation of autophagic vacuoles. ATG7 is also involved in the phosphatidylethanolamine modification of LC3 during the formation of autophagesome. More importantly, Atg7 knockout mice are more susceptible to liver cancer compared with healthy mice [7], suggesting that the protein is involved in the autophage-regulated tumor suppression. Moreover, ATG7 is also essential for the cancer therapy-induced autophagy. Mice with reduced Atg7 expression diaplays markedly decreased immune responses to chemotherapy [8]. On the other hand, suppressed autophage by Atg7 silencing will lead to increased sensitivity of tumor cells to chemo- or radio-therapy, suggesting a cytoprotective effect of therapy-induced autophagy [9].
Apoptosis, the type I PCD, is a highly regulated and controlled process that is closely associated with the occurrence and development of cancer. Caspases (cysteine-aspartic proteases) are a family of protease enzymes that are essential in classical apoptotic pathways [10]. As two different forms of programmed cell death, autophage and apoptosis are both important in cellular growth, as well as the occurrence and progression of various diseases [11]. Nevertheless, the association between autophage and apoptosis has not been fully clarified. Generally, autophagy can protect cells by preventing them from undergoing apoptosis. However, autophagy can also trigger apoptosis under certain conditions [12]. Therefore, the investigation of their association and related signal pathway is essential for the identification of novel therqpeutic targets. This study explored the effect of different autophagy levels on the apoptosis and proliferation of clear cell renal cell carcinoma (ccRCC) 786-O cells in order to clarify the relationship between autophage and apoptosis.
Materials and methods
Cell culture
Human renal cell carcinoma 786-O cells were purchased from the Chinese Academy of Sciences Library (CCCAS, Shanghai) and maintained in high glucose RPMI-1640 medium (HyClone company) containing 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 mg/mL streptomycin at 37°C in an incubator with 5% CO2. Cells at the exponential phase were used for subsequent experiments.
Main reagents and instruments
Thiazolyl blue tetrazolium blue (MTT) and dimethyl sulfoxide (DMSO) were purchased from Sigma. AnnexinV-FITC/7AAD apoptosis staining kit was purchased from BD Biosciences (San Diego, CA, USA). BCA-100 protein quantification kit was purchased from Dongsheng Biotech. (Shenyang, China). Rabbit anti-human Caspase-3, Caspase-4, Caspase-8 and Caspase-9 polyclonal antibodies were purchased from Proteintech (Chicago, IL, USA). Mouse anti-human β-actin monoclonal antibody, HRP-labeled goat anti-rabbit IgG, and HRP-labeled goat anti-mouse IgG were purchased from TransGen (Beijing, China). ECL luminous color reagent was purchased from Millipore (Temecula, CA, USA).
Construct of recombinant lentivirus
Human autophagy related gene Atg7 was cloned into pLenti6.3-MCS-IRES-GFP vector (Invitrogen, Beijing, China). Empty vector control was also established. Sequence of Atg7 shRNA, 5’-CCGGAAGGAGTCACAGCTCTTCCTTCTCGAGAAGGAAGAGCTGTGACTCCTTTTTTTG-3’, was cloned into pLVshRNA-mCherry(2A)-puro vector (Invitrogen). Sequence of RNA scramble was also cloned to construct vector control. All vectors were respectively transfected into packaging cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacture’s instructions. Lentiviruses were purified and concentrated. The obtained virus, empty control, Atg7-knockdown virus, and RNA scramble control was named.
Establishment of stable Atg7-overexpressing, Atg7-knockdown cells
To obtain stable transfectants, 786-O cells were transfected with the appropriate lentivirus (10 μL) using Lipofectamine 2000 reagent and incubated at 37°C, 5% CO2. Medium was replaced every 24 h. After 96 h, cells were selected in complete growth medium containing 4.0 µg/mL Blasticidin (Invitrogen) or 2.0 µg/mL Puromycin (Invitrogen). The selected Atg7-expressing cells, empty control, Atg7-knockdown cells, and scramble control cells were named as pLenti6.3-ATG7, pLenti6.3-GFP, sh-ATG7-2 lv, and sh-scramb-con lv, respectively.
Flow cytometry
pLenti6.3-ATG7, pLenti6.3-GFP, sh-ATG7-2 lv, and sh-scramb-con lv cells were cultured in complete medium with or without 100 µM inhibitor Z-VAD-FMK at 37°C, 5% CO2 for 24 h. Cells at exponential phase were trypsinized and washed with PBS. Cells were then mixed with binding buffer and treated with 5 ul of Annexin V-FITC for 15 min in dark. 7AAD (5 ul) was added. Cells were incubated for 5 min and detected by a flow cytometer within 1 h to detect the apoptosis in each group. Triplicate samples were prepared for each group.
MTT assay
Cell proliferation was determined by MTT assay. Briefly, cells in each group were cultured in complete medium with or without inhibitor Z-VAD-FMK at 37°C, 5% CO2 for 24 h and treated with 20 μl of 5 mg/ml MTT for 4 h. The medium was discarded and DMSO was then added to solubilize the formazan crystals. The absorbance (OD) at 590 nm was measured using a microplate autoreader (Bio-Tek Instruments, Vermont, USA). Triplicate samples were prepared for each group.
Western blot
Cells at the exponential phase were collected and mixed with RIPA lysis buffer. Total protein was extracted and quantified using a BCA kit according to the manufacture’s instruction. Equal amounts of total protein (30 μg) were separated by SDS-PAGE electrophoresis and transferred to polyvinylidene difluoride membranes. The membrane was blocked in TBS buffer containing 5% skim milk at room temperature for 2 h, and incubated with caspase-3, caspase-4, caspase-8 and caspase-9 and β-actin antibodies overnight at overnight at 4°C. The membrane was washed and incubated with HRP-labeled secondary antibodies at 37°C for 1 h. Protein signal was then detected using the ECL detection system. The intensity of bands was detected by a Molecular Imager® ChemiDocTM XRS System (Bio-Rad Laboratories). The gray value of bands was analyzed by Image Lab 2.0 software (Bio-Rad Laboratories).
Statistical analysis
All data were expressed as mean ± standard deviation and analyzed using SPSS 17.0 (IBM SPSS, Chicago, IL, USA). Difference among groups was compared by ANOVA. Difference between groups was compared by t-tests. P values smaller than 0.05 are considered statistically significant.
Results
Effects of autophagy on the apoptosis of 786-O cells
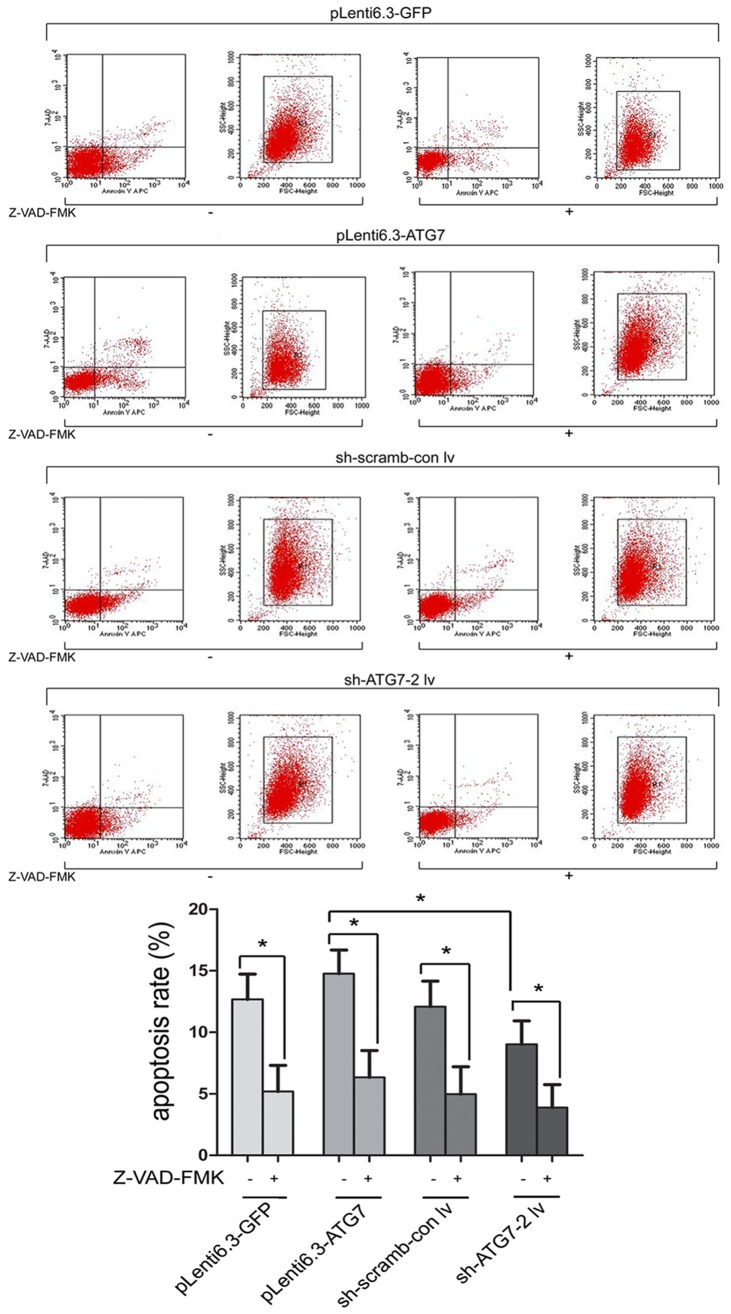
As shown in Figure 1, flow cytometry demonstrated that the apoptosis rate of pLenti6.3-ATG7 cultured in medium without apoptosis inhibitor Z-VAD-FMK was significantly higher than that of sh-ATG7-2 lv cells (P < 0.05). No significant difference in the apoptosis rate was observed among other groups. When cells were treated with Z-VAD-FMK, the apoptosis rate was significantly decreased compared with untreated cells (P < 0.05). However, no significant difference in the apoptosis rate was detected among cell groups with different level of autophage, suggesting that the level of autophage did not affect the apoptosis of cells, and autophage and apoptosis were independent processes of PCD.
Figure 1.
Flow cytometry comparing the apoptosis rate in different cell groups. *, P < 0.05.
Effects of autophagy on the proliferation of 786-O cells
Although the apoptosis rate of cells treated with Z-VAD-FMK was significantly decreased compared with untreated cells (Figure 1), they exhibited similar cell viability (P > 0.05, Figure 2). When cell apoptosis were inhibited by Z-VAD-FMK, the cell viability in pLenti6.3-ATG7 group was significantly reduced compared with 786-O control (P < 0.05), whereas the cell viability in sh-ATG7-2 lv was significantly enhanced (P < 0.05, Figure 2), indicating that cell proliferation was closely associated with the level of autophage. Moreover, the Z-VAD-FMK treatment induced significant changes in apoptosis rate in all cell groups, but only slightly changed the cell proliferation (Figure 3), further suggesting that the difference in cell viability among different cell groups was caused by different level of autophage instead of apoptosis, and autophage was the primary process of PCD in 786-O cells.
Figure 2.
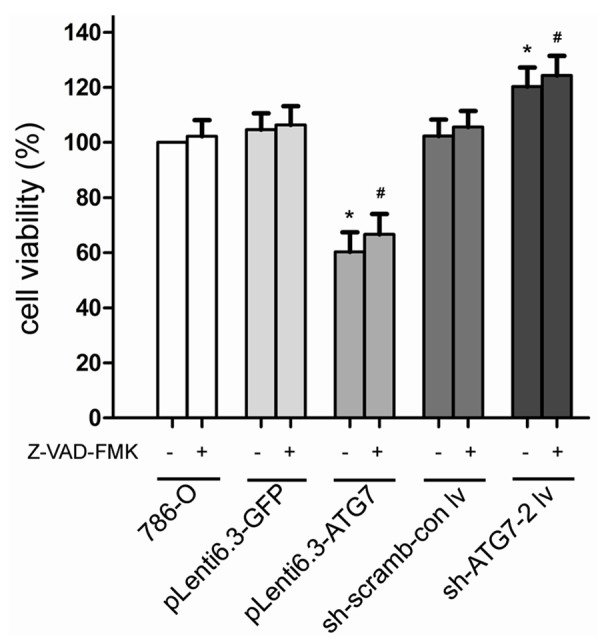
Comparison of cell viability of different cell groups. *, P < 0.05 compared with 786-O cells, and #, P < 0.05 compared with 786-O cells treated with Z-VAD-FMK.
Figure 3.
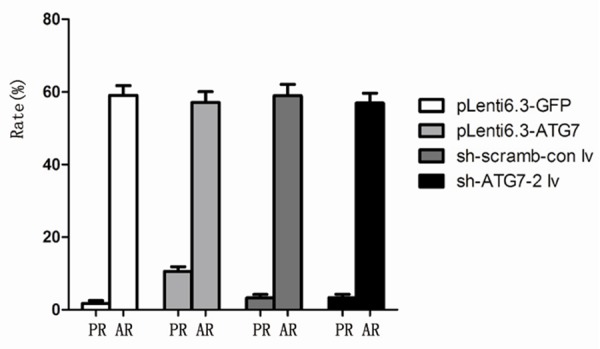
Comparison of proliferation rate (PR) and apoptosis rate (AR) of different cell groups.
Effects of autophagy on apoptotic signaling pathway proteins
As shown in Figure 4, the highest expression of caspase3, 4, 8 and 9, the apoptotic signaling pathway proteins, was detected in pLenti6.3-ATG7 group, followed by pLenti6.3-GFP or sh-scramb-con lv groups. The lowest expression of caspase proteins was observed in sh-ATG7-2 lv group. The expression of caspase proteins in pLenti6.3-ATG7 was significantly higher compared with sh-ATG7-2 lv group (P < 0.05). These results suggested that autophage was closely associated with the apoptotic signaling pathway proteins, and a crosstalk might exist between autophage and apoptotic pathways.
Figure 4.
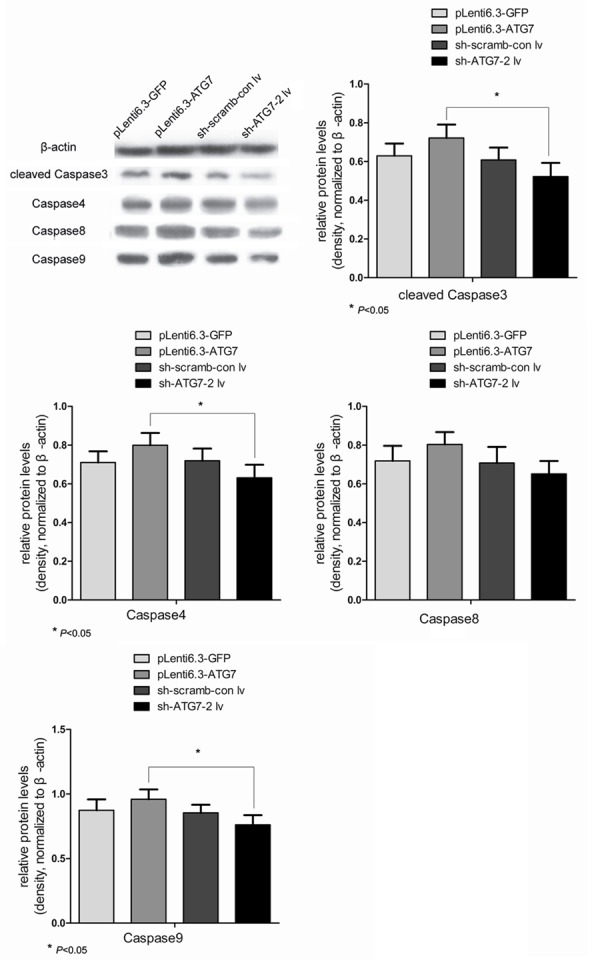
Western blot analyses comparing the relative expression of key apoptotic proteins (cleaved caspase-3, caspase-4, -8 and -9) in different cell groups. *, P < 0.05.
Discussion
Currently, the relationship between autophage and apoptosis has not been fully clarified. Studies have suggested that autophage and apoptosis coordinately regulate cell death. They may occur simultaneously in cells. For instance, TNF-related apoptosis inducing ligand (TRAIL) may induce both autophage and caspase-dependent apoptosis of human breast epithelial MCF-10A cells [13]. It has also been found that arsenic trioxide induces not only apoptosis but also autophagic cell death in leukemia cell lines [14]. Apoptosis and autophage are both activated in the treatment of Kaposi’s sarcoma [15]. Alternatively, cells start to undergo autophage or apoptosis once the other form of CPD is deactivated. When apoptosis in neurons is inhibited, autophage becomes the primary CPD [16]. Nevertheless, studies have also found that autophage induces the process of apoptosis, and thus is required for the latter. In breast cancer cells, autophage occurs prior to apoptosis, and the autophage inhibitor 3-MA can suppress both autophage and apoptosis [17]. Damage regulated autophagy modulator (DRAM) may promote apoptosis through mediating autophage, whereas its deactivation suppresses autophage, leading to reduced apoptosis [18]. In addition, some studies have suggested a mutual antagonism between autophagy and apoptosis. Bauvy et al. have considered depolarized mitochondrial degradation by autophagy as a protective mechanism against apoptosis in human colon cancer HT-29 cells [19]. The inhibition of autophage can also enhance the sensitivity of breast cancer cells to radiotherapy [20]. In this study, we found that the level of autophage did not affect the apoptosis of cells, indicating that autophage and apoptosis were independent processes of PCD in ccRCC 786-O cells. Moreover, the apoptosis rate in pLenti6.3-ATG7 group was significantly increased compared with sh-ATG7-2 lv group (P < 0.05), which suggested that some of the cells with high autophage level might start the process of apoptosis. Autophage therefore promoted the occurrence of apoptosis.
We further compared the cell viability of cells treated with apoptosis inhibitor Z-VAD-FMK in order to investigate the primary PCD in 786-O cells. It was found that although the apoptosis rate of cells treated with Z-VAD-FMK was significantly decreased compared with untreated cells, they exhibited similar cell viability. The Z-VAD-FMK treatment induced significant changes in apoptosis rate in all cell groups, but only slightly changed the cell proliferation. Moreover, the cell viability in pLenti6.3-ATG7 group was significantly reduced compared with 786-O control, whereas the cell viability in sh-ATG7-2 lv was significantly enhanced. These results suggested that cell proliferation of 786-O cells was mainly affected by the autophage level instead of apoptosis, and autophage was the primary type of PCD in 786-O cells. To our best knowledge, the current study is the first report on the main type of PCD in ccRCC.
Autophagy and apoptosis may occur simultaneously or successively in cells. Therefore, the investigation on the interplay between the two may provide novel therapeutic targets in the treatment of several diseases, especially cancer. There are three classical apoptotic pathways including death receptor, mitochondrial and endoplasmic reticulum pathway [21,22]. In this study, we further compared the expression of key apoptotic pathway proteins (caspase-3, -4, -8 and -9) in cells with different levels of autophage. The expression of caspase proteins in pLenti6.3-ATG7 was significantly higher compared with sh-ATG7-2 lv group (P < 0.05), indicating a close association between autophage and these apoptotic signaling pathway proteins, and thereby an interplay between autophage and apoptosis through the classical apoptotic pathways.
In summary, autophagy and apoptosis are independent processes of PCD in human ccRCC 786-O cells. Autophagy is the main type of PCD and may be closely associated with apoptosis through the classical death receptor, mitochondria and endoplasmic reticulum apoptosis pathway.
Acknowledgements
This study was supported by the Key Research Project at the Second Affiliated Hospital of Xi’an Jiaotong University (YJ(ZD)201318) and the International Forefront and Independent Innovation Foundation (xjj2015012).
Disclosure of conflict of interest
None.
References
- 1.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuya N, Liang XH, Levin B. Autophagy and cancer. In: Klionsky DJ, editor. Landes Bioscience. Texas, USA: Georgetown; 2004. pp. 244–253. [Google Scholar]
- 6.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–61. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 7.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, Rello-Varona S, Tailler M, Menger L, Vacchelli E, Galluzzi L, Ghiringhelli F, di Virgilio F, Zitvogel L, Kroemer G. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–7. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- 9.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–37. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasmahapatra G, Almenara JA, Grant S. Flavopiridol and histone deacetylase inhibitors promote mitochondrial injury and cell death in human leukemia cells that overexpress Bcl-2. Mol Pharmacol. 2006;69:288–98. doi: 10.1124/mol.105.016154. [DOI] [PubMed] [Google Scholar]
- 11.Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–98. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills KR, Reginato M, Debnath J, Queenan B, Brugge JS. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc Natl Acad Sci U S A. 2004;101:3438–43. doi: 10.1073/pnas.0400443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian W, Liu J, Jin J, Ni W, Xu W. Arsenic trioxide induces not only apoptosis but also autophagic cell death in leukemia cell lines via up-regulation of Beclin-1. Leuk Res. 2007;31:329–39. doi: 10.1016/j.leukres.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 15.Basciani S, Vona R, Matarrese P, Ascione B, Mariani S, Cauda R, Gnessi L, Malorni W, Straface E, Lucia MB. Imatinib interferes with survival of multi drug resistant Kaposi’s sarcoma cells. FEBS Lett. 2007;581:5897–903. doi: 10.1016/j.febslet.2007.11.069. [DOI] [PubMed] [Google Scholar]
- 16.Van Doorn WG, Beers EP, Dangl JL, Franklin-Tong VE, Gallois P, Hara-Nishimura I, Jones AM, Kawai-Yamada M, Lam E, Mundy J, Mur LA, Petersen M, Smertenko A, Taliansky M, Van Breusegem F, Wolpert T, Woltering E, Zhivotovsky B, Bozhkov PV. Morphological classification of plant cell deaths. Cell Death Differ. 2011;18:1241–6. doi: 10.1038/cdd.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Q, Tashiro S, Onodera S, Minami M, Ikejima T. Autophagy preceded apoptosis in oridonin-treated human breast cancer MCF-7 cells. Biol Pharm Bull. 2007;30:859–64. doi: 10.1248/bpb.30.859. [DOI] [PubMed] [Google Scholar]
- 18.Criollo A, Dessen P, Kroemer G. DRAM: a phylogenetically ancient regulator of autophagy. Cell Cycle. 2009;8:2319–20. doi: 10.4161/cc.8.15.9153. [DOI] [PubMed] [Google Scholar]
- 19.Bauvy C, Gane P, Arico S, Codogno P, Ogier-Denis E. Autophagy delays sulindac sulfide-induced apoptosis in the human intestinal colon cancer cell line HT-29. Exp Cell Res. 2001;268:139–49. doi: 10.1006/excr.2001.5285. [DOI] [PubMed] [Google Scholar]
- 20.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–10. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 21.Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281:1305–8. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura K, Bossy-Wetzel E, Burns K, Fadel MP, Lozyk M, Goping IS, Opas M, Bleackley RC, Green DR, Michalak M. Changes in endoplasmic reticulum luminal environment affect cell sensitivity to apoptosis. J Cell Biol. 2000;150:731–40. doi: 10.1083/jcb.150.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]