Abstract
This study aimed to explore the role and mechanism of lncRNA small nucleolar RNA host gene 12 (SNHG12) in the development of prostate cancer (PCa). The expression of SNHG12 in the serum of PCa patients as well as PCa cells was determined, and then we investigated whether SNHG12 could act as a competing endogenous RNA (ceRNA) to mediate the development of PCa. Furthermore, the association between SNHG12 and activation of the PI3K/AKT/mTOR pathway was explored. SNHG12 expression was up-regulated in the serum of PCa patients as well as PCa cells. High expression of SNHG12 resulted in a poor prognosis of PCa patients. Moreover, suppression of SNHG12 inhibited viability and promoted apoptosis and autophagy of LNCaP cells. Furthermore, SNHG12 was found to act as a ceRNA to regulate the expression of Cyclin E1 (CCNE1) by sponging miR-195. Lastly, suppression of SNHG12 inhibited the activation of PI3K/AKT/mTOR pathway. Our results revealed that up-regulation of SNHG12 promoted the viability and inhibited apoptosis and autophagy of PCa cells by regulating CCNE1 expression by sponging miR-195. Moreover, activation of PI3K/AKT/mTOR pathway is a key downstream mechanism regulating SNHG12-mediated the development of PCa. Our findings provide an experimental basis for targeted therapy of PCa.
Keywords: Prostate cancer, competing endogenous RNA, SNHG12, miR-195
Introduction
Prostate cancer (PCa) is one of the most prevalent cancers among men worldwide [1]. The incidence and morbidity of PCa have been increasing rapidly in the past decade in China [2]. Moreover, a high proportion (13.3%-26%) of bone metastasis is revealed in Chinese patients with PCa in the initial diagnosis [3-5], resulting in the poor prognosis of PCa. Despite extensive efforts, rare biomarkers for PCa have been introduced in clinical practice. Therefore, exploring effective biomarkers for the PCa diagnosis has a great significance.
Long noncoding RNAs (lncRNAs) are a class of non-coding transcripts longer than 200 nucleotides. Aberrant expression of lncRNAs is widely involved in the physiological and pathological processes of various human cancers [6,7], including PCa [8,9]. Recently, lncRNA small nucleolar RNA host gene 12 (SNHG12) is reported to promote the proliferation and migration of human osteosarcoma cells by upregulating angiomotin gene expression [10]. Lan et al. demonstrated that SNHG12 could promote tumorigenesis and metastasis hepatocellular carcinoma through functioning as an endogenous sponge for miR-199a/b-5p to target MLK3 expression [11]. In addition to these studies, SNHG12 is also identified to be involved in the pathogenesis of various cancers, including non-small cell lung cancer [12], nasopharyngeal carcinoma [13], gastric cancer [14], papillary thyroid carcinoma [15], and cervical cancer [16]. However, whether SNHG12 plays a key role in PCa development has not been reported, let alone the underlying mechanism.
Growing evidence has supported that lncRNAs can function as competing endogenous RNAs (ceRNAs) to sponge miRNAs, thus playing a regulatory role in many diseases including cancers [17-19]. In this study, we detected the expression of SNHG12 in the serum of PCa patients as well as PCa cells, and then investigated whether SNHG12 could act as a ceRNA to mediate the development of PCa. Furthermore, the association between SNHG12 and activation of PI3K/AKT/mTOR pathway was explored. Our findings will help to the deepen understanding of the key mechanism underlying PCa.
Materials and methods
Patients
This study was approved by the ethics committee of our hospital. Prior to the study, all patients gave informed consent for research.
From March 2014 to March 2018, 56 PCa patients with the average age of 60.3 ± 6.8 years old and 45 patients with benign prostatic hyperplasia (BPH) with the average age of 59.9 ± 8.8 years old were enrolled. All the PCa patients were diagnosed with adenocarcinoma of the prostate and their grade was assessed according to Gleason score. The patients received any treatment before recruitment and had other diseases, such as diabetes, cardiovascular disease, kidney disease or various malignancies were excluded. The clinical features of PCa patients were collected, including age, Gleason score, clinical stage, bone metastasis, disease recurrence and serum PSA. Meanwhile, 37 healthy volunteers with average age of 58.4 ± 8.5 years old were recruited as the control group. There was no statistical difference in the age of the three groups.
Peripheral venous blood samples were collected from subjects who had underwent over 10 h fasting and 8 h water deprivation. Moreover, all enrolled subjects were prohibited heavy drinking and greasy and high-protein food within 3 days prior to drawing blood. After centrifugation with a rate of 4500 rpm/h for 15 min, blood were collected and then kept at -80°C.
All PCa patients received follow-ups. The last time of follow-up was May 1st, 2018. The survival time of patients was defined from the operation date to the death date. The patients died from tumor metastasis and recurrences excluding other severe diseases and accidental death. If the patients were still alive at the last time of follow-up, the data was considered as censored data. The lost patients were processed as the last time of encounter data. Patients’ prognosis was assessed according to overall survival.
Cell lines
Human PCa cell lines, including 22RV1, Du145, LNCaP, MDaPCa2b, and a non-malignant cell line (RWPE1) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and then cultured according to ATCC protocol.
Transient transfection
SiRNA for SNHG12 (si-SNHG12#1 or si-SNHG12#2) and that for Cyclin E1 (CCNE1) (100 nM, GenePharma, China) was introduced into LNCaP cells to suppress the expression of SNHG12 and CCNE1, respectively; for overexpression of SNHG12 and CCNE1, the sequence of SNHG12 and CCNE1 was respectively inserted into the plasmid pcDNA3.1 (GenePharma) to construct the overexpression vector pcDNA-SNHG12 or pcDNA-CCNE1. Si-NC was the negative control for si-SNHG12 and empty vector pcDNA3.1 was the negative control for pcDNA-SNHG12. The miR-195 mimic (50 nM), mimic control (50 nM), and miR-195 inhibitor (150 nM), and inhibitor NC (150 nM) (GenePharma) were also introduced into LNCaP cells to regulate the expression of miR-195. Cell transfection was then performed using Lipofectamine® 2000 (Invitrogen, Carlsbad, CA, USA) that was diluted into Opti-MEM (Reduced Serum Media, Gibico). After 48 h of transfection, cells were harvested.
Real-time quantitative PCR (qPCR)
Total RNA was extracted from patients’ serum and cells using Trizol reagent (Invitrogen), followed by performing the reverse-transcription reactions using an M-MLV Reverse Transcriptase kit (Invitrogen). Subsequently, real-time qPCR was carried out following a standard protocol of SYBR Green PCR kit (Toyobo, Osaka, Japan) on a thermocycler (Rotor-Gene RG-3000A; Corbett Life Science, Sidney, Australia). The expression levels of miRNAs and RNAs were respectively normalized to U6 and β-actin, and quantitated by 2-ΔΔCt method.
Dual-luciferase reporter assay and RNA binding protein immunoprecipitation (RIP)
The pMIR-REPORT-SNHG12-wt/mut and pMIR-REPORT-CCNE1-3’UTR-wt/mut (Obio, Shanghai, China) were constructed and then transfected into LNCaP cells, along with miR-195 mimic or mimic control. After 48 h of transfection, luciferase activity of each group was detected using Dual-Luciferase Reporter Assay System (E1910, Promega, WI, USA).
RIP assay was carried out following the manufacturer’s instructions of RNA-Binding Protein Immunoprecipitation Kit (17e700, Millipore, USA). Notably, AntiAgo2 (Millipore, USA) was applied to the enrichment of SNHG12 and miR-195. Normal mouse Anti-IgG (Millipore, USA) served as a negative control.
Analysis of cell viability by MTT assay
Approximately 2×103 LNCaP cells were plated into a 96-well plate. At various times following treatments, MTT (20 μl of 5 mg/mL, Sigma-Aldrich, St. Louis, USA) was added into each well to incubate cells for 4 h at 37°C. Subsequently, 150 μl of dimethyl sulfoxide was added to dissolve the formazan precipitates. The absorbance (470 nm) of each well was measured with a MRX II absorbance reader (DYNEX Technologies, Chantilly, Virginia, USA).
Detection of apoptotic cells by flow cytometry
At various times following treatments, the cells were harvested and resuspended in 100 μl binding buffer at a concentration of 1×106 cells/ml. According to the manufacturer’s instructions of Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA), cells were then double-stained with Annexin V and PI. The apoptotic cells were then detected by the BD LSRII Flow Cytometer System with FACSDiva Software within 1 h.
Western blot
The LNCaP cells were collected and the total protein was extracted by lysing with cell lysis buffer (Beyotime, Haimen, China). The proteins (30 μg per lane) were separated on 12% SDS-polyacrylamide gels and subsequently transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The primary antibodies to pro-caspase-3, cleaved-caspase-3, Bax, Bcl-2, pro-caspase-9, cleaved-caspase-9, p62, Beclin-1, LC3II, LC3I, CCNE1, PI3K, p-PI3K, AKT, p-AKT, PTEN, mTOR, p-mTOR and β-actin (Abcam, Cambridge, UK) were diluted in 1:1000 and then used for incubating LNCaP cells overnight at 4°C. β-actin was used as the control. After incubation with the recommended secondary antibodies, the protein signals were revealed using an Odyssey Infrared Imaging System (LI-COR, Lincoln, NE, USA).
Statistical analysis
The statistical analysis was performed using SPSS 20.0 (SPSS Inc, Chicago, IL, USA). The measurement data were expressed by mean ± standard deviation. The comparison between two groups was carried out by Student’s t test, while the multi-group comparisons were analyzed by one-way ANOVA and following LSD-t test. The survival analysis performed by Kaplan-Meier method and Log-rank test was adopted to compare the differences between high- and low-SNHG12 expression levels. P < 0.05 was considered significant with a two-sided test.
Results
The expression levels of SNHG12 in PCa, BPH and healthy group
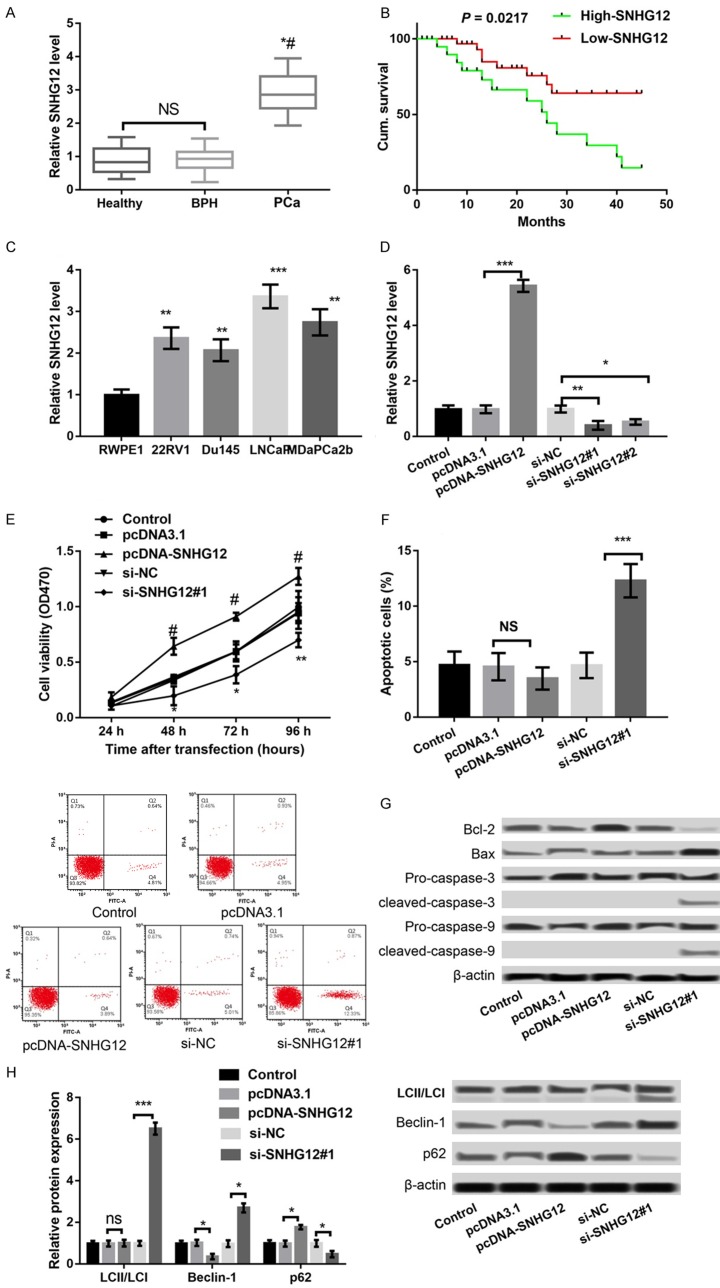
To investigate the role of SNHG12 in PCa, we detected the expression levels of SNHG12 in the serum of PCa patients, BPH patients, and healthy controls. As shown in Figure 1A, SNHG12 expression in the serum of PCa patients was significantly higher than that in BPH patients or healthy controls (P < 0.05), and there was no significant difference in SNHG12 expression between BPH patients and healthy controls. According to the mean expression levels of SNHG12, 34 of the 56 PCa patients were classified into the high expression group and the remaining 22 were in the low expression group. Further survival analysis revealed that the overall survival of the low-SNHG12 expression group was significantly higher than that in the high-SNHG12 expression group (P = 0.0217), indicating that the poor prognosis of PCa patients was associated with the high expression of SNHG12 (Figure 1B). Furthermore, the correlation of SNHG12 expression with clinical features of PCa patients was further explored. As shown in Table 1, SNHG12 expression was significantly correlated to the Gleason score, clinical stage, bone metastasis, disease recurrence and serum PSA (P < 0.01), but had nothing to do with patients’ age.
Figure 1.
LncRNA SNHG12 was up-regulated in the serum of prostate cancer (PCa) patients and correlated to the poor prognosis of PCa patients. A: qPCR showed the expression of SNHG12 in the serum of PCa patients, BPH patients, and healthy controls. *P < 0.05 compared to healthy controls, and #P < 0.05 compared to BPH patients. B: Survival analysis revealed the correlation between SNHG12 expression and the poor prognosis of PCa patients. C-H. LncRNA SNHG12 was up-regulated in PCa cells and dysregulation of SNHG12 regulated LNCaP cell viability, apoptosis and autophagy. C: qPCR showed the expression of SNHG12 in four PCa cell lines, including 22RV1, Du145, LNCaP, MDaPCa2b, as well as in a non-malignant cell line RWPE1. D: qPCR showed the expression of SNHG12 in LNCaP cells after transfection with pcDNA-SNHG12, pcDNA3.1, si-SNHG12#1, si-SNHG12#2 and si-NC. E: MTT assay showed LNCaP cell viability in different transfected groups. F: Flow cytometry showed LNCaP cell apoptosis in different transfected groups. G: Western blot showed the expression of apoptosis-related proteins in different transfected groups. H: Western blot showed the expression of autophagy markers in different transfected groups. *P < 0.05, **P < 0.01, and ***P < 0.001 compared to their respective controls.
Table 1.
The association between SNHG12 expression and clinicopathological characteristics of prostate cancer patients
| Characteristics | Cases (N=56) | SNHG12 expression | P value | ||
|---|---|---|---|---|---|
|
| |||||
| Low (N=34) | High (N=22) | ||||
| Age | < 60 | 16 | 9 | 7 | 0.235 |
| ≥ 60 | 40 | 25 | 15 | ||
| Gleason score | ≤ 7 | 19 | 16 | 3 | 0.004 |
| > 7 | 37 | 18 | 19 | ||
| Clinical stage | A | 6 | 5 | 1 | < 0.001 |
| B | 8 | 5 | 3 | ||
| C | 28 | 19 | 9 | ||
| D | 14 | 5 | 9 | ||
| Bone metastasis | Yes | 14 | 3 | 11 | <0.001 |
| No | 42 | 31 | 11 | ||
| Recurrence | Yes | 16 | 8 | 8 | < 0.001 |
| No | 40 | 26 | 14 | ||
| Serum PSA (μg/L) | ≤ 10 | 13 | 10 | 3 | < 0.001 |
| > 10 | 43 | 24 | 19 | ||
Effects of SNHG12 expression on cell viability, apoptosis and autophagy
We further determined the expression of SNHG12 in PCa cells. The results showed that SNHG12 expression was significantly increased in four PCa cell lines, including 22RV1, Du145, LNCaP, MDaPCa2b, in relation to that in a non-malignant cell line RWPE1 (P < 0.01, Figure 1C). LNCaP cells were selected for subsequent experiments due to the highest expression levels of SNHG12 among the four PCa cell lines. Subsequently, SNHG12 expression was markedly overexpressed in pcDNA-SNHG12-transfected LNCaP cells relative to that in pcDNA3.1-transfected LNCaP cells (P < 0.001), and dramatically suppressed in both si-SNHG12#1- and si-SNHG12#2-transfected LNCaP cells compared to that in si-NC-transfected LNCaP cells (P < 0.05) (Figure 1D). Because the inhibitory effects of si-SNHG12#1 was stronger than si-SNHG12#2, si-SNHG12#1 was used for following experiments. Subsequently, the results of MTT assay showed that overexpression of SNHG12 significantly promoted LNCaP cell viability, whereas suppression of SNHG12 had an opposite effect (P < 0.05, Figure 1E). The results of flow cytometry disclosed that suppression of SNHG12 significantly promoted the apoptosis of LNCaP cells (P < 0.01, Figure 1F); consistently, suppression of SNHG12 resulted in the increased expression of Bax/Bcl-2 ratio, cleaved/pro-caspase-3 and cleaved/pro-caspase-9 (Figure 1G); however, overexpression of SNHG12 did not exhibit opposite effects on cell apoptosis and the expression of apoptosis-related proteins. Furthermore, the expression levels of autophagy markers were detected. The results showed that suppression of SNHG12 obviously promoted the expression of Bclin-1 and LC3II/LC3I but markedly decreased the p62 expression (P < 0.05), indicating that suppression of SNHG12 promoted cell autophagy (Figure 1H). However, overexpression of SNHG12 had opposite effects on the expression of Bclin-1 and p62 (P < 0.05) (Figure 1H).
Association between SNHG12 and miR-195 in LNCaP cells
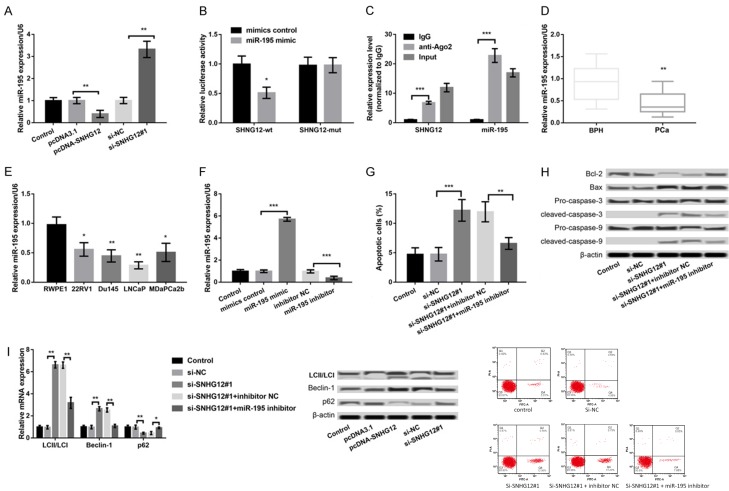
In previous studies, SNHG12 has been shown to regulate the tumorigenesis in glioma and osteosarcoma through sponging miR-195 [20,21]. Therefore, we also investigated the association between SNHG12 and miR-195 in LNCaP cells. As presented in Figure 2A, the expression of miR-195 in pcDNA-SNHG12-transfected LNCaP cells was remarkably lower than that in pcDNA3.1-transfected LNCaP cells (P < 0.01), while the miR-195 expression in si-SNHG12#1-transfected LNCaP cells was obviously higher than that in si-NC-transfected LNCaP cells (P < 0.01). These data indicated a negative regulatory relationship between SNHG12 and miR-195. In addition, the results of dual-luciferase reporter assay showed that the luciferase activity of SNHG12-wt but not SNHG12-mut was significantly decreased after co-transfection with miR-195 mimic (P < 0.05, Figure 2B). Furthermore, the results of RIP confirmed that SNHG12 and miR-195 were in the same RNA-induced silencing complex (RISC) (Figure 2C).
Figure 2.
Association between SNHG12 and miR-195 in LNCaP cells. A: qPCR showed the expression of miR-195 in LNCaP cells after transfection with pcDNA-SNHG12, pcDNA3.1, si-SNHG12#1, and si-NC. B: Dual-luciferase reporter assay showed the luciferase activity of SNHG12-wt and SNHG12-mut after co-transfection with miR-195 mimic or mimic control. C: RIP confirmed that SNHG12 and miR-195 were in the same RNA-induced silencing complex. D-I. SNHG12 regulated apoptosis and autophagy of LNCaP cells through sponging miR-195. D: qPCR showed the expression of miR-195 in PCa and BPH patients. E: qPCR showed the expression of miR-195 in four PCa cell lines and a non-malignant cell line RWPE1. F: qPCR showed the expression of SNHG12 in LNCaP cells after transfection with miR-195 mimic, mimic control, miR-195 inhibitor, and inhibitor control. G: Flow cytometry showed LNCaP cell apoptosis in different transfected groups. H: Western blot showed the expression of apoptosis-related proteins in different transfected groups. I: qPCR and western blot showed the expression of autophagy markers in different transfected groups. *P < 0.05, **P < 0.01, and ***P < 0.001 compared to their respective controls.
SNHG12 regulated apoptosis and autophagy of LNCaP cells through sponging miR-195
The expression levels of miR-195 in the serum of PCa patients and PCa cells were also measured. As shown in Figure 2D, miR-195 expression was significantly decreased in the serum of PCa patients compared to that in BPH patients (P < 0.01). Moreover, obvious down-regulation of SNHG12 expression was also revealed in the four PCa cell lines, including 22RV1, Du145, LNCaP, and MDaPCa2b, in comparison with that in a non-malignant cell line RWPE1 (P < 0.05, Figure 2E). The miR-195 expression was subsequently overexpressed in LNCaP cells by transfection with miR-195 mimic and inhibited by transfection with miR-195 inhibitor, and the transfection efficiency was confirmed by qPCR (P < 0.001, Figure 2F). To further reveal whether SNHG12 regulated the PCa development via regulating miR-195, LNCaP cells were co-transfected with si-SNHG12#1 and miR-195 inhibitor. The results showed that the effects of SNHG12 suppression on cell apoptosis (P < 0.01, Figure 2G) and the expression of apoptosis-related proteins (Figure 2H) and autophagy-markers (Figure 2I) in LNCaP cells were significantly reversed after suppression of SNHG12 and inhibition of miR-195 at the same time.
CCNE1 was verified as a functional target of miR-195
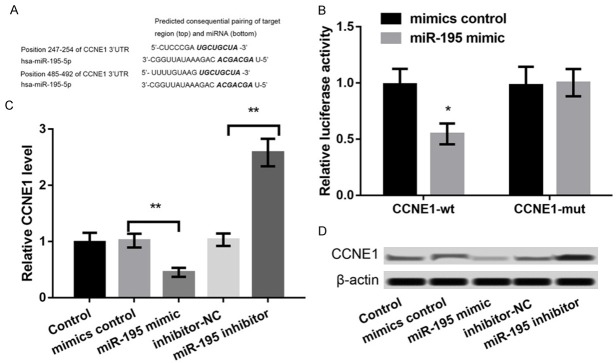
To explore the regulatory mechanism of miR-195, the targets of miR-195 were predicted. Based on the information of TargetScan software, CCNE1 was identified as a potential target of miR-195 (Figure 3A). The results of dual-luciferase reporter assay showed that the luciferase activity of CCNE1-wt but not CCNE1-mut was remarkably inhibited after co-transfection with miR-195 mimic (P < 0.05, Figure 3B). Furthermore, the expression of CCNE1 in miR-195 mimic-transfected LNCaP cells was markedly down-regulated in relation to that in mimic NC-transfected LNCaP cells (P < 0.01), while the CCNE1 expression in miR-195 inhibitor-transfected LNCaP cells was up-regulated compared to that in inhibitor-NC-transfected LNCaP cells (P < 0.01) (Figure 3C, 3D). These data indicated that CCNE1 was negatively regulated by miR-195.
Figure 3.
CCNE1 was verified as a functional target of miR-195. A: TargetScan software predicted the binding sequence of CCNE1 and miR-195. B: Dual-luciferase reporter assay showed that the luciferase activity of CCNE1-wt and CCNE1-mut co-transfection with miR-195 mimic or mimic control. C, D: qPCR and western blot showed the expression of CCNE1 in LNCaP cells after transfection with miR-195 mimic, mimic control, miR-195 inhibitor, and inhibitor control. *P < 0.05, and **P < 0.01 compared to their respective controls.
miR-195 regulated the apoptosis and autophagy of LNCaP cells by targeting CCNE1
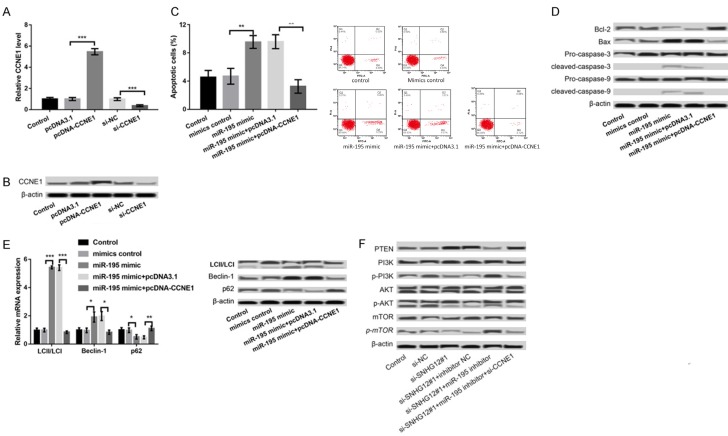
The expression of CCNE1 was further overexpressed and suppressed in LNCaP cells. As presented in Figure 4A, 4B, the expression of CCNE1 was significantly increased in pcDNA-CCNE1 group compared to that in pcDNA3.1 group (P < 0.001) and dramatically decreased in si-CCNE1 group relative to that in si-NC group (P < 0.001), indicating that the transfection efficiency was high. In addition, we found that overexpression of miR-195 promoted the apoptosis (P < 0.01, Figure 4C, 4D), and autophagy (Figure 4E) of LNCaP cells, which were significantly reversed after overexpression of miR-195 and CCNE1 concurrently (Figure 4C-E).
Figure 4.
miR-195 regulated the apoptosis and autophagy of LNCaP cells by targeting CCNE1. A, B: qPCR and western blot showed the expression of CCNE1 in LNCaP cells after transfection with pcDNA-CCNE1, pcDNA3.1, si-CCNE1, and si-NC. C: Flow cytometry showed LNCaP cell apoptosis in different transfected groups. D: Western blot showed the expression of apoptosis-related proteins in different transfected groups. E: qPCR and western blot showed the expression of autophagy markers in different transfected groups. F: The expression of PI3K/AKT/mTOR pathway-related proteins after suppression of SNHG12, miR-195 and/or CCNE1. *P < 0.05, **P < 0.01, and ***P < 0.001 compared to their respective controls.
The association of SNHG12/miR-195/CCNE1 axis and activation of PI3K/AKT/mTOR pathway
To explore the downstream regulatory mechanism of SNHG12/miR-195/CCNE1 axis, the expression of PI3K/AKT/mTOR pathway-related proteins was explored after dysregulation of SNHG12, miR-195, and/or CCNE1. The results showed that suppression of SNHG12 resulted in the increased expression of PTEN and the decreased expression of p-PI3K, p-AKT, and p-mTOR, indicating that suppression of SNHG12 inhibited the activation of PI3K/AKT/mTOR pathway (Figure 4F). Moreover, concurrent suppression of SNHG12 and miR-195 could reverse the inhibitory effect of suppression of SNHG12 alone on the activation of PI3K/AKT/mTOR pathway, which was further reversed after suppression of SNHG12, miR-195 and CCNE1 at the same time (Figure 4F).
Discussion
LncRNAs are shown to participate in a wide range of cellular and pathologic processes. Especially in PCa, a growing number of lncRNAs have been identified to regulate tumor occurrence and progression, such as MALAT1 [22], GAS5 [23], NEAT1 [24], and PCAT-1 [25]. In the present study, we found that SNHG12 expression was up-regulated in the serum of PCa patients as well as PCa cells, and suppression of SNHG12 inhibited viability and promoted apoptosis and autophagy of LNCaP cells. Moreover, SNHG12 was found to act as a ceRNA to regulate CCNE1 expression by sponging miR-195. Furthermore, suppression of SNHG12 inhibited the activation of PI3K/AKT/mTOR pathway.
Consistent with previous findings [15,16], we also found that SNHG12 expression was up-regulated in the serum of PCa patients as well as PCa cells, and the high expression of SNHG12 is correlated to the poor prognosis of PCa patients. Moreover, suppression of SNHG12 promoted apoptosis and autophagy of LNCaP cells. Apoptosis is a programmed cell death, which maintains the balance of survival and death in living organisms [26,27]. Autophagy is also an evolutionarily conserved catabolic process that plays either pro-death or pro-survival role in living organisms according to the cell type and strength of specific stimuli [28,29]. Based on our results, we speculate that SNHG12 may play an oncogenic role in PCa. In line with previous studies [20,21], we also found that SNHG12 could sponge miR-195. It has been confirmed that miR-195 plays a tumor suppressive role in PCa cells [30]. Zhang et al. also revealed that down-regulation of miR-195 contributed to the progression of PCa [31]. Cai et al. confirmed that miR-195 could suppress PCs cell proliferation and angiogenesis [32]. In this study, there was a negative regulatory relationship between SNHG12 and miR-195, suggesting that SNHG12 may contribute to PCa via negative regulation of miR-195. Furthermore, CCNE1 was verified as a functional target of miR-195. A previous study has shown that overexpression of CCNE1 is related to poor survival in ovarian cancer and CCNE1-targeted therapy may be a promising strategy for ovarian cancer patients with CCNE1 amplification [33]. CCNE1 is also confirmed to play an oncogenic role in other cancers [34,35]. Our results showed that miR-195 regulated the apoptosis and autophagy of LNCaP cells by targeting CCNE1. Although the role of CCNE1 in PCa has not been fully investigated, our results suggest that CCNE1 may be a downstream target of SNHG12/miR-195 to mediate the development of PCa.
Furthermore, PI3K/AKT/mTOR pathway is widely implicated in the regulation of cellular processes, such as cell proliferation and apoptosis [36,37]. PI3K/AKT/mTOR signaling is found to be upregulated in 30-50% of PCa and deregulation of the PI3K pathway is increasingly involved in the carcinogenesis of PCa [38,39]. In addition, PI3K/AKT/mTOR pathway inhibitors are shown to have an ability of enhancing radiosensitivity in radioresistant prostate cancer cells via regulating apoptosis and autophagy [40]. In this study, suppression of SNHG12 inhibited the activation of PI3K/AKT/mTOR pathway, which was reversed by concurrent suppression of SNHG12 and miR-195. Based on the important role of PI3K/Akt/mTOR pathway in PCa, we speculate that PI3K/Akt/mTOR pathway is a downstream mechanism of SNHG12 in regulating PCa development.
In conclusion, our results revealed that up-regulation of SNHG12 promoted the viability and inhibited apoptosis and autophagy of PCa cells via regulating CCNE1 expression by sponging miR-195. Moreover, activation of PI3K/AKT/mTOR pathway is a key downstream mechanism regulating SNHG12-mediated the development of PCa. Our findings provide an experimental basis for targeted therapy of PCa.
Acknowledgements
This work was supported by National Natural Science Foundation of China, Grant No: 81472454 and 81572082, and Medical and Bethune program of Jilin University, Grant No: 2015318, and the department of science and technology agency project of Jinlin Province, Grant No: 2150414015GH.
Disclosure of conflict of interest
None.
References
- 1.Kgatle MM, Kalla AA, Islam MM, Sathekge M, Moorad R. Prostate cancer: epigenetic alterations, risk factors, and therapy. Prostate Cancer. 2016;2016:5653862. doi: 10.1155/2016/5653862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27:2–12. doi: 10.3978/j.issn.1000-9604.2015.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yiwei W, Jianming G, Lei X, Naiqing Z, Zhibing X, Hang W, Yanjun Z, Shuai J, Nianqin Y, Yuanfeng Y. Should bone scan be performed in Chinese prostate cancer patients at the time of diagnosis? Urol Int. 2013;91:160–4. doi: 10.1159/000348330. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Dhakal IB, Zhao Z, Li L. Trends in mortality from cancers of the breast, colon, prostate, esophagus, and stomach in East Asia: role of nutrition transition. Eur J Cancer Prev. 2012;21:480–9. doi: 10.1097/CEJ.0b013e328351c732. [DOI] [PubMed] [Google Scholar]
- 6.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 7.Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer. 2016;15:39. doi: 10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitobe Y, Takayama KI, Horie-Inoue K, Inoue S. Prostate cancer-associated lncRNAs. Cancer Lett. 2018;418:159–166. doi: 10.1016/j.canlet.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Smolle MA, Bauernhofer T, Pummer K, Calin GA, Pichler M. Current insights into long non-coding RNAs (LncRNAs) in prostate cancer. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18020473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruan W, Wang P, Feng S, Xue Y, Li Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumour Biol. 2016;37:4065–73. doi: 10.1007/s13277-015-4256-7. [DOI] [PubMed] [Google Scholar]
- 11.Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. 2017;36:11. doi: 10.1186/s13046-016-0486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. 2017;8:84086. doi: 10.18632/oncotarget.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu ZB, Tang C, Jin X, Liu SH, Wen W. Increased expression of lncRNA SNHG12 predicts a poor prognosis of nasopharyngeal carcinoma and regulates cell proliferation and metastasis by modulating Notch signal pathway. Cancer Biomark. 2018;23:603–613. doi: 10.3233/CBM-181873. [DOI] [PubMed] [Google Scholar]
- 14.Yang B, Cai W, Chen B. LncRNA SNHG12 regulated the proliferation of gastric carcinoma cell BGC-823 by targeting microRNA-199a/b-5p. Eur Rev Med Pharmacol Sci. 2018;22:1297–1306. doi: 10.26355/eurrev_201803_14471. [DOI] [PubMed] [Google Scholar]
- 15.Ding S, Qu W, Jiao Y, Zhang J, Zhang C, Dang S. LncRNA SNHG12 promotes the proliferation and metastasis of papillary thyroid carcinoma cells through regulating wnt/β-catenin signaling pathway. Cancer Biomark. 2018;22:217–226. doi: 10.3233/CBM-170777. [DOI] [PubMed] [Google Scholar]
- 16.Jin XJ, Chen XJ, Zhang ZF, Hu WS, Ou RY, Li S, Xue JS, Chen LL, Hu Y, Zhu H. Long noncoding RNA SNHG12 promotes the progression of cervical cancer via modulating miR-125b/STAT3 axis. J Cell Physiol. 2019;234:6624–6632. doi: 10.1002/jcp.27403. [DOI] [PubMed] [Google Scholar]
- 17.Huang G, Wu X, Li S, Xu X, Zhu H, Chen X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Scientific Reports. 2016;6:26524. doi: 10.1038/srep26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Mejias A, Tay Y. Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J Hematol Oncol. 2015;8:30. doi: 10.1186/s13045-015-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kartha RV, Subbaya S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Zheng J, Xue Y, Qu C, Chen J, Wang Z, Li Z, Zhang L, Liu Y. Inhibition of TDP43-mediated SNHG12-miR-195-SOX5 feedback loop impeded malignant biological behaviors of glioma cells. Mol Ther Nucleic Acids. 2018;10:142–158. doi: 10.1016/j.omtn.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. 2018;495:1822. doi: 10.1016/j.bbrc.2017.12.047. [DOI] [PubMed] [Google Scholar]
- 22.Wang D, Ding L, Wang L, Zhao Y, Sun Z, Karnes RJ, Zhang J, Huang H. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. 2015;6:41045. doi: 10.18632/oncotarget.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pickard M, Mourtada-Maarabouni M, Williams G. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832:1613–23. doi: 10.1016/j.bbadis.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, Evans JR, Knudsen KE, Paulsen MT, Ljungman M. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassan M, Watari H, Abualmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014:150845. doi: 10.1155/2014/150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Sankari SL, Masthan KM, Babu NA, Bhattacharjee T, Elumalai M. Apoptosis in cancer--an update. Asian Pac J Cancer Prev. 2012;13:4873–8. doi: 10.7314/apjcp.2012.13.10.4873. [DOI] [PubMed] [Google Scholar]
- 28.Maycotte P, Thorburn A. Autophagy and cancer therapy. Cancer Biol Ther. 2011;11:127–37. doi: 10.4161/cbt.11.2.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janku F, Mcconkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–39. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 30.Cai C, Chen QB, Han ZD, Zhang YQ, He HC, Chen JH, Chen YR, Yang SB, Wu YD, Zeng YR, Qin GQ, Liang YX, Dai QS, Jiang FN, Wu SL, Zeng GH, Zhong WD, Wu CL. miR-195 inhibits tumor progression by targeting RPS6KB1 in human prostate cancer. Clin Cancer Res. 2015;21:4922–34. doi: 10.1158/1078-0432.CCR-15-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Tao T, Liu C, Guan H, Huang Y, Xu B, Chen M. Downregulation of miR-195 promotes prostate cancer progression by targeting HMGA1. Oncol Rep. 2016;36:376–82. doi: 10.3892/or.2016.4797. [DOI] [PubMed] [Google Scholar]
- 32.Cai C, He H, Duan X, Wu W, Mai Z, Zhang T, Fan J, Deng T, Zhong W, Liu Y. miR-195 inhibits cell proliferation and angiogenesis in human prostate cancer by downregulating PRR11 expression. Oncol Rep. 2018;39:1658–1670. doi: 10.3892/or.2018.6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama N, Nakayama K, Shamima Y, Ishikawa M, Katagiri A, Iida K, Miyazaki K. Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer. 2010;116:2621–34. doi: 10.1002/cncr.24987. [DOI] [PubMed] [Google Scholar]
- 34.Matsushita R, Seki N, Chiyomaru T, Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S, Itesako T. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br J Cancer. 2015;113:282–9. doi: 10.1038/bjc.2015.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Hu S, Zhang X, Wang L, Zhang X, Yan B, Zhao J, Yang A, Zhang R. MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2014;443:1078–84. doi: 10.1016/j.bbrc.2013.12.095. [DOI] [PubMed] [Google Scholar]
- 36.Karar J, Maity A. PI3K/AKT/mTOR pathway in angiogenesis. Front Mol Neurosci. 2011;4:51. doi: 10.3389/fnmol.2011.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang BH, Liu LZ. PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochim Biophys Acta. 2008;1784:150–158. doi: 10.1016/j.bbapap.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Sarker D, Reid AH, Yap TA, de Bono JS. Targeting the PI3K/AKT pathway for the treatment of prostate cancer. Clin Cancer Res. 2009;15:4799–4805. doi: 10.1158/1078-0432.CCR-08-0125. [DOI] [PubMed] [Google Scholar]
- 39.Morgan TM, Koreckij TD, Corey E. Targeted therapy for advanced prostate cancer: inhibition of the PI3K/Akt/mTOR pathway. Curr Cancer Drug Targets. 2009;9:237–49. doi: 10.2174/156800909787580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang L, Graham PH, Hao J, Ni J, Bucci J, Cozzi PJ, Kearsley JH, Li Y. PI3K/Akt/mTOR pathway inhibitors enhance radiosensitivity in radioresistant prostate cancer cells through inducing apoptosis, reducing autophagy, suppressing NHEJ and HR repair pathways. Cell Death Dis. 2014;5:e1437. doi: 10.1038/cddis.2014.415. [DOI] [PMC free article] [PubMed] [Google Scholar]