Abstract
Background: Hepatocellular carcinoma (HCC) is the fifth most frequently diagnosed cancer worldwide and the second most frequent cause of cancer death. The aim of this study is to investigate the early diagnostic value of a panel of peripheral blood exosomal micro-RNAs (miRNAs) in patients with HCC compared with patients with Hepatitis B virus (HBV) and hepatocirrhosis. Patients and methods: Blood samples from 72 patients with HCC, 72 patients with hepatocirrhosis and 72 patients with HBV were obtained at Beijing Friendship Hospital, Capital Medical University. The miRNA expression levels were detected by real-time polymerase chain reaction (RT-PCR). Areas under curve (AUCs) were used to compare diagnostic values of plasmic and exosomal miRNAs. Results: We screened plasmic and exosomal solutions of 3 HCC, 3 cirrhosis and 3 HBV patients to perform miRNA microarray analysis. Three distinctly differential microRNAs including miRNA-26a, miRNA-29c, and miRNA-21 were selected to perform further evaluation. First, we found that the expressions of miRNA-26a, miRNA-29c, and miRNA-21 were significantly lower in patients with HCC compared with cirrhotic and HBV group in both exosomes and plasma. Second, we found miRNA-26a, miRNA-29c, and miRNA-21 were significantly down-regulated in HCC tumor tissues compared with normal tissues. Thirdly, we found miRNAs in exosomes had better diagnostic value for patients with HCC compared with plasmic miRNAs among different groups. Conclusions: In conclusion, we found that the expression of miRNA-26a, miRNA-29c, and miRNA-21 were significantly lower in patients with HCC, and we confirmed miRNA-26a, miRNA-29c, and miRNA-21 could be identified as independent diagnostic biomarkers for patients with HCC.
Keywords: miRNAs, exosomes, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is a frequently diagnosed malignant tumor and the second most frequent cause of cancer death worldwide [1], with the highest incidence in Asian countries and increasing in western countries [2]. Despite application of surgical resection, chemotherapy, radiotherapy and molecular targeting therapy, tumor recurrence is common in patients with HCC. There are several factors associated with the prognosis of HCC, including the completeness of tumor resections, alpha-fetoprotein (AFP) levels, tumor size, number of tumor lesions and distant metastases [3,4]. The poor prognosis of patients with HCC is attributed to the lack of effective modalities of early diagnosis. Only 30% to 40% of patients are candidates for a potential radical liver resection at the time of diagnosis. Patients with Hepatitis B Virus (HBV) and hepatocirrhosis are much more frequent in eastern countries, especially in China [5]. Less-invasive diagnostic methods, such as fecal occult blood testing (FOBT) and serum AFP screening in blood are of limited value owning to poor sensitivity and specificity in distinguishing HCC, hepatocirrhosis, and HBV. Therefore, discovery of an effective and reliable tool distinguishing patients with HCC, hepatocirrhosis, and HBV would play a pivotal role in early diagnosis and prognostic improvement of patients with HCC.
In recent years, interest in the function of exosomes has increased, particularly in cancer research. Exosomes are small, lipid bilayer vesicles (30-100 nm) from multiple vesicles (MVBs), the luminal membrane composed of the release of the fusion with the cell membrane, provide protection including RNAs and protein membranes [6-9]. New evidence suggests that tumor-derived exosomes are involved in tumor growth, angiogenesis, tumorigenesis, tumor immune escape, resistance and metastasis [10,11].
MicroRNAs (miRNAs) are the most stable nucleic acid molecules in the body and only about 19 to 23 nucleotides act as regulating gene expression at the post-transcriptional level through complex miRNA-mRNA interactions [12]. Recently, miRNAs of serological origin have been reported as novel markers for diagnosing HCC in many studies, which provides a simple procedure for early diagnosis of HCC [13,14]. Many studies have demonstrated significant differences in miRNAs expression between HCC and para-carcinoma tissues [15-17]. Moreover, emerging evidence shows that the quantification of tumor-derived exosomes plays a decisive role in the dialogue between peripheral blood and exosome-mediated miRNA transduction of human tumors and their microenvironment [18]. Taking account to liver diseases, miRNAs derived from exosomes have already been used as non-invasive diagnostic markers for hepatitis rating and classification [19].
In the npresent study, we aim to investigate the early diagnostic value of a panel of exosomal miRNAs in patients with HCC from the peripheral blood, compared with patients with HBV and hepatocirrhosis.
Materials and methods
Patients
Blood samples from 72 patients with HCC, 72 patients with hepatocirrhosis and 72 patients with HBV were obtained at Beijing Friendship Hospital, Capital Medical University. The tissues of HCC were immediately frozen in liquid nitrogen after surgical removal and stored at -80°C until use. HCC diagnosis was based on WHO criteria. Tumor staging was determined according to the sixth edition of the tumor-node-metastasis (TNM) classification of the International Union against Cancer. The characteristics of patients are shown in Table 1. The study was approved by the Research Ethics Committee of Beijing Friendship Hospital, Capital Medical University. Informed consent was obtained from all patients.
Table 1.
Patient and tumor characteristics (N = 216)
| Variable | HBV group | Cirrhosis group | HCC group | P value |
|---|---|---|---|---|
| Case, n | 72 | 72 | 72 | |
| Age | 60.5 ± 5.8 | 61.0 ± 7.6 | 61.8 ± 6.1 | 0.321 |
| Sex | 0.854 | |||
| Female | 21 | 23 | 20 | |
| Male | 51 | 49 | 52 | |
| HBsAg | 0.001 | |||
| Positive | 72 | 46 | 53 | |
| Negative | 0 | 26 | 19 | |
| HBeAg | 0.001 | |||
| Positive | 62 | 34 | 42 | |
| Negative | 10 | 38 | 30 | |
| Liver cirrhosis | ||||
| Yes | 0 | 72 | 61 | 0.001 |
| No | 72 | 0 | 11 | |
| TBL (umol/l) | 12.5 ± 8.3 | 15.1 ± 7.3 | 16.1 ± 8.2 | 0.025 |
| ALB (g/dl) | 39.4 ± 6.6 | 38.9 ± 6.5 | 37.9 ± 4.6 | 0.016 |
| ALT (U/L) | 25.7 ± 14.1 | 50.4 ± 30.2 | 79.4 ± 66.5 | 0.001 |
| Anti-virus therapy | 0.762 | |||
| Yes | 16 | 20 | 18 | |
| No | 56 | 52 | 54 | |
| AFP at diagnosis (ng/ml) | 0.001 | |||
| ≤ 400 | 72 | 64 | 52 | |
| > 400 | 0 | 8 | 20 | |
| Tumor size (cm) | - | |||
| > 5 cm | - | - | 35 | |
| ≤ 5 cm | - | - | 37 | |
| Microvascular invasion | - | |||
| Yes | - | - | 30 | |
| No | - | - | 42 | |
| TNM staging | - | |||
| I | - | - | 20 | |
| II | - | - | 25 | |
| III-IV | - | - | 27 | |
| Metastases | - | |||
| Yes | - | - | 20 | |
| No | - | - | 52 |
Abbreviations: TBL: total bilirubin; ALB: albumin; ALT: alanine aminotransferase; AFP, alpha-fetoprotein; HBV, Hepatitis B Virus.
Cell lines and culture conditions
Human HCC cell lines (SMMC-7721, Huh-7 and Hep3B cells) were purchased from the Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). SMMC-7721, Huh-7 and Hep3B cell lines were cultured in RPMI-1640 Medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin.
RNA extraction
RNA was isolated from 200 μl serum using mirVana Paris Kit (Ambion, Austin, TX, USA) according to the manufacturer’s protocol. To allow for normalization of sample-to-sample variation in RNA extraction procedures, synthetic C. elegans miRNA cel-miR-39 (5 nM/L, 5 μl RiboBio, Guangzhou, China) was spiked into each denatured sample after combining the serum sample with denaturing solution (Ambion, Austin, TX, USA). Total RNA was extracted from specimens stored in the -80°C refrigerator using the RNA Micro Kit (Ambion, Austin, TX, USA). RNA was eluted with 100 μl of RNase-free water and stored at -80°C for further use. The ultraviolet spectrophotometer was used to evaluate the concentration and purity of the total RNA.
Extraction of exosomes from peripheral blood
Whole blood was centrifuged at 3000*g for 15 min to remove cells or cell debris, the supernatant liquid was then placed into a centrifuge tube, added with 63 μl of ExoQuick reagent per 250 μl of serum and allowed to stand at 4°C for 30 min. In a 4°C environment, the mixture was centrifuged at 1500*g for 30 min (exosomes precipitated at the bottom of the centrifuge tube). Supernatant was aspirated completely and centrifuged at 1500*g, 4°C for 5 min. Supernatant was aspirated completely (during which there should be no shaking of the centrifuge tube), completely dissolved and precipitated with 20 μl of 1 * PBS and stored at -20°C.
Transmission electron microscopy
The isolated exosome fraction was dissolved in HEPES buffer, and a drop of the suspension was placed on a sheet. A carbon-coated copper grid was floated on the drop for 10 s. The grid was then removed, and excess liquid was drained from the edge of the grid using a piece of clean filter paper. The grid was touched onto a drop of 2% uranyl acetate or phosphotungstic acid, pH 7.0, for B5s, and excess liquid was removed. The grid was allowed to dry for several minutes and was then examined using a JEM1400 microscope [20].
miRNA microarray analysis
Sample processing: RNAs were quantified with NanoDrop ND-1000, and the integrity of RNAs was examined by agarose gel electrophoresis. miRNA microarray selection: we used the Affymetrix miRNA 4.0 array, which contained 30424 (including rodent) miRNA from the latest miRBase database.
RNA labeling and hybridization: we used the FlashTag Biotin HSR RNA Labeling Kit to label the miRNA, then performed ELOSA QC assay. RNAs were eluted through the Affymetrix GeneChip 450 System, and sequentially hybridized with the Affymetrix GeneChip 645 System. Then the hybridized sequences were observed using the Affymetrix GeneChip 7G Microarray Scanner, and the observed images were input into the Affymetrix Expression Console Software. Data were expressed using the Robust Multichip Average (RMA + DABG) in the Affymetrix Expression Console Software, and CHP files were collected after calibration. Transcriptome Analysis Console Software was used for further analysis of data. The genes were divided into six grades, and different gene expression levels were presented as Fold Change. Finally, clustering analysis was performed on gene expression of each sample.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The amplification of miRNA was performed using the specific primers of reverse transcription (RT) and polymerase chain reaction (PCR) from Bulge-Loop™ miRNA qRT-PCR Primer Set (RiboBio, Guangzhou, China) as previously described [21]. The quantification of PCR product was evaluated by the level of fluorescence emitted by SYBR Green (SYBR® Premix Ex Taq™ II, TaKaRa). RT and PCR were performed as previously described [21,22]. RT reactions were carried out at 42°C for 60 min followed by 70°C for 10 min. The qRT-PCR was run on a LightCycler® 480 Real-Time PCR System (Roche Diagnostics, Mannheim, Germany) in 384-well plates at 95°C for 20 sec followed by 40 cycles of 95°C for 10 sec, 60°C for 20 sec and then 70°C for 10 sec. Melting analysis was added finally to evaluate the specificity of PCR products. The expression of miRNAs in tissue specimens, serum samples, and exosomes were calculated using the comparative 2-ΔΔCt method. The let-7a was used as an internal control because it has been reported to be a reliable endogenous control for analysis of miRNA by RT-PCR in humans [23,24]. The raw data are presented as the relative quantity of target miRNA, normalized with respect to let-7a and compared with a reference sample.
Western blotting
Cells were harvested and protein was extracted from cells as previously described [25]. The protein concentration was determined using a protein assay kit (Bio-Rad, Hercules, CA, USA) and samples were separated in SDS polyacrylamide gels, with various concentrations depending on the molecular weight of the protein under investigation. After probing with a primary antibody, the membrane was incubated with a secondary antibody labeled with either IRDye 800CW or IRDye 680. Finally, signal intensity was determined using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA).
Diagnosis and treatment
After a detailed history and a complete physical examination, the hepatitis B and C serology, liver function test and tumor marker examination which included alpha-fetoprotein (AFP), carbohydrate antigen 19-9 (CA19-9), and carcinoembryonic antigen (CEA) was routinely performed. Other routine investigations were chest X-ray, upper gastrointestinal endoscopy, abdominal ultrasound, contrast-enhanced computerized tomography (CT) and/or magnetic resonance imaging (MRI). A clinical diagnosis of HCC was based on the criteria of the American Association for the Study of Liver Diseases (AASLD) [26].
The type of partial hepatectomy carried out was based on the tumor size, number, location, presence/absence of cirrhosis and estimated volume of future liver remnant. As far as possible, anatomical liver resection was carried out basing on Couinaud’s liver segments, sectors and hemilivers. Histopathologic study of the resected specimens was carried out independently by three pathologists who came to a consensus by discussion if there was any discrepancy.
Statistical analysis
Continuous variables were expressed as mean ± SD (standard deviation) and compared using a two-tailed unpaired Student’s t test; categorical variables were compared using χ2 or Fisher analysis. The predictive performances of plasma and exosomal miRNAs were measured using the area under receiver operating characteristic (ROC) curve (AUC). AUCs were also used to compare plasmic and exosomal miRNAs using the Hanley and McNeil method [27]. miRNA panel was further analyzed by logistic regression model for the differentiation between the HCC and HBV groups. Statistical analyses were conducted with the SPSS for Windows version 18.0 release (SPSS, Inc., Chicago, IL) and ROC curve analysis were computed using MedCalcV.11.0.3.0 (MedCalc software, Mariakerke, Belgium). A value of P < 0.05 was considered significant in all analyses.
Results
Characterization of exosomes released from cancer cells
Exosomes can be actively released from a variety of cell types including cancer cells. To determine whether microRNA-carrying exosomes can be released from hepatocarcinoma cell lines, we incubated three cell lines: SMMC-7721, Huh-7 and Hep3B cells, in exosome-free medium made from exosome-free FBS. The exosomes in the conditioned media were isolated by a serial centrifugations and filtration. The harvested exosomes were then resuspended in PBS or other agents depending upon further use. To determine the relative purity of isolated exosomes and their morphology, the exosome pellets were resuspended in PBS and then examined by transmission electron microscopy (TEM). To further determine the nature of the isolated exosomes, we used the exosome-specific marker CD63 which had been used to provide a quantification of the exosomes present in cell culture supernatants.
Characteristics of the patients
The characteristics of patients with HCC, cirrhotic, and HBV groups enrolled in this study are shown in Table 1. There was no significant difference among HCC, cirrhotic and control groups in the distribution of age or gender (P > 0.05).
Profiling of plasmic and exosomal miRNAs for HCC detection
In this study, we collected plasmic and exosomal solutions of 3 patients with HCC, 3 cirrhotic patients and 3 patients with HBV to perform miRNA microarray analysis. The results revealed 38 significantly differential miRNAs expressed among different exosomal groups, which were much higher than 26 differential miRNAs in the plasmic groups. miRNAs showed more than 1.5 fold altered expression between all 3 pooled HCC samples and the control pool samples were selected as candidates. We next performed qRT-PCR assay to confirm the expression of 24 candidate miRNAs in 30 HCC patients, 30 cirrhotic patients and 30 patients with HBV. 3 out of the 24 miRNAs (miRNA-26a, miRNA-29c and miRNA-21) showed significant differences in both plasma and exosome levels among patients with HCC, cirrhosis, and HBV groups. The differential expression of the three miRNAs in the 72 samples in both plasma and exosomes levels among HCC, cirrhosis, and patients with HBV groups were shown in Figure 1.
Figure 1.
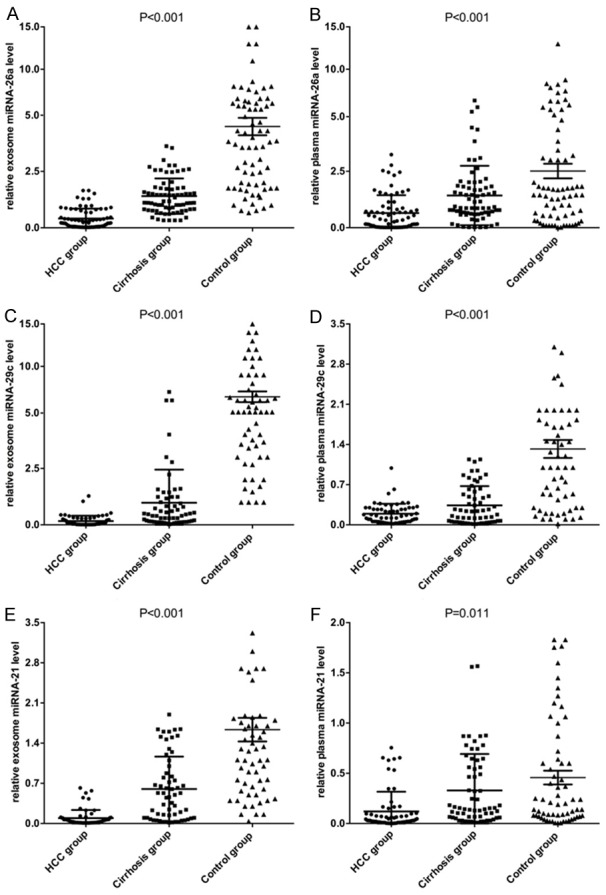
Comparing expression levels of miRNA-26a (A, B), miRNA-29c (C, D) and miRNA-21 (E, F) in both plasma and exosomes among three groups.
Expression of serum miRNAs in tissue samples
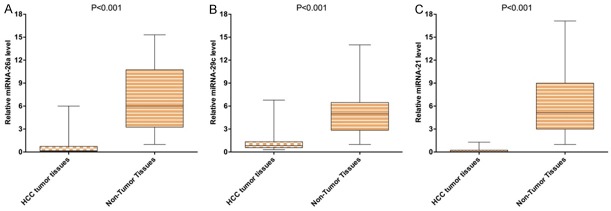
The three miRNAs identified in both plasma and exosome levels were further detected by qRT-PCR in an additional 10 pairs of tissue samples. Consistent with our previous results of plasma and exosomes levels, the expression of miRNA-26a, miRNA-29c and miRNA-21 were found to be significantly down-regulated in HCC tumor tissues compared to those in normal tissues (Figure 2).
Figure 2.
Comparing expression levels of miRNA-26a (A), miRNA-29c (B), and miRNA-21 (C) in HCC tumor tissues with non-tumor tissues.
Diagnostic values of candidate miRNAs
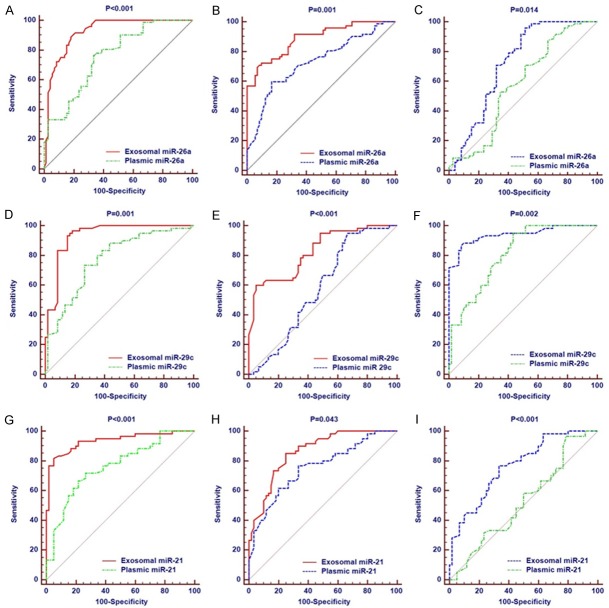
We generated ROC curves to evaluate the performance of the three miRNAs in discriminating the HCC patients from patients with HBV. The AUC for plasmic miRNA-26a was 0.757 (95% CI: 0.679 to 0.825), which was significantly smaller than the AUC (0.922, 95% CI: 0.865 to 0.960) of exosomal miRNA-26a (P < 0.001, Figure 3A). When we compared the HCC group with cirrhotic group, the AUC for plasmic miRNA-26a (0.728, 95% CI: 0.648 to 0.799) was significantly smaller than the AUC (0.887, 95% CI: 0.824 to 0.934) of exosomal miRNA-26a (P < 0.001, Figure 3B). We also explored the diagnostic value of exosomal miRNA-26a in discriminating the cirrhotic patients from patients with HBV. The AUC for plasmic miRNA-26a (0.577, 95% CI: 0.492 to 0.659) was significantly smaller than the AUC (0.722, 95% CI: 0.641 to 0.793) of exosomal miRNA-26a (P < 0.001, Figure 3C). Consistent with the results of miRNA-26a, the other 2 miRNAs (miRNA-29c and miRNA-21) showed significantly predictive value in discriminating the patients with HCC, cirrhosis, and HBV. The AUCs for plasmic miRNA-29c and miRNA-21 were significantly different from those of exosomal miRNA-29c and miRNA-21, respectively (Figure 3D-I).
Figure 3.
Comparison of diagnostic significance of miRNA-26a for all patients. A: Diagnostic value of plasmic and exosomal miRNA-26a in distinguishing HCC and HBV groups; B: Diagnostic value of plasmic and exosomal miRNA-26a in distinguishing HCC and hepatocirrhosis groups; C: Diagnostic value of plasmic and exosomal miRNA-26a in distinguishing hepatocirrhosis and HBV groups; D: Diagnostic value of plasmic and exosomal miRNA-29c in distinguishing HCC and HBV groups; E: Diagnostic value of plasmic and exosomal miRNA-29c in distinguishing HCC and hepatocirrhosis groups; F: Diagnostic value of plasmic and exosomal miRNA-29c in distinguishing hepatocirrhosis and HBV groups; G: Diagnostic value of plasmic and exosomal miRNA-21 in distinguishing HCC and HBV groups; H: Diagnostic value of plasmic and exosomal miRNA-21 in distinguishing HCC and hepatocirrhosis groups; I: Diagnostic value of plasmic and exosomal miRNA-21 in distinguishing hepatocirrhosis and HBV groups.
Multivariate logistic regression analysis identified exosomal miRNA-26a, miRNA-29c, and miRNA-21 as independent diagnostic markers for HCC
Considering that the exosomal expression of miRNA-26a, miRNA-29c and miRNA-21 were much better than the plasmic expression of these three miRNAs in terms of diagnostic specificity and sensitivity for HCC, we further performed multivariate logistic regression analysis including the variables of miRNA-26a, miRNA-29c, and miRNA-21 (Table 2). We found that these three miRNAs had independent diagnostic values in patients with HCC after multivariable adjustment.
Table 2.
Logistic analysis and diagnostic performance of miRNAs in patients with HCC compared with HBV group
| MicroRNA group | AUC | Univariate | Multivariate | ||
|---|---|---|---|---|---|
|
| |||||
| P value | HR | 95% CI | P value | ||
| AFP at diagnosis | 0.712 | 0.002 | 1.152 | 0.962-1.457 | 0.743 |
| ALT (U/L) | 0.689 | 0.012 | 1.021 | 0.583-1.522 | 0.552 |
| Liver cirrhosis | 0.677 | 0.023 | 0.788 | 0.367-1.263 | 0.156 |
| miRNA-26a | 0.922 | 0.001 | 0.134 | 0.024-0.241 | 0.001 |
| miRNA-29c | 0.934 | 0.001 | 0.104 | 0.016-0.832 | 0.004 |
| miRNA-21 | 0.935 | 0.001 | 0.084 | 0.027-0.932 | 0.012 |
Abbreviations: CI, confidence interval; AUC, area under the receiver operating characteristic curve; HCC, hepatocellular carcinoma.
Discussion
Although the prognosis of patients with HCC has improved in recent years as multiple therapeutic modalities had been applied, the overall survival is still poor [28,29]. Novel potential diagnosis and therapies for HCC are urgently required. A structured approach and early diagnosis of HCC are crucial for patients to prevent tumor progression and improve overall outcome.
Many studies have shown that exosomes in the urine, pleural fluid, and ascites fluid have a variety of biologic functions, including intracellular antigen presentation, communication, and transmission of signals and RNA and miRNA transfer [30,31]. Evidence has indicated that circulating miRNAs were involved in several pathophysiologic processes and related to cancer [32,33]. Moreover, miRNAs in malignant cells play important roles in the regulation of gene expression and could be identified as potential therapy targets [34]. Recent study showed that miRNAs such as-miR-125b, miR-15a, miR-122 and-miR-146b-5p might be novel biomarkers for hepatocellular carcinoma gene therapy [35].
A large number of exosomes can pass out of tumor cells, which is due to the impact of hypoxia secretion, changes in the internal environment and other factors. These exosomes are not easily degraded in the intercellular space or in the peripheral blood due to plasma membrane protection. With respect to patients with HCC, exosomal miR-21 recently has been reported as a novel biomarker for HCC diagnosis [36]. In this study, we compared the expression levels of miRNAs derived from exosomes and plasmas among different groups, and investigated their sensitivity and specificity for early diagnosis of patients with HCC.
Many studies have reported that the miR-26 level is dysregulated in various cancers [37-39]. To date, many oncogenes have been identified as miR-26a targets, which are involved in a variety of biologic pathways including cell proliferation, invasion, differentiation, angiogenesis, and energy metabolism [40-42]. miR-26a plays a dual role in tumorigenesis as a tumor suppressor and oncogene [43,44].
The miRNA-29 family includes miR-29a and mir-29b-1/2 and miR-29c. This group of miRNAs has been extensively studied and has been shown to be downregulated in many cancers, such as stomach, peripheral nerve sheath tumors, esophageal squamous cell carcinoma, melanoma, and breast cancer [45]. Research indicates that miR-29 participates in the processes of apoptosis, proliferation and epithelial-mesenchymal transition (EMT) in cancer.
In the present study, we screened plasmas and exosomes solutions of 3 HCC patients, 3 cirrhotic patients and 3 HBV patients to perform miRNA microarray analysis. Based on some preliminary studies and our results from microarray analysis, three distinctly differential miRNAs including miRNA-26a, miRNA-29c, and miRNA-21 were selected to perform further evaluation. First, we found expression of miRNA-26a, miRNA-29c, and miRNA-21 were significantly lower in the HCC group compared with control and cirrhotic group in both exosomes and plasma. Second, we found miRNA-26a, miRNA-29c, and miRNA-21 were significantly under-expressed in HCC tumor tissues compared with adjacent tissues. Third, we found miRNAs in exosomes had better diagnostic significance for patients with HCC compared with plasmic miRNAs among different groups.
Recently, Wang et al. [36] demonstrated that miR-21 was enriched in serum exosomes which provided increased sensitivity of detection compared to whole serum. The potential mechanism was that miR-21 expression is down-regulated by tumor suppressor phosphatase and tensin homolog (PTEN), resulting in the production of unsaturated fatty acids in hepatocytes. The value of exosomal miR-21 served as a potential biomarker for HCC diagnosis was rarely reported. In this study, miR-21 was significantly downregulated in serum and exosomes of HCC patients, in contrast with results reported previously. The sample size of all these studies including ours was too small and the results were not validated. Further studies are needed to confirm the expression levels of all miRNAs reported in this study.
There are several limitations of the present study: first of all, the sample size is too small in this study, and further studies with larger sample size are needed to confirm the present results. Secondly, since patients with HCC, cirrhosis and HBV received various therapies, whether different therapy modalities influenced the optimal specificity and sensitivity of these three miRNAs are also needed future confirmation.
In conclusion, we found expression of miRNA-26a, miRNA-29c, and miRNA-21 was significantly lower in patients with HCC compared with cirrhotic and HBV group, and we confirmed miRNA-26a, miRNA-29c and miRNA-21 could be identified as independent diagnostic biomarkers for patients with HCC.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (No. 81600453), Beijing Nova Program (No. Z171100001117114), the Military Medical Science and Technology Project of Youth Development (No. 15QNP084), the Doctorial Innovation Fund of Chinese PLA Medical School (No. B14015) and the 302 Hospital Foundation (No. QNPY2015007).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Tateishi R, Yoshida H, Matsuyama Y, Mine N, Kondo Y, Omata M. Diagnostic accuracy of tumor markers for hepatocellular carcinoma: a systematic review. Hepatol Int. 2008;2:17–30. doi: 10.1007/s12072-007-9038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nathan H, Schulick RD, Choti MA, Pawlik TM. Predictors of survival after resection of early hepatocellular carcinoma. Ann Surg. 2009;249:799–805. doi: 10.1097/SLA.0b013e3181a38eb5. [DOI] [PubMed] [Google Scholar]
- 5.Qin Y, Zhong Y, Ma T, Zhang J, Yang G, Guan F, Li Z, Li B. A pilot study of salivary N-glycome in HBV-induced chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Glycoconj J. 2017;34:523–535. doi: 10.1007/s10719-017-9768-5. [DOI] [PubMed] [Google Scholar]
- 6.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83. doi: 10.1186/s13045-015-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo W, Gao Y, Li N, Shao F, Wang C, Wang P, Yang Z, Li R, He J. Exosomes: new players in cancer (Review) Oncol Rep. 2017;38:665–675. doi: 10.3892/or.2017.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–15. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SIJ, Wurdinger T, Pegtel DM, van Rheenen J. In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046–1057. doi: 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le MT, Hamar P, Guo C, Basar E, Perdigão-Henriques R, Balaj L, Lieberman J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124:5109–28. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naidu S, Magee P, Garofalo M. MiRNA-based therapeutic intervention of cancer. J Hematol Oncol. 2015;8:68. doi: 10.1186/s13045-015-0162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–8. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Kharaziha P, Ceder S, Li Q, Panaretakis T. Tumor cell-derived exosomes: a message in a bottle. Biochim Biophys Acta. 2012;1826:103–11. doi: 10.1016/j.bbcan.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Shimoda M, Khokha R. Proteolytic factors in exosomes. Proteomics. 2013;13:1624–36. doi: 10.1002/pmic.201200458. [DOI] [PubMed] [Google Scholar]
- 16.Mu W, Rana S, Zoller M. Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia. 2013;15:875–87. doi: 10.1593/neo.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson MC, Azorsa DO. High-throughput RNAi screening for the identification of novel targets. Methods Mol Biol. 2013;986:89–95. doi: 10.1007/978-1-62703-311-4_6. [DOI] [PubMed] [Google Scholar]
- 18.Challagundla KB, Fanini F, Vannini I, Wise P, Murtadha M, Malinconico L, Cimmino A, Fabbri M. microRNAs in the tumor microenvironment: solving the riddle for a better diagnostics. Expert Rev Mol Diagn. 2014;14:565–74. doi: 10.1586/14737159.2014.922879. [DOI] [PubMed] [Google Scholar]
- 19.Murakami Y, Toyoda H, Tanahashi T, Tanaka J, Kumada T, Yoshioka Y, Kosaka N, Ochiya T, Taguchi YH. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS One. 2012;7:e48366. doi: 10.1371/journal.pone.0048366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M, Baba H. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159–1167. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 21.Zhao DS, Chen Y, Jiang H, Lu JP, Zhang G, Geng J, Zhang Q, Shen JH, Zhou X, Zhu W, Shan QJ. Serum miR-210 and miR-30a expressions tend to revert to fetal levels in Chinese adult patients with chronic heart failure. Cardiovasc Pathol. 2013;22:444–50. doi: 10.1016/j.carpath.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Du Y, Zhu M, Zhou X, Huang Z, Zhu J, Xu J, Cheng G, Shu Y, Liu P, Zhu W, Wang T. miR-20a enhances cisplatin resistance of human gastric cancer cell line by targeting NFKBIB. Tumour Biol. 2016;37:1261–9. doi: 10.1007/s13277-015-3921-1. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Zhang L, Liu F, Xiang G, Jiang D, Pu X. Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma. Dis Markers. 2015;2015:893594. doi: 10.1155/2015/893594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie X, Jiang C, Peng Z, Liu B, Hu W, Wang Y, Lin M, Lu M, Kuang M. Local recurrence after radiofrequency ablation of hepatocellular carcinoma: treatment choice and outcome. J Gastrointest Surg. 2015;19:1466–75. doi: 10.1007/s11605-015-2850-z. [DOI] [PubMed] [Google Scholar]
- 25.Takaoka Y, Shimizu Y, Hasegawa H, Ouchi Y, Qiao S, Nagahara M, Ichihara M, Lee JD, Adachi K, Hamaguchi M, Iwamoto T. Forced expression of miR-143 represses ERK5/c-Myc and p68/p72 signaling in concert with miR-145 in gut tumors of Apc(Min) mice. PLoS One. 2012;7:e42137. doi: 10.1371/journal.pone.0042137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology (Baltimore, Md.) 2005;42:1208–1236. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 27.Hanley JA. Receiver operating characteristic (ROC) methodology: the state of the art. Crit Rev Diagn Imaging. 1989;29:307–35. [PubMed] [Google Scholar]
- 28.Borzio M, Dionigi E, Vitale A, Rossini A, Marignani M, Fornari F, Vicari S, De Sio I, Farinati F, Bertolini E, Oliveri F, Leandro G, Francica G, Mitra M, Omazzi B, Boccia S, Salmi A, Toldi A, Sacco R. Management and prognosis of HCC in the elderly: results of an in-field multicenter cohort study. Liver Int. 2017;37:1184–1192. doi: 10.1111/liv.13392. [DOI] [PubMed] [Google Scholar]
- 29.Chang YJ, Chung KP, Chang YJ, Chen LJ. Long-term survival of patients undergoing liver resection for very large hepatocellular carcinomas. Br J Surg. 2016;103:1513–20. doi: 10.1002/bjs.10196. [DOI] [PubMed] [Google Scholar]
- 30.Raj DA, Fiume I, Capasso G, Pocsfalvi G. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. 2012;81:1263–72. doi: 10.1038/ki.2012.25. [DOI] [PubMed] [Google Scholar]
- 31.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–87. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 32.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–7. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang HH, Wei PL, Hung CS, Wu CT, Wang W, Huang MT, Chang YJ. MicroRNA-200a/b influenced the therapeutic effects of curcumin in hepatocellular carcinoma (HCC) cells. Tumour Biol. 2013;34:3209–18. doi: 10.1007/s13277-013-0891-z. [DOI] [PubMed] [Google Scholar]
- 35.Ding W, Yang H, Gong S, Shi W, Xiao J, Gu J, Wang Y, He B. Candidate miRNAs and pathogenesis investigation for hepatocellular carcinoma based on bioinformatics analysis. Oncol Lett. 2017;13:3409–3414. doi: 10.3892/ol.2017.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. 2014;2014:864894. doi: 10.1155/2014/864894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng M, Tang HL, Lu XH, Liu MY, Lu XM, Gu YX, Liu JF, He ZM. miR-26a suppresses tumor growth and metastasis by targeting FGF9 in gastric cancer. PLoS One. 2013;8:e72662. doi: 10.1371/journal.pone.0072662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian X, Zhao P, Li W, Shi ZM, Wang L, Xu Q, Wang M, Liu N, Liu LZ, Jiang BH. MicroRNA-26a promotes tumor growth and angiogenesis in glioma by directly targeting prohibitin. CNS Neurosci Ther. 2013;19:804–12. doi: 10.1111/cns.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu P, Tang H, Chen B, He Z, Deng M, Wu M, Liu X, Yang L, Ye F, Xie X. miR-26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett. 2015;357:384–392. doi: 10.1016/j.canlet.2014.11.050. [DOI] [PubMed] [Google Scholar]
- 40.Zhang B, Liu XX, He JR, Zhou CX, Guo M, He M, Li MF, Chen GQ, Zhao Q. Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis. 2011;32:2–9. doi: 10.1093/carcin/bgq209. [DOI] [PubMed] [Google Scholar]
- 41.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chai ZT, Kong J, Zhu XD, Zhang YY, Lu L, Zhou JM, Wang LR, Zhang KZ, Zhang QB, Ao JY, Wang M, Wu WZ, Wang L, Tang ZY, Sun HC. MicroRNA-26a inhibits angiogenesis by down-regulating VEGFA through the PIK3C2alpha/Akt/HIF-1alpha pathway in hepatocellular carcinoma. PLoS One. 2013;8:e77957. doi: 10.1371/journal.pone.0077957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang JR, Zeng C, Zhuang SM. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 2012;40:4615–25. doi: 10.1093/nar/gkr1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu B, Wu X, Liu B, Wang C, Liu Y, Zhou Q, Xu K. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim Biophys Acta. 2012;1822:1692–704. doi: 10.1016/j.bbadis.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 45.Sandhu R, Rivenbark AG, Mackler RM, Livasy CA, Coleman WB. Dysregulation of microRNA expression drives aberrant DNA hypermethylation in basal-like breast cancer. Int J Oncol. 2014;44:563–72. doi: 10.3892/ijo.2013.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]