Abstract
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide. microRNAs (miRNAs) have been reported to play essential roles in HCC progression and radiosensitivity. However, the effect of miR-106a on HCC progression and radiosensitivity as well as its mechanism remain largely unknown. The expressions of miR-106a and F-box and WD repeat domain containing 7 (FBXW7) were measured by quantitative real-time polymerase chain reaction (qRT-PCR) or western blot, respectively. Cell migration and invasion were analyzed by trans-well assay. The radiosensitivity was investigated by colony formation and western blot. The interaction between miR-106a and FBXW7 was explored by luciferase activity and RNA immunoprecipitation (RIP) analyses. Then miR-106a expression was elevated in HCC tissues and cells and associated with tumor stage as well as overall survival. Knockdown of miR-106a impeded cell migration and invasion but contributed to irradiation-induced inhibition of survival and increase of phosphorylation of histone in Serine 139 (γ-H2AX) protein in HCC cells. Moreover, FBXW7 was indicated as a target of miR-106a and negatively correlated with miR-106a. Besides, interference of FBXW7 reversed the regulatory effect of miR-106a abrogation on migration, invasion and radiosensitivity in HCC cells. The results showed down-regulation of miR-106a suppressed migration and invasion and increased radiosensitivity of HCC cells by targeting FBXW7, providing a novel avenue for HCC treatment.
Keywords: Hepatocellular carcinoma, radiosensitivity, migration, invasion, miR-106a, FBXW7
Introduction
Hepatocellular carcinoma (HCC) is the most common liver cancer with high mortality and incidence all over the world [1]. Many treatments have been used for HCC, such as chemotherapeutics, radiotherapy, antibody therapy, targeted therapy and immunotherapy [2]. Despite great advances in radiotherapy development, the efficacy and safety of radiation therapy for HCC remain a huge challenge because of the resistance [3]. Thus, a novel strategy for enhancing radiosensitivity is needed for treatment of HCC.
microRNAs (miRNAs) are a class of endogenous noncoding RNAs, which play important roles in therapeutics and diagnosis of HCC [4]. miRNAs have been reported to have essential roles in regulating proliferation, invasion, metastasis and drug resistance by varying mechanisms in HCC [5]. For example, miR-19a-3p contributes to metastasis and chemoresistance by regulating phosphatase and tensin homolog (PTEN)/protein kinase B (AKT) pathway in HCC [6]. miR-203 promotes cell radiosensitivity by targeting B-cell-specific Moloney leukemia virus insertion site 1 (Bmi-1) in HCC [7]. As for miR-106a, it is suggested to promote proliferation, migration and invasion in prostate cancer and cervical cancer [8,9]. Moreover, miR-106a has been reported to inhibit chemosensitivity in colorectal cancer and non-small cell lung cancer [10,11]. A previous study suggests that miR-106a is associated with sensitivity of cancer cells to radiation by regulating lipopolysaccharide-induced tumor necrosis factor-alpha factor (LITAF) and ataxia telangiectasia mutated (ATM) in prostate cancer [12]. Notably, miR-106a has been reported to be highly expressed and serve as biomarker for diagnosis and prognosis of HCC [13]. However, the role of miR-106a in radiosensitivity as well as its mechanism remain poorly understood.
F-box and WD repeat domain containing 7 (FBXW7), a member of the F-box protein family, has been reported to function as a tumor suppressor in various human cancers [14]. Accruing efforts suggest that FBXW7 suppresses cell migration and drug-resistance in breast cancer, cholangiocarcinoma and non-small-cell lung cancer cells [15-17]. Notably, the former findings indicate that FBXW7 is implicated in prognosis and progression of HCC [18,19]. However, there is no direct evidence in support of the relationship between miR-106a and FBXW7. Intriguingly, bioinformatics analysis showed the potential binding sites of miR-106a and FBXW7. Hence, we hypothesized FBXW7 might participate in miR-106a-mediated progress in HCC. In this study, we explored the effect of miR-106a on migration, invasion, and radiosensitivity as well as the interaction between miR-106a and FBXW7.
Materials and methods
Tissues samples
A total of 29 paired tumor and peri-tumor tissues were collected from patients with HCC and then immediately stored at -80°C until used. All patients without chemotherapy, radiotherapy or other therapy were enrolled from Chongqing General Hospital in this study. Written informed consent was obtained from all participants and this study was approved by the Research Ethics Committee of Chongqing General Hospital. The patients were classified into early stage (I-II, n=11) and advanced stage (III-IV, n=18) according to tumor-mode-metastasis (TNM) stage. The low miR-106a expression group (n=14) and high expression group (n=15) were classified according to the mean value of miR-106a expression level in HCC patients. The overall survival rate was analyzed in patients after the follow-up.
Cell culture and transfection
The human normal liver cell line (LO2) and HCC cell lines (MHCC97-H, SMMC-7721, Hep3B and HepG2) cells were purchased from American Tissue Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA) at 37°C in a humidified atmosphere with 5% CO2.
miR-106a mimic (miR-106a), miRNA negative control (miR-NC), miR-106a inhibitor (anti-miR-106a), inhibitor negative control (anti-miR-NC), small interfering RNA (siRNA) against FBXW7 (si-FBXW7) and siRNA negative control (si-NC) were synthesized by Genepharma (Shanghai, China). Cell transfection was performed in SMMC-7721 and HepG2 cells for 48 h by using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from tissues or cells by using TRIzol reagent (Invitrogen) and then reverse transcribed into complementary DNA (cDNA) using TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) or M-MLV Reverse Transcription Kit (Thermo Fisher, Wilmington, DE, USA) according to the manufacturer’s instructions. The cDNA was diluted and used for qRT-PCR using SYBR green (Applied Biosystems) with the following amplification protocol: 95°C for 5 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. Every sample was prepared in triplicate and the experiment was repeated three times. The expressions of miR-106a and FBXW7 were calculated using 2-ΔΔCt method with U6 small RNA or GAPDH as endogenous control, respectively [20]. The primers were list as follows: miR-106a (Forward, 5’-GCGGCGGAAAAGTGCTTACAGTG-3’; Reverse, 5’-ATCCAGTGCAGGGTCCGAGG-3’), U6 (Forward, 5’-GTGCTCGCTTCGGCAGCA CATATAC-3’; Reverse, 5’-AAAAATATGGAACGCTCACGAATTTG-3’), FBXW7 (Forward, 5’-ACTGGAAAGTGACTCTGGGA-3’; Reverse, 5’-TACTGGGGCTAGGCAAACAA-3’), GAPDH (Forward, 5’-AACGGATTTGGTCGTATTGGG-3’; Reverse, 5’-TCGCTCCTGGAAGATGGTGAT-3’).
Trans-well assay
Transwell assay was conducted to analyze the migrated and invasive abilities of cells. For cell migration, transfected SMMC-7721 and HepG2 cells (1 × 104 cells/well) in 100 μl serum-free DMEM medium were placed in the upper chambers (Costar, Corning, NY, USA), and 500 μl DMEM medium containing 10% FBS was added to the lower chambers. Cells remaining on the top surface were removed with a cotton swab after the incubation for 24 h, while migrated cells through the membranes were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet (Sigma, St. Louis, MO, USA). The stained cells were counted from five random fields under a microscope (Olympus, Tokyo, Japan). For cell invasion, the experiment was performed following the same approach using transwell chambers pre-treated with Matrigel (BD, San Jose, CA, USA).
Irradiation treatment
Transfected SMMC-7721 and HepG2 cells were treated with different doses of irradiation (0, 2, 4, 6 and 8 Gy) using the x-ray apparatus (Rad Source Technologies, Alpharetta, GA, USA) with a dose rate of 200 cGy/min. At 24 h after irradiation, the expression of miR-106a and survival fraction were measured in transfected cells. The survival fraction was analyzed by colony formation. After incubation for 14 d, treated cells were fixed with methanol (Sigma) for 30 min and then stained with 0.01% crystal violet for 15 min. The colony formation was analyzed under a microscope and survival fraction was calculated by normalizing to the control cells. For measurement of cells sensitivity of radiotherapy, the expression of phosphorylation of histone in Serine 139 (γ-H2AX) protein was measured in transfected cells at different times (0, 12 and 24 h) after irradiation of 6 Gy.
Western blot
Treated cells were washed with PBS and then incubated with RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China). Total protein in supernatant was obtained by centrifugation at 12,000 × g for 20 min at 4°C and then quantified by BCA protein assay kit (Beyotime Biotechnology), followed by denatured at 98°C for 10 min. Subsequently, equal amounts of proteins were separated by SDS-PAGE gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat milk for 1 h at room temperature, and then incubated with primary antibodies against γ-H2AX (ab11174, 1:2000 dilution, Abcam, Cambridge, UK), FBXW7 (ab109617, 1:1000 dilution, Abcam) or GAPDH (ab9485, 1:2500 dilution, Abcam) overnight at 4°C and horseradish peroxidase (HRP)-conjugated secondary antibody (ab6721, 1:10000 dilution, Abcam) for 2 h at room temperature. GAPDH was used as loading control in this study. The protein signals were visualized using enhanced chemiluminescence (ECL) chromogenic substrate (Beyotime Biotechnology).
Luciferase activity assay
The potential targets of miR-106a were predicted by starBase. The 3’ untranslated regions (3’-UTR) sequences of FBXW7 containing wild-type (WT) or mutant-type (MUT) binding sites of miR-106a were amplified and then cloned into the pGL3 vectors (Promega, Madison, WI, USA) to synthesize luciferase reporter vectors (FBXW7-WT or FBXW7-MUT), respectively. SMMC-7721 and HepG2 cells were co-transfected with 20 ng FBXW7-WT or FBXW7-MUT, 15 ng control vector and 40 nM miR-106a or miR-NC using Lipofectamine 2000 according to the manufacturer’s protocols. After the transfection for 48 h, cells were collected for luciferase activity assay with luciferase assay kit (Promega) according to the manufacturer’s instructions.
RNA immunoprecipitation (RIP)
RIP assay was performed by using RNA-binding protein immunoprecipitation kit (Millipore) according to the manufacturer’s protocols. In brief, SMMC-7721 and HepG2 cells transfected with miR-106a or miR-NC were lysed in RIP buffer containing magnetic beads bound with Ago2 antibody or IgG. The enrichment of FBXW7 immunoprecipitated on beads were detected by qRT-PCR.
Statistical analysis
The data are presented as the mean ± standard deviation (SD) from three independent experiments. The overall survival curve of patients was generated by the Kaplan-Meier method. The relationship between the abundance of miR-106a and FBXW7 was analyzed by Spearman rank correlation. Student’s t test or one-way analysis of variance (ANOVA) was performed to analyze statistical differences between groups by using GraphPad Prism 5 (GraphPad Inc., La Jolla, CA, USA). P<0.05 was regarded as statistically significant.
Results
The expression of miR-106a was enhanced in HCC
To explore the potential role of miR-106a in HCC progression, its expression was first measured in HCC tissues and cells. Compared with peri-tumor samples, HCC tumor tissues displayed high expression of miR-106a (Figure 1A). Moreover, tissues at advanced stage (III-IV) showed higher abundance of miR-106a than that in early stage (I-II) (Figure 1B). In addition, high expression of miR-106a was associated with poor overall survival of patients (P=0.04) (Figure 1C). The abundance of miR-106a was significantly enhanced in HCC cells (MHCC97-H, SMMC-7721, Hep3B and HepG2) compared with that in LO2 cells (Figure 1D). Thus, SMMC-7721 and HepG2 cells with higher expression of miR-106a were used for further experiments.
Figure 1.
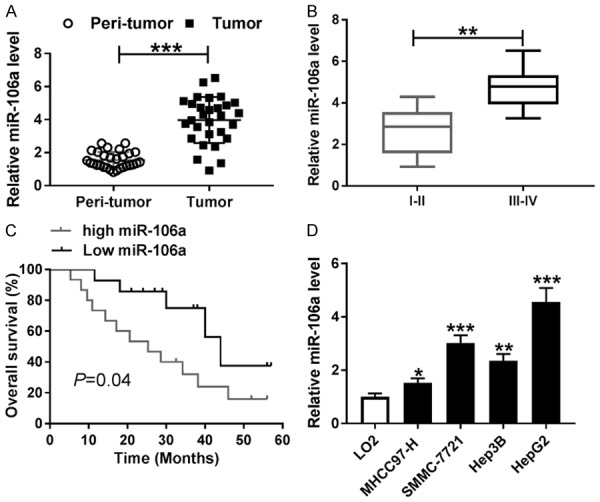
miR-106a expression is up-regulated in HCC tissues and cells. A. The expression of miR-106a was measured in tumor and peri-tumor tissues by qRT-PCR. B. The level of miR-106a was detected in tumor tissues of patients at early (I-II) or advanced (III-IV) stage by qRT-PCR. C. Overall survival of patients was analyzed according to miR-106a level. D. The abundance of miR-106a was examined in HCC cells by qRT-PCR. *P<0.05, **P<0.01, ***P<0.001.
Abrogation of miR-106a inhibited migration and invasion in HCC cells
To investigate the effect of miR-106a on cell migration and invasion, SMMC-7721 and HepG2 cells were transfected with anti-miR-106a or anti-miR-NC. As a result, the abundance of miR-106a was effectively reduced in SMMC-7721 and HepG2 cells transfected with anti-miR-106a compared with that in anti-miR-NC group (Figure 2A). Moreover, transwell assay showed that knockdown of miR-106a significantly impaired the migrated ability of SMMC-7721 and HepG2 cells (Figure 2B). Similarly, cell invasion was also greatly impeded in SMMC-7721 and HepG2 cells transfected with anti-miR-106a (Figure 2C).
Figure 2.
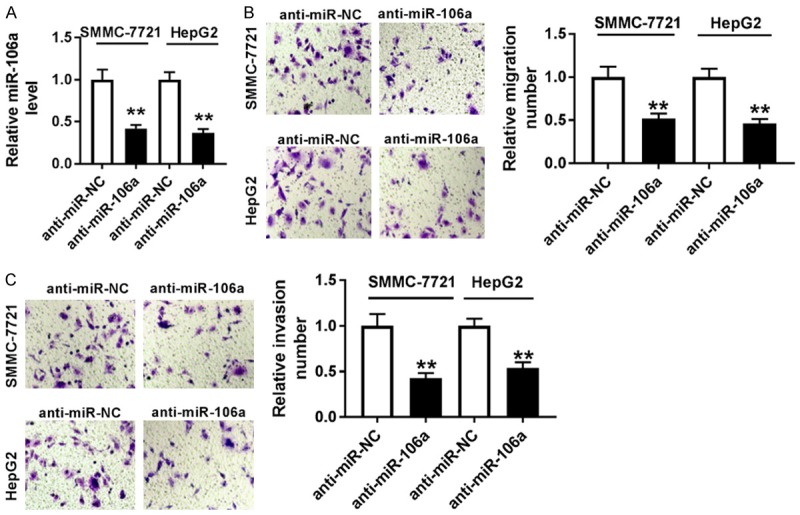
Knockdown of miR-106a inhibited migration and invasion in HCC cells. A. The expression of miR-106a was measured in SMMC-7721 and HepG2 cells transfected with anti-miR-106a or anti-miR-NC by qRT-PCR. B and C. Cell migration and invasion were analyzed in SMMC-7721 and HepG2 cells transfected with anti-miR-106a or anti-miR-NC by transwell assay. **P<0.01.
Inhibition of miR-106a enhanced radiosensitivity in HCC cells
To evaluate the effect of miR-106a on radiosensitivity of HCC cells, SMMC-7721 and HepG2 cells were treated with various doses of irradiation. After treatment of irradiation for 24 h, the expression of miR-106a was significantly inhibited in SMMC-7721 and HepG2 cells in a dose-dependent manner (Figure 3A and 3B). Moreover, inhibition of miR-106s led to obvious loss of colony formation in SMMC-7721 and HepG2 cells after exposure of different doses of irradiation (Figure 3C and 3D). Also, treatment of 6 Gy irradiation resulted in an increase of γ-H2AX protein abundance in SMMC-7721 and HepG2 cells in a time-dependent manner, which was exacerbated by exhaustion of miR-106a (Figure 3E and 3F).
Figure 3.
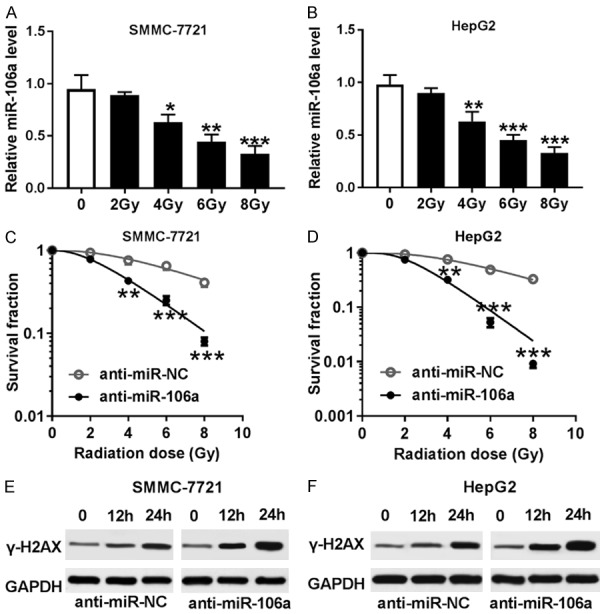
Inhibition of miR-106a enhanced radiosensitivity of HCC cells. A and B. The expression of miR-106a was detected in SMMC-7721 and HepG2 cells after various doses of irradiation for 24 h by qRT-PCR. C and D. The survival fraction was analyzed in SMMC-7721 and HepG2 cells transfected with anti-miR-106a or anti-miR-NC after various doses of irradiation for 24 h by colony formation. E and F. The abundance of γ-H2AX was examined in SMMC-7721 and HepG2 cells transfected with anti-miR-106a or anti-miR-NC after 6 Gy irradiation for different times by western blot. *P<0.05, **P<0.01, ***P<0.001.
FBXW7 was a target of miR-106a
To explore the potential mechanism that allows miR-106a participating in HCC progression, the promising target was probed by starBase. The binding sites of miR-106a and FBXW7 were shown in Figure 4A, and we conducted the WT or MUT luciferase reporter vector. Results showed that overexpression of miR-106a significantly repressed luciferase activity in SMMC-7721 and HepG2 cells transfected with FBXW7-WT, while its efficacy was lost in terms of the FBXW7-MUT group (Figure 4B). Moreover, RIP assay was conducted in SMMC-7721 and HepG2 cells transfected with miR-106a or miR-NC. Up-regulation of miR-106a induced higher abundance of FBXW7 by Ago2 RIP in SMMC-7721 and HepG2 cells, whereas there was little enrichment in IgG group (Figure 4C).
Figure 4.
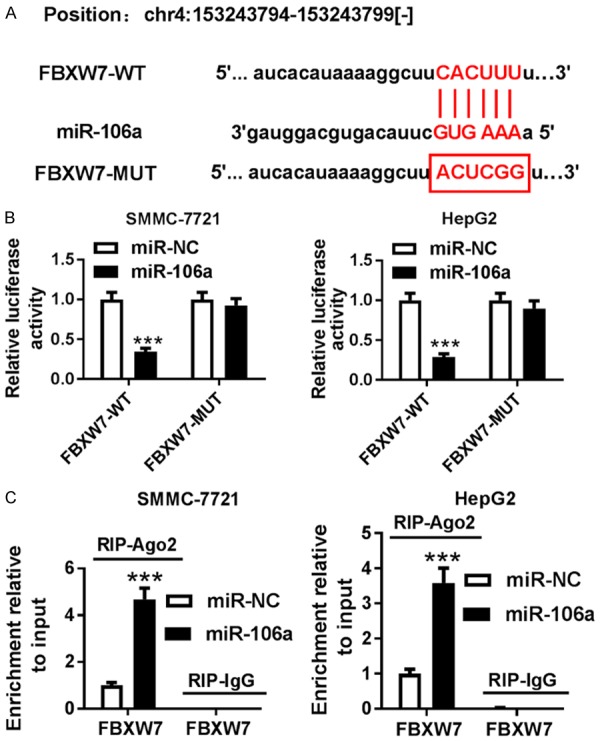
FBXW7 is a direct target of miR-106a. A. The putative binding sites of miR-106a and FBXW7 were predicted by starBase. B. Luciferase activity was analyzed in SMMC-7721 and HepG2 cells co-transfected with FBXW7-WT or FBXW7-MUT and miR-106a or miR-NC. C. The enrichment of FBXW7 was measured in SMMC-7721 and HepG2 cells transfected with miR-106a or miR-NC after Ago2 or IgG RIP by qRT-PCR. ***P<0.001.
FBXW7 was down-regulated and negatively regulated by miR-106a
Seeing that FBXW7 was targeted by miR-106a, its expression was measured in HCC. The expression of FBXW7 mRNA was abnormally reduced in HCC tumor tissues compared with that in peri-tumor samples (Figure 5A). Meanwhile, FBXW7 mRNA level was significantly decreased in HCC cells (Figure 5B). Spearman rank correlation assay showed that the expression of FBXW7 was negatively correlated with miR-106a level in HCC tissues (R=-0.546, P=0.012) (Figure 5C). Furthermore, the effect of miR-106a on FBXW7 protein expression was analyzed in SMMC-7721 and HepG2 cells. Results showed that addition of miR-106a greatly inhibited the FBXW7 protein level, while its depletion induced FBXW7 expression (Figure 5D and 5E).
Figure 5.
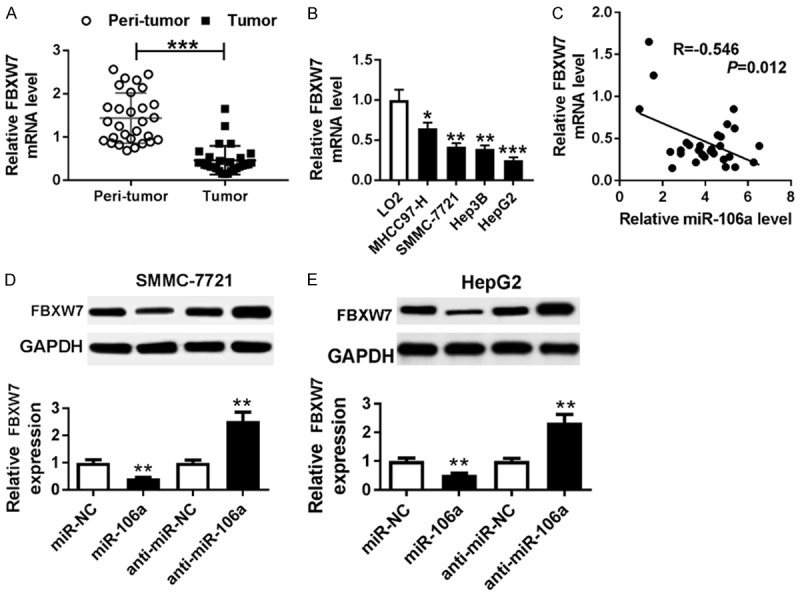
FBXW7 was down-regulated in HCC and negatively regulated by miR-106a. A and B. The expression of FBXW7 was measured in HCC tissues and cells by qRT-PCR. C. The relationship between miR-106a and FBXW7 mRNA levels was analyzed by Spearman rank correlation. D and E. The expression of FBXW7 protein was detected in SMMC-7721 and HepG2 cells transfected with miR-106a, miR-NC, anti-miR-106a, or anti-miR-NC by western blot. *P<0.05, **P<0.01, ***P<0.001.
FBXW7 was required for miR-106a-mediated progression of HCC
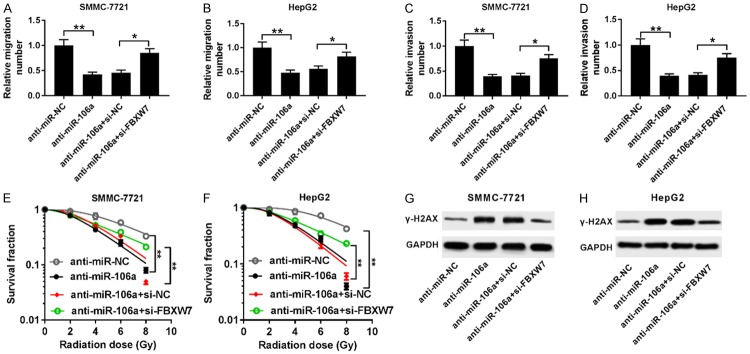
To explore whether FBXW7 was implicated in miR-106a-mediated migration, invasion and radiosensitivity of HCC cells, SMMC-7721 and HepG2 cells were transfected with anti-miR-NC, anti-miR-106a, anti-miR-106a+si-NC or anti-miR-106a+si-FBXW7. Interference with FBXW7 reversed miR-106a knockdown-mediated inhibition of migration in SMMC-7721 and HepG2 cells (Figure 6A and 6B). Similarly, FBXW7 exhaustion protected cell invasion from miR-106a depletion in SMMC-7721 and HepG2 cells (Figure 6C and 6D). Moreover, knockdown of FBXW7 attenuated the inhibitory effect of miR-106a deficiency on survival fraction in irradiation-treated SMMC-7721 and HepG2 cells (Figure 6E and 6F). Finally, down-regulation of FBXW7 ablated silencing of miR-106a-induced expression of γ-H2AX protein in irradiation-treated SMMC-7721 and HepG2 cells (Figure 6G and 6H).
Figure 6.
miR-106a regulated migration, invasion, and radiosensitivity of HCC cells by targeting FBXW7. A-D. Cell migration and invasion were measured in SMMC-7721 and HepG2 cells transfected with anti-miR-NC, anti-miR-106a, anti-miR-106a+si-NC or anti-miR-106a+si-FBXW7 by trans-well assay. E and F. The survival fraction was analyzed in SMMC-7721 and HepG2 cells transfected with anti-miR-NC, anti-miR-106a, anti-miR-106a+si-NC or anti-miR-106a+si-FBXW7 after various doses of irradiation for 24 h by colony formation. G and H. The abundance of γ-H2AX was examined in SMMC-7721 and HepG2 cells transfected with anti-miR-NC, anti-miR-106a, anti-miR-106a+si-NC, or anti-miR-106a+si-FBXW7 after 6 Gy irradiation for different times by western blot. *P<0.05, **P<0.01.
Discussion
The available evidence has indicated that miRNAs have important impact on tumor development, apoptosis, metastasis, and progression of HCC [21]. In this study, we found that miR-106a expression was enhanced in HCC tissues and cells, which is also in agreement with a former study [13]. Here we first provide the view that miR-106a knockdown enhanced radiosensitivity of HCC and validated the interaction between miR-106a and FBXW7.
miR-106a-5p, as a dominant strand of miR-106a, has been reported to facilitate proliferation, migration, and invasion in HCC [22,23]. Moreover, previous work suggested that miR-106a promoted proliferation and invasion in HCC cells [13]. Similarly, we also found that knockdown of miR-106a suppressed cell migration and invasion in HCC cells. Radiotherapy is one of the main strategies for HCC treatment [24]. Furthermore, miR-106a was indicated to increase radioresistance in prostate cancer [12]. In the present study, results showed that depletion of miR-106a enhanced radiosensitivity of HCC cells, which also suggested the association between miR-106a and radioresistance. However, the mechanism allows miR-106a participating in progression and radiosensitivity of HCC remains largely unclear. Functional miRNAs are known to regulate target expressions in many conditions. A number of investigators have reported various targets of miR-106a in various cells. For instance, miR-106a promoted tumor growth, migration and invasion by regulating Kruppel-like factor 4 (KLF4) expression in gastric cancer [25]. In addition, miR-106a knockdown induced cell apoptosis but reduced migration and invasion by targeting B-cell lymphoma 2-like protein 11 (BCL2L11) in human endometrial adenocarcinoma [26]. Also, PTEN was indicated as a target of miR-106a, which was associated with proliferation and metastasis in prostate cancer [8]. In this study, we first validated the interaction between miR-106a and FBXW7 in HCC cells by luciferase activity and RIP assays.
Silencing of FBXW7 was reported to enhance cell invasion through regulating epithelial-mesenchymal transition in HCC [27]. Moreover, FBXW7 suppressed proliferation and tumorigenesis in HCC [28,29]. Here we found that FBXW7 mRNA expression was decreased in HCC tissues and cells, which is also consistent with former finding [30]. Additionally, knockdown of FBXW7 has been reported to induce migration and invasion by Notch1 pathway in HCC [31]. Similarly, we also revealed that inhibition of FBXW7 reversed depletion of miR-106a-mediated suppression of migration and invasion in HCC. Besides, we also indicated that knockdown of FBXW7 attenuated the promotional role of miR-106a abrogation on radiosensitivity of HCC cells. In vivo experiments are responsible for further understanding the mechanism involved in progression and radiosensitivity of HCC [32,33]. Hence, the role of miR-106a is needed to be explored using a xenograft model in vivo in future.
In conclusion, miR-106a expression was up-regulated and FBXW7 mRNA level was down-regulated in HCC tissues and cells. Knockdown of miR-106a inhibited cell migration and invasion but promoted radiosensitivity in HCC cells. Moreover, FBXW7 was regarded as a functional target of miR-106a and its interference reversed the regulatory effect of miR-106a abrogation on migration, invasion and radiosensitivity in HCC cells. Collectively, inhibition of miR-106a suppressed migration and invasion and enhanced radiosensitivity of HCC cells by targeting FBXW7, providing a novel biomarker for HCC treatment.
Disclosure of conflict of interest
None.
References
- 1.Llovet J, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 2.Dutta R, Mahato R. Recent advances in hepatocellular carcinoma therapy. Pharmacol Ther. 2017;173:106–117. doi: 10.1016/j.pharmthera.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalogeridi M, Zygogianni A, Kyrgias G, Kouvaris J, Chatziioannou S, Kelekis N, Kouloulias V. Role of radiotherapy in the management of hepatocellular carcinoma: a systematic review. World J Hepatol. 2015;7:101–112. doi: 10.4254/wjh.v7.i1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang N, Ekanem N, Sakyi C, Ray S. Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv Drug Deliv Rev. 2015;81:62–74. doi: 10.1016/j.addr.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, Cai F, Ma L, Yu Y. The role of MicroRNAs in hepatocellular carcinoma. J Cancer. 2018;9:3557–3569. doi: 10.7150/jca.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X, Yu X, Liu T, Zhu H, Shi X, Bilegsaikhan E, Guo H, Song G, Weng S, Huang X, Dong L, Janssen H, Shen X, Zhu J. microRNA-19a-3p promotes tumor metastasis and chemoresistance through the PTEN/Akt pathway in hepatocellular carcinoma. Biomed Pharmacother. 2018;105:1147–1154. doi: 10.1016/j.biopha.2018.06.097. [DOI] [PubMed] [Google Scholar]
- 7.Shao Y, Zhang D, Li X, Yang J, Chen L, Ning Z, Xu Y, Deng G, Tao M, Zhu Y, Jiang J. MicroRNA-203 increases cell radiosensitivity via directly targeting bmi-1 in hepatocellular carcinoma. Mol Pharm. 2018;15:3205–3215. doi: 10.1021/acs.molpharmaceut.8b00302. [DOI] [PubMed] [Google Scholar]
- 8.Luo B, Kang N, Chen Y, Liu L, Zhang Y. Oncogene miR-106a promotes proliferation and metastasis of prostate cancer cells by directly targeting PTEN in vivo and in vitro. Minerva Med. 2018;109:24–30. doi: 10.23736/S0026-4806.17.05342-3. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Zhou Q, Tao L, Yu C. MicroRNA-106a promotes cell migration and invasion by targeting tissue inhibitor of matrix metalloproteinase 2 in cervical cancer. Oncol Rep. 2017;38:1774–1782. doi: 10.3892/or.2017.5832. [DOI] [PubMed] [Google Scholar]
- 10.Qin Y, Chen X, Liu Z, Tian X, Huo Z. miR-106a reduces 5-fluorouracil (5-FU) sensitivity of colorectal cancer by targeting dual-specificity phosphatases 2 (DUSP2) Med Sci Monit. 2018;24:4944–4951. doi: 10.12659/MSM.910016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Y, Sun C, Zhang L, Pan Y. Clinical significance of miRNA-106a in non-small cell lung cancer patients who received cisplatin combined with gemcitabine chemotherapy. Cancer Biol Med. 2018;15:157–164. doi: 10.20892/j.issn.2095-3941.2017.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoey C, Ray J, Jeon J, Huang X, Taeb S, Ylanko J, Andrews D, Boutros P, Liu S. miRNA-106a and prostate cancer radioresistance: a novel role for LITAF in ATM regulation. Mol Oncol. 2018;12:1324–1341. doi: 10.1002/1878-0261.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue X, Zhao Y, Wang X, Qin L, Hu R. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120:135–142. doi: 10.1002/jcb.27165. [DOI] [PubMed] [Google Scholar]
- 14.Yeh C, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17:115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang G, Shi W, Fang H, Zhang X. miR-27a promotes human breast cancer cell migration by inducing EMT in a FBXW7-dependent manner. Mol Med Rep. 2018;18:5417–5426. doi: 10.3892/mmr.2018.9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori A, Masuda K, Ohtsuka H, Shijo M, Ariake K, Fukase K, Sakata N, Mizuma M, Morikawa T, Hayashi H, Nakagawa K, Motoi F, Naitoh T, Fujishima F, Unno M. FBXW7 modulates malignant potential and cisplatin-induced apoptosis in cholangiocarcinoma through NOTCH1 and MCL1. Cancer Sci. 2018;109:3883–3895. doi: 10.1111/cas.13829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao G, Li Y, Wang M, Li X, Qin S, Sun X, Liang R, Zhang B, Du N, Xu C, Ren H, Liu D. FBXW7 suppresses epithelial-mesenchymal transition and chemo-resistance of non-small-cell lung cancer cells by targeting snai1 for ubiquitin-dependent degradation. Cell Prolif. 2018;51:e12473. doi: 10.1111/cpr.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tu K, Yang W, Li C, Zheng X, Lu Z, Guo C, Yao Y, Liu Q. Fbxw7 is an independent prognostic marker and induces apoptosis and growth arrest by regulating YAP abundance in hepatocellular carcinoma. Mol Cancer. 2014;13:110. doi: 10.1186/1476-4598-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C, Dou C, Xu M, Liu Q, Tu K. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16:123. doi: 10.1186/s12943-017-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Chen E, Xu X, Liu R, Liu T. Small but heavy role: micrornas in hepatocellular carcinoma progression. Biomed Res Int. 2018;2018:6784607. doi: 10.1155/2018/6784607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B, Cai H, Zheng R, Yang S, Zhou Z, Tu J. Long non-coding RNA 657 suppresses hepatocellular carcinoma cell growth by acting as a molecular sponge of miR-106a-5p to regulate PTEN expression. Int J Biochem Cell Biol. 2017;92:34–42. doi: 10.1016/j.biocel.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Wu J, Huang J, Wang W, Xu J, Yin M, Cheng N, Yin J. Long non-coding RNA Fer-1-like protein 4 acts as a tumor suppressor via miR-106a-5p and predicts good prognosis in hepatocellular carcinoma. Cancer Biomark. 2017;20:55–65. doi: 10.3233/CBM-170090. [DOI] [PubMed] [Google Scholar]
- 24.Ohri N, Dawson L, Krishnan S, Seong J, Cheng J, Sarin S, Kinkhabwala M, Ahmed M, Vikram B, Coleman C, Guha C. Radiotherapy for hepatocellular carcinoma: new indications and directions for future study. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu M, Zhang N, Lu X, He S. Negative regulation of kruppel-like factor 4 on microRNA-106a at upstream transcriptional level and the role in gastric cancer metastasis. Dig Dis Sci. 2018;63:2604–2616. doi: 10.1007/s10620-018-5143-z. [DOI] [PubMed] [Google Scholar]
- 26.Tang W, Li J, Liu H, Zhou F, Liu M. MiR-106a promotes tumor growth, migration, and invasion by targeting in human endometrial adenocarcinoma. Am J Transl Res. 2017;9:4984–4993. [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J, Zhang W, Gao F, Liu Y, Chen Z, Cheng L, Xie S, Zheng S. FBW7 increases chemosensitivity in hepatocellular carcinoma cells through suppression of epithelial-mesenchymal transition. Hepatobiliary Pancreat Dis Int. 2014;13:184–91. doi: 10.1016/s1499-3872(14)60029-1. [DOI] [PubMed] [Google Scholar]
- 28.Tang B, Lei B, Qi G, Liang X, Tang F, Yuan S, Wang Z, Yu S, He S. MicroRNA-155-3p promotes hepatocellular carcinoma formation by suppressing FBXW7 expression. J Exp Clin Cancer Res. 2016;35:93. doi: 10.1186/s13046-016-0371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Wang H, Wang J, Huang S, Zhang W. STAT1 inhibits human hepatocellular carcinoma cell growth through induction of p53 and Fbxw7. Cancer Cell Int. 2015;15:111. doi: 10.1186/s12935-015-0253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imura S, Tovuu L, Utsunomiya T, Morine Y, Ikemoto T, Arakawa Y, Kanamoto M, Iwahashi S, Saito Y, Takasu C, Yamada S, Ishikawa D, Bando Y, Shimada M. Role of Fbxw7 expression in hepatocellular carcinoma and adjacent non-tumor liver tissue. J Gastroenterol Hepatol. 2014;29:1822–1829. doi: 10.1111/jgh.12623. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Zhang J, Zhou L, Sun W, Zheng Z, Lu P, Gao Y, Yang X, Zhang Z, Tao K, Dou K. Fbxw7 regulates hepatocellular carcinoma migration and invasion via Notch1 signaling pathway. Int J Oncol. 2015;47:231–243. doi: 10.3892/ijo.2015.2981. [DOI] [PubMed] [Google Scholar]
- 32.Cun Y, Dai N, Xiong C, Li M, Sui J, Qian C, Li Z, Wang D. Silencing of APE1 enhances sensitivity of human hepatocellular carcinoma cells to radiotherapy in vitro and in a xenograft model. PLoS One. 2013;8:e55313. doi: 10.1371/journal.pone.0055313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Jin Y, Lyu Y, Tang X, Zhang Y, Chen J, Zheng D, Liang Y. Lupeol enhances radiosensitivity of human hepatocellular carcinoma cell line SMMC-7721 in vitro and in vivo. Int J Radiat Biol. 2015;91:202–208. doi: 10.3109/09553002.2015.966209. [DOI] [PubMed] [Google Scholar]