Abstract
MicroRNAs play important roles in the initiation and progression of acute myeloid leukemia (AML). This study aimed to detect serum miR-203 expression levels in AML and explore its potential clinical significance. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was performed to measure the serum miR-203 levels in 134 patients with AML and 70 healthy controls. The results demonstrated that serum miR-203 expression was significantly reduced in AML patients compared with healthy controls. Receiver operating characteristic curve (ROC) analysis revealed miR-203 could distinguish AML cases from normal controls. Low serum miR-203 levels were associated with worse clinical features, as well as poorer overall survival and relapse free survival of AML patients. Moreover, multivariate analysis confirmed low serum miR-203 expression to be an independent unfavorable prognostic predictor for AML. The bioinformatics analysis showed that the downstream genes and pathways of miR-203 was closely associated with tumorigenesis. Downregulation of miR-203 in AML cell lines upregulated the expression levels of oncogenic promoters such as CREB1, SRC and HDAC1. Thus, these findings demonstrated that serum miR-203 might be a promising biomarker for the diagnosis and prognosis of AML.
Keywords: Serum miR-203, acute myeloid leukemia, diagnosis, prognosis
Introduction
Acute myeloid leukemia (AML) is the most frequent type of hematopoietic malignancy with proliferation of myeloblasts in the bone marrow. Although the survival rates of AML have been improved over the past decades, the relapse risk remains high in AML and a large proportion of cases still fail to achieve 5-year survival [1-3]. In addition, the prognosis of AML is variable and ranges from a few-weeks’ survival to a cure; thus it is very difficult to evaluate the clinical outcome of AML patients [4,5]. Therefore, identification of a novel biomarker would help diagnosis and prognosis prediction of AML.
MicroRNAs (miRNAs) are a class of small, non-coding RNAs that negatively regulate gene expression at the post-transcriptional level by binding to 3’-untranslated regions of target messenger RNA (mRNA), leading to degradation of mRNA and/or translational inhibition [6,7]. miRNAs are actively involved in regulating multiple processes of carcinogenesis, includng cell growth, apoptosis, differentiation, invasion, and proliferation [8,9]. Many aberrantly expressed miRNAs have been reported to be present in serum/plasma in a stable and reproducible fashion and are identified as useful tools for diagnosis and prognosis of AML. For instance, the expression of miR-328 [10], miR-96 [11], miR-215 [12] is reduced in AML cases and downregulation of these miRNAs were negatively associated with unfavorable clinical outcome. However, elevated expression of miR-99a [13], and miR-210 [14] is a common event in patients with AML.
miR-203 has been widely reported to have tumor-suppressive function and modulate the expression of multiple oncogenes such as Src, BCR/ABL, and survivin. In AML, Zhang et al revealed miR-203 was greatly decreased in CD34+ AML cells compared to CD34- cells and controls. In addition, upregulation of miR-203 significantly inhibited proliferation, self-renewal and prompted apoptosis in vivo, and suppressed tumorigenesis in xenograft tumor tissues. Both Bmi-1 and Survivin were identified as its direct and functional target genes [15]. However, the clinical significance of serum miR-203 in AML remains uncertain. In this study, we investigate serum miR-203 levels in AML patients and assess its potential clinical significance.
Materials and methods
Cell culture and transfections
The AML cell lines THP-1 and K-562 were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA). The miR-203 inhibitor and non-targeting control (scrambled miRNA) were obtained from Sigma. Lipofectamine RNAiMAX (Invitrogen) was used to transfect AML cells with 50 nmol/L of the miR-203 inhibitor or scrambled miRNA based on the manufacturer’s instructions.
Patient samples
The study was approved by the Ethics Committee of The Second Hospital of Shanxi Medical University. A total of 134 cases diagnosed with de novo AML (non-M3) were enrolled. According to the French-America-British (FAB) classification, 7 patients had AML M0, 40 had M1, 52 had M2, 17 had M4, 15 had M5, and 3 had M7. A control group of 70 healthy volunteers was recruited and none of them had any clinical symptoms of cancer or other diseases. AML complete remission (CR) was defined as a normocellular BM containing less than 5% blasts and normalization of the peripheral blood counts at one month after starting induction therapy. Details of clinical features of all patients are provided in Table 1. Prior informed consent was obtained from all participants.
Table 1.
Correlation between miR-203 expression and clinicopathologic parameters
| Characteristics | Cases | Serum miR-203 level | P | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Gender | NS | |||
| Men | 79 | 41 | 38 | |
| Women | 55 | 29 | 26 | |
| Age | NS | |||
| <55 | 82 | 42 | 40 | |
| ≥55 | 52 | 28 | 24 | |
| Bone marrow blasts | NS | |||
| <50% | 61 | 28 | 33 | |
| ≥50% | 73 | 42 | 31 | |
| WBC | NS | |||
| <10 | 40 | 17 | 23 | |
| ≥10 | 94 | 53 | 41 | |
| Complete Remission | 0.0178 | |||
| Yes | 57 | 23 | 34 | |
| No | 77 | 47 | 30 | |
| Cytogenetics | 0.0166 | |||
| Favorable | 38 | 13 | 25 | |
| Intermediate | 65 | 36 | 29 | |
| Unfavorable | 31 | 21 | 10 | |
| FAB Classification | 0.0039 | |||
| M0 | 7 | 2 | 5 | |
| M1 | 40 | 12 | 28 | |
| M2 | 52 | 35 | 17 | |
| M4 | 17 | 8 | 9 | |
| M5 | 15 | 11 | 4 | |
| M7 | 3 | 2 | 1 | |
NS, no significant; WBC, white blood cells; FAB, French-American-British.
Up to 5 ml whole blood was withdrawn from each participant and collected in EDTA-K2 tubes. Subsequently, the supernatant was isolated from blood samples by centrifuging at 1200 g for 10 min, then centrifuged at 12000 g for 10 min at 4°C, and transferred to cryotubes and stored at -80°C.
RNA extraction, quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the serum using a QIAamp RNA Blood kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s protocols. The quantity and concentration of RNA were spectrophotometrically assessed by measuring absorbance at A260/280. Reverse transcription reactions were carried out with the Prime-Script RT reagent kit (TaKaRa, Dalian, China). Quantitative RT-PCR was run on an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using miScript SYBR green PCR kit (Qiagen, Hilden, Germany). The relative miR-203 expression was calculated by the comparative 2-ΔΔCt method using cer-miR-39 as the endogenous control. Triplicates were performed for all qRT-PCR reactions.
Bioinformatic analysis of the downstream genes of miR-203
The targeted genes of miR-203 were obtained from TargetScan7.2 (http://www.targetscan.org/vert_71/). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/). STRING online database (https://string-db.org/) was used for the protein-protein interaction (PPI) analysis. The central nodes with most connections were chosen for validation in AML cell lines.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software (GraphPad Software, San Diego, CA, USA). Overall survival (OS) was defined as the time from diagnosis of AML to any cause of death. Relapse-free survival (RFS) was defined as the time from CR to relapse or death. The Mann-Whitney U test or Kruskal-Wallis test were used to assess a significant difference in serum miR-203 expression between two groups or among multiple groups respectively. Chi-square analysis was used to evaluate the difference of categorical variables. The receiver operating characteristic (ROC) curve was constructed to determine the diagnosis value of serum miR-203 level. Kaplan-Meier method was used to estimate the relationship between miR-203 expression and OS and RFS. Multivariate survival analysis was performed using the Cox proportional hazards model. Differences were considered statistically significant when P<0.05.
Results
Reduced serum miR-203 expression in AML patients
miR-203 levels were detected in blood samples from all participants by qRT-PCR. As shown in Figure 1A, serum miR-203 expression was greatly decreased in AML patients compared to that in healthy controls. In addition, serum miR-203 levels in all AML subtypes significantly downregulated compared to healthy controls, but no significant difference in serum miR-203 expression was observed among AML subtypes (M0, M1, M2, M4, M5 and M7) (P>0.05, Figure 1B).
Figure 1.
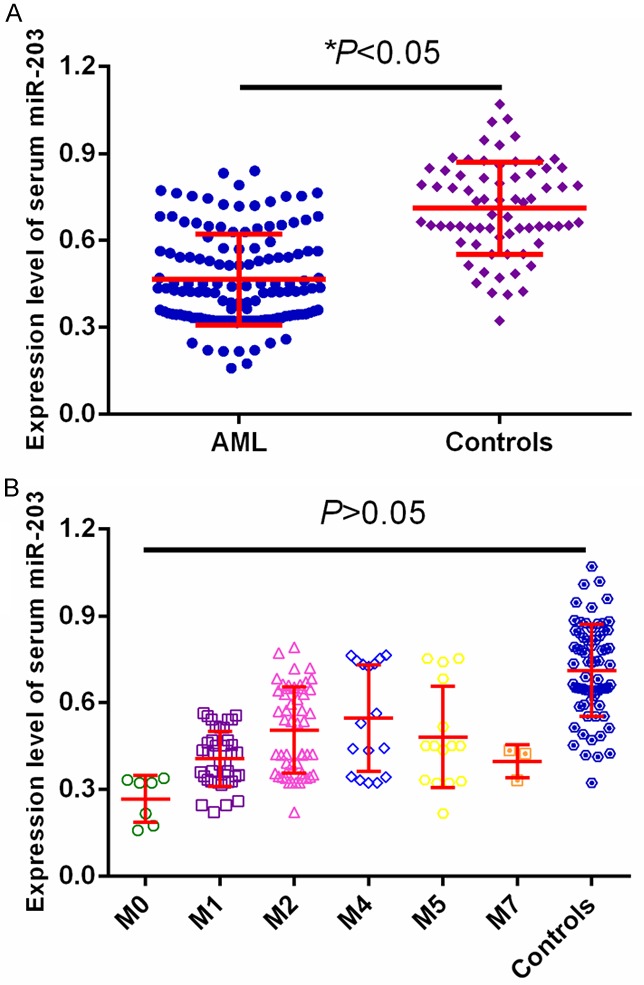
A. miR-203 expression was significantly lower in AML than in controls. B. miR-203 expression was significantly lower in different AML subtypes than in controls.
The diagnostic value of serum miR-203 for AML
ROC curve analysis identified serum miR-203 as a reliable biomarker for discriminating AML subjects from controls with the area under the ROC curve (AUC) of 0.853, and the sensitivity and specificity were 77.61% and 81.43%, respectively (Figure 2). Among all AML patients, 57 cases achieved CR. We compared the miR-203 levels in paired blood samples of these patients and found miR-203 expression was significantly upregulated when CR was achieved after chemotherapy (P<0.05, Figure 3).
Figure 2.
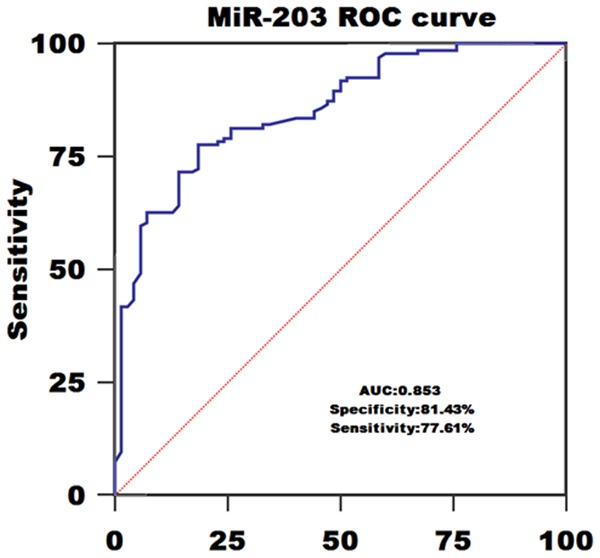
ROC curve of using serum miR-203 levels to diagnose AML.
Figure 3.
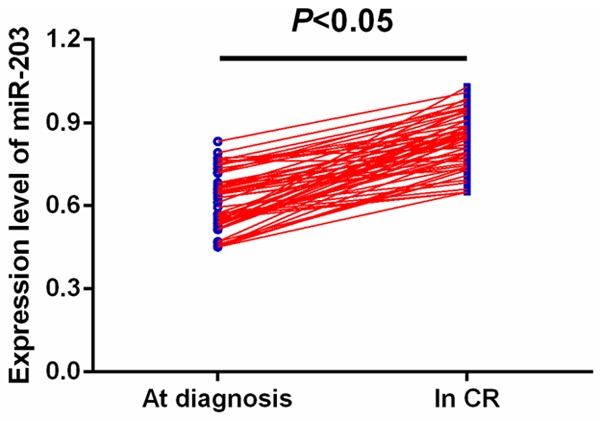
Comparison of miR-203 levels in 57 paired serum samples before and after CR.
Correlation between serum miR-203 expression and clinicopathologic factors
Table 1 depicts the correlation between serum miR-203 levels and various clinical factors. All AML patients were divided into high and low serum miR-203 group based on the median fold-change values. Serum miR-203 expression was significantly associated with cytogenetics (P=0.0166), complete remission (P=0.0178), and FAB classification (P=0.0039). However, other clinical characteristics including gender, age, bone marrow blasts and white blood cells (WBC) were not directly correlated to serum miR-203 expression (all P>0.05).
Correlation between serum miR-203 expression and prognosis of AML
Kaplan-Meier analysis revealed that AML patients with low serum miR-203 levels had shorter OS and RFS rates than those with high serum miR-203 levels (P=0.016, Figure 4A; P=0.012, Figure 4B, respectively).
Figure 4.
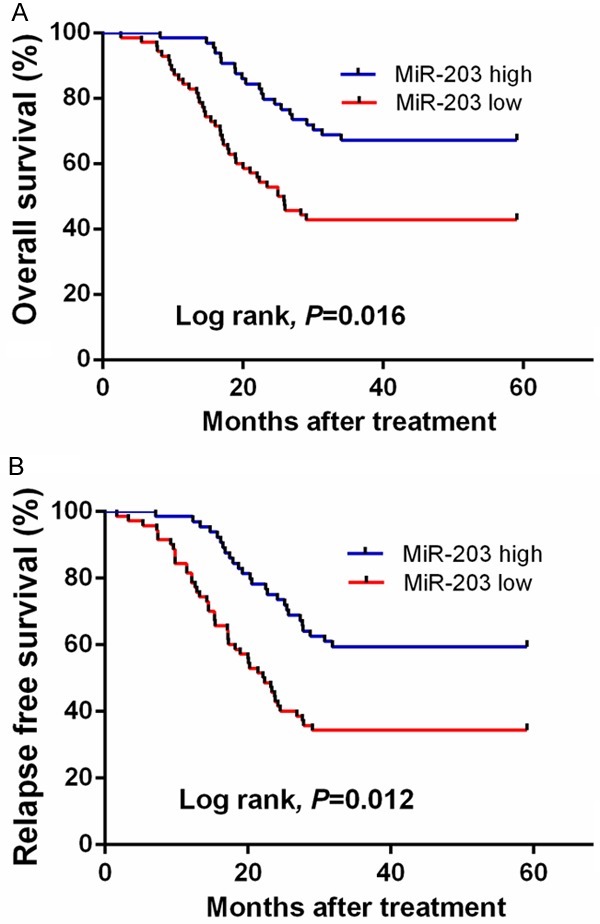
A. Kaplan-Meier curve of OS of all AML patients stratified by serum miR-203 levels. B. Kaplan-Meier curve of RFS of all AML patients stratified by serum miR-203 levels.
Multivariate survival analysis demonstrated that serum miR-203 level (OS: OR=3.35, 95% CI=1.78-5.06, P=0.002; RFS: OR=3.02, 95% CI=1.52-4.73, P=0.003), cytogenetics (OS: OR=2.67, 95% CI=1.34-4.15, P=0.005; RFS: OR=2.27, 95% CI=1.07-3.55, P=0.008), and FAB classification (OS: OR=2.86, 95% CI=1.43-4.35, P=0.004; RFS: OR=2.36, 95% CI=1.14-3.69, P=0.008) were prognostic markers for OS and RFS (Table 2).
Table 2.
Multivariable analysis of the impact of variables on OS and RFS in 134 AML patients
| Variable | Overall survival | Relapse free survival | ||
|---|---|---|---|---|
|
|
|
|||
| OR (95% CI) | P | OR (95% CI) | P | |
| miR-203 level | 3.35 (1.78-5.06) | 0.002 | 3.02 (1.52-4.73) | 0.003 |
| Cytogenetics | 2.67 (1.34-4.15) | 0.005 | 2.27 (1.07-3.55) | 0.008 |
| FAB Classification | 2.86 (1.43-4.35) | 0.004 | 2.36 (1.14-3.69) | 0.008 |
Bioinformatics analysis
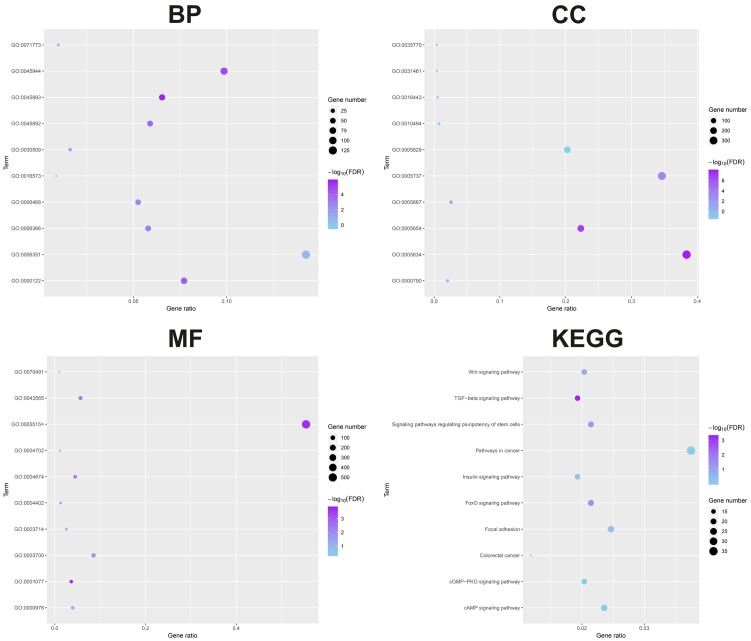
Bioinformatic analysis showed that GO:005893~positive regulation of transcription, DNA-templated, GO:0045944~positive regulation of transcription from RNA polymerase II promoter and GO:0000122~negative regulation of transcription from RNA polymerase II promoter were the top biologic processes. GO:0005634~nucleus, GO:0005654~nucleoplasm, GO:0005737~cytoplasm, were the top enriched cellular components. GO:0001077~transcritional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding, GO:0005515~protein binding and GO:0043565~sequence-specific DNA binding were the top enriched in molecular function. TGF-beta signaling pathway, FoxO signaling pathway, Signaling pathways regulating pluripotency of stem cells, Wnt signaling pathway, and Insulin signaling pathway were the top enriched KEGG pathways (Figure 5). We first identified the central nodes of the PPI network from the downstream targets (connections ≥20) (Figure 6A). Our qPCR results showed that the expression levels of CREB1, SRC, and HDAC1 were significantly higher in AML cell lines subjected to miR-203 inhibitor transfection compared to the controls (Figure 6B).
Figure 5.
GO and KEGG analysis of the downstream genes of miR-203.
Figure 6.
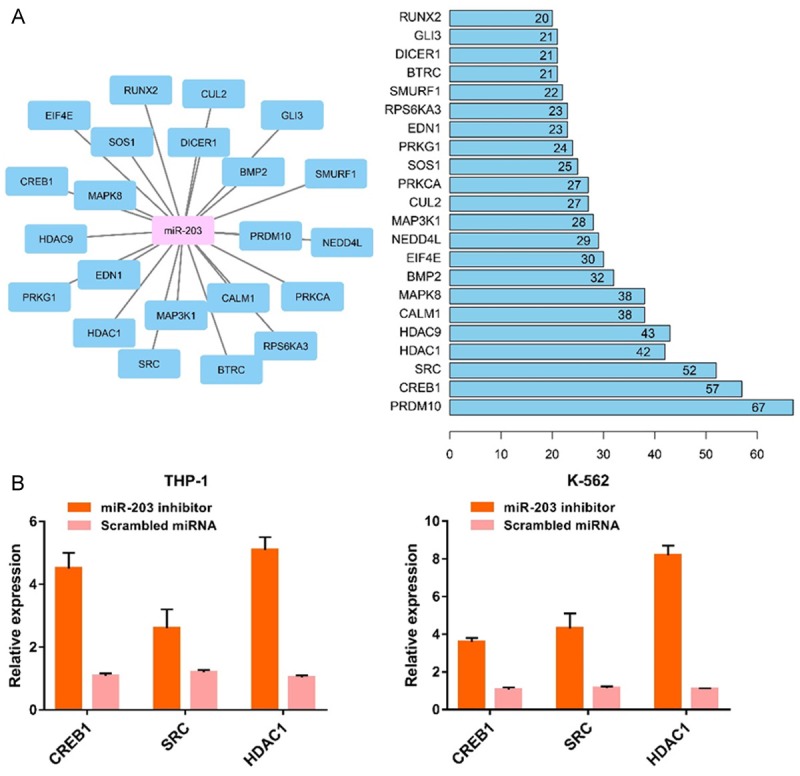
The central nodes of the PPI network (A). The expression levels of CREB1, SRC and HDAC1 were significantly higher in AML cells subjected to miR-203 suppression (B).
Discussion
miRNAs are reliably detectable in plasma/serum and their expression has been used as a biomarker for various types of cancer. Our study demonstrated that serum miR-203 expression was significantly lower in AML cases than that in healthy controls. The ROC curve analysis showed that serum miR-203 was useful as a potential tumor marker for diagnosis of AML. In addition, serum miR-203 expression was closely associated with cytogenetics, complete remission and FAB classification. Serum miR-203 levels were increased significantly when the patients obtained a CR. Moreover, AML patients with low serum miR-203 expression had shorter survival, and multivariate analysis showed that serum miR-203 expression was an independent indicator of OS/RFS. Bioinformatics analysis showed that the downstream genes and pathways of miR-203 were closely associated with tumorigenesis. Downregulation of miR-203 in AML cell lines upregulated the expression levels of oncogenic promoters such as CREB1, SRC and HDAC1. These results suggested that low serum miR-203 is an adverse prognostic factor in AML and might serve as a useful diagnostic and prognostic biomarker. However, the limitation of this study is the small sample size, and our results require further validation with large cohorts.
In line with our findings, miR-203 had been reported to exert a tumor suppressive role in multiple cancers. For instance, the expression level of miR-203 was decreased in bladder cancer tissues, and upregulation of miR-203 induced apoptosis of cancer cells and suppressed cell proliferation. Moreover, in vitro and in vivo evidence identified bcl-w as its downstream target [16]. Similarly, Zhang et al verified that deletion of serum miR-203 was found in patients with bladder cancer, and reduced serum miR-203 predicted poorer survival. Ectopic expression of miR-203 suppressed bladder cancer tumorigenic potential and enhanced cisplatin cytotoxicity by regulating Bcl-w and survivin [17]. In lung cancer, overexpression of miR-203 greatly reduced cancer cell proliferation, and migration and stimulated apoptosis via degrading LIN28B [18], PKCα [19] and SRC [20]. In osteosarcoma, miR-203 levels were significantly decreased in cancer cell lines and tissues. Restoration of miR-203 markedly inhibited cancer cell growth, invasion, migration, and suppressed mesenchymal-to-epithelial reversion transition (MErT) through targeting RAB22A [21] or TBK1 [22]. Also, miR-203 expression was dramatically down-regulated in the tissues and cell lines of cervical cancer. Upregulation of miR-203 greatly suppressed tumorigenicity and angiogenesis in vivo by silencing VEGFA expression [23]. miR-203 overexpression was inversely correlated with lymph node metastasis [24]. Zhao et al demonstrated that miR-203 was downregulated in ovarian cancer tissues. Enforced miR-203 expression could greatly attenuate cell proliferation, invasion and migration, and inhibit epithelial-mesenchymal transition by targeting Snai2 [25]. In prostate cancer (PC), a reduction in miR-203 expression was found in bone metastatic PC, PC tissues and cell lines. Furthermore, miR-203 overexpression markedly suppressed cell growth, migration and invasion in vitro and in vivo through the repression of ZEB2, Bmi, survivin and Rap1A [26,27]. In gastric cancer, Chu and colleagues reported low miR-203 expression predicted poor prognosis of patients, and either loss of PIBF1 or miR-203 upregulation restrained cell proliferation in vitro and inhibited tumorigenicity in vivo [28]. In hepatocellular carcinoma (HCC), reduced miR-203 levels were observed in HCC tissues and associated with aggressive clinical variables. miR-203 overexpression resulted in the inhibition of the proliferation and lung metastasis of hepatic residual HCC [29,30]. Moreover, miR-203 downregulation was found in non-small cell lung cancer tissues. In vitro [31] and in vivo [32] evidence showed that its overexpression strongly inhibited the carcinogenesis by targeting Bmi1 and RGS17. In glioblastoma (GBM), miR-203 was downregulated in GBM tissues and cell lines. Elevated miR-203 expression decreased cell viability and growth through Robo1/ERK/MMP-9 signaling [33]. In head and neck squamous cell carcinoma, high miR-203 expression was shown to inhibit cell invasion, promoted mesenchymal-epithelial transition and negatively correlated with poor clinical outcome [34].
More interestingly, the tumor-suppressive role of miR-203 remains controversial in some cancer types. In breast cancer (BC), significantly lower miR-203 expression was detected in metastatic BC cells and cell lines, and ectopic miR-203 expression could inhibit cell invasion, migration, and lung metastatic colonization [35,36]. In contrast, miR-203 might have an oncogenic activity because higher miR-203 levels were detected in BC tissues and the MCF-7 cell line, and miR-203 knockdown decreased colony formation, and transformation, and sensitized MCF-7 cells to cisplatin [37,38]. Furthermore, Miao et al revealed elevated miR-203 expression repressed cancer cell migration, invasion and epithelial to mesenchymal transition by targeting caveolin-1 in pancreatic cancer [39]. However, Greither and colleagues demonstrated high miR-203 expression was an independent indicator of shorter survival in patients with pancreatic ductal adenocarcinomas, indicating miR-203 might be an oncogenic miRNA [40]. Therefore, miR-203 might have different regulatory roles during the initiation and progression in some kinds of tumors.
In conclusion, we have demonstrated that low serum miR-203 expression is associated with aggressive clinical features and poor survival of AML. Therefore, serum miR-203 might be a promising marker for the diagnosis and prognosis of AML.
Acknowledgements
The present study was supported by the Natural Science Foundation of Shanxi Province (no. 201801D121330).
Disclosure of conflict of interest
None.
References
- 1.Löwenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 2.Tallman MS, Gilliland DG, Rowe JM. Drug therapy for acute myeloid leukemia. Blood. 2005;106:1154–1163. doi: 10.1182/blood-2005-01-0178. [DOI] [PubMed] [Google Scholar]
- 3.Gregory TK, Wald D, Chen Y, Vermaat JM, Xiong Y, Tse W. Molecular prognostic markers for adult acute myeloid leukemia with normal cytogenetics. J Hematol Oncol. 2009;2:23. doi: 10.1186/1756-8722-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thol F, Ganser A. Molecular pathogenesis of acute myeloid leukemia: a diverse disease with new perspectives. Front Med China. 2010;4:356–362. doi: 10.1007/s11684-010-0220-5. [DOI] [PubMed] [Google Scholar]
- 5.Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013;88:318–327. doi: 10.1002/ajh.23404. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Chen R, Zhang Y, Fan W, Xiao F, Yan X. Low expression of circulating microRNA-328 is associated with poor prognosis in patients with acute myeloid leukemia. Diagn Pathol. 2015;10:109. doi: 10.1186/s13000-015-0345-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Lu Q, Zhu J, Fu J, Chen YX. Prognostic value of miR-96 in patients with acute myeloid leukemia. Diagn Pathol. 2014;9:76. doi: 10.1186/1746-1596-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YX, Zhang TJ, Yang DQ, Yao DM, Yang L, Zhou JD, Deng ZQ, Ma JC, Guo H, Wen XM, Lin J, Qian J. Reduced miR-215 expression predicts poor prognosis in patients with acute myeloid leukemia. Jpn J Clin Oncol. 2016;46:350–356. doi: 10.1093/jjco/hyv204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Si X, Zhang X, Hao X, Li Y, Chen Z, Ding Y, Shi H, Bai J, Gao Y, Cheng T, Yang FC, Zhou Y. Upregulation of miR-99a is associated with poor prognosis of acute myeloid leukemia and promotes myeloid leukemia cell expansion. Oncotarget. 2016;7:78095–78109. doi: 10.18632/oncotarget.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X, Chen L, Yan X, Li Y, Xiong Y, Zhou X. Overexpression of miR-210 is associated with poor prognosis of acute myeloid leukemia. Med Sci Monit. 2015;21:3427–3433. doi: 10.12659/MSM.894812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Zhou SY, Yan HZ, Xu DD, Chen HX, Wang XY, Wang X, Liu YT, Zhang L, Wang S, Zhou PJ, Fu WY, Ruan BB, Ma DL, Wang Y, Liu QY, Ren Z, Liu Z, Zhang R, Wang YF. MiR-203 inhibits proliferation and self-renewal of leukemia stem cells by targeting survivin and Bmi-1. Sci Rep. 2016;6:19995. doi: 10.1038/srep19995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bo J, Yang G, Huo K, Jiang H, Zhang L, Liu D, Huang Y. MicroRNA-203 suppresses bladder cancer development by repressing bcl-w expression. FEBS J. 2011;278:786–792. doi: 10.1111/j.1742-4658.2010.07997.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Zhang Y, Liu X, Fang A, Li P, Li Z, Liu T, Yang Y, Du L, Wang C. MicroRNA-203 is a prognostic indicator in bladder cancer and enhances chemosensitivity to cisplatin via apoptosis by targeting bcl-w and survivin. PLoS One. 2015;10:e0143441. doi: 10.1371/journal.pone.0143441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Liang H, Liao Z, Wang Y, Hu X, Chen X, Xu L, Hu Z. MiR-203 enhances let-7 biogenesis by targeting LIN28B to suppress tumor growth in lung cancer. Sci Rep. 2017;7:42680. doi: 10.1038/srep42680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Wang X, Liang H, Wang T, Yan X, Cao M, Wang N, Zhang S, Zen K, Zhang C, Chen X. MiR-203 inhibits cell proliferation and migration of lung cancer cells by targeting PKCα. PLoS One. 2013;8:e73985. doi: 10.1371/journal.pone.0073985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang N, Liang H, Zhou Y, Wang C, Zhang S, Pan Y, Wang Y, Yan X, Zhang J, Zhang CY, Zen K, Li D, Chen X. MiR-203 suppresses the proliferation and migration and promotes the apoptosis of lung cancer cells by targeting SRC. PLoS One. 2014;9:e105570. doi: 10.1371/journal.pone.0105570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang D, Liu G, Wang K. MiR-203 acts as a tumor suppressor gene in osteosarcoma by regulating RAB22A. PLoS One. 2015;10:e0132225. doi: 10.1371/journal.pone.0132225. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Liu S, Feng P. MiR-203 determines poor outcome and suppresses tumor growth by targeting TBK1 in osteosarcoma. Cell Physiol Biochem. 2015;37:1956–1966. doi: 10.1159/000438556. [DOI] [PubMed] [Google Scholar]
- 23.Zhu X, Er K, Mao C, Yan Q, Xu H, Zhang Y, Zhu J, Cui F, Zhao W, Shi H. MiR-203 suppresses tumor growth and angiogenesis by targeting VEGFA in cervical cancer. Cell Physiol Biochem. 2013;32:64–73. doi: 10.1159/000350125. [DOI] [PubMed] [Google Scholar]
- 24.Zhao S, Yao DS, Chen JY, Ding N. Aberrant expression of miR-20a and miR-203 in cervical cancer. Asian Pac J Cancer Prev. 2013;14:2289–2293. doi: 10.7314/apjcp.2013.14.4.2289. [DOI] [PubMed] [Google Scholar]
- 25.Zhao G, Guo Y, Chen Z, Wang Y, Yang C, Dudas A, Du Z, Liu W, Zou Y, Szabo E, Lee SC, Sims M, Gu W, Tillmanns T, Pfeffer LM, Tigyi G, Yue J. MiR-203 functions as a tumor suppressor by inhibiting epithelial to mesenchymal transition in ovarian cancer. J Cancer Sci Ther. 2015;7:34–43. doi: 10.4172/1948-5956.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang J, Bian C, Wang H, Huang S, Wu D. MiR-203 down-regulates Rap1A and suppresses cell proliferation, adhesion and invasion in prostate cancer. J Exp Clin Cancer Res. 2015;34:8. doi: 10.1186/s13046-015-0125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V, Chen Y, Deng G, Tanaka Y, Dahiya R. Regulatory role of mir-203 in prostate cancer progression and metastasis. Clin Cancer Res. 2011;17:5287–5298. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
- 28.Chu SJ, Wang G, Zhang PF, Zhang R, Huang YX, Lu YM, Da W, Sun Q, Zhang J, Zhu JS. MicroRNA-203 suppresses gastric cancer growth by targeting PIBF1/Akt signaling. J Exp Clin Cancer Res. 2016;35:47. doi: 10.1186/s13046-016-0323-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Liu Y, Ren F, Rong M, Luo Y, Dang Y, Chen G. Association between underexpression of microrna-203 and clinicopathological significance in hepatocellular carcinoma tissues. Cancer Cell Int. 2015;15:62. doi: 10.1186/s12935-015-0214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng XB, Chen XB, Xu LL, Zhang M, Feng L, Yi PS, Tang JW, Xu MQ. MiR-203 inhibits augmented proliferation and metastasis of hepatocellular carcinoma residual in the promoted regenerating liver. Cancer Sci. 2017;108:338–346. doi: 10.1111/cas.13167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen T, Xu C, Chen J, Ding C, Xu Z, Li C, Zhao J. MicroRNA-203 inhibits cellular proliferation and invasion by targeting Bmi1 in non-small cell lung cancer. Oncol Lett. 2015;9:2639–2646. doi: 10.3892/ol.2015.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi Y, Jin Q, Liu X, Xu L, He X, Shen Y, Zhou Q, Zhang J, Jin M. MiR-203 inhibits cell proliferation, invasion, and migration of non-small-cell lung cancer by downregulating RGS17. Cancer Sci. 2017;108:2366–2372. doi: 10.1111/cas.13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dontula R, Dinasarapu A, Chetty C, Pannuru P, Herbert E, Ozer H, Lakka SS. MicroRNA 203 modulates glioma cell migration via Robo1/ERK/MMP-9 signaling. Genes Cancer. 2013;4:285–296. doi: 10.1177/1947601913500141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obayashi M, Yoshida M, Tsunematsu T, Ogawa I, Sasahira T, Kuniyasu H, Imoto I, Abiko Y, Xu D, Fukunaga S, Tahara H, Kudo Y, Nagao T, Takata T. MicroRNA-203 suppresses invasion and epithelial-mesenchymal transition induction via targeting NUAK1 in head and neck cancer. Oncotarget. 2016;7:8223–8239. doi: 10.18632/oncotarget.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding X, Park SI, McCauley LK, Wang CY. Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J Biol Chem. 2013;288:10241–10253. doi: 10.1074/jbc.M112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z, Dong JT. Epigenetic silencing of miR-203 upregulates SNAI2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer. 2011;2:782–791. doi: 10.1177/1947601911429743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ru P, Steele R, Hsueh EC, Ray RB. Anti-miR-203 upregulates SOCS3 expression in breast cancer cells and enhances cisplatin chemosensitivity. Genes Cancer. 2011;2:720–727. doi: 10.1177/1947601911425832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He S, Zhang G, Dong H, Ma M, Sun Q. MiR-203 facilitates tumor growth and metastasis by targeting fibroblast growth factor 2 in breast cancer. Onco Targets Ther. 2016;9:6203–6210. doi: 10.2147/OTT.S108712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miao L, Xiong X, Lin Y, Cheng Y, Lu J, Zhang J, Cheng N. MiR-203 inhibits tumor cell migration and invasion via caveolin-1 in pancreatic cancer cells. Oncol Lett. 2014;7:658–662. doi: 10.3892/ol.2014.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greither T, Grochola LF, Udelnow A, Lautenschläger C, Würl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]