Abstract
Purpose: Whether patients with stage IA-IIA lung adenocarcinoma require conventional chemotherapy is still a controversy. An ideal metastasis risk prediction model in lung adenocarcinoma is valuable for determining the prognosis and giving timely, individualized treatment. Results: Analyzing the clinical cases of 153 lung adenocarcinoma patients using an χ2 test, Kaplan-Meier survival curves, and a multivariate logistic regression analysis, we selected the most valuable factors for determining metastasis and constructed metastasis prediction models. We confirmed the importance of the tumor markers (CEA, NSE) and a molecular marker (CAMKII) as independent prognostic factors in lung adenocarcinoma. The result of a five-year survival status was significantly associated with CAMKII and CEA (P < 0.05). A nomogram was created using CEA, NSE, CYFRA 21-1, and CAMKII to estimate the metastasis probability for individuals, specifically, 78 stage I lung adenocarcinoma patients were used to verify the effectiveness of the nomogram. Using machine learning, LASSO selected the subset of variables that minimized the predictive error of the outcome, including CEA, NSE, CYFRA 21-1, CAMKII, tumor size, histologic type, lymph node status, smoking, and age. A ten-fold cross-validation showed the average accuracy of this model was 86.208%, with an area under the curve of 0.857, a sensitivity value of 0.840, and a specificity value of 0.873. Conclusion: Using both complementary methods, the predictive models illustrated that the combination of tumor markers and a key molecule to predict the prognosis of lung adenocarcinoma patients in early stages is valuable. The postoperative transfer rate of stage I patients can be effectively predicted by these complementary methods.
Keywords: Nomogram, machine learning, CAMKII, serum tumor markers, risk of metastasis
Introduction
Lung carcinoma is the most common human malignant disease and the leading cause of cancer-related death in the world, with almost 1.6 million people dying from lung cancer annually [1]. Particularly, lung adenocarcinoma (ADC) is a histological type whose incidence is increasing year by year [2]. Despite recent advances in the target chemo- or radio-methods available for application, the 5-year survival rate remains poor. Even among early-stage patients, the mortality risk remains high, with a high rate of relapse and metastasis [3]. Staging according to the tumor, node, and metastasis (TNM) system has been widely used to estimate the outcomes of patients with adenocarcinoma in current clinical practice. The US National Comprehensive Cancer Network guidelines recommend that chemotherapy be selected for patients with high-risk factors for stage IA-IIA lung adenocarcinoma. However, there is no definite prognostic biomarker for non-small-cell lung cancer yet [4]. Whether patients with stage IA-IIA lung adenocarcinoma should receive conventional chemotherapy is still controversial. Metastasis is the leading cause of treatment failure and cancer-associated mortalities in lung adenocarcinoma. Therefore, the construction of an ideal metastasis risk prediction model in adenocarcinoma patients is valuable for guiding the judgment of prognosis and giving individualized treatment on time.
Tumor markers are easy to obtain and detect in patients’ serum and have a wide range of clinical applications. Some markers are considered to be a convenient supplementary method for disease diagnosis or post-therapy surveillance, such as prostate-specific antigen (PSA) for prostate cancer. For lung cancer diagnosis, the related tumor markers comprise carcinoembryonic antigen (CEA), cytokeratin 19 fragments (CYFRA 21-1), neuron-specific enolase (NSE), total prostate-specific antigen (TPSA), and squamous cell carcinoma antigen (SCC) [5,6]. Recent studies have reported that an increase in these tumor markers is associated with metastasis and poor prognosis [7,8]. Clinically, the combined detection of multiple markers is the most common method.
Ca2+/calmodulin-dependent protein kinase II (CAMKII), a molecular marker, is a serine/threonine-specific protein kinase that responds to Ca2+ fluctuations [9]. Recent studies have demonstrated that high levels of CAMKII play a critical role in the regulation of the proliferation, differentiation and survival of various cancer cells, expressed in several cancers such as breast, prostate and colon cancer [10-12]. Increasing evidence has suggested that the role of CAMKII as a biomarker in cancer diagnosis and therapy should be explored in future research [13]. We found the expression of CAMKII to be closely associated with tumor metastasis in lung adenocarcinoma. As an independent prognostic factor, the importance of CAMKII was confirmed by machine learning as well.
A nomogram is a meaningful and well-accepted tool for predicting cancer prognosis because it provides intuitive and convenient information on the probability of clinical events based on the ability to combine statistically significant features and quantified risk in a graphical form [14]. The capability of the nomogram to precisely estimate the outcome for individuals would be helpful for clinicians in arriving at a treatment decision. Recent studies focus on single factors for building survival models, such as age, sex or TNM stage [15]. We combined tumor markers and molecular markers for the prediction of poor prognosis with a nomogram. The method of machine learning was then applied to evaluate the metastasis risk.
Machine learning is a field of computer science which combines the study of pattern recognition and computational learning theory to construct algorithms that can learn from data and make predictions on outcomes as well as uncover hidden insights [16]. It is already widely employed in biological domains such as genomics, proteomics, microarrays, systems biology, evolution, and text mining [17]. Compared with human intuition and standard statistics, the use of intensively computational approaches such as machine learning is part of a growing trend toward personalized, predictive medicine.
In our study, we evaluated the prognostic value of the traditional clinicopathological factors, leading tumor markers and expression of major proteins. We aimed to focus on the investigation of patients with early-stage lung adenocarcinoma that will have distant metastasis based on these factors, using nomograms and machine learning.
Materials and methods
Patients
As shown in the diagram of patient selection in Figure 1, in a database of 787 lung cancer patients undergoing curative resection at the Tianjin Medical University Cancer Institute and Hospital from June 1, 2011 to December 31, 2011, 223 patients were histologically diagnosed with lung adenocarcinoma. One hundred and sixty-two cases of 180 patients underwent EGFR mutation detection among the 223 lung adenocarcinoma patients who were followed up. Excluding patients who had no pre-therapy tumor marker examination, 153 patients with full panels of tumor marker data (CEA, TPSA, SCC, CYFRA 21-1, and NSE) were included in the final analysis. Seventy-eight patients diagnosed with stage I lung adenocarcinoma from January to April 2013 also had their diagnoses verified by machine learning. These patients had negative margins after surgery. All characteristics including age, gender, smoking, primary tumor size, histological tumor type, TNM stage, and development of lymphatic and distant metastases were determined and made up the patients’ clinical and pathological data. None of the patients had received chemotherapy or radiotherapy before their operations. The follow-up periods ranged from 64 to 84 months and ended on October 31, 2018. The protocols of this study were approved by the hospital’s Protection of Human Subjects Committee.
Figure 1.

Diagram of patient selection and study design.
Tumor marker measurement
Subjects had 5 ml of venous blood drawn with heparin anticoagulation in a fasting state before surgery or any other treatments. The serum was separated by centrifugation at 3000 rpm. The levels of the tumor markers (CEA, TPSA, SCC, NSE, and CYFRA 21-1) in the serum were measured by electrochemiluminescence using Roche COBAS6000 and Roche’s supporting reagents. The upper limits of the reference concentration recommended by the manufacturers of CEA, TPSA, SCC, CYFRA 21-1 and NSE were 5 µg/ml, 80 µg/ml, 1.5 µg/ml, 3.3 µg/ml, 15.2 µg/ml, respectively. Laboratory quality control guaranteed the validity of the results. All assays were blind to clinical information.
Immunohistochemistry
With approval from the Ethics Committee, lung cancer tissue samples were obtained from 162 patients undergoing surgical resection with a histologic diagnosis of lung cancer at the Tianjin Cancer Institute and Hospital. The paraffin-embedded tissues cut into 4 µm sections were deparaffinized by sequential washing with xylene, graded ethanol, and phosphate-buffered saline. The tissues were incubated overnight at 4°C with rabbit anti-CAMKII (Santa Cruz Biotechnology). The slides were treated with a broad-spectrum secondary antibody and then treated with dimethyl-aminoazobenzene. The expression of CAMKII was analyzed only histologically in neoplastic epithelial cells. Immunoreactivity was semiquantitatively scored according to the estimated percentage of positive tumor cells as previously described; staining intensity was scored 1 (negative) and 2 (positive). The immunoreactivity percentage was scored on a scale from 0 to 3 (0 for no positive cells, 1 for < 25% of cells being positive, 2 for 25-50% of cells being positive, and 3 for > 50% of cells being positive). A final immunoreactive score, also known as the staining index, was calculated between 0 and 6 by multiplying the percentage of positive cells by the staining intensity score. A total score of 0-3 indicates a negative expression of protein, and a total score ≥ 4 indicates a positive expression protein. The processed sections were examined using an Olympus BX51 microscope and the resulting images were captured using the AnalySIS program.
Construction of nomogram and model validation
Nomograms were used to assign 98 stage II-IV cases among 153 lung adenocarcinoma patients diagnosed in 2011 as a training cohort and 78 stage I lung adenocarcinoma patients as a validation cohort in addition. Univariate and multivariate analyses were used to screen for significant prognostic factors, including tumor markers, molecular markers, and clinical and pathological features. Only predictors with a P-value < 0.05 were incorporated into the nomogram. The resulting multivariate log-rank regression model was used to calculate the risk score and build the final nomogram prognostic model. In the validation cohort, the predictive ability of the nomogram was measured using a receiver operating characteristic (ROC) curve analysis. All statistical analyses were carried out using R software (version 3.1.0) and SPSS version 22.0 (IBM Corp., Armonk, NY, USA). A value of P < 0.05 was considered statistical significant.
Machine learning
For the model constructed, the random forest machine learning scheme was employed for the classification. The least absolute shrinkage and selection operator (LASSO) was used to pre-identify the most predictive features before the classification experiments. Ten-fold cross-validation was performed to identify the model classification performance. Classification performance was evaluated by the area under the AUC, accuracy, sensitivity and specificity. The model was constructed using Matlab version R2017a (MathWorks Inc., Natick, Massachusetts, USA).
Statistical analysis
The paired χ2-test for continuous variables and the chi-square test for categorical variables were used to compare two groups. Logistic regression models were used to estimate the odds ratio (OR) and the 95% confidence interval (CI) and to identify independent prognostic variables for 5-year distant metastasis. Kaplan-Meier survival curves and a multivariate Cox regression analysis were used to analyze mortality at 5 years. All the other statistical tests were performed using R (version 3.1.0) and SPSS software (version 22.0). A value of P < 0.05 was considered statistically significant.
Results
General characteristics of the study population
The general clinicopathological characteristics of the adenocarcinoma patients are shown in Table 1. The median age of all patients was 58 years, with 23 (15%) cases being younger than 50 years. About half of the adenocarcinoma tissues obtained from surgery were smaller than 3 cm. Comparatively, 19 patients (12.4%) were diagnosed with TNM stage IV, 62.1% cases were in the early stage (55 TNM stage I cases and 40 cases in TNM stage II). Fifty-six cases of 153 adenocarcinoma patients were diagnosed with EGFR mutation, comprising approximately 36.6% of all tested cases. As shown in the table regarding the general characteristics, 63 patients used to smoke, and adenocarcinoma patients comprised half of the men and women in our study. While highly differentiated histological type tissues accounted for 70% of our cases (56 with acinar predominant and 51 with lepidic predominant types), the poorly differentiated cases comprised only 18.9% with 17 solid predominant and 12 micropapillary predominant. The upper limits of reference concentration recommended by the manufacturers of CEA, TPSA, SCC, CYFRA 21-1, and NSE were 5 µg/ml, 80 µg/ml, 1.5 µg/ml, 3.3 µg/ml, 15.2 µg/ml, respectively. According to these limits, 63 (41.2%) CEA, 29 (19%) TPSA, 9 (5.9%) SCC, 47 (30.7%) CYFRA 21-1, and 24 (15.7%) NSE were higher than the normal value.
Table 1.
General characteristics of the ADC patients
| Factors | Patient Number (%) | |
|---|---|---|
| Age (year) | Median (Range) | 58 (52.9, 47.1) |
| < 50 | 23 (15) | |
| ≥ 50 | 130 (85) | |
| Gender | Male | 77 (50.3) |
| Female | 76 (49.7) | |
| Smoking | Never | 90 (58.8) |
| Former or current | 63 (41.2) | |
| Tumor Size (cm) | Median (Range) | 3 (49,51) |
| ≤ 3 | 75 (49) | |
| > 3 | 78 (51) | |
| Histological type | Acinar predominant | 56 (36.6) |
| Lepidic predominant | 51 (33.3) | |
| Papilary predominant | 17 (11.1) | |
| Solid predominant | 17 (11.1) | |
| Micropapilary predominant | 12 (7.8) | |
| TNM | TNMI | 55 (35.9) |
| TNMII | 40 (26.2) | |
| TNMIII | 39 (25.5) | |
| TNMIV | 19 (12.4) | |
| NSE | Median (Range) | 11.85 (50.3, 49.7) |
| ≤ 15.2 ug/ml | 129 (84.3) | |
| > 15.2 ug/ml | 24 (15.7) | |
| TPSA | Median (Range) | 45.41 (50.3, 49.7) |
| ≤ 80 ug/ml | 124 (81) | |
| > 80 ug/ml | 29 (19) | |
| SCC | Median (Range) | 0.7 (56.9, 43.1) |
| ≤ 1.5 ug/ml | 144 (94.1) | |
| > 1.5 ug/ml | 9 (5.9) | |
| CEA | Median (Range) | 3.87 (50.65, 49.35) |
| ≤ 5 ug/ml | 90 (58.8) | |
| > 5 ug/ml | 63 (41.2) | |
| Cyfra 21-1 | Median (Range) | 2.53 (50.3, 49.7) |
| ≤ 3.3 ug/ml | 106 (69.3) | |
| > 3.3 ug/ml | 47 (30.7) | |
| CAMKII | Low | 63 (41.2) |
| High | 90 (58.8) | |
| EGFR | Mutation | 56 (36.6) |
| Wild | 97 (63.4) |
The association of CAMKII expression with the clinicopathological features of the lung adenocarcinoma cases
In order to reveal the clinical significance of CAMKII protein expression levels in the lung adenocarcinoma tissues, we selected 153 human lung adenocarcinoma cases, and subjected them to an χ2 test. Among the 153 lung adenocarcinoma samples, 90 (58.8%) showed positive CAMKII expression (Figure 2A). The relationships between the CAMKII expressions and each of the cases with their clinicopathological parameters are summarized in Table 2. We found that the expression of CAMKII was not significantly associated with patient age or gender, tumor size or smoking. However, the histological type (P < 0.05), TNM stage (P < 0.05), lymphatic and distant metastasis (P < 0.05) were strongly close to the expression of CAMKII. The solid predominant and micropapillary predominant types are more malignant than others; we found high-level expressions of CAMKII (82.4% and 91.7% respectively) in these two types (Figure 2B). The patients with positive CAMKII expressions were more vulnerable to metastasis compared to those with negative CAMKII expressions. Interestingly, the expression of CAMKII was associated with EGFR mutation (P < 0.05) and might provide a novel target for adenocarcinoma treatment. Thus, upregulated CAMKII expression has the potential to be developed as a biomarker for malignant phenotypes of lung adenocarcinoma.
Figure 2.
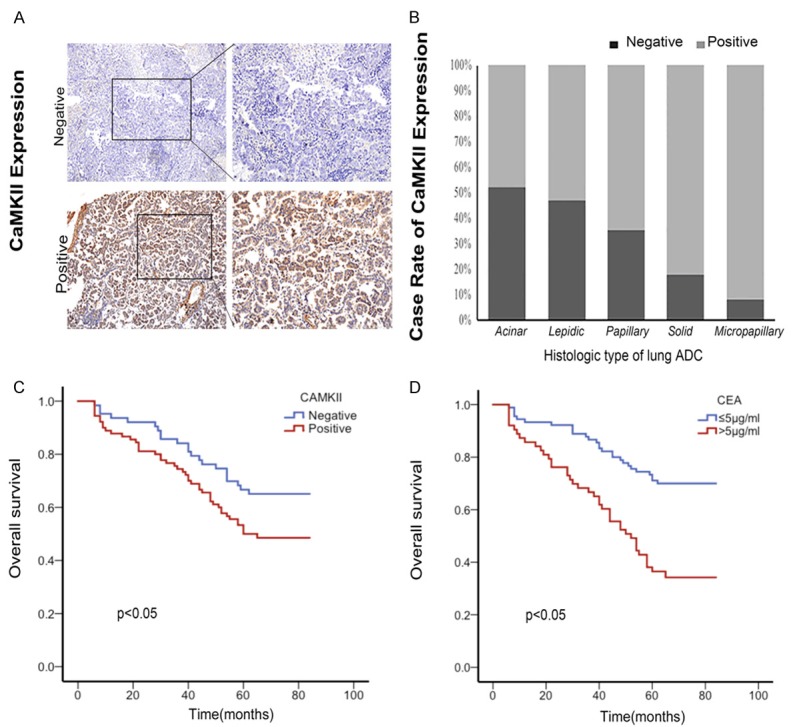
The expression of CAMKII and Kaplan-Meier estimates of overall survival. A. Representative IHC images showing the expression of CaMKII in lung ADC in micropapillary. 200*. B. The case rate expression of CaMKII in different histological types of ADC. Along with the ADC deteriorate, the expression of CAMKII was upregulated. Especially, the CAMKII expression in micropapillary predominant samples was totally positive. C. Positive CaMKII expression predicts a poor prognosis in patients with ADC. Kaplan-Meier curves displaying the overall survival of patients with negative CAMKII expression vs positive CaMKII expression (P < 0.05, Log-rank Test). D. Higher CEA (> 5 μg/ml) predicts poor prognosis in patients with ADC. Kaplan-Meier curves displaying the overall survival of patients with higher CEA vs positive lower CEA (P < 0.05, Log-rank Test).
Table 2.
The relationship between CAMKII expression and the clinicopathological characteristics of lung ADC
| Variable | Total (%) | CAMK2 expression | X2 | P | |
|---|---|---|---|---|---|
|
| |||||
| Low | High | ||||
| Age | |||||
| < 50 | 23 (15) | 11 (47.8) | 12 (52.2) | 0.494 | 0.499 |
| ≥ 50 | 130 (85) | 52 (40) | 78 (60) | ||
| Sex | |||||
| Male | 77 (50.3) | 32 (41.6) | 45 (58.4) | 0.009 | 1.000 |
| Female | 76 (49.7) | 31 (40.8) | 45 (59.2) | ||
| Smoking | |||||
| Never smoker | 90 (58.8) | 35 (38.9) | 55 (61.1) | 0.472 | 0.509 |
| Former or current smoker | 63 (41.2) | 28 (44.4) | 35 (55.6) | ||
| Tumor Size (cm) | |||||
| ≥ 3 | 78 (51) | 30 (38.5) | 48 (61.5) | 0.484 | 0.514 |
| < 3 | 75 (49) | 33 (44) | 42 (56) | ||
| Histological type | |||||
| Acinar predominant | 56 (36.6) | 29 (51.8) | 27 (48.2) | ||
| Lepidic predominant | 51 (33.3) | 24 (47.1) | 27 (52.9) | ||
| Papilary predominant | 17 (11.1) | 6 (35.3) | 11 (64.7) | 12.65 | 0.013* |
| Solid predominant | 17 (11.1) | 3 (17.6) | 14 (82.4) | ||
| Micropapilary predominant | 12 (7.8) | 1 (8.3) | 11 (91.7) | ||
| TNM stage | |||||
| TNMI | 55 (35.9) | 31 (56.4) | 24 (43.6) | ||
| TNMII | 40 (26.1) | 15 (37.5) | 25 (62.5) | 8.937 | 0.030* |
| TNM III | 39 (25.5) | 12 (30.8) | 27 (69.2) | ||
| TNM IV | 19 (12.4) | 5 (26.3) | 14 (73.7) | ||
| Lymphatic Metastasis | |||||
| Present | 66 (43.1) | 19 (28.8) | 47 (71.2) | 7.355 | 0.008* |
| Absent | 87 (56.9) | 44 (50.6) | 43 (49.4) | ||
| Distant Metastasis | |||||
| Present | 49 (32) | 14 (28.6) | 35 (71.4) | 4.729 | 0.035* |
| Absent | 104 (68) | 49 (47.1) | 55 (52.9) | ||
| EGFR | |||||
| Mutation | 56 (36.6) | 17 (30.4) | 39 (69.6) | 4.269 | 0.042* |
| Wild | 97 (63.4) | 46 (47.4) | 51 (52.6) | ||
Significantly different.
Multivariate analyses for independent prognostic factor identification
We observed a significant correlation between the occurrence of distant metastasis and CEA, NSE, lymph metastasis and CAMKII expression with R software (version 3.1.0). For CAMKII, the OR of distant metastasis increased 0.135 in the positive expression group (95% CI: 1.000-1.135, P < 0.05). Compared with the negative groups, the higher NSE groups reflected an increased risk of poor prognosis (OR=0.22, 95% CI: 0.711-0.931, P=0.003). There was a significant difference between the negative CEA groups and the positive CEA groups. The distant metastasis OR of the positive CEA groups rose by 0.236 (95% CI: 1.073-1.309, P < 0.001). As expected, the occurrence of lymph node metastasis was an independent predictor of poor prognosis (OR=0.407, 95% CI: 1.868-2.275, P < 0.001). Thus, Table 3 confirmed the importance of CEA, NSE, and CAMKII as independent prognostic factors for predicting poor prognosis in lung adenocarcinoma.
Table 3.
Multivariate logistic regression analyses for distant metastasis in Lung ADC
| Variable | Multivariate analysis | |
|---|---|---|
|
| ||
| HR (95% CI) | P value | |
| Sex, male versus female | 0.194 (0.869-1.063) | 0.440 |
| Age, < 50 versus ≥ 50 years | 0.238 (0.832-1.070) | 0.365 |
| Tumor sizes, ≤ 3 versus > 3 cm | 0.244 (0.835-1.079) | 0.421 |
| TPSA, ≤ 80 µg/ml versus > 80 µg/ml | 0.247 (0.886-1.133) | 0.966 |
| SCC, ≤ 1.5 µg/ml versus > 1.5 µg/ml | 0.476 (0.981-1.457) | 0.076 |
| NSE, ≤ 15.2 µg/ml versus > 15.2 µg/ml | 0.22 (0.711-0.931) | 0.003* |
| CEA, ≤ 5 µg/ml versus > 5 µg/ml | 0.236 (1.073-1.309) | 0.001* |
| Cyfra 21-1, ≤ 3.3 µg/ml versus > 3.3 µg/ml | 0.217 (0.873-1.090) | 0.659 |
| EGFR, wild versus mutation | 0.192 (0.896-1.088) | 0.795 |
| Histologic style | 0.068 (0.992-1.060) | 0.140 |
| Lymph metastasis, present versus absent | 0.407 (1.868-2.275) | < 0.001* |
| Smoking, never versus former or current | 0.23 (0.996-1.226) | 0.057 |
| CAMKII, low versus high | 0.135 (1.000-1.135) | 0.049* |
Significantly different.
CAMKII and CEA levels correlated with 5-year survival
Kaplan-Meier survival curves and a multivariate Cox regression analysis were used to analyze mortality at 5 years using SPSS version 22.0. Kaplan-Meier survival curves, as shown in Figure 2C and 2D, indicated the result of five-year survival status is significantly associated with CAMKII (P < 0.05) and CEA (P < 0.001). Kaplan-Meier and log-rank test analyses suggested that lung adenocarcinoma patients with positive CAMKII expression levels have shorter overall survival (OS) and higher metastasis rates than those with negative CAMKII expression levels. These findings suggest that CAMKII generally reflected a poor prognosis in lung adenocarcinoma. Higher CEA levels were closely correlated with lower 5-year survival in lung adenocarcinoma cases. A multivariate Cox regression analysis demonstrated that the higher baseline CEA level (> 5 ng/ml) remained independently associated with poorer OS (HR=2.076, 95% CI: 1.262-3.415, P=0.004). As expected, lymph node metastasis was closely related to the 5-year survival rate (HR=6.514, 95% CI: 3.663-11.586, P < 0.001).
Nomogram development and model validation
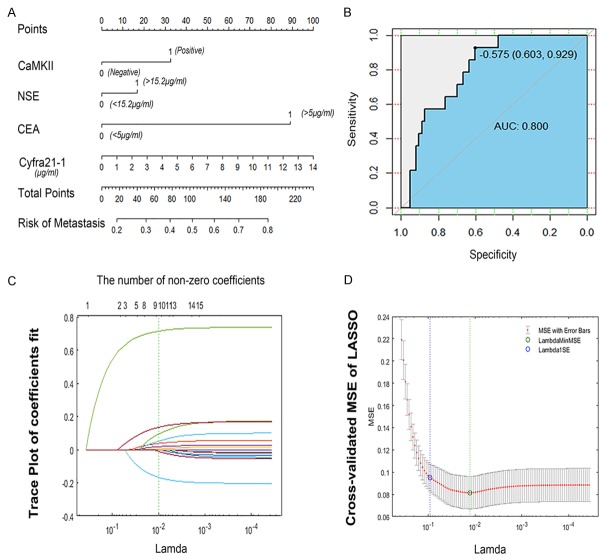
The nomogram was used to assign 98 stage II-IV cases of 153 lung adenocarcinoma patients diagnosed in 2011 as the training cohort and 78 stage I patients as a validation cohort in addition. Univariate and multivariate analysis were used to screen for the significant prognostic factors, including tumor markers, molecular markers, clinical and pathological features in the training cohort (Table 3). A nomogram that fed the significant prognostic factors (CEA, CYFRA 21-1, and CAMKII) was established. The resulting multivariate log-rank regression model was used to calculate the risk score and build the final nomogram prognostic model (Figure 3A). Obviously, the CEA level made a huge contribution to a poor prognosis. In the validation cohort, the predictive ability of the nomogram was measured using an ROC analysis (Figure 3B). With stage IA lung adenocarcinoma patients validated, the accuracy of the nomogram was 64.94%, with an AUC value of 0.800, a sensitivity value of 0.929, and a specificity value of 0.603. The validation cohort illustrated that the nomogram was suited for predicting the poor prognosis of early stage lung adenocarcinoma patients. For example, a pathologically diagnosed patient with stage I lung adenocarcinoma after surgery has a negative CEA, NSE and CAMKII, and a CYFRA 21-1 value of 2.59. Therefore, we draw a line perpendicular to this scale down until it meets the “risk of metastasis” axis; his total points are 18. The cross point is approximately 0.2, which means the probability of distant metastasis in this patient is about 20%.
Figure 3.
The nomogram of predicting the risk of metastasis and feature selection using the least absolute shrinkage and selection operator (LASSO) binary logistic regression model. A. The construction of a nomogram of predicting metastasis risk in lung ADC, including CaMKII, NSE, CEA, a value of Cyfra 21-1. In the nomogram, each variable value is assigned a score, and the final sum of the scores is projected to the corresponding probability of metastasis. B. The ROC curve of the nomogram, the AUC was 0.800, with a sensitivity value of 0.929 and specificity value of 0.603. C. LASSO coefficient profiles of the 15 features. A vertical line was drawn at the value selected using 10-fold cross-validation, where optimal 1 resulted in 9 nonzero coefficients. D. Tuning parameter (Lambda) selection in the LASSO model used 10-fold cross-validation via minimum criteria. The green circle and dotted line locate the Lambda with the minimum cross-validation mean squared error (MSE).
Prediction of distant metastasis via machine learning
The random forest machine learning scheme was employed for the classification. LASSO was used to identify the most predictive features before the classification experiments. LASSO utilizes both variable selection and regularization to select the subset of variables that minimizes predictive error of the outcome, including the expression of CAMKII, Cyfra 21-1, NSE, CEA, tumor size, histologic type, lymph node status, smoking status and age (Figure 3C). A ten-fold cross-validation was performed to identify the model classification performance (Figure 3D). Classification performance was evaluated by AUC, accuracy, sensitivity, and specificity. The average accuracy of this model was 86.208%, with the AUC value of 0.857, a sensitivity value of 0.840 and a specificity value of 0.873. The model was constructed using Matlab (version R2017a).
Discussion
Lung carcinoma is the leading cause of mortality among all cancers worldwide. Moreover, metastasis is the leading cause of treatment failure and cancer-associated mortality in lung carcinoma, especially in lung adenocarcinoma. Approximately 15% of stage I lung adenocarcinoma patients experience distant metastasis after surgery [3]. However, whether stage IA-IIA lung adenocarcinoma patients require conventional chemotherapy is still controversial. That means this some patients with lung adenocarcinoma who may be transferred may not receive timely prevention and treatment. On the other hand, some patients with early low-risk lung adenocarcinoma may receive radiotherapy and chemotherapy. For the poor prognosis, there is lack of an intuitive and significant method to predict and assess whether early-stage patients require therapy or not. In our study, we used molecular markers, tumor markers, and clinical and pathological factors associated with tumor progression to build a machine learning model and a nomogram which was able to predict a poor prognosis after surgery. Therefore, our predictive survival model provides a useful and objective adjunct to current staging criteria that incorporates the heterogeneity existing in the biology of lung adenocarcinoma. Hence, this needs to be kept in mind when interpreting our results.
With immunohistochemistry and data analysis, our findings indicated that CAMKII expression was associated with histology type (P < 0.05), lymphatic metastasis (P < 0.05) and distant metastasis (P < 0.05) and TNM stage (P < 0.05). The interesting finding that CAMKII expression was related to EGFR mutations (P < 0.05) provides a novel idea for patients with EGFR tolerance. A multivariable analysis confirmed the importance of CAMKII as an independent prognostic molecular marker for predicting distant metastasis in lung adenocarcinoma. Kaplan-Meier analyses and a log-rank test suggested that lung adenocarcinoma patients with positive CAMKII expression levels have shorter OS and higher metastasis rates than those with negative CAMKII expression levels. Machine learning also confirms the importance of CAMKII for transfer prediction. Recent data from several groups have highlighted that CAMKII plays an important role in the regulation of cancer progression and therapy response, such as breast, prostate, and colon cancer. Wang et al. found that the activation of CaMKII significantly increased cell motility and the capacity of wound healing in prostate cancer cell lines [18]. The rate of wound closure was decreased by 80% after the inhibition of CaMKII. Kim et al. suggested that the promoter methylations of CaMKIIβ can be used as a biomarker for the diagnosis of breast cancer [19]. Mamaeva et al. showed that the expression profile of CaMKII isoforms is tissue-specific and could be used as a biomarker to distinguish the origins of cancer cells [20]. This molecular marker was also the focus of our previous experiments. All the data demonstrated the importance of CAMKII expression added to the diagnostic features and prognosis model.
In our multivariate Cox regression analysis, the higher levels of CEA and NSE were significant factors for predicting distant metastasis, but only CEA was found to be an independent predictor of survival in lung adenocarcinoma patients. In 98 stage II-IV cases of these lung adenocarcinoma patients used as a training cohort, CEA, CYFRA 21-1 and CAMKII constituted a nomogram to predict the poor prognosis. Additionally, using 78 stage I patients for validation certificated that the nomogram also suited early-stage lung adenocarcinoma cases. In the construction of the nomogram, the result showed that CEA played an important role in the predictive model of lung adenocarcinoma metastasis. Moreover, CAMKII and CYFRA 21-1, as significant factors, added on to the model. Lymph node metastasis is also a central indicator, but it was deleted because the validation group deviated from the training group. Recent studies reported that increasing tumor markers and some clinical features associated with metastasis and poor prognosis and even some of these studies have established related models. Grunnet considered that the serum level of CEA carries prognostic and predictive information of risk of recurrence and of death in NSCLC independent of the treatment or study design [21]. Jingbo Wang et al. reported that elevated CEA served as an unfavorable determinant of OS, increased the NSE level and was predictive of poor distant metastasis-free survival [22]. They created a nomogram integrating KPS, TNM stage, CEA and CYFRA 21-1 in a total of 224 non-small-cell lung cancer patients. Niels Lyhne Christensen et al. found that COPD, high-risk alcohol intake, low nutritional status, and the number of cigarettes smoked after diagnosis were associated with death within one year among recently diagnosed Danish stage I lung cancer patients [23]. In the process of modeling, we also screened the above indicators. Our statistics on the tumor markers are also consistent with these reports. Furthermore, we only used data regarding stage I lung adenocarcinoma patients for validation and more factors in order to screen for more meaningful indicators. The construction of nomogram could offer institutional data for clinicians to distinguish high-risk patients in the early stages who require chemotherapy.
Since a model for the joint prediction of molecular and tumor markers has never been proposed, and due to the bias of the sample distribution, in order to further study the synergy between these factors, we chose the method of machine learning for further exploration. Additionally, we built a LASSO model for predicting the poor prognosis of lung adenocarcinoma patients by machine learning, including CAMKII expression, major tumor markers (CEA and NSE) and traditional clinicopathological factors (tumor size, lymph node metastasis, smoking status, histology type, and age). In this study, the random forest machine learning scheme was employed for the classification. Moreover, using 10-fold cross-validation to identify the model classification performance, the average accuracy of this model was 86.208%, with an AUC value of 0.857, a sensitivity value of 0.840 and a specificity value of 0.873. Machine learning is a technology that learns from a set of examples (training sets) to perform tasks so that it can perform with a completely new data set. The simulation model based on actual cases can give insights about the overall future of that population [17]. In recent years, more reports show that machine learning is widely used in the prediction of disease prognosis with image data, and also with gene-sequencing [24,25]. In addition, some studies have used machine learning to hunt for the differences in prediction methods [26,27]. However, a combination of protein markers, serum tumor markers, clinical and pathological features has rarely been utilized with machine learning. Our study focused on the clinical factors combined with serum tumor markers, molecular markers and clinicopathological features. The more comprehensive coverage content brought into play machine learning and showed that the joint application of different types of features has a higher sensitivity and specificity for predicting the prognosis of lung adenocarcinoma.
In conclusion, this retrospective study constructed two kinds of novel predictive models of poor prognosis in lung adenocarcinoma patients with nomogram and machine learning. Both complementary models confirmed that a combination of molecular markers (CAMKII) and tumor markers can be used to predict metastasis in patients with lung adenocarcinoma. The higher risk tumor metastasis patients could benefit maximally from postoperative adjuvant therapy at the early stages. Each of these two methods has its own advantages. From the above results, it is clear that machine learning methods can be used to substantially (15-25%) improve the accuracy of predicting cancer metastasis susceptibility and recurrence. Undeniably, the nomogram could be more suitable for widespread application because of its simplicity and intuitiveness. Compared with previous reports, our model has more comprehensive information which included a novel molecular marker, tumor markers, and clinical and pathological factors. However, it must be acknowledged that this study is a retrospective study with a limited sample size. The models also still require more clinical cases to verify and improve some imaging features.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant no. 81402420), the Tianjin Municipal Health Bureau Science and Technology Foundation (grant no. 16KG125), and the Tianjin Natural Science Foundation (grant no. 15JCQNJC12400). The authors are grateful for the valuable assistance they received from Dr. Guo Yuhong and Liu Zhijuan.
Disclosure of conflict of interest
None.
References
- 1.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 2.Song MA, Benowitz NL, Berman M, Brasky TM, Cummings KM, Hatsukami DK, Marian C, O’Connor R, Rees VW, Woroszylo C, Shields PG. Cigarette filter ventilation and its relationship to increasing rates of lung adenocarcinoma. J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer. 2016;16:525–537. doi: 10.1038/nrc.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L, Teng J, Zhang L, Cong P, Yao Y, Sun G, Liu Z. The combination of the tumor markers suggests the histological diagnosis of lung cancer. Biomed Res Int. 2017;2017:2013989. doi: 10.1155/2017/2013989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamura K, Takayama K, Izumi M, Harada T, Furuyama K, Nakanishi Y. Diagnostic value of CEA and CYFRA 21-1 tumor markers in primary lung cancer. Lung Cancer. 2013;80:45–49. doi: 10.1016/j.lungcan.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Liu D, Li L, Pu D, Zhou P, Jing Y, Yu H, Wang Y, Zhu Y, He Y, Li Y, Zhao S, Qiu Z, Li W. The important role of circulating Cyfra 21-1 in metastasis diagnosis and prognostic value compared with carcinoembryonic antigen and neuron-specific enolase in lung cancer patients. BMC Cancer. 2017;17:96. doi: 10.1186/s12885-017-3070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Q, Zhang P, Wu R, Lu K, Zhou H. Identifying the best marker combination in cEA, CA125, CY211, NSE, and SCC for lung cancer screening by combining ROC curve and logistic regression analyses: is it feasible? Dis Markers. 2018;2018:2082840. doi: 10.1155/2018/2082840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo E, Salzano M, De Falco V, Mian C, Barollo S, Secondo A, Bifulco M, Vitale M. Calcium/calmodulin-dependent protein kinase ii and its endogenous inhibitor alpha in medullary thyroid cancer. Clin Cancer Res. 2014;20:1513–1520. doi: 10.1158/1078-0432.CCR-13-1683. [DOI] [PubMed] [Google Scholar]
- 10.Britschgi A, Bill A, Brinkhaus H, Rothwell C, Clay I, Duss S, Rebhan M, Raman P, Guy CT, Wetzel K, George E, Popa MO, Lilley S, Choudhury H, Gosling M, Wang L, Fitzgerald S, Borawski J, Baffoe J, Labow M, Gaither LA, Bentires-Alj M. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc Natl Acad Sci U S A. 2013;110:E1026–1034. doi: 10.1073/pnas.1217072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu G, Cheng CJ, Lin SC, Lee YC, Frigo DE, Yu-Lee LY, Gallick GE, Titus MA, Nutt LK, Lin SH. Organelle-derived acetyl-CoA promotes prostate cancer cell survival, migration, and metastasis via activation of calmodulin kinase II. Cancer Res. 2018;78:2490–2502. doi: 10.1158/0008-5472.CAN-17-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W, An P, Quan XJ, Zhang J, Zhou ZY, Zou LP, Luo HS. Ca(2+)/calmodulin-dependent protein kinase II regulates colon cancer proliferation and migration via ERK1/2 and p38 pathways. World J Gastroenterol. 2017;23:6111–6118. doi: 10.3748/wjg.v23.i33.6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi M, Evans H, Gilchrist J, Mayhew J, Hoffman A, Pearsall EA, Jankowski H, Brzozowski JS, Skelding KA. Phosphorylation of calcium/calmodulin-stimulated protein kinase II at T286 enhances invasion and migration of human breast cancer cells. Sci Rep. 2016;6:33132. doi: 10.1038/srep33132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16:e173–180. doi: 10.1016/S1470-2045(14)71116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang W, Zhang L, Jiang G, Wang Q, Liu L, Liu D, Wang Z, Zhu Z, Deng Q, Xiong X, Shao W, Shi X, He J. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J. Clin. Oncol. 2015;33:861–869. doi: 10.1200/JCO.2014.56.6661. [DOI] [PubMed] [Google Scholar]
- 16.Cruz JA, Wishart DS. Applications of machine learning in cancer prediction and prognosis. Cancer Inform. 2007;2:59–77. [PMC free article] [PubMed] [Google Scholar]
- 17.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Li N, Liu X, Zheng Y, Cao X. A novel endogenous human CaMKII inhibitory protein suppresses tumor growth by inducing cell cycle arrest via p27 stabilization. J Biol Chem. 2008;283:11565–11574. doi: 10.1074/jbc.M800436200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Kim JH, Kim TW, Kim SJ. Downregulation of ARFGEF1 and CAMK2B by promoter hypermethylation in breast cancer cells. BMB Rep. 2011;44:523–528. doi: 10.5483/bmbrep.2011.44.8.523. [DOI] [PubMed] [Google Scholar]
- 20.Mamaeva OA, Kim J, Feng G, McDonald JM. Calcium/calmodulin-dependent kinase II regulates notch-1 signaling in prostate cancer cells. J Cell Biochem. 2009;106:25–32. doi: 10.1002/jcb.21973. [DOI] [PubMed] [Google Scholar]
- 21.Grunnet M, Sorensen JB. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer. 2012;76:138–143. doi: 10.1016/j.lungcan.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Jiang W, Zhang T, Liu L, Bi N, Wang X, Hui Z, Liang J, Lv J, Zhou Z, Xiao Z, Feng Q, Chen D, Yin W, Wang L. Increased CYFRA 21-1, CEA and NSE are prognostic of poor outcome for locally advanced squamous cell carcinoma in lung: a nomogram and recursive partitioning risk stratification analysis. Transl Oncol. 2018;11:999–1006. doi: 10.1016/j.tranon.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen NL, Lokke A, Dalton SO, Christensen J, Rasmussen TR. Smoking, alcohol, and nutritional status in relation to one-year mortality in Danish stage I lung cancer patients. Lung Cancer. 2018;124:40–44. doi: 10.1016/j.lungcan.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 24.Hao X, Luo H, Krawczyk M, Wei W, Wang W, Wang J, Flagg K, Hou J, Zhang H, Yi S, Jafari M, Lin D, Chung C, Caughey BA, Li G, Dhar D, Shi W, Zheng L, Hou R, Zhu J, Zhao L, Fu X, Zhang E, Zhang C, Zhu JK, Karin M, Xu RH, Zhang K. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A. 2017;114:7414–7419. doi: 10.1073/pnas.1703577114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dallora AL, Eivazzadeh S, Mendes E, Berglund J, Anderberg P. Machine learning and microsimulation techniques on the prognosis of dementia: a systematic literature review. PLoS One. 2017;12:e0179804. doi: 10.1371/journal.pone.0179804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ten Haaf K, Jeon J, Tammemägi MC, Han SS, Kong CY, Plevritis SK, Feuer EJ, de Koning HJ, Steyerberg EW, Meza R. Risk prediction models for selection of lung cancer screening candidates: a retrospective validation study. PLoS Med. 2017;14:e1002277. doi: 10.1371/journal.pmed.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]