Abstract
Human Papillomavirus (HPV) 16 infection has led to clinical disorders and is considered one of the important causes of human cervical cancer. Recently, microRNAs (miRNAs) have been proven to play an important role in many viral infections through regulating the Type I IFN immune response. However, reports concerning the role of miRNAs in HPV 16 infection are unclear. The aim of this study was to identify and evaluate the potential functions of miRNAs in HPV 16 replication and reveal the detailed mechanism for regulating IFN immune response. Using microarray and qRT-PCR assays, microRNA-221 (miR-221) was found to be significantly up-regulated in the serum samples from patients with HPV 16 infection, as well as in HPV 16-positive cervical cancer cells. miR-221 overexpression inhibited, while miR-221 knockdown facilitated HPV 16 E1-E2 mediated DNA replication in vitro. Moreover, overexpression of miR-221 was associated with upregulation of IFN-α and IFN-β at mRNA and protein levels in infected cells. Conversely, IFN-α and IFN-β mRNA or protein expression was significantly downregulated during inhibition of miR-221. Subsequently, we demonstrated that upregulation of miR-221 promoted the expression of representative interferon stimulated genes (ISGs) such as myxovirus protein A (MxA), 2’,5’-oligoadenylate synthetases (OAS) and murine IFN-stimulated gene 15 (ISG15). In contrast, miR-221 inhibition significantly decreased ISGs expression. Furthermore, we found that suppressor of cytokine signaling 1 (SOCS1), a suppressor of interferon signaling pathway, was a direct target of miR-221 and overexpression of SOCS1 reversed the effects of miR-221 on the IFN-I response and HPV 16 E1-E2 mediated DNA replication. Collectively, the findings provide new evidence that miR-221 could inhibit HPV 16 E1-E2 mediated DNA replication through the SOCS1/Type I IFN signaling pathway suggesting it may be a novel anti-HPV therapeutic target.
Keywords: Human papillomavirus 16, microRNA-221, DNA replication, SOCS1/Type I IFN signaling pathway
Introduction
Human cervical cancer (CC) is a malignant neoplasm and the 4th most common cancer in women, which results in an estimated 500,000 cases every year [1,2]. Many studies reveal that human papillomavirus (HPV) infection is the most common cause of cervical cancer [3]. Consequently, reduction of HPV infection could be benefit for treatment of cervical cancer.
MicroRNAs (miRNAs) are a conserved group of small noncoding RNAs comprising of 18-22 nucleotides, that regulate gene expression at post-transcription level [4-6]. Recently, miRNAs have been implicated in response to virus infection. For example, dengue virus (DENV) induced miR-146a has been shown to intensify viral replication partially through impairment of IFN-β by targeting TNFR-associated factor 6 (TRAF6) [7]. Another study showed that miR-130a has been shown to increase virus replication of HCV through downregulating IFN-induced transmembrane protein 1 (IFITM1) protein levels [8,9]. As with HPV 16, differential expression of cellular microRNAs in HPV 16 transfected cells was observed including miR-125a-5p, miR-129-3p, miR-363, and others [10]. However, the functions of those differently expressed miRNAs during HPV 16 infection are still not completely understood. Therefore, identification of more miRNAs and elucidation of their functional mechanisms may provide better control strategies for the treatment of HPV 16 infection.
In the present study, the differentially expressed miRNAs in serums of patients with HPV 16 infection were tested by using microarray technology, and then the biological functions of the aimed miRNAs on HPV 16 E1-E2 mediated DNA replication and Type I IFN response were examined, which will provide potential mechanisms and new targets for the treatment of HPV 16 infection.
Materials and methods
Samples
In total, 32 serum samples were collected from January 2015 to January 2016 in Department of Sexually Transmitted Disease Institute, Shanghai Skin Disease Hospital, including 16 from HPV 16 infected patients and 16 healthy persons. All patients were diagnosed by pathological tests and imaging examinations. Human papillomavirus (HPV) infection was detected by highly sensitive polymerase chain reaction (PCR) techniques. This study has been approved by the ethics committee of Shanghai Skin Disease Hospital.
Microarray
Total RNA was isolated from 3 paired serum samples by miRNeasy isolation kit (Qiagen, Milan, Italy) and Cy3- or Cy5-labeled cDNAs were hybridized on the miRCURYTM LNA Array (v.16.0) according to the manufacturer’s instruction. After washing, the slides were scanned using an Axon GenePix 4000 B microarray scanner (Axon Instruments, Foster City, CA, USA). Scanned images were then imported into the GenePix Pro6.0 program (Axon Instruments) for grid alignment and data extraction. Replicated miRNAs were averaged, and miRNAs with intensities ≥ 50 in all samples were used to calculate a normalization factor. Expressed data were normalized by median normalization. After normalization, the miRNAs that were significantly differentially expressed were identified by Volcano plot filtering. Finally, the expression data were subjected to hierarchical clustering and subsequently depicted in a heat map format using GeneSpring GX, version 7.3 (Agilent Technologies, California, United Stages). The most differentially expressed miRNAs were confirmed by quantitative reverse transcription polymerase chain reaction (qRT-PCR).
qRT-PCR
miRNA was extracted using a miRNeasy mini kit (Qiagen, USA) and total RNA was extracted with the TRIzol reagent (Invitrogen, USA) according to the manufacturer’s protocol. For miRNA reverse transcription, cDNA was synthesized using a universal tag by using a miScript II RT kit (Qiagen, USA). For mRNA reverse transcription, cDNA was synthesized using the PrimeScriptTM First Strand cDNA Synthesis (Takara Bio, China). Real-time PCR for miRNA and mRNA were performed using a standard protocol from the SYBR Green PCR kit (Toyobo, Osaka, Japan) on an ABI 7900HT Fast Real-Time PCR System (Life Technologies, USA). Relative quantification was determined by normalization to U6 or GAPDH. The primers for qRT-PCR analysis were as follows: miR-221 forward: 5’-GTTGGTGGGAGCTACATTGTCTGC-3’; miR-221 reverse: 5’-GTGTCGTGGACTCGGCAATTC-3’; U6 forward: 5’-TGCGGGTGCTCGCTTCGCAGC-3’; U6 reverse: 5’-CCAGTGCAGGGTCCGAGGT-3’; IFN-α forward: 5’-GCCTGAAGGACAGACATGACTTT-3’, IFN-α reverse: 5’-GGATGGTTTGAGCCTTTTGG-3’; IFN-β forward: 5’-AAACTCATGAGCAGTCTGCA-3’, IFN-β reverse: 5’-AGGAGATCTTCAGTTTCGGAGG-3’; OAS forward: 5’-AGGTGGTAAAGGGTGGCT-3’, OAS reverse: 5’-TGCTTGACTAGGCGGATG-3’ MxA forward: 5’-GGGAAGGTGAAGGTCGGAGT-3’, MxA reverse: 5’-TTGAGGTCAATGAAGGGGTCA-3’; murine IFN-stimulated gene 15 (ISG15) forward: 5’-GGTGTCCGTGACTAACTCCAT-3’, ISG15 reverse: 5’-TGGAAAGGGTAAGACCGTCCT-3’; SOCS1 forward: 5’-CTGGGATGCCGTGTTATTT-3’, SOCS1 reverse: 5’-TAGGAGGTGCGAGTTCAGGT-3’; GAPDH forward: 5’-AGGTCGGTGTGAACGGATTTG-3’, GAPDH reverse: 5’-TGTAGACCATGTAGTTGAGGTCA-3’. The PCR amplification protocol was as follows: an initial 95°C for 5 min and 40 cycles of 94°C for 15 s, 55°C for 30 s, and 70°C for 30 s. The qRT-PCR assays were performed in triplicate and the change in expression level was calculated using the 2-ΔΔCt method.
Cell culture and plasmids
Human cervical squamous cell carcinoma cell lines SiHa (1-2 copies of HPV16 per cell), HeLa (HPV18 positive), and C33A [11] (HPV negative) were included. At the same time, 293 cell line was used as a control group. All cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in Dulbecco Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA) containing 5% or 10% fetal bovine serum (Gibco, Grand Island, NY, USA) at 37°C in a humidified atmosphere of 95% air and 5% CO2. Plasmids used in this study such as HPV16-E2, hemagglutinin-E1 (HAE1), pOri plasmid are already described [12,13].
Transfection
The miR-221 mimics, mimic negative control (mimics NC), miR-221 inhibitor, and inhibitor NC were bought from GenePharm (Shanghai, China). The coding domain sequences of SOCS1 mRNA were amplified by PCR, and inserted into pcDNA 3.0 vector to enhance its expression (Invitrogen, Grand Island, NY, USA), named as pcDNA-SOCS1. Transfections of the miRNAs were performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions.
DNA replication assay
SiHa cells (5 × 105 cells) were plated into 100 mm2 tissue culture dishes for 24 h, and cells were then transfected using the calcium phosphate method using indicated plasmids. The cells were washed the following day with PBS and replenished with fresh media and allowed to grow for 48 h. The cells were then harvested using HIRT solution (0.6% SDS, 10 mM EDTA) and low molecular weight DNA extracted and processed for quantitative PCR as previously reported [12,14].
ELISA assay
IFN-β and IFN-α in supernatant were measured by ELISA kit (both from PBL Biomedical Laboratories, Piscataway, NJ, USA) according to manufacture. SiHa cells were transfected with miR-221 inhibitor, inhibitor NC or miR-221 mimics and mimics NC. At 24 h post-transfection, SiHa cells were transfected with E1/E2/pOri plasmids, and cell culture supernatants were collected 24 h after transfection to perform ELISA assay.
Target genes prediction of miR-221
The target genes of miR-221 were predicted by bioinformatics analysis. The analysis was performed by TargetScan 5.1 (http://targetscan.org/), miRanda (http://microRNA.org) and PicTar5 (http://pictar.mdcberlin.de/).
Luciferase reporter assay
The 3’-UTR of SOCS1 and the fragment of SOCS1 3’-UTR mutant were individually inserted into the pGL3 control vector (Promega Corporation, Madison, WI, USA) to construct wt SOCS1-3’-UTR vector and mutant SOCS1-3’-UTR vector, respectively. For reporter assay, 293 cells were co-transfected with wild-type (mutant) reporter plasmid and miR-221 mimics, miR-221 inhibitor using Lipofectamine 2000 (Invitrogen). 48 hours after transfection, the dual-luciferase reporter assay system (Promega, Shanghai, the People’s Republic of China) were used to measure the luciferase activity according to the manufacturer’s instructions (Promega, Madison, WI, USA). All experiments were performed in triplicate.
Western blot
After 48 h transfection, total protein was extracted from cells using radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology, Shanghai, China). Cell lysates were centrifuged at 12,000 × g for 20 min at 4°C, and the protein concentrations of supernatant were determined using a BCA protein assay reagent kit (Beyotime). Then, the supernatant lysates were run on 8% SDS-polyacrylamide gels (40 μg/lane), and proteins were transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare, Freiburg, DE) by electroblotting. Primary antibodies against SOCS1 (Cell Signaling Technology, 1:1,000 dilution) and β-actin (Santa Cruz Biotechnology, 1:2000 dilution) were probed with proteins on the membrane at 4°C overnight. After incubating with secondary antibodies (1:10000, Cell Signaling Technology, Danvers, MA), Bands were detected by enhanced chemiluminescence (ECL) kit (GE Healthcare, Freiburg, DE). The intensity of the bands of interest was analyzed by ImageJ software (Rawak Software, Inc. Munich, Germany).
Statistical analysis
Statistical analysis was performed using the SPSS program (version 18.0; SPSS, Chicago, IL, USA). Data are presented as mean ± S.D. Student’s t-test or one-way ANOVA was used to analyze the difference among/between sample groups. P ≤ 0.05 was considered significant.
Results
miR-221 expression was upregulated by HPV 16 infection
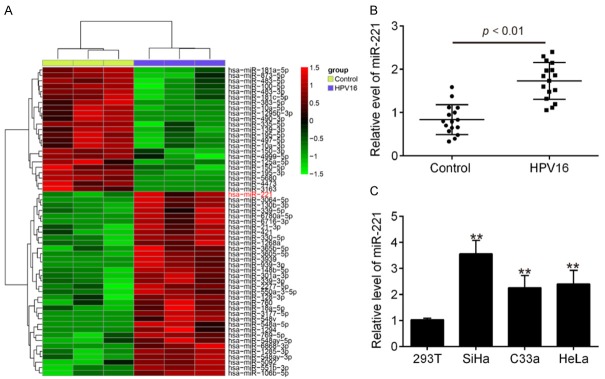
To determine the potential involvement of miRNAs in HPV 16 infection, we performed miRNA microarray profiling to determine miRNA levels in serum from 3 HPV infected patients and 3 healthy persons. Our data revealed that compared with the control group, 23 miRNAs were upregulated and 34 miRNAs were downregulated in the HPV group (Figure 1A). Among the differentially expressed miRNAs, miR-221 was one of the most upregulated miRNAs and many studies have shown that miR-221 has been found to be highly expressed in response to virus infection, such as hepatitis C virus (HCV) [15] and human immunodeficiency virus type 1 (HIV-1) [16], and inhibited virus replication by promoting the Type I IFN response [17]. However, the function and mechanism of miR-221 in HPV 16 infection have not been characterized. Thus, we chose miR-221 for further analysis. Next, the expression levels of miR-221 in 32 serum samples from 16 HPV infected patients and 16 healthy persons were validated by qRT-PCR. Consistent with the array data, the level of miR-221 expression in serums of HPV infected patients was significantly higher than those of healthy persons (Figure 1B). Moreover, the expression of miR-221 was higher in human cervical squamous cell carcinoma cell lines SiHa, HeLa and C33A compared with that in 293 cells, especially in SiHa cells. Collectively, these results indicate that miR-221 is upregulated after HPV 16 infection and it may be involved in the host-virus response of HPV 16 infection.
Figure 1.
miR-221 expression is upregulated by HPV infection. A. Heatmap of normalized expression levels of miRNAs in serum samples from HPV 16 infected patients (n = 3) and healthy persons (n = 3). Green indicates low expression levels; red indicates high expression levels. B. The expression of miR-221 was detected by qRT-PCR in serum samples from HPV 16 infected patients (n = 16) and healthy persons (n = 16). P < 0.01 vs. Control group. C. The expression of miR-221 was detected by qRT-PCR in human cervical squamous cell carcinoma cell lines SiHa, HeLa and C33A and 293 cells used as control. Data represent the mean ± SD of three independent experiments. **P < 0.01 vs. 293.
The effects of miR-221 on HPV 16 E1-E2 mediated DNA replication
To examine whether miR-221 can influence HPV 16 E1-E2 mediated DNA replication, we overexpressed or knocked down the expression of miR-221 in SiHa cells using miR-221 mimics or inhibitor. As shown in Figure 2A, 2C, miR-221 mimics or inhibitor effectively overexpressed or inhibited the expression of miR-221. Subsequently, SiHa cells were transfected with miR-221 mimics or inhibitor for 24 h, followed treatment using the calcium phosphate protocol with E2, E1 and pOri (a plasmid containing the HPV16 origin of replication) plasmids [13,18]. Then, real time PCR was applied to quantify the levels of viral DNA replication. As shown in Figure 2B, 2D, we found that miR-221 overexpression inhibited HPV 16 E1-E2 mediated DNA replication, while miR-221 knockdown facilitated it in vitro. Together, these results indicate that upregulation of miR-221 inhibits HPV 16 E1-E2 mediated DNA replication in vitro.
Figure 2.
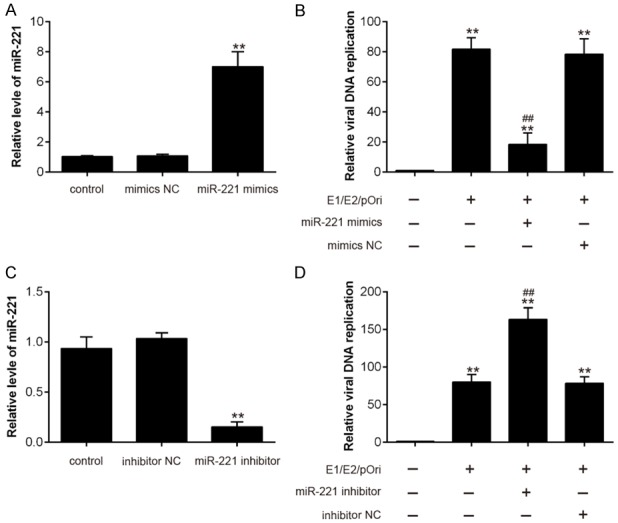
The effects of miR-221 on HPV 16 E1-E2 mediated DNA replication. A, C. The efficiency of miR-221 mimics and inhibitor was confirmed through qRT-PCR assay. B, D. SiHa cells were transfected with miR-221 mimics or inhibitor for 24 h, followed by treatment using the calcium phosphate protocol with E2, E1 and pOri (a plasmid containing the HPV16 origin of replication) plasmids. Then, real time PCR was applied to quantify the levels of HPV 16 E1-E2 mediated DNA replication. Data represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 vs. mimics NC or inhibitor NC; ##P < 0.01 vs. E1/E2/pOri.
miR-221 positively regulate HPV 16 induced IFN-I production
A previous study revealed that miR-221 could positively regulate type I IFN in HCV infection [17]. It is probable that miR-221 may be involved in the HPV 16-triggered response in host cells. To further determine whether HPV 16-induced miR-221 upregulation could affect HPV 16-triggered immune response in host cells, we investigated the role of miR-221 in type I IFN production after HPV 16 infection. As shown in Figure 3A, miR-221 overexpression enhanced IFN-α and IFN-β mRNA expression in E1/E2/pOri transfected SiHa cells. Similarly, the protein levels of IFN-α and IFN-β were significantly increased in E1/E2/pOri transfected cells that overexpressed miR-221 (Figure 3B). Conversely, IFN-α and IFN-β mRNA or protein expression was significantly reduced during inhibition of miR-221 (Figure 3C, 3D). All data indicate that miR-221 may positively regulate the production of type I IFN in HPV 16 infection.
Figure 3.
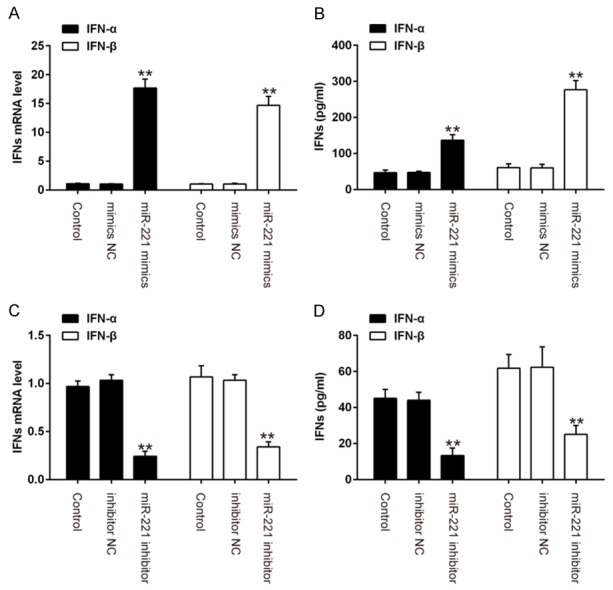
miR-221 positively regulates HPV 16 induced IFN-I production. SiHa cells were transfected with miR-221 mimics or inhibitor for 24 h, followed by treatment using the calcium phosphate protocol with E2, E1 and pOri (a plasmid containing the HPV16 origin of replication) plasmids. A, C. Real-time PCR was performed to measure IFN-α and IFN β mRNA expression. B, D. The protein expression levels of IFN-α and IFN β weredetermined by ELISA. Data represent the mean ± SD of three independent experiments. **P < 0.01 vs. mimics NC or inhibitor NC.
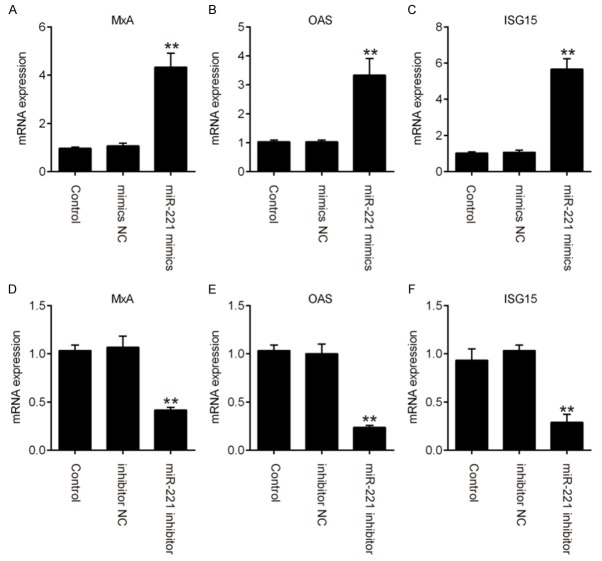
miR-221 positively regulates ISG expression
It has been reported that the antiviral effectors interferon stimulated genes (ISGs) induced by the JAK/STAT pathway play critical roles in the antiviral responses of type I IFN [19,20]. Thus, we sought to determine whether the expression of ISGs was influenced by miR-221 expression. The results of qRT-PCR showed that overexpression of miR-221 promoted the expressions of ISGs like MxA, OAS1 and ISG15 (Figure 4A-C). In contrast, miR-221 inhibition obviously reduced the expressions of these ISGs (Figure 4D-F). These data suggest that miR-221 positively regulates the expressions of ISGs during HPV 16 infection.
Figure 4.
miR-221 positively regulates ISGs expression. SiHa cells were transfected with miR-221 mimics or inhibitor for 24 h, followed treatment using the calcium phosphate protocol with E2, E1 and pOri plasmids. Expression level of MxA (A, D), OAS (B, E) and ISG15 (C, F) were examined by qRT-PCR assay. Data represent the mean ± SD of three independent experiments. **P < 0.01 vs. mimics NC or inhibitor NC.
SOCS1 was a direct target of miR-221
To illustrate the possible mechanism by which miR-221 functions in the regulation of HPV 16 E1-E2 mediated DNA replication and Type I IFN immune response, the potential targets of miR-221 were screened by TargetScan, miRanda and PicTar databases. According to bioinformatic analysis, we focused on suppressor of cytokine signaling-1 (SOCS1), a suppressor of interferon signaling pathway. As shown in Figure 5A, the RNA sequence alignment showed that the 3’-UTR of SOCS1 mRNA contained a complementary site for the seed region of miR-221. To validate whether SOCS1 is the target of miR-221, 3’UTR fragment of SOCS1 containing miR-221 binding sequence were subcloned downstream of a luciferase reporter vector. The reporters were co-transfected with either miR-221 mimic/inhibitor or with negative control (NC) mimic/inhibitor into 293 cells, and luciferase activity was then measured. We observed that overexpression of miR-221 decreased relative luciferase activity in the presence of the wild-type 3’-UTR, whereas knockdown of miR-221 increased the relative luciferase activity (Figure 5B). Similarly, we observed that the luciferase activity did not change significantly when the targeted sequence of SOCS1 was mutated in the miR-221-binding site. To further confirm that SOCS1 was negatively regulated by miR-221, the levels of SOCS1 mRNA and protein expressions were analyzed by qRT-PCR and western blot analysis. We found that SOCS1 levels were significantly downregulated after miR-221 mimics transfection, while transfection with miR-221 inhibitor enhanced SOCS1 expression at mRNA and protein levels (Figure 5C, 5D). Together, these results showed that miR-221 could regulate the expressions of human SOCS1 by directly targeting the 3’-UTR of SOCS1 mRNA.
Figure 5.
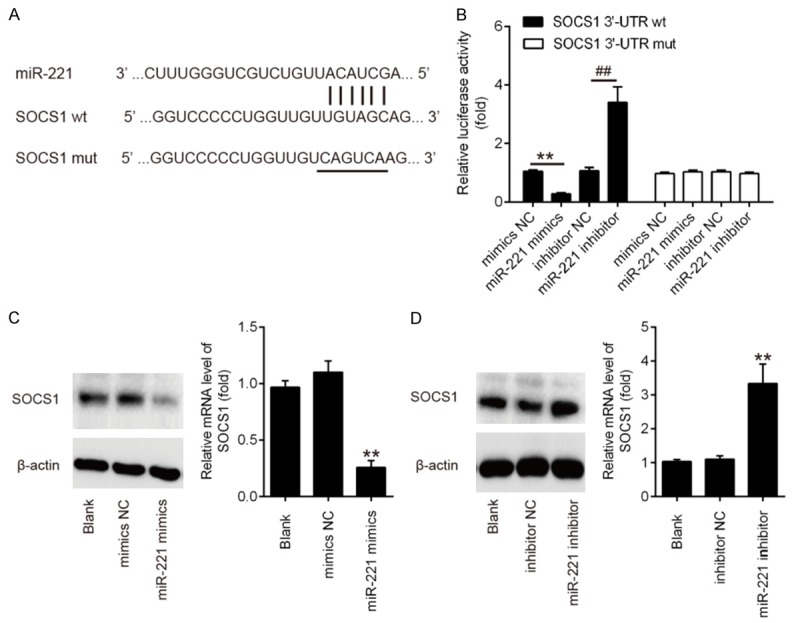
SOCS1 is a direct target of miR-221. A. Schematic diagram of the predicted target sites of miR-221 in SOCS1 3’-UTRs. The predicted target sites are underlined and mutated as indicated. B. Luciferase assay of 293 cells co-transfected with firefly luciferase constructs containing the SOCS1 wild-type or mutated 3’-UTRs and miR-221 mimic, mimic NC, miR-221 inhibitor or inhibitor NC, as indicated (n = 3). Data represent the mean ± SD of three independent experiments. **P < 0.01 vs. mimic NC, ##P < 0.01 vs. inhibitor NC. C, D. The expressions of SOCS1 mRNA and protein after transfection with miR-221 mimic or miR-221 inhibitor were measured by qRT-PCR and western blot. Data represent the mean ± SD of three independent experiments. **P < 0.01 vs. mimics NC or inhibitor NC.
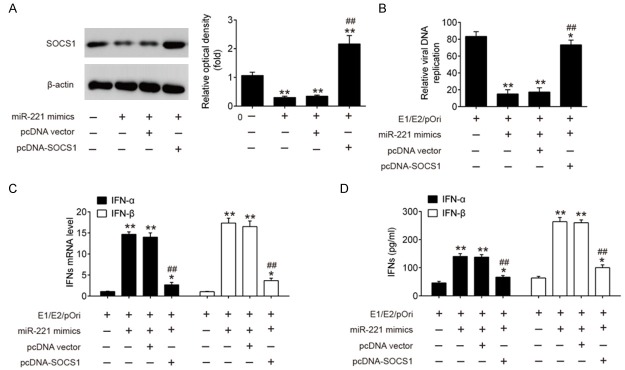
Overexpression of miR-221 inhibited HPV 16 E1-E2 mediated DNA replication by targeting SOCS1
In order to further confirm whether SOCS1 mediates the inhibitory effect of miR-221 overexpression on the HPV 16 E1-E2 mediated DNA replication, we co-transfected miR-221 mimics and pcDNA SOCS1 into SiHa cells, followed by treatment of E1/E2/pOri plasmids. As shown in Figure 6A, the protein level of SOCS1 was greatly decreased in E1/E2/pOri transfected SiHa cells by transfection of miR-221 mimics, whereas co-transfection of pcDNA-SOCS1 significantly reversed this effect. Consistently, HPV 16 E1-E2 mediated DNA replication in E1/E2/pOri transfected SiHa cells was dramatically reduced by miR-221 overexpression. However, the antiviral effect was obviously impaired by transfection of pcDNA-SOCS1 (Figure 6B). Moreover, the induction of IFN α and IFN β by miR-221 mimics at both mRNA level and protein levels was significantly reduced by transfection of pcDNA-SOCS1 (Figure 6C, 6D). Our findings suggest that the antiviral function of miR-221 during HPV 16 infection is mainly through targeting SOCS1 and subsequently promoting type I IFN.
Figure 6.
Overexpression of miR-221 inhibited HPV 16 E1-E2 mediated DNA replication by targeting SOCS1. SiHa cells were co-transfected with miR-221 mimics and pcDNA-SOCS1 for 24 h, followed treatment using the calcium phosphate protocol with E2, E1 and pOri (a plasmid containing the HPV16 origin of replication) plasmids. A. The protein expression of SOCS1 was measured by western blot. B. Real time PCR was applied to quantify the levels of HPV 16 E1-E2 mediated DNA replication. C, D. Real-time PCR and ELISA assay were performed to measure IFN-α and IFN-β expression. Data represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 vs. E1/E2/pOri; ##P < 0.01 vs. E1/E2/pOri + miR-221 mimics + pcDNA vector.
Discussion
In the present study, we first found that the expression of miR-221 was significantly upregulated in HPV infected cell lines and serum in HPV infected patients. We have also demonstrated, in vitro, that overexpression of miR-221 inhibited HPV 16 E1-E2 mediated DNA replication through positively regulating IFN-I production and ISGs expression. Further work revealed that miR-221 works mainly through targeting SOCS1 to promote IFN-I immune response, and thus inhibited HPV 16 E1-E2 mediated DNA replication. On this basis, miR-221 may serve as a potential therapeutic target for HPV 16 infected patients.
Increasing evidence has indicated that miRNAs are involved in the complicated regulation of host-pathogens interactions and emerge as important regulators of immune response [21-23]. Recently, a large number of studies have investigated miRNA expression profiles in host cell or viruses and have reported it to play an important role in virus replication [24-26]. For example, miR-373 was found to be upregulated after HSV-1 infection and promote HSV-1 replication in Hela cell lines by suppressing IRF1 [27]. He et al. showed that miR-182 inhibited HCMV replication by targeting FOXO3 in neural cells [28]. Moreover, Li et al. showed that miR-9-5p suppresses Enterovirus 71 (EV71) replication through retinoic acid-induced gene 1 (RIG-I)-mediated innate immune response [29]. Liu et al. found that miR-26b could inhibit vesicular stomatitis virus (VSV) replication through positively regulating type-I IFN signaling [30]. In the current study, we found that the expression of miR-221 was significantly increased in serum of HPV 16 infected patients and HPV 16 infected cell lines. miR-221 has also been confirmed to play fundamental roles in the replication of HCV [31], HIV [32] and avian leukosis virus subgroup J (ALV-J) [33]. However, little previous research attention has been paid to the functions of miR-221 in HPV 16 infection. Our further studies showed that overexpression of miR-221 led to decreased HPV 16 E1-E2 mediated DNA replication and increased IFN-α and IFN-β expressions, as well as increased ISGs expression. However, knockdown of miR-221 has an opposite result. All data suggest that that overexpression of miR-221 may affect HPV 16 E1-E2 mediated DNA replication through promoting a type I IFN response.
How did miR-221 inhibit HPV 16 E1-E2 mediated DNA replication? Based on the database and double-luciferase reporter assay, the results showed that miR-221 can directly target SOCS1, which is a negative regulator of type I IFN signaling pathway [34]. Thus, miR-221 may rely on adjusting SOCS1/IFN I signaling pathway to influence the HPV 16 E1-E2 mediated DNA replication. SOCS1 belongs to members of the SOCS family, and acts as a suppressor of type I IFN function against several phylogenetically distinct viruses infections, such as herpes simplex virus, influenza A virus and hepatitis C virus [35,36]. For example, Shao et al. found that SOCS1 negatively regulated the antiviral effect of IFN in HCV infection and the upregulation of classical ISGs by IFN is reduced when SOCS1 is overexpressed [37]. Zheng et al. showed that respiratory syncytial virus (RSV) infection upregulated the expression of SOCS1, which utilized a feedback loop to inhibit the type I interferon dependent antiviral signaling pathway [38]. In our study, SOCS1 was proven to be a direct target of miR-221 and SOCS1 was negatively regulated by miR-221, which is consistent with a previous report [39]. Given the important role of SOCS1 in the Type I IFN response, it is reasonable for us to infer that miR-221 suppress SOCS1 and thus decreases HPV 16 E1-E2 mediated DNA replication. As expected, enhanced SOCS1 expression by pcDNA-SOCS1 transfection reversed the anti-virus effects and type I IFN production of miR-221 overexpression. These data indicate that miR-221 suppresses HPV 16 E1-E2 mediated DNA replication through regulating the SOCS1/type I IFN signaling pathway.
In conclusion, this study demonstrated for the first time that miR-221 is up-regulated during HPV 16 infection and overexpression of miR-221 inhibited HPV 16 E1-E2 mediated DNA replication in SiHa cell lines by activating the SOCS1/Type I IFN response. These findings provide new evidence regarding the role of host miRNAs in HPV 16 infection and suggest that miR-221 is a novel promising anti-HPV 16 therapeutic target.
Disclosure of conflict of interest
None.
References
- 1.Arbyn M, Castellsague X, de Sanjose S, Bruni L, Saraiya M, Bray F, Ferlay J. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 2.Yang BH, Bray FI, Parkin DM, Sellors JW, Zhang ZF. Cervical cancer as a priority for prevention in different world regions: an evaluation using years of life lost. Int J Cancer. 2004;109:418–424. doi: 10.1002/ijc.11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Wu S, He L, Li Y, Wang T, Feng L, Jiang L, Zhang P, Huang X. miR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. J Infect. 2013;67:329–341. doi: 10.1016/j.jinf.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Bhanja Chowdhury J, Shrivastava S, Steele R, Di Bisceglie AM, Ray R, Ray RB. Hepatitis C virus infection modulates expression of interferon stimulatory gene IFITM1 by upregulating miR-130A. J Virol. 2012;86:10221–10225. doi: 10.1128/JVI.00882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Duan X, Li Y, Liu B, McGilvray I, Chen L. MicroRNA-130a inhibits HCV replication by restoring the innate immune response. J Viral Hepat. 2014;21:121–128. doi: 10.1111/jvh.12131. [DOI] [PubMed] [Google Scholar]
- 10.Dreher A, Rossing M, Kaczkowski B, Andersen DK, Larsen TJ, Christophersen MK, Nielsen FC, Norrild B. Differential expression of cellular microRNAs in HPV 11, -16, and -45 transfected cells. Biochem Biophys Res Commun. 2011;412:20–25. doi: 10.1016/j.bbrc.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Wei L, Gravitt PE, Song H, Maldonado AM, Ozbun MA. Nitric oxide induces early viral transcription coincident with increased DNA damage and mutation rates in human papillomavirus-infected cells. Cancer Res. 2009;69:4878–4884. doi: 10.1158/0008-5472.CAN-08-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor ER, Morgan IM. A novel technique with enhanced detection and quantitation of HPV-16 E1- and E2-mediated DNA replication. Virology. 2003;315:103–109. doi: 10.1016/s0042-6822(03)00588-9. [DOI] [PubMed] [Google Scholar]
- 13.Donaldson MM, Mackintosh LJ, Bodily JM, Dornan ES, Laimins LA, Morgan IM. An interaction between human papillomavirus 16 E2 and TopBP1 is required for optimum viral DNA replication and episomal genome establishment. J Virol. 2012;86:12806–12815. doi: 10.1128/JVI.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan IM, Taylor ER. Detection and quantitation of HPV DNA replication by Southern blotting and real-time PCR. Methods Mol Med. 2005;119:349–362. doi: 10.1385/1-59259-982-6:349. [DOI] [PubMed] [Google Scholar]
- 15.Braconi C, Valeri N, Gasparini P, Huang N, Taccioli C, Nuovo G, Suzuki T, Croce CM, Patel T. Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clin Cancer Res. 2010;16:957–966. doi: 10.1158/1078-0432.CCR-09-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fowler L, Conceicao V, Perera SS, Gupta P, Chew CB, Dyer WB, Saksena NK. First evidence for the disease-stage, cell-type, and virus specificity of micrornas during human immunodeficiency virus type-1 infection. Med Sci (Basel) 2016;4 doi: 10.3390/medsci4020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu G, Yang F, Ding CL, Wang J, Zhao P, Wang W, Ren H. MiR-221 accentuates IFNs anti-HCV effect by downregulating SOCS1 and SOCS3. Virology. 2014;462-463:343–350. doi: 10.1016/j.virol.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 18.Gagnon D, Joubert S, Senechal H, Fradet-Turcotte A, Torre S, Archambault J. Proteasomal degradation of the papillomavirus E2 protein is inhibited by overexpression of bromodomain-containing protein 4. J Virol. 2009;83:4127–4139. doi: 10.1128/JVI.02468-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo XK, Zhang Q, Gao L, Li N, Chen XX, Feng WH. Increasing expression of microRNA 181 inhibits porcine reproductive and respiratory syndrome virus replication and has implications for controlling virus infection. J Virol. 2013;87:1159–1171. doi: 10.1128/JVI.02386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169–175. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen L, Wu E, Ye X, Gao GF, Wang F, Chen Y, Duan Z, Meng S. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1) -modulated P53 activity. Hepatology. 2012;55:730–741. doi: 10.1002/hep.24809. [DOI] [PubMed] [Google Scholar]
- 24.Ru J, Sun H, Fan H, Wang C, Li Y, Liu M, Tang H. MiR-23a facilitates the replication of HSV-1 through the suppression of interferon regulatory factor 1. PLoS One. 2014;9:e114021. doi: 10.1371/journal.pone.0114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo H, Liu H, Mitchelson K, Rao H, Luo M, Xie L, Sun Y, Zhang L, Lu Y, Liu R, Ren A, Liu S, Zhou S, Zhu J, Zhou Y, Huang A, Wei L, Guo Y, Cheng J. MicroRNAs-372/373 promote the expression of hepatitis B virus through the targeting of nuclear factor I/B. Hepatology. 2011;54:808–819. doi: 10.1002/hep.24441. [DOI] [PubMed] [Google Scholar]
- 26.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 27.Xie Y, He S, Wang J. MicroRNA-373 facilitates HSV-1 replication through suppression of type I IFN response by targeting IRF1. Biomed Pharmacother. 2018;97:1409–1416. doi: 10.1016/j.biopha.2017.11.071. [DOI] [PubMed] [Google Scholar]
- 28.He X, Teng J, Cui C, Li D, Wen L. MicroRNA-182 inhibits HCMV replication through activation of type I IFN response by targeting FOXO3 in neural cells. Exp Cell Res. 2018;369:197–207. doi: 10.1016/j.yexcr.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 29.Li B, Zheng J. MicroR-9-5p suppresses EV71 replication through targeting NFkappaB of the RIG-I-mediated innate immune response. FEBS Open Bio. 2018;8:1457–1470. doi: 10.1002/2211-5463.12490. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Liu C, Zhang L, Xu R, Zheng H. miR-26b inhibits virus replication through positively regulating interferon signaling. Viral Immunol. 2018;31:676–682. doi: 10.1089/vim.2018.0067. [DOI] [PubMed] [Google Scholar]
- 31.Ding CL, Xu G, Ren H, Zhao LJ, Zhao P, Qi ZT, Wang W. HCV infection induces the upregulation of miR-221 in NF-kappaB dependent manner. Virus Res. 2015;196:135–139. doi: 10.1016/j.virusres.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Lodge R, Ferreira Barbosa JA, Lombard-Vadnais F, Gilmore JC, Deshiere A, Gosselin A, Wiche Salinas TR, Bego MG, Power C, Routy JP, Ancuta P, Tremblay MJ, Cohen EA. Host microRNAs-221 and -222 inhibit HIV-1 entry in macrophages by targeting the CD4 viral receptor. Cell Rep. 2017;21:141–153. doi: 10.1016/j.celrep.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 33.Ren C, Yu M, Zhang Y, Fan M, Chang F, Xing L, Liu Y, Wang Y, Qi X, Liu C, Zhang Y, Cui H, Li K, Gao L, Pan Q, Wang X, Gao Y. Avian leukosis virus subgroup J promotes cell proliferation and cell cycle progression through miR-221 by targeting CDKN1B. Virology. 2018;519:121–130. doi: 10.1016/j.virol.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Skjesol A, Liebe T, Iliev DB, Thomassen EI, Tollersrud LG, Sobhkhez M, Lindenskov Joensen L, Secombes CJ, Jorgensen JB. Functional conservation of suppressors of cytokine signaling proteins between teleosts and mammals: atlantic salmon SOCS1 binds to JAK/STAT family members and suppresses type I and II IFN signaling. Dev Comp Immunol. 2014;45:177–189. doi: 10.1016/j.dci.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Akhtar LN, Benveniste EN. Viral exploitation of host SOCS protein functions. J Virol. 2011;85:1912–1921. doi: 10.1128/JVI.01857-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pothlichet J, Chignard M, Si-Tahar M. Cutting edge: innate immune response triggered by influenza A virus is negatively regulated by SOCS1 and SOCS3 through a RIG-I/IFNAR1-dependent pathway. J Immunol. 2008;180:2034–2038. doi: 10.4049/jimmunol.180.4.2034. [DOI] [PubMed] [Google Scholar]
- 37.Shao RX, Zhang L, Hong Z, Goto K, Cheng D, Chen WC, Jilg N, Kumthip K, Fusco DN, Peng LF, Chung RT. SOCS1 abrogates IFN’s antiviral effect on hepatitis C virus replication. Antiviral Res. 2013;97:101–107. doi: 10.1016/j.antiviral.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng J, Yang P, Tang Y, Pan Z, Zhao D. Respiratory syncytial virus Nonstructural proteins upregulate SOCS1 and SOCS3 in the different manner from endogenous IFN signaling. J Immunol Res. 2015;2015:738547. doi: 10.1155/2015/738547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI, Blazar BR, Zeng Y, Zhou X. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117:4293–4303. doi: 10.1182/blood-2010-12-322503. [DOI] [PMC free article] [PubMed] [Google Scholar]