Abstract
Background: Abnormal proliferation of PASMCs is the main phenotype of pulmonary arterial hypertension (PAH). MicroRNAs (miRNAs) were reported to participate in regulating the progression of PAH. Here, we aimed to investigate the impact of miR-107 on proliferation and migration of PASMCs and potential mechanism. Methods: MTT assay was carried out to examine the cell viability of PASMCs. PASMC migration ability was verified through Transwell assay. RT-qPCR was performed to detect the expression of miR-107 and NOR1. Western blot was conducted to detect the expression of cell proliferation markers Ki-67, p27 and Cyclin D1, as well as NOR1. Bioinformatics analysis was conducted to verify whether the 3’-untranslated region (3’-UTR) of NOR1 contains a binding site for miR-107, and luciferase reporter assay and RNA immunoprecipitation (RIP) were employed to confirm the relationship between miR-107 and NOR1. Results: Platelet-derived growth factor (PDGF)-BB promoted the cell viability and migration of PASMCs, and suppressed miR-107 expression in a time-dependent and concentration-dependent manner. Introduction of miR-107 inhibited the promotion of proliferation and migration of PASMCs stimulated by PDGF-BB, while loss of miR-107 facilitated PDGF-BB-induced promoted effects. NOR1 was identified as a downstream gene of miR-107 and down-regulated by miR-107. Knockout of NOR1 also repressed the promotion of proliferation and migration of PASMCs stimulated by PDGF-BB. Additionally, restoration of NOR1 attenuated the inhibition of miR-107 on the cell viability and migration ability of PASMCs. Conclusion: miR-107 inhibits PDGF-BB-induced PASMCs proliferation and migration through targeting NOR1.
Keywords: PDGF-BB, PASMCs, miR-107, NOR1, proliferation, migration
Introduction
Pulmonary arterial hypertension (PAH) is a peculiar disease featured with vasoconstriction and remodeling of the pulmonary arteries (PAs), causing increased arterial pressure which eventually leads to right heart failure [1]. It was estimated that the prevalence of PAH is 15 per million in the world [2]. Hyper-proliferation of PASMCs is the main initial malignant phenotypes during vascular remodeling process of the lungs of PAH patients, suggesting a vital role of this cell type in the pathogenesis of PAH [3,4].
The platelet-derived growth factor (PDGF) family consists of 5 different dimeric isoforms, including PDGF-AA, PDGF-AB, PDGF-BB, PDGF-CC, and PDGF-DD [5]. PDGF contributes to the proliferation and migration of PASMCs, and has been regarded as a novel potential therapeutic target in PAH [6]. Researchers verified that PDGF stimulates proliferation of PASMCs by up-regulating TRPC6 expression, a member of the short transient receptor potential channel gene subfamily [7]. Here, we made efforts to explore other potential mechanisms.
MicroRNA is a single-stranded, non-coding RNA approximately 22 nucleotides in length, key regulator for gene expression, that leads to mRNA degradation, translation inhibition, or mRNA cleavage by binding to the 3’-UTR of target mRNAs [8,9]. Former reports indicated that several miRNAs have essential functions in the proliferation and (or) apoptosis of human PASMCs, such as miR-34a [10], let-7g [11], miR-200c [12], miR-222 [13], miR-1281 [14] and miR-214 [15]. Also, miR-107 and miR-103, belong to the same family, which differ only at one nucleotide close to their 3’ends and are located on different human chromosomes [16]. It has been documented that miR-103/107 could repress tumor angiogenesis, contribute to adipogenesis, suppress cell proliferation, regulate lipid and control insulin sensitivity [17]. Li et al. demonstrated that miR-107 expression was diminished in PDGF-stimulated human aortic smooth muscle cells (SMCs) [18]. Little is known about the role of miR-107 in PASMCs.
The oxidored-nitro domain-containing protein 1 (NOR1) gene (also named as organic solute carrier partner 1, OSCP1), was initially extracted from nasopharyngeal carcinoma (NPC) [19]. Belonging to the ligand-independent NR4A subfamily, NOR1 is implicated in cell proliferation, differentiation, and apoptosis, and functions as a key transcriptional regulator of SMC proliferation [20]. NOR1 also participates in the metabolism reprogramming in NPC cells, regulates oxidative stress and autophagy apoptosis crosstalk, mediates tumor cell adaptation to hypoxia, and suppresses epithelial-to-mesenchymal transition (EMT) and metastasis through FOXA1-HDAC2/slug axis, serving as a novel tumor suppressor [21].
In the current study, we investigated the effects of miR-107 and NOR1 on the proliferation and migration abilities of PDGF-BB-pretreated PASMCs, and explore a potential mechanism.
Materials and methods
Cell culture and PDGF-BB treatment
Normal human pulmonary artery smooth muscle cells (PASMCs) were purchased from Lonza (Walkersville, MD, USA), and the cells were maintained in SmGM-2 medium (Lonza) containing 5% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), penicillin (100 U/mL, Sigma-Aldrich, St. Louis, MO, USA), and streptomycin (100 mg/mL, Sigma-Aldrich) in a humidified incubator at 37°C. 293T cell line was obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% FBS at 37°C.
PASMCs were divided into five groups, including Vehicle group, 1 ng/mL PDGF-BB (Thermo Fisher Scientific) group, 10 ng/mL PDGF-BB group, 20 ng/mL PDGF-BB group and 40 ng/mL PDGF-BB group, which were incubated at 37°C.
Reagent and transfection
miR-107 mimics (miR-107), miR-NC mimics (miR-NC), miR-107 inhibitor (Anti-miR-107), miR-NC inhibitors (Anti-miR-NC), si-NOR1, si-NC (Scramble), pcDNA3.1-NOR1 (NOR1) and pcDNA3.1 vector (vector) were purchased from (GenePharma Co. Ltd. Shanghai, China). Above nucleotides or plasmids were transfected into PASMCs using LipofectamineTM 2000 reagent (Invitrogen, Carlsbad, CA, USA) for 48 h, referring to the manufacturer’s instructions.
MTT assay
2 × 103 PASMCs were seeded in 96-well plates (Corning Costar, Corning, NY, USA) and incubated for 24 h, 48 h and 72 h at 37°C. Then 20 µL MTT (5 mg/mL, Sigma-Aldrich) was added into each well, and incubation was continued for another 4 h at 37°C. After removing supernatant, 150 μL dimethyl sulfoxide (DMSO, Sigma-Aldrich) was added into each well. The absorbance at 490 nm of each well was measured using a microplate reader (Thermo Labsystems, Waltham, MA, USA). All experiments were performed three times.
Transwell migration assay
The migration assay was performed using millicell chambers (8 μm pores; Millipore, Billerica, MA, USA). 5 × 104 PASMCs were suspended in 100 μL serum-free SmGM-2 medium and added into the upper chamber, and the lower compartment of each chamber contained 500 µL SmGM-2 with 10% FBS as the chemoattractant. The chamber was then incubated at 37°C for 24 h. Cells attached to the lower surface of the filter were fixed and stained with 0.1% crystal violet (Sigma-Aldrich), and we counted the mean number of six random areas under a light microscope (Nikon E100; Nikon Corp, Japan) at 200 × magnification.
RT-qPCR assay
miRNAs were isolated using mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA), and the miR expression was analyzed with a miRNA-specific TaqMan MiRNA Assay Kit (Applied Biosystems, Foster City, CA). As for mRNA expression, total RNA was first isolated with TRIzol reagent (TaKaRa Bio, Inc., Otsu, Japan), then 500 ng total RNA was reverse-transcribed to produce cDNA using a Prime ScriptTM RT Reagent Kit (TaKaRa). qPCR was performed with SYBR® Premix Ex TaqTM II (TaKaRa) on ABI Prism7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). U6 and GAPDH were used as the internal reference for miR-107 and NOR1, respectively. All used primers were as follows: miR-107, 5’-AGCAGCA-TTGTACAGGGCTATCA-3’ (sense) and 5’-AAGGCGAGACGCACATTCTT-3’ (anti-sense); U6, 5’-GCTTCGGCAGCACATATACTAAAAT-3’ (sense) and 5’-CGCTTCACGAATTTGCGTGTCAT-3’ (anti-sense); NOR1, 5’-ATAGTCTGAAAGGGAGGAGAGG-3 (sense) and 5’-ATCATGCAGATTGGAGGAGAAG-3’ (anti-sense); GAPDH, 5’-ACCCACTCCTCCACCTTTG-3’ (sense) and 5’-CACCACCCTGTTGCTGTAG-3’ (anti-sense). The relative expression level of miR-107 and NOR1 were calculated using the threshold cycle 2-ΔΔCt method. All the real-time PCR analyses were conducted in triplicate.
Western blot assay
Total cellular proteins were isolated by a Protein Extraction Kit (Bio-Rad, Hercules, CA, USA) and quantified using a bicinchoninic acid assay (BCA) protein assay kit (Beyotime Biotechnology, Haimen, China). Thirty micrograms protein samples were resolved by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels, then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore) for western blot analysis. After being blocked with 5% fat-free milk at room temperature for 1 h, the membranes were washed with TBST and incubated with primary antibodies against Ki-67 (1:1000 dilution), p27 (1:1000 dilution), Cyclin D1 (1:1000 dilution), NOR1 (1:1000 dilution) and GAPDH (1:2000 dilution) overnight at 4°C. After being washed with TBST for three times, the membranes were then incubated with corresponding secondary antibody (1:2000 dilution) at room temperature for 2 h. The protein bands were visualized by an enhanced chemiluminescence kit (ECL kit, GE Healthcare Life Sciences, Piscataway, NJ, USA) and the band intensity was quantified using Image Lab Software (Bio-Rad). All antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA).
Luciferase reporter assay
To predict the potential target genes of miR-107, the online bioinformatics software TargetScanHuman (http://www.targetscan.org/vert_72/) was utilized and identified NOR1 as a downstream target of miR-107. In order to further confirm the target interaction between miR-107 and NOR1, we conducted luciferase reporter assay in 96-well plates with a Dual-Luciferase Reporter Assay System (Promega Corp., Madison, WI, USA). The fragment of the 3’-UTR of NOR1 for miR-107 was amplified and cloned into PGL3 luciferase promoter vector (pGL3-empty, Promega) to synthesize luciferase reporter gene plasmid PGL3-NOR1-wt. Similarly, the mutant of NOR1 vector (PGL3-NOR1-mut) was synthesized. 293T cells were co-transfected with PGL3-NOR1-wt or PGL3-NOR1-mut, with miR-107 or miR-NC using LipofectamineTM 2000 reagent. 48 h later, cells were harvested to analyze the luciferase activity using the above-mentioned System.
RNA immunoprecipitation (RIP)
RIP was conducted with Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the protocol of manufacture and the AGO2 antibody. 293T cell pellets were collected and lysed in RIP lysis buffer (Sigma-Aldrich). Then the supernatant incubated with protein A/G magnetic beads with human anti-Argonaute2 (Ago2) antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or IgG (Cell Signaling Technology). The RNA RNAs were purified with TRIzol and analyzed by RT-qPCR assay to examine miR-107 and NOR1 in the precipitates.
Statistical analysis
The GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA) was applied for statistical analysis. All data in the present study are presented as mean ± SD (standard deviation). The differences between groups were analyzed by Student’s t test. A P-value less than 0.05 was considered significant.
Results
PDGF-BB treatment elevated the cell viability and migration ability of PASMCs
To validate the effects of PDGF-BB on the cell viability and migration ability of PASMCs, MTT assay and Transwell assay were conducted, respectively. As shown in Figure 1A, the cell viability of PASMCs treated with various concentrations of PDGF-BB was evaluated. MTT assay revealed that the cell viabilities of PASMCs treated with PDGF-BB were all elevated, compared with those in cells treated with vehicle. PDGF-BB at lower concentrations (1 ng/mL, 10 ng/mL and 20 ng/mL) promoted the cell viability of PASMCs in a concentration-dependent way, while the cell viability of PASMCs in 40 ng/mL PDGF-BB group was lower than that in 20 ng/mL PDGF-BB group. PDGF-BB treatment also enhanced the migration ability of PASMCs, which was confirmed by Transwell assay. Similarly, PDGF-BB at lower concentrations (1 ng/mL, 10 ng/mL and 20 ng/mL) elevated the migratory cell number of PASMCs in a concentration-dependent way, whereas the migratory cell number of PASMCs in 40 ng/mL PDGF-BB group was lower than that in 20 ng/mL PDGF-BB group (Figure 1B).
Figure 1.
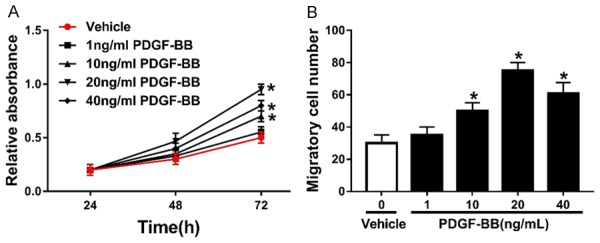
PDGF-BB treatment elevates the cell viability and migration ability of PASMCs. A. The cell viability of PASMCs treated with 1 ng/mL, 10 ng/mL, 20 ng/mL and 40 ng/mL PDGF-BB. B. The migratory cell number of PASMCs treated with 1 ng/mL, 10 ng/mL, 20 ng/mL and 40 ng/mL PDGF-BB. *P<0.05 compared to PASMCs treated with Vehicle.
miR-107 was down-regulated in PDGF-BB-pretreated PASMCs
RT-qPCR assay was employed to measure the miR-107 expression in PDGF-BB-pretreated PASMCs. Obviously, the miR-107 expression in PASMCs treated with PDGF-BB was lower than that in cells treated with Vehicle. PDGF-BB treatment inhibited miR-107 expression in a concentration-dependent manner (Figure 2A). As 20 ng/mL PDGF-BB treatment time increased, the expression level of miR-107 gradually decreased, and PDGF-BB treatment inhibited miR-107 expression in a time-dependent manner (Figure 2B).
Figure 2.
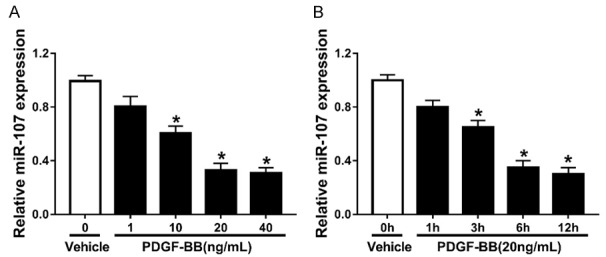
miR-107 is down-regulated in PDGF-BB-pretreated PASMCs. A. The miR-107 expression in PASMCs treated with 1 ng/mL, 10 ng/mL, 20 ng/mL and 40 ng/mL PDGF-BB. B. The miR-107 expression in PASMCs treated with 20 ng/mL PDGF-BB for 1 h, 3 h, 6 h and 12 h. *P<0.05 compared to PASMCs treated with Vehicle.
miR-107 repressed proliferation and migration of PASMCs promoted by PDGF-BB
To determine the effects of miR-107 on the proliferation of PASMCs, a series of introduction or knockout assays were conducted. We first transfected PASMCs with different doses of miR-107 mimics. Following that, RT-qPCR assay was performed to validate transfection efficiency. We could conclude that miR-107 mimics facilitated miR-107 expression in a dose-dependent way (Figure 3A). MTT assay was used to detect the impact of miR-107 on the cell viability of PASMCs pre-treated with PDGF-BB and revealed that up-regulation of miR-107 suppressed the cell viability of PASMCs and weakened the promotion of PDGF-BB on the viability of PASMCs (Figure 3B). As shown in Figure 3C, up-regulation of miR-107 reduced the protein levels of Ki-67 and Cyclin D1, and elevated p27 expression. Introduction of miR-107 attenuated the impact of PDGF-BB on the expression of the three. Next, we down-regulated miR-107 in PASMCs by transfection with Anti-miR-NC (Figure 3D). MTT assay showed that knockout of miR-107 contributed to the viability of PASMCs, and promoted the elevation of viability of PASMCs triggered by PDGF-BB (Figure 3E). Transwell assay was conducted to detect the migratory cell number of PASMCs and indicated that miR-107 repressed the migration ability of PASMCs and mitigated the promotion of PDGF-BB on the migration ability of PASMCs (Figure 3F).
Figure 3.
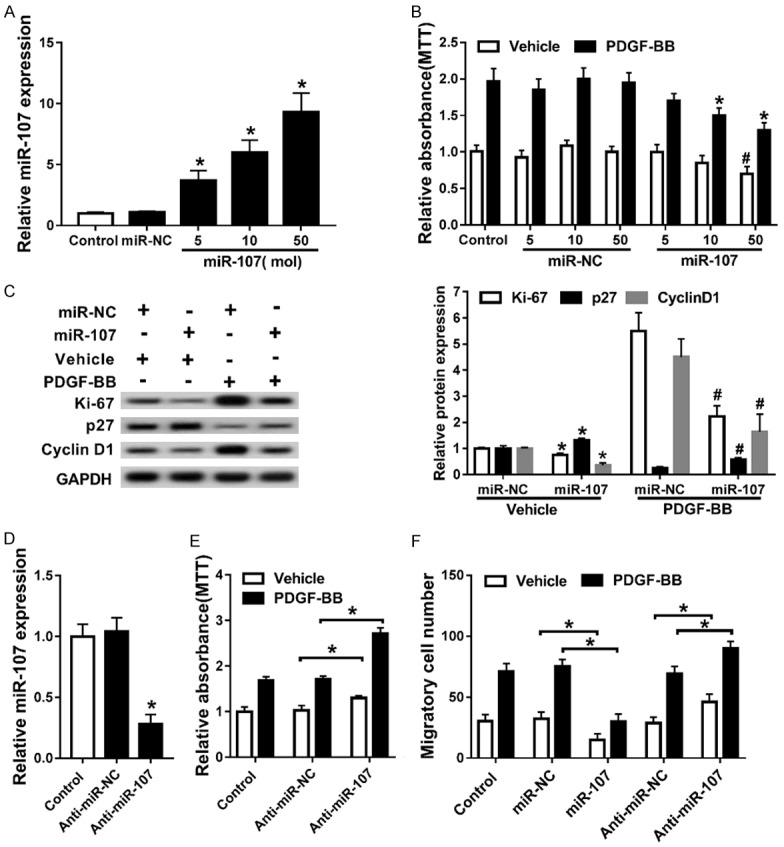
miR-107 repressed proliferation and migration of PASMCs promoted by PDGF-BB. A. The miR-107 expression in PASMCs transfected with different doses of miR-107 mimics. *P<0.05 compared to PASMCs transfected with miR-NC. B. The cell viability of PASMCs co-treated with PDGF-BB and miR-107 mimics or miR-NC. *P<0.05 compared to PASMCs co-treated with PDGF-BB and 5 mol miR-107 mimics. #P<0.05 compared to PASMCs co-treated with Vehicle and 50 mol miR-NC. C. Western blot assay for proteins Ki-67, p27, and Cyclin D1. *P<0.05 compared to PASMCs co-treated with Vehicle and miR-NC. #P<0.05 compared to PASMCs co-treated with PDGF-BB and miR-NC. D. The miR-107 expression in PASMCs transfected with Anti-miR-107 or Anti-miR-NC. *P<0.05 compared to PASMCs transfected with Anti-miR-NC. E. The cell viability of PASMCs co-treated with PDGF-BB and Anti-miR-107 or Anti-miR-NC. *P<0.05 compared to PASMCs co-treated with Vehicle and Anti-miR-NC. *P<0.05 compared to PASMCs co-treated with PDGF-BB and Anti-miR-NC. F. The migratory cell number of PASMCs. *P<0.05 compared to PASMCs co-treated with Vehicle and miR-NC. *P<0.05 compared to PASMCs co-treated with PDGF-BB and miR-NC. *P<0.05 compared to PASMCs co-treated with Vehicle and Anti-miR-NC. *P<0.05 compared to PASMCs co-treated with PDGF-BB and Anti-miR-NC.
NOR1 is a target of miR-107
Online software TargetScanHuman predicted the potential targets of miR-107 and identified NOR1 as a downstream target of miR-107. The predicted binding site of miR-107 on the 3’-UTR of NOR1 was marked in Figure 4A. Following luciferase reporter gene assay was applied to verify that whether miR-107 could bind to 3’-UTR of NOR1. The luciferase activity of PASMCs co-transfected with NOR1-wt and miR-107 mimics was much lower than that in cells co-transfected with NOR1-wt and miR-NC, while no luciferase activity change was observed in NOR1-mut-transfected PASMCs (Figure 4B). Subsequent RIP further confirmed the target relationship between miR-107 and NOR1 (Figure 4C). Strikingly, NOR1 was counter-regulated by miR-107 (Figure 4D).
Figure 4.
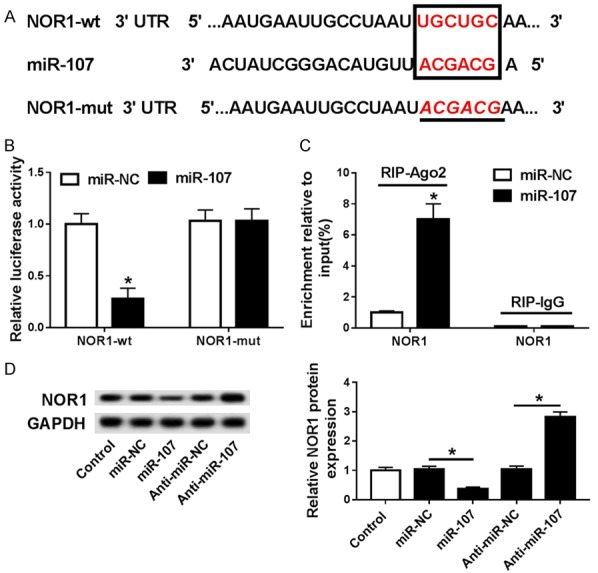
NOR1 is a target of miR-107. A. The predicted binding site and the mutant sites of miR-107 on the 3’-UTR of NOR1 mRNA are marked in red. B. The luciferase activity of PASMCs that were co-transfected with NOR1-wt or NOR1-mut and miR-107 mimics or miR-NC. *P<0.05 compared to PASMCs co-transfected with NOR1-wt and miR-NC. C. RIP was conducted, and expression of NOR1 was determined. *P<0.05 compared to PASMCs co-treated with RIP-Ago2 antibody and miR-NC. D. Western blot assay for NOR1. *P<0.05 compared to PASMCs transfected with miR-NC. *P<0.05 compared to PASMCs transfected with Anti-miR-NC.
PDGF-BB promoted the proliferation of PASMCs by down-regulating miR-107 and thereby up-regulating NOR1
PDGF-BB treatment facilitated the mRNA expression level of NOR1 in PASMCs, compared with cells treated with Vehicle (Figure 5A). We constructed PASMCs with NOR1 down-regulated through transfected with siNOR1 (Figure 5B). MTT assay suggested that knockout of NOR1 weakened the elevation of the cell viability of PASMCs induced by PDGF-BB (Figure 5C). Western blot indicated that down-regulation of NOR1 attenuated the regulation of PDGF-BB on the protein levels of Ki-67, p27 and Cyclin D1 (Figure 5D). Down-regulation of NOR1 suppressed the migration ability of PASMCs stimulated by PDGF-BB (Figure 5E). Restoration of NOR1 rescued the inhibition of overexpression of miR-107 on the cell viability of PASMCs (Figure 5F). Restoration of NOR1 also rescued the miR-107 up-regulation-induced suppression of the migration ability of PASMCs (Figure 5G).
Figure 5.
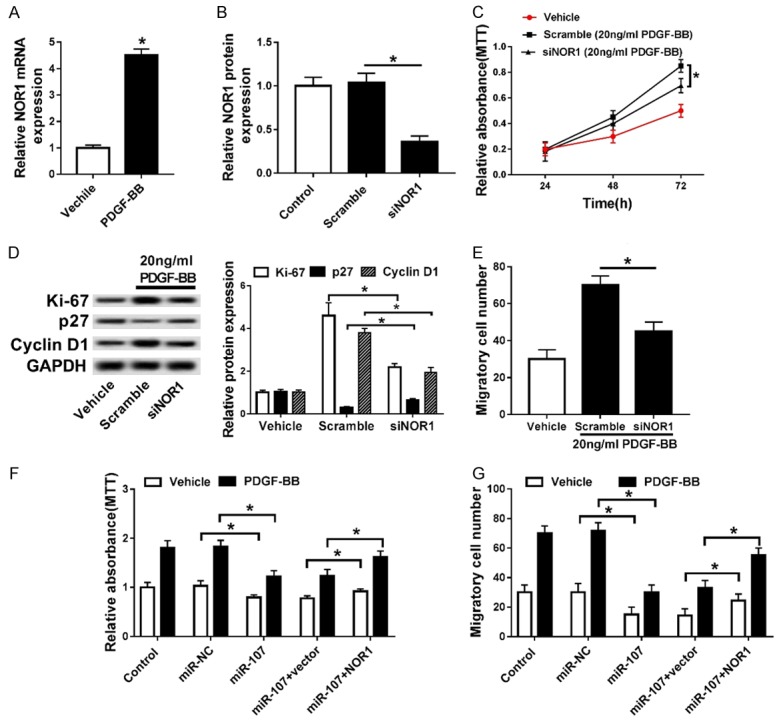
PDGF-BB promoted the proliferation of PASMCs by down-regulating miR-107 and thereby up-regulating NOR1. A. The mRNA expression of NOR1 in PASMCs treated with PDGF-BB. *P<0.05 compared to PASMCs treated with Vehicle. B. The protein level of NOR1 in PASMCs transfected with siNOR1. *P<0.05 compared to PASMCs transfected with Scramble. C. MTT assay for PASMCs co-treated with siNOR1 and PDGF-BB. *P<0.05 compared to PASMCs co-treated with Scramble and PDGF-BB. D. Western blot assay for Ki-67, p27 and Cyclin D1. *P<0.05 compared to PASMCs transfected with Scramble. E. The migrated cell number of PASMCs co-treated with siNOR1 and PDGF-BB. *P<0.05 compared to PASMCs co-treated with Scramble and PDGF-BB. F, G. The cell viability and migrated cell number of PASMCs co-transfected with miR-107 mimics and pcDNA3.1-NOR1 or vector. *P<0.05 compared to PASMCs co-treated with Vehicle and miR-NC. *P<0.05 compared to PASMCs co-treated with PDGF-BB and miR-NC. *P<0.05 compared to PASMCs co-treated with Vehicle, miR-107, and vector. *P<0.05 compared to PASMCs co-treated with PDGF-BB, miR-107 and vector.
Discussion
During the last two decades, PAH has gained increasing widespread attention. In Asia, PAH is still a lethal disease despite modern treatment [22]. Remodeling of PA is a significant feature of PAH, and excessive proliferation and migration of PASMCs is regarded as a primary cause [23]. Therefore, there is an urgent need for a better understanding of the molecular pathogenesis of PAH.
A previous study showed that PDGF-BB elevates human PASMC proliferation and survival, which is probably regulated by the JNK pathway [24]. Chen and his colleagues demonstrated that the up-regulation of miR-376b induced by PDGF-BB mediated the down-regulation of BMPR2, which led to expression change of its target genes and promoted the proliferation of PASMCs through multi-omics analysis [25]. Neuroblastoma suppressor of tumorigenicity 1 (NBL1) inhibited PDGF-BB-induced proliferation of human PASMCs, and the underlying mechanism is probably related to the reduced cyclin D1-CDK4 activity and raised p27 by decreasing the phosphorylation of p27 through the repression of PDGFRβ-p38MAPK signaling cascade [26]. In the current study, we found that PDGF-BB treatment promoted the cell viability and migration of PASMCs in a time-dependent and a concentration-dependent manner, which was in keeping with previous studies.
The proliferation of PASMCs can be meditated by several miRNAs. miR-34a was significantly down-regulated in hypoxic lung tissue, pulmonary artery, and PASMCs. miR-34a impeded the proliferation of PASMCs, and has potential to be used as a treatment target in PAH [10]. Yuan et al. demonstrated that miR-200c triggered cell proliferation and restrained cell apoptosis in PASMCs treated with endothelin-1 in vitro, which supplies a potential molecular basis for miR-200c regulation in the progression of PAH [12]. miR-222 had a pro-proliferation effect on PASMCs, at least partially by targeting p27 and TIMP3. Hence, inhibition of miR-222 in PASMCs may be a potential therapy strategy for PAH [13]. Additionally, Li et al. proposed a novel PDGFBB-respondent pathway, phosphatidylinositol 3-kinase (PI3K)/DNA methyltransferase 1(DNMT1)/miR-1281/histone deacetylase 4 (HDAC4), that plays a crucial role in PASMC proliferation and migration [14]. In our study, we found that miR-107 was down-regulated in PDGF-BB-pretreated PASMCs, which was similar to a former study [18]. Previously, miR-107 was reported to regulate adipose insulin sensitivity, and promote endoplasmic reticulum stress-induced apoptosis through targeting the Wnt3a/β-catenin/ATF6 pathway in preadipocytes [17,27]. The study conducted by Wang et al. suggested that the expression of miR-107 was significantly decreased in patients with Alzheimer’s disease (AD) at the early stage, and miR-107 probably participates in promoting AD progression via regulation of BACE1 [28]. Besides, miR-107 and its paralogs are down-regulated in certain tumors and supply potential therapeutic targets [29]. For example, Takahashi and his colleagues demonstrated that miR-107 can induce cell cycle arrest in human non-small cell lung cancer cell lines [30]. miR-107 also induced cell cycle arrest and inhibited invasion in gastric cancer cells by directly targeting cyclin-dependent kinase 6 (CDK6) [31]. Thus we investigated the role of PDGF-BB on miR-107 expression, and found that PDGF-BB inhibited miR-107 expression in a concentration-dependent and time-dependent manner. Then we further investigated the interaction between PDGF-BB and miR-107, and observed that miR-107 repressed proliferation and migration of PASMCs stimulated by PDGF-BB.
Dual luciferase reporter gene assays, RT-qPCR, and western blotting were performed to validate that NOR1 was a direct target of miR-107 in human PASMCs, and NOR1 was negatively-regulated by miR-107. NOR1 was identified as a direct target of miR-638 in human vascular smooth muscle cells, and down-regulation of NOR1 was vital for miR-638-mediated suppressor effects on PDGF-induced cyclin D1 expression, cell proliferation, and migration in human aortic smooth muscle cells [18]. Nomiyama et al. demonstrated that NOR1 targeted cyclin D1 and the lack of NOR1 reduced neointimal formation in response to vascular injury [32]. The research performed by Gizard et al. indicated that NOR1 induced the activation of the S Phase Kinase-associated Protein 2 (Skp2)-p27 pathway in response to mitogenic stimulation of vascular smooth muscle cells (VSMCs) and during neointima formation [33]. Wang et al. concluded that the upregulation of NOR1 is related to hypoxia-induced pulmonary vascular remodeling in chronic obstructive pulmonary disease (COPD) by contributing to human PASMCs proliferation [34]. In our study, NOR1 was up-regulated by PDGF-BB, and knockout of NOR1 attenuated the PDGF-BB-mediated proliferation and migration of PASMCs. Recuperation of NOR1 weakened the miR-107 up-regulation-induced inhibition of proliferation and migration of PASMCs.
In summary, we observed that PDGF-BB treatment promoted the cell viability and migration of PASMCs in a time-dependent and a concentration-dependent manner. PDGF-BB treatment suppressed the expression level of miR-107 in a concentration-dependent and time-dependent way. Up-regulation of miR-107 repressed proliferation and migration of PASMCs promoted by PDGF-BB, while loss of miR-107 deepened the promoted effects induced by PDGF-BB. NOR1 is a direct target of miR-107 and is counter-regulated by miR-107. Knockout of NOR1 weakened the elevation of the proliferation and migration ability of PASMCs induced by PDGF-BB. Restoration of NOR1 attenuated the inhibition of miR-107 on the cell viability and migration ability of PASMCs. In conclusion, miR-107 inhibits PDGF-BB-induced PASMCs proliferation and migration through targeting NOR1.
Disclosure of conflict of interest
None.
References
- 1.Encyclopædia Britannica: Encyclopædia Britannica Online. 2009. Leaf-nosed bat. [Google Scholar]
- 2.Hoeper MM, Humbert M, Souza R, Idrees M, Kawut SM, Sliwa-Hahnle K, Jing ZC, Gibbs JS. A global view of pulmonary hypertension. Lancet Respir Med. 2016;4:306–322. doi: 10.1016/S2213-2600(15)00543-3. [DOI] [PubMed] [Google Scholar]
- 3.Sutendra G, Michelakis ED. The metabolic basis of pulmonary arterial hypertension. Cell Metab. 2014;19:558–573. doi: 10.1016/j.cmet.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Schermuly RT, Ghofrani HA, Wilkins MR, Grimminger F. Mechanisms of disease: pulmonary arterial hypertension. Nat Rev Cardiol. 2011;8:443–455. doi: 10.1038/nrcardio.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredriksson L, Li H, Eriksson U. The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004;15:197–204. doi: 10.1016/j.cytogfr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Perros F, Montani D, Dorfmuller P, Durand-Gasselin I, Tcherakian C, Le Pavec J, Mazmanian M, Fadel E, Mussot S, Mercier O, Herve P, Emilie D, Eddahibi S, Simonneau G, Souza R, Humbert M. Platelet-derived growth factor expression and function in idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med. 2008;178:81–88. doi: 10.1164/rccm.200707-1037OC. [DOI] [PubMed] [Google Scholar]
- 7.Yu Y, Sweeney M, Zhang S, Platoshyn O, Landsberg J, Rothman A, Yuan JX. PDGF stimulates pulmonary vascular smooth muscle cell proliferation by upregulating TRPC6 expression. Am J Physiol Cell Physiol. 2003;284:C316–330. doi: 10.1152/ajpcell.00125.2002. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- 10.Wang P, Xu J, Hou Z, Wang F, Song Y, Wang J, Zhu H, Jin H. miRNA-34a promotes proliferation of human pulmonary artery smooth muscle cells by targeting PDGFRA. Cell Prolif. 2016;49:484–493. doi: 10.1111/cpr.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang WF, Xiong YW, Zhu TT, Xiong AZ, Bao HH, Cheng XS. MicroRNA let-7g inhibited hypoxia-induced proliferation of PASMCs via G0/G1 cell cycle arrest by targeting c-myc. Life Sci. 2017;170:9–15. doi: 10.1016/j.lfs.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 12.Yuan C, Xu M, Rong R, Mei Y, Cai W, Li L, Xue Y, Zhu B, Sun K, Han L. miR-200c regulates endothelin-1 induced PASMCs abnormal proliferation and apoptosis. IUBMB Life. 2017;69:877–886. doi: 10.1002/iub.1686. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Bei Y, Shen S, Zhang J, Lu Y, Xiao J, Li X. MicroRNA-222 promotes the proliferation of pulmonary arterial smooth muscle cells by targeting P27 and TIMP3. Cell Physiol Biochem. 2017;43:282–292. doi: 10.1159/000480371. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Li L, Qian Z, Lin B, Chen J, Luo Y, Qu J, Raj JU, Gou D. Phosphatidylinositol 3-Kinase-DNA methyltransferase 1-miR-1281-Histone deacetylase 4 regulatory axis mediates platelet-derived growth factor-induced proliferation and migration of pulmonary artery smooth muscle cells. J Am Heart Assoc. 2018;7 doi: 10.1161/JAHA.117.007572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahoo S, Meijles DN, Al Ghouleh I, Tandon M, Cifuentes-Pagano E, Sembrat J, Rojas M, Goncharova E, Pagano PJ. MEF2C-MYOCD and leiomodin1 suppression by miRNA-214 promotes smooth muscle cell phenotype switching in pulmonary arterial hypertension. PLoS One. 2016;11:e0153780. doi: 10.1371/journal.pone.0153780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, Parenti AR, Daidone MG, Bicciato S, Piccolo S. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Wu S, Muhammad S, Ren Q, Sun C. miR-103/107 promote ER stress-mediated apoptosis via targeting the Wnt3a/beta-catenin/ATF6 pathway in preadipocytes. J Lipid Res. 2018;59:843–853. doi: 10.1194/jlr.M082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P, Liu Y, Yi B, Wang G, You X, Zhao X, Summer R, Qin Y, Sun J. MicroRNA-638 is highly expressed in human vascular smooth muscle cells and inhibits PDGF-BB-induced cell proliferation and migration through targeting orphan nuclear receptor NOR1. Cardiovasc Res. 2013;99:185–193. doi: 10.1093/cvr/cvt082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie X, Zhang B, Li X, Xiang J, Xiao B, Ma J, Zhou M, Zhu S, Lu H, Gui R, Shen S, Li G. Cloning, expression, and mutation analysis of NOR1, a novel human gene down-regulated in HNE1 nasopharyngeal carcinoma cell line. J Cancer Res Clin Oncol. 2003;129:410–414. doi: 10.1007/s00432-003-0451-9. [DOI] [PubMed] [Google Scholar]
- 20.Nomiyama T, Nakamachi T, Gizard F, Heywood EB, Jones KL, Ohkura N, Kawamori R, Conneely OM, Bruemmer D. The NR4A orphan nuclear receptor NOR1 is induced by platelet-derived growth factor and mediates vascular smooth muscle cell proliferation. J Biol Chem. 2006;281:33467–33476. doi: 10.1074/jbc.M603436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi M, Yang J, Li W, Li X, Xiong W, McCarthy JB, Li G, Xiang B. The NOR1/OSCP1 proteins in cancer: from epigenetic silencing to functional characterization of a novel tumor suppressor. J Cancer. 2017;8:626–635. doi: 10.7150/jca.17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds PN. Pulmonary arterial hypertension: in Asia, as elsewhere, still a lethal disease despite modern treatment. Respirology. 2019;24:99–100. doi: 10.1111/resp.13438. [DOI] [PubMed] [Google Scholar]
- 23.Stenmark KR, Fagan KA, Frid MG. Hypoxia-induced pulmonary vascular remodeling: cellular and molecular mechanisms. Circ Res. 2006;99:675–691. doi: 10.1161/01.RES.0000243584.45145.3f. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Lv W, Piao H, Chu X, Wang H. Role of platelet-derived growth factor-BB (PDGF-BB) in human pulmonary artery smooth muscle cell proliferation. J Recept Signal Transduct Res. 2014;34:254–260. doi: 10.3109/10799893.2014.908915. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Cui X, Qian Z, Li Y, Kang K, Qu J, Li L, Gou D. Multi-omics analysis reveals regulators of the response to PDGF-BB treatment in pulmonary artery smooth muscle cells. BMC Genomics. 2016;17:781. doi: 10.1186/s12864-016-3122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui C, Zhang H, Guo LN, Zhang X, Meng L, Pan X, Wei Y. Inhibitory effect of NBL1 on PDGF-BB-induced human PASMC proliferation through blockade of PDGFbeta-p38MAPK pathway. Biosci Rep. 2016;36:e00374. doi: 10.1042/BSR20160199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 28.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng L, Xie Y, Zhang H, Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol. 2012;29:856–863. doi: 10.1007/s12032-011-9823-1. [DOI] [PubMed] [Google Scholar]
- 32.Nomiyama T, Zhao Y, Gizard F, Findeisen HM, Heywood EB, Jones KL, Conneely OM, Bruemmer D. Deficiency of the NR4A neuron-derived orphan receptor-1 attenuates neointima formation after vascular injury. Circulation. 2009;119:577–586. doi: 10.1161/CIRCULATIONAHA.108.822056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gizard F, Zhao Y, Findeisen HM, Qing H, Cohn D, Heywood EB, Jones KL, Nomiyama T, Bruemmer D. Transcriptional regulation of S phase kinase-associated protein 2 by NR4A orphan nuclear receptor NOR1 in vascular smooth muscle cells. J Biol Chem. 2011;286:35485–35493. doi: 10.1074/jbc.M111.295840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang CG, Li C, Lei W, Jiang JH, Huang JA, Zeng DX. The association of neuron-derived orphan receptor 1 with pulmonary vascular remodeling in COPD patients. Int J Chron Obstruct Pulmon Dis. 2018;13:1177–1186. doi: 10.2147/COPD.S151820. [DOI] [PMC free article] [PubMed] [Google Scholar]


