Abstract
Netrin 4 (NTN4) is downregulated in breast cancer (BC) and can inhibit the migration of BC cells. miRNAs dysregulation plays prominent roles in BC tumorigenesis. However, the function of miR-17-5p, its relationship with NTN4 and its underlying functional mechanism in BC are unclear and were investigated in the current study. Compared with normal breast samples, miR-17-5p was upregulated in BC specimens in The Cancer Genome Atlas (TCGA). A clinical analysis based on TCGA showed that miR-17-5p expression correlated with BC tumor stage, lymph node status, estrogen receptor, and progesterone receptor status. A wound-healing assay and Transwell assay implied that miR-17-5p upregulation promotes BC cell migration and invasion. Reverse transcription-quantitative PCR and ELISA showed that NTN4 mRNA and protein were both downregulated after miR-17-5p was overexpressed in Hs578T cells, whereas miR-17-5p inhibition had the opposite effect in MCF-7 cells. We also performed a dual-fluorescent reporter assay, the results of which demonstrated that miR-17-5p represses NTN4 expression by directly targeting the 3’ untranslated region of NTN4 mRNA. In summary, miR-17-5p considerably promotes BC cell migration by suppressing NTN4 expression, and may therefore offer a potential therapeutic target for BC.
Keywords: Breast cancer (BC), NTN4, miR-17-5p, migration
Introduction
Breast cancer (BC) is the second commonest cause of cancer-related death in females [1-3]. In the past three decades, the treatment and early diagnosis of BC have improved markedly. The overall 5-year survival rate of BC patients has shown an encouraging trend [1]. However, Globocan estimated that 187,000 people in China were first diagnosed with BC in 2012, and approximately 48,000 of them died [2,4]. Metastasis is the major challenge for BC treatment, although many significant advances have been made in the study of cancer metastasis.
The netrin family of proteins functions in the modulation of both neuronal axon development and angiogenesis [5]. Netrin 4 (NTN4) is a secreted molecule, a novel member of this family [6], which is generally reduced in tumors but is upregulated in cancerous effusions or at the invasive edges of solid tumors [7,8]. In our previous study, we demonstrated that NTN4 acts as a BC suppressor and inhibits the metastasis of BC cells by downregulating epithelial-mesenchymal transition (EMT)-related biomarkers [9].
microRNAs (miRNAs), a class of 19-25 nucleotide small noncoding RNAs, can bind to the 3’ untranslated regions (3’-UTRs) of the mRNAs thus inhibiting the expression of the corresponding proteins [10,11]. So far, several published studies have suggested that a series of miRNAs are abnormally expressed in BC, and may modulate cell proliferation, apoptosis, migration, and invasion [12-14]. starBase [15] was used to predict the miRNAs that may target NTN4, with six upregulated miRNAs identified. A correlation analysis of the most relevant miRNA, miR-17-5p, was performed. Several studies have reported that miR-17-5p plays pivotal roles in BC, acting as an onco-miR to promote BC cell proliferation, and could have utility as a biomarker predicting BC recurrence [16,17]. However, how miR-17-5p influences BC cell invasiveness and the molecular mechanism by which it regulates NTN4 have not been determined.
In this study, we investigated the function of miR-17-5p, a differentially expressed miRNA associated with NTN4, in BC cell invasion, and the possible mechanism of this function. We found that miR-17-5p acts as an onco-miR, repressing NTN4 expression by binding to the 3’-UTR of NTN4 mRNA and thus promoting BC cell metastasis.
Materials and methods
Cell culture and transfection
Hs578T, MCF-7, MDA-MB-231, and T-47D, the four human BC cell lines used in this research, were obtained from the Cell Bank of China Academy of Sciences (Shanghai, China). All cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, Utah, USA) supplemented with 1% penicillin-streptomycin (HyClone), and 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) in a humidified incubator (37°C) containing 5% CO2. The cells (2.0 × 105) were seeded in each well of six-well plates, and then cultured for 24 h before transfection. The cells were transfected with miR-17-5p mimic (miR-17 mimic) or miR-17-5p inhibitor (miR-17 inhibitor), with Lipofectamine® 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
RT-qPCR analysis of mRNA and miRNA
Total cellular RNA (including miRNA) was purified with TRIzol (Invitrogen). RT-qPCR was used to evaluate the level of NTN4 mRNA, using the One-Step TB GreenTM PrimeScriptTM RT-PCR Kit II (TaKaRa Bio, Shiga, Japan), according to the manufacturer’s instruction. β-actin was used as the reference gene for the normalization of NTN4. The primers used in this assay were: NTN4 forward primer 5’-GTACTTTGCGACTAACTGCTCC-3’, and reverse primer 5’-TCCAGTGCATGGAAAAGGACT-3’; and β-actin forward primer 5’-ACCCACACTGTGCCCATCTAC-3’, reverse primer 5’-TCGGTGAGGATCTCATGAGGTA-3’.
To quantify miR-17-5p, the Mir-XTM miRNA First-Strand Synthesis Kit (Clontech, CA, USA) was used to convert miRNAs into cDNA; SYBR® Premix Ex TaqTM II (TaKaRa Bio) was then used for qPCR, performed on Light Cycler® 480 System (Roche Diagnostics, Switzerland). miR-17-5p was standardized to the value of U6 snRNA with 2-ΔΔCt method. The miR-17-5p primers were synthesized by RiboBio (RiboBio Co., Ltd, Guangzhou, China).
Colony formation assays
After transfection for 24 h, 103 single cells were seeded into each well of six-well plates, and cultured in 2 ml of DMEM that included 10% FBS for 10 days or until colonies could be seen with the naked eye. Then 95% ethanol was used as the stationary liquid for the colonies. The colonies were then fixed for 15 min, and stained for another 15 min with 0.1% crystal violet solution. The ImageJ software (National Institutes of Health, USA) was used to count numbers of clones.
Wound-healing assay
The wound-healing assay was used to detect the horizontal migration capacity of BC cells. In a scratch assay, the cell monolayer was wounded with 200 μl pipette tips after transfection for 24 h, and then the same site on the monolayer was photographed at 0, 24, 48 h under a microscope (Olympus CKX41, Tokyo, Japan). The gap size was measured with the ImageJ software and the percentage migration was calculated based on the size of the wound at 0 h.
Transwell migration and invasion assay
The Transwell assay, with Transwell inserts (8 μm, 24-well plate; Corning, USA), was used to analyze the migration and invasion capabilities of BC cells. Cells (6 × 104) suspended in DMEM without serum were added to the upper chamber, DMEM containing 10% FBS was added to the lower chamber, and the cells were cultured for 24 h to evaluate their vertical migration. For the invasion assay, the chamber membrane was covered with Matrigel (BD Biosciences, San Jose, CA, USA) and cells were incubated in the same environment for 48 h. Cells that had migrated to or invaded the lower chamber were treated with 95% ethanol, 0.1% crystal violet dye for 15 min respectively, and counted in five random fields.
Elisa
NTN4 protein concentration was measured with an ELISA kit (CSB-E11900h; CUSABIO, China, https://www.cusabio.com/), according to manufacturer’s instructions, as described previously [9]. The results were detected with a plate reader (Anthos Fluido 2010), as the absorbance at 450 nm. A standard curve was constructed with the software Curve Expert 1.4 and the NTN4 concentrations were then calculated.
Target prediction for miR-17-5p and NTN4
Two different online databases, miRDB (http://mirdb.org/index.html) [18] and TargetScan (http://www.targetscan.org/) [19], were used to predict the possible target sites of miR-17-5p within NTN4 mRNA. The detailed protocols can be obtained at the relevant websites.
Dual-luciferase reporter assay
The pmiR-RB-Report vector (RiboBio) was used in the dual-luciferase reporter assay, with the target region of miR-17-5p (sequence 5’-TTCCTTGTATAAAGCACTTTA-3’) inserted in and was designated the wild-type-NTN4-expressing plasmid (h-NTN4-WT). The control plasmid containing a mutant sequence (5’-TTCCTTGTATAAACGTGAAAA-3’) was designated the mutant-NTN4-expressing plasmid (h-NTN4-MUT). MCF-7 cells were plated in a 24-well plate (1.0 × 105 cells per well), and cultured for 24 h. The MCF-7 cells were then cotransfected with 300 ng of h-NTN4-WT or h-NTN4-MUT and 50 nM miR-17 mimic or negative control using 3 μl of Lipofectamine® 3000 per well. After transfection for 48 h, the Dual-Luciferase Reporter System (Promega, Madison, WI, USA) was used to detect the Renilla and firefly luciferase activities, according to the instructions of the manufacturer.
Statistical analysis
All statistical analyses were performed with the SPSS 20.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prime 7.00 (La Jolla, CA, USA). The paired Student’s t test and unpaired Student’s t test were used to analyze the gene expression differences between two groups. The relationship between miR-17-5p and NTN4 expression was analyzed with Pearson’s correlation analysis. miR-17-5p expression and the clinical characteristics were analyzed with a χ2 test. Kaplan-Meier method were performed for the survival analysis, and log-rank test was used to calculate the P-values. Data are presented as the means ± standard deviations (SD) of at least three independent experiments. P < 0.05 was considered significant.
Results
Upregulation of miR-17-5p in malignant tissues and sera of BC patients
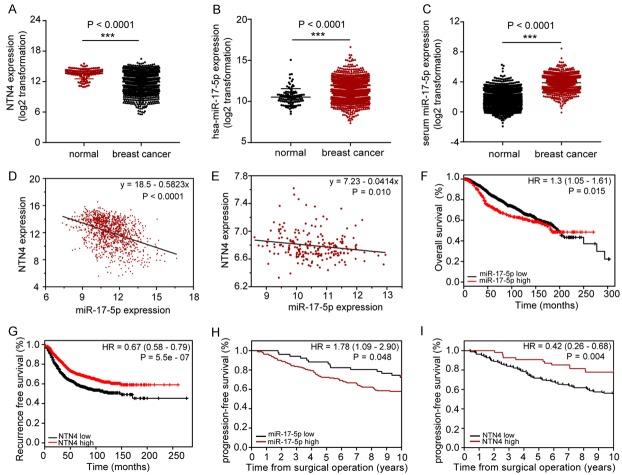
The levels of NTN4 and miR-17-5p in BC were analyzed with data from the public database The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/). The results showed that NTN4 mRNA expression was significantly lower in BC tissues than in normal breast samples (Figure 1A), whereas miR-17-5p expression was significantly higher in the BC tissues (Figure 1B). We also analyzed the serum miR-17-5p levels of the BC patients and control subjects without cancer using data from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Compared with the noncancer samples, miR-17-5p was elevated in the sera of BC patients (P < 0.001) (Figure 1C), which is consistent with the expression trend in tissues. We also analyzed the relationship between miR-17-5p and NTN4 expression in the TCGA and GEO database (GSE22220) (Figure 1D, 1E). The results suggested a significant negative association between miR-17-5p and NTN4 expression in BC.
Figure 1.
Markedly negative correlation between miR-17-5p and NTN4 mRNA in BC lesions in a database analysis. A. An analysis of NTN4 mRNA expression in 113 normal breast tissues and 1109 BC tissues in the TCGA database. B. miR-17-5p expression in 104 normal breast specimens and 1103 BC samples from the data in TCGA. C. Serum miR-17-5p expression in GSE73002, including 2686 noncancer controls and 1280 BC samples, ***P < 0.001. D, E. Correlations between miR-17-5p and NTN4 mRNA in TCGA (1066 pairs of samples, P < 0.0001) and GSE22220 (207 pairs of tissues, P = 0.010). F, G. Survival analysis of miR-17-5p and NTN4 in BC with a Kaplan-Meier plotter. H, I. Progression-free survival of miR-17-5p and NTN4 in GSE22220.
Survival analysis and clinicopathologic characteristic analysis of miR-17-5p in BC patients
Kaplan-Meier plotter (http://www.kmplot.com/mirpower) was used to analyze the influence of miR-17-5p expression on BC prognosis. The results suggested that low-miR-17-5p-expressing BC patients displayed better prognoses (P = 0.015) (Figure 1F), whereas patients with high-NTN4 expression had prolonged recurrence-free survival (RFS) (P = 5.5e-07) (Figure 1G). A similar result was obtained with the GSE22220 dataset (Figure 1H, 1I).
We also analyzed data from TCGA to determine whether miR-17-5p expression in BC patients was strongly associated with their clinicopathologic features. miR-17-5p expression was found to be significantly correlated with tumor stage (P = 0.002), lymph node status (P = 0.001), estrogen receptor (ER) status (P < 0.001) and progesterone receptor (PR) status (P < 0.001), but not significantly with other characteristics (Table 1). These results strongly indicate that miR-17-5p participates in the progression and prognosis of BC.
Table 1.
Correlation between miR-17-5p expression in BC and clinical characteristics in TCGA
| miR-17-5p-low (%) | miR-17-5p-high (%) | P value | |
|---|---|---|---|
| Age | |||
| < 40 | 37 (50.7) | 36 (49.3) | |
| 40-49 | 125 (57.3) | 93 (42.7) | |
| 50-59 | 137 (48.9) | 143 (51.1) | |
| 60-69 | 145 (51.8) | 135 (48.2) | |
| ≥ 70 | 124 (55.1) | 101 (44.9) | 0.378 |
| Tumor stage | |||
| T1 | 158 (56.4) | 122 (43.6) | |
| T2 | 287 (46.3) | 333 (53.7) | |
| T3 | 63 (46.7) | 72 (53.3) | |
| T4 | 28 (70.0) | 12 (30.0) | 0.002 |
| Lymph node | |||
| Negative | 226 (44.4) | 283 (55.6) | |
| Positive | 311 (54.7) | 258 (45.3) | 0.001 |
| Stage | |||
| I | 95 (52.2) | 87 (47.8) | |
| II | 285 (46.8) | 324 (53.2) | |
| III | 126 (51.6) | 118 (48.4) | |
| IV | 13 (65.0) | 7 (35.0) | |
| V | 10 (76.9) | 3 (23.1) | 0.072 |
| ER status | |||
| Negative | 64 (27.6) | 168 (72.4) | |
| Positive | 458 (57.6) | 337 (42.4) | 0.000 |
| PR status | |||
| Negative | 118 (35.2) | 217 (64.8) | |
| Positive | 406 (59.0) | 282 (41.0) | 0.000 |
| Her2 status | |||
| Negative | 287 (51.8) | 267 (48.2) | |
| Positive | 88 (55.3) | 71 (44.7) | 0.431 |
miR-17-5p significantly promotes the clonogenicity of BC cells
The levels of miR-17-5p in Hs578T, MDA-MB-231, MCF-7, and T-47D cells were evaluated using RT-qPCR. miR-17-5p was expressed most strongly in MCF-7 cells, and significantly lower in Hs578T cells than in the other cell types. The level of NTN4 was lowest in the MCF-7 cells and second highest in Hs578T cells (Figure 2A). Therefore, MCF-7 and Hs578T cells were chosen as the experimental cells. To explore the biological roles of miR-17-5p, we transfected Hs578T and MCF-7 cells with miR-17 mimic or miR-17 inhibitor to overexpress and knockdown miR-17-5p, respectively. After the transfection, miR-17-5p was expressed 67.7-fold more strongly in the Hs578T cells and its expression in the MCF-7 cells was knocked down by 68.7% (Figure 2B, 2C).
Figure 2.
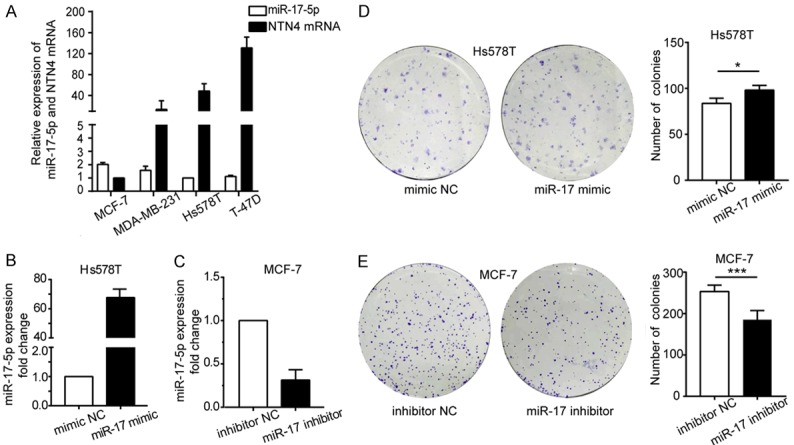
Validation of miR-17-5p transfection. A. miR-17-5p and NTN4 mRNA expression in four BC cell lines, determined with RT-qPCR. B, C. Fold change in miR-17-5p expression after cell transfection with miR-17 mimic, miR-17 inhibitor, or their matching controls. ‘Mimic NC’ denotes mimic-transfected negative control, and ‘inhibitor NC’ denotes inhibitor-transfected negative control. D, E. Influence of miR-17-5p on colony formation. D. miR-17-5p upregulation in Hs578T cells promoted colony formation. E. Reduced miR-17-5p in MCF-7 cells suppressed colony formation. Colony numbers are quantified in the right bar graphs. *P < 0.05, ***P < 0.001.
The capacity to form colonies can reflect the tumorigenicity of cells, so we used a colony formation assay to determine whether the cellular miR-17-5p affects the clonogenicity of BC cells. After Hs578T cells were transfected with the miR-17 mimic and MCF-7 cells were transfected with miR-17 inhibitor, the Hs578T cells generated significantly more colonies than the mimic-transfected normal control cells (NC; P = 0.032) (Figure 2D), whereas the MCF-7 cells produced substantially fewer colonies than the inhibitor-transfected NC (P < 0.001) (Figure 2E). These results imply that miR-17-5p promotes colony formation in BC cells.
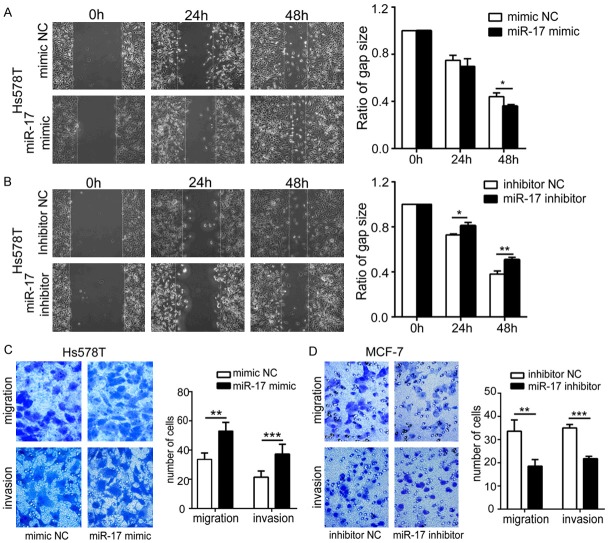
miR-17-5p promotes the migration and invasiveness of BC cells
We also studied whether miR-17-5p expression influences the cellular migration and invasiveness of BC, which affect tumor metastasis and overall survival. A scratch assay was performed in Hs578T cells, because of their high migration capacity. miR-17-5p overexpression accelerated scratch healing in the Hs578T cells (Figure 3A) and its depletion reduced cell migration (Figure 3B). In the Transwell migration assays, Hs578T cells overexpressing miR-17-5p migrated more than the NC cells (P < 0.001) (Figure 3C, upper panel), whereas miR-17-5p depletion in the MCF-7 cells strongly diminished their migration compared with that of the corresponding control (Figure 3D, upper panel). The results of the migration assay are consistent with those of the cell invasion test (Figure 3C and 3D, lower panel) and suggest that miR-17-5p increases the cellular metastatic and invasive capacities of BC.
Figure 3.
Upregulation or downregulation of miR-17-5p affects the migration and invasion capacities of BC cells. A, B. Effects of miR-17-5p on wound-healing assay in Hs578T cells. A. miR-17-5p overexpression promoted wound healing in Hs578T cells P = 0.046) at 48 h. B. Reduced miR-17-5p suppressed wound healing in Hs578T cells at 24 h (P = 0.032) and 48 h (P = 0.010). C, D. Effects of miR-17-5p evaluated with a Transwell assay. C. miR-17-5p overexpression increased Hs578T cells migration and invasion. D. Inhibition of miR-17-5p inhibited MCF-7 cells migration and invasion. Bar graphs on the right show the quantification of the left panels. *P < 0.05, **P < 0.01, ***P < 0.001.
NTN4 mRNA is the target of miR-17-5p
To study whether the function of miR-17-5p in promoting BC cell migration directly involves NTN4, RT-qPCR and ELISA were used after miR-17-5p transfection to evaluate the changes in NTN4 expression. The levels of NTN4 mRNA and protein were downregulated in the miR-17-5p-overexpressing Hs578T cells, and their expression was elevated in miR-17-5p-reduced MCF-7 cells (Figure 4A). This indicates that miR-17-5p reduces NTN4 expression, which is consistent with the interactions predicted with the databases.
Figure 4.
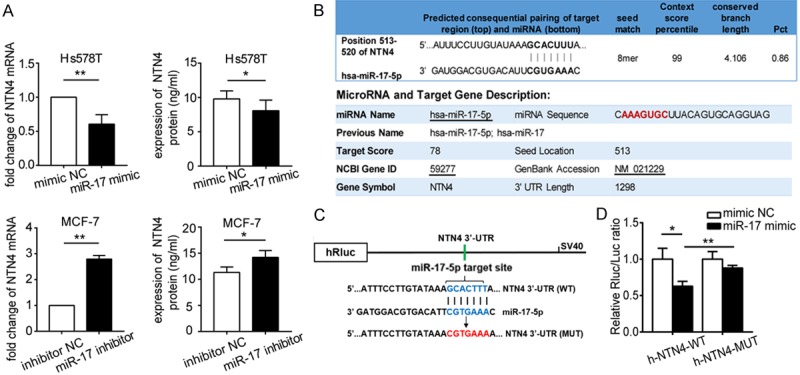
miR-17-5p directly binds to NTN4 mRNA. A. Expression of NTN4 mRNA (left panel) and protein (right panel) changed after the transfection of BC cells with miR-17 mimic or miR-17 inhibitor. B. Binding site of miR-17-5p within NTN4 mRNA predicted with TargetScan and miRDB. C. h-NTN4-WT and h-NTN4-MUT were constructed in the pmiR-RB-REPORT vector, with wild-type and mutated miR-17-5p binding sites inserted, respectively, downstream from the luciferase reporter. D. In the luciferase reporter assay, miR-17-5p significantly reduced the luciferase activity expressed from h-NTN4-WT (P < 0.05) but not that from h-NTN4-MUT (P > 0.05). *P < 0.05, **P < 0.01.
Bioinformatic tools based on different algorithms were used to identify the possible target sites of miR-17-5p within NTN4 mRNA. We used two different miRNA databases, TargetScan and miRDB, to maximize the accuracy of the prediction. The results showed that a sequence within the 3’-UTR of NTN4 mRNA was predicted to be a binding site of miR-17-5p. The binding site is located at nucleotides 513-520, with a high target score (Figure 4B). Therefore, luciferase reporter vectors were constructed (Figure 4C), and dual-luciferase reporter assays were conducted in MCF-7 cells. As suggested in Figure 4D, miR-17-5p significantly dampened the luciferase activity of h-NTN4-WT, and the suppression of NTN4 expression by miR-17-5p was attenuated when the putative binding site of miR-17-5p in the NTN4 3’-UTR was mutated. Therefore, miR-17-5p directly suppresses NTN4 expression in human BC by targeting the 3’-UTR of NTN4 mRNA.
Discussion
Metastasis is a severe manifestation of BC, and metastasis is the main reason for the high mortality observed in BC patients [20]. In a previous study, we demonstrated that NTN4 has a protective function in BC by reducing the migration and invasion of BC cells [9]. However, its function in human BC is not fully understood. Accumulating evidence indicates that miRNAs play vital roles in regulating the occurrence and progression of many tumors [12,13]. miRNAs are detectable in plasma or serum, and are therefore expected to have utility as novel biomarkers of cancer prognosis [20].
In this study, we analyzed data from BC patients in the TCGA and GEO databases and found that miR-17-5p was clearly highly expressed in both tissue and serum samples from BC patients. NTN4 was significantly reduced in BC tissues, which correlated significantly negatively with miR-17-5p expression. We also investigated the functions of miR-17-5p in BC cell migration and the molecular mechanism by which it regulates NTN4 expression.
Many studies have shown that NTN4 acts as a tumor suppressor in various cancers, including BC and cervical cancer [9,21]. Through the analysis of data from the Kaplan-Meier plotter database and GEO datasets, we found that BC patients with high-NTN4-expressing tumors had markedly longer RFS than those with low-NTN4-expressing tumors. Nevertheless, a survival analysis of BC patients showed the patients with high-miR-17-5p tumors tended to show poor overall survival. Other clinical characteristics implied that BC patients strongly expressing miR-17-5p are more likely to have a higher tumor stage and negative ER status, which is consistent with the results of many studies that have shown that ER-negative tumors indicate poorer survival than ER-positive tumors [22]. All the results together indicate that miR-17-5p promotes tumor progression.
For the functional experiment, we selected two experimental cell lines, MCF-7 cells, because they express relatively high miR-17-5p and low NTN4 levels, and Hs578T cells, with low miR-17-5p and high NTN4 levels. Our results indicated that miR-17-5p overexpression markedly increased the cellular metastatic and invasive abilities of BC. Therefore, we infer that miR-17-5p is associated with BC metastasis, and contrary to the normal roles of NTN4. We also explored the possible mechanisms underlying the cancer-promoting effects of miR-17-5p in BC. After miR-17-5p depletion, both NTN4 mRNA and protein were clearly increased, whereas miR-17-5p upregulation had the opposite effect. Because the expression of NTN4 can be directly regulated by miR-17-5p, and miRNAs usually modulate their target gene expression by binding to the 3’-UTR of their mRNAs, we predicted the target site of miR-17-5p in the NTN4 mRNA with online tools. We then confirmed this with a dual-luciferase reporter assay, which showed that miR-17-5p modulates NTN4 expression by binding directly to the NTN4 mRNA 3’-UTR. However, the relative fluorescence of the group in which miR-17-5p and h-NTN4-WT were cotransfected was also quenched, so we suspect that miR-17-5p may have other binding sites in the 3’-UTR of NTN4 mRNA. A further study is required to investigate this possibility.
Importantly, the results of this study provide the first evidence that miR-17-5p directly modulates NTN4 expression and promotes the metastasis and invasion of BC cells. We have confirmed the miR-17-5p is negatively associated with NTN4 by investigating their expression and functions, both in database analyses and experimentally. However, because our findings were mainly obtained in databases and with cell-based experiments, further investigation of the role of miR-17-5p in BC progress is still required.
Acknowledgements
This work was supported by the Public Technology Research Project of Zhejiang Province (LGF18H200006) and the Medicines Health Technology Plan Project of Zhejiang Province (2018PY073).
Disclosure of conflict of interest
None.
References
- 1.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 4.Wang F, Yu ZG. Current status of breast cancer prevention in China. Chronic Dis Transl Med. 2015;1:2–8. doi: 10.1016/j.cdtm.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson BD, Ii M, Park KW, Suli A, Sorensen LK, Larrieu-Lahargue F, Urness LD, Suh W, Asai J, Kock GA, Thorne T, Silver M, Thomas KR, Chien CB, Losordo DW, Li DY. Netrins promote developmental and therapeutic angiogenesis. Science. 2006;313:640–644. doi: 10.1126/science.1124704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch M, Murrell JR, Hunter DD, Olson PF, Jin W, Keene DR, Brunken WJ, Burgeson RE. A novel member of the netrin family, β-netrin, shares homology with the β chain of laminin: identification, expression, and functional characterization. J Cell Biol. 2000;151:221–234. doi: 10.1083/jcb.151.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan Y, Leszczynska M, Konstantinovsky S, Trope CG, Reich R, Davidson B. Netrin-4 is upregulated in breast carcinoma effusions compared to corresponding solid tumors. Diagn Cytopathol. 2011;39:562–566. doi: 10.1002/dc.21424. [DOI] [PubMed] [Google Scholar]
- 8.Latil A, Chene L, Cochant-Priollet B, Mangin P, Fournier G, Berthon P, Cussenot O. Quantification of expression of netrins, slits and their receptors in human prostate tumors. Int J Cancer. 2003;103:306–315. doi: 10.1002/ijc.10821. [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Yan Q, Wang Y, Dong X. NTN4 is associated with breast cancer metastasis via regulation of EMT-related biomarkers. Oncol Rep. 2017;37:449–457. doi: 10.3892/or.2016.5239. [DOI] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Wu Q, Luo G, Yang Z, Zhu F, An Y, Shi Y, Fan D. miR-17-5p promotes proliferation by targeting SOCS6 in gastric cancer cells. FEBS Lett. 2014;588:2055–2062. doi: 10.1016/j.febslet.2014.04.036. [DOI] [PubMed] [Google Scholar]
- 12.Bao S, Wang X, Wang Z, Yang J, Liu F, Yin C. MicroRNA-30 mediates cell invasion and metastasis in breast cancer. Biochem Cell Biol. 2018;96:825–831. doi: 10.1139/bcb-2018-0032. [DOI] [PubMed] [Google Scholar]
- 13.Ren Z, Yang T, Ding J, Liu W, Meng X, Zhang P, Liu K, Wang P. MiR-520d-3p antitumor activity in human breast cancer via post-transcriptional regulation of spindle and kinetochore associated 2 expression. Am J Transl Res. 2018;10:1097–1108. [PMC free article] [PubMed] [Google Scholar]
- 14.You F, Luan H, Sun D, Cui T, Ding P, Tang H, Sun D. miRNA-106a promotes breast cancer cell proliferation, clonogenicity, migration, and invasion through inhibiting apoptosis and chemosensitivity. DNA Cell Biol. 2018;38:198–207. doi: 10.1089/dna.2018.4282. [DOI] [PubMed] [Google Scholar]
- 15.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang F, Li Y, Xu L, Zhu Y, Gao H, Zhen L, Fang L. miR-17 as a diagnostic biomarker regulates cell proliferation in breast cancer. Onco Targets Ther. 2017;10:543–550. doi: 10.2147/OTT.S127723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Li J, Dai L, Zheng J, Yi Z, Chen L. MiR-17-5p may serve as a novel predictor for breast cancer recurrence. Cancer Biomark. 2018;22:721–726. doi: 10.3233/CBM-181228. [DOI] [PubMed] [Google Scholar]
- 18.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGuire A, Brown JA, Kerin MJ. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34:145–155. doi: 10.1007/s10555-015-9551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Zheng F, Yu G, Yin Y, Lu Q. miR-196a targets netrin 4 and regulates cell proliferation and migration of cervical cancer cells. Biochem Biophys Res Commun. 2013;440:582–588. doi: 10.1016/j.bbrc.2013.09.142. [DOI] [PubMed] [Google Scholar]
- 22.Ryu JM, Choi HJ, Kim I, Lee SK, Yu J, Kim JE, Kang BI, Lee JE, Nam SJ, Kim SW. Only estrogen receptor “positive” is not enough to predict the prognosis of breast cancer. Breast Cancer Res Treat. 2018;172:627–636. doi: 10.1007/s10549-018-4948-y. [DOI] [PubMed] [Google Scholar]