Abstract
The role of microRNA-107 (miR-107) as a tumor suppressor has been explored in different types of human cancer. However, the expression and role of miR-107 in oral squamous cell carcinoma (OSCC) remains elusive. Here, the expression of miR-107 in OSCC cell lines was explored by reverse transcription-quantitative polymerase chain reaction. Online prediction algorithms and luciferase activity reporter assay were conducted to validate the targets of miR-107. Cell counting kit-8 assay and wound-healing assay were performed to analyze the functions of miR-107 on OSCC cells. These results indicated that miR-107 had reduced expression in OSCC cells. Overexpression of miR-107 inhibits OSCC cell proliferation and migration. It was found that miR-107 directly targets TP53 regulated inhibitor of apoptosis 1 (TRIAP1) to regulate gene expression. Together, these results demonstrated that miR-107 also plays a tumor suppressive role by inhibiting proliferation and migration of OSCC cells by targeting TRIAP1. Our finding provides novel insights into the mechanism of OSCC progression.
Keywords: miR-107, TRIAP1, oral squamous cell carcinoma, tumor suppressive miRNA, cell behavior
Introduction
Oral squamous cell carcinoma (OSCC) has a high incidence worldwide and accounts for approximately 95% of all oral cancer [1]. The risk factors for OSCC include tobacco exposure, alcohol usage, and the infection of human papilloma virus [2,3]. Despite improvements in treatment measures for OSCC, the overall survival of OSCC remains undesirable [4]. Therefore, it is urgent to investigate the mechanisms related to OSCC progression.
The initiation and progression of cancer is a complex process and is characterized by abnormal cell status and expression of many cancer-related genes [5]. Previously, most attention has been put into the abnormal expression of protein-coding genes and thus generated multiple novel anti-cancer treatment methods [6]. Over the past decades, attention has been focused on non-coding RNAs (ncRNAs) including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), small nucleolar RNAs (snoRNAs) as approximately 66% of all human genes are regulated by non-coding RNAs [7-9]. These ncRNAs were found to serve crucial roles in regulating cellular progression [7,8].
miRNAs acts as a gene modulator mainly through binding the 3’-untranslated region (3’-UTR) [10]. miR-107 is reported to be abnormally expressed in human cancers [11-13]. It was found that miR-107 mimic transfection repressed non-small cell lung cancer cell (NSCLC) proliferation through targeting inhibitor of nuclear factor kappa B kinase subunit gamma [11]. Importantly, the overexpression of miR-107 could sensitive NSCLC cells to parthenolide [11]. miR-107 was revealed to be decreased in gastric cancer [12]. Moreover, miR-107 overexpression was shown to inhibit cell proliferation and metastasis by targeting BDNF through the PI3K/AKT pathway [12]. These results indicated a tumor suppressive role of miR-107 in NSCLC and gastric cancer. On the contrary, miR-107 expression was revealed to be overexpressed in pancreatic ductal adenocarcinoma and associated with poor clinicopathologic parameters and prognosis, suggesting the oncogenic role of miR-107 [13]. In OSCC, many miRNAs have been identified as biomarkers for cancer diagnosis, treatment, or prognosis prediction [14-17]. However, there is no study to date investigating the role of miR-107 in OSCC.
In this study, we analyzed the expression level of miR-107 in OSCC cell lines. We analyzed the connection of miR-107 and TP53 regulated inhibitor of apoptosis 1 (TRIAP1) through bioinformatic analysis and western blot. Moreover, the biological functions of miR-107 and TRIAP1 in OSCC progression were explored in OSCC cells.
Materials and methods
Cell line and culture
Human normal oral epithelial keratinocytes (hNOK) and OSCC cell lines (CAL-27 and OSC-4) were obtained from the Cell Bank of Chinese Academy of Science (Shanghai, China). These cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen) in a 37°C humidified incubator containing 5% CO2.
Transfection protocol
miR-107 mimic and miR-107 negative control (miR-NC) were purchased from Genechem (Shanghai, China). TRIAP1 expression vector (pTRIAP1) and empty vector (pcDNA3.1) were purchased from GenScript (Nanjing, China). Lipofectamine 2000 (Invitrogen) was employed for miRNA or expression vector transfection following the manufacturer’s protocol.
Bioinformatic analysis
The TargetScan and miRDB online prediction algorithms were utilized for miR-107 target prediction. Among all the predicted targets, TRIAP1 was selected for further investigation.
Dual-luciferase activity reporter assay
Wild-type 3’-UTR of TRIAP1 (TRIAP1-WT) and mutated 3’-UTR of TRIAP1 (TRIAP1-MUT) were cloned into pMIR-REPORT (Promega, Madison, WI, USA). Cells were seeded in 6-well plates and co-transfected with TRIAP1-WT or TRIAP1-MUT and miR-107 mimic and miR-NC using Lipofectamine 2000. Dual Luciferase-Reporter System (Promega) was employed to measure relative luciferase activity and Renilla activity was used as internal control.
RNA extraction and quantitative real-time PCR (qRT-PCR)
TRIzol (Invitrogen) reagent was used to extract total RNA according to the manufacturer’s instructions. First-strand complementary DNA (cDNA) was synthesized by TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). qRT-PCR reaction was conducted using SYBR Green Mix (Takara, Dalian, China) on the ABI 7500 system (Applied Biosystems) with the following procedure: 1 cycle of 95°C for 10 min; 40 cycles of 95°C for 1 min, 63°C for 2 min, 72°C for 1 min; final 1 cycle 72°C for 10 min. The primer sequences were as follows: miR-107 forward, 5’-AGCAGCATTGTACAGGGCTATCA-3’, reverse, 5’-GCGAGCACAGAATTAATACGAC-3’; U6 snRNA forward, 5’-AGAGCCTGTGGTGTCCG-3’, reverse, 5’-CATCTTCAAAGCACTTCCCT-3’. U6 snRNA was used as internal control to normalize the levels of miR-107.
Western blot
Protease inhibitor in combination with RIPA lysis buffer (Beyotime, Haimen, Jiangsu, China) was employed to isolate total proteins from cells. Protein concentration was measured by BCA Protein Assay Kit (Beyotime). Protein samples were isolated in 10% sodium dodecyl sulfate-polyacrylamide gels and transferred to PVDF membranes (Beyotime), then membranes were blocked with fat-free milk. All membranes were incubated for 1 h with a buffer at room temperature. Subsequently, primary antibodies utilized to incubate with the membranes (anti-TRIAP1: cat. LS-C346398-50; LifeSpan Biosciences, Inc., Seattle, WA, USA, anti-GAPDH: cat. ab181602; Abcam, Cambridge, MA, USA) at 4°C for overnight. The membranes were further incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (cat. ab6721; Abcam) at room temperature for 2 h. Bands were visualized using BeyoECL kit (Beyotime).
Cell viability assay
Cell Counting Kit-8 (CCK-8) assay (Beyotime) was used to assess cell proliferation based on the manufacturer’s protocols. Briefly, 3,000 cells were seeded into 96-well plates and incubated to the indicated time before the addition of 10 μl CCK-8 reagent. After further incubation for 2 h, the absorbance was measured at 450 nm with a microplate reader (Thermo Fisher Scientific, Inc.).
Wound-healing assay
Wound healing assay was used to assess cell migration. A 10 μl pipette tip was used to create a wound at confluent cells, and then incubated in the above mentioned medium containing Mitomycin C (5 μg/ml). Distance migrated was quantitated by taking pictures at 0 and 24 h.
Statistical analysis
Data are presented as the mean ± standard deviation (SD) after analyzing by SPSS 19 (SPSS, Inc., Chicago, IL, USA). Differences between two groups were analyzed using Student’s t-test, while analysis of variance and Tukey post-hoc test was utilized for comparisons of more than two groups. P-values less than 0.05 were considered significant.
Results
miR-107 expression was reduced in OSCC
The expression levels of miR-107 in OSCC cell lines were analyzed by qRT-PCR. Results revealed that miR-107 expression was significantly reduced in OSCC cell lines (CAL-27 and OSC-4) compared with the hNOK cell line (Figure 1).
Figure 1.
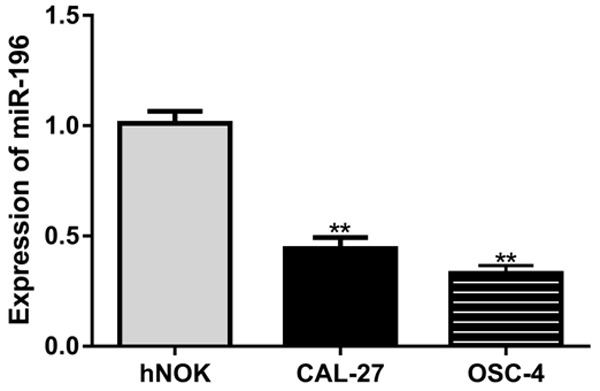
miR-107 expression was significantly reduced in OSCC cell lines (CAL-27 and OSC-4) compared with the hNOK cell line. miR-107: microRNA-107; OSCC: oral squamous cell carcinoma.
miR-107 regulates OSCC cell proliferation and migration
To investigate the influence of miR-107 on OSCC, cells were transfected with miR-107 mimic and miR-NC. qRT-PCR results revealed that miR-107 mimic transfection significantly enhanced the levels of miR-107 in OSCC cells (Figure 2A). Furthermore, CCK-8 assay revealed that the proliferation of OSCC cells was impaired in the miR-107 mimic group (Figure 2B). In a wound-healing assay, the migration of OSCC cell lines was inhibited by miR-107 mimic as compared with miR-NC (Figure 2C).
Figure 2.
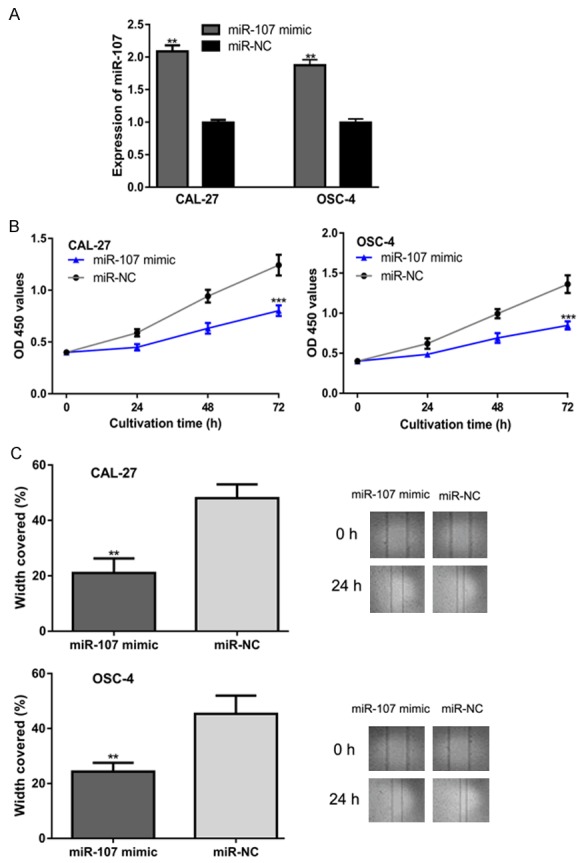
miR-107 overexpression inhibits OSCC cell proliferation and migration. (A) miR-107 expression, (B) Cell proliferation, and (C) Cell migration in OSCC cells transfected with synthetic miRNAs. miR-107: microRNA-107; OSCC: oral squamous cell carcinoma; miR-NC: negative control miRNA.
TRIAP1 was a direct target of miR-107
A putative binding site in the 3’-UTR of TRIAP1 was predicted by TargetScan and miRDB (Figure 3A). To confirm the interaction between miR-107 and 3’-UTR of TRIAP1, luciferase activity reporter assay was conducted. It was found that miR-107 mimic transfection significantly inhibited the luciferase activity of cells transfected with TRIAP1-WT but not TRIAP1-MUT (Figure 3B). TRIAP1 expression in OSCC cells transfected with miR-107 mimic or miR-NC was further analyzed by western blot. It was found that TRIAP1 expression was reduced by miR-107 mimic (Figure 3C).
Figure 3.
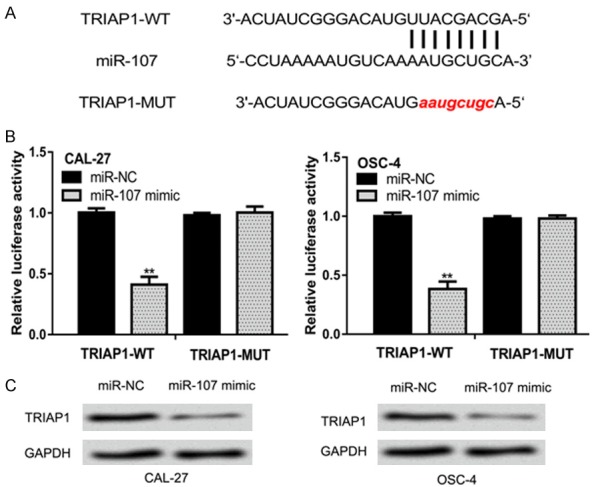
miR-107 regulates TRIAP1 expression by 3’-UTR binding. A. Binding region between miR-107 and the 3’-UTR of TRIAP1. B. Luciferase activity in cells transfected with luciferase reporter plasmids and synthetic miRNAs. C. TRIAP1 expression in cells transfected with synthetic miRNAs. miR-107: microRNA-107; OSCC: oral squamous cell carcinoma; miR-NC: negative control miRNA; UTR: untranslated region; WT: wild-type; MUT: mutant; TRIAP1: TP53 regulated inhibitor of apoptosis 1.
Co-transfection of TRIAP1 in cells with miR-107 mimic restores cell proliferation and migration
To further investigate whether the roles of miR-107 on OSCC cell proliferation and migration were mediated by targeting TRIAP1, overexpression vector for TRIAP1 was co-transfected with miR-107 mimic into the OSCC cells. Western blot showed pTRIAP1 transfection increased the levels of TRIAP1 and reversed the effect of miR-107 mimic on TRIAP1 expression (Figure 4A). CCK-8 assay showed the overexpression of TRIAP1 enhanced OSCC cell proliferation (Figure 4B). Wound-healing assay revealed that introduction of pTRIAP1 promoted cell migration (Figure 4C). Importantly, cell proliferation and migration abilities were downregulated by the miR-107 mimic but were restored after pTRIAP1 transfection (Figure 4B and 4C).
Figure 4.
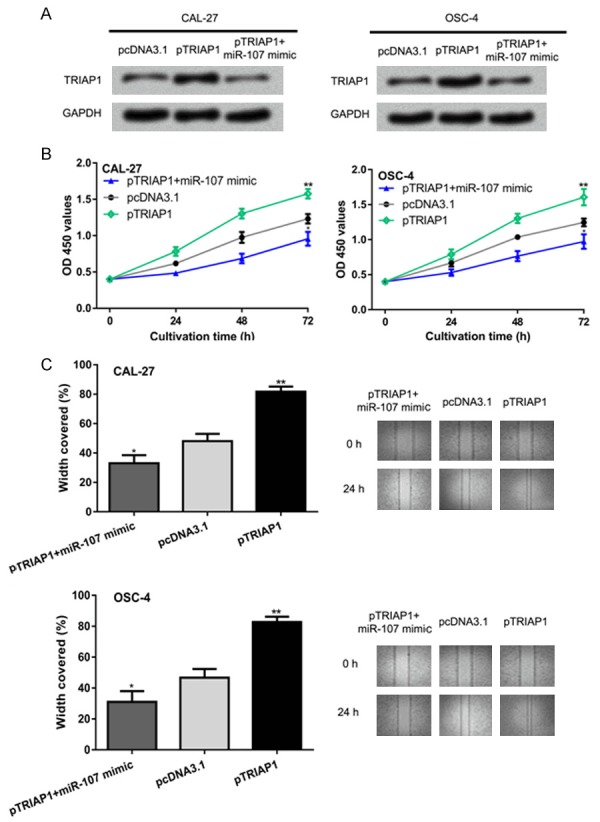
miR-107 regulates OSCC cell proliferation and migration by targeting TRIAP1. (A) TRIAP1 expression, (B) cell proliferation, and (C) cell migration in OSCC cells transfected with pTRIAP1, pcDNA3.1, or pTRIAP1 and miR-107 mimic. miR-107: microRNA-107; OSCC: oral squamous cell carcinoma; TRIAP1: TP53 regulated inhibitor of apoptosis 1.
Discussion
The progression of human cancer is due to the accumulated genetic and epigenetic changes, which can impair the function of signaling pathways [5]. miRNAs have been reported to be important regulators for OSCC carcinogenesis [14-17]. For instance, miRNA-375 was reported to inhibit growth and enhance radiosensitivity of OSCC cells by regulating the expression of insulin like growth factor 1 receptor [14]. Moreover, low miR-375 expression was correlated with poor survival of OSCC patients, indicating miR-375 may be a target for OSCC treatment [14]. miR-182-5p directly targeted CAMK2NA to promote OSCC cell growth [15]. In an animal model, it was found that injection of the miR-182-5p inhibitor-transfected OSCC cell lines in mice increased tumor size and weight [15]. In addition, miR-139-5p was reported to inhibit OSCC cell events by inhibiting HOXA9 expression [16]. Moreover, the overexpression of miR-133a-3p was able to inhibit OSCC cell migration and proliferation through targeting COL1A1 [17].
In this study, we demonstrated that miR-107 expression was markedly reduced in OSCC cell lines compared with the normal cell line. Previous studies showed that miR-107 was able to regulate multiple cancer cell behaviors [12,13]. We therefore investigated the functions of miR-107 on OSCC cell proliferation and migration through synthetic miRNA transfection. We found overexpression of miR-107 significantly inhibited OSCC cell proliferation and migration. These data revealed a tumor suppressive role of miR-107 in OSCC.
We further investigated the potential targets of miR-107 using bioinformatic tools and found TRIAP1 was the putative downstream target of miR-107. TRIAP1 is a conserved protein containing 76 amino acids and contributes to the reduction of cell death [18]. Moreover, it was found that TRIAP1 silencing induces late apoptosis through the APAF1/Caspase 9 pathway in a multiple myeloma cell line [19]. In ovarian cancer, TRIAP1 expression was revealed to be regulated by miR-18a to inhibit cell proliferation [20]. In this work, TRIAP1 was verified as a direct target of miR-107 through bioinformatic analysis and western blot analysis. In addition, we verified that overexpression of TRIAP1 counteracted the effects of miR-107 on OSCC cell proliferation and migration. The limitation of this study was that we only investigated the function of miR-107 in OSCC cells. Therefore, further studies should be conducted to verify the effects of miR-107 in animal model and explore the clinical significance of miR-107 in humans.
Taken together, our study revealed miR-107 expression was downregulated in OSCC cell lines, and the overexpression of mR-107 inhibited cell proliferation and migration in part through regulating the expression of TRIAP1. To the best of knowledge, this is the first report to demonstrate the tumor suppressive role of miR-107 through targeting TRIAP1. Our study will advance our understanding of mechanisms underlying the tumorigenesis of OSCC.
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Moore SR, Johnson NW, Pierce AM, Wilson DF. The epidemiology of mouth cancer: a review of global incidence. Oral Dis. 2000;6:65–74. doi: 10.1111/j.1601-0825.2000.tb00104.x. [DOI] [PubMed] [Google Scholar]
- 3.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, Shah KV, Sidransky D. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 4.Fukumoto I, Hanazawa T, Kinoshita T, Kikkawa N, Koshizuka K, Goto Y, Nishikawa R, Chiyomaru T, Enokida H, Nakagawa M, Okamoto Y, Seki N. MicroRNA expression signature of oral squamous cell carcinoma: functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br J Cancer. 2015;112:891–900. doi: 10.1038/bjc.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Tsimberidou AM. Targeted therapy in cancer. Cancer Chemother Pharmacol. 2015;76:1113–1132. doi: 10.1007/s00280-015-2861-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Momen-Heravi F, Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134–143. doi: 10.1016/j.cellsig.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474:4219–4251. doi: 10.1042/BCJ20170079. [DOI] [PubMed] [Google Scholar]
- 9.Kersey PJ, Allen JE, Allot A, Barba M, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Grabmueller C, Kumar N, Liu Z, Maurel T, Moore B, McDowall MD, Maheswari U, Naamati G, Newman V, Ong CK, Paulini M, Pedro H, Perry E, Russell M, Sparrow H, Tapanari E, Taylor K, Vullo A, Williams G, Zadissia A, Olson A, Stein J, Wei S, Tello-Ruiz M, Ware D, Luciani A, Potter S, Finn RD, Urban M, Hammond-Kosack KE, Bolser DM, De Silva N, Howe KL, Langridge N, Maslen G, Staines DM, Yates A. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018;46:D802–D808. doi: 10.1093/nar/gkx1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Moeng S, Seo HA, Hwang CY, Cipolla GA, Lee DJ, Kuh HJ, Park JK. MicroRNA-107 targets IKBKG and sensitizes A549 cells to parthenolide. Anticancer Res. 2018;38:6309–6316. doi: 10.21873/anticanres.12987. [DOI] [PubMed] [Google Scholar]
- 12.Cheng F, Yang Z, Huang F, Yin L, Yan G, Gong G. microRNA-107 inhibits gastric cancer cell proliferation and metastasis by targeting PI3K/AKT pathway. Microb Pathog. 2018;121:110–114. doi: 10.1016/j.micpath.2018.04.060. [DOI] [PubMed] [Google Scholar]
- 13.Xiong J, Wang D, Wei A, Lu H, Tan C, Li A, Tang J, Wang Y, He S, Liu X, Hu W. Deregulated expression of miR-107 inhibits metastasis of PDAC through inhibition PI3K/Akt signaling via caveolin-1 and PTEN. Exp Cell Res. 2017;361:316–323. doi: 10.1016/j.yexcr.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B, Li Y, Hou D, Shi Q, Yang S, Li Q. MicroRNA-375 inhibits growth and enhances radiosensitivity in oral squamous cell carcinoma by targeting insulin like growth factor 1 receptor. Cell Physiol Biochem. 2017;42:2105–2117. doi: 10.1159/000479913. [DOI] [PubMed] [Google Scholar]
- 15.Li N, Nan CC, Zhong XY, Weng JQ, Fan HD, Sun HP, Tang S, Shi L, Huang SX. miR-182-5p promotes growth in oral squamous cell carcinoma by inhibiting CAMK2N1. Cell Physiol Biochem. 2018;49:1329–1341. doi: 10.1159/000493411. [DOI] [PubMed] [Google Scholar]
- 16.He B, Lin X, Tian F, Yu W, Qiao B. MiR-133a-3p inhibits oral squamous cell carcinoma (OSCC) proliferation and invasion by suppressing COL1A1. J Cell Biochem. 2018;119:338–346. doi: 10.1002/jcb.26182. [DOI] [PubMed] [Google Scholar]
- 17.Wang K, Jin J, Ma T, Zhai H. MiR-139-5p inhibits the tumorigenesis and progression of oral squamous carcinoma cells by targeting HOXA9. J Cell Mol Med. 2017;21:3730–3740. doi: 10.1111/jcmm.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 19.Fook-Alves VL, de Oliveira MB, Zanatta DB, Strauss BE, Colleoni GW. TP53 Regulated Inhibitor of Apoptosis 1 (TRIAP1) stable silencing increases late apoptosis by upregulation of caspase 9 and APAF1 in RPMI8226 multiple myeloma cell line. Biochim Biophys Acta. 2016;1862:1105–1110. doi: 10.1016/j.bbadis.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Liu P, Qi X, Bian C, Yang F, Lin X, Zhou S, Xie C, Zhao X, Yi T. MicroRNA-18a inhibits ovarian cancer growth via directly targeting TRIAP1 and IPMK. Oncol Lett. 2017;13:4039–4046. doi: 10.3892/ol.2017.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]


