Abstract
Alterations in colonic mucus secretion are linked to the induction and maintenance of inflammation during inflammatory bowel disease (IBD) and its progression to colorectal cancer (CRC). MUC1, a multifunctional glycoprotein, is the best studied cell surface mucin in mouse models of IBD and CRC. However, little information on MUC1 expression and localization in different types of pathologic human colon mucosa is available. In this work, expression and subcellular localization of MUC1 in different types of diseased human colon mucosa from a cohort of Tunisian patients is analyzed and correlated with the type of disorder. Colon tissue samples were obtained from 39 cases of CRC and 18 cases of IBD. 13 cases of normal adjacent colon mucosa tissues served as controls. Biopsies were subjected to immunohistochemical analysis of MUC1 expression. Signals were quantified densitometrically and characterized with regard to tissue and intracellular distribution. Results were then correlated with the different types of colon disorder. Immunohistochemical investigation of MUC1 in a cohort of inflammatory bowel diseases and colorectal cancer showed a significant divergence in the expression of MUC1 in terms intensity (18.96% ± 0.55 vs 27.26% ± 1.24 respectively; P=0.005) and localization between the two types of lesions (30.76% vs 70.96% respectively; P=0.0199). Our findings show divergent characteristic patterns for MUC1 expression and localization in different types of pathologic alterations of the colon mucosa. These results are of potential diagnostic and predictive clinical value.
Keywords: Mucin-1, MUC1, inflammatory bowel disease, IBD, colorectal cancer, CRC
Introduction
Both benign and malignant disorders with severe clinical implications and overlapping symptoms frequently affect the colonic mucosa. Importantly, inflammatory diseases of the colon may favor the development of cancer. It is, thus, important to differentiate between categories of colonic malady at a preferably early stage to optimize appropriate treatment.
The term inflammatory bowel disease (IBD) includes mainly two clinically defined entities. Crohn’s disease (CD) and ulcerative colitis (UC) refer to chronic remittent or progressive inflammatory disorders that concern the intestinal tract [1]. The risk of IBD is influenced by life style; prevalence is considerably higher e.g in Europe compared to North Africa. As a consequence of changing life style in developing regions, IBD cases are expected to progressively rise in the near future [2,3]. Notably, patients with IBD have a clearly increased risk to develop colorectal carcinoma (CRC), a cancer with a high rate of mortality [4,5].
Until now, the accepted etiology of IBD combines environmental and genetic factors which contribute to the loss of intestinal integrity and an excessive immune response to the commensal microbiota [6]. Proteins of the mucin family, which form the major component of the protective mucus layer, constitute the only physical and biochemical barrier between microbiota and immune cells. Both secreted and transmembrane mucins are closely involved in the maintenance of intestinal homeostasis [7,8]. Formation of leaks in the epithelial barrier structural and/or quantity alterations of mucin plays key roles in the initiation and progression of IBD and also in its progression to cancer [9,10]. Among mucins, MUC1 has attracted particular interest as a potential target in diagnostics and future immunotherapy in both IBD and CRC [11]. Full length MUC1 is a large glycosylated type I transmembrane protein 200-500 Kda in size expressed on glandular or luminal epithelial cells of serval organs including colonic mucosa [12].
The extracellular domain contains a variable number of tandem repeat regions (VNTR), composed of 20-200 tandem repeat (TR) units, each 20 amino acids in length and rich in prolines and O-glycosylated serines and threonines. It undergoes autoproteolytic cleavage resulting in the generation of two subunits which remain non-covalently linked [13]. On normal epithelia, MUC1 is heavily glycosylated and expressed at low levels. In contrast, it is strongly over-expressed and markedly hypo-glycosylated in various epithelial cancers such as carcinomas of the pancreas, breast, prostate and bladder [14]. Importantly, these features of MUC1 form have been correlated with poor prognosis in these cancers [15].
Hence, we analyzed MUC1 expression and localization in different types of lesioned mucosa: in a cohort of Tunisian patients affected by IBD or CRC with the aim of associating MUC1 abundance and distribution with disease characteristics.
Material and methods
Patients and tissues
Seventy paraffin-embedded tissues from colon-diseased patients collected by the department of pathology of the Institute of Cancer Salah Azaeiz of Tunis were included into the study. The patient cohort comprised 18 cases of IBD (mean age 31; 8 males, 10 females), 39 cases of CRC (mean age 54; 17 males, 22 females) and 13 (control) cases without pathologic findings. Histomorphological assessment of samples was performed by certified pathologists on the basis of corresponding hematoxylin and eosin-stained slides.
All clinicopathologic and personal information was anonymized and detached from any personal identifiers. The study was conducted in concordance with local ethical regulations.
Antibodies
Rabbit polyclonal antibody (H-295) (Santa Cruz, Dallas, TX, USA), recognizing amino acid residues 961-1255 at the C-terminus of human MUC1 was employed for immunohistochemical detection of MUC1. Anti-rabbit Polymer-HRP-IgG provided with the NovoLinkTMPolymer detection system (Novocastra Laboratories Ltd, Newcastle, UK) was used as secondary antibody.
Immunohistochemistry
Hematoxylin-eosin staining and immunohistochemical analysis was performed as described [16]. Briefly, tissues were fixed for 24 h, dehydrated and embedded in paraffin before 4 μm sections were processed employing the NovoLinkTM Polymer Detection Systems (Novocastra Laboratories Ltd.) method. Following deparaffination and rehydration, antigens were retrieved by incubation of slides in 0.1 M citric acid. Endogenous peroxidase activity was then quenched by incubation with Peroxidase Block (3-4% (v/v) hydrogen peroxide) and non-specific binding of primary antibody was prevented by incubation of sections with Protein Block (0.4% Casein in phosphate-buffered saline, with stabilizers, surfactant, and 0.2% Bronidox L as a preservative) (Novocastra).
Tissue sections were incubated with anti-MUC1 serum overnight at 4°C at a dilution of 1:400 followed by incubations with Post Primary Block (Rabbit anti mouse IgG (<10 μg/ml) in 10% (v/v) animal serum in Tris-buffered saline/0.09% ProClinTM 950 (Novocastra Laboratories Ltd.) for 30 min, NovoLinkTM Polymer (Anti-rabbit Poly-HRP-IgG (<25 μg/mL) containing 10% (v/v) animal serum in Tris-buffered saline/0.09% ProClinTM 950 (Novocastra Laboratories Ltd.) for 30 min) and 3, 3’-diaminobenzidine (DAB) working solution (Novocastra Laboratories Ltd.) for 5 min to assess peroxidase activity.
Slides were counterstained with hematoxylin and visualized using an Olympus CX41 microscope equipped with 10 ×, 40 × and 100 × objective. Colon tissues of each type were incubated with 50 μl blocking peptides (Santa Cruz Biotechnology, Dallas, TX, USA) in place of primary antibody for control purposes.
For quantification of antigen abundance, staining intensities (optical densities) within 6 randomly chosen surface areas (microscopic fields, 40 × objective) per slide were determined using the color deconvolution plugin algorithm in ImageJ FIJI (created by J. W. Rasband at the US National Institutes of Health). From the average values for each tissue type, the means (± SEM) were calculated [16,17].
Statistical analysis
The statistical significance was assessed by Fisher exactitude, T-test and the one-way ANOVA test (post hoc: Tukey’s multiple compress tests) using the computer program GraphPad PRISMA 7.0. For all statistical tests, P<0.05 was considered significant.
Fisher exact test was used to test the significance of the proportion of each variable, T-test to compare means of 2 groups, and Tukey’s multiple compress to compare the means of more than 2 groups.
Results
MUC1 expression in diseased colon mucosa
We examined expression level and localization of MUC1 in clinical specimens from patients with different types of pathological alterations within the colon mucosa by immunohistochemistry. The studied patient cohort (n=70) consisted of 13 (healthy) controls, 18 cases of IBD (among them 8 samples classified as CD, 8 as UC and 2 as unspecific inflammation) and 39 cases of CRC. The samples were subjected to qualitative and quantitative assessment of MUC1 expression by immunohistochemistry.
Figure 1 shows a typical example of anti-MUC1 stained sections from each of the three groups, visualized at two different magnifications. Strong or very strong apical and luminal staining was observed in the glandular epithelial cells of both inflamed and tumorous mucosa, while in normal mucosa immunoreactivity was very small. IBD and CRC samples showed different subcellular distribution of MUC1. While in IBD samples, immunostaining preferentially localized to apical membranes, in CRC tissues MUC1 staining mostly showed a diffuse cytoplasmic localization.
Figure 1.
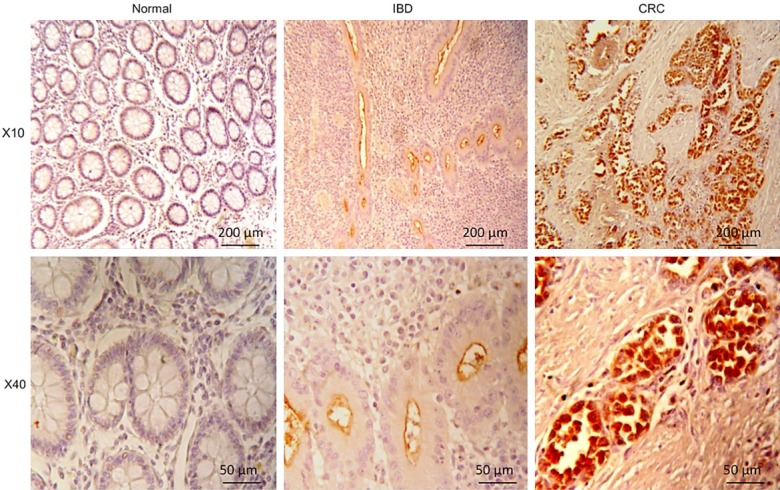
Immunohistochemical staining for MUC1 in normal and lesional human colon mucosa. Representative thin sections from healthy individuals (“normal”) and IBD and CRC patients were stained with anti-MUC1 serum and probed with peroxidase-coupled secondary antibody as described in Material and Methods. For visualization, a × 10 objective (top panel) and a × 40 (bottom panel) were employed. Scale bars represent 200 μm or 50 μm, respectively.
Correlation of MUC1 expression with different types of colon mucosa disorder
We performed a quantitative analysis of MUC1 immunochemical reactivity in relation to the type of colon disease. Out of each studied groups of patient samples (normal, IBD and CRC), six sections were selected at random. In each section, the staining intensity was determined in six microscopic fields. MUC1 expression after microscopic examination (defined as “reactive”) in normal, inflamed, and tumorous tissues were 23% (3/13), 72% (13/18) and 79% (31/39) respectively. The correlation of MUC1 expression with colon musosa disease was highly significant (P=0.0004). We then determined optical densities of immunostain in the three biopsy groups and found mean MUC1 expression markedly increased in inflamed compared to normal colon tissue (18.96% ± 0.55 vs 1.72% ± 1.58). The highest mean level of MUC1 expression was observed in the CRC specimens (27.26% ± 1.24). The rise in MUC1 expression in relation to healthy tissue was highly significant for both types of disease P=0.0025 and P<0.0001, respectively). Also, the distinction between IBD and CRC in terms of MUC1 expression was significant (P=0.005) (Figure 2A).
Figure 2.
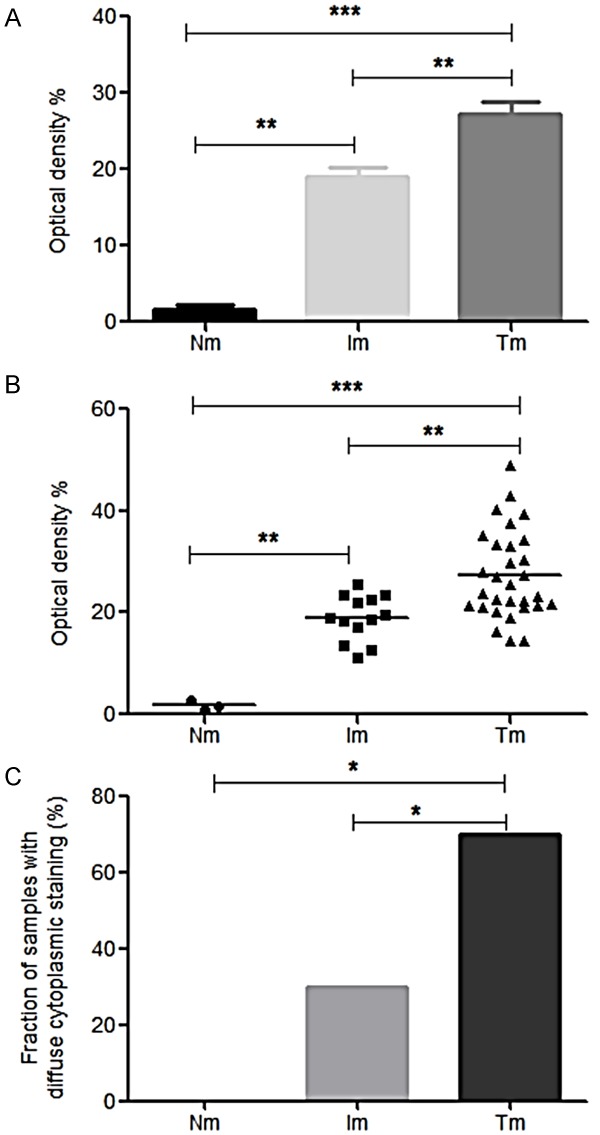
Quantitative assessment of MUC1 expression in healthy and pathologic colon mucosa. (A) Quantification of MUC1 expression in normal (“Nm”), IBD (“Im”) and tumorous CRC (“Tm”) biopsies by determination of mean optical intensity. From each biopsy of the respective groups, staining intensities (optical densities) for 6 arbitrarily selected microscopic fields, (× 40 objective) were determined as detailed in the Material and Methods section. Error bars represent standard deviations (± SEM). Significance was determined by the Tukey’s multiple comparisons test (GraphPad PRISMA 7.0) computer program) **: P ≤ 0.01, ****: P ≤ 0.0001. Average optical densities were included only for samples,positive here mean presenting immunoreactivity under microscope (B) Distribution of MUC1 immunostaining intensity within the investigated groups of normal and diseased colon mucosa specimens (designation of groups, methodology and statistics as in (A)). Bars represent the respective median values. (C) Analysis of subcellular distribution of MUC1 in the groups of colon mucosa specimens under study. The percentage of staining in the cytoplasmic compartment was determined for all specimens investigated by microscopic estimation and expressed as percentage of total cells. Significance was determined by Fisher exactitude test, *: P ≤ 0.05.
The distribution of individual MUC1 expression levels within the IBD group was relatively homogeneous (median 18.82), whereas the CRC group showed a far more heterogeneous distribution (median 24.60). The difference between the two groups was significant (P=0.005) (Figure 2B).
Next we further characterized the subcellular distribution of MUC1 expression and determined the percentage of cytoplasmic stain in all three groups under investigation. The percentage of cytoplasmic MUC1 abundance was 30.76% within the IBD group and 70.96% within the CRC group. This difference between the two altered colon mucosa groups was significant (P=0.0199) (Figure 2C).
Correlation of MUC1 expression with the tumor grade
The finding that CRC samples showed a particular heterogeneity with regard to MUC1 expression prompted us to study in how far the level of MUC1 expression was associated with cancer characteristics. The 39 CRC tissues included within the study were categorized with regard to clinicopathologic parameters (Figure 3A). For most of the parameters, the relatively small number of samples combined with the dominance of individual subgroups did not allow for a meaningful correlation of MUC1 expression with cancer specifications. However, consideration of the tumor grade in connection with MUC1 expression yielded a significant correlation: Comparison of low grade (G1) with high grade G2/G3 tumors showed significantly stronger MUC1 expression in high grade CRC tissue (31.16% ± 2.9 vs 24.28% ± 1.37; P=0.0433) (Figure 3B).
Figure 3.
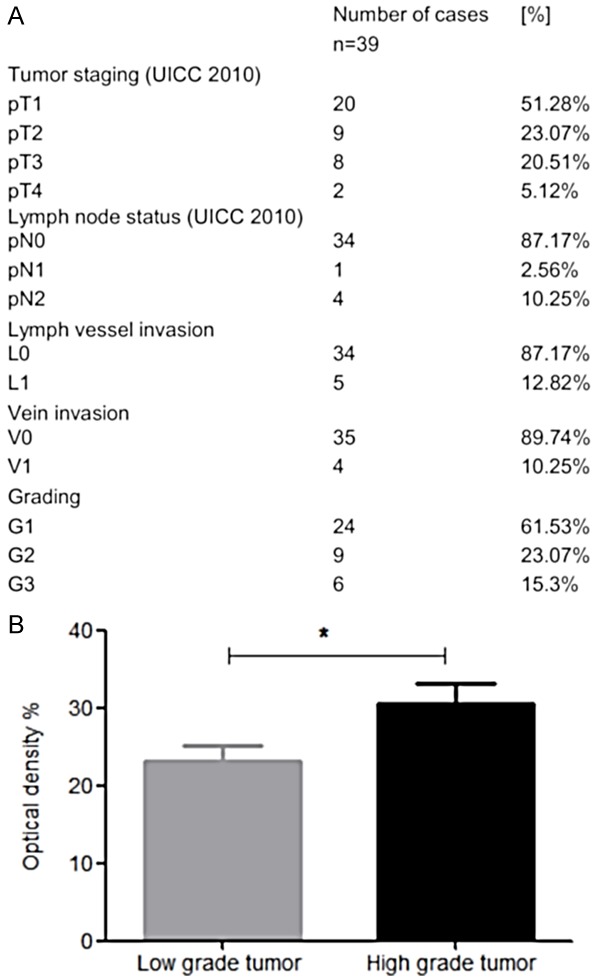
Association of MUC1 expression levels with CRC tumor grade. (A) Clinicopathologic parameters of CRC samples investigated in this study. (B) Comparison of MUC1 expression intensity in biopsies obtained from low grade and high grade CRC tumors. “Low grade tumor” refers to the G1 cases (n=24), “High grade tumor” to the combined G2 and G3 cases (n=15) from (A). Acquisition and presentation of data and statistics is as in Figure 2A; *: P ≤ 0.05.
Discussion
The finding that expression levels and subcellular distribution of MUC1 are significantly distinct between colon mucosa from healthy individuals and from IBD and CRC patients, respectively supports the notion of a crucial role of MUC1 in inflammatory and cancerous conditions of the colon. Our estimation of MUC1 expression in CRC cases is in consistent with earlier reports. There are, however, also contradicting publications [18,19]. This considerable heterogeneity in interpretation is perhaps attributable to the properties of antibodies used in different studies, i.e. their respective recognized structures of the antigen. MUC1 is a huge and dynamic molecule that exists in different variants, e.g. in hyper- or hypoglycosylated forms or as a full size or processed protein [12]. The used antibody in this study recognizes amino acid 961-1255 at the C terminus of human MUC1. This section of the MUC1-C subunit is involved in function.
Most of the previous studies on MUC1 function in cancer dealt with MUC1-N, since this subunit was described as overexpressed and aberrantly glycosylated in breast cancer. Based on these findings, early work on targeting MUC1 by drugs focused on MUC1-N. Only recently, MUC1-C was recognized as the oncogenic subunit and, thus, has been addressed as a potential target [20].
With regard to IBD, studies in various mouse models have been performed to characterize the involvement of MUC1 in the pathogenesis of these diseases. In IL-10(-/-) mice crossed to human MUC1-transgenic mice, MUC1 expression was shown to accelerate the disease and its progression to CRC [21]. Abnormal MUC1 expression in an IBD mouse model was also detected in pancreas where it may cause associated pancreatitis [22].
Regardless of these and other revelations, very few data describing the expression of MUC1 in cases of UC and CD are available. Our data show a significant upregulation in MUC1 expression in these disease states compared to healthy biopsies, which is in agreement with the two previous reports [23,24].
Our study shows that MUC1 expression is significantly stronger in CRC compared to IBD, supporting the concept of a gradual development from chronically inflamed tissue to cancer through mutual, cytokine and chemokine driven interactions of epithelial cells with the microenvironment [25,26]. Although the two types of microenvironment in IBD and CRC appear similar and involve activities of comparable effectors and mediators, qualitative differences obviously exist [27,28]. Several studies have revealed that the activities of pro-inflammatory cytokines such Interleukin (IL)-1, IL-6 and Tumor Necrosis Factor-(TNF-) α strongly influence the expression of MUC1 [29,30]. On the other hand, MUC1 was also shown to foster CRC development by inducing pro-inflammatory cytokines in a mouse disease model [31,32].
A further interesting aspect is the localization of MUC1. In this study, apical localization of was observed in almost all MUC1-positive samples including normal mucosa. In contrast, diffuse cytoplasmic expression was seen only in the two groups of lesional mucosa, indicating an association between cytoplasmic localization and cellular stress. Notably, the localization of MUC1 to the cytoplasmic compartment was found more related to malignant cells than to cells showing inflammatory alterations, hinting at a functional role of MUC1 in the course of tissue transformation to cancerous characteristics.
Lastly, our study revealed a significant association of the MUC1 expression level with the grade of CRC. An increase in MUC1 intensity was noticed in undifferentiated tumor cells which is consistent with some previous reports [33,34]. The differentiation grade of tumor is a major feature in classifying solid malignancies [35]. Potential future therapies may aim at convert undifferentiated cells back to “normal” [36].
Acknowledgements
This work was supported by funding from the German Federal Ministry of Education and Research (BMBF) and the Tunisian Ministry for Higher Education and Scientific Research (MESRS) cooperation.
Disclosure of conflict of interest
None.
References
- 1.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12:720–27. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 3.M’Koma AE. Inflammatory bowel disease: an expanding global health problem. Clin Med Insights Gastroenterol. 2013;6:33–47. doi: 10.4137/CGast.S12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorsteinsdottir S, Gudjonsson T, Nielsen OH, Vainer B, Seidelin JB. Pathogenesis and biomarkers of carcinogenesis in ulcerative colitis. Nat Rev Gastroenterol Hepatol. 2011;8:395–404. doi: 10.1038/nrgastro.2011.96. [DOI] [PubMed] [Google Scholar]
- 5.Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–14. e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 6.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goll R, Van Beelen Granlund A. Intestinal barrier homeostasis in inflammatory bowel disease. Scand J Gastroenterol. 2015;50:3–12. doi: 10.3109/00365521.2014.971425. [DOI] [PubMed] [Google Scholar]
- 8.Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 2010;1:254–68. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michielan A, D’Incà R. Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediators Inflamm. 2015;2015:628157. doi: 10.1155/2015/628157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng YH, Hasnain SZ, Florin THJ, McGuckin MA. Mucins in inflammatory bowel diseases and colorectal cancer. J Gastroenterol Hepatol. 2012;27:28–38. doi: 10.1111/j.1440-1746.2011.06909.x. [DOI] [PubMed] [Google Scholar]
- 11.Beatty PL, Narayanan S, Gariépy J, Ranganathan S, Finn OJ. Vaccine against MUC1 antigen expressed in inflammatory bowel disease and cancer lessens colonic inflammation and prevents progression to colitis-associated colon cancer. Cancer Prev Res (Phila) 2010;3:438–46. doi: 10.1158/1940-6207.CAPR-09-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332–42. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levitin F, Stern O, Weiss M, Gil-Henn C, Ziv R, Prokocimer Z, Smorodinsky NI, Rubinstein DB, Wreschner DH. The MUC1 SEA module is a self-cleaving domain. J Biol Chem. 2005;280:33374–86. doi: 10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- 14.Lau SK, Weiss LM, Chu PG. Differential Expression of MUC1, MUC2, and MUC5AC in Carcinomas of various sites: an immunohistochemical study. Am J Clin Pathol. 2004;122:61–9. doi: 10.1309/9R66-73QE-C06D-86Y4. [DOI] [PubMed] [Google Scholar]
- 15.Xu F, Liu F, Zhao H, An G, Feng G. Prognostic significance of mucin antigen MUC1 in various human epithelial cancers. Medicine (Baltimore) 2015;94:e2286–96. doi: 10.1097/MD.0000000000002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben Jemaa A, Bouraoui Y, Sallami S, Banasr A, Ben Rais N, Ouertani L, Nouira Y, Horchani A, Oueslati R. Co-expression and impact of prostate specific membrane antigen and prostate specific antigen in prostatic pathologies. J Exp Clin Cancer Res CR. 2010;29:171–80. doi: 10.1186/1756-9966-29-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2011;23:291–99. [PubMed] [Google Scholar]
- 18.Wang HS, Wang LH. The expression and significance of Gal-3 and MUC1 in colorectal cancer and colon cancer. Onco Targets Ther. 2015;8:1893–998. doi: 10.2147/OTT.S83502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldus SE, Mönig SP, Hanisch FG, Zirbes TK, Flucke U, Oelert S, Zilkens G, Madejczik B, Thiele J, Schneider PM, Hölscher AH, Dienes HP. Comparative evaluation of the prognostic value of MUC1, MUC2, sialyl-Lewis(a) and sialyl-Lewis(x) antigens in colorectal adenocarcinoma. Histopathology. 2002;40:440–9. doi: 10.1046/j.1365-2559.2002.01389.x. [DOI] [PubMed] [Google Scholar]
- 20.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–85. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beatty PL, Plevy SE, Sepulveda AR, Finn OJ. Cutting edge: transgenic expression of human MUC1 in IL-10-/- mice accelerates inflammatory bowel disease and progression to colon cancer. J Immunol. 2007;179:735–9. doi: 10.4049/jimmunol.179.2.735. [DOI] [PubMed] [Google Scholar]
- 22.Kadayakkara DK, Beatty PL, Turner MS, Janjic JM, Ahrens ET, Finn OJ. Inflammation driven by overexpression of the hypoglycosylated abnormal mucin 1 (MUC1) links inflammatory bowel disease and pancreatitis. Pancreas. 2010;39:510–5. doi: 10.1097/MPA.0b013e3181bd6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furr AE, Ranganathan S, Finn OJ. Aberrant expression of MUC1 mucin in pediatric inflammatory bowel disease. Pediatr Dev Pathol. 2010;13:24–31. doi: 10.2350/08-06-0479.1. [DOI] [PubMed] [Google Scholar]
- 24.Campbell BJ, Yu LG, Rhodes JM. Altered glycosylation in inflammatory bowel disease: a possible role in cancer development. Glycoconj J. 2001;18:851–8. doi: 10.1023/a:1022240107040. [DOI] [PubMed] [Google Scholar]
- 25.Fernandes JV, Cobucci RN, Jatobá CA, Fernandes TA, de Azevedo JW, de Araújo JM. The role of the mediators of inflammation in cancer development. Pathol Oncol Res. 2015;21:527–34. doi: 10.1007/s12253-015-9913-z. [DOI] [PubMed] [Google Scholar]
- 26.West NR, McCuaig S, Franchini F, Powrie F. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol. 2015;15:615–29. doi: 10.1038/nri3896. [DOI] [PubMed] [Google Scholar]
- 27.Rahat MA, Shakya J. Parallel aspects of the microenvironment in cancer and autoimmune disease. Mediators Inflamm. 2016;2016:4375120. doi: 10.1155/2016/4375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francescone R, Hou V, Grivennikov SI. Cytokines, IBD and colitis-associated cancer. Inflamm Bowel Dis. 2015;21:409–18. doi: 10.1097/MIB.0000000000000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albertsmeyer AC, Kakkassery V, Spurr-Michaud S, Beeks O, Gipson IK. Effect of pro-inflammatory mediators on membrane-associated mucins expressed by human ocular surface epithelial cells. Exp Eye Res. 2010;90:444–51. doi: 10.1016/j.exer.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li YY, Hsieh LL, Tang RP, Liao SK, Yeh KY. Interleukin-6 (IL-6) released by macrophages induces IL-6 secretion in the human colon cancer HT-29 cell line. Hum Immunol. 2009;70:151–58. doi: 10.1016/j.humimm.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Cascio S, Faylo J, Sciurba J, Xue J, Finn OJ. MUC1 promotes an inflammatory microenvironment aggravating colitis-associated tumorigenesis in mice through up-regulation of pro-inflammatory cytokines. J Immunol. 2016;196(Suppl 1):73–7. [Google Scholar]
- 32.Yu XW, Rong W, Xu FL, Xu GY, Sun YR, Feng MY. Expression and clinical significance of Mucin and E-cadherin in colorectal tumors. Ai Zheng. 2007;26:1204–10. [PubMed] [Google Scholar]
- 33.Kesari MV, Gaopande VL, Joshi AR, Babanagare SV, Gogate BP, Khadilkar AV. Immunohistochemical study of MUC1, MUC2 and MUC5AC in colorectal carcinoma and review of literature. Indian J Gastroenterol. 2015;34:63–7. doi: 10.1007/s12664-015-0534-y. [DOI] [PubMed] [Google Scholar]
- 34.Yu XW, Rong W, Xu FL, Xu GY, Sun YR, Feng MY. Expression and clinical significance of Mucin and E-cadherin in colorectal tumors. Ai Zheng. 2007;26:1204–10. [PubMed] [Google Scholar]
- 35.Jögi A, Vaapil M, Johansson M, Påhlman S. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Ups J Med Sci. 2012;117:217–24. doi: 10.3109/03009734.2012.659294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan M, Liu Q. Differentiation therapy: a promising strategy for cancer treatment. Chin J Cancer. 2016;35:1–3. doi: 10.1186/s40880-015-0059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]


