Abstract
Background: Staphylococcus aureus is a highly prevalent respiratory pathogen in cystic fibrosis (CF). It is unclear how this organism establishes chronic infections in CF airways. We hypothesized that S. aureus isolates from patients with CF would share common virulence properties that enable chronic infection. Methods: 77 S. aureus isolates were obtained from 45 de-identified patients with CF at the University of Iowa. We assessed isolates phenotypically and used genotyping assays to determine the presence or absence of 18 superantigens (SAgs). Results: We observed phenotypic diversity among S. aureus isolates from patients with CF. Genotypic analysis for SAgs revealed 79.8% of CF clinical isolates carried all six members of the enterotoxin gene cluster (EGC). MRSA and MSSA isolates had similar prevalence of SAgs. We additionally observed that EGC SAgs were prevalent in S. aureus isolated from two geographically distinct CF centers. Conclusions: S. aureus SAgs belonging to the EGC are highly prevalent in CF clinical isolates. The greater prevalence in these SAgs in CF airway specimens compared to skin isolates suggests that these toxins confer selective advantage in the CF airway.
Keywords: cystic fibrosis, Staphylococcus aureus, superantigen, enterotoxin gene cluster, MRSA
1. Introduction
Cystic fibrosis (CF) is a common lethal genetic disease, which results in chronic airway infections, irreversible bronchiectasis, and respiratory failure. Staphylococcus aureus is the most prevalent bacterial pathogen in children with CF [1], and is present in ≈70% of all individuals with CF in the United States. Although Pseudomonas aeruginosa is the predominant pathogen in older patients, S. aureus is the most common bacterial species in patients with CF under age 24 [2]. Unlike P. aeruginosa, which has multiple effective treatments to eradicate early infections and control chronic infections [3,4,5], S. aureus infections can be difficult to control with antibiotics [6]. CFTR modulator drugs may help prevent incident infections with S. aureus, but they are unlikely to eliminate chronic S. aureus infections [7,8]. Understanding how S. aureus infects and persists in the CF airway is critically important, as these infections may increase the risk of subsequent disease progression [9,10].
We hypothesized that S. aureus isolates in the CF airway would share common virulence properties. Some readily visible phenotypes such as hemolysis, pigmentation, and protease secretion could enable S. aureus to elude host defenses. People with CF are commonly treated with antibiotics; resistance to antibiotics may be occur under the selective pressure of antibiotic exposure. Another potential mechanism enabling S. aureus to establish chronic infection is the secretion of toxins that misdirect the immune response. S. aureus produces a large number of secreted toxins that may be critical for establishing infections [11]. These include 18 unique superantigens (SAgs), secreted toxins that bind both the T cell receptor and major histocompatibility complex molecules on antigen presenting cells [12,13].
Some SAgs are well known for their roles in acute infection. In the extreme example of toxic shock syndrome, the SAg TSST-1 cross-links T cells and antigen presenting cells, stimulating massive cytokine release and blocking the immune system from developing lasting immunity [12,13]. By contrast, the enterotoxin gene cluster (EGC), an element encoding six staphylococcal enterotoxin (SE) and SE-like SAgs G, I, M, N, O, and U, is generally associated with long term mucosal colonization. The EGC is present in between 50% to 70% of isolates from individuals with nasal carriage of S. aureus [14,15]. These EGC toxins can stimulate T cell proliferation [16], yet neutralizing antibody response to these toxins is surprisingly poor [17]. While these toxins have been associated with asymptomatic colonization, experimental studies in rabbits show that EGC SAgs may play crucial for infections such as endocarditis [18].
It is not clear what role SAgs play in CF respiratory infections. EGC SAgs are prevalent in clinical isolates of S. aureus; a recent European study showed that 57% of CF isolates harbored at least one gene belonging to the EGC [19]. This is similar to the prevalence of EGC in isolates from a cohort of patients with atopic dermatitis in the United States and Europe [20,21]. Our study had two goals: To determine whether these SAgs are as prevalent in CF isolates in the United States, and to determine if the superantigens were associated with methicillin resistance, which is common in the United States and has been linked to worse outcomes [10,22].
2. Materials and Methods
Ethics Statements: All bacterial isolates examined in this study were de-identified when they were supplied to the research team. The University of Iowa Institutional Review Board (IRB) approved specimen collection after obtaining informed consent under approval numbers 200311016 and 200803708.
Sources of Clinical Isolates: University of Iowa Cystic Fibrosis Biobank. 77 de-identified S. aureus clinical isolates were obtained from the University of Iowa Hospitals and Clinics clinical laboratory following CF clinic visits made between 12 December 2011 and 20 July 2012. Specimens were obtained from both adult and pediatric patients.
CF Biospecimen Registry (CFBR) at Emory and Children’s Center for Cystic Fibrosis: 20 S. aureus isolates from people with CF were collected between 1 January 2012 and 31 December 2013. These human subject samples were provided by the CF Biospecimen Registry at the Children’s Healthcare of Atlanta and Emory University CF Discovery Core courtesy of Arlene Stecenko. The Emory University IRB has approved collecting and banking of these specimens after obtaining informed consent.
The Geisel School of Medicine at Dartmouth University: The Hogan laboratory at the Geisel School of Medicine at Dartmouth University graciously provided 12 deidentified S. aureus isolates from the Dartmouth CF Translational Research Core. These isolates were obtained from adult patients with CF between 1 January 2015 and 31 December 2017 with support by the CF Foundation RDP grant STANTO15R0.
Bacterial Phenotypes: Clinical isolates of S. aureus were streaked onto tryptic soy agar (TSA) and blood agar to examine colony size, color, and hemolysis pattern. We streaked colonies onto milk agar to score for secreted protease. Beta-toxin was scored by partial lysis on sheep blood agar; alpha-toxin by complete lysis on rabbit blood agar. Oxacillin resistance was determined by growth on Mueller–Hinton agar with 4% NaCl in the presence or absence of 6 µg/mL oxacillin at 33–35 °C. We examined for chloramphenicol, tetracycline, or erythromycin resistance by presence or absence of growth with 10 µg/mL of each antibiotic.
Superantigen Testing: Clinical isolates of S. aureus were tested for the presence or absence of SAg genes using PCR of genomic DNA preparations following a published protocol with appropriate positive and negative controls for each of the SAgs [23]. PCR primers are listed in supplemental materials.
Statistical Analysis: We determined the prevalence of SAgs as the number of subjects positive for a given SAg divided by the total number of subjects analyzed. In subgroup analysis, we compared SAg prevalence in subjects with a single culture vs. those with multiple cultures using Fisher’s exact test. We compared the proportions of MRSA and MSSA isolates that were positive for each of the SAgs using Fisher’s exact test. To measure the strength of the association, we calculated an odds ratio to determine the increase in odds that the individual SAg would be present in MRSA compared to MSSA. Odds ratios were calculated using conditional maximum likelihood estimate with the fisher.test command in R. P < 0.05 was considered statistically significant. We did not adjust for multiple comparisons. To determine whether MRSA and MSSA have distinct complements of SAgs, we performed unsupervised hierarchical clustering of the University of Iowa Biobank isolates based on the presence of SAg genes. We used R Studio version 0.98.1085 or SAS version 9.4 for statistical testing.
3. Results
3.1. S. aureus Specimens From Patients With CF Are Heterogeneous in Phenotypic Appearance
We obtained 77 clinical isolates of S. aureus from adult and pediatric patients with CF at the University of Iowa. These specimens were obtained from N = 45 patients in visits between 12 December 2011 and 20 July 2012. The median age of these patients was 15.75 years as of the date of their last culture (IQR 8.34–26.89, range 5.27–58.66). Between 1 and 7 specimens were obtained per subject (Supplemental Figure S1), with 28 subjects having a single culture. 44 of these specimens were from sputum samples and 33 were from oropharyngeal swabs.
3.1.1. Colony Morphology
In chronic airway infections, CF pathogens like Pseudomonas aeruginosa diversify through genetic mutations [24]. However, if phenotypes are required for survival in the CF airway, these features may be found with increased frequency. Therefore, we examined the S. aureus isolates from patients with CF for colony phenotypes (Table 1). S. aureus normally expresses staphyloxanthin, a golden pigment that protects against host-derived oxidants [25]. We found that many isolates were hypopigmented: 13 were white, 37 were yellow, and 27 were gold.
Table 1.
Phenotypes of S. aureus isolated from individuals with cystic fibrosis.
| Characteristic | Number of Isolates (Total = 77) | % |
|---|---|---|
| Clinical Source | ||
| Sputum | 44 | 57.1% |
| Throat culture | 33 | 42.9% |
| Antibiotic Resistance | ||
| Oxacillin | 28 | 36.4% |
| Chloramphenicol | 0 | 0.0% |
| Tetracycline | 12 | 15.6% |
| Erythromycin | 75 | 97.4% |
| Hemolysis | ||
| Complete, rabbit blood agar ( α-toxin) | 63 | 81.8% |
| Partial, sheep blood agar (β-toxin) | 41 | 53.2% |
| Color | ||
| White | 13 | 16.9% |
| Yellow | 37 | 48.1% |
| Gold | 27 | 35.1% |
| Secreted Protease | ||
| Not detected | 41 | 53.2% |
| Faint | 13 | 16.9% |
| Present | 23 | 29.9% |
Protease secretion is considered a virulence factor in skin infections [26], and protease production could be damaging to airways. Therefore, we tested for secreted protease by examining milk agar plates for zones of clearance. 23 of the isolates had distinct zones of clearance consistent with protease secretion, 13 had small or faint zones of clearance, and 41 isolates exhibited no clearance of milk agar in this assay. There was a strong correlation between hypopigmentation and protease activity. Protease activity, as determined by clearance of milk agar, was detected in 85% of white colonies but 7.5% of gold colonies (P < 0.001).
Another characteristic of S. aureus is hemolysis, a phenotype that is linked to alpha and beta hemolysin toxins. Previous studies of bacteria deficient in alpha toxin reveal its importance in cellular and animal models of CF [27,28]. We tested for the activity of these toxins by hemolysis patterns on sheep and rabbit blood agar plates. Alpha toxin (encoded by hla) activity was observed in the majority of specimens, whereas beta-toxin (hlb) activity was observed in 40% of specimens.
3.1.2. Antibiotic Resistance
CF pathogens are under selective pressure from antibiotic treatment. We determined MRSA status of these isolates by growth on Mueller–Hinton agar in the presence of 6 µg/mL of oxacillin. 28 isolates derived from 19 individuals were phenotypically resistant to oxacillin. 49 isolates from 30 individuals were methicillin-susceptible S. aureus (MSSA). Four subjects had isolates of both MRSA and MSSA. Because macrolides and tetracyclines are commonly prescribed to patients with CF [2], we hypothesized that the S. aureus isolates would be resistant to these antibiotic classes, but remain susceptible to antibiotics that are not routinely given. Each isolate was grown on TSA containing either erythromycin, tetracycline, or chloramphenicol. Chronic azithromycin is routinely prescribed at the University of Iowa CF center [8]. The vast majority of isolates from the University of Iowa (75/77) exhibited erythromycin resistance, but tetracycline resistance was less common (12/77). Within the University of Iowa collection, the two isolates susceptible to erythromycin were obtained as oropharyngeal cultures.
3.2. High Prevalence of Enterotoxin Gene Cluster Genes in S. aureus Isolated From Patients With Cystic Fibrosis
S. aureus encodes a variety of secreted toxins, including bacterial superantigens (SAgs). We hypothesized that there would be similar heterogeneity in S. aureus secreted toxins. Using previously described methods [23], we assessed for the presence of 18 unique SAgs. Among these toxins, the most common were genes belonging to the enterotoxin gene cluster (EGC), including seg, sel-i, sel-m, sel-n, sel-o, and sel-u. These genes were highly prevalent in isolates of S. aureus from patients with CF. All six EGC genes were identified in 37 of the 45 patients examined. 97.8% of patients within this cohort grew S. aureus that encoded at least one member of the EGC (Table 2). The genes encoding EGC toxins were significantly more prevalent in CF specimens compared to the classic S. aureus SAg toxic shock syndrome toxin-1(TSST-1; gene tstH), which was present in 11.7% of isolates.
Table 2.
Prevalence of S. aureus superantigen genes detected from individuals with cystic fibrosis.
| Toxin | Iowa Subjects with CF Total = 45 |
Single Culture Total = 28 |
Multiple Cultures Total = 17 |
P † | |||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| sea | 19 | 42.2% | 8 | 28.6% | 11 | 64.7% | 0.03 |
| seb | 2 | 4.4% | 1 | 3.6% | 1 | 5.9% | 1.00 |
| sec | 4 | 8.9% | 2 | 7.1% | 2 | 11.8% | 0.63 |
| sed | 6 | 13.3% | 2 | 7.1% | 4 | 23.5% | 0.18 |
| see | 3 | 6.7% | 0 | 0.0% | 3 | 17.6% | 0.05 |
| seg | 42 | 93.3% | 25 | 89.3% | 17 | 100.0% | 0.28 |
| sel-h | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | - |
| sel-i | 42 | 93.3% | 25 | 89.3% | 17 | 100.0% | 0.28 |
| sel-k | 4 | 8.9% | 2 | 7.1% | 2 | 11.8% | 0.63 |
| sel-l | 3 | 6.7% | 1 | 3.6% | 2 | 11.8% | 0.55 |
| sel-m | 39 | 86.7% | 23 | 82.1% | 16 | 94.1% | 0.38 |
| sel-n | 41 | 91.1% | 25 | 89.3% | 16 | 94.1% | 1.00 |
| sel-o | 43 | 95.6% | 26 | 92.9% | 17 | 100.0% | 0.52 |
| sel-p | 30 | 66.7% | 15 | 53.6% | 15 | 88.2% | 0.02 |
| sel-q | 15 | 33.3% | 8 | 28.6% | 7 | 41.2% | 0.52 |
| sel-u | 41 | 91.1% | 24 | 85.7% | 17 | 100.0% | 0.28 |
| sel-x | 39 | 86.7% | 23 | 82.1% | 16 | 94.1% | 0.38 |
| tstH | 8 | 17.8% | 3 | 10.7% | 5 | 29.4% | 0.23 |
| ≥1 of egc | 44 | 97.8% | 27 | 96.4% | 17 | 100.0% | 1.00 |
| 6 of egc | 37 | 82.2% | 21 | 75.0% | 16 | 94.1% | 0.13 |
†P values compare subjects with single culture to subjects with multiple cultures using Fisher’s exact test. Bold font indicates genes belonging to the enterotoxin gene cluster.
Because some subjects within the University of Iowa cohort had multiple cultures, there may be greater opportunities to identify specific bacterial genes within these subjects. Therefore, we compared the prevalence of each toxin in subjects with multiple cultures vs. those with one culture. We identified sea and more frequently in patients with repeated cultures. Genes encoding EGC toxins were highly prevalent in both groups. We observed no statistically significant differences in age or culture source between groups.
3.2.1. EGC Prevalence in MRSA and MSSA
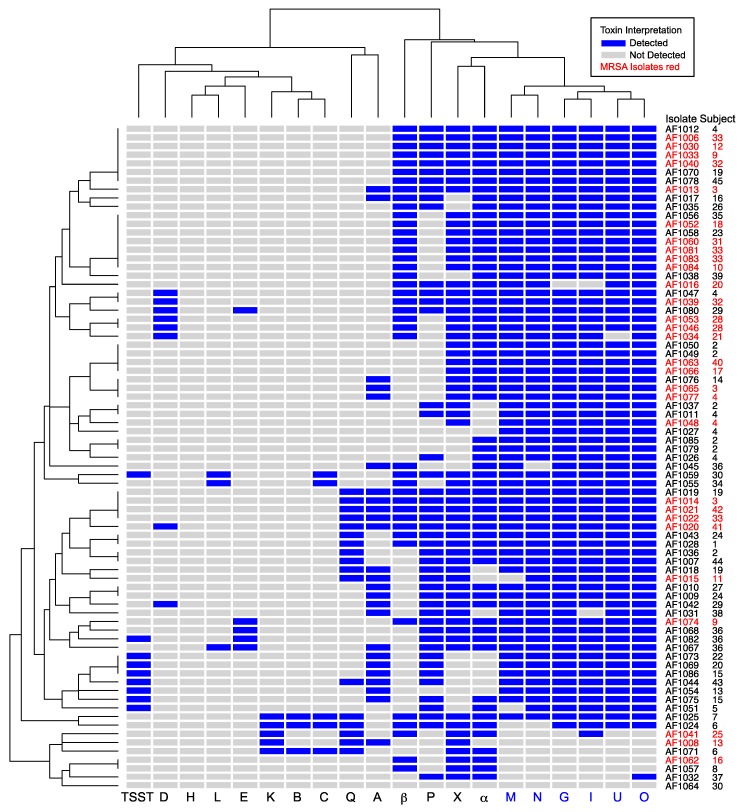
We hypothesized that the genes encoding secreted toxins may be associated with either methicillin susceptibility or resistance. To determine whether MRSA isolates had specific toxin signature(s), we performed hierarchical clustering based on the presence or absence of toxins (Figure 1). MRSA isolates were distributed widely in this analysis and often shared the same toxin profile as MSSA isolates. We separately tested whether individual toxins were associated with MRSA or MSSA (Table 3). None of the MRSA isolates were positive for tstH, consistent with previous observations that TSST-1 is generally associated with MSSA [29]. MSSA was more likely than MRSA to be positive for sel-p (P = 0.03). While sel-p and tstH were more common in MSSA, sel-x was more common in MRSA. However, no combination of SAgs was perfectly predictive of methicillin resistance, and genes encoding EGC toxins were prevalent in both MSSA and MRSA isolates.
Figure 1.
Unsupervised hierarchical clustering of cystic fibrosis (CF) clinical isolates based on presence of S. aureus toxin genes. Isolate numbers and subjects are indicated at right. Toxin genes are on the bottom margin, with members of the enterotoxin gene cluster (EGC) in blue. Letters A, B, C, D, E, and G are staphylococcal enterotoxin (SE) superantigens characterized as causing emesis after oral administration. Letters H, I, K, L, M–Q, and X are SE-like superantigens. TSST is toxic shock syndrome toxin-1 superantigen. α and β are cytotoxins. Blue shading represents toxin presence. MRSA isolates are indicated with red font. Dendrograms at left and top show relatedness of the isolates and toxins, respectively. The EGC was prevalent in both MRSA and MSSA isolates.
Table 3.
Association of S. aureus toxin genes with methicillin susceptibility or resistance.
|
S. aureus Total = 77 |
MSSA Total = 49 |
MRSA Total = 28 |
OR * | P † | ||||
|---|---|---|---|---|---|---|---|---|
| Toxin | N | % | N | % | N | % | ||
| sea | 25 | 32.5% | 17 | 34.7% | 8 | 28.6% | 0.76 | 0.62 |
| seb | 3 | 3.9% | 3 | 6.1% | 0 | 0.0% | - | 0.30 |
| sec | 5 | 6.5% | 5 | 10.2% | 0 | 0.0% | - | 0.15 |
| sed | 8 | 10.4% | 3 | 6.1% | 5 | 17.9% | 3.28 | 0.13 |
| see | 5 | 6.5% | 4 | 8.2% | 1 | 3.6% | 0.42 | 0.65 |
| seg | 69 | 89.6% | 45 | 91.8% | 24 | 85.7% | 0.54 | 0.45 |
| sel-h | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% | - | - |
| sel-i | 69 | 89.6% | 44 | 89.8% | 25 | 89.3% | 0.95 | 1.00 |
| sel-k | 5 | 6.5% | 3 | 6.1% | 2 | 7.1% | 1.18 | 1.00 |
| sel-l | 3 | 3.9% | 3 | 6.1% | 0 | 0.0% | - | 0.30 |
| sel-m | 67 | 87.0% | 42 | 85.7% | 25 | 89.3% | 1.38 | 0.74 |
| sel-n | 68 | 88.3% | 43 | 87.8% | 25 | 89.3% | 1.16 | 1.00 |
| sel-o | 71 | 92.2% | 46 | 93.9% | 25 | 89.3% | 0.55 | 0.66 |
| sel-p | 46 | 59.7% | 34 | 69.4% | 12 | 42.9% | 0.34 | 0.03 |
| sel-q | 17 | 22.1% | 11 | 22.4% | 6 | 21.4% | 0.94 | 1.00 |
| sel-u | 69 | 89.6% | 45 | 91.8% | 24 | 85.7% | 0.54 | 0.45 |
| sel-x | 61 | 79.2% | 33 | 67.3% | 28 | 100.0% | - | 0.0003 |
| tstH | 9 | 11.7% | 9 | 18.4% | 0 | 0.0% | - | 0.02 |
| hla | 63 | 81.8% | 37 | 75.5% | 26 | 92.9% | 4.15 | 0.07 |
| hlb | 41 | 53.2% | 19 | 38.8% | 22 | 78.6% | 5.65 | 0.001 |
* OR = Odds ratio, values > 1 indicate increased odds of the toxin being encoded in MRSA versus methicillin-susceptible S. aureus (MSSA). †P values calculated by Fisher’s exact test. Bold font indicates genes belonging to the enterotoxin gene cluster.
3.2.2. EGC Prevalence in S. aureus From Other U.S. CF Centers
We considered the possibility that the high prevalence of S. aureus encoding EGC was due to geographic sampling. To address this possibility, we obtained 12 S. aureus isolates from adults with CF at Dartmouth University and 20 S. aureus isolates from Emory University. We genotyped these isolates for the same set of toxins (Table 4). Notably, there were no significant differences between isolates obtained in Iowa compared to Dartmouth or Emory, suggesting that the high prevalence of the EGC is not related to geographic sampling of one region of the United States.
Table 4.
S. aureus toxin genes identified in CF clinical isolates from geographically separate regions. Bold font indicates genes belonging to the enterotoxin gene cluster.
| Toxin | Iowa Total = 77 |
Emory Total = 20 |
Dartmouth Total = 12 |
|||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| sea | 25 | 32.5% | 0 | 0.0% | 2 | 16.7% |
| seb | 3 | 3.9% | 0 | 0.0% | 0 | 0.0% |
| sec | 5 | 6.5% | 1 | 5.0% | 0 | 0.0% |
| sed | 8 | 10.4% | 3 | 15.0% | 0 | 0.0% |
| see | 5 | 6.5% | 4 | 20.0% | 0 | 0.0% |
| seg | 69 | 89.6% | 16 | 80.0% | 10 | 83.3% |
| sel-h | 0 | 0.0% | 0 | 0.0% | 0 | 0.0% |
| sel-i | 69 | 89.6% | 16 | 80.0% | 10 | 83.3% |
| sel-k | 5 | 6.5% | 1 | 5.0% | 2 | 16.7% |
| sel-l | 3 | 3.9% | 1 | 5.0% | 0 | 0.0% |
| sel-m | 67 | 87.0% | 15 | 75.0% | 9 | 75.0% |
| sel-n | 68 | 88.3% | 16 | 80.0% | 10 | 83.3% |
| sel-o | 71 | 92.2% | 15 | 75.0% | 9 | 75.0% |
| sel-p | 46 | 59.7% | 5 | 25.0% | 0 | 0.0% |
| sel-q | 17 | 22.1% | 2 | 10.0% | 2 | 16.7% |
| sel-u | 69 | 89.6% | 16 | 80.0% | 10 | 83.3% |
| sel-x | 61 | 79.2% | 19 | 95.0% | 11 | 91.7% |
| tstH | 9 | 11.7% | 1 | 5.0% | 2 | 16.7% |
4. Discussion
S. aureus isolates from patients with CF displayed heterogeneity of color and protease secretion. However, the majority of these diverse S. aureus isolates encoded the EGC. The heterogeneity of S. aureus colony phenotypes suggests that putative virulence factors such as staphyloxanthin and protease may not be under strong selective pressure to remain in the CF airway. By contrast, alpha-toxin mediated hemolysis was routinely observed. Chronic antibiotic exposure represents a strong selective pressure; most of the isolates from the University of Iowa were resistant to erythromycin. The high prevalence of EGC toxins suggests that S. aureus is under pressure to maintain these genes during infection of the CF airway.
We considered the possibility that high prevalence of EGC was related to geographic exposure. Using three geographically distinct collections of S. aureus isolated from U.S. patients with cystic fibrosis, we found that EGC toxins had similarly high prevalence. This is similar to a recent study of S. aureus isolates from Europe patients with CF, in which EGC toxins were present in ≈57% of isolates [19]. The EGC prevalence within this U.S. collection is even higher, suggesting the continued emergence of strains encoding this locus, possibly through the spread of one or more clones. S. aureus clonality is common in CF. In the European study, 5 spa types accounted for 25.6% of all S. aureus isolates. However, EGC-positive S. aureus isolates were not limited to a single clonal group [19].
Most of the CF isolates in the current study were obtained in 2011 and 2012, a time period similar to when specimens were collected for a study of atopic dermatitis that used the same genotyping methodology [21]. We compared the prevalence of each of the SAgs in CF versus atopic dermatitis. All of the SAgs belonging to the EGC (seg, sel-i, sel-m, sel-n, sel-o, and sel-u) were significantly more prevalent in CF as compared to atopic dermatitis collection. Among subjects with atopic dermatitis, three distinct genotypes of S. aureus were apparent. The first of these S. aureus genotypes, which nearly always encoded all EGC toxins, appears highly similar to the prevailing S. aureus within the CF population. Longitudinal studies are needed to determine whether or not the EGC associates with persistence of S. aureus in diseases like CF and atopic dermatitis.
In comparing the prevalence of the EGC in these CF isolates to S. aureus isolates derived from other anatomic sites, we find that CF has a uniquely high prevalence for this group of SAg toxins. The EGC is more prevalent in CF than in atopic dermatitis, diabetic foot ulcer, and significantly more prevalent than in vaginal mucosa [30] and in patients with menstrual toxic shock syndrome (TSS) [12]. The enrichment of EGC-positive isolates in these CF airway isolates suggests that the EGC may confer selective advantage for S. aureus strains in adapting to its role as a chronic pathogen in the airway. Typically, members of the EGC are considered colonization SAgs, due to low-level production [31].
In striking contrast to the CF, S. aureus isolated from acute inflammatory infections such as TSS and post-influenza necrotizing pneumonia have very high prevalence of tstH and produce SAgs in higher concentration [12,18]. We observed that a minority of CF clinical isolates encode tstH; it is unknown what effect this SAg may have on progression of CF lung disease.
It is unclear whether patients with CF develop intact immune responses to S. aureus [32,33]. Compared to P. aeruginosa, patients with CF may have attenuated antibody production [33]. We hypothesize that this could be a consequence of immune misdirection by S. aureus SAgs. Moreover, these SAgs could facilitate increased inflammation, an important factor in CF disease progression. Future studies should examine the immune response to these prevalent S. aureus SAgs.
S. aureus SAgs represent a possible target for vaccination. Given the high prevalence of EGC SAgs in CF, future attempts at immunizing patients with CF against S. aureus may use these antigens as vaccine targets. Notably, a recent study has shown that immunization of rabbits against the SAgs TSST-1 and SEC, and the cytotoxin α-toxin protected 87/88 animals after intra-pulmonary challenge [34]. This vaccination strategy depended on formation of cross-protective neutralizing antibodies. For example, antibodies raised against SEC protect against both SEB and the EGC SAg SEl-U. That study also suggested that vaccination against toxoids may be a more effective strategy than vaccination against cell surface S. aureus virulence factors. In keeping with this notion, a group has recently performed a first-in-humans vaccine trial against the TSST-1 toxoid [35].
4.1. Advantages
This study establishes that enterotoxin gene cluster members are highly prevalent in S. aureus isolates in American patients with CF, independent of methicillin resistance. Because we used consistent methodology for genotyping, we can compare respiratory isolates of S. aureus from patients with CF to cutaneous isolates from patients with atopic dermatitis that were obtained at a similar time. This comparison reveals that genes encoding EGC toxins are enriched in respiratory isolates. We have also confirmed the high prevalence of EGC toxins using S. aureus isolates from geographically distinct CF centers.
4.2. Limitations
Because these data are cross-sectional, it is unknown how long these strains have been present in the CF airway. Many of these patients were sputum-producing. Thus, these infections may represent chronic infection rather than initial infection. We are unable to correlate the presence or absence of secreted toxins with pulmonary outcomes such as FEV1, since the specimens are de-identified. Although many strains encode secreted toxins, we cannot determine whether these genes are actively expressed in the CF airway. Because of de-identification, we are also unable to determine whether there is adaptive host response to presence or absence of these toxins. We intend to address these limitations in future studies with a larger number of fully identified specimens.
5. Conclusions
S. aureus remains a prevalent pathogen in CF. Improvements in the prevention and treatment of S. aureus infection remain a major goal in this disease. This study reveals the SAg ECG gene cluster is highly prevalent in S. aureus CF isolates, revealing a potential vaccination target for this organism in CF.
Acknowledgments
We acknowledge Timothy Starner for assistance with University of Iowa CF Biobank isolates, Arlene Stecenko for assistance with Emory University isolates, and Deborah Hogan and Alex Crocker for assistance with Dartmouth isolates.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/12/1036/s1, Supplemental Methods: Genotyping assay. Figure S1: Frequency of repeated cultures within University of Iowa Biobank.
Author Contributions
Conceptualization, A.J.F. and P.M.S.; methodology, A.J.F., S.H.K., S.B.S., D.H.L., P.D.A., A.R.H.; formal analysis, A.J.F., S.H.K., D.H.L., P.M.S.; resources, S.B.S. and D.H.L.; data curation, A.J.F., S.H.K., S.B.S., P.M.S.; writing—original draft preparation, A.J.F.; writing—review and editing, A.J.F., D.H.L., S.B.S., P.M.S.; visualization, A.J.F.; supervision, A.J.F., P.M.S.; project administration, A.J.F. and P.M.S.; funding acquisition, A.J.F. and P.M.S.
Funding
A.J.F. was supported in part by the University of Iowa Department of Pediatrics, the Cystic Fibrosis Foundation CFF FISCHE16I0, and NHLBI K08 HL136927. S.H.K. and P.M.S. were supported by University of Iowa start-up funds. D.H.L. is supported by CFF Post-doc to Faculty Transition Award LIMOLI18F5 and University of Iowa start-up funds.
Conflicts of Interest
P.M.S. has received patent protection regarding S. aureus secreted toxins, including methods to induce immune response to S. aureus with novel toxoids. The remaining authors have no financial conflicts of interest. The funders of this study had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Salsgiver E.L., Fink A.K., Knapp E.A., LiPuma J.J., Olivier K.N., Marshall B.C., Saiman L. Changing epidemiology of the respiratory bacteriology of patients with cystic fibrosis. Chest. 2016;149:390–400. doi: 10.1378/chest.15-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cystic Fibrosis Foundation 2016 Patient Registry Annual Data Report. [(accessed on 4 November 2019)];2017 Available online: https://www.cff.org/research/researcher-resources/patient-registry/2016-patient-registry-annual-data-report.pdf.
- 3.Ramsey B.W., Pepe M.S., Quan J.M., Otto K.L., Montgomery A.B., Williams-Warren J., Vasiljev K.M., Borowitz D., Bowman C.M., Marshall B.C., et al. Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. Cystic fibrosis inhaled tobramycin study group. N. Engl. J. Med. 1999;340:23–30. doi: 10.1056/NEJM199901073400104. [DOI] [PubMed] [Google Scholar]
- 4.Gibson R.L., Emerson J., McNamara S., Burns J.L., Rosenfeld M., Yunker A., Hamblett N., Accurso F., Dovey M., Hiatt P., et al. Significant microbiological effect of inhaled tobramycin in young children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 2003;167:841–849. doi: 10.1164/rccm.200208-855OC. [DOI] [PubMed] [Google Scholar]
- 5.Tiddens H.A., De Boeck K., Clancy J.P., Fayon M., Arets H.G.M., Bresnik M., Derchak A., Lewis S.A., Oermann C.M. Open label study of inhaled aztreonam for Pseudomonas eradication in children with cystic fibrosis: The alpine study. J. Cyst. Fibros. 2015;14:111–119. doi: 10.1016/j.jcf.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Dezube R., Jennings M.T., Rykiel M., Diener-West M., Boyle M.P., Chmiel J.F., Dasenbrook E.C. Eradication of persistent methicillin-resistant Staphylococcus aureus infection in cystic fibrosis. J. Cyst. Fibros. 2018 doi: 10.1016/j.jcf.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Heltshe S.L., Mayer-Hamblett N., Burns J.L., Khan U., Baines A., Ramsey B.W., Rowe S.M. Pseudomonas aeruginosa in cystic fibrosis patients with G551D-CFTR treated with ivacaftor. Clin. Infect. Dis. 2015;60:703–712. doi: 10.1093/cid/ciu944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh S.B., McLearn-Montz A.J., Milavetz F., Gates L.K., Fox C., Murry L.T., Sabus A., Porterfield H.S., Fischer A.J. Pathogen acquisition in patients with cystic fibrosis receiving ivacaftor or lumacaftor/ivacaftor. Pediatr. Pulmonol. 2019 doi: 10.1002/ppul.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caudri D., Turkovic L., Ng J., de Klerk N.H., Rosenow T., Hall G.L., Ranganathan S.C., Sly P.D., Stick S.M. The association between Staphylococcus aureus and subsequent bronchiectasis in children with cystic fibrosis. J. Cyst. Fibros. 2018;17:462–469. doi: 10.1016/j.jcf.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Dasenbrook E.C., Merlo C.A., Diener-West M., Lechtzin N., Boyle M.P. Persistent methicillin-resistant Staphylococcus aureus and rate of FEV1 decline in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2008;178:814–821. doi: 10.1164/rccm.200802-327OC. [DOI] [PubMed] [Google Scholar]
- 11.Dinges M.M., Orwin P.M., Schlievert P.M. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 2000;13:16–34. doi: 10.1128/CMR.13.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spaulding A.R., Salgado-Pabon W., Kohler P.L., Horswill A.R., Leung D.Y., Schlievert P.M. Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 2013;26:422–447. doi: 10.1128/CMR.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormick J.K., Yarwood J.M., Schlievert P.M. Toxic shock syndrome and bacterial superantigens: An update. Annu. Rev. Microbiol. 2001;55:77–104. doi: 10.1146/annurev.micro.55.1.77. [DOI] [PubMed] [Google Scholar]
- 14.Jarraud S., Peyrat M.A., Lim A., Tristan A., Bes M., Mougel C., Etienne J., Vandenesch F., Bonneville M., Lina G. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 2001;166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- 15.Becker K., Friedrich A.W., Peters G., von Eiff C. Systematic survey on the prevalence of genes coding for staphylococcal enterotoxins SELM, SELO, and SELN. Mol. Nutr. Food Res. 2004;48:488–495. doi: 10.1002/mnfr.200400044. [DOI] [PubMed] [Google Scholar]
- 16.Grumann D., Scharf S.S., Holtfreter S., Kohler C., Steil L., Engelmann S., Hecker M., Volker U., Broker B.M. Immune cell activation by enterotoxin gene cluster (egc)-encoded and non-egc superantigens from Staphylococcus aureus. J. Immunol. 2008;181:5054–5061. doi: 10.4049/jimmunol.181.7.5054. [DOI] [PubMed] [Google Scholar]
- 17.Holtfreter S., Bauer K., Thomas D., Feig C., Lorenz V., Roschack K., Friebe E., Selleng K., Lovenich S., Greve T., et al. egc-Encoded superantigens from Staphylococcus aureus are neutralized by human sera much less efficiently than are classical staphylococcal enterotoxins or toxic shock syndrome toxin. Infect. Immun. 2004;72:4061–4071. doi: 10.1128/IAI.72.7.4061-4071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stach C.S., Vu B.G., Merriman J.A., Herrera A., Cahill M.P., Schlievert P.M., Salgado-Pabon W. Novel tissue level effects of the Staphylococcus aureus enterotoxin gene cluster are essential for infective endocarditis. PLoS ONE. 2016;11:e0154762. doi: 10.1371/journal.pone.0154762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garbacz K., Piechowicz L., Podkowik M., Mroczkowska A., Empel J., Bania J. Emergence and spread of worldwide Staphylococcus aureus clones among cystic fibrosis patients. Infect. Drug Res. 2018;11:247–255. doi: 10.2147/IDR.S153427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mempel M., Lina G., Hojka M., Schnopp C., Seidl H.P., Schafer T., Ring J., Vandenesch F., Abeck D. High prevalence of superantigens associated with the egc locus in Staphylococcus aureus isolates from patients with atopic eczema. Eur. J. Clin. Microbiol. Infect. Dis. 2003;22:306–309. doi: 10.1007/s10096-003-0928-0. [DOI] [PubMed] [Google Scholar]
- 21.Merriman J.A., Mueller E.A., Cahill M.P., Beck L.A., Paller A.S., Hanifin J.M., Ong P.Y., Schneider L., Babineau D.C., David G., et al. Temporal and racial differences associated with atopic dermatitis Staphylococcus aureus and encoded virulence factors. mSphere. 2016;1 doi: 10.1128/mSphere.00295-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dasenbrook E.C., Checkley W., Merlo C.A., Konstan M.W., Lechtzin N., Boyle M.P. Association between respiratory tract methicillin-resistant Staphylococcus aureus and survival in cystic fibrosis. JAMA J. Am. Med. Assoc. 2010;303:2386–2392. doi: 10.1001/jama.2010.791. [DOI] [PubMed] [Google Scholar]
- 23.Salgado-Pabon W., Case-Cook L.C., Schlievert P.M. Molecular analysis of staphylococcal superantigens. Methods Mol. Biol. 2014;1085:169–185. doi: 10.1007/978-1-62703-664-1_10. [DOI] [PubMed] [Google Scholar]
- 24.Smith E.E., Buckley D.G., Wu Z., Saenphimmachak C., Hoffman L.R., D’Argenio D.A., Miller S.I., Ramsey B.W., Speert D.P., Moskowitz S.M., et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clauditz A., Resch A., Wieland K.P., Peschel A., Gotz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 2006;74:4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolar S.L., Ibarra J.A., Rivera F.E., Mootz J.M., Davenport J.E., Stevens S.M., Horswill A.R., Shaw L.N. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. MicrobiologyOpen. 2013;2:18–34. doi: 10.1002/mbo3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarry T.M., Memmi G., Cheung A.L. The expression of alpha-haemolysin is required for Staphylococcus aureus phagosomal escape after internalization in CFT-1 cells. Cell. Microbiol. 2008;10:1801–1814. doi: 10.1111/j.1462-5822.2008.01166.x. [DOI] [PubMed] [Google Scholar]
- 28.Keitsch S., Riethmuller J., Soddemann M., Sehl C., Wilker B., Edwards M.J., Caldwell C.C., Fraunholz M., Gulbins E., Becker K.A. Pulmonary infection of cystic fibrosis mice with Staphylococcus aureus requires expression of alpha-toxin. Biol. Chem. 2018;399:1203–1213. doi: 10.1515/hsz-2018-0161. [DOI] [PubMed] [Google Scholar]
- 29.Schlievert P.M., Strandberg K.L., Lin Y.C., Peterson M.L., Leung D.Y. Secreted virulence factor comparison between methicillin-resistant and methicillin-sensitive Staphylococcus aureus, and its relevance to atopic dermatitis. J. Allergy Clin. Immunol. 2010;125:39–49. doi: 10.1016/j.jaci.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vu B.G., Stach C.S., Salgado-Pabon W., Diekema D.J., Gardner S.E., Schlievert P.M. Superantigens of Staphylococcus aureus from patients with diabetic foot ulcers. J. Infect. Dis. 2014;210:1920–1927. doi: 10.1093/infdis/jiu350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roetzer A., Gruener C.S., Haller G., Beyerly J., Model N., Eibl M.M. Enterotoxin gene cluster-encoded sei and seln from Staphylococcus aureus isolates are crucial for the induction of human blood cell proliferation and pathogenicity in rabbits. Toxins. 2016;8:314. doi: 10.3390/toxins8110314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hollsing A.E., Granstrom M., Strandvik B. Prospective study of serum staphylococcal antibodies in cystic fibrosis. Arch. Dis. Child. 1987;62:905–911. doi: 10.1136/adc.62.9.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moss R.B., Hsu Y.P., Lewiston N.J. 125I-Clq-binding and specific antibodies as indicators of pulmonary disease activity in cystic fibrosis. J. Pediatr. 1981;99:215–222. doi: 10.1016/S0022-3476(81)80453-2. [DOI] [PubMed] [Google Scholar]
- 34.Spaulding A.R., Salgado-Pabon W., Merriman J.A., Stach C.S., Ji Y., Gillman A.N., Peterson M.L., Schlievert P.M. Vaccination against Staphylococcus aureus pneumonia. J. Infect. Dis. 2014;209:1955–1962. doi: 10.1093/infdis/jit823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roetzer A., Jilma B., Eibl M.M. Vaccine against toxic shock syndrome in a first-in-man clinical trial. Expert Rev. Vaccines. 2017;16:81–83. doi: 10.1080/14760584.2017.1268921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.