Abstract.
Rotavirus is the leading cause of childhood deaths due to diarrhea. Although existing oral rotavirus vaccines are highly efficacious in high-income countries, these vaccines have been demonstrated to have decreased efficacy in low- and middle-income countries. A possible explanation for decreased efficacy is the impact of gut microbiota on the enteric immune system’s response to vaccination. We analyzed the gut microbiome of 50 children enrolled in a prospective study evaluating response to oral pentavalent rotavirus vaccination (RV5) to assess associations between relative abundance of bacterial taxa and seroconversion following vaccination. Stool samples were taken before the first RV5 dose, and microbiome composition characterized using 16S rRNA amplicon sequencing and Quantitative Insights Into Microbial Ecology software. Relative abundance of bacterial taxa between seroconverters following the first RV5 dose, those with ≥ 4-fold increase in rotavirus-specific IgA titers, and nonseroconverters were compared using the Wilcoxon–Mann–Whitney test. We identified no significant differences in microbiome composition between infants who did and did not respond to vaccination. Infants who responded to vaccination tended to have higher abundance of Proteobacteria and Eggerthella, whereas those who did not respond had higher abundance of Fusobacteria and Enterobacteriaceae; however, these differences were not statistically significant following a multiple comparison correction. This study suggests a limited impact of gut microbial taxa on response to oral rotavirus vaccination among infants; however, additional research is needed to improve our understanding of the impact of gut microbiome on vaccine response, toward a goal of improving vaccine efficacy and rotavirus prevention.
INTRODUCTION
Diarrhea is the second leading cause of death among children younger than 5 years, resulting in more than 800,000 deaths annually.1 More than 90% of deaths due to diarrheal disease occur in low-income countries. Rotavirus is the leading cause of diarrheal-related deaths worldwide, accounting for over 215,000 deaths each year, primarily in low- and middle-income countries (LMICs).2,3 Although rotavirus vaccines (RV) have been highly efficacious in high-income countries, numerous studies have demonstrated decreased efficacy in LMICs.4–12 This presents a major challenge in addressing global childhood mortality caused by diarrheal disease.
The reason for decreased RV efficacy in LMICs is not entirely understood. One possible explanation is the impact that the developing gut microbiota has on the enteric immune response to this oral vaccine. Development of the human gut microbiota begins after birth and plays an important role in maturity of the enteric and systemic immune system.13 Commensal bacteria influence anti-inflammatory properties of many immune cells,14 induce regulatory T cells,15 and affect the overall maturation of the immune system.16 Moreover, a number of factors influence the composition of an infant’s gut microbiome, including mode of delivery (vaginal versus cesarean),17,18 breastfeeding,19 and exposure to antibiotics.20 Environmental enteropathy (EE), an intestinal condition characterized by increased intestinal permeability and chronic inflammation due to repeated pathogenic exposure, is associated with impaired enteric immunity and may also limit response to RV vaccination.21
The impact of the gut microbiota on response to orally delivered vaccination has been investigated with regards to both oral polio vaccine (OPV) and RV.22 One recent study found an association between high relative abundance of Bifidobacterium spp. and increased OPV response,23 whereas another found no statistically significant associations between bacterial microbiota abundance (at the phylum or genus levels) following false discovery rate (FDR) correction.24 In addition, abundance of Bifidobacterium longum subspecies infantis in early infancy was positively associated with the T-cell responses induced by BCG, TT, and hepatitis B vaccines.25 Response to rotavirus vaccination in a trial of Ghanaian children found response to RV to be positively associated with the Bacilli class (Streptococcus bovis) and negatively associated with bacteria in the Bacteroidetes phylum (Bacteroides and Prevotella).26 Another study among Pakistani infants found Clostridium cluster XI and Proteobacteria to be positively associated with response at the phylum level and Serratia and Escherichia coli to be positively associated with response at the genus level.27 Evaluation of immune response in gnotobiotic pigs transplanted with either healthy or unhealthy human gut microbiota (defined by their concentrations of biomarkers of environmental enteric dysfunction) found stronger adaptive immune response among pigs transplanted with healthy gut microbiota.28 Furthermore, microbiota ablation in mice, via the administration of antibiotics or use of germ-free mice, delayed and initiated a more robust adaptive immune response, indicating interplay between commensal bacteria and host immunity.29
To date, the published studies that have evaluated the association between infant microbiome composition and response to oral rotavirus vaccines have all investigated the monovalent RV (RotaRix, GlaxoSmithKline, Brentford, United Kingdom), made from a live attenuated human strain of rotavirus. The aim of this study was to evaluate the relationship between gut microbiome community structure and response to oral pentavalent RV (RotaTeq, RV5, Merck, Kenilworth, NJ), made from reassortment of bovine and human rotavirus strains, among Nicaraguan children. We hypothesized that the relative abundance of different phyla or genera of bacteria isolated from participants’ stool samples will be associated with vaccine response to RV5.
METHODS
Study design and participants.
The study enrolled 50 children from a prospective study of RV5 immunogenicity. Participants were recruited between September and November 2014 from records of recent deliveries and pregnancies from the Perla Maria and Subtiava Health Sectors in Leon, Nicaragua. Eligibility criteria, recruitment methodology, and sample collection have been previously reported.30 Blood samples were obtained immediately before immunization and 4 weeks after the first RV5 dose to assess seroconversion. All participants received their first dose of RV5 at 2 months of age, following the Nicaraguan National Immunization Schedule of three doses at 2, 4, and 6 months. The study was approved by the Institutional Review Boards of the National Autonomous University of Nicaragua, Leon (UNAN-Leon) (IRB # 110), and the University of North Carolina at Chapel Hill (IRB # 14-1136).
Relative abundances of bacterial taxa were estimated using infant’s stool. Stool samples were collected from the infant’s diaper at 1–3 days before receipt of RV5. The samples were then diluted 20-fold, homogenized in sterile pre-reduced anaerobic 0.1 M potassium phosphate buffer saline (pH 7.2), snap-frozen in liquid nitrogen, and stored at −80°C until laboratory analysis.
Laboratory methods.
A composite EE score was calculated using the concentration of four fecal biomarkers indicative of presence of EE, including alpha-1 antitrypsin, neopterin, myeloperoxidase, and calprotectin. Biomarker concentration was determined using ELISA and composite EE scores ranged from 0 to 11. Participants with an EE score in the upper quartile were categorized as high EE, whereas all others were categorized as low EE. Seroconversion following the first dose of RV5 was defined as ≥ 4-fold increase in rotavirus-specific IgA titers. Environmental enteropathy score calculation and determination of titers through the use of ELISA was conducted as previously described.30
16S rRNA amplicon sequencing.
Fecal samples were transferred to a 2-mL tube containing 200 mg of ≤ 106 μm glass beads (Sigma, St. Louis, MO) and 0.3 mL of Qiagen ATL buffer (Valencia, CA), supplemented with 20 mg/mL lysozyme (Thermo Fisher Scientific, Grand Island, NY). The suspension was incubated at 37°C for 1 hour with occasional agitation. Subsequently, the suspension was supplemented with 600IU of Qiagen proteinase K and incubated at 60°C for 1 hour. Finally, 0.3 mL of Qiagen AL buffer was added and a final incubation at 70°C for 10 minutes was carried out. Bead beating was then used for 3 minutes in a Qiagen TissueLyser II at 30 Hz. After a brief centrifugation, supernatants were aspirated and transferred to a new tube containing 0.3 mL of ethanol. DNA was purified using a standard on-column purification method with Qiagen buffers AW1 and AW2 as washing agents, and eluted in 10 mM Tris (pH 8.0).
Total DNA from fecal samples was amplified using primers targeting the V4 hypervariable region of the bacterial 16S rRNA gene31 containing overhang adaptor sequences compatible with Illumina sequencing platform. Master mixes contained 12.5 ng of total DNA, 0.2 µm of each primer, and 2x KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Wilmington, MA). Library preparation was performed as previously described.32 The normalized DNA library pool was denatured with NaOH, diluted with hybridization buffer, and heat-denatured before loading on the MiSeq reagent cartridge (Illumina, San Diego, CA) and on the MiSeq instrument (Illumina). The average sequencing depth was 200,000–250,000 per sample. Automated cluster generation and paired-end sequencing with dual reads were performed according to the manufacturer’s instructions.
Bioinformatics analysis and data visualization.
Sequencing output from the Illumina MiSeq platform were converted to FASTQ format and demultiplexed using Illumina Bcl2Fastq 2.18.0.12.33 The resulting paired-end reads were joined using the Quantitative Insights Into Microbial Ecology (QIIME) 1.9.034 invocation of fastq-join35 with the default parameters. Index and linker primer sequences were trimmed and the reads were subsequently filtered for quality removing any read where the percentage of quality scores below the quality threshold of 24 fell below 70%. Quality control of both raw and processed sequencing reads was verified by FastQC.36
Sequences were clustered into operational taxonomic units (OTUs) based on the de novo OTU picking algorithm using the QIIME implementation of UCLUST37 at a similarity threshold of 97%. operational taxonomic units identified as chimeric by VSEARCH 38 of the ChimeraSlayer “gold” reference database39 and those composed of a single read (singletons) were eliminated. The remaining OTUs were assigned taxonomic identifiers with respect to the Greengenes database,40 their sequences were aligned using template alignment through PyNAST,41 and a phylogenetic tree was built with FastTree 2.1.3.42
Alpha diversity with respect to PD whole tree, Shannon index, Chao1, and observed species number metrics was estimated using QIIME at a rarefaction depth of 1,000 sequences per subsample. Beta diversity estimates were calculated within QIIME using weighted and unweighted UniFrac distances43,44 between samples at a subsampling depth of 1,000. Results were summarized and visualized through principal coordinate analysis as implemented in QIIME. Default values were used with all QIIME processing.
PhyloToAST,45 was used to calculate the mean relative abundance (MRA) per sample from a normalized BIOM table in JSON format using the normalize_table. py and biom.convert commands, respectively. The taxonomy table from the assign_taxonomy QIIME script was condensed and trimmed to include sequences that map to unique OTUs using the otu_condense.py and filter_rep_set.py commands. The condensed representative sequence set was imported into Geneious46 software for MUSCLE alignment and tree building using the HKY option and exported in NEWICK format. The Interactive Tree of Life (iTol) tree and table were generated using iTol.py command, with the normalized OTU json table as an input file. The variable “seroconversion” from the mapping file was used to generate the iTol table. The iTol (itol.embl.de)47,48 was used for visualization and annotation of phylogenetic trees.
Statistical analysis.
Participants’ baseline characteristics were calculated and reported as frequencies for categorical variables and group means for continuous variables. Baseline differences among categorical variables were assessed using the Fisher’s exact test, whereas differences among continuous variables were assessed using the Students t-test. Differences in relative abundance of each bacteria genera and phyla were statistically assessed between groups (seroconverters versus non-seroconverters) using the Wilcoxon–Mann–Whitney test. For bacterial taxa included in Enterobacteriaceae, Erysipelotrichaceae, and Xanthomonadaceae, comparisons were limited to the family level, as the 16s rRNA sequence approach used could not distinguish genera below this level. Given the large number of bacteria taxa included in the analysis, 10 phyla and 147 genera, statistical significance was assessed using the Benjamini–Hochberg FDR correction. An FDR correction was only applied to comparisons of relative abundance of bacterial taxa, and not to the comparison of participant characteristics. Statistical testing was completed using SAS software version 9.3 and 9.4 (SAS, Cary, NC).
RESULTS
Baseline characteristics.
Stool samples were collected from 50 children before vaccination in the prospective study; however, five children were excluded for analysis because of the mother’s refusal to provide all required biological samples (three participants), moving out of the study area (one participant), or incomplete receipt of vaccine (vomiting after vaccine administration) (one participant). All children included in the study were 2 months of age at the time of baseline characteristic assessment and vaccine administration.
Vaccine seroconversion occurred after the first RV5 dose in 25 of 45 participants (55.6%). Among the overall study population, 42% were female, 89% had been breastfed the previous day, 98% had municipally piped water in the home, 84% had indoor sanitation, 71% had non-dirt floors in the home, 69% had animal(s) living in the home, and 7% had experienced a diarrhea episode before receiving the first RV5 dose. Interestingly, 45% of non-seroconverters were identified as high EE, whereas only 16% of seroconverters had high EE (P = 0.05), which was the only statistically significant difference in the comparison of characteristics between seroconverters and non-seroconverters (Table 1).
Table 1.
Population characteristics
| Variable | Seroconverters (25 infants), number (%) or mean (SD) | Non-seroconverters (20 infants), number (%) or mean (SD) | P-value* |
|---|---|---|---|
| Gender (% female) | 11 (44%) | 8 (40%) | 1.00 |
| Breastfed yesterday (% yes) | 23 (92%) | 17 (85%) | 0.64 |
| Exclusively breastfed (% yes) | 9 (36%) | 4 (20%) | 0.32 |
| Maternal education (% with any secondary education) | 18 (72%) | 14 (70%) | 1.00 |
| Diarrhea episode before receipt of first rotavirus vaccine dose | 1 (4%) | 2 (10%) | 0.58 |
| High EE Score (% yes)† | 4 (16%) | 9 (45%) | 0.05 |
| Birth weight (g, SD) | 3,137.6 (313.1) | 3,046.5 (331.6) | 0.35 |
| Household characteristics | |||
| Household water source (% municipal piped water) | 25 (100) | 19 (95%) | 0.44 |
| Household sanitation (% indoor toilet) | 23 (92) | 15 (75%) | 0.21 |
| Floor type (% non-dirt floor) | 20 (80) | 12 (60%) | 0.19 |
| Animal(s) present in the home (% yes) | 17 (68) | 14 (70%) | 1.00 |
EE = environmental enteropathy. Baseline characteristics assessed at study enrollment.
* P-values assessed using Fisher’s exact test for dichotomous variables and difference of distributions for continuous variables.
† Children categorized as high EE had scores in the upper quartile.
Microbiome composition.
Comparisons between seroconverters and non-seroconverters did not yield statistically significant differences between Shannon diversity (3.70 versus 3.68 [P = 0.91]) and species richness scores (80.97 versus 90.13 [P = 0.24]). Also, based on PCoA, PERMANOVA (P = 0.88), and ANOSIM (P = 0.48) analyses, the relationship between seroconversion and microbiome composition was not significant.
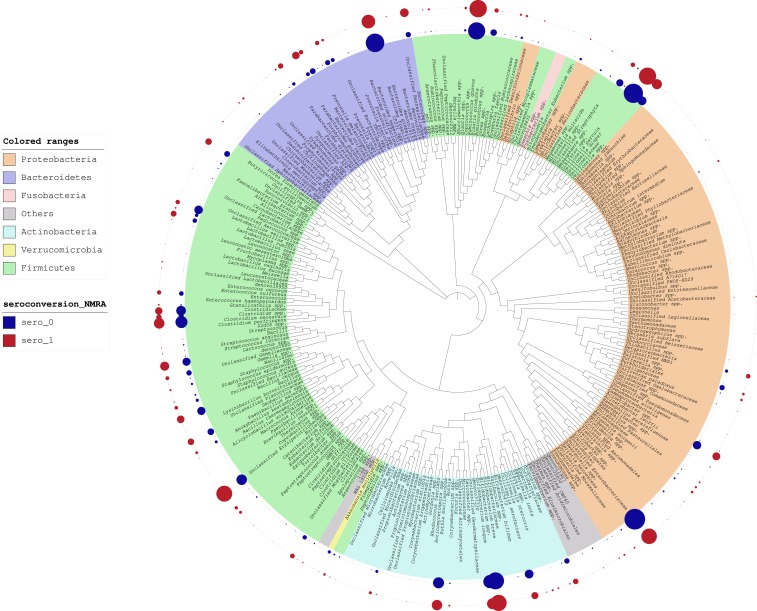
Upon inspection of the iTol table (Figure 1), the microbiome composition of fecal samples, represented as normalized MRA (NMRA),49 we identified apparent differences in NMRA between seroconverters and non-seroconverters; however, it is important to note that these differences do not reflect formal statistical comparisons. We observed a higher relative abundance of Bacteroides fragilis in the non-seroconverters; however, relative abundance of B. caccae, which is also a member of the B. fragilis group, was higher in the seroconverters. A notably higher abundance of bacteria belonging to the Enterobacteriaceae family was also observed among non-seroconverters. In addition, we observed a higher abundance of beneficial microbes, including Lactobacillus spp., B. longum, and Bifidobacterium bifidum, in non-seroconverters. Among the non-seroconverters, we observed an increase in NMRA of Akkermansia muciniphila. Seroconverters exhibited a higher NMRA of a potentially pathogenic bacterium, Eggerthella lenta.50
Figure 1.
Interactive Tree of Life analysis of relative abundance of bacteria comparing RV seroconverters vs. non-seroconverters. Relative abundance is depicted as the size of circles aligned with the noted bacterial taxa. Relative abundance of bacterial taxa among non-seroconverters and seroconverters is reflected in the inner blue and red rings, respectively.
Seroconverters had a lower relative abundance of Proteobacteria (P = 0.059, FDR adjusted P = 0.32) compared with non-seroconverters, although this result did not attain statistical significance. In addition, seroconverters were more likely to have a higher relative abundance of Fusobacteria (P = 0.064, FDR adjusted P = 0.32). Seroconverters were more likely to have a lower relative abundance of family Enterobacteriaceae (P = 0.0299, FDR adjusted P = 1) and higher relative abundance of genus Eggerthella (P = 0.0208, FDR adjusted P = 1) (Table 2). However, these comparisons did not reach statistical significance after FDR correction. The Wilcoxon–Mann–Whitney analysis did not account for confounding, and analyses to adjust for confounding were not conducted because of limited sample size.
Table 2.
Selected associations between relative abundance of bacteria and seroresponse to RV5 vaccination*
| Bacteria | Responders, MRA (SD) | Nonresponders, MRA (SD) | Wilcoxon–Mann–Whitney 2-sided P-value | FDR-corrected P-value |
|---|---|---|---|---|
| Phylum | ||||
| Proteobacteria | 0.040022 (0.032079) | 0.092023 (0.108095) | 0.06 | 0.31 |
| Fusobacteria | 0.004647 (0.021920) | 0.000009 (0.000026) | 0.06 | 0.31 |
| Actinobacteria | 0.174979 (0.167543) | 0.141047 (0.156533) | 0.31 | 0.85 |
| Thermi | 0.000009 (0.000031) | 0.000012 (0.000053) | 0.34 | 0.85 |
| Tenericutes | 0.000001 (0.000007) | 0.000000 (0.000000) | 0.56 | 0.92 |
| TM7 | 0.000113 (0.000405) | 0.000106 (0.000308) | 0.63 | 0.92 |
| Cyanobacteria | 0.000003 (0.000010) | 0.000004 (0.000013) | 0.67 | 0.92 |
| Verrucomicrobia | 0.000077 (0.000318) | 0.002436 (0.006906) | 0.83 | 0.92 |
| Firmicutes | 0.502032 (0.239870) | 0.482842 (0.268181) | 0.83 | 0.92 |
| Bacteroidetes | 0.277153 (0.244999) | 0.280203 (0.273953) | 0.97 | 0.97 |
| Genera | ||||
| Eggerthella | 0.000139 (0.000312) | 0.000007 (0.000020) | 0.02 | 1.00 |
| Enterobacteriaceae† | 0.015892 (0.020168) | 0.036539 (0.045746) | 0.03 | 1.00 |
| Megamonas | 0.011465 (0.057317) | 0.000794 (0.003480) | 0.05 | 1.00 |
| Roseburia | 0.000012 (0.000029) | 0.000000 (0.000000) | 0.06 | 1.00 |
| Fusobacterium | 0.004644 (0.021921) | 0.000008 (0.000026) | 0.06 | 1.00 |
| Parabacteroides | 0.017832 (0.036384) | 0.004744 (0.013960) | 0.07 | 1.00 |
| Erysipelotrichaceae† | 0.062857 (0.118074) | 0.009887 (0.017497) | 0.09 | 1.00 |
| Xanthomonadaceae† | 0.000036 (0.000128) | 0.000002 (0.000005) | 0.09 | 1.00 |
| Sutterella | 0.005797 (0.018688) | 0.015933 (0.031513) | 0.10 | 1.00 |
FDR = false discovery rate; MRA = mean relative abundance.
* Includes Wilcoxon–Mann–Whitney results for all phyla but only selected genera that had a P-value of < 0.10 after applying a FDR correction.
† The 16s rRNA amplicon sequencing approach used could not distinguish below the family level for Enterobacteriaceae, Erysipelotrichaceae, and Xanthomonadaceae.
DISCUSSION
We examined the microbiome composition among 45 Nicaraguan infants who did versus those who did not seroconvert following the first dose of RV5. Pre- and post-vaccination rotavirus-specific antibody titers were assessed to dichotomously categorize seroresponse and then relative abundance of bacteria in the gut were compared with identify taxa that were associated with response.
This study is the first to analyze associations between gut microbiome composition and response to RV5. Prior analyses included infants vaccinated with monovalent RV, which may have implications regarding the comparability of findings. Our results provide information from an additional world region on the association between the gut microbiome and response to oral RV. Our findings agree with an examination of the microbiota among children in India and Ghana, which also showed minimal difference in the microbiome between infants who did versus did not seroconvert.26,51
At the phylum level, Proteobacteria tended to have greater abundance among infants who did not seroconvert to the rotavirus vaccine, but this difference was not statistically significant. Although this conflicts with the results of a previous study conducted among Pakistani infants that found higher levels of Proteobacteria among seroconverters, those results were similarly not significant after applying an FDR correction.27 A higher relative abundance of the enteropathogen-rich phylum Proteobacteria among RV nonresponders is biologically relevant, as increased abundance of Proteobacteria has been associated with lower response to other oral and parenteral vaccines because of lowered humoral and cellular immunity.23,52
Our analysis at the phyla and genus level did not identify differences in bacterial taxa after adjustment for multiple comparisons, which may reflect the sample and effect sizes. Of note, the family Enterobacteriaceae tended to be higher among infants who did not seroconvert to the vaccine as compared with seroconverters. Enterobacteriaceae is a family of Gram-negative bacteria in the phylum Proteobacteria that includes several pathogens, including species of Salmonella, Shigella, and diarrheagenic E. coli. Differences in abundance of Enterobacteriaceae may be the drivers of observed differences among Proteobacteria at the phylum level. The 16S rRNA amplicon sequencing approach does not typically provide identification down to the species level to distinguish whether these specific pathogens differed between groups. Interestingly, Parker, et al.51 found that at the time of the first dose of monovalent RV, seroconverters were more likely to have an enteropathogen, as detected by TaqMan Array Cards, although no individual comparisons of enteropathogens were found to be statistically significant in their study.
Through inspection of the iTol table, we identified potential differences in the NMRA between seroconverters and non-seroconverters with regards to A. muciniphila, which was more abundant in non-seroconverters. There is increasing evidence that this bacterium has probiotic properties with a role in improving gut barrier integrity and insulin resistance.53,54 We also found that the genus Eggerthella of the family Coriobacteriaceae had greater abundance among seroconverters than non-seroconverters. Eggerthella is a Gram-positive anaerobic bacillus that has been implicated as a cause of bacteremia in patients with abdominal pathology. Finally, we observed higher abundance of B. fragilis in non-seroconverters, an anaerobic pathogen which can cause intra-abdominal infections and bacteremia in humans.55
One limitation of this study was the sample size, which paired with a small effect size resulted in insufficient power to detect differences between seroconverters and non-seroconverters after correction for multiple comparisons. Furthermore, the sample size limitation restricted controlling for potential confounding factors, including participant demographics or other characteristics (EE score, breastfeeding, delivery mode, and nutritional status of both mother and infant). As we have reported previously, having a high EE score was associated with poor RV5 response.30 Our analysis may have also benefitted from a full assessment of enteric infections, which have been demonstrated to interfere with response to oral vaccines22; however, the enteric virome was not assessed as part of our study. Prior analyses overcame this problem through 1:1 matching of responders to nonresponders on relevant variables, which was not possible in this cohort. An additional potential limitation is that the infant microbiome composition was characterized using a single stool sample collected at enrollment. The use of 16S metagenomics, as opposed to whole genome sequencing, may have limited the ability to identify important differences in microbiome composition. Given the numerous factors that contribute to gut microbiome composition, it is possible the snapshot characterization of each participant’s microbiome may not be a perfect representation of the true bacterial composition present during the time that vaccine response is mounted. Longitudinal assessment of microbiome composition over a period leading up to vaccination may provide a more accurate characterization of microbiota composition.
Our study identified no significant differences in the microbiome composition between RV seroconverters and non-seroconverters. The observed differences should be considered in future research on the impact of gut microbiota on RV response. Additional research into the contribution of gut microbiome to vaccine response may provide an explanation for differing performance of rotavirus vaccines around the world, potentially creating a path for further improvements to vaccine efficacy and effectiveness.
REFERENCES
- 1.Liu L, et al. Child Health Epidemiology Reference Group of WHO, Unicef , 2012. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet 379: 2151–2161. [DOI] [PubMed] [Google Scholar]
- 2.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD; World Health Organization-Coordinated Global Rotavirus Surveillance Network , 2016. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin Infect Dis 62 (Suppl 2): S96–S105. [DOI] [PubMed] [Google Scholar]
- 3.Kotloff KL, et al. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, GEMS): a prospective, case-control study. Lancet 382: 209–222. [DOI] [PubMed] [Google Scholar]
- 4.Vesikari T, et al. 2006. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med 354: 23–33. [DOI] [PubMed] [Google Scholar]
- 5.Armah GE, et al. 2010. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet 376: 606–614. [DOI] [PubMed] [Google Scholar]
- 6.Zaman K, et al. 2010. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. Lancet 376: 615–623. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz-Palacios GM, et al. 2006. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med 354: 11–22. [DOI] [PubMed] [Google Scholar]
- 8.Vesikari T, Karvonen A, Prymula R, Schuster V, Tejedor JC, Cohen R, Meurice F, Han HH, Damaso S, Bouckenooghe A, 2007. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. Lancet 370: 1757–1763. [DOI] [PubMed] [Google Scholar]
- 9.Madhi SA, et al. 2010. Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med 362: 289–298. [DOI] [PubMed] [Google Scholar]
- 10.Linhares AC, et al. 2008. Efficacy and safety of an oral live attenuated human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in Latin American infants: a randomised, double-blind, placebo-controlled phase III study. Lancet 371: 1181–1189. [DOI] [PubMed] [Google Scholar]
- 11.Bhandari N, et al. 2014. Efficacy of a monovalent human-bovine (116E) rotavirus vaccine in Indian infants: a randomised, double-blind, placebo-controlled trial. Lancet 383: 2136–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isanaka S, et al. 2017. Efficacy of a low-cost, heat-stable oral rotavirus vaccine in Niger. N Engl J Med 376: 1121–1130. [DOI] [PubMed] [Google Scholar]
- 13.Murgas Torrazza R, Neu J, 2011. The developing intestinal microbiome and its relationship to health and disease in the neonate. J Perinatol 31 (Suppl 1): S29–S34. [DOI] [PubMed] [Google Scholar]
- 14.Brestoff JR, Artis D, 2013. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol 14: 676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atarashi K, et al. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL, 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118. [DOI] [PubMed] [Google Scholar]
- 17.Neu J, Rushing J, 2011. Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clin Perinatol 38: 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey PK, Verma P, Kumar H, Bavdekar A, Patole MS, Shouche YS, 2012. Comparative analysis of fecal microflora of healthy full-term Indian infants born with different methods of delivery (vaginal vs cesarean): Acinetobacter sp. prevalence in vaginally born infants. J Biosci 37: 989–998. [DOI] [PubMed] [Google Scholar]
- 19.O’Sullivan A, Farver M, Smilowitz JT, 2015. The influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutr Metab Insights 8: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimura KE, Slusher NA, Cabana MD, Lynch SV, 2010. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther 8: 435–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korpe PS, Petri WA, Jr., 2012. Environmental enteropathy: critical implications of a poorly understood condition. Trends Mol Med 18: 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker EP, Ramani S, Lopman BA, Church JA, Iturriza-Gomara M, Prendergast AJ, Grassly NC, 2018. Causes of impaired oral vaccine efficacy in developing countries. Future Microbiol 13: 97–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB, 2014. Stool microbiota and vaccine responses of infants. Pediatrics 134: e362–e372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Praharaj I, et al. 2019. Influence of nonpolio enteroviruses and the bacterial gut microbiota on oral poliovirus vaccine response: a study from south India. J Infect Dis 219: 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huda MN, Ahmad SM, Alam MJ, Khanam A, Kalanetra KM, Taft DH, Raqib R, Underwood MA, Mills DA, Stephensen CB, 2019. Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics 143: e20181489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris VC, et al. 2017. Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural Ghana. J Infect Dis 215: 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris V, et al. 2017. Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes 9:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Twitchell EL, et al. 2016. Modeling human enteric dysbiosis and rotavirus immunity in gnotobiotic pigs. Gut Pathog 8: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uchiyama R, Chassaing B, Zhang B, Gewirtz AT, 2014. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J Infect Dis 210: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker-Dreps S, Vilchez S, Bucardo F, Twitchell E, Choi WS, Hudgens MG, Perez J, Yuan L, 2017. The association between fecal biomarkers of environmental enteropathy and rotavirus vaccine response in Nicaraguan infants. Pediatr Infect Dis J 36: 412–416. [DOI] [PubMed] [Google Scholar]
- 31.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R, 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108 (Suppl 1): 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allali I, et al. 2017. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol 17: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Illumina Inc. , 2016. Bcl2Fastq 2.18.0.12. San Diego, CA. [Google Scholar]
- 34.Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aronesty E, 2013. Comparison of sequencing utility programs. Open Bioinform J 7: 1–8. [Google Scholar]
- 36.Babraham Institute , 2014. FastQC 0.11.2. Cambridge, United Kingdom. [Google Scholar]
- 37.Edgar RC, 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- 38.Rognes T, Flouri T, Nichols B, Quince C, Mahe F, 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4: e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas BJ, et al. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL, 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R, 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price MN, Dehal PS, Arkin AP, 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozupone C, Hamady M, Knight R, 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinf. 7: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lozupone C, Knight R, 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dabdoub SM, Fellows ML, Paropkari AD, Mason MR, Huja SS, Tsigarida AA, Kumar PS, 2016. PhyloToAST: bioinformatics tools for species-level analysis and visualization of complex microbial datasets. Sci Rep 6: 29123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kearse M, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letunic I, Bork P, 2007. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128. [DOI] [PubMed] [Google Scholar]
- 48.Letunic I, Bork P, 2011. Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Letunic I, Bork P, 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gardiner BJ, Tai AY, Kotsanas D, Francis MJ, Roberts SA, Ballard SA, Junckerstorff RK, Korman TM, 2015. Clinical and microbiological characteristics of Eggerthella lenta bacteremia. J Clin Microbiol 53: 626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parker EPK, et al. 2018. Influence of the intestinal microbiota on the immunogenicity of oral rotavirus vaccine given to infants in south India. Vaccine 36: 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimmermann P, Curtis N, 2018. The influence of the intestinal microbiome on vaccine responses. Vaccine 36: 4433–4439. [DOI] [PubMed] [Google Scholar]
- 53.Naito Y, Uchiyama K, Takagi T, 2018. A next-generation beneficial microbe: Akkermansia muciniphila. J Clin Biochem Nutr 63: 33–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chelakkot C, et al. 2018. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med 50: e450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aldridge KE, Ashcraft D, O’Brien M, Sanders CV, 2003. Bacteremia due to Bacteroides fragilis group: distribution of species, beta-lactamase production, and antimicrobial susceptibility patterns. Antimicrob Agents Chemother 47: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]