Abstract
Prostate cancer (PCa) is the most frequent urological malignancy in men worldwide. DNA methylation has an essential role in the etiology and pathogenesis of PCa. The purpose of the present study was to identify the aberrantly methylated-differentially expressed genes and to determine their potential roles in PCa. The important node genes identified were screened by integrated analysis. Gene expression microarrays and gene methylation microarrays were downloaded and aberrantly methylated-differentially expressed genes were obtained. Enrichment analysis and protein-protein interactions (PPI) were obtained, their interactive and visual networks were created, and the node genes in the PPI network were validated. A total of 105 hypomethylation-high expression genes and 561 hypermethylation-low expression genes along with their biological processes were identified. The top 10 node genes obtained from the PPI network were identified for each of the two gene groups. The methylation and gene expression status of node genes in TCGA database, GEPIA tool, and the HPA database were generally consistent with those of our results. In conclusion, the present study identified 20 aberrantly methylated-differentially expressed genes in PCa by combining bioinformatics analyses of gene expression and gene methylation microarrays, and concurrently, the survival of these genes was analyzed. Notably, methylation is a reversible biological process, which makes it of great biological significance for the diagnosis and treatment of prostate cancer using bioinformatics technology to determine abnormal methylation gene markers. The present study provided novel therapeutic targets for the treatment of PCa.
Keywords: PCa, methylation, DEGs, bioinformatics, node genes
Introduction
Globally, prostate cancer (PCa) is the second most frequent cancer. It is the fifth leading cause of cancer-related deaths in males, and is the most frequently diagnosed cancer among men in more than half of all countries (1). It has been established that age, race, and family history are risk factors associated with PCa (2). Previous studies have implicated the initiation and development of PCa as a multistep complex process, driven by changes in the expression of a large number of genes with epigenetic alterations influencing the expression of crucial genes (3,4).
Among the epigenetic modifications, methylation, phosphorylation, acetylation, and ubiquitination have been identified (5,6). A study by De Carvalho et al (7) revealed that DNA methylation can genetically alter gene expression without a change in the DNA sequence. Hypermethylation of a promoter may downregulate gene expression and influence the progression of human cancer (8). Recently, studies have revealed that DNA methylation can identify invasive lesions and silence tumor suppressor genes in PCa, providing a new direction for the treatment of PCa (9,10).
Bioinformatics analysis based on high-throughput platform microarray technology has been extensively used to predict biomarkers of cancers over the last few decades (11–13). Numerous gene expression microarrays have been used to identify potential target genes and their functions in PCa (14–16). However, the aforementioned studies focused on gene expression microarrays, the number of which is limited, preventing the accurate identification of target genes and their functions in PCa. Therefore, an approved approach includes the combination of gene expression and gene methylation microarray data.
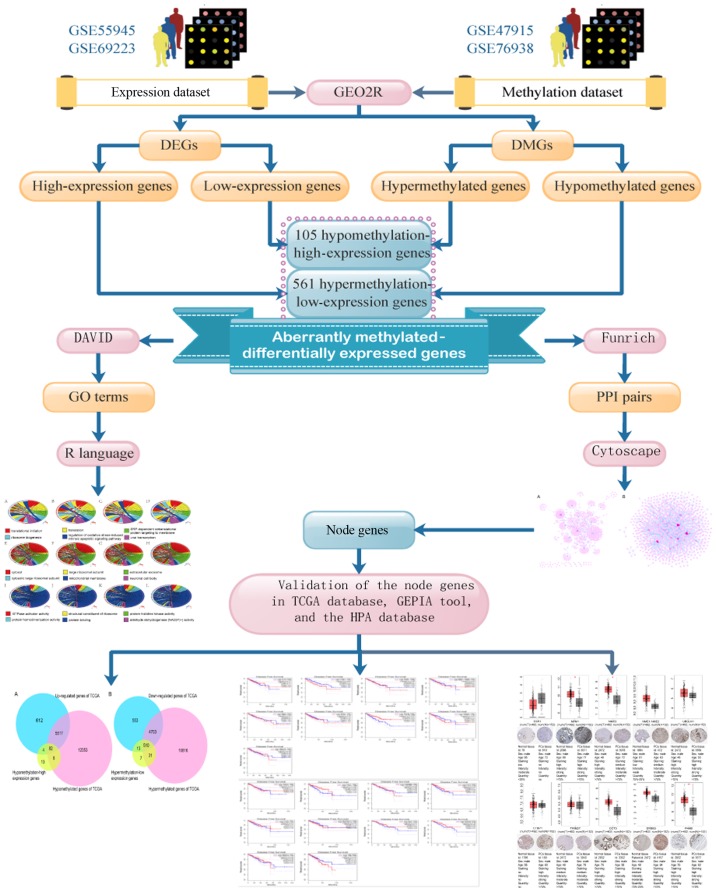
The purpose of this study was to identify aberrantly methylated-differentially expressed genes based on gene expression and gene methylation microarray datasets. The important node genes were screened by integrated analysis with the goal of identifying a novel therapeutic target for the treatment of PCa. The screening steps for determining the aberrantly methylated-differentially expressed genes in PCa are summarized in Fig. 1.
Figure 1.
Flow chart of aberrantly methylated-differentially expressed genes in prostate cancer. DEGs, differentially expressed genes; DMGs, differentially methylated genes; GO, Gene Ontology; PPI, protein-protein interactions; DAVID, Database for Annotation, Visualization, and Integrated Discovery; TCGA, The Cancer Genome Atlas; GEPIA, Gene Expression Profiling Interactive Analysis; HPA, Human Protein Atlas.
Materials and methods
Data sources
In the present study, the raw data were selected from the Gene Expression Omnibus (GEO), which is an international public repository that can be found on the National Center for Biotechnology Information (NCBI) home page (https://www.ncbi.nlm.nih.gov/geo/). Microarray gene expression data found at accession GSE55945 involved data from 13 PCa samples and eight normal samples, and accession GSE69223 encompassed 15 PCa samples and 15 normal samples, with the platform GPL570 of the two datasets ([HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array). Methylation profile data in GSE47915 comprised four PCa samples and four normal samples, while GSE76938 contained 73 PCa samples and 63 normal samples. The platform of both datasets (GSE47915 and GSE76938) was based on GPL13534 (Illumina HumanMethylation450 BeadChip).
Data processing
The raw data analysis was carried out using GEO2R, which can separately screen differentially methylated genes (DMGs) and differentially expressed genes (DEGs) between normal and cancer prostate sample datasets (17). DMGs and DEGs were obtained using the criteria|t|>2 and P<0.05. The intersection of DMGs and DEGs was derived using the FunRich Venn function (http://www.funrich.org) (18), followed by obtaining the hypomethylation-high expression genes and hypermethylation-low expression genes.
Gene ontology (GO) term enrichment analysis
The GO terms, including the hypomethylation-high expression genes and hypermethylation-low expression genes, were enriched using the Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.niaid.nih.gov), and P-values <0.05 were considered statistically significant. The chord plots from the GO results were created using R language with ggplot2 and GOplot packages (19).
Construction of PPI networks
Protein-protein interactions (PPI) are critical events in signaling pathways, especially when interpreting the molecular mechanisms of cellular activities during carcinogenesis. The PPI relationships of the hypomethylation-high expression genes and hypermethylation-low expression genes were obtained by FunRich, and their interactive and visual networks were created using Cytoscape v3.5.0 software (https://cytoscape.org/) (20).
Node gene validation
Gene expression profiling data (HTSeq-FPKM) and methylation sequencing data (Illumina Human Methylation 450) were downloaded from the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). DMGs and DEGs were obtained using the criteria |t|>2 and P<0.05. The hypomethylated-high expression genes and hypermethylated-low expression genes obtained from the GEO database were respectively validated in TCGA database, and the methylation and gene expression status of the top ten node genes were also validated. Subsequently, the expression status, translational levels and the mRNA levels of the top ten node genes were validated using the Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn) and The Human Protein Atlas database (HPA; http://www.proteinatlas.org/), respectively. In addition, the Disease-Free Survival (RFS) method of the GEPIA online tool was used to analyze 20 aberrantly-methylated genes including hypomethylated-high expression genes and hypermethylated-low expression genes. Group cutoff was set as Quartile (Cutoff-High (%) was 75% and Cutoff-Low (%) was 25%). The confidence interval (CU) was 95%. High and low expression genes were respectively represented in red and blue colour (21,22).
Results
Methylated DEGs in PCa screening results
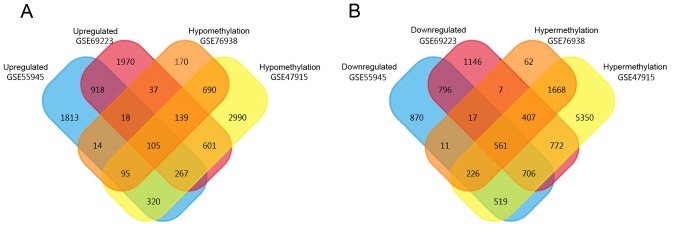
A total of 3,550 high-expression and 3,706 low-expression genes were obtained from GSE55945, while 4,055 high-expression and 4,412 low-expression genes were obtained from GSE69223. In addition, 10,209 hypermethylated and 5,207 hypomethylated genes were obtained from GSE47915 and 2,959 hypermethylated and 1,268 hypomethylated genes were obtained from GSE76938 by GEO2R. Using the FunRich Venn function, 105 hypomethylation-high expression genes and 561 hypermethylation-low expression genes were identified, as revealed in Fig. 2.
Figure 2.
Identification of aberrantly methylated-differentially expressed genes in prostate cancer. (A) Hypomethylation-high expression genes. (B) Hypermethylation-low expression genes.
GO term enrichment analysis
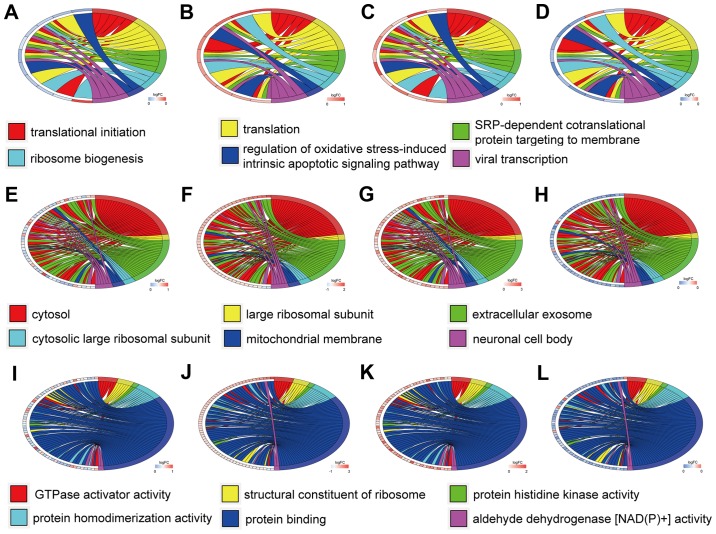
The top six GO annotation results of aberrantly methylated-differentially expressed genes are summarized in Table I. The ontology enrichment analysis of hypomethylation-high expression genes is presented in Fig. 3 in the form of cluster chord diagrams. The diagrams of cluster chord A-D, E-F and I-L respectively show the biological processes, cellular components and molecular functions of GO analysis. The four columns of chord diagrams include the first column including A, E and I, the second column including B, F and J, the third column including C, G and K, and the fourth column including D, H and L respectively enriched the GO of GSE47915, GSE55945, GSE69223 and GSE769938 datasets. Similarly, Fig. 4 reveals the enrichment of hypermethylation-low expression genes. Among the hypomethylation-high expression genes, the biological processes (BP) were mainly associated with translational initiation, translation, SRP-dependent co-translational protein targeting to membrane, ribosome biogenesis, regulation of oxidative stress-induced intrinsic apoptotic signaling pathways, and viral transcription. The major cell components (CC) included: Cytosol, large ribosomal subunit, extracellular exosome, cytosolic large ribosomal subunit, mitochondrial membrane, and neuronal cell body, with the most important being the cytosol. The molecular functions (MF) were primarily focused on GTPase activator activity, ribosome structure, protein histidine kinase activity, protein homodimerization activity, protein binding, and aldehyde dehydrogenase [NAD(P)+] activity.
Table I.
GO analysis of aberrantly methylated-differentially expressed genes in prostate cancer.
| A, Hypomethylation and high expression | ||||
|---|---|---|---|---|
| GO analysis | Term | Gene count | Percentage (%) | P-value |
| BP | Translational initiation | 5 | 4.76 | 5.15×10−3 |
| BP | Translation | 6 | 5.71 | 9.21×10−3 |
| BP | SRP-dependent co-translational protein targeting to membrane | 4 | 3.81 | 1.21×10−2 |
| BP | Ribosome biogenesis | 3 | 2.86 | 1.43×10−2 |
| BP | Regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 2 | 1.90 | 1.51×10−2 |
| BP | Viral transcription | 4 | 3.81 | 1.92×10−2 |
| CC | Cytosol | 34 | 32.38 | 1.94×10−5 |
| CC | Large ribosomal subunit | 3 | 2.86 | 2.07×10−3 |
| CC | Extracellular exosome | 25 | 23.81 | 3.56×10−3 |
| CC | Cytosolic large ribosomal subunit | 4 | 3.81 | 4.49×10−3 |
| CC | Mitochondrial membrane | 4 | 3.81 | 1.10×10−2 |
| CC | Neuronal cell body | 6 | 5.71 | 1.91×10−2 |
| MF | GTPase activator activity | 7 | 6.67 | 2.67×10−3 |
| MF | Structural constituent of ribosome | 6 | 5.71 | 5.00×10−3 |
| MF | Protein histidine kinase activity | 2 | 1.90 | 9.93×10−3 |
| MF | Protein homodimerization activity | 10 | 9.52 | 1.03×10−2 |
| MF | Protein binding | 55 | 52.38 | 1.56×10−2 |
| MF | Aldehyde dehydrogenase [NAD(P)+] activity | 2 | 1.90 | 2.95×10−2 |
| B, Hypermethylation and low expression | ||||
| BP | Cell adhesion | 39 | 6.96 | 2.49×10−8 |
| BP | Extracellular matrix organization | 23 | 4.11 | 1.46×10−7 |
| BP | Response to hypoxia | 18 | 3.21 | 2.09×10−5 |
| BP | Muscle contraction | 14 | 2.50 | 2.24×10−5 |
| BP | Positive regulation of cell migration | 18 | 3.21 | 4.97×10−5 |
| BP | Regulation of phosphatidylinositol 3-kinase signaling | 11 | 1.96 | 1.25×10−4 |
| CC | Focal adhesion | 53 | 9.46 | 6.76×10−20 |
| CC | Z disc | 22 | 3.93 | 3.87×10−11 |
| CC | Proteinaceous extracellular matrix | 32 | 5.71 | 1.08×10−10 |
| CC | Extracellular matrix | 30 | 5.36 | 2.00×10−8 |
| CC | Actin cytoskeleton | 25 | 4.46 | 3.70×10−8 |
| CC | Cytoskeleton | 32 | 5.71 | 2.49×10−7 |
| MF | Actin binding | 28 | 5.00 | 1.03×10−7 |
| MF | Integrin binding | 15 | 2.68 | 3.34×10−6 |
| MF | Collagen binding | 11 | 1.96 | 1.15×10−5 |
| MF | Actin filament binding | 15 | 2.68 | 4.73×10−5 |
| MF | Signal transducer activity | 17 | 3.04 | 4.92×10−4 |
| MF | Scaffold protein binding | 8 | 1.43 | 5.61×10−4 |
GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
Figure 3.
Gene Ontology analysis of the hypomethylation-high expression genes. Chord plot revealing a circular dendrogram of the clustering of the expression profiles. Cluster revealing genes and their (A-D) assigned biological process, (E-H) cellular component and (I-L) molecular function terms are connected by ribbons. Blue to red on the left side of the chord plot represent logFC. Each color from red to purple on the right side of the chord plot represents a different term. A, E and I reveal the expression in the methylated GSE47915 dataset. B, F and J reveal the expression in the GSE55945 dataset. C, G and K reveal the expression in the GSE69223 dataset. D, H and L reveal the expression in the methylated GSE76938 dataset.
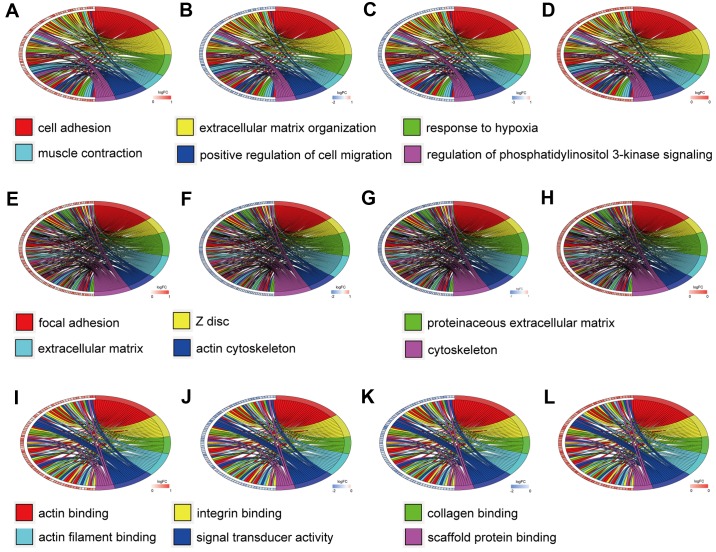
Figure 4.
Gene Ontology analysis of the hypermethylation-low expression genes. Chord plot revealing a circular dendrogram of the clustering of the expression profiles. Cluster revealing genes and their assigned (A-D) biological process, (E-H) cellular component and (I-L) molecular function terms are connected by ribbons. Blue to red on the left side of the chord plot represent logFC. Each color from red to purple on the right side of the chord plot represents a different term. A, E and I reveal the expression in the methylated GSE47915 dataset. B, F and J reveal the expression in the GSE55945 dataset. C, G and K reveal the expression in the GSE69223 dataset. D, H and L reveal the expression in the methylated GSE76938 dataset.
For the hypermethylation-low expression genes, cell adhesion, extracellular matrix organization, response to hypoxia, muscle contraction, positive regulation of cell migration, and regulation of phosphatidylinositol 3-kinase signaling were the predominant BP. The CC of these genes were primarily distributed in focal adhesions, Z discs, proteinaceous extracellular matrices, other extracellular matrices, and the cytoskeleton. The MF mainly included actin binding, integrin binding, collagen binding, actin filament binding, signal transducer activity, and scaffold protein binding.
Selection of PPI network node genes
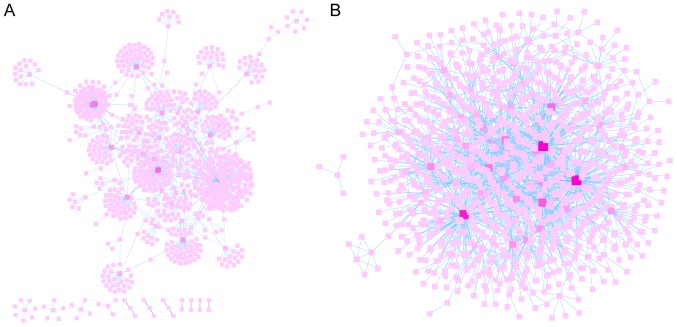
FunRich was used to predict the relationships between genes and proteins of the hypomethylation-high expression and hypermethylation-low expression gene groups. Subsequently, their interaction networks were visualized by Cytoscape v3.5.0 software as revealed Fig. 5A and B. Cytoscape is a software focused on open source web visualization and analysis. Its core is to provide the basic functional layout and query network, and is based on the combination of basic data into a visual network. Derived from systems biology, Cytoscape is used to integrate biomolecular interaction networks with high-throughput gene expression data and other molecular state information. Its most powerful function is for the analysis of large-scale PPIs, protein-DNA and genetic interactions (21). The top ten node genes in the hypomethylation-high expression and hypermethylation-low expression gene groups are listed separately in Tables II and III, based on the degree of distribution of the nodes.
Figure 5.
PPI network of the aberrantly methylated-differentially expressed genes. (A) PPI network of the hypomethylation-high expression genes. (B) PPI network of the hypermethylation-low expression genes. The distribution degree of nodes is indicated by the color of the square blocks, and the darkness of the color indicates the significance of the nodes. PPI, protein-protein interaction.
Table II.
Top ten node genes in the hypomethylation-high expression genes.
| Name | Degree |
|---|---|
| ESR1 | 234 |
| NPM1 | 81 |
| NME2 | 77 |
| NME1-NME2 | 64 |
| UBQLN1 | 47 |
| CTBP1 | 45 |
| TRIM27 | 41 |
| CCT3 | 40 |
| ERBB3 | 35 |
| P4HB | 33 |
Table III.
Top ten node genes in the hypermethylation-low expression genes.
| Name | Degree |
|---|---|
| CREBBP | 160 |
| SMAD3 | 159 |
| MCC | 142 |
| VIM | 92 |
| ATXN1 | 79 |
| CAV1 | 68 |
| FLNA | 68 |
| MAP3K14 | 55 |
| PRKCB | 53 |
| FHL2 | 52 |
Validation of the node genes
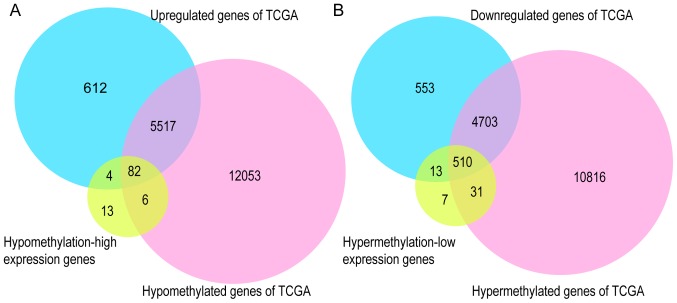
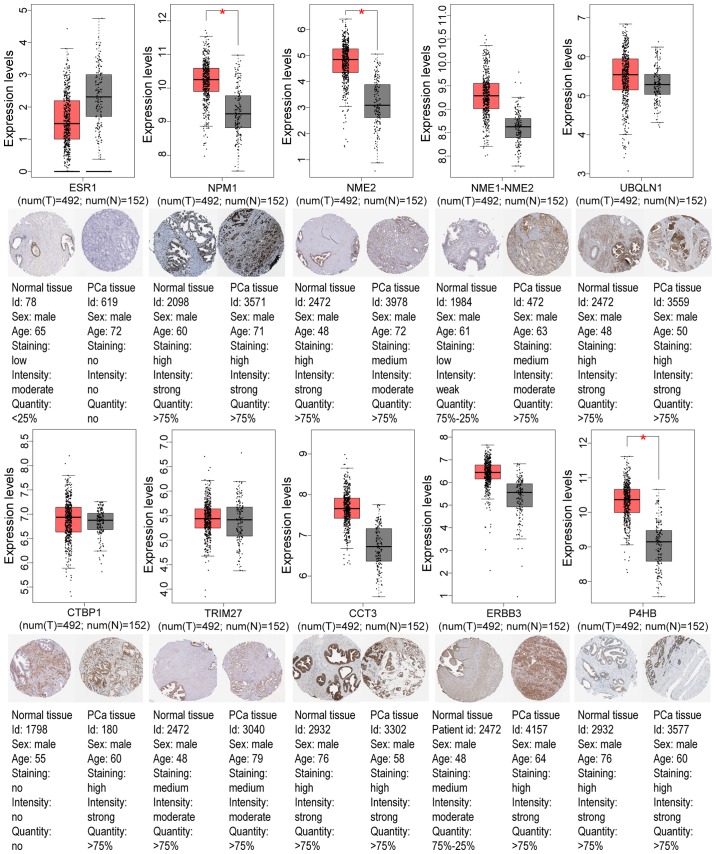
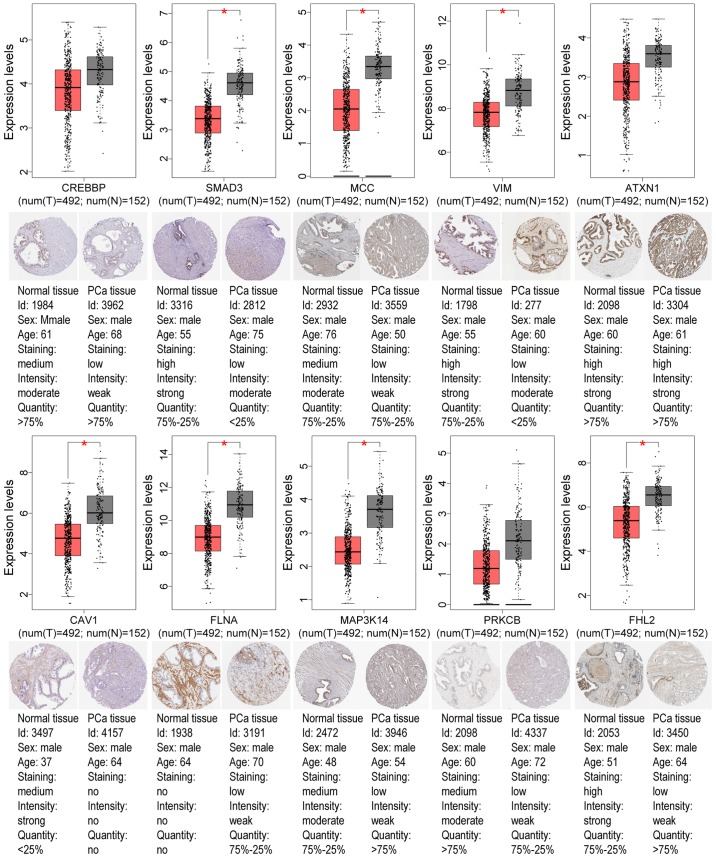
To further validate the present results, TCGA database, the GEPIA tool, and the HPA database were employed. Venn diagram analysis of the GEO database and TCGA database is presented in Fig. 6, from which it can be observed that most of the aberrantly methylated and expressed genes in GEO were contained within TCGA dataset. The methylation and gene expression status of the top ten node genes were similar between GEO and TCGA databases, as revealed in Tables IV and V. Using the GEPIA tool and HPA database, the expression of node genes in tumor and normal prostate samples were further verified and the immunohistochemical staining combined with patient information, node gene expression, and the mRNA levels, respectively, were obtained, as revealed in Figs. 7 and 8 and Tables II and III. In Figs. 7 and 8, immunohistochemical images and data were generated by the online GEPIA tool, log2(TPM+1) for log-scale was used to express gene expression on the y-axis. Red and gray were set as the colors of tumor and normal data sets respectively. The size of jitter across the box was as 0.4. It was revealed that the expression of node genes was generally consistent with previous results.
Figure 6.
Validation of the aberrantly methylated-differentially expressed genes in the TCGA database. (A) Hypomethylation-high expression genes. (B) Hypermethylation-low expression genes. TCGA, The Cancer Genome Atlas.
Table IV.
Validation of the top ten hypomethylation-high expression genes in TCGA database.
| Gene | Methylation status | P-value | Expression status | P-value |
|---|---|---|---|---|
| ESR1 | Hypomethylation | 3.70×10−143 | Upregulated | 1.20×10−1 |
| NPM1 | Hypomethylation | 5.51×10−119 | Upregulated | 3.12×10−15 |
| NME2 | Hypomethylation | 2.76×10−119 | Upregulated | 1.71×10−5 |
| NME1-NME2 | Hypomethylation | 6.35×10−132 | Upregulated | 6.43×10−9 |
| UBQLN1 | Hypomethylation | 4.24×10−117 | Upregulated | 8.87×10−1 |
| CTBP1 | Hypomethylation | 1.85×10−126 | Upregulated | 2.01×10−7 |
| TRIM27 | Hypomethylation | 5.11×10−129 | Upregulated | 5.39×10−27 |
| CCT3 | Hypomethylation | 1.92×10−129 | Upregulated | 2.40×10−18 |
| ERBB3 | Hypomethylation | 1.46×10−121 | Upregulated | 2.89×10−18 |
| P4HB | Hypomethylation | 2.12×10−119 | Upregulated | 4.29×10−18 |
TCGA, The Cancer Genome Atlas.
Table V.
Validation of the top ten hypermethylation-low expression genes in TCGA database.
| Gene | Methylation status | P-value | Expression status | P-value |
|---|---|---|---|---|
| CREBBP | Hypermethylation | 1.06×10−123 | Downregulated | 1.64×10−1 |
| SMAD3 | Hypermethylation | 6.27×10−121 | Downregulated | 1.34×10−10 |
| MCC | Hypermethylation | 2.39×10−126 | Downregulated | 2.13×10−20 |
| VIM | Hypermethylation | 1.11×10−134 | Downregulated | 1.26×10−5 |
| ATXN1 | Hypermethylation | 3.54×10−134 | Downregulated | 2.13×10−4 |
| CAV1 | Hypermethylation | 1.31×10−133 | Downregulated | 7.54×10−26 |
| FLNA | Hypermethylation | 8.64×10−126 | Downregulated | 1.12×10−15 |
| MAP3K14 | Hypermethylation | 2.39×10−123 | Downregulated | 1.79×10−7 |
| PRKCB | Hypermethylation | 1.24×10−133 | Downregulated | 6.56×10−20 |
| FHL2 | Hypermethylation | 1.74×10−123 | Downregulated | 1.71×10−12 |
TCGA, The Cancer Genome Atlas.
Figure 7.
Validation of the top ten hypomethylation-high expression genes. The boxplots reveal the expression levels of the top ten hypomethylation-high expression genes in PRAD tissues (n=492) and their corresponding normal samples (n=152) based on The Cancer Genome Atlas and Gene Expression Profiling Interactive Analysis databases. To express gene expression, on the y-axis, the log2(TPM+1) for log-scale was used. The red color was set for the tumor dataset. The red and gray boxes represent prostate tumor and normal tissues, respectively. The size of jitter across the box was set as 0.4. The staining patterns revealed immunohistochemical staining of these genes between PRAD samples and normal prostate samples. *P<0.05. PRAD, prostate adenocarcinoma; PCa, prostate cancer.
Figure 8.
Validation of the top ten hypermethylation-low expression genes. The boxplots reveal the expression levels of the top ten hypermethylation-low expression genes in PRAD tissues (n=492) and their corresponding normal samples (n=152) based on The Cancer Genome Atlas and Gene Expression Profiling Interactive Analysis databases. The red and gray boxes represent prostate tumor and normal tissues, respectively. The staining patterns revealed the immunohistochemical staining of these genes between PRAD samples and normal prostate samples. *P<0.05. PRAD, prostate adenocarcinoma; PCa, prostate cancer.
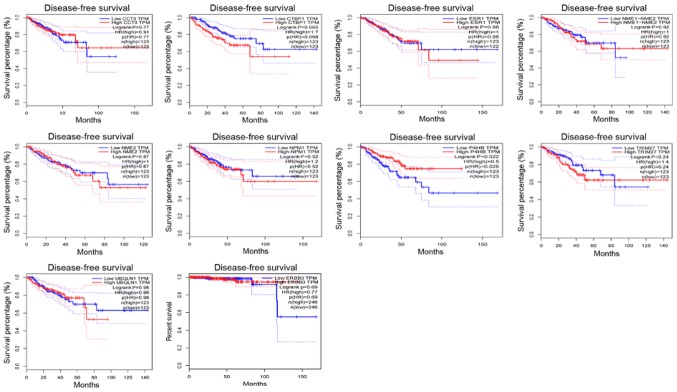
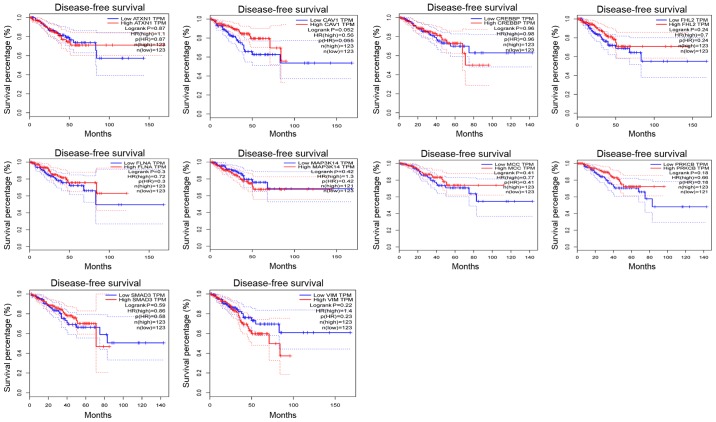
Survival analysis of the node genes
In addition, the Disease-Free Survival (RFS) method of GEPIA online tool was used to analyze 20 aberrantly-methylated genes. Group Cutoff was set as Quartile (Cutoff-High (%) was 75% and Cutoff-Low (%) was 25%). The CI was 95%. Hypomethylated-high expression genes (Fig. 9) and hypermethylated-low expression genes (Fig. 10) were respectively represented in red and blue colour. It was revealed that the survival analysis of the P4HB gene exhibited a statistically significant difference in survival between cancer and normal groups (log-rank P-value 0.022).
Figure 9.
Kaplan-Meier survival curves analysis of hypomethylation-high expression node genes. Kaplan-Meier curves were used to analyze the association of the signature genes with clinical outcomes.
Figure 10.
Kaplan-Meier survival curves analysis of the top 10 hypermethylation-low expression node genes. Kaplan-Meier curves were used to analyze the association of the genes signature with clinical outcomes.
P4HB is a key protein of disulfide isomerase (protein di-sulphideisomerase, PDI), which is also referred to as a post-translational modifications collagen synthase, that is involved in antioxidant and detoxification reactions. Its encoding gene is on chromosome 17 q25 (22,23). P4HB was revealed to be highly expressed in glioblastoma and liver cancer (24,25). Notably, Xia et al (25) revealed that high expression of P4HB in liver cancer tissues was associated with poor survival.
Discussion
In recent years, DNA methylation is a major part of epigenetic modification and plays an important role in maintaining chromosome stability and gene expression in mammals. It refers to the process of transferring methyl groups to specific bases catalyzed by DNA methyl transfer enzymes (DNMTs) using s-adenosine methionine (SAM) as the methyl donor. Increasing studies (26) have revealed that aberrant DNA methylation is intimately associated with tumorigenesis. It was revealed that the overall methylation level of DNA in tumor cells was lower than that in normal cells, however some specific gene CpG islands were hypermethylated (27). DNA hypomethylation activates proto-oncogenes and abnormal proliferation of cancer cells. However, hypermethylation of CpG islands in the promoter region can inhibit gene expression and inactivate tumor suppressor genes, thus promoting the occurrence and development of tumors. Therefore, methylation is considered to be another mechanism of tumorigenesis (28). DNA methylation is not only involved in the regulation of the cell cycle, proliferation, apoptosis and metastasis, but also in the regulation of drug resistance and intracellular signal transduction pathway of tumor cells (29).
At present, some progress has been made in genetics and in the molecular pathogenesis of PCa, however effective diagnosis and treatment of PCa still require further advances (30). In the present study, the gene expression and methylation datasets of PCa were analyzed using a variety of online tools. A total of 105 hypomethylation-high expression genes and 561 hypermethylation-low expression genes were obtained.
GO annotations of the hypomethylation-high expression gene group in PCa predominantly included translational initiation, translation, SRP-dependent co-translational protein targeting to the membrane, ribosome biogenesis, regulation of oxidative stress-induced intrinsic apoptotic signaling pathways, viral transcription, GTPase activator activity, structural constituents of ribosomes, protein histidine kinase activity, protein homodimerization activity, protein binding, and aldehyde dehydrogenase [NAD(P)+] activity. Multiple translation initiation factors were regularly magnified or diminished in tumors, promoting proliferation, survival, angiogenesis, and metastasis (31,32). Ribosome biogenesis is a process closely correlated to cell growth and proliferation, which are upregulated in the vast majority of cancers including PCa. This leads to downregulation of the expression and activity of p53, and thus promotes tumorigenesis (33,34). As a GTPase activator, DOCK4 can promote intercellular adhesion by activating Rap GTPase (35).
According to the PPI network for the hypomethylation-high expression gene group constructed by Cytoscape, the node degree of each gene was obtained and the top 10 node genes were selected, including ESR1, NPM1, NME2, NME1-NME2, UBQLN1, CTBP1, TRIM27, CCT3, ERBB3, and P4HB. Toy et al (36) suggested that activation of the ESR1 ligand binding domain mutations had different effects on the efficacy of estrogen receptor antagonists. Nucleophosmin (NPM1) is generally overexpressed, mutated, and rearranged in cancer and has been revealed to be overexpressed in PCa (37,38). In addition, NPM1 was revealed to participate in the progression, invasion, and metastasis of tumors in high-grade serous ovarian adenocarcinoma (39). Underexpression of NME2 can inhibit the metastasis of lung cancer and other studies concluded that hypomethylation and high expression of NME2 can cause apoptosis of the mouse testes cells (40,41). UBQLN1 was revealed to be overexpressed in gastric cancer. Reduced UBQLN1 expression can cause enhanced cell migration and invasion, actin cytoskeleton rearrangements, and stimulation of the epithelial-mesenchymal transition (EMT) (42,43). Scholars have revealed that CTBP1 is carcinogenic in PCa and other adenomas, and overexpression of CTBP1 is thought to be involved in cell survival, proliferation, migration, invasion, and EMT (44–46). TRIM27 was initially considered to be part of a fusion gene RET (rearranged during transfection), which is a proto-oncogene that is upregulated in multitudinal cancers, including PCa (47,48). CCT3 was significantly correlated with cancer cell proliferation and was revealed to be upregulated in osteosarcoma and hepatocellular carcinoma (49–51). ERBB3 is the only gene identified whose impaired kinase domain may be involved in the progression and invasion of PCa (52,53). P4HB was overexpressed in colon cancer and downregulation of expression may promote cancer cell apoptosis (54).
For the hypermethylation-low expression gene group in this PCa study, the BP revealed from the GO annotations were associated with cell adhesion, extracellular matrix organization, response to hypoxia, muscle contraction, positive regulation of cell migration, and regulation of phosphatidylinositol 3-kinase signaling. Loss of cell adhesion is one of the critical steps in tumor progression (55). Several studies have demonstrated that the extracellular matrix, hypoxia, cell migration, and the phosphatidylinositol 3-kinase signaling pathway may play crucial roles in cancer metastasis (56–58). Furthermore, the phosphatidylinositol 3-kinase signaling pathway is involved in tumorigenesis and progression (58). The molecular functions have primarily focused on actin, integrin, collagen, scaffold protein, and actin filament binding, as well as signal transducer activity. Previous studies have revealed that actin-binding proteins have inhibitory roles in PCa cell growth and metastasis (59,60). Integrins play a pivotal role in cell adhesion by connecting the cytoskeleton and extracellular matrix, promoting tumor cell proliferation, metastasis, and invasion (61,62).
In the present study, the top 10 hypermethylation-low expression node genes that were obtained from the PPI network included CREBBP, SMAD3, MCC, VIM, ATXN1, CAV1, FLNA, MAP3K14, PRKCB, and FHL2. There is evidence that CREBBP is downregulated in PCa and is a tumor suppressor gene in small-cell lung cancer and other neuroendocrine tumors (63,64). Smad3/Sox5/Twist1 was reported to promote EMT and cancer progression in PCa (65). MCC, a candidate tumor suppressor gene, was revealed to be often silenced by hypermethylation of its promoter in colorectal cancer (66). Several studies have revealed that the EMT marker, VIM, was positively associated with bone metastasis in PCa, and its promoter is often hypermethylated in cervical cancer cells (67,68). It has been reported that downregulation of ATXN1 induced EMT in cervical cancer, but its role in PCa remains to be elucidated (69). Studies have revealed that CAV1 can promote growth and metastasis of PCa, and the hypermethylation of its promoter could make CAV1 a tumor suppressor gene in PCa (70–72). The results of FLNA expression are consistent with those of down-regulation in PCa, indicating that FLNA may play a critical role as a negative regulator in PCa (73). MAP3K14, also known as NIK, is a regulatory component of the mitochondrial division machinery and has a dominant effect on cancer cell invasion (74). PRKCB is a member of the PKC family, and when its expression is downregulated, cell apoptosis and suppression of tumorigenesis occur (75,76). A previous study reported that FHL2 could be considered as a PCa biomarker (77).
GEPIA is a TCGA database-based online tool that can be used to study the prognostic effects of genes in various cancers. Survival analysis was conducted on 20 aberrantly-methylated genes which exhibit potential as diagnostic markers of PCa using GEPIA. Notably, the expression of P4HB was the only one among the 20 genes that was significantly correlated with survival. P4HB is found to be highly expressed in glioblastoma and liver cancer, decreasing the apoptosis of tumor cells (24,25). Xia et al (25) have revealed that high expression of P4HB in liver cancer tissues was associated with poor survival. However, there is also evidence suggesting that the low expression of P4HB could lead to proteasome inhibition-induced autophagy (78), since the boundary between apoptosis and autophagy has always been controversial due to tumor heterogeneity (79). Based on the present results revealing that the patients with hypermethylation and low expression of P4HB had poor prognosis, it was hypothesized that hypermethylation of P4HB in prostate cancer would activate autophagy of prostate cancer cells and promote the tumor progression. Further experiments are required to verify the hypothesis.
To validate the present results, TCGA database, the GEPIA tool, and the HPA database were employed to determine node gene methylation and gene expression status, and the results were generally consistent with our previous results, revealing the reliability of this study. However, there are still several limitations in the present study, including the focus on data mining and analysis without experimental confirmation. In addition, the methylation status of the node genes was only verified in TCGA database. Therefore, further experiments are required to verify our results.
In conclusion, the present study identified 20 aberrantly methylated-differentially expressed genes that may be used as biomarkers in PCa. The bioinformatics approach included the analysis of both gene expression and gene methylation microarrays, providing a novel and practical approach for the diagnosis, treatment, and prognosis of PCa. However, based on the limitations of the present study, additional molecular biologal experiments are required to confirm these findings.
It is anticipated that further study on the relationship between DNA methylation and histone codes and their mutual regulatory mechanism could provide new breakthroughs in the gene regulation of tumor genesis. Prostate cancer has high morbidity and mortality, however its pathogenesis has not been fully elucidated, and its therapeutic effect is not satisfactory. The study of the relationship between DNA methylation and histone codes is crucial to improve understanding of some signaling pathways which widely participate in cell physiology and pathology, making subtle adjustments to the complex signaling network, yet there have been few studies conducted on its clinical application. With the deeper understanding of the role of DNA methylation in tumor and the increasing number of high-throughput data of DNA methylation in tumors, bioinformatics technology has laid the foundation for the early diagnosis of tumors, the search for molecular markers for prognosis evaluation and new tumor therapeutic targets.
In the present study, high throughput microarray data of biological databases was mined by bioinformatics technology, to the best of our knowledge for the first time, to determine the aberrantly-methylated biomarkers of prostate cancer. The strategy applied was the selection of the intersecting high methylation and low expression genes as biological markers. The low methylation and high expression genes were analyzed by the same procedure. The markers were further enriched to display their related signaling pathways. Thus, the carcinogenic mechanism of prostate cancer could be clarified, to provide theories for clinical research and treatment.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- PCa
prostate cancer
- DEGs
differentially expressed genes
- GEO
Gene Expression Omnibus
- PPI
protein-protein interaction
- TCGA
The Cancer Genome Atlas
- GEPIA
Gene Expression Profiling Interactive Analysis
- HPA
Human Protein Atlas
- NCBI
National Center for Biotechnology Information
- DMGs
differentially methylated genes
- GO
Gene Ontology
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- BP
biological processes
- EMT
epithelial-mesenchymal transition
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZQ conceived and designed the present study. LW and BW collected, extracted and analyzed the data. LW and BW wrote the manuscript. LW and ZQ reviewed the final manuscript. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 4.Nephew KP, Huang TH. Epigenetic gene silencing in cancer initiation and progression. Cancer Lett. 2003;190:125–133. doi: 10.1016/S0304-3835(02)00511-6. [DOI] [PubMed] [Google Scholar]
- 5.Nakao M. Epigenetics: Interaction of DNA methylation and chromatin. Gene. 2001;278:25–31. doi: 10.1016/S0378-1119(01)00721-1. [DOI] [PubMed] [Google Scholar]
- 6.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 9.Mundbjerg K, Chopra S, Alemozaffar M, Duymich C, Lakshminarasimhan R, Nichols PW, Aron M, Siegmund KD, Ukimura O, Aron M, et al. Identifying aggressive prostate cancer foci using a DNA methylation classifier. Genome Biol. 2017;18:3. doi: 10.1186/s13059-016-1129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia D, Wang D, Kim SH, Katoh H, DuBois RN. Prostaglandin E2 promotes intestinal tumor growth via DNA methylation. Nat Med. 2012;18:224–226. doi: 10.1038/nm.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iizuka N, Oka M, Yamada-Okabe H, Nishida M, Maeda Y, Mori N, Takao T, Tamesa T, Tangoku A, Tabuchi H, et al. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361:923–929. doi: 10.1016/S0140-6736(03)12775-4. [DOI] [PubMed] [Google Scholar]
- 12.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 13.Verma M, Khoury MJ, Ioannidis JP. Opportunities and challenges for selected emerging technologies in cancer epidemiology: Mitochondrial, epigenomic, metabolomic, and telomerase profiling. Cancer Epidemiol Biomarkers Prev. 2013;22:189–200. doi: 10.1158/1055-9965.EPI-12-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu W, Ding Z. Identification of key genes in prostate cancer gene expression profile by bioinformatics. Andrologia. 2019;51:e13169. doi: 10.1111/and.13169. [DOI] [PubMed] [Google Scholar]
- 15.Ye Y, Li SL, Wang SY. Construction and analysis of mRNA, miRNA, lncRNA, and TF regulatory networks reveal the key genes associated with prostate cancer. PLoS One. 2018;13:e0198055. doi: 10.1371/journal.pone.0198055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Z, Tang F, Lu Z, Huang Y, Lei H, Li Z, Zeng G. Analysis of differentially expressed genes, clinical value and biological pathways in prostate cancer. Am J Transl Res. 2018;10:1444–1456. [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41(Database Issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 19.Lin P, He RQ, Dang YW, Wen DY, Ma J, He Y, Chen G, Yang H. An autophagy-related gene expression signature for survival prediction in multiple cohorts of hepatocellular carcinoma patients. Oncotarget. 2018;9:17368–17395. doi: 10.18632/oncotarget.24089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris JH, Wu A, Yamashita RA, Marchler-Bauer A, Ferrin TE. cddApp: A Cytoscape app for accessing the NCBI conserved domain database. Bioinformatics. 2015;31:134–136. doi: 10.1093/bioinformatics/btu605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galligan JJ, Petersen DR. The human protein disulfide isomerase gene family. Hum Genomics. 2012;6:6. doi: 10.1186/1479-7364-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pajunen L, Jones TA, Goddard A, Sheer D, Solomon E, Pihlajaniemi T, Kivirikko KI. Regional assignment of the human gene coding for a multifunctional polypeptide (P4HB) acting as the beta-subunit of prolyl 4-hydroxylase and the enzyme protein disulfide isomerase to 17q25. Cytogenet Cell Genet. 1991;56:165–168. doi: 10.1159/000133078. [DOI] [PubMed] [Google Scholar]
- 24.Goplen D, Wang J, Enger PØ, Tysnes BB, Terzis AJ, Laerum OD, Bjerkvig R. Protein disulfide isomerase expression is related to the invasive properties of malignant glioma. Cancer Res. 2006;66:9895–9902. doi: 10.1158/0008-5472.CAN-05-4589. [DOI] [PubMed] [Google Scholar]
- 25.Xia W, Zhuang J, Wang G, Ni J, Wang J, Ye Y. P4HB promotes HCC tumorigenesis through downregulation of GRP78 and subsequent upregulation of epithelial-to-mesenchymal transition. Oncotarget. 2017;8:8512–8521. doi: 10.18632/oncotarget.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. 2010;17:2141–2151. doi: 10.2174/092986710791299966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrlich M. DNA methylation in cancer: Too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 28.Davis CD, Uthus EO. DNA methylation, cancer susceptibility, and nutrient interactions. Exp Biol Med (Maywood) 2004;229:988–995. doi: 10.1177/153537020422901002. [DOI] [PubMed] [Google Scholar]
- 29.Toyota M, Itoh F, Imai K. DNA methylation and gastrointestinal malignancies: Functional consequences and clinical implications. J Gastroenterol. 2000;35:727–734. doi: 10.1007/s005350070030. [DOI] [PubMed] [Google Scholar]
- 30.Bjurlin MA, Taneja SS. Prostate cancer. Urol Clin North Am. 2017;44:xv–xvi. doi: 10.1016/S0094-0143(17)30088-5. [DOI] [PubMed] [Google Scholar]
- 31.Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol. 2013;5(pii):a012336. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu J, Cargnello M, Topisirovic I, Pelletier J. Translation initiation factors: Reprogramming protein synthesis in cancer. Trends Cell Biol. 2016;26:918–933. doi: 10.1016/j.tcb.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Ray S, Johnston R, Campbell DC, Nugent S, McDade SS, Waugh D, Panov KI. Androgens and estrogens stimulate ribosome biogenesis in prostate and breast cancer cells in receptor dependent manner. Gene. 2013;526:46–53. doi: 10.1016/j.gene.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 34.Derenzini M, Montanaro L, Trerè D. Ribosome biogenesis and cancer. Acta Histochem. 2017;119:190–197. doi: 10.1016/j.acthis.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Yajnik V, Paulding C, Sordella R, McClatchey AI, Saito M, Wahrer DC, Reynolds P, Bell DW, Lake R, van den Heuvel S, et al. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–684. doi: 10.1016/S0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 36.Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AM, Smith A, Wilson J, Morrow C, Wong WL, et al. Activating ESR1 mutations differentially affect the efficacy of ER antagonists. Cancer Discov. 2017;7:277–287. doi: 10.1158/2159-8290.CD-15-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Box JK, Paquet N, Adams MN, Boucher D, Bolderson E, O'Byrne KJ, Richard DJ. Nucleophosmin: From structure and function to disease development. BMC Mol Biol. 2016;17:19. doi: 10.1186/s12867-016-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Destouches D, Sader M, Terry S, Marchand C, Maillé P, Soyeux P, Carpentier G, Semprez F, Céraline J, Allory Y, et al. Implication of NPM1 phosphorylation and preclinical evaluation of the nucleoprotein antagonist N6L in prostate cancer. Oncotarget. 2016;7:69397–69411. doi: 10.18632/oncotarget.8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan X, Wen L, Li Y, Lou L, Liu W, Zhang J. The expression profile and prognostic value of APE/Ref-1 and NPM1 in high-grade serous ovarian adenocarcinoma. APMIS. 2017;125:857–862. doi: 10.1111/apm.12733. [DOI] [PubMed] [Google Scholar]
- 40.Thakur RK, Yadav VK, Kumar A, Singh A, Pal K, Hoeppner L, Saha D, Purohit G, Basundra R, Kar A, et al. Non-metastatic 2 (NME2)-mediated suppression of lung cancer metastasis involves transcriptional regulation of key cell adhesion factor vinculin. Nucleic Acids Res. 2014;42:11589–11600. doi: 10.1093/nar/gku860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu Y, Xu W, Nie D, Zhang D, Dai J, Zhao X, Zhang M, Wang Z, Chen Z, Qiao Z. Nicotine induces Nme2-mediated apoptosis in mouse testes. Biochem Biophys Res Commun. 2016;472:573–579. doi: 10.1016/j.bbrc.2016.03.044. [DOI] [PubMed] [Google Scholar]
- 42.Bao J, Jiang X, Zhu X, Dai G, Dou R, Liu X, Sheng H, Liang Z, Yu H. Clinical significance of ubiquilin 1 in gastric cancer. Medicine. 2018;97:e9701. doi: 10.1097/MD.0000000000009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah PP, Lockwood WW, Saurabh K, Kurlawala Z, Shannon SP, Waigel S, Zacharias W, Beverly LJ. Ubiquilin1 represses migration and epithelial-to-mesenchymal transition of human non-small cell lung cancer cells. Oncogene. 2015;34:1709–1717. doi: 10.1038/onc.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porretti J, Dalton GN, Massillo C, Scalise GD, Farré PL, Elble R, Gerez EN, Accialini P, Cabanillas AM, Gardner K, et al. CLCA2 epigenetic regulation by CTBP1, HDACs, ZEB1, EP300 and miR-196b-5p impacts prostate cancer cell adhesion and EMT in metabolic syndrome disease. Int J Cancer. 2018;143:897–906. doi: 10.1002/ijc.31379. [DOI] [PubMed] [Google Scholar]
- 45.Phelps RA, Chidester S, Dehghanizadeh S, Phelps J, Sandoval IT, Rai K, Broadbent T, Sarkar S, Burt RW, Jones DA. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–634. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blevins MA, Huang M, Zhao R. The role of CtBP1 in oncogenic processes and its potential as a therapeutic target. Mol Cancer Ther. 2017;16:981–990. doi: 10.1158/1535-7163.MCT-16-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma Y, Wei Z, Bast RC, Jr, Wang Z, Li Y, Gao M, Liu Y, Wang X, Guo C, Zhang L, Wang X. Downregulation of TRIM27 expression inhibits the proliferation of ovarian cancer cells in vitro and in vivo. Lab Invest. 2016;96:37–48. doi: 10.1038/labinvest.2015.132. [DOI] [PubMed] [Google Scholar]
- 48.Shaikhibrahim Z, Lindstrot A, Ochsenfahrt J, Fuchs K, Wernert N. Epigenetics-related genes in prostate cancer: Expression profile in prostate cancer tissues, androgen-sensitive and -insensitive cell lines. Int J Mol Med. 2013;31:21–25. doi: 10.3892/ijmm.2012.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Wang Y, Wei Y, Wu J, Zhang P, Shen S, Saiyin H, Wumaier R, Yang X, Wang C, Yu L. Molecular chaperone CCT3 supports proper mitotic progression and cell proliferation in hepatocellular carcinoma cells. Cancer Lett. 2016;372:101–109. doi: 10.1016/j.canlet.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 50.Xiong Y, Wu S, Du Q, Wang A, Wang Z. Integrated analysis of gene expression and genomic aberration data in osteosarcoma (OS) Cancer Gene Ther. 2015;22:524–529. doi: 10.1038/cgt.2015.48. [DOI] [PubMed] [Google Scholar]
- 51.Cui X, Hu ZP, Li Z, Gao PJ, Zhu JY. Overexpression of chaperonin containing TCP1, subunit 3 predicts poor prognosis in hepatocellular carcinoma. World J Gastroenterol. 2015;21:8588–8604. doi: 10.3748/wjg.v21.i28.8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaiswal BS, Kljavin NM, Stawiski EW, Chan E, Parikh C, Durinck S, Chaudhuri S, Pujara K, Guillory J, Edgar KA, et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell. 2013;23:603–617. doi: 10.1016/j.ccr.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 53.Koumakpayi IH, Le Page C, Delvoye N, Saad F, Mes-Masson AM. Macropinocytosis inhibitors and Arf6 regulate ErbB3 nuclear localization in prostate cancer cells. Mol Carcinog. 2011;50:901–912. doi: 10.1002/mc.20766. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Y, Yang J, Zhang Q, Xu Q, Lu L, Wang J, Xia W. P4HB knockdown induces human HT29 colon cancer cell apoptosis through the generation of reactive oxygen species and inactivation of STAT3 signaling. Mol Med Rep. 2019;19:231–237. doi: 10.3892/mmr.2018.9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Bras GF, Taubenslag KJ, Andl CD. The regulation of cell-cell adhesion during epithelial-mesenchymal transition, motility and tumor progression. Cell Adh Migr. 2012;6:365–373. doi: 10.4161/cam.21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacquemet G, Hamidi H, Ivaska J. Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr Opin Cell Biol. 2015;36:23–31. doi: 10.1016/j.ceb.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 58.Liu X, Xu Y, Zhou Q, Chen M, Zhang Y, Liang H, Zhao J, Zhong W, Wang M. PI3K in cancer: Its structure, activation modes and role in shaping tumor microenvironment. Future Oncol. 2018;14:665–674. doi: 10.2217/fon-2017-0588. [DOI] [PubMed] [Google Scholar]
- 59.Haffner MC, Esopi DM, Chaux A, Gürel M, Ghosh S, Vaghasia AM, Tsai H, Kim K, Castagna N, Lam H, et al. AIM1 is an actin-binding protein that suppresses cell migration and micrometastatic dissemination. Nat Commun. 2017;8:142. doi: 10.1038/s41467-017-00084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanaja DK, Grossmann ME, Cheville JC, Gazi MH, Gong A, Zhang JS, Ajtai K, Burghardt TP, Young CY. PDLIM4, an actin binding protein, suppresses prostate cancer cell growth. Cancer Invest. 2009;27:264–272. doi: 10.1080/07357900802406319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramovs V, Te Molder L, Sonnenberg A. The opposing roles of laminin-binding integrins in cancer. Matrix Biol. 2017;57-58:213–243. doi: 10.1016/j.matbio.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 62.Desgrosellier JS, Cheresh DA. Integrins in cancer: Biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jia D, Augert A, Kim DW, Eastwood E, Wu N, Ibrahim AH, Kim KB, Dunn CT, Pillai SPS, Gazdar AF, et al. Crebbp loss drives small cell lung cancer and increases sensitivity to HDAC inhibition. Cancer Discov. 2018;8:1422–1437. doi: 10.1158/2159-8290.CD-18-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaikhibrahim Z, Lindstrot A, Buettner R, Wernert N. Analysis of laser-microdissected prostate cancer tissues reveals potential tumor markers. Int J Mol Med. 2011;28:605–611. doi: 10.3892/ijmm.2011.746. [DOI] [PubMed] [Google Scholar]
- 65.Hu J, Tian J, Zhu S, Sun L, Yu J, Tian H, Dong Q, Luo Q, Jiang N, Niu Y, Shang Z. Sox5 contributes to prostate cancer metastasis and is a master regulator of TGF-β-induced epithelial mesenchymal transition through controlling Twist1 expression. Br J Cancer. 2018;118:88–97. doi: 10.1038/bjc.2017.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohonen-Corish MR, Sigglekow ND, Susanto J, Chapuis PH, Bokey EL, Dent OF, Chan C, Lin BP, Seng TJ, Laird PW, et al. Promoter methylation of the mutated in colorectal cancer gene is a frequent early event in colorectal cancer. Oncogene. 2007;26:4435–4441. doi: 10.1038/sj.onc.1210210. [DOI] [PubMed] [Google Scholar]
- 67.Lang SH, Hyde C, Reid IN, Hitchcock IS, Hart CA, Bryden AA, Villette JM, Stower MJ, Maitland NJ. Enhanced expression of vimentin in motile prostate cell lines and in poorly differentiated and metastatic prostate carcinoma. Prostate. 2002;52:253–263. doi: 10.1002/pros.10088. [DOI] [PubMed] [Google Scholar]
- 68.Jung S, Yi L, Kim J, Jeong D, Oh T, Kim CH, Kim CJ, Shin J, An S, Lee MS. The role of vimentin as a methylation biomarker for early diagnosis of cervical cancer. Mol Cells. 2011;31:405–411. doi: 10.1007/s10059-011-0229-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kang AR, An HT, Ko J, Kang S. Ataxin-1 regulates epithelial-mesenchymal transition of cervical cancer cells. Oncotarget. 2017;8:18248–18259. doi: 10.18632/oncotarget.15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li L, Yang G, Ebara S, Satoh T, Nasu Y, Timme TL, Ren C, Wang J, Tahir SA, Thompson TC. Caveolin-1 mediates testosterone-stimulated survival/clonal growth and promotes metastatic activities in prostate cancer cells. Cancer Res. 2001;61:4386–4392. [PubMed] [Google Scholar]
- 71.Tahir SA, Yang G, Goltsov AA, Watanabe M, Tabata K, Addai J, Fattah el MA, Kadmon D, Thompson TC. Tumor cell-secreted caveolin-1 has proangiogenic activities in prostate cancer. Cancer Res. 2008;68:731–739. doi: 10.1158/0008-5472.CAN-07-2668. [DOI] [PubMed] [Google Scholar]
- 72.Cui J, Rohr LR, Swanson G, Speights VO, Maxwell T, Brothman AR. Hypermethylation of the caveolin-1 gene promoter in prostate cancer. Prostate. 2001;46:249–256. doi: 10.1002/1097-0045(20010215)46:3<249::AID-PROS1030>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 73.Sun GG, Lu YF, Zhang J, Hu WN. Filamin A regulates MMP-9 expression and suppresses prostate cancer cell migration and invasion. Tumour Biol. 2014;35:3819–3826. doi: 10.1007/s13277-013-1504-6. [DOI] [PubMed] [Google Scholar]
- 74.Jung JU, Ravi S, Lee DW, McFadden K, Kamradt ML, Toussaint LG, Sitcheran R. NIK/MAP3K14 regulates mitochondrial dynamics and trafficking to promote cell invasion. Curr Biol. 2016;26:3288–3302. doi: 10.1016/j.cub.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hagiwara K, Ito H, Murate T, Miyata Y, Ohashi H, Nagai H. PROX1 overexpression inhibits protein kinase C beta II transcription through promoter DNA methylation. Genes Chromosomes Cancer. 2012;51:1024–1036. doi: 10.1002/gcc.21985. [DOI] [PubMed] [Google Scholar]
- 76.Surdez D, Benetkiewicz M, Perrin V, Han ZY, Pierron G, Ballet S, Lamoureux F, Rédini F, Decouvelaere AV, Daudigeos-Dubus E, et al. Targeting the EWSR1-FLI1 oncogene-induced protein kinase PKC-β abolishes ewing sarcoma growth. Cancer Res. 2012;72:4494–4503. doi: 10.1158/0008-5472.CAN-12-0371. [DOI] [PubMed] [Google Scholar]
- 77.Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- 78.Rui YN, Xu Z, Chen Z, Zhang S. The GST-BHMT assay reveals a distinct mechanism underlying proteasome inhibition-induced macroautophagy in mammalian cells. Autophagy. 2015;11:812–832. doi: 10.1080/15548627.2015.1034402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.