Abstract
Toll-like receptors (TLRs) are the most widely studied pattern recognition receptors. Mounting evidence suggests an important association between TLRs and the occurrence and development of breast cancer. Thus, targeting these receptors may be a potential strategy for breast cancer treatment. The current study analyzed the data of 1,215 patients with breast cancer obtained from The Cancer Genome Atlas (TCGA) database. It was observed that, in addition to TLR6, TLR7 and TLR8, the expression of the remaining TLRs in breast cancer tissues was lower than that in normal tissues. In addition, TLR3 and TLR9 displayed significantly different expression levels in ER-/PR-negative breast cancer compared with the control tissues, while TLR5 expression was significantly reduced in HER2-enriched breast cancer. Furthermore, TLR10 exhibited lower expression levels in advanced stages of the disease as compared with that observed in earlier stages. Survival analysis revealed that the expression of TLR4 and TLR7 had a significant impact on survival, and higher expression levels suggested worse prognosis. Finally, the expression levels of TLR1, TLR2, TLR4, TLR5, TLR6 and TLR10 were correlated with those of the inflammatory cytokines interleukin-1β and tumor necrosis factor-α, while the expression levels of TLR3, TLR7, TLR8 and TLR9 were correlated with those of interferon-β and C-X-C motif chemokine ligand 10. Taken together, the current study results suggest that TLR expression may serve as a biomarker of cancer pathogenesis and progression, and may provide new insights for the treatment of breast cancer through the regulation and targeting of TLRs.
Keywords: Toll-like receptors, breast cancer, survival, inflammatory cytokines, the cancer genome atlas
Introduction
Toll-like receptors (TLRs) are well-conserved pattern-recognition receptors that are primarily expressed in human epithelial and immune cells (1,2). The main function of TLRs is to promote the synthesis and release of inflammatory cytokines and chemokines, thus triggering the inflammatory response (3,4). To date, a total of 10 TLRs, namely TLR1-TLR10, have been identified in humans. Previous studies have demonstrated that TLRs are also expressed in numerous tumor cells, and serve key roles in tumorigenesis, development and metastasis (5,6). Research on how to effectively inhibit the expression and activation of TLRs in order to reduce the production and release of the corresponding inflammatory factors, and ultimately inhibit the proliferation of cancer cells has been receiving increasing attention.
Breast cancer is one of the most common malignant tumors worldwide, and is the most prevalent type of cancer and second leading cause of cancer-associated mortality in women (7). In China, breast cancer is the second most common type of cancer, second only to lung cancer (8). Breast cancer is an intrinsically heterogeneous disease with different biological characteristics and clinical outcomes. Common immunohistochemistry markers, such as estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2), together with traditional clinicopathological features, including tumor size, tumor grade and nodal involvement, are used to predict the outcome and treatment response in breast cancer (9). Sørlie et al (10) have proposed five subgroups of breast cancer based on gene expression profiling using DNA microarrays, including Luminal A, Luminal B, HER2 overexpressing, basal-like and normal-like breast cancer. Luminal type breast cancer (both A and B, also known as Lum A/B) is the most common type, accounting for ~70% of patients with breast cancer. Basal-like type breast cancer lacks ER, PR and HER2 receptors (triple-negative breast cancer), and has the worst prognosis (11,12).
Although the association between TLRs and breast cancer has not been thoroughly investigated, previous studies suggested the presence of an important link between TLRs and breast cancer. It has been reported that the expression of TLR2 was ~10-fold lower in the more malignant MDA-MB-231 cells as compared with that in the less malignant MCF7 cells. Activation of TLR2 results in the activation of nuclear factor-κB (NF-κB) and upregulation of interleukin (IL)-6, transforming growth factor-β, vascular endothelial growth factor and matrix metalloproteinase 9 (13). Salaun et al (14) reported that, among 194 cases of patients with breast cancer, 36–45% of breast cancer cells expressed TLR3, while treatment with poly(A:U) reduced the risk of recurrence and metastasis in patients with TLR3-positive breast cancer. In addition, Haricharan and Brown (15) observed that TLR4 activation in TP53-mutant breast cancer regulated the proliferation of cancer cells by promoting the secretion of pro-growth cytokines. TLR4 expression has also been demonstrated to be strongly associated with clinical indicators in metastatic ductal carcinoma (16). Cai et al (17) further reported that 80% of 75 cases of breast cancer expressed TLR5, the majority of which were high-grade ductal carcinomas. TLR5 was also overexpressed in 256 breast carcinomas specimens, and was correlated with lymph node metastasis and cancer grade (18). Furthermore, Berger et al (19) studied frozen breast specimens from 124 female patients with breast cancer and found that TLR9 mRNA expression was positively correlated with tumor grade, suggesting that TLR9 may be a molecular marker for poorly differentiated breast cancer. Another study reported that activation of TLR9 in MDA-MB-231 cells by CpG-ODN increased in vitro invasion (20), suggesting that TLR9 is involved in tumor progression and metastasis. These previous studies collectively suggest that TLR signaling may serve an important role in regulating the growth, metastasis and apoptosis of breast cancer cells.
In the current study, mRNA expression and clinical data of 1,215 patients with breast cancer were obtained from The Cancer Genome Atlas (TCGA) database to analyze the expression patterns of TLRs in different sample types, tumor subtypes and tumor stages. Furthermore, cytokines downstream of the TLR signaling pathway were studied, and survival analysis was performed to investigate the effect of TLR expression on the outcome and prognosis of patients with breast cancer. By investigating the role of TLRs in the development of breast cancer, the present study provides new insights for the diagnosis and treatment of breast cancer.
Materials and methods
Patients and TCGA data retrieval
The data of patients with breast cancer, including mRNA expression and corresponding clinical information, were retrieved from TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), which is an open access, publicly available database. Clinical characteristics of the patients included in the current study are listed in Table I. The TCGA gene expression profile was measured using the Illumina HiSeq 2000 RNA Sequencing System (HiSeqV2; Illumina, Inc., San Diego, CA, USA). The RSEM (also known as RNA-Seq by Expectation-Maximization) normalized count was used as the gene level expression estimates in the present study. Intrinsic tumor subtype, tumor stage information and overall survival time were also extracted from TCGA data portal. Patients without detailed clinicopathological data, such as age, gender, race, histological subtype, tumor-node- metastasis (TNM) stage or overall survival were excluded from the present study. The study met the ethics and policies provided by TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/history/policies).
Table I.
Clinical characteristics of breast cancer patients included in The Cancer Genome Atlas database.
| Characteristic | Value | Percentage (%) |
|---|---|---|
| Age at diagnosis (years) | 57.98 | − |
| Follow up (years)a | 1.65 | − |
| Ethnicity | ||
| Caucasian | 757 | 60.99 |
| Black or African American | 183 | 14.75 |
| Asian | 61 | 4.92 |
| American Indian or Alaska native | 1 | 0.08 |
| NA | 239 | 19.26 |
| Survival | ||
| Alive at last follow-up or succumbed to unrelated cause | 816 | 65.75 |
| Disease-specific mortality | 135 | 10.88 |
| NA | 290 | 23.37 |
| Tumor grade (Nottingham) (37) | ||
| I | 133 | 10.72 |
| II | 446 | 35.94 |
| III | 175 | 14.10 |
| IV | 15 | 1.21 |
| X | 10 | 0.81 |
| NA | 462 | 37.23 |
| Subtype | ||
| Normal | 113 | 9.11 |
| Luminal A | 434 | 34.97 |
| Luminal B | 194 | 15.63 |
| HER2-enriched | 67 | 5.40 |
| Basal-like | 142 | 11.44 |
| Not classified | 291 | 23.45 |
Presented as the median. NA, not available; HER2, human epidermal growth factor receptor 2.
Statistical analysis
The analysis focused on 1,215 breast cancer cases included in TCGA database. The data were expressed as the mean ± standard deviation. Paired or unpaired Student's t-test, and one-way analysis of variance functions of GraphPad Prism software (version 6; GraphPad Software, Inc., San Diego, CA, USA) were used to evaluate the significance of differential expression levels of candidate genes among different samples, intrinsic tumor subtypes and tumor stages. In addition, Spearman correlation and linear regression analysis were used to examine the association of TLR expression with various cytokines [IL-1β, IL-6, IL-8, necrosis factor α (TNF-α), interferon (IFN)-α, IFN-β and C-X-C motif chemokine 10 (CXCL10)], with 0<r<1 indicating that two variables were changing in the same direction in a correlative manner. Survival analysis was conducted using SPSS software (version 22.0; IBM Corp., Armonk, NY, USA). Patients were grouped based on the mRNA expression of TLRs, with the upper 50% and the lower 50% representing the high and low expression groups, respectively. Survival curves were estimated by the Kaplan-Meier method, and the log-rank test was used to compare the overall survival curves between groups. Only mortality cases as a result of breast cancer were considered in the analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of TLRs in breast cancer cases included in TCGA database
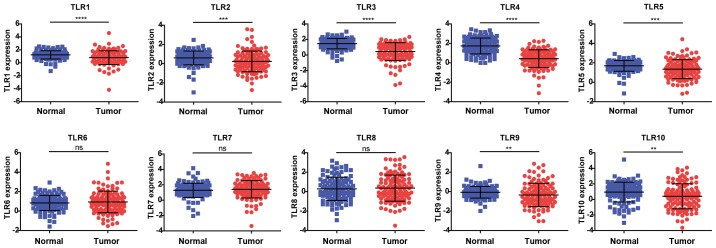
The publicly available TCGA database, composed of 2.5 petabytes of data describing tumor tissues and matched normal tissues from >11,000 patients, was used in the present study. The expression levels of TLR1-TLR10 were initially analyzed using TCGA breast cancer database in order to compare normal and tumor tissues. Compared with the normal control tissues, the expression levels of TLR1, TLR2, TLR3, TLR4, TLR5, TLR9 and TLR10 were significantly decreased in breast cancer tissues. In addition, the expression levels of TLR6, TLR7 and TLR8 were slightly increased in breast cancer tissues, although statistical significance was not reached (Fig. 1).
Figure 1.
TLR expression levels in breast cancer. The raw expression data were exported from The Cancer Genome Atlas breast cancer database, including 113 tumor samples (red) and matched normal tissues (blue). Data are expressed as the mean ± standard deviation. Paired Student's t-test was used to evaluate the statistical significance of differential mRNA expression levels of candidate genes between normal and tumor groups. **P<0.01, ***P<0.001 and ****P<0.0001. TLR, Toll-like receptor; ns, no significant difference.
Expression levels of TLRs in various subtypes and stages of breast cancer
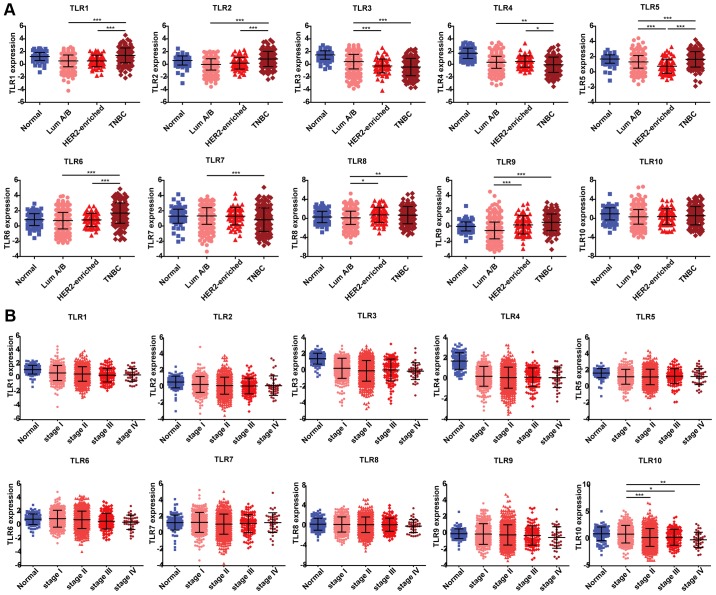
The expression of TLRs in different subtypes of breast cancer was subsequently analyzed. TLR1, TLR2 and TLR6 displayed higher expression levels in the triple-negative breast cancer subtype as compared with those in Lum A/B and HER2-enriched subtypes. The expression of TLR3 exhibited a gradient decline among normal tissues and all the three subtypes; however, the difference between the HER2-enriched and triple-negative subtypes was not statistically significant. The TLR4 and TLR7 expression levels were lower in the triple-negative subtype as compared with the other two subtypes. TLR5 was the only gene that exhibited a statistically significant difference in terms of expression among all the three subtypes, with the lowest expression observed in the HER2-enriched subtype, followed by the Lum A/B and triple-negative subtypes. Furthermore, the expression of TLR8 was reduced in the Lum A/B subtype compared with that exhibited in the other two subtypes, although this difference was not significant. TLR9 expression increased gradually among the three subtypes; however, similarly to TLR3, there was no statistically significant difference between the HER2-enriched and triple-negative subtypes. By contrast, TLR10 expression did not differ among the three subtypes of breast cancer (Fig. 2A). These results suggest that TLR3 and TLR9 may be useful biomarkers of ER-/PR-negative breast cancer (HER2-enriched and triple-negative subtypes), while TLR5 may serve an important role in HER2-enriched breast cancer.
Figure 2.
TLR expression among different subtypes and stages of breast cancer. (A) Subtype information was exported from TCGA breast cancer database, including 113, 628, 67 and 142 samples from normal tissues, Luminal A/B, HER2-enriched and triple-negative subtypes, respectively. (B) TNM stage information was exported from TCGA breast cancer database, including 113, 279, 622, 131 and 40 samples from normal tissues, stage I, II, III and IV, respectively. Data are expressed as the mean ± standard deviation. One-way analysis of variance was used to evaluate the statistical significance of differential mRNA expression levels of candidate genes among groups. *P<0.05, **P<0.01 and ***P<0.001. TLR, Toll-like receptor; TCGA, The Cancer Genome Atlas; ns, no significant difference.
Next, the expression levels of TLRs in various stages of cancer progression were analyzed based on the data obtained from TCGA database. Breast cancer cases were staged according to the TNM staging system, which uses the size and extension of the primary tumor, its lymphatic involvement and the presence of metastases to classify the progression of all solid tumors (https://www.cancer.gov/types/breast/patient/adult/breast-treatment-pdq). It was observed that the expression levels of TLRs varied in different stages of breast cancer (Fig. 2B). Consistent with our earlier results (Fig. 1), the expression levels of TLR1, TLR3 and TLR4 were significantly reduced in all stages relative to normal tissues, whereas TLR2, TLR5 and TLR10 exhibited lower expression in certain stages compared with that in normal tissues. TLR10 was the only gene whose expression varied significantly between stage I and II–IV breast cancer, with its expression decreasing as the cancer stage advanced. These results suggest that TLR10 may serve as a biomarker of breast cancer progression. By contrast, the expression levels of TLR6, TLR7, TLR8 and TLR9 did not differ among different disease stages or in comparison with those found in normal tissues.
Survival analysis
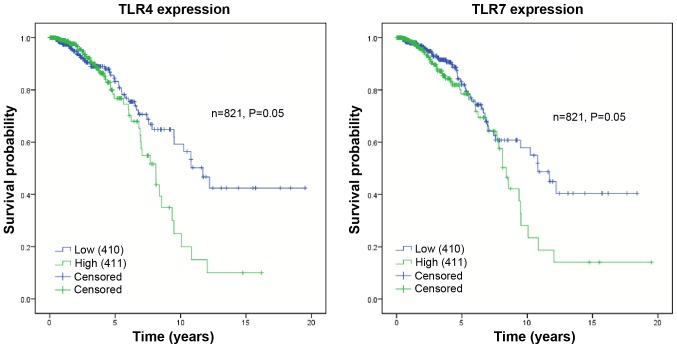
The current study used overall survival data of breast cancer patients (OS ≥90 days) from TCGA database to evaluate the effect of TLR expression on patient prognosis. The patient survival status was described as 0 or 1, where 0 represented mortality and 1 represented censoring (patients who were lost, or succumbed to other causes, or were alive at the end of follow-up). The samples were divided into two groups based on TLR expression levels, including the low and high expression groups. Fig. 3 illustrates the association between patient survival and the expression of TLR4 or TLR7. The mean survival time following the initial diagnosis of patients with low TLR4 expression was 12.38 years, while that of patients with high expression was 8.07 years (P=0.05). Furthermore, the mean survival time of patients with low TLR7 expression was 11.66 years, while that of patients with high expression was 8.96 years (P=0.05). The remaining TLRs exhibited no significant association between expression levels and survival (Table II). Overall, high TLR4 and TLR7 expression may be associated with poor prognosis.
Figure 3.
Kaplan-Meier curves for overall survival stratified by TLR4 and TLR7 mRNA expression. The overall survival data was exported from The Cancer Genome Atlas breast cancer database, including 410 and 411 samples with low (blue lines) and high (green lines) TLR expression, respectively. Only cases exhibiting cancer-associated mortality were included in this analysis. The symbol + denotes censored observations. The log-rank test was used to compare the overall survival curves between groups. TLR, Toll-like receptor.
Table II.
Analysis of overall survival in relation to TLR expression.
| 95% confidence interval | ||||||
|---|---|---|---|---|---|---|
| Gene | Expression | Mean survival (years) | Lower | Upper | χ2 | P-value |
| TLR1 | Low | 11.25 | 9.65 | 12.85 | 0.15 | 0.70 |
| High | 9.62 | 7.91 | 11.33 | |||
| TLR2 | Low | 10.59 | 9.04 | 12.14 | 0.41 | 0.52 |
| High | 10.64 | 8.83 | 12.46 | |||
| TLR3 | Low | 10.70 | 9.00 | 12.39 | 0.63 | 0.43 |
| High | 10.66 | 8.95 | 12.36 | |||
| TLR4 | Low | 12.38 | 10.71 | 14.06 | 4.00 | 0.05 |
| High | 8.07 | 6.94 | 9.20 | |||
| TLR5 | Low | 10.17 | 8.33 | 12.00 | 0.00 | 0.99 |
| High | 10.68 | 9.25 | 12.11 | |||
| TLR6 | Low | 11.20 | 9.64 | 12.76 | 1.98 | 0.16 |
| High | 9.93 | 8.08 | 11.77 | |||
| TLR7 | Low | 11.66 | 7.32 | 10.60 | 3.92 | 0.05 |
| High | 8.96 | 10.15 | 13.18 | |||
| TLR8 | Low | 11.77 | 10.15 | 13.40 | 1.90 | 0.17 |
| High | 8.44 | 7.18 | 9.70 | |||
| TLR9 | Low | 11.05 | 9.19 | 12.90 | 0.02 | 0.89 |
| High | 10.22 | 8.78 | 11.67 | |||
| TLR10 | Low | 11.19 | 9.48 | 12.90 | 0.02 | 0.89 |
| High | 9.62 | 8.30 | 10.93 | |||
TLR, Toll-like receptor.
Correlation of inflammatory cytokines and chemokines with TLRs
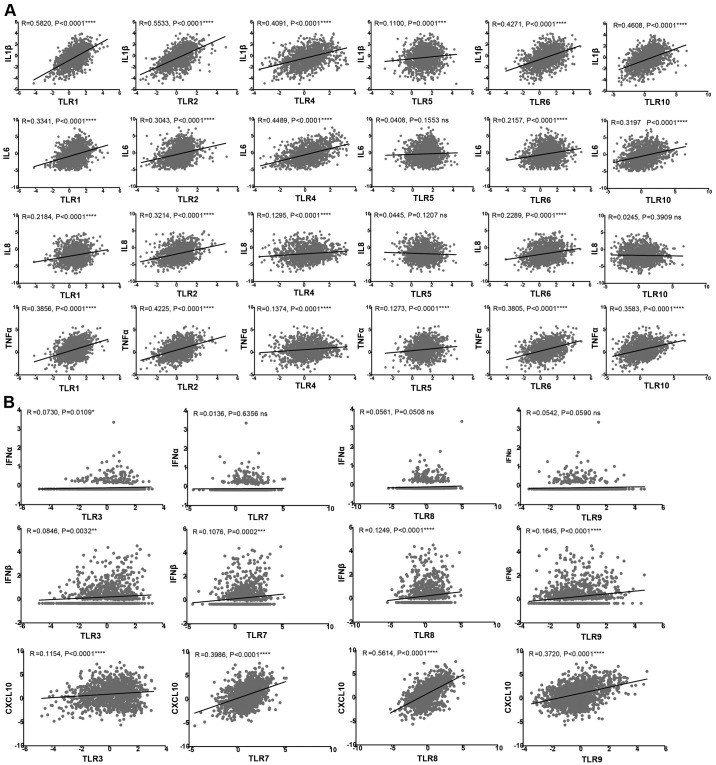
TLR expression in the tumor microenvironment has been reported to be associated with cancer progression and to be involved in inflammation (21). The activation of TLRs expressed in tumor cells initiates signaling cascades that mediate the release of cytokines, chemokines, pro-angiogenic mediators and growth factors, leading to tumor survival and progression (22). Due to the different locations of TLRs in cancer cells, different downstream signaling pathways are activated, resulting in the production of different cytokines. In the present study, the TLR family was divided into two groups: The first group included TLR1, TLR2, TLR4, TLR5, TLR6 and TLR10, which are expressed on the cell surface, while the second group comprised TLR3, TLR7, TLR8 and TLR9, which are present in intracellular vesicles (23). A total of seven representative cytokines and chemokines downstream of TLR signaling pathways were then selected to analyze their correlation with TLR expression.
Fig. 4A shows the correlation of inflammatory cytokines IL-1β, IL-6, IL-8 and tumor necrosis factor α (TNF-α) with the cell surface receptors TLR1, TLR2, TLR4, TLR5, TLR6 and TLR10. The expression levels of TLR1, TLR2, TLR4 and TLR6 were found to be directly correlated with these four cytokines. By contrast, the expression of TLR5 was only correlated with IL-1β and TNF-α, while TLR10 was correlated with IL-1β, IL-6 and TNF-α. As shown in Fig. 4B, the expression levels of the four intracellular receptors (namely TLR3, TLR7, TLR8 and TLR9) was positively correlated with IFN-β and CXCL10. TLR3 was also correlated with IFN-α, whereas TLR7, TLR8 and TLR9 did not exhibit significant correlations with IFN-α.
Figure 4.
(A) Cell surface TLR and (B) intracellular TLR expression correlation with cytokines. The raw expression data were exported from The Cancer Genome Atlas breast cancer database. The R and P-values are indicated in the graphs. Linear regression and association analyses were conducted by GraphPad Prism 6 software. TLR, Toll-like receptor; ns, no significant difference; IL, interleukin; TNF-α, tumor necrosis factor α; IFN, interferon; CXCL10, C-X-C motif chemokine ligand 10.
Discussion
Breast cancer is a major public health issue for women worldwide. The occurrence and development of breast cancer are affected by complex environmental and genetic factors. Numerous studies have suggested that TLRs, which are normally associated with immunity and inflammation, may be involved in the progression and prognosis of breast cancer. TLRs are highly expressed in breast cancer cells, and activation of these receptors can induce aggressive tumor behavior, cell proliferation, cell invasion, cell migration and metastasis (24). The ‘cross-talk’ between TLRs and various other signaling pathways in breast cancer constitutes a markedly complicated signaling network system to promote secretion of inflammatory cytokines and chemokines. These inflammatory mediators, in turn, can promote tumor proliferation and apoptosis resistance, thus participating in the development and progression of breast cancer (25). Therefore, TLRs may be potential drug targets for breast cancer treatment.
The current study demonstrated that the expression levels of the majority of TLRs (except for TLR6-TLR9) were downregulated compared with those in normal tissues; these results differ from those reported in previous studies (13,16,17,26–28). This discrepancy may be due to the source of the model system from which the data were derived. Previous studies have mainly focused on the expression of TLRs in animal models or breast cancer cell lines, whereas the present study utilized clinical data from TCGA database. In the present study, although TLRs were found to be generally downregulated in tumor tissues, certain tumor subtypes exhibited higher expression compared with normal tissues. For instance, TLR6 and TLR9 expression was significantly increased in triple-negative subtype in comparison with that in normal tissues. Thus, it is speculated that the possible reasons for the discrepancy of the results between the present and previous studies may be the size and source of samples, or the selection of tumor subtypes. The present study involved a large number of samples (1,215 cases), and different sample sources, subtypes, races, genders and ages were included.
It has previously been reported that TLR3 sequence variants may reduce 82% of breast cancer risk among African-American women (29), and the activation of TLR3 was associated with a significant decrease in the risk of metastatic relapse in 194 patients with breast cancer, suggesting an association between clinical outcome and TLR3 expression (26). This is consistent with the results of the present study, which revealed lower levels of TLR3 expression in more malignant subtypes. It was also demonstrated that high level of TLR9 was strongly associated with ER-negative subtypes (HER2-enriched and triple-negative), which was also verified by previous studies. For instance, Berger et al (19) reported that TLR9 expression promoted cell migration, cell invasion and aggressive tumor behavior in an ER-negative breast cancer cell line. In addition, Jukkola-Vuorinen et al (30) demonstrated increased TLR9 expression in ER-negative versus ER-positive breast cancer samples via immunofluorescence. The current study also revealed that low expression of TLR5 was significantly associated with HER2-enriched breast cancer, although further research is required to confirm this association.
Multiple studies have focused on the effect of TLRs on breast cancer prognosis. González-Reyes et al (27) revealed that high expression of TLR4 was associated with a large tumor size, distant metastasis and recurrence upon investigating tumors from 74 patients with breast cancer. In addition, TLR4 was expressed in a functional form in patients with ER-/PR-negative breast cancer and was correlated with a decreased survival (31). Lu et al (32) demonstrated that activation of TLR7 was associated with a significant regression of spontaneous breast cancer in mice, suggesting a better prognosis. In the present study, high expression levels of TLR4 and TLR7 were revealed to have a close association with shortened survival and worse prognosis. However, no significant differences in survival were observed between high and low expression levels of other TLRs, which may be due to the short follow-up period of patients included in TCGA database (<5 years).
TLR signaling pathways promote survival, proliferation and apoptosis of cancer cells, as well as IFN, cytokine and chemokine production. For instance, TLR1 and TLR2 mediated by the PI3K-Akt pathway, or TLR4, TLR5 and TLR6 mediated by the NF-κB pathway lead to increased levels of several pro-inflammatory cytokines (including TNF-α, IL-1β, IL-6 and IL-12) and chemokines [including IL-8, C-C motif chemokine ligand 3 (CCL3), CCL4 and CCL5], thus causing inflammation. TLR3, TLR7, TLR8 and TLR9 mediated by MyD88-dependent/independent pathways induce the production of co-stimulatory molecules [such as cluster of differentiation 40 (CD40), CD80 and CD86], inflammatory cytokines (including IFN-α and IFN-β) and chemokines (including CXCL9, CXCL10 and CXCL11), thus promoting antibacterial and antiviral effects (33–36). In line with this, the present study revealed positive correlations between TLRs and the downstream signaling molecules IL-1β, IL-6, IL-8, TNF-α, IFN-α, IFN-1β and CXCL10 in breast cancer. These results suggest that TLR-mediated signaling serves an important role in the regulation of tumorigenesis. It may be useful to investigate how various TLR pathways affect the different breast tumor cell types and their potential roles in breast cancer development in future studies.
In conclusion, the current study analyzed the expression profile of TLRs and identified that most TLRs were downregulated in breast cancer. TLR3, TLR5 and TLR9 were associated with specific subtypes, and TLR10 was related to tumor stages, suggesting that TLRs have profound effects on breast cancer incidence and progression. TLRs and their downstream inflammatory cytokines serve important roles in innate immunity and are also key regulatory factors in tumor development. Silencing or activating TLRs may be an effective therapeutic target of breast cancer. Therefore, studying the expression patterns of TLRs and the interaction of their signaling pathways in breast cancer will help to explore the clinical applications of anti-tumor therapy and to provide new insights for breast cancer treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Shandong Provincial Natural Science Foundation of China (grant nos. ZR2011HZ004 and ZR2017BC073) and the Project of Shandong Province Higher Educational Science and Technology Program (grant nos. J10LC21 and J16LE07).
Availability of data and materials
The datasets used and analyzed during the current study are available from TCGA database (http://cancergenome.nih.gov/).
Authors' contributions
SS and CX analyzed the data and drafted the manuscript. XF, YZ and HL contributed to the interpretation of data and revised the manuscript. GY and WW conceived and designed the study. All authors have reviewed and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mifsud EJ, Tan AC, Jackson DC. TLR Agonists as modulators of the innate immune response and their potential as agents against infectious disease. Front Immunol. 2014;5:79. doi: 10.3389/fimmu.2014.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: A cell biological perspective. Annu Rev Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palm E, Demirel I, Bengtsson T, Khalaf H. The role of Toll-like and protease-activated receptors in the expression of cytokines by gingival fibroblasts stimulated with the periodontal pathogen Porphyromonas gingivalis. Cytokine. 2015;76:424–432. doi: 10.1016/j.cyto.2015.08.263. [DOI] [PubMed] [Google Scholar]
- 4.Johnston DG, Corr SC. Toll-like receptor signalling and the control of intestinal barrier function. Methods Mol Biol. 2016;1390:287–300. doi: 10.1007/978-1-4939-3335-8_18. [DOI] [PubMed] [Google Scholar]
- 5.Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nat Rev Cancer. 2009;9:57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 6.Yu L, Chen S. Toll-like receptors expressed in tumor cells: Targets for therapy. Cancer Immunol Immunother. 2008;57:1271–1278. doi: 10.1007/s00262-008-0459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 8.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 9.Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J, Shi B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929–2243. [PMC free article] [PubMed] [Google Scholar]
- 10.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 12.Yadav BS, Chanana P, Jhamb S. Biomarkers in triple negative breast cancer: A review. World J Clin Oncol. 2015;6:252–263. doi: 10.5306/wjco.v6.i6.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie W, Wang Y, Huang Y, Yang H, Wang J, Hu Z. Toll-like receptor 2 mediates invasion via activating NF-kappaB in MDA-MB-231 breast cancer cells. Biochem Biophys Res Commun. 2009;379:1027–1032. doi: 10.1016/j.bbrc.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Salaun B, Zitvogel L, Asselin-Paturel C, Morel Y, Chemin K, Dubois C, Massacrier C, Conforti R, Chenard MP, Sabourin JC, et al. TLR3 as a biomarker for the therapeutic efficacy of double-stranded RNA in breast cancer. Cancer Res. 2011;71:1607–1614. doi: 10.1158/0008-5472.CAN-10-3490. [DOI] [PubMed] [Google Scholar]
- 15.Haricharan S, Brown P. TLR4 has a TP53-dependent dual role in regulating breast cancer cell growth. Proc Natl Acad Sci USA. 2015;112:E3216–E3225. doi: 10.1073/pnas.1420811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehsan N, Murad S, Ashiq T, Mansoor MU, Gul S, Khalid S, Younas M. Significant correlation of TLR4 expression with the clinicopathological features of invasive ductal carcinoma of the breast. Tumour Biol. 2013;34:1053–1059. doi: 10.1007/s13277-013-0645-y. [DOI] [PubMed] [Google Scholar]
- 17.Cai Z, Sanchez A, Shi Z, Zhang T, Liu M, Zhang D. Activation of Toll-like receptor 5 on breast cancer cells by flagellin suppresses cell proliferation and tumor growth. Cancer Res. 2011;71:2466–2475. doi: 10.1158/0008-5472.CAN-10-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shuang C, Weiguang Y, Zhenkun F, Yike H, Jiankun Y, Jing X, Xinghan L, Yue L, Dalin L. Toll-like receptor 5 gene polymorphism is associated with breast cancer susceptibility. Oncotarget. 2017;8:88622–88629. doi: 10.18632/oncotarget.20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger R, Fiegl H, Goebel G, Obexer P, Ausserlechner M, Doppler W, Hauser-Kronberger C, Reitsamer R, Egle D, Reimer D, et al. Toll-like receptor 9 expression in breast and ovarian cancer is associated with poorly differentiated tumors. Cancer Sci. 2010;101:1059–1066. doi: 10.1111/j.1349-7006.2010.01491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merrell MA, Ilvesaro JM, Lehtonen N, Sorsa T, Gehrs B, Rosenthal E, Chen D, Shackley B, Harris KW, Selander KS. Toll-like receptor 9 agonists promote cellular invasion by increasing matrix metalloproteinase activity. Mol Cancer Res. 2006;4:437–447. doi: 10.1158/1541-7786.MCR-06-0007. [DOI] [PubMed] [Google Scholar]
- 21.Ridnour LA, Cheng RY, Switzer CH, Heinecke JL, Ambs S, Glynn S, Young HA, Trinchieri G, Wink DA. Molecular pathways: Toll-like receptors in the tumor microenvironment-poor prognosis or new therapeutic opportunity. Clin Cancer Res. 2013;19:1340–1346. doi: 10.1158/1078-0432.CCR-12-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato Y, Goto Y, Narita N, Hoon DS. Cancer cells expressing Toll-like receptors and the tumor microenvironment. Cancer Microenviron. 2009;2(Suppl 1):S205–S214. doi: 10.1007/s12307-009-0022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Smith C, Yin H. Targeting Toll-like receptors with small molecule agents. Chem Soc Rev. 2013;42:4859–4866. doi: 10.1039/c3cs60039d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidd LC, Rogers EN, Yeyeodu ST, Jones DZ, Kimbro KS. Contribution of Toll-like receptor signaling pathways to breast tumorigenesis and treatment. Breast Cancer (Dove Med Press) 2013;5:43–51. doi: 10.2147/BCTT.S29172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green TL, Santos MF, Ejaeidi AA, Craft BS, Lewis RE, Cruse JM. Toll-like receptor (TLR) expression of immune system cells from metastatic breast cancer patients with circulating tumor cells. Exp Mol Pathol. 2014;97:44–48. doi: 10.1016/j.yexmp.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Amarante MK, de Oliveira KB, Guembarovski RL, da Silva do Amaral Herrera AC, Guembarovski AL, Sobrinho WJ, Voltarelli JC, Watanabe MA. Toll-like receptor 3: implications for proinflammatory microenvironment in human breast cancer. Mol Biol Rep. 2012;39:11087–11092. doi: 10.1007/s11033-012-2013-1. [DOI] [PubMed] [Google Scholar]
- 27.González-Reyes S, Marín L, González L, González LO, del Casar JM, Lamelas ML, González-Quintana JM, Vizoso FJ. Study of TLR3, TLR4 and TLR9 in breast carcinomas and their association with metastasis. BMC Cancer. 2010;10:665. doi: 10.1186/1471-2407-10-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Zhou H, Feng P, Zhou X, Wen H, Xie X, Shen H, Zhu X. Reduced expression of Toll-like receptor 4 inhibits human breast cancer cells proliferation and inflammatory cytokines secretion. J Exp Clin Cancer Res. 2010;29:92. doi: 10.1186/1756-9966-29-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeyeodu ST, Kidd LR, Oprea-Ilies GM, Burns BG, Vancleave TT, Shim JY, Kimbro KS. IRAK4 and TLR3 sequence variants may alter breast cancer risk among African-American women. Front Immunol. 2013;4:338. doi: 10.3389/fimmu.2013.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jukkola-Vuorinen A, Rahko E, Vuopala KS, Desmond R, Lehenkari PP, Harris KW, Selander KS. Toll-like receptor-9 expression is inversely correlated with estrogen receptor status in breast cancer. J Innate Immun. 2009;1:59–68. doi: 10.1159/000151602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mehmeti M, Allaoui R, Bergenfelz C, Saal LH, Ethier SP, Johansson ME, Jirström K, Leandersson K. Expression of functional toll like receptor 4 in estrogen receptor/progesterone receptor-negative breast cancer. Breast Cancer Res. 2015;17:130. doi: 10.1186/s13058-015-0640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu H, Wagner WM, Gad E, Yang Y, Duan H, Amon LM, Van Denend N, Larson ER, Chang A, Tufvesson H, Disis ML. Treatment failure of a TLR-7 agonist occurs due to self-regulation of acute inflammation and can be overcome by IL-10 blockade. J Immunol. 2010;184:5360–5367. doi: 10.4049/jimmunol.0902997. [DOI] [PubMed] [Google Scholar]
- 33.Bhatelia K, Singh K, Singh R. TLRs: Linking inflammation and breast cancer. Cell Signal. 2014;26:2350–2357. doi: 10.1016/j.cellsig.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 34.Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, Sandercock AM, Robinson CV, Latz E, Gay NJ. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gay NJ, Gangloff M, O'Neill LA. What the Myddosome structure tells us about the initiation of innate immunity. Trends Immunol. 2011;32:104–109. doi: 10.1016/j.it.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, et al. Breast cancer prognostic classification in the molecular era: The role of histological grade. Breast Cancer Res. 2010;12:207. doi: 10.1186/bcr2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from TCGA database (http://cancergenome.nih.gov/).