Abstract
Triple-negative breast cancer (TNBC) accounts for the worst prognosis of all types of breast cancers due to a high risk of recurrence and a lack of targeted therapeutic options. Extensive effort is required to identify novel targets for TNBC. In the present study, a robust rank aggregation (RRA) analysis based on genome-wide gene expression datasets involving TNBC patients from the Gene Expression Omnibus (GEO) database was performed to identify key genes associated with TNBC. A total of 194 highly ranked differentially expressed genes (DEGs) were identified in TNBC vs. non-TNBC. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) enrichment analysis was utilized to explore the biological functions of the identified genes. These DEGs were mainly involved in the biological processes termed positive regulation of transcription from RNA polymerase II promoter, negative regulation of apoptotic process, response to drug, response to estradiol and negative regulation of cell growth. Genes were mainly involved in the KEGG pathway termed estrogen signaling pathway. The aberrant expression of several randomly selected DEGs were further validated in cell lines, clinical tissues and The Cancer Genome Atlas (TCGA) cohort. Furthermore, all the top-ranked DEGs underwent survival analysis using TCGA database, of which overexpression of 4 genes (FABP7, ART3, CT83, and TTYH1) were positively correlated to the life expectancy (P<0.05) of TNBC patients. In addition, a model consisting of two genes (FABP7 and CT83) was identified to be significantly associated with the overall survival (OS) of TNBC patients by means of Cox regression, Kaplan-Meier, and receiver operating characteristic (ROC) analyses. In conclusion, the present study identified a number of key genes as potential biomarkers involved in TNBC, which provide novel insights into the tumorigenesis of TNBC at the gene level and may serve as independent prognostic factors for TNBC prognosis.
Keywords: triple-negative breast cancer, robust rank aggregation, biomarker, prognostic risk model, differentially expressed gene
Introduction
In the United States, breast cancer is the most common cancer among women, accounting for 30% of all new cancer diagnoses in women, and is the second leading cause of cancer-related deaths of over 40,000 women each year (1). Triple-negative breast cancer (TNBC), a subtype defined clinically by the lack of estrogen receptor alpha (ERα), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2), is composed of 15 to 20% of all types of breast cancers and is associated with highly aggressive biological characteristics (2). TNBC frequently presents in younger patients, and often ends with poor clinical outcome with high rates of recurrence and mortality due in part to lack of therapeutic options beyond chemotherapy (3,4). However, although TNBC exhibits an excellent clinical response to neoadjuvant chemotherapy (NCT) or conventional postoperative chemotherapy, it often becomes refractory (5,6). It is reported that >50% of patients will develop recurrent disease with residual disease after chemotherapy (7). For these reasons, there is an urgent need to reveal the molecular mechanism and identify novel targeted therapeutics strategies for TNBC patients.
Recent evidence highlights that TNBC is a complex disease characterized by molecular and phenotypic heterogeneity compared with other types (8), and can be further subdivided into four to six distinct molecular subtypes for their unique expression signatures and ontologies, including luminal androgen receptor, basal-like and mesenchymal subtypes (9,10). Currently, high-throughput genome-wide gene expression datasets are available freely in public databases, such as Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) database. By means of comprehensive gene expression analysis, genome-wide analysis of these public high-throughput data can provide insight into the molecular mechanism underlying those complex diseases with different patterns. However, due to limited sample size and heterogeneous characteristics, the results of individual gene expression analysis are often inconsistent or even discrepant among each study, and may not convincingly predict the functional gene networks involved in disease. Therefore, a comprehensive integrated analysis of this enormous volume of data to get credible outcomes is necessary.
The robust rank aggregation (RRA) analysis, a rigorous and well-accepted approach designed specifically for comparison of several ranked gene lists, can be used to integrate multiple genome-wide gene expression datasets and identify key genes most likely associated in several diseases (11,12). By means of comparison of several ranked gene lists in an unbiased manner, RRA is particularly suitable for identification of statistically significant genes when high-throughput sequencing experiments are conducted by different platforms covering different sets of gene probes (11,13). At present, there has been no attempt to integrate these genome-wide gene expression datasets regarding TNBC vs. non-TNBC using the RRA method.
In the present study, RRA analysis based on four high-throughput genome-wide gene profiling datasets was first performed in the GEO database to identify key genes involved in TNBC. The differentially expressed genes (DEGs) were firstly identified by comparing the gene expression profiles between TNBC and non-TNBC samples. These genes then underwent gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis to explore the biological functions. Subsequently, the top-ranked DEGs in cell lines, clinical tissues and TCGA cohorts were further randomly validated. Finally, the potential clinical significance of these genes and a two-gene signature acting as novel molecular prognostic markers were examined in TCGA cohorts using the Kaplan-Meier method and ROC analysis. The present study aimed to provide insights into TNBC pathogenesis and identify some novel potential biomarkers for TNBC.
Materials and methods
Dataset search and eligibility criteria
To identify genome-wide gene expression datasets in TNBC patients, the widely used Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo/) database was searched.
The search terms were based on a combination of Triple-Negative Breast Cancer and TNBC. The search results and relevant datasets were filtrated according to the following eligibility criteria: i) Genome-wide expression profiling designed for comparison between TNBC and non-TNBC or involving TNBC and non-TNBC using high-throughput array or next generation sequencing were included; ii) databases using only cell lines or peripheral blood of patients were excluded; iii) the total number of samples should not be <15; iv) there should not be <10 differentially expressed genes identified in each gene expression dataset; and v) the raw data should be provided in selected databases which could be used for reanalysis. Studies that did not meet the aforementioned eligibility criteria were excluded.
RRA analysis
The gene expression profiling was firstly annotated using the annotation document of corresponding platforms, followed with normalization of the expression data using ‘limma’ package of Bioconductor (http://www.bioconductor.org/packages/release/bioc/html/limma.html). Subsequently, the ranked lists of both upregulated and downregulated genes in each dataset were generated according to their fold changes. Finally, an R package of ‘Robust Rank Aggregation’ was utilized for the integrated analysis of these ranked gene lists (12). P-values were calculated for each gene, indicating the possibility of ranking high in the final gene list. In addition, a Bonferroni correction was also used to reduce false-positive results.
Functional enrichment analysis
To reveal the possible functions of these DEGs identified by RRA method with Bonferroni's adjusted P<0.05 and logarithmic fold changes (logFCs)>1, GO functional enrichment analysis and KEGG pathway enrichment analysis were performed using ‘GO.db’ (http://www.bioconductor.org/packages/release/data/annotation/html/GO.db.html) and ‘org.Hs.eg.db’ (http://www.bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html) packages in Bioconductor. The top enriched ontological or pathway terms with Bonferroni's adjusted P<0.05 were selected.
Cell culture
The TNBC and non-TNBC cell lines, MDA-MB-231 and MCF-7, were purchased from the Cell Bank of the Chinese Scientific Academy. These two cell lines were maintained in Roswell Park Memorial Institute (RMPI)-1640 medium (Gibco; Life Technologies; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum (FBS; Biological Industries) at 37°C and 5% CO2.
Validation of the selected DEGs using quantitative real-time PCR
To validate the results of RRA analysis, immunohistochemically diagnosed fresh samples were collected from 20 TNBC patients and 20 non-TNBC patients by experienced surgeons at the First Affiliated Hospital of Zhejiang University. The written informed consents were provided from all patients, and the study was approved by the Institute Ethics Committee of the hospital. After being surgically resected, the samples were stored in sterile tubes and frozen immediately in liquid nitrogen.
Total RNA of the cell lines and clinical samples was extracted using Qiagen RNeasy Kit (QIAGEN, Inc.) and then reverse-transcribed into complementary DNA (cDNA) using a cDNA synthesis kit (Takara Biotechnology Co., Ltd.), at 37°C for 15 min and 85°C for 5 sec, according to the manufacturer's instructions. Subsequently, six genes randomly selected from the identified 40 top-ranked DEGs through RRA analysis were used to validate the expression by RT-qPCR using the SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.). The 20-µl system included 10 µl SYBR Premix Ex Taq, 1 µl each primer, 2 µl cDNA and 6 µl double distilled water. PCR was performed as follows: 10 sec at 95°C for 1 cycle, then 5 sec at 95°C and 20 sec at 60°C for 39 cycles. GAPDH was used as the reference gene, and the relative expression of each gene to GAPDH. The specific primers used in the present study were as follows: ADP-ribosyltransferase 3 (ART3) forward, 5′-GCAACCATGATTCTAGTGGACA-3′ and reverse, 5′-CTTTAGCAGTTGGGGAACGTAT-3′; fatty acid binding protein 7 (FABP7) forward, 5′-GGCTTTGCCACTAGGCAGG-3′ and reverse, 5′-TGACCACTTTGTCTCCTTCTTGA-3′; HORMA domain containing 1 (HORMAD1) forward, 5′-GCCCAGTTGCAGAGGACTC-3′ and reverse, 5′-TCTTGTTCCATAAGCGCATTCT-3′; trefoil factor1 (TFF1) forward, 5′-CCCCGTGAAAGACAGAATTGT-3′ and reverse, 5′-GGTGTCGTCGAAACAGCAG-3′; anterior gradient 2 (AGR2) forward, 5′-GTCAGCATTCTTGCTCCTTGT-3′ and reverse, 5′-GGGTCGAGAGTCCTTTGTGTC-3′; and forkhead box A1 (FOXA1) forward, 5-GCAATACTCGCCTTACGGCT-3′ and reverse, 5′-TACACACCTTGGTAGTACGCC-3′. All experiments were performed in triplicate.
TCGA database
The mRNA expression data and corresponding clinical information were downloaded from TCGA data portal (https://genome-cancer.ucsc.edu/). After extracting the ER, PR and HER2 information of each sample, a total of 99 TNBC cases and 558 non-TNBC cases were identified. Cases with uncertain status (equivocal, indeterminate or unknown) of any ER, PR or HER2 were excluded. All the top-ranked DEGs underwent survival analysis using Kaplan-Meier survival curve analysis. Differences were considered significant with a P-value <0.05.
Statistical analyses
All data in the present study were analyzed using the R statistical package (R version 3.4.4) unless otherwise stated. The univariate Cox proportional hazard analysis was firstly conducted in the TCGA cohort to identify genes significantly (P-value <0.05) correlated with TNBC patient survival as candidate genes. Then, these genes were further screened to construct a gene signature using multivariate Cox regression analysis. Two genes from the candidate genes were selected to construct a risk score formula by which a prognostic risk score for each patient was calculated. Subsequently, these patients were further separated into ‘low-risk’ and ‘high-risk’ groups using the median risk score as the cutoff point. Kaplan-Meier survival curve analysis was then applied to compare the differences between the two groups for survival time. Finally, ROC analysis was conducted and visualized by SPSS 24.0 (SPSS, Inc.).
Results
Characteristics of included microarray datasets
After thoroughly searching in the Gene Expression Omnibus (GEO) database according to the eligibility criteria, a total of 4 genome-wide gene expression datasets involving TNBC and non-TNBC were finally included. The characteristics of these 4 datasets are summarized in Table I including GSE number, involved participants and detection platforms. Among these datasets, the number of TNBC patients ranged from 14 to 198, while the number of non-TNBC subjects ranged from 5 to 67. Finally, the pooled dataset included 251 TNBC samples and 158 non-TNBC samples. Various microarray platforms were used in the studies including GPL570 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL570), GPL10558 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL10558) and GPL6244 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL6244).
Table I.
Summary of 4 genome-wide gene expression datasets including TNBC and non-TNBC tissues.
| Dataset ID | GSE number | Samples | Platform |
|---|---|---|---|
| 1 | GSE76275 | 198 TNBC and 67 non-TNBC | GPL570 |
| 2 | GSE36693 | 21 TNBC and 66 non-TNBC | GPL10558 |
| 3 | GSE27447 | 14 TNBC and 5 non-TNBC | GPL6244 |
| 4 | GSE3744 | 18 TNBC and 20 non-TNBC | GPL570 |
TNBC, triple-negative breast cancer.
RRA analysis
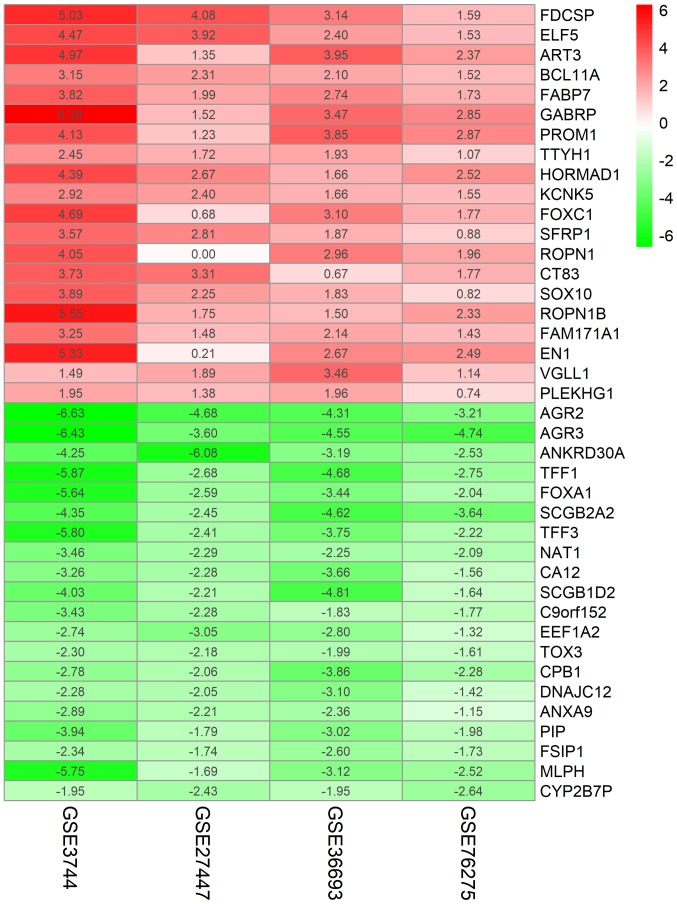
To identify credible aberrantly expressed genes involved in TNBC vs. non-TNBC, an integrated analysis was performed using the R package ‘Robust Rank Aggregation’, and upregulated and downregulated ranked gene lists were successfully generated. A total of 194 highly ranked differentially expressed genes (DEGs) with P-values <0.05 were identified in TNBC vs. non-TNBC. Table SI exhibited the top 100 significant DEGs in TNBC compared with non-TNBC samples. As revealed in Fig. 1, the top 20 upregulated and downregulated genes expressed consistently across all profiling were identified by the RRA analysis.
Figure 1.
Top 20 upregulated and downregulated genes in TNBC vs. non-TNBC. The red indicates increased gene expression, whereas the green indicates decreased gene expression; the number exhibited in the figure indicates the logarithmic fold change of genes in each dataset. TNBC, triple-negative breast cancer.
Functional analysis of DEGs
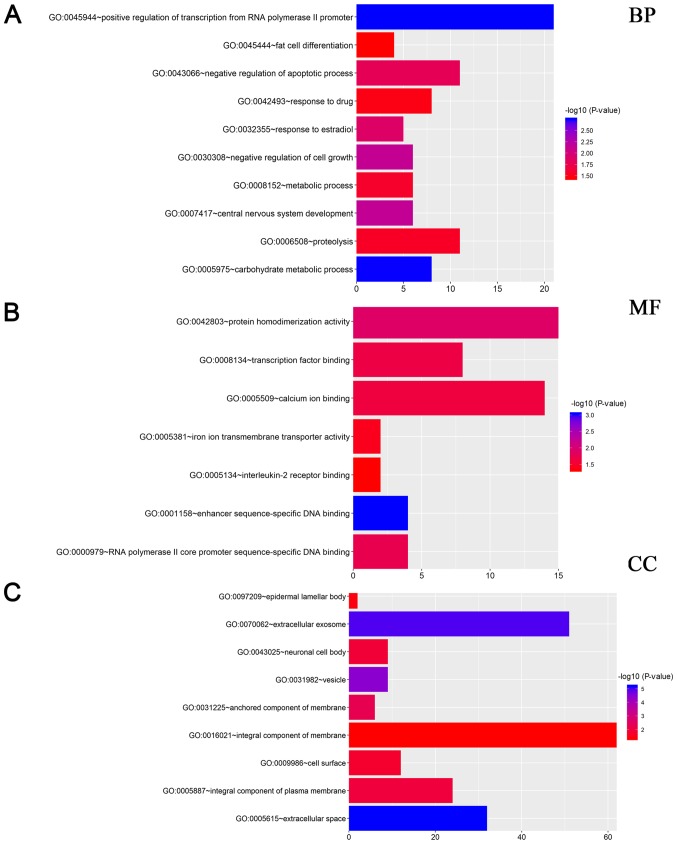
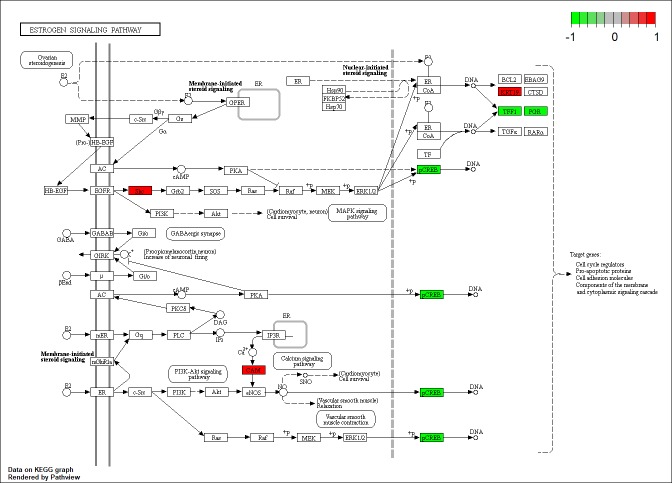
To explore the systematic features and biological functions of the identified DEGs, GO term enrichment analysis and KEGG pathway analysis, were performed. For GO annotation, cellular component (CC), molecular function (MF) and participation in biological processes (BP) were included. In the BP analysis, it was revealed that DEGs were significantly enriched in terms of ‘positive regulation of transcription from RNA polymerase II promoter’ (Fig. 2A). Moreover, as revealed in Fig. 2A, DEGs were also involved in negative regulation of apoptotic process, response to drug, and response to estradiol, indicating that these gene were closely associated with TNBC. For MF analysis, DEGs were significantly related with protein homodimerization activity, transcription factor binding and calcium ion binding (Fig. 2B). CC analysis further revealed that these genes were mainly localized in extracellular exosome, integral component of membrane, and extracellular space (Fig. 2C). Furthermore, as revealed in Fig. 3, KEGG pathway annotation revealed that genes were significantly enriched in estrogen signaling pathway, which was closely related with non-TNBC.
Figure 2.
GO analysis of DEGs identified by the RRA method. (A) Enriched BP of these genes; (B) enriched MF of these genes; (C) enriched CC of these genes. DEGs, differentially expressed genes; RRA robust rank aggregation; BP, biological processes; MF, molecular functions; CC, cellular components.
Figure 3.
Differentially expressed genes in the estrogen signaling pathway. Red, upregulated genes; green, downregulated genes.
Validation of mRNA by RT-qPCR
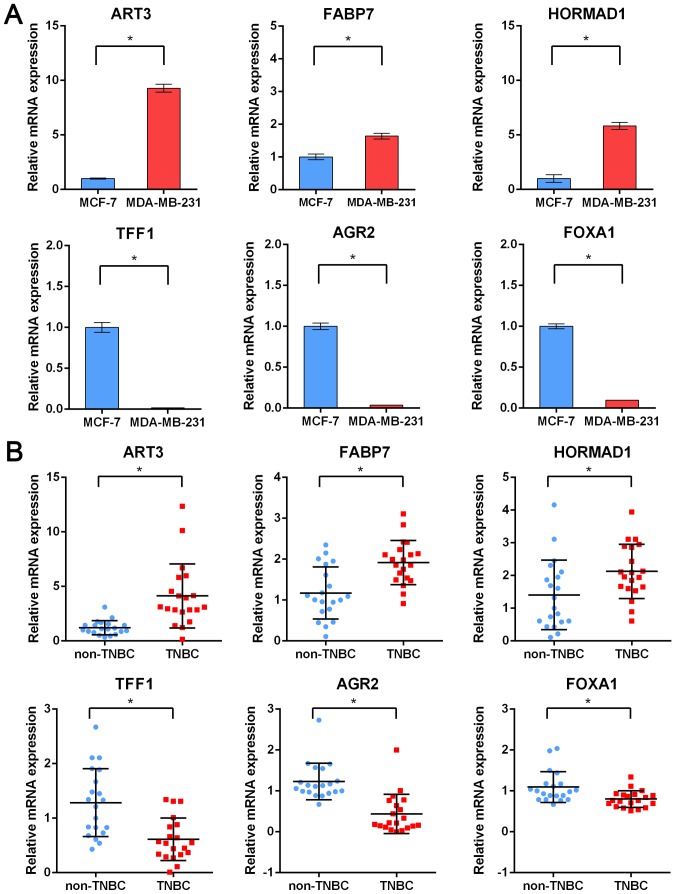
According to the results analyzed by bioinformatics, the expression of six genes randomly selected in top upregulated and downregulated DEGs revealed in the RRA analysis was then validated. Two cell lines, MDA-MB-231 and MCF-7 belonging to TNBC and non-TNBC respectively, were selected to determine the expression of randomly selected key genes including ART3, FABP7, HORMAD1, TFF1, AGR2 and FOXA1. As revealed in Fig. 4A, the expression of ART3, FABP7 and HORMAD1 was significantly upregulated whereas the expression of TFF1, AGR2 and FOXA1 was significantly downregulated in the TNBC cell line, which was consistent with the results in the RRA analysis. Next, 20 fresh clinical tissues originating from TNBC or non-TNBC patients were used to further validate these six randomly selected genes. Consistent with the results obtained with the cell lines, it was revealed that ART3, FABP7 and HORMAD1 were respectively upregulated in TNBC tissues compared to non-TNBC breast tissues, whereas TFF1, AGR2 and FOXA1 were downregulated analogously (Fig. 4B). Furthermore, similar results were also obtained in the TCGA validation cohort including 99 TNBC samples and 558 non-TNBC samples (Fig. S1). Collectively, these findings indicated that reliable analysis results were obtained from the RRA method and these top DEGs may serve as key regulators of TNBC.
Figure 4.
Validation of six randomly selected DEGs through RT-qPCR. (A) Expression of ART3, FABP7, HORMAD1, TFF1, AGR2 and FOXA1 in TNBC cell lines compared with non-TNBC cell lines. (B) Expression of ART3, FABP7, HORMAD1, TFF1, AGR2 and FOXA1 in TNBC patients compared with non-TNBC patients. *P<0.05. DEGs, differentially expressed genes; ART3, ADP-ribosyltransferase 3; FABP7, fatty acid binding protein 7; HORMAD1, HORMA domain containing 1; TFF1, trefoil factor1; AGR2, anterior gradient 2; FOXA1, forkhead box A1; TNBC, triple-negative breast cancer.
Association between DEGs and clinical outcome in TNBC
To explore the prognostic values of DEGs in TNBC patients, the breast cancer data including gene expression and clinical outcome were extracted and analyzed from TCGA, which has vigorous criteria for sample collection and processing (14). Next, the association between the identified top DEGs and overall survival (OS) of 99 TNBC patients was assessed using the Kaplan-Meier survival curve analysis. Notably, there were 4 genes, including FABP7, ART3, CT83 and TTYH1, which were positively correlated to the life expectancy (Fig. S2), indicating that these genes may serve as prognostic biomarkers.
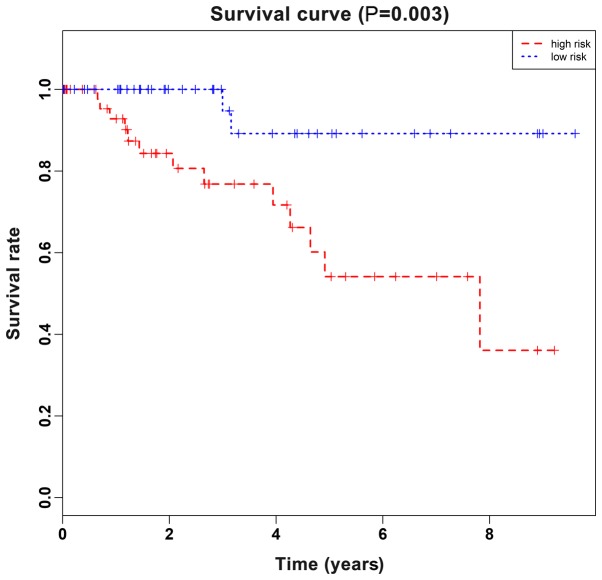
To further identify key genes in the top 40 DEGs associated with OS of TNBC patients, univariate Cox proportional hazard regression analysis (data not shown) was performed using gene expression as variables in the TCGA dataset. The results revealed, 5 genes that were significantly associated with the OS of TNBC patients with P-values <0.05. Subsequently, multivariate Cox regression with stepwise regression (data not shown) was performed and screened for these 5 genes. In consequence, a hazard ratio model consisting of 2 genes, including FABP7 and CT83, was identified as the optimum prognostic model for predicting the prognosis of breast cancer patients. Notably, the risk scoring formula of these 3 genes was obtained as follows: Risk score=−0.1256×FABP7-0.119×CT83. TNBC patients in TCGA dataset were divided into a high-risk group or low-risk group using the median of risk score as the cutoff point, which calculated by the aforementioned formula. As revealed in Fig. 5, the patients in the high-risk group suffered a significantly (P<0.05) worse prognosis compared with the low-risk group. Collectively, these results demonstrated that the two-gene signature could well differentiate high-risk patients from low-risk patients, which hinted its potential clinical application value in the prognostic prediction of TNBC patients.
Figure 5.
Kaplan-Meier estimates of the OS of TNBC patients in TCGA cohort using a two-gene signature. A log-rank test was conducted to evaluate the survival differences between the two curves. OS, overall survival; TNBC, triple-negative breast cancer; TCGA, The Cancer Genome Atlas.
Evaluation of the predictive performance using ROC analysis
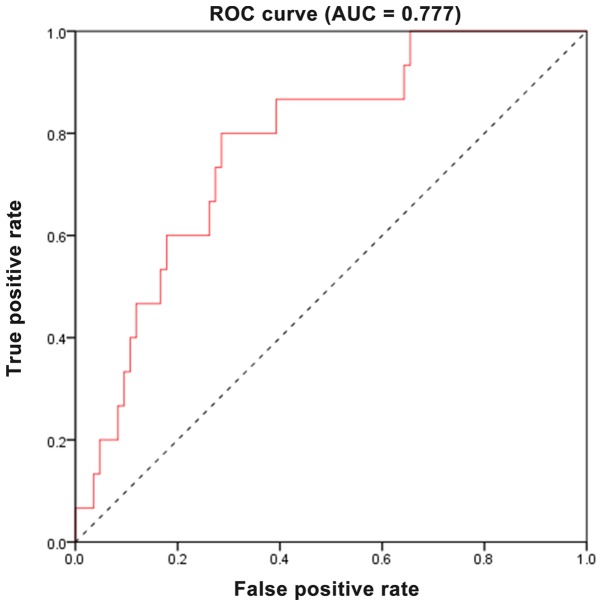
To further evaluate the predictive accuracy of the two-gene signature, the sensitivity and specificity of the two-gene signature in predicting prognosis were assessed using ROC analysis. As revealed in Fig. 6, the area under the curve (AUC) was 0.777, revealing that the two-gene signature had a relative high sensitivity and specificity. Consequently, along with the aforementioned results, the two-gene signature may serve as a potential marker to predict the prognostic survival of patients with TNBC, possessing significant clinical application value.
Figure 6.
ROC curve analysis reveals the sensitivity and specificity of a two-gene signature in predicting the OS of patients. ROC, receiver operating characteristic; OS, overall survival.
Discussion
TNBC, a subtype of breast cancer comprising 15–20% of breast cancers, is a highly aggressive cancer with poor prognosis due to its tendency for recurrence and metastasis (9,15–17). Its pathogenesis and underlying mechanisms are still unclear. In the present study, various genome-wide gene expression datasets were integrated using the RRA method for the purpose of exploring the underlying mechanism of TNBC at a systems level. According to inclusion criteria, four genome-wide datasets were finally downloaded from the GEO database, which involved a total of 251 TNBC and 158 non-TNBC patients. By means of integrated analysis, a large cohort of significantly or downregulated genes were identified, some of which has been documented to be strongly associated with estrogen sensitivity in breast cancer, such as ELF5 (18,19), TFF1 (20,21), and TFF3 (22,23). However, there are also some genes identified to be novel TNBC gene signatures, and their underlying mechanisms in TNBC are still poorly understood, which require further exploration in a future study.
Subsequently, GO annotation and KEGG pathway analysis were performed to elucidate the significance of these identified aberrantly expressed genes. The GO analysis indicated that these genes were significantly enriched in the following GO terms: Positive regulation of transcription from RNA polymerase II promoter, negative regulation of apoptotic process, response to drug, and response to estradiol. This indicated that DEGs exerted their biological function by catalyzing the process of transcription. In addition, the enriched GO terms also suggested that DEGs were closely related to the biological behavior of TNBC, since it has been well documented that TNBC exhibits a distinctly aggressive nature with higher rates of relapse, resistance to endocrine therapy (ET) and sensitivity to cytotoxic chemotherapy (24–26). Moreover, the KEGG pathway analysis revealed that DEGs were significantly enriched in estrogen signaling pathway, which is of importance to luminal breast cancer with positive estrogen receptor (27).
Next, further confirmation of the DEGs analyzed from the RRA method was performed using genes selected at random. RT-PCR data revealed that ART3, FABP7 and HORMAD1 were significantly upregulated whereas TFF1, AGR2 and FOXA1 were significantly downregulated in both cell lines and clinical samples. Notably, all these genes were confirmed to be associated with tumorigenesis. Previous investigation indicated that overexpression of ART3 promoted TNBC via activation of Akt and ERK pathways (28). FABP7 has been reported to act as an oncogene in TNBC, and the FABP7/RXRβ pathway was revealed to promote cell proliferation in TNBC (29). HORMAD1 has been demonstrated to contribute to homologous recombination (HR) deficiency in TNBC and be associated with response to platinum-based chemotherapy in this disease (30). A study by Fritzsche et al (31) demonstrated that AGR2 was positively correlated with improved outcomes in breast cancer. In addition, AGR2 was recently revealed to be linked with FOXA1, and the FOXA1/ERα/AGR2 signaling axis may be utilized as a therapeutic target for the treatment of breast cancer (32). Furthermore, deficiency of TFF1, a small cysteine-rich acidic secreted protein, was demonstrated to be involved in tumorigenesis of gastric cancer (33) and breast cancer (34). These results, collectively, indicated that integrated analysis could provide reliable results and these DEGs may play pivotal roles in tumorigenesis and development in breast cancer.
In order to determine whether these identified top 40 DEGs were related to the life expectancy of patients with TNBC, all the top DEGs underwent survival analysis using TCGA database. Using Kaplan-Meier survival curve analysis, 4 genes (FABP7, ART3, CT83 and TTYH1) were identified to be positively correlated to life expectancy. Moreover, a two-gene signature, including FABP7 and CT83, was predicted to be significantly associated with the OS of breast cancer patients using univariate Cox analysis followed with multivariate Cox analysis. By calculating risk score according to a formula, the two-gene signature performed well in differentiating low-risk and high-risk groups, which exhibited significant prognostic differences. Finally, ROC analysis was performed and a relatively high AUC of 0.777 was demonstrated, revealing that the two-gene signature could act as an independent predictor of survival of patients with TNBC.
To sum up, the present study provides an integrated analysis of gene expression profiles of TNBC compared with non-TNBC and identifies a number of key genes involved in the pathogenesis of TNBC. It is inferred from the present research that these DEGs may regulate the initiation and progression of TNBC in various ways. Some of these key genes are novel and their precise roles in TNBC remain unclear. The present findings in this study may provide insights into the pathogenesis of TNBC at a systems level, and also identify some alternative biomarkers and potential therapeutic targets for TNBC patients. Continuous studies on these key genes should be performed to elucidate their detailed role in TNBC.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by grant 2017KY019 from the Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (China).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GZ, WL and QS conceived and designed the present study. GZ, WL, QS and KY performed the experiments and analyzed the data. YZ interpreted the data and wrote the manuscript. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Zhejiang University (Zhejiang, China). Written informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 3.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 4.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 5.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: Disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 6.Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 7.von Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich J, Huober J, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 8.Byler S, Goldgar S, Heerboth S, Leary M, Housman G, Moulton K, Sarkar S. Genetic and epigenetic aspects of breast cancer progression and therapy. Anticancer Res. 2014;34:1071–1077. [PubMed] [Google Scholar]
- 9.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Võsa U, Kolde R, Vilo J, Metspalu A, Annilo T. Comprehensive meta-analysis of microRNA expression using a robust rank aggregation approach. Methods Mol Biol. 2014;1182:361–373. doi: 10.1007/978-1-4939-1062-5_28. [DOI] [PubMed] [Google Scholar]
- 12.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi KQ, Lin Z, Chen XJ, Song M, Wang YQ, Cai YJ, Yang NB, Zheng MH, Dong JZ, Zhang L, Chen YP. Hepatocellular carcinoma associated microRNA expression signature: Integrated bioinformatics analysis, experimental validation and clinical significance. Oncotarget. 2015;6:25093–25108. doi: 10.18632/oncotarget.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin L, Andersen JN, Futreal PA. Cancer genomics: From discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 15.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 17.Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer. 2009;9(Suppl 2):S73–S81. doi: 10.3816/CBC.2009.s.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalyuga M, Gallego-Ortega D, Lee HJ, Roden DL, Cowley MJ, Caldon CE, Stone A, Allerdice SL, Valdes-Mora F, Launchbury R, et al. ELF5 suppresses estrogen sensitivity and underpins the acquisition of antiestrogen resistance in luminal breast cancer. PLoS Biol. 2012;10:e1001461. doi: 10.1371/journal.pbio.1001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furth PA, Nakles RE, Millman S, Diaz-Cruz ES, Cabrera MC. Signal transducer and activator of transcription 5 as a key signaling pathway in normal mammary gland developmental biology and breast cancer. Breast Cancer Res. 2011;13:220. doi: 10.1186/bcr2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L, Yan T, Jiang Y, Di G, Shen Z, Shao Z, Lu J. Prognostic and predictive value of TFF1 for adjuvant endocrine therapy in Chinese women with early ER positive breast cancer: Comparing aromatase inhibitors with tamoxifen. Breast. 2011;20:15–20. doi: 10.1016/j.breast.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Prest SJ, May FE, Westley BR. The estrogen-regulated protein, TFF1, stimulates migration of human breast cancer cells. FASEB J. 2002;16:592–594. doi: 10.1096/fj.01-0498fje. [DOI] [PubMed] [Google Scholar]
- 22.May FE, Westley BR. TFF3 is a valuable predictive biomarker of endocrine response in metastatic breast cancer. Endocr Relat Cancer. 2015;22:465–479. doi: 10.1530/ERC-15-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishibashi Y, Ohtsu H, Ikemura M, Kikuchi Y, Niwa T, Nishioka K, Uchida Y, Miura H, Aikou S, Gunji T, et al. Serum TFF1 and TFF3 but not TFF2 are higher in women with breast cancer than in women without breast cancer. Sci Rep. 2017;7:4846. doi: 10.1038/s41598-017-05129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abramson VG, Lehmann BD, Ballinger TJ, Pietenpol JA. Subtyping of triple-negative breast cancer: Implications for therapy. Cancer. 2015;121:8–16. doi: 10.1002/cncr.28914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Podo F, Buydens LM, Degani H, Hilhorst R, Klipp E, Gribbestad IS, Van Huffel S, van Laarhoven HW, Luts J, Monleon D, et al. Triple-negative breast cancer: Present challenges and new perspectives. Mol Oncol. 2010;4:209–229. doi: 10.1016/j.molonc.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 27.Iwamoto T, Bianchini G, Booser D, Qi Y, Coutant C, Shiang CY, Santarpia L, Matsuoka J, Hortobagyi GN, Symmans WF, et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J Natl Cancer Inst. 2011;103:264–272. doi: 10.1093/jnci/djq524. [DOI] [PubMed] [Google Scholar]
- 28.Tan L, Song X, Sun X, Wang N, Qu Y, Sun Z. ART3 regulates triple-negative breast cancer cell function via activation of Akt and ERK pathways. Oncotarget. 2016;7:46589–46602. doi: 10.18632/oncotarget.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu RZ, Graham K, Glubrecht DD, Lai R, Mackey JR, Godbout R. A fatty acid-binding protein 7/RXRβ pathway enhances survival and proliferation in triple-negative breast cancer. J Pathol. 2012;228:310–321. doi: 10.1002/path.4001. [DOI] [PubMed] [Google Scholar]
- 30.Watkins J, Weekes D, Shah V, Gazinska P, Joshi S, Sidhu B, Gillett C, Pinder S, Vanoli F, Jasin M, et al. Genomic complexity profiling reveals that HORMAD1 overexpression contributes to homologous recombination deficiency in triple-negative breast cancers. Cancer Discov. 2015;5:488–505. doi: 10.1158/2159-8290.CD-14-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritzsche FR, Dahl E, Pahl S, Burkhardt M, Luo J, Mayordomo E, Gansukh T, Dankof A, Knuechel R, Denkert C, et al. Prognostic relevance of AGR2 expression in breast cancer. Clin Cancer Res. 2006;12:1728–1734. doi: 10.1158/1078-0432.CCR-05-2057. [DOI] [PubMed] [Google Scholar]
- 32.Wright TM, Wardell SE, Jasper JS, Stice JP, Safi R, Nelson ER, McDonnell DP. Delineation of a FOXA1/ERalpha/AGR2 regulatory loop that is dysregulated in endocrine therapy-resistant breast cancer. Mol Cancer Res. 2014;12:1829–1839. doi: 10.1158/1541-7786.MCR-14-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soutto M, Peng D, Katsha A, Chen Z, Piazuelo MB, Washington MK, Belkhiri A, Correa P, El-Rifai W. Activation of β-catenin signalling by TFF1 loss promotes cell proliferation and gastric tumorigenesis. Gut. 2015;64:1028–1039. doi: 10.1136/gutjnl-2014-307191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buache E, Etique N, Alpy F, Stoll I, Muckensturm M, Reina-San-Martin B, Chenard MP, Tomasetto C, Rio MC. Deficiency in trefoil factor 1 (TFF1) increases tumorigenicity of human breast cancer cells and mammary tumor development in TFF1-knockout mice. Oncogene. 2011;30:3261–3273. doi: 10.1038/onc.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.