Abstract
Histone deacetylase 9 (HDAC9) is involved in a variety of malignant tumors, and leads to malignant tumor development and poor prognosis. However, the association between HDAC9 expression, and the prognosis and clinicopathological features of patients with pancreatic ductal adenocarcinoma (PDAC) remains unclear. The present study used reverse transcription-quantitative PCR, western blotting and immunohistochemistry to detect the expression level of HDAC9 in PDAC tumors and cell lines. The Kaplan-Meier method and Pearson's χ2 test were applied to evaluate the prognostic impact of HDAC9. The present study investigated the effect of HDAC9 on the biological function of PDAC cells. The present results indicated that HDAC9 was highly expressed in PDAC tissue and PDAC cell lines (P<0.05). HDAC9 expression level in tumor tissues was negatively associated with tumor size (P=0.026), T stage (P=0.014) and N stage (P=0.004). Kaplan-Meier analysis suggested that patients with high HDAC9 had shorter recurrence-free survival (RFS; P=0.017) and disease-specific survival (DSS; P=0.022). Moreover, the present results suggested that T stage, N stage and HDAC9 expression level were independent predictive factors for RFS and DSS in patients with PDAC. In addition, silencing HDAC9 significantly inhibited the proliferation and migration of PDAC cells. The present results indicated that high expression levels of HDAC9 were associated with tumor progression and poor prognosis; thus, HDAC9 may serve as a prognostic predictor of PDAC.
Keywords: histone deacetylase 9, prognosis, pancreatic ductal adenocarcinoma
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is among the most chemoresistant and lethal types of cancer due to its multiple mechanisms of pathogenesis and the heterogeneity of its genetic mutations (1,2). Both PDAC and its treatment significantly affect patient quality of life (2,3). The only potentially curative treatment for patients with PDAC is radical resection; however, only 15–20% of patients are candidate for radical resection at the time of diagnosis due to the early metastasis and late presentation of PDAC (3,4). If a patient presents as borderline resectable, tumor cells have only invaded the local blood vessels, and neoadjuvant therapy combined with surgical resection is the main treatment (1). For patients with distant metastasis, comprehensive treatment based on chemotherapy is the only viable option (5). However, patients with PDAC usually have a poor response to chemotherapy and radiotherapy (5). In addition, targeted therapy based on molecular pathways has significantly improved survival in numerous types of cancer, but improved prognosis in patients with PDAC is still limited (6). There is an urgent need to understand the molecular mechanisms underlying PDAC development, which may help identify new biomarkers for PDAC detection and improve current treatment strategies.
Histone deacetylases (HDACs) are a class of enzymes that control the transcription of histones and non-histones via deacetylation (7). Dysregulated expression and mutation of HDACs have been found in various diseases and are related to carcinogenesis (8). Therefore, HDACs are potential therapeutic targets for some neoplasms. Previous studies have demonstrated that HDAC inhibitors can induce cell cycle arrest, apoptosis and tumor cell differentiation (9,10). Several HDAC inhibitors have been studied in patients with solid neoplasms or other malignancies, and are considered potential anticancer drug candidates (11,12). HDAC is a unique family member of class IIa HDACs with tissue-specific expression (13). HDAC can regulate a variety of biological processes, including T-regulatory cell function, muscle differentiation, neuronal disorders, development and tumorigenesis (14). Due to its unique biological function, HDAC9 can selectively encode multiple protein isoforms (15). Recent studies have also demonstrated that HDAC9 has differing oncogenic effects in various malignancies, including oral squamous cell carcinoma, breast cancer and retinoblastoma (10,15,16).
To the best of our knowledge, the role of HDAC9 in the development and progression of the PDAC has not been previously studied. The present study investigated the expression levels of HDAC9 in PDAC using reverse transcription-quantitative PCR (RT-qPCR), western blotting and immunohistochemistry (IHC). In addition, the present study evaluated associations between HDAC9 expression levels, and the clinicopathologic parameters and prognosis of patients with PDAC.
Materials and methods
Human tissue specimens and clinicopathological information
A tissue microarray containing 97 PDAC specimens, PDAC tumor tissue and paired adjacent tissue were collected from 97 patients with primary PDAC (age range, 21–77 years; sex, 60 males and 37 females) who underwent surgical resection from September 2011 to December 2013 at The Second Affiliated Hospital of Chongqing Medical University. All patients had been diagnosed with typical PDAC by the pathologist after surgery. TNM stage and clinical stage were evaluated according to the American Joint Committee on Cancer manual (17). The present study was approved by the Ethics Committee of The Second Affiliated Hospital of Chongqing Medical University and was conducted in accordance with the Declaration of Helsinki. An informed consent document was signed by all patients.
Cell culture and transfection
PDAC cell lines (CFPAC-1, HPAC, SW1990 and Capan-2), and pancreatic ductal epithelial cells (HPDE) were obtained from the Institute of Biochemistry and Cell Biology (Chinese Academy of Sciences). All cell lines were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), and were incubated at 37°C with 5% CO2. A total of 3×105 CFPAC-1 cells were transfected with 50 nM/ul siRNA (si) against HDAC9 (si-HDAC9) or negative control (si-NC). The sequences for the siRNAs were as follows: si-HDAC9 forward, 5′-AACGCCGGAGCUUUCACGTAT-3′ and reverse, 5′-GCGTTCAAUCAUGGUGGCACUTT-3′; and si-NC forward, 5′-AACUCCAAATGUTTCUCGATT-3′ and reverse, 5′-GAAUCACGCCCUAAGTTAATT-3′. The cells were transiently transfected using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc), and the transfection effect was maintained for >72 h.
Western blotting
Total protein was extracted from 1×106 CFPAC-1, HPAC, SW1990, Capan-2 and HPDE cell lines, and 100 mg PDAC and adjacent tissues using RIPA lysis buffer (Beyotime Institute of Biotechnology). Total protein was quantified using the Bradford protein assay (Bio-Rad Laboratories, Inc.) with a Nanodrop spectrophotometer and 25 µg protein/lane was separated by SDS-PAGE on a 10% gel. After blocking with 5% non-fat powdered milk at room temperature for 1 h, the PVDF membranes were probed at 4°C overnight using the following primary antibodies: Anti-HDAC9 (1:1,000; cat. no. ab59718; Abcam), anti-Bax (1:5,000; cat. no. ab32503; Abcam), anti-Bcl-2 (1:500, cat. no. ab182858; Abcam), anti-KI67 (1:1,000; cat. no. ab15580; Abcam) and anti-GAPDH (1:5,000; cat. no. ab181602; Abcam). Subsequently, the PVDF membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (1:5,000; cat. no. ab6721; Abcam) at room temperature for 1 h. Protein bands were visualized using enhanced chemiluminescence solution (EMD Millipore) and a ChemiDoc Imaging System (Bio-Rad Laboratories, Inc.). Protein expression was quantified using Quantity One version 4.6.6 software (Bio-Rad Laboratories, Inc.), with GAPDH as the loading control.
RT-qPCR
Total RNA from 1×106 CFPAC-1, HPAC, SW1990, Capan-2 and HPDE cell lines, and 70 mg PDAC and paired adjacent tissues was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A PrimeScript RT kit (Takara Bio, Inc.) was used for RT of cDNA. The temperature protocol for RT was as follows: 32°C for 10 min, followed by 42°C for 30 min and 75°C for 10 min. qPCR was performed using SYBR® Premix Ex Taq™ II (Takara Bio, Inc.) using a LightCycler system (Roche Molecular Systems, Inc.). The following primer sequences were used for the qPCR: HDAC9 forward, 5′-GAACTCTAAGCCAGATGGGG-3′ and reverse, 5′-GCCCACAGGAACTTCTGACT-3′; and GAPDH forward, 5′-TTCCAGCCTTCCTTCCTGGG-3′ and reverse, 5′-TTGCGCTCAGGAGGAGCAAT-3′. The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 2 min; 40 cycles of 95°C for 15 sec, 60°C for 34 sec and 72°C for 30 sec. The mRNA expression levels of HDAC9 were quantified using the 2−ΔΔCq method (18) and expression levels were normalized to the internal reference gene GAPDH.
IHC
IHC of HDAC9 expression levels was performed using PDAC tissue microarrays. PDAC and paired adjacent tissues were fixed in 10% neutral formalin solution at room temperature for 24 h. Subsequently, paraffin-embedded tissue-array sections (4 µm) were dried at 80°C for 24 h, de-paraffinized in xylene I for 15 min and xylene II for 15 min, and then rehydrated in graded ethanol (100% ethanol for 5 min, 95% ethanol for 5 min, 80% ethanol for 5 min and 75% ethanol for 5 min). To block the endogenous peroxidase activity, the sections were incubated in 3% H2O2 for 30 min at room temperature. After washing with 0.01 M PBS three times, sections were incubated for 15 min at room temperature with 5% goat serum (OriGene Technologies, Inc.) to block non-specific binding, followed by incubation with a rabbit monoclonal anti-HDAC9 antibody (1:500; cat. no. ab109446; Abcam) at 4°C overnight. The sections were then incubated with an anti-rabbit secondary IgG antibody (1:5,000; cat. no. TA140003; OriGene Technologies, Inc.) at 37°C for 30 min. After washing with PBS, the signal was visualized using diaminobenzidine (Wuhan Boster Biological Technology, Ltd.), and counterstaining was performed with hematoxylin for 2 min at room temperature. The histopathological examination was performed using an Olympus DP70 light microscope (magnification, ×200; Olympus Corporation). Finally, HDAC9 immunostaining was scored and examined by two independent assessors, who were blinded to the clinicopathological data.
Scoring systems for IHC staining
The staining intensity score and the proportion of HDAC9 positive cells were evaluated by the pathologist as follows: Staining intensity, i) negative=0; ii) weakly stained=1; iii) moderately stained=2; and iv) strongly stained=3. Staining extent: i) none=0; ii) 1–20%=1; iii) 21–40%=2; iv) 41–60%=3; v) 61–80%=4; and vi) 81–100%=5. The final immunoreactive score (IRS) of HDAC9 expression level was calculated by multiplying the staining intensity score with the staining extent score. IRS was dichotomised using X-tile software version 3.4.7 software (Yale School of Medicine), which is a useful bio-informatics tool for outcome-based cut-point optimization (19). IRS ≤7.5 was designated as low expression, while IRS >7.5 was designated as high expression.
Cell Counting Kit-8 assay (CCK-8)
A total of 2×103 CFPAC-1 cells/well were transfected with si-HDAC9 or si-NC for 48 h and cultured in a 96-well plate for 24, 48 and 72 h. Cell proliferation analysis was performed using the CCK-8 assay (Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocol.
Wound healing assay
A total of 3×105 CFPAC-1 cells were transfected with si-HDAC9 or si-NC for 48 h and seeded into 6-well plates. When the cell density reached 70–80%, all cell lines were cultured in RPMI-1640 medium containing 0% FBS and were incubated at 37°C with 5% CO2 for 24 h. The cell monolayer was scratched with a pipette tip (size, 10 ml) to generate three scratch wounds and then rinsed twice with PBS to remove non-adherent cells. Cells were visualized and counted using a light microscope (magnification, ×200). The distance between scratches was measured at 0, 24 and 48 h. The cell migration rate (%) was calculated using the following equation: [(Original gap distance-current gap distance)/original gap distance] ×100.
5-Ethynyl-2′-deoxyuridine (EdU) assay
A total of 1×103 CFPAC-1 cells were transfected for 48 h and cultured in a 96-well plate. Cells were incubated with 50 µM EdU, 100 µl 1X ApolloR reaction cocktail (cat. no. 100T; Guangzhou RiboBio Co., Ltd.) and 100 µl 1X Hoechst 33342 for 30 min at 37°C. Cell proliferation was analyzed by counting the mean number of cells in three fields for each sample using a fluorescence microscope (magnification, ×100).
Tumorigenesis assay
A total of 6 BALB/c-nu mice (age, 5 weeks; sex, male; weight, 20–22 g) were purchased from The Shanghai Experimental Animal Center and housed in a sterile room at The Animal Center of Chongqing Medical University at 25°C and 40–70% humidity, with a 12-h light/dark cycle and free access to food and water. The study was approved by Ethics Committee of Chongqing Medical University (approval no. IACUC-20180117021). All animal experiments were performed in accordance with the institutional guidelines, and the method of euthanasia was cervical dislocation (when the heart stopped completely, the mouse was determined as dead). Body weight loss >20% was assumed to be a humane endpoint for euthanasia. Xenograft tumors were generated by subcutaneously injecting 3×106 PDAC cells into the left hip flanks of the mice (n=3 per group; 2 groups; each mouse was inoculated with a single tumor site of PDAC cells). Tumor volume was calculated according to the following formula: Volume=(length × width2)/2. Then, 28 days after injection, the mice were sacrificed, and tumors were collected for analysis. The tumor experiments were ended when tumor diameters were <20 mm (the maximum tumor volume was 523 mm3).
Statistical analysis
Data are presented as the mean ± SD and each cell experiment was repeated three times. Statistical data were analyzed using SPSS version 23.0 software (IBM Corp.) and GraphPad Prism version 7.0 software (GraphPad Software, Inc.). A paired t-test was used to evaluate the significance of HDAC9 expression level between PDAC tissues and adjacent normal tissues. Association analyses of clinicopathological factors was performed with Pearson's χ2 test. The Kaplan-Meier method was used to plot survival curves and calculate survival probabilities for overall survival. The statistical significance of survival curves was determined using the log-rank test. Univariate and multivariate analyses (Cox regression analysis) were applied to identify the clinicopathological features, and covariates with P<0.05 in univariate analysis were further analyzed by multivariate analysis. Statistical differences were analyzed by one-way ANOVA, followed by Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of HDAC9 protein in PDAC
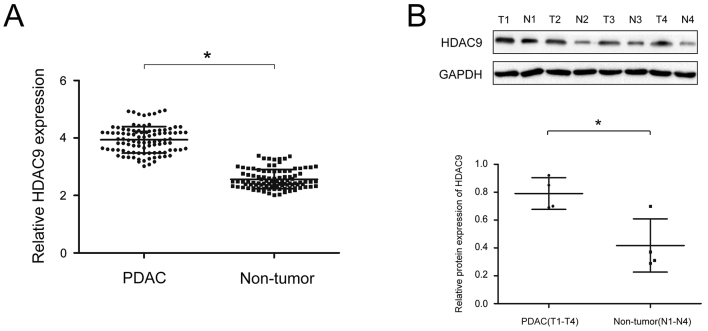
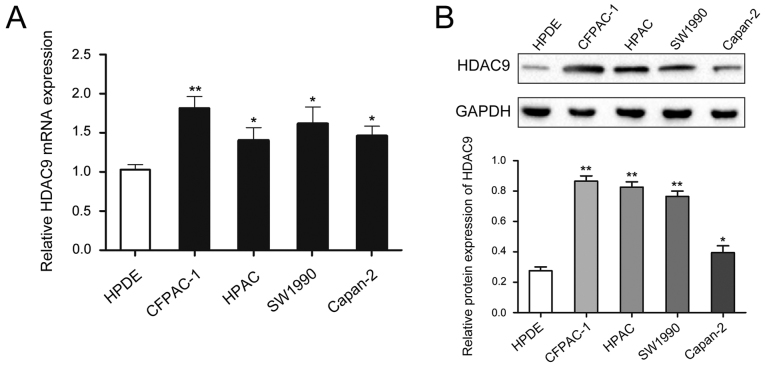
To analyze the expression of HDAC9 protein in PDAC, RT-qPCR and western blot analyses were performed on PDAC and paired adjacent tissue. The expression of HDAC9 was significantly higher in PDAC tissues compared with paired adjacent tissues (Fig. 1A and B). Moreover, the expression level of HDAC9 was higher in PDAC cell lines compared with a pancreatic ductal epithelial cell line (Fig. 2).
Figure 1.
Expression levels of HDAC9 in PDAC. (A) Relative expression levels of HDAC9 between 97 PDAC tissue samples and paired adjacent tissue samples as determined via reverse transcription-quantitative PCR analysis. (B) Western blot analysis of HDAC9 protein expression in PDAC tissue samples and paired adjacent tissue. *P<0.05. PDAC, pancreatic ductal adenocarcinoma; T, cancer tissues; N, paired adjacent tissue samples; HDAC9, histone deacetylase 9.
Figure 2.
Expression levels of HDAC9 in PDAC cell lines. (A) Relative expression levels of HDAC9 in PDAC cell lines and a pancreatic ductal epithelial cell line. (B) HDAC9 protein expression in PDAC cell lines and a pancreatic ductal epithelial cell line. *P<0.05, **P<0.01 vs. HPDE. PDAC, pancreatic ductal adenocarcinoma; HDAC9, histone deacetylase 9.
HDAC9 is associated with clinicopathological features of patients with PDAC
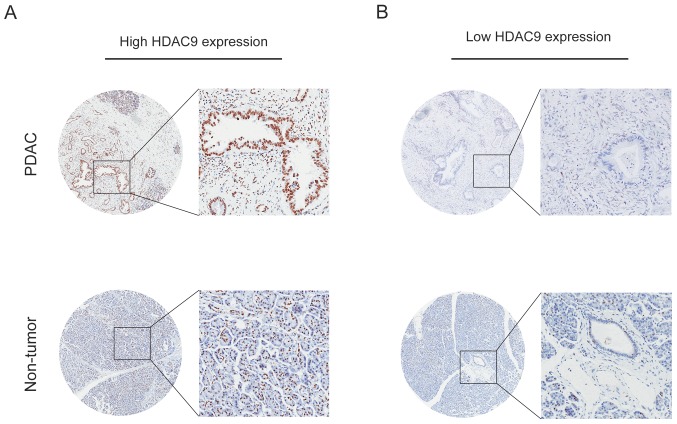
The expression level of HDAC9 in PDAC was analyzed by IHC, and the results indicated that high HDAC9 expression was observed in 64.0% of PDAC samples (62/97). Furthermore, positive staining for HDAC9 was primarily localized in the nuclei of tumor and normal pancreatic cells (Fig. 3).
Figure 3.
Immunohistochemical staining analysis of HDAC9 protein expression in PDAC. (A) High HDAC9 staining in PDAC tissue samples and paired adjacent tissue samples. (B) Low HDAC9 staining in in PDAC tissue samples and paired adjacent tissue samples. Magnifications, ×50 and ×200. PDAC, pancreatic ductal adenocarcinoma; HDAC9, histone deacetylase 9.
The present study analyzed the association between HDAC9 expression level and the clinicopathological characteristics of patients with PDAC. High HDAC9 expression level was identified to be positively associated with tumor size (P=0.026), T stage (P=0.014) and N stage (P=0.004; Table I). There was no significant association between HDAC9 expression level and age, sex, pathological grade, M stage, clinical stage or diabetes.
Table I.
Association of HDAC9 expression with clinicopathological features of pancreatic ductal adenocarcinoma.
| Histone deacetylase 9 expression | ||||
|---|---|---|---|---|
| Variables | Cases | Low (n=35) | High (n=62) | P-value |
| Age, years | 0.295 | |||
| <65 | 49 | 15 | 34 | |
| ≥65 | 48 | 20 | 28 | |
| Sex | 0.665 | |||
| Male | 60 | 23 | 37 | |
| Female | 37 | 12 | 25 | |
| Tumor size, cm | 0.026 | |||
| ≤5 | 74 | 22 | 52 | |
| >5 | 23 | 13 | 10 | |
| Pathological grade | 0.498 | |||
| I–II | 66 | 22 | 44 | |
| III–IV | 31 | 13 | 18 | |
| T stage | 0.014 | |||
| T1 | 14 | 10 | 4 | |
| T2 | 64 | 15 | 49 | |
| T3 | 19 | 10 | 9 | |
| N stage | 0.004 | |||
| N0 | 36 | 20 | 16 | |
| N1 | 61 | 15 | 46 | |
| M stage | 0.551 | |||
| M0 | 94 | 35 | 59 | |
| M1 | 3 | 0 | 3 | |
| Clinical stage | 0.504 | |||
| 1 | 35 | 10 | 25 | |
| 2 | 51 | 18 | 33 | |
| 3 | 8 | 5 | 3 | |
| 4 | 3 | 2 | 1 | |
| Diabetes | 0.295 | |||
| No | 48 | 20 | 28 | |
| Yes | 49 | 15 | 34 | |
T, tumor; N, lymph node involvement; M, metastasis.
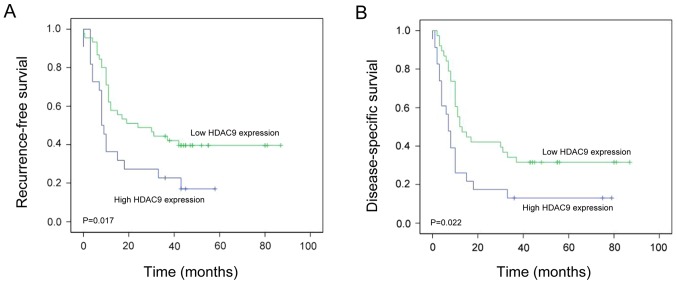
Effect of HDAC9 expression on recurrence-free survival (RFS) in PDAC
Kaplan-Meier survival analysis revealed that patients with PDAC exhibiting high HDAC9 expression levels have significantly shorter RFS compared with patients with low HDAC9 expression levels (P=0.017; Fig. 4A). To determine whether HDAC9 expression was an independent predictive factor for RFS in PDAC, Cox proportional hazards regression analysis was conducted. The univariate analysis indicated that T stage (HR=1.684; P=0.029), N stage (HR=1.448; P=0.013) and HDAC9 expression level (HR=1.647; P=0.018) were significantly associated with RFS in PDAC (Table II). The multivariate analysis suggested that T stage (HR=1.537; P=0.021), N stage (HR=1.597; P=0.019) and HDAC9 expression (HR=1.739; P=0.011) were independent prognostic factors for RFS in PDAC (Table II).
Figure 4.
RFS and DSS in patients with high or low expression of HDAC9 in PDAC. (A) Kaplan-Meier analysis of RFS in patients with PDAC exhibiting high or low HDAC9 expression. (B) Kaplan-Meier analysis of DSS in patients with PDAC exhibiting high or low HDAC9 expression. HDAC9, histone deacetylase 9; RFS, recurrence-free survival; DSS, disease-specific survival; PDAC, pancreatic ductal adenocarcinoma.
Table II.
Univariate and multivariate analysis of different prognostic variables and recurrence-free survival in pancreatic ductal adenocarcinoma.
| Univariate analysis model | Multivariate analysis model | ||||
|---|---|---|---|---|---|
| Variables | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex | 0.948 (0.540–1.684) | 0.308 | |||
| Male | 60 | ||||
| Female | 37 | ||||
| Age, years | 0.648 (0.608–1.840) | 0.584 | |||
| <65 | 49 | ||||
| ≥65 | 48 | ||||
| Tumor size, cm | 0.608 (0.771–2.017) | 0.309 | |||
| ≤5 | 74 | ||||
| >5 | 23 | ||||
| Pathological grade | 1.308 (0.667–2.554) | 0.706 | |||
| I/II | 66 | ||||
| III/IV | 31 | ||||
| T stage | 1.684 (0.846–2.647) | 0.029 | 1.537 (0.647–2.394) | 0.021 | |
| T1 | 14 | ||||
| T2 | 64 | ||||
| T3 | 19 | ||||
| N stage | 1.448 (0.670–2.298) | 0.013 | 1.597 (0.849–2.937) | 0.019 | |
| N0 | 36 | ||||
| N1 | 61 | ||||
| M stage | 1.075 (0.834–2.840) | 0.384 | |||
| M0 | 94 | ||||
| M1 | 3 | ||||
| Clinical stage | 0.572 (0.637–1.846) | 0.608 | |||
| 1 | 35 | ||||
| 2 | 51 | ||||
| 3 | 8 | ||||
| 4 | 3 | ||||
| Diabetes | 1.087 (1.557–2.947) | 0.284 | |||
| Presence | 52 | ||||
| Absence | 53 | ||||
| Histone deacetylase 9 expression | 1.647 (0.71–2.127) | 0.018 | 1.739 (1.067–3.337) | 0.011 | |
| High | 62 | ||||
| Low | 35 | ||||
T, tumor; N, lymph node involvement; M, metastasis.
Effect of HDAC9 expression level on disease-specific survival (DSS) in PDAC
Kaplan-Meier survival analysis revealed that patients with PDAC exhibiting high HDAC9 expression levels had significantly shorter DSS compared with those with low HDAC9 expression levels (P=0.022; Fig. 4B). The univariate analysis suggested that T stage (HR=1.509; P=0.015), N stage (HR=1.493; P=0.025) and HDAC9 expression levels (HR=1.210; P=0.010) were significantly associated with DSS in PDAC (Table III). The multivariate analysis indicated that T stage (HR=1.805; P=0.011), N stage (HR=1.690; P=0.022) and HDAC9 expression levels (HR=1.394; P=0.008) were independent prognostic factors for DSS in PDAC (Table III).
Table III.
Univariate and multivariate analysis of different prognostic variables and disease-specific survival in pancreatic ductal adenocarcinoma.
| Univariate analysis model | Multivariate analysis model | ||||
|---|---|---|---|---|---|
| Variables | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex | 0.647 (0.428–1.553) | 0.648 | |||
| Male | 60 | ||||
| Female | 37 | ||||
| Age, years | 0.480 (0.571–1.669) | 0.806 | |||
| <65 | 49 | ||||
| ≥65 | 48 | ||||
| Tumor size, cm | 0.940 (0.807–1.694) | 0.638 | |||
| ≤5 | 74 | ||||
| >5 | 23 | ||||
| Pathological grade | 1.249 (0.947–2.048) | 0.634 | |||
| I/II | 66 | ||||
| III/IV | 31 | ||||
| T stage | 1.509 (1.178–3.642) | 0.015 | 1.805 (1.049–2.648) | 0.011 | |
| T1 | 14 | ||||
| T2 | 64 | ||||
| T3 | 19 | ||||
| N stage | 1.493 (0.477–1.947) | 0.025 | 1.690 (0.644–2.684) | 0.022 | |
| N0 | 36 | ||||
| N1 | 61 | ||||
| M stage | 0.948 (1.067–2.785) | 0.673 | |||
| M0 | 94 | ||||
| M1 | 3 | ||||
| Clinical stage | 1.545 (0.884–2.153) | 0.735 | |||
| 1 | 35 | ||||
| 2 | 51 | ||||
| 3 | 8 | ||||
| 4 | 3 | ||||
| Diabetes | 1.604 (1.241–3.517) | 0.570 | |||
| Presence | 52 | ||||
| Absence | 53 | ||||
| Histone deacetylase 9 expression | 1.210 (0.879–1.846) | 0.010 | 1.394 (0.957–2.047) | 0.008 | |
| High | 62 | ||||
| Low | 35 | ||||
T, tumor; N, lymph node involvement; M, metastasis.
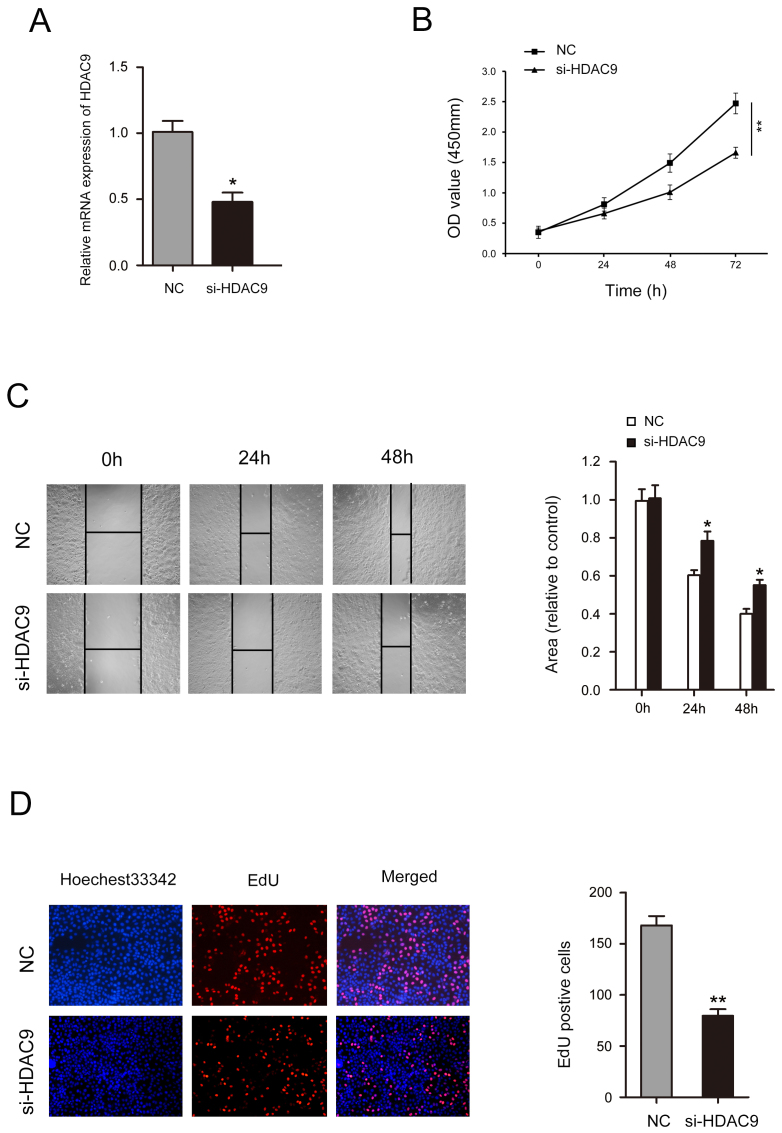
Decreased HDAC9 expression inhibits the proliferation and migration of CFPAC-1 cells
After treatment with si-HDAC9, the expression level of HDAC9 was significantly decreased (Fig. 5A). A CCK-8 assay identified that decreased HDAC9 expression significantly inhibited cell proliferation in CFPAC-1 cells (Fig. 5B). Wound healing assay results indicated that CFPAC-1 cell mobility was significantly decreased following HDAC9 knockdown (Fig. 5C). The number of CFPAC-1 cells incorporating EdU in the HDAC9 knockdown group was significantly decreased compared with the control group (Fig. 5D).
Figure 5.
Low expression of HDAC9 inhibits proliferation and migration in CFPAC-1 cells. (A) Reverse transcription-quantitative PCR analysis of the expression of HDAC9 in CFPAC-1 cells transfected with si-HDAC9 or NC. (B) Cell Counting Kit-8 assay was used to investigate the effects of HDAC9 silencing on the proliferative ability of CFPAC-1 cells. (C) Wound healing assay was performed to investigate the effects of HDAC9 silencing on the migration of CFPAC-1 cells. (D) Cell proliferation was assessed using EdU in CFPAC-1 cells after HDAC9 silencing. Each experiment was repeated ≥3 times. *P<0.05, **P<0.01 vs. NC. EdU, 5-ethynyl-2′-deoxyuridine; HDAC9, histone deacetylase 9; OD, optical density; NC, negative control; siRNA, small interfering RNA.
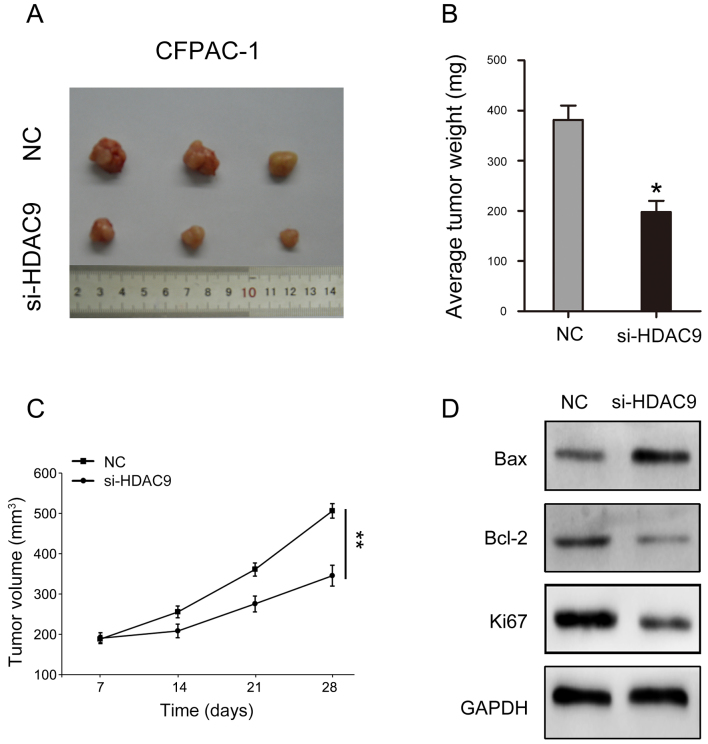
Decreased HDAC9 expression suppresses tumorigenicity in PDAC
The present study investigated the influence of HDAC9 in vivo. The results indicated that reducing the expression of HDAC9 may significantly inhibit the volume and weight of tumors formed by CFPAC-1 cells (Fig. 6A-C). In addition, key molecules involved in cell proliferation and apoptosis were analyzed. The present results suggested that HDAC9 silencing decreased Bcl-2 and Ki67 expression levels, and increased Bax expression (Fig. 6D).
Figure 6.
Low expression of HDAC9 inhibits tumorigenicity of PDAC in vivo. (A) CFPAC-1 cells transfected with si-HDAC9 were subcutaneously inoculated into mice to form tumors. (B) Tumor weight was recorded in each group. (C) Tumor volume was recorded in each group. (D) Protein expression of Bax, Bcl-2 and Ki67, after transfection with si-HDAC9 in CFPAC-1 cells. *P<0.05, **P<0.01 vs. NC. HDAC9, histone deacetylase 9; PDAC, pancreatic ductal adenocarcinoma; NC, negative control; siRNA, small interfering RNA.
Discussion
HDACs are proteases that serve an important role in the remodeling of chromatin structure and epigenetic regulation of gene expression (20). HDACs can repress transcription by deacetylating regulatory transcription factors, and are involved in the development and progression of various diseases, including cancer (10). As a class II HDAC, HDAC9 has been studied in several malignancies, but its biological activities in different tumors remain unclear (14). A previous study has shown that HDAC9 is highly expressed in childhood acute lymphoblastic leukemia and that its expression level is negatively associated with the 5-year disease-free survival of patients (21). The overexpression of HDAC9 in breast cancer, retinoblastoma, medulloblastoma and oral squamous cell carcinoma significantly promotes tumor cell proliferation and reduces programmed cell death (9,22–24). In contrast, a previous study of lung cancer found that the protein expression levels of HDAC9 are markedly lower in lung cancer compared with normal epithelial cells (25). This previous study also showed that HDAC9 attenuates the growth of lung cancer cells in vitro and may have tumor suppressor effects in lung cancer (25).
The role of HDAC9 in tumorigenesis is complex, having dual roles in different malignancies (26). Previous studies have reported that HDAC9 appears to be a pro-oncogenic factor in certain malignancies including glioblastoma, leiomyosarcoma and rhabdoid sarcomas (8,27,28). However, HDAC9 appears to be a tumor suppressor in other cancer types, such as non-small cell lung cancer (25). There have been few studies on the role of HDAC9 in PDAC. To the best of our knowledge, the only study on HDAC9 in PDAC showed that HDAC9 is highly expressed in subtypes of PDAC with high microvessel density and endothelial cell abundance (29); however, this previous study only reported that the prognosis of patients with high microvessel density was improved and did not investigate how HDAC9 may influence these findings. The present study investigated the expression of HDAC9 in PDAC and its effect on the malignant progression of patients with PDAC. The present results suggested that HDAC9 was highly expressed in PDAC, and patients with high expression of HDAC9 had a worse prognosis. In addition, high expression levels of HDAC9 were associated with tumor size, T stage and N stage. Moreover, HDAC9 expression level and clinical stage appeared to be independent risk factors for RFS and DSS in PDAC. The present results indicated that silencing HDAC9 significantly inhibited the proliferation and migration of PDAC cells. The present study used si-HDAC9 to reduce the expression levels of HDAC9. The present results suggested that after si-HDAC9 transfection, cell proliferation was inhibited at 48 h, and had a significant effect on cell proliferation at 72 h. Moreover, in vivo results suggested that key molecules Bax, Bcl-2 and Ki67, which regulate cell proliferation and apoptosis, are affected by HDAC9. Decreasing the expression level of HDAC9 can inhibit the expression of Bcl-2 and Ki67, and promote the expression of Bax, thereby inhibiting the proliferation and migration of pancreatic cancer cells (30). Although the present study has limitations, future research should investigate the specific molecular mechanism of HDAC9 to understand the role of HDAC9 in the malignant progression of PDAC.
In summary, the present study suggested that expression levels of HDAC9 were higher in patients with PDAC. In addition, HDAC9 may be associated with RFS and DSS in PDAC. The present univariate and multivariate analysis suggested that high HDAC9 expression level and clinical stage predicted poor prognosis, and that HDAC9 expression level was an independent prognostic factor for DSS in PDAC. Therefore, HDAC9 may serve as a new diagnostic target and facilitate the treatment of PDAC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the subject of Yongchuan Hospital affiliated to Chongqing Medical University (grant no. YJQN201422).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HL and JPG designed the study. HL and JPG conducted the research. XCL and HPL collected and analyzed the data. HL, XCL and HPL cultured cells, collected reference articles and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients provided informed consent and the study was approved by the Ethics Committee of Chongqing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Adamska A, Elaskalani O, Emmanouilidi A, Kim M, Abdol Razak NB, Metharom P, Falasca M. Molecular and cellular mechanisms of chemoresistance in pancreatic cancer. Adv Biol Regul. 2018;68:77–87. doi: 10.1016/j.jbior.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Kong F, Kong X, Du Y, Chen Y, Deng X, Zhu J, Du J, Li L, Jia Z, Xie D, et al. STK33 promotes growth and progression of pancreatic cancer as a critical downstream mediator of HIF1a. Cancer Res. 2017;77:6851–6862. doi: 10.1158/0008-5472.CAN-17-0067. [DOI] [PubMed] [Google Scholar]
- 3.Raptis DA, Fessas C, Belasyse-Smith P, Kurzawinski TR. Clinical presentation and waiting time targets do not affect prognosis in patients with pancreatic cancer. Surgeon. 2010;8:239–246. doi: 10.1016/j.surge.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Zhou B, Sun C, Hu X, Zhan H, Zou H, Feng Y, Qiu F, Zhang S, Wu L, Zhang B. MicroRNA-195 suppresses the progression of pancreatic cancer by targeting DCLK1. Cell Physiol Biochem. 2017;44:1867–1881. doi: 10.1159/000485876. [DOI] [PubMed] [Google Scholar]
- 5.Garrido-Laguna I, Hidalgo M. Pancreatic cancer: From state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12:319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- 6.Adamska A, Domenichini A, Falasca M. Pancreatic ductal adenocarcinoma: Current and evolving therapies. Int J Mol Sci. 2017;18(pii):E1338. doi: 10.3390/ijms18071338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/bj20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang R, Wu Y, Wang M, Sun Z, Zou J, Zhang Y, Cui H. HDAC9 promotes glioblastoma growth via TAZ-mediated EGFR pathway activation. Oncotarget. 2015;6:7644–7656. doi: 10.18632/oncotarget.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Wu D, Xia F, Xian H, Zhu X, Cui H, Huang Z. Downregulation of HDAC9 inhibits cell proliferation and tumor formation by inducing cell cycle arrest in retinoblastoma. Biochem Biophys Res Commun. 2016;473:600–606. doi: 10.1016/j.bbrc.2016.03.129. [DOI] [PubMed] [Google Scholar]
- 10.Rastogi B, Raut SK, Panda NK, Rattan V, Radotra BD, Khullar M. Overexpression of HDAC9 promotes oral squamous cell carcinoma growth, regulates cell cycle progression, and inhibits apoptosis. Mol Cell Biochem. 2016;415:183–196. doi: 10.1007/s11010-016-2690-5. [DOI] [PubMed] [Google Scholar]
- 11.Lakshmaiah KC, Jacob LA, Aparna S, Lokanatha D, Saldanha SC. Epigenetic therapy of cancer with histone deacetylase inhibitors. J Cancer Res Ther. 2014;10:469–478. doi: 10.4103/0973-1482.137937. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Wallach J, Duane S, Wang Y, Wu J, Wang J, Adejare A, Ma H. Developing selective histone deacetylases (HDACs) inhibitors through ebselen and analogs. Drug Des Devel Ther. 2017;11:1369–1382. doi: 10.2147/DDDT.S124977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Z, Peng L, Radhakrishnan R, Seto E. Histone deacetylase 9 (HDAC9) regulates the functions of the ATDC (TRIM29) protein. J Biol Chem. 2010;285:39329–39338. doi: 10.1074/jbc.M110.179333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrie K, Guidez F, Howell L, Healy L, Waxman S, Greaves M, Zelent A. The histone deacetylase 9 gene encodes multiple protein isoforms. J Biol Chem. 2003;278:16059–16072. doi: 10.1074/jbc.M212935200. [DOI] [PubMed] [Google Scholar]
- 15.Salgado E, Bian X, Feng A, Shim H, Liang Z. HDAC9 overexpression confers invasive and angiogenic potential to triple negative breast cancer cells via modulating microRNA-206. Biochem Biophys Res Commun. 2018;503:1087–1091. doi: 10.1016/j.bbrc.2018.06.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Q, He W, Chen L, Yang Y, Shi K, You Z. MicroRNA-101-3p inhibits proliferation in retinoblastoma cells by targeting EZH2 and HDAC9. Exp Ther Med. 2018;16:1663–1670. doi: 10.3892/etm.2018.6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 20.Weichert W, Röske A, Gekeler V, Beckers T, Ebert MP, Pross M, Dietel M, Denkert C, Röcken C. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: A retrospective analysis. Lancet Oncol. 2008;9:139–148. doi: 10.1016/S1470-2045(08)70004-4. [DOI] [PubMed] [Google Scholar]
- 21.Moreno DA, Scrideli CA, Cortez MA, de Paula Queiroz R, Valera ET, da Silva Silveira V, Yunes JA, Brandalise SR, Tone LG. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br J Haematol. 2010;150:665–673. doi: 10.1111/j.1365-2141.2010.08301.x. [DOI] [PubMed] [Google Scholar]
- 22.Lapierre M, Linares A, Dalvai M, Duraffourd C, Bonnet S, Boulahtouf A, Rodriguez C, Jalaguier S, Assou S, Orsetti B, et al. Histone deacetylase 9 regulates breast cancer cell proliferation and the response to histone deacetylase inhibitors. Oncotarget. 2016;7:19693–19708. doi: 10.18632/oncotarget.7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P, Deubzer HE, Lodrini M, Taylor MD, von Deimling A, et al. HDAC5 and HDAC9 in medulloblastoma: Novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16:3240–3252. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 24.Cha JD, Kim HJ, Cha IH. Genetic alterations in oral squamous cell carcinoma progression detected by combining array-based comparative genomic hybridization and multiplex ligation-dependent probe amplification. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2011;111:594–607. doi: 10.1016/j.tripleo.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Okudela K, Mitsui H, Suzuki T, Woo T, Tateishi Y, Umeda S, Saito Y, Tajiri M, Masuda M, Ohashi K. Expression of HDAC9 in lung cancer-potential role in lung carcinogenesis. Int J Clin Exp Pathol. 2014;7:213–220. [PMC free article] [PubMed] [Google Scholar]
- 26.Gore J, Craven KE, Wilson JL, Cote GA, Cheng M, Nguyen HV, Cramer HM, Sherman S, Korc M. TCGA data and patient-derived orthotopic xenografts highlight pancreatic cancer-associated angiogenesis. Oncotarget. 2015;6:7504–7521. doi: 10.18632/oncotarget.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Giorgio E, Franforte E, Cefalu S, Rossi S, Dei Tos AP, Brenca M, Polano M, Maestro R, Paluvai H, Picco R, Brancolini C. The co-existence of transcriptional activator and transcriptional repressor MEF2 complexes influences tumor aggressiveness. PLoS Genet. 2017;13:e1006752. doi: 10.1371/journal.pgen.1006752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahali B, Yu J, Marquez SB, Thompson KW, Liang SY, Lu L, Reisman D. The silencing of the SWI/SNF subunit and anticancer gene BRM in Rhabdoid tumors. Oncotarget. 2014;5:3316–3332. doi: 10.18632/oncotarget.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craven KE, Gore J, Wilson JL, Korc M. Angiogenic gene signature in human pancreatic cancer correlates with TGF-beta and inflammatory transcriptomes. Oncotarget. 2016;7:323–341. doi: 10.18632/oncotarget.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du H, Xu Q, Xiao S, Wu Z, Gong J, Liu C, Ren G, Wu H. MicroRNA-424-5p acts as a potential biomarker and inhibits proliferation and invasion in hepatocellular carcinoma by targeting TRIM29. Life Sci. 2019;224:1–11. doi: 10.1016/j.lfs.2019.03.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.