Abstract
Tailed, double-stranded DNA bacteriophages provide a well-characterized model system for the study of viral assembly, especially for herpesviruses and adenoviruses. A wealth of genetic, structural, and biochemical work has allowed for the development of assembly models and an understanding of the DNA packaging process. The portal complex is an essential player in all aspects of bacteriophage and herpesvirus assembly. Despite having low sequence similarity, portal structures across bacteriophages share the portal fold and maintain a conserved function. Due to their dynamic role, portal proteins are surprisingly plastic, and their conformations change for each stage of assembly. Because the maturation process is dependent on the portal protein, researchers have been working to validate this protein as a potential antiviral drug target. Here we review recent work on the role of portal complexes in capsid assembly, including DNA packaging, as well as portal ring assembly and incorporation and analysis of portal structures.
Keywords: portal fold, DNA packaging, bacteriophage assembly, virus assembly
INTRODUCTION
Bacteriophages have been used and researched for over a century. In the 1950s, bacteriophages were used to perform experiments that laid the foundation for the central dogma of modern molecular biology. More recent innovations have focused on the role phages can play in human health. This includes the use of phages for vaccines, either for antigenic display or for delivery of DNA vaccines to elicit a greater immune response than traditional vaccination methods. Furthermore, exploration of the human microbiome has prompted interest in the role phages play in intestinal health and diseases. Additionally, as antibiotic resistance continues to rise, phage therapy is garnering increased interest. Phages have successfully been used to combat resistant bacterial infections once all other conventional therapeutics failed (1, 2).
Well-characterized phages also provide a platform to understand pathogenic human viruses. Bacteriophages work well as model systems for double-stranded DNA (dsDNA) viruses such as herpesviruses and adenoviruses. The general assembly mechanisms follow the same pathway, and the major proteins—including coat, scaffolding, and portal proteins—have significant structural similarities and biochemical properties. Here we discuss progress in understanding the structure and function of viral portal proteins in the assembly and infection cycle of bacteriophages as well as herpresviruses.
BACTERIOPHAGE ASSEMBLY
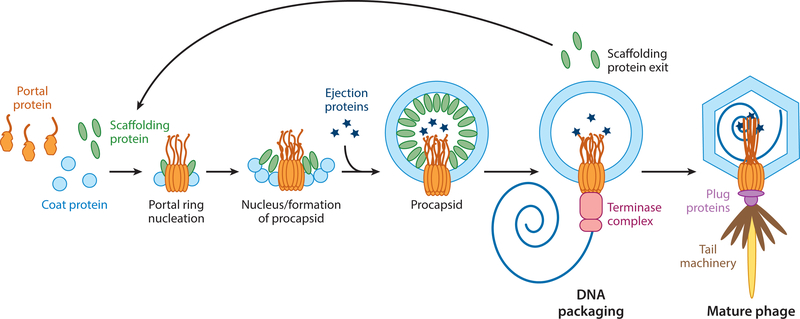
The assembly mechanisms of several dsDNA bacteriophages and herpesviruses have been studied in depth. The general pathway involves three major proteins: the coat, scaffolding (or delta domains attached to the coat protein), and portal proteins. Scaffolding proteins copolymerize with the coat proteins into an icosahedral procapsid or prohead made up of hexamers and pentamers, which contain a dodecameric portal protein replacing a pentamer at a single vertex (Figure 1). In P22, the scaffolding protein coassembles with the coat protein on a growing edge of the procapsid, whereas in T4 and herpesviruses, the coat protein assembles around a preformed scaffolding protein core. In all cases, the scaffolding protein is inside the procapsid (3–13). During maturation, the scaffolding protein exits the procapsid and is either degraded (as with phages lambda, T4, and HK97 and herpesviruses) or recycled for further rounds of assembly (as in P22 and Phi29), making room for DNA encapsidation (13–17). In P22, the scaffolding protein exits intact through holes in the coat protein lattice (9). In all bacteriophages, terminase proteins package the genome into the capsid until the DNA reaches liquid crystalline density, generating more than 50 pN of pressure (18–21). During the maturation process, the head expands, taking on a more angular shape and increasing the volume (19, 22). Once packaging is complete, the terminase proteins dissociate from the portal complex to allow for the addition of plug and tail proteins (23).
Figure 1.
Bacteriophage P22 assembly pathway. Coat monomers, scaffolding protein, and portal monomers make up a nucleation complex, forming the portal ring around which the coat protein shell can be assembled by scaffolding protein. The ejection proteins are incorporated into the procapsid prior to DNA packaging by terminase proteins. Pressure from DNA induces a conformational change in the capsid and portal protein facilitating the dissociation of the terminase complex and addition of the tail machinery.
In the majority of phages, DNA packaging into procapsids begins with the small subunit of the terminase complex (TerS) recognizing the concatemeric viral DNA. Then, the large terminase (TerL) assembles onto the TerS:DNA complex and then the portal complex in the procapsid. The TerL cuts the DNA and packages it in an ATP-dependent process (24–26). To ensure packaging of a full genome into each capsid, these phages use one of two strategies. In headful phages, such as P22, T4, and SPP1, an initial nonspecific cut is made near a recognition (pac) site. All following cuts are nonspecific and occur following packaging of just over 100% of the genome (until the capsid head is full). The DNA in headful phages contains redundancy in the terminal ends to ensure that each capsid contains the entire genome. In cohesive end site (cos) phages, such as lambda, HK97, P2, and herpesviruses, nicks are produced at specific sites (cosN) such that a full genome length is packaged between the cut sites (25, 27). Bacteriophage Phi29, as well as adenoviruses, produces genome-length units of DNA and therefore does not require a cleavage event, although Phi29 does require a NoRC-associated RNA (pRNA) molecule for packaging (25, 28, 29). DNA packaging is a multistep process that couples ATP hydrolysis to translocation of dsDNA inside an empty capsid (20, 21, 30, 31). Recent advances in single-molecule techniques have allowed for further analysis of DNA packaging mechanisms and rates in vitro, which are further discussed in this review. After release of the newly generated phages or viruses from infected cells, each must infect a new host. The genome is ejected into the new host cell via the portal protein. Thus, the portal complex is the conduit for DNA entry into and exit from the virus.
PORTAL PROTEIN: THE ESSENTIAL PLAYER IN PROCAPSID ASSEMBLY
The portal protein complex plays a unique role in tailed dsDNA bacteriophage assembly and herpesviruses. It is the nucleator for procapsid assembly in some bacteriophages, such as lambda and T4, and in herpesviruses (7, 32–34). In contrast, the absence of portal protein does not change the initiation of assembly or assembly kinetics in P22 and SPP1 (35, 36). Although procapsid-like particles can form or be in vitro assembled with only coat and scaffolding proteins, these procapsids cannot package DNA or mature into infectious viruses (3, 4, 36, 37). Only phages with portal incorporated at a single pentameric vertex are able to package the correct amount of DNA and are viable. In several bacteriophages, including T4, SPP1, and Phi29, the incorporation of the portal complex promotes the formation of capsids with the correct morphology (7, 36–39). In P22, the presence of excess portal has been linked to the formation of T = 4 particles and aberrant spirals, indicating that portal concentration plays a significant role in procapsid assembly (40). Across the bacteriophages, in addition to regulating procapsid assembly, portal proteins act as DNA sensors that facilitate both packaging and release of the genome (41–47). The portal complex is, therefore, indispensable as it plays a direct role in every step of phage assembly.
In recent years, there has been increased attention to bacteriophage and dsDNA virus portal complexes due to their unique role in capsid assembly and potential as a drug target. In this review, we discuss what is known about bacteriophage portal complexes, as well as their homologs in herpesviruses and adenoviruses. We focus on portal complex assembly and incorporation as well as the portal structure and its plasticity during DNA packaging and ejection. In each section, we use P22 as a model to compare and contrast with other bacteriophages as well as herpesviruses. To avoid overlap with other recent reviews, we recommend consulting References 23,25, and 48 for a more detailed overview of bacteriophage DNA packaging and the tail machinery.
PORTAL ASSEMBLY AND INCORPORATION
Portal Ring Incorporation
Before portal can act as a conduit for DNA, it must first be assembled from monomers and incorporated into a single vertex of the procapsid. In several bacteriophages as well as Herpes simplex virus (HSV-1), scaffolding protein is thought to interact with the portal complex to facilitate incorporation (32,36, 49–52). In bacteriophage P22, progeny from infections with phages that have an amber mutation in the scaffolding protein gene lack the portal protein and lead to aberrant coat protein structures (49). Interestingly, correctly sized P22 procapsids can form in the absence of the portal complex and still encapsulate the internal ejection proteins, although these particles cannot package DNA to mature into infectious virions (9, 10, 40, 53–55). In vitro work has been used to show that in order for the P22 portal to be incorporated into procapsids, it must be present with coat protein monomers and scaffolding protein at the initiation of assembly, although excess portal induces the formation of petite procapsids and aberrant structures (33, 40, 56). Building capsids around a nucleation site that includes the portal complex, as well as scaffolding protein and potentially the major capsid protein, provides one explanation for the inclusion of portal at a single vertex.
Structural evidence from cryo-electron microscopy (cryoEM) also suggests that P22 portal and scaffolding protein interact. In a recent asymmetric cryoEM reconstruction of the P22 portal complex incorporated into a procapsid (8.7 A), density below the portal wing domain was attributed to the helix-turn-helix domains of 10 scaffolding proteins, although biochemical evidence suggests that P22 scaffolding protein interacts more strongly with portal monomers than portal rings (56, 57).
Scaffolding protein is also important for portal incorporation and procapsid morphology in other phages. In phage lambda, amber mutations were used to show that portal and scaffolding protein interact prior to coat protein assembly (32). Soluble complexes of HSV-1 portal (UL6) and scaffolding protein (UL26.5) can be used to initiate capsid assembly upon the addition of coat protein. SPP1 portal (gp6) forms a stable complex with coat (gp13) and scaffolding (gp11) proteins prior to capsid formation (33, 36, 52). In both P22 and Phi29, assembly reactions containing coat protein, scaffolding protein, and the portal complex are too rapid to detect intermediates or sequential binding (17). Scaffolding protein in Phi29 has been shown to bind portal rings in vitro, and coat protein interacts with portal rings only if scaffolding protein is also present (17, 38, 56). The exact order of binding events and mechanism of portal incorporation differ between phages, but it is likely that scaffolding protein is required for portal incorporation in most phages.
The portal complex also plays a critical role in procapsid morphology. In phage T4, although the portal is not necessary for core (scaffolding and internal proteins) assembly, inclusion of the portal protein regulates capsid dimensions during procapsid assembly (6, 7, 37, 39, 58). Thus, interactions between the portal and the scaffolding protein core may be necessary to form procapsids of the correct size in T4. Despite not forming a scaffolding protein core for portal to dock onto, SPP1 portal protein presence also regulates the formation of T = 7 procapsids (36). The Phi29 portal complex also has an impact on phage morphology. Capsids assembled in vivo from scaffolding and coat proteins in the absence of portal were irregularly shaped and sized (38). In bacteriophages, the formation of functional procapsids requires the interaction of coat, scaffolding (or a delta domain in the case of HK97), and portal proteins.
Portal Ring Assembly
Scaffolding protein not only is required for the incorporation of the portal complex but also is necessary for the assembly of the dodecameric rings from monomers. In P22, infections with phages containing an amber mutation in the coat protein gene lead to formation of exclusively portal monomers, which is thought to be due to scaffolding protein autoregulation (16, 59). Full-length P22 scaffolding protein and, to a lesser extent, coat protein have been shown to oligomerize assembly-competent portal rings. In addition, in vitro complementation experiments with P22 scaffolding protein truncation mutants identified the region from amino acids 229–238 as being required for portal incorporation in vivo (56, 60). Temperature-sensitive scaffolding protein mutants Y214W and S242F are unable to recruit portal, indicating that specific scaffolding protein residues may be responsible for incorporating portal into the growing procapsid (61).
Further genetic evidence for portal-scaffolding interactions in P22 includes the scaffolding protein escape mutant, G287E, which corrects head assembly in the presence of excess portal, but is correlated with lower incorporation in conditions with normal levels of portal expression (62). Scaffolding protein C-terminus mutations also affect scaffolding protein-coat protein interactions, potentially leading to irregularities in portal incorporation (60, 62). Weak affinity chromatography experiments with P22 portal, coat, and scaffolding proteins also demonstrate that scaffolding protein interacts preferentially with portal monomers as opposed to portal rings, lending further evidence to the role of scaffolding protein in the oligomerization of portal rings (56). In P22, scaffolding protein messenger RNA (mRNA) may provide a docking site upon which assembly can be initiated (63). Scaffolding protein binds its own mRNA to autoregulate its translation, which could create a locally high concentration of scaffolding protein to assemble the portal complex prior to coat polymerization.
Mutational evidence has also indicated that coat protein may play a role in portal ring assembly and ensuring the incorporation of only one portal in P22. In in vitro assembly experiments, coat protein facilitated the assembly of portal rings, albeit to a much lesser extent than scaffolding protein (56). The A285T mutation in the coat protein I-domain enables the incorporation of two portals (64). Furthermore, suppressor searches revealed sites in the I-domain that enable incorporation of cold-sensitive portal mutants (64, 65). The current model for P22 assembly involves the formation of a nucleation complex consisting of scaffolding, coat, and portal protein monomers that oligomerize the portal ring around which the coat protein can assemble (Figure 1).
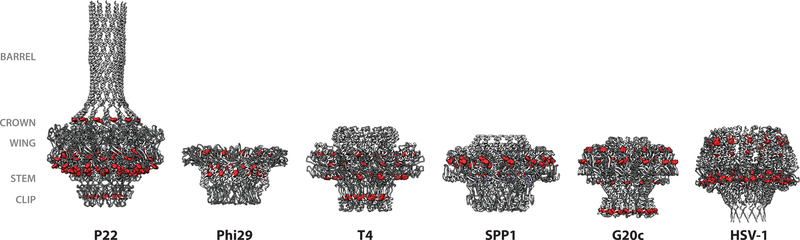
Portal ring assembly across bacteriophages and herpesviruses share similarities with P22, including the importance of scaffolding protein to portal ring formation. In phage lambda, the scaffolding protein GpNu3 has been implicated in the formation of portal (gpB) rings (14). In HSV-1 scaffolding protein (UL26.5), residues 143–151 are required for portal incorporation. The residues YYPGE in this region may be responsible as they are well conserved across alpha-herpesviruses (66, 67). On the HSV-1 portal protein, tryptophan residues in portal are critical to portal-scaffolding protein interactions as well as incorporation (68). It is possible that scaffolding protein-portal binding is driven by hydrophobic interactions. Some of these tryptophan residues are conserved in P22 as well, where they form a ring encircling the wing domain. Phi29 also has a belt of hydrophobic residues in the portal’s wing domain, suggesting that exposed hydrophobic residues on the portal wing domain may have a conserved function (43) Both SPP1 and the thermophilic bacteriophage G20c, a close relative of P23–45, as well as HSV-1, also feature tryptophan residues in the portal wing domain (Figure 2). In vitro experiments in Phi29 demonstrated scaffolding protein-portal ring binding; crosslinking evidence was used to develop a docking model to show six scaffolding protein dimers bound to the dodecameric portal wing domain (38, 51, 69). This evidence suggests that scaffolding proteins in several phages rely on similar hydrophobic interactions with the portal wing domain. However, the interactions are phage specific, as in vitro experiments using T4 portal with Phi29 coat and scaffolding proteins demonstrated that proteins from different phages did not bind (17).
Figure 2.
The tryptophan belt (red spheres) shown in portal proteins from P22 (5JJ3), Phi29 (1FOU), T4 (3JA7), SPP1 (2JES), G20c (4ZJN), and HSV-1 (6OD7). The portal complexes shown are all in the mature virion conformation. The well-conserved ring of tryptophan residues in the portal wing domains are important for portal interactions with scaffolding protein and ring assembly.
Other factors within the cellular environment may facilitate the assembly and incorporation of portal rings into procapsids. In T4, a phage-encoded protein (gp40) is thought to provide a membrane anchor for the assembly of the portal and core complex. Mutations in the 5’ upstream noncoding region of the portal protein can bypass the need for gp40 and increase the intracellular concentration of portal (34,70). This perhaps indicates that a high local concentration of the portal protein, facilitated by a chaperone protein, is crucial for ring assembly. In Phi29, in vitro portal incorporation during procapsid assembly can be achieved with the addition of 6% polyethylene glycol as a molecular crowding agent (50). The portal complex in Phi29 is also able to bind both RNA and DNA, as shown by filter binding assays. Although there is no evidence that this aids in complex oligomerization or incorporation into procapsids, it does mirror the idea that P22 mRNA could enhance portal ring assembly (71).
While work has been done to understand portal ring assembly and incorporation in bacteriophages, a significant amount of detail has yet to be explored. For example, the effect of scaffolding protein dimers on portal ring assembly has not been investigated, despite widely accepted evidence that P22, Phi29, and SPP1 scaffolding proteins are active for coat assembly as dimers or tetramers (51,72–74).
PORTAL PROTEIN STRUCTURES
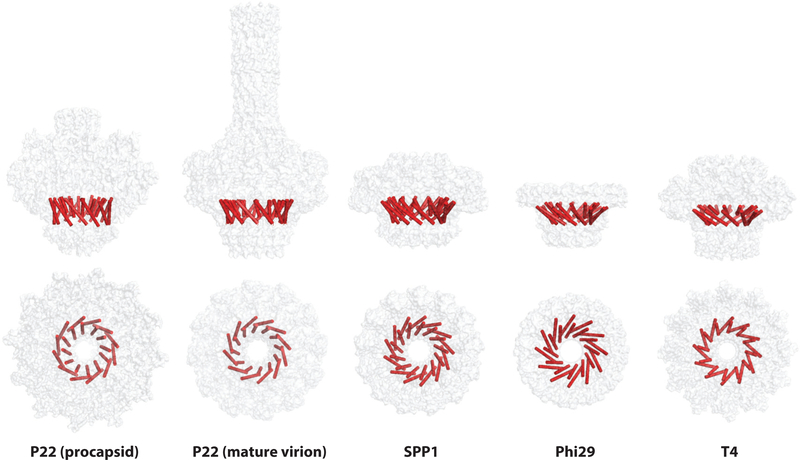
A common key component of bacteriophage, herpesvirus, and adenovirus assemblies is the presence of a single portal complex through which DNA is packaged and ejected. Portal complex structures have been studied using a range of techniques, including cryoEM, X-ray crystallography, and mass spectrometry. Each portal ring is composed of 12 monomers arranged around a central pore (also referred to as a channel or tunnel), although portal complex masses vary greatly from 400 kDa in Phi29 to 1 MDa in P22 (75). There is also some variability in the oligomeric states of purified portal complexes. In P22, mass spectrometry and electron microscopy studies have shown that purified in vitro assembled portal rings consisted of 11- and 12-mers, while Phi29 and SPP1 portals are 12-mers or 13-mers (56, 76). Image analysis and three-dimensional reconstruction of HSV-1 portal negative-stain and cryo-electron micrographs were used to determine that the oligomeric state lies between 11-mers and 14-mers, but the dodecameric form is likely incorporated into procapsids (77). In bacteriophages, the portal complex is always dodecameric in capsids (59, 76, 78–83).
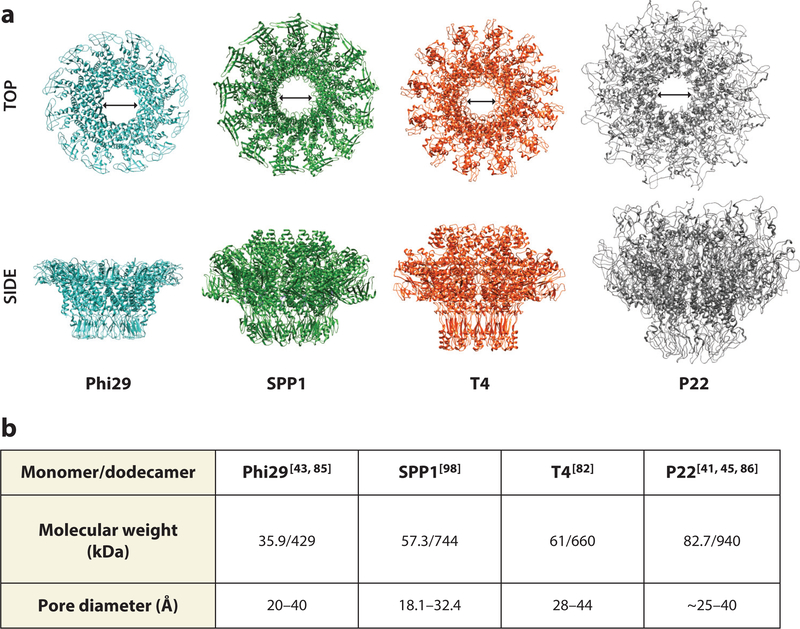
In addition to 12-fold symmetry, each portal complex features a central pore or channel. During packaging and ejection, the central pore of the portal complexes has a minimum diameter of 28 Å and is large enough to accommodate dsDNA (20 Å diameter) as well as some proteins that are packaged into procapsids (41, 84). Across bacteriophages, the internal diameters of the central channels do not vary as widely as the overall sizes of the portal complexes, ranging from approximately 18 Å to 44 Å across the entire length of the pore (41, 43, 77, 84–86) (Figure 3). In P22, the only phage for which the structure of portal protein was determined in the procapsid and mature virion conformations (45, 84), the diameter of the DNA channel decreases in the procapsid portal from ~40 Å to ~25 Å (Figure 4c–f). This suggests that residues lining the channel can make direct contact with DNA during packaging in the procapsid portal. Thus, the channel size may be relatively constant across bacteriophages due to the conserved nature of its function, with variations that depend more on the assembly state of the oligomer than the type of portal ring.
Figure 3.
(a) Portal ring structures from Phi29 (1FOU), SPP1 (2JES), T4 (3JA7), and P22 (5JJ3) from side and top views (arrows indicate internal pore diameter). (b) Chart summarizing the molecular weight and pore diameters of the portal complexes.
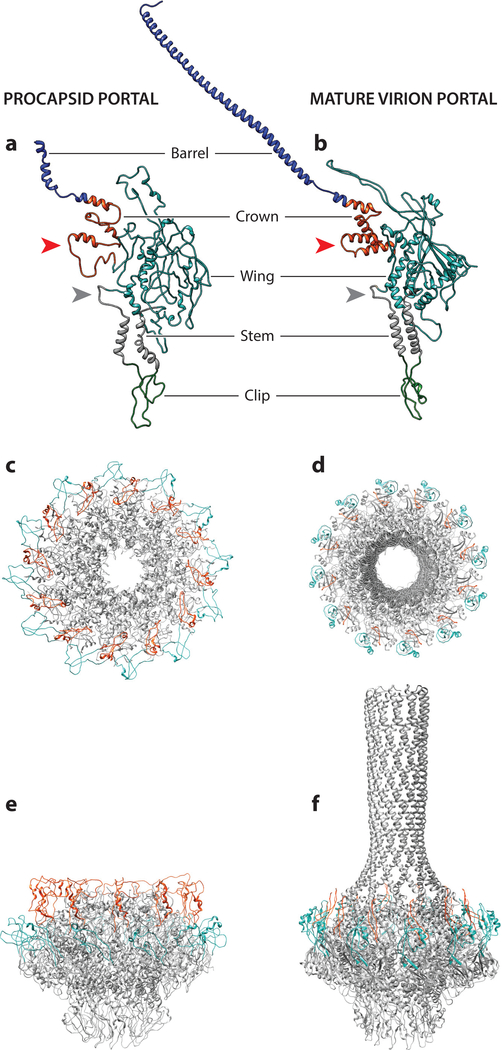
Figure 4.
P22 procapsid (left panel) and mature virion (right panel) portal structures. The monomers (a,b) have been colored by domain: barrel (blue), crown (red), wing (cyan), stem (gray), and clip (green). The tunnel (*) and channel (**) loops have been denoted. The top (c,d) and side (e,f) views of the procapsid and mature virion portal structures are shown with the hammer (red) and trigger loops (cyan) highlighted.
In short-tailed Podoviridae P22, Sf6 and T7 and the tailless phage phiX174 ejection proteins are expelled through the portal pore, along with DNA, to facilitate entry into the host cell by forming a tube that spans the bacterial membrane (87–90). In P22, the tail needle punctures the host cell’s outer membrane, and prior to DNA ejection, the ejection proteins (gp7, gp16, and gp20) exit the capsid to form subsequent parts of the transmembrane channel spanning to the inner membrane (88). In bacteriophage N4, astonishingly, a 3,500-amino acid phage-encoded DNA-dependent RNA polymerase is packaged into the capsid and ejected into the host through the portal during infection. The diameter of the channel is likely to accommodate only a folded globular protein less than 10 kDa, suggesting that most of these proteins must unfold during ejection and then refold to perform their function in the infection process. Despite variation in overall size and low sequence homology, portal structures show remarkable structural similarity, leading to the definition of the portal fold. Between the portal proteins from P22, T4, SPP1, and Phi29, there is less than 12% sequence identity, yet each portal protomer consists of a clip (also called a stalk), a stem, and a wing domain, while some structures have an additional crown domain (82, 91) (Figure 4a,b).The portal complex from P22, as well as many P22-related Podoviridae, is unique in that it also has an additional 200-Å alpha helical barrel domain that juts into the internal compartment of the capsid (41, 84).
The Clip Domain
The clip domains, which protrude out to the capsid exterior, comprised an α/β fold (23, 82, 84, 85, 91, 92). There is some structural variation in the clip region between phages, with some featuring more loop regions than others. In P22, this region of the portal protein has been shown to interact with the coat protein, as observed in an asymmetric cryoEM reconstruction (57). This region is also responsible for the large terminase binding during DNA packaging, as well as subsequent plug proteins and tail machinery. The plug proteins function to close the portal channel to prevent the accidental release of the packaged DNA as well as interact with proteins in the tail machinery (23, 82, 92, 93). The clip regions in the dodecamer impart additional stability to the overall structure (23,82,92).
In a comparison of SPP1, Phi29, and T4 portal proteins, there is significant structural homology as well as some conserved residues in the clip region, which have been implicated in terminase binding and DNA packaging. Mutations in the SPP1 clip region inhibited DNA packaging (94). The T4 portal clip region binds to the C terminus of the large terminase in order to package DNA. Mutations to the charged residues in the large terminase resulted in a loss of binding to the portal complex, indicating the importance of electrostatic interactions both within portal and for interprotein interactions (82, 94). In Phi29, the clip region also comprises the binding site for the pRNA, which forms a hexameric ring around the base of the portal and facilitates DNA packaging (29, 43, 85). Due to its protrusion from the capsid interior, the clip region provides a docking site for DNA packaging complex and the tail machinery. Changes in the conformation of the P22 clip region are implicated in the binding of TerL to the procapsid state of the portal and release of TerL once DNA has been packaged (45). The recent 4.3Å cryoEM reconstruction of HSV-1 portal shows five tentacle helices extending from the outer face of the portal complex, which are proposed to be part of the clip domain. These helices, punctuated with disordered residues, are thought to be flexible and allow the portal to contact both the pentameric terminase protein and the portal cap. The flexibility of the structure in the clip likely allows for the intriguing symmetry mismatch between the 12-mer portal ring and the 5-mer terminase protein (95).
The portal is a surprisingly plastic assembly, and structural signals can be transmitted through the domains. In HSV-1, a predicted leucine zipper motif from residues 422–443 in the clip domain of the portal is important both for portal ring formation and, indirectly, for recruitment of the terminase complex (95). Mutations to this region demonstrate that a hydrophobic interface may stabilize monomer-monomer interactions. Perturbations may alter the portal structure in ways that inhibit binding of the terminase proteins (96, 97). A putative leucine zipper has also been predicted from residues 600–650 in the P22 portal barrel domain, but its role has not been investigated (84).
In addition to providing docking sites for other proteins, the portion of the clip region facing the inner channel forms the entrance to the channel. Charged residues on these loops, such as the well-conserved channel loop (Figure 4), have been shown to interact with DNA. Negatively charged aspartate and glutamate residues at the channel entrance and on the channel loop are required for DNA translocation in several phages, including SPP1, HK97, Phi29, and T4. These residues may play a role in keeping DNA centered in the channel (82, 98). Rings of positively charged residues such as arginine and lysine are also implicated in DNA binding in several phages, including P22, Phi29, andT4, as well as HSV-1. Positive charges within the channel likely prevent backsliding during DNA translocation (84). The clip domain may also be important for maintaining monomer-monomer interactions. In Phi29 there are several hydrophobic contacts between adjacent monomers (43, 85). Thus, the electrostatic profile and buried hydrophobic residues of the clip domain are important for the functions of this domain including binding DNA packaging and tail machinery as well as increasing dodecamer stability (82, 84, 85, 94, 99).
The Stem Domain
Connected to the clip region is the stem domain, which comprised two well-conserved helices that extend into the wing domain (43, 85) (Figures 4a,b and 5). In the predicted herpes virus portal structure, these helices are also conserved (100). The 24 stem helices form the central channel in the dodecameric portal complex and have greater compression in the immature procapsid portal of P22 than in the mature virus conformation. This is reflected by a change in the angle that each helix makes relative to the 12-fold portal axis that increases from 20° to 30° in the procapsid versus mature virion portal (45, 95). The conserved channel loop in the stem domain has been shown to be important for T4 DNA packaging and was hypothesized to be necessary for DNA translocation in SPP1 (98,101). The central channel of the portal rings is typically electronegative but contains conserved basic residues involved in DNA binding (82, 91, 101). In T7 and Phi29, rings of conserved lysine residues in the stem domain are important for DNA translocation and likely interact with the phosphate backbone (43, 83). In P22, five glutamate rings within the portal channel are thought to be important in keeping the DNA centered during packaging (84, 91). Likewise, HSV-1 features two rings of positively charged arginine residues in the stem domain, likely for DNA interaction, while the channel facing beta-clip features asparagine and glutamine residues in the electronegative portion of the pore, similar to P22 (95).
Figure 5.
Portal complexes from P22, SPP1, Phi29, and T4 side and top views shown as gray spheres, with central helices highlighted in red. The core helices are important for oligomerization as well as DNA ejection and the conformational changes required for the maturation and ejection processes.
The Wing Domain
The adjoining wing domain shows greater structural variation between different portal proteins, although the core helices, which are suspected of aiding in intersubunit interactions, are structurally conserved in phages as well as herpesviruses (100) (Figure 5). The portal wing domains also likely contain the putative coat-binding region. In the T4 portal, the wing domain N-terminal 73 residues are predicted to comprise the coat-binding domain of the portal complex. Deletion of the N-terminal portal residues prevents head assembly in T4. Thus, these wing domain residues are predicted to be important for coat binding and the initiation of assembly (82, 101). The N terminus, which is not resolved by cryoEM, is thought to be disordered and reside in a pocket between the clip domain and the adjacent hexamers (82). N-terminal loop regions in the stem and wing domains of the P22 portal, from residues 41–59 and 191–217, have been proposed as the coat-binding domain as they change conformation upon capsid maturation (41, 56). A recent cryoEM reconstruction of the thermophilic bacteriophage P23–45 portal vertex in both the procapsid and expanded capsid forms also shows the portal N terminus in the wing domain interacting with the surrounding coat protein, likely through electrostatic interactions (102).
The wing domain comprises a significant portion of the portal protein monomer-monomer interface. Hydrogen bonding and electrostatic interactions likely stabilize the subunits, where net positive and net negative faces of the monomers border each other (82, 84, 85). In P22, positively charged lysine and arginine residues on one subunit interact with negatively charged aspartate and glutamate residues of the neighboring subunit. Similar monomer-monomer interactions have been shown in Phi29 (43,85). Further evidence for the electrostatic stabilization of the oligomer is demonstrated by salt dependence and ethylenediaminetetraacetic acid requirement for dodecamer assembly, which has been shown in both P22 and T4 (82, 84).
P22 and HSV-1 feature cysteine residues in the portal wing domain that are important for portal ring formation. In P22, the mutant C283S portal forms hyperstable monomers; in HSV-1, C166A and C254A are both ring assembly incompetent, indicating that disulfide bonds may also play a role in portal ring stabilization (103, 104). The portal rings are highly stable once formed, and in Phi29, they require a strong denaturant, such as guanidine hydrochloride, in order to denature and disassemble the oligomer (71).
The wing domain is also important for binding other proteins. The outer regions of portal complexes tend to be charged at the top portion of the wing/crown and clip domains, which may be important for contacts with the ejection proteins, terminase proteins, and plug proteins/tail machinery (82, 84, 85, 94, 99). This domain also contains a typtophan belt in P22, T4, Phi29, and HSV-1 portal proteins (Figure 3), which strengthens the hydrophobic core and could play a role in scaffolding protein binding during incorporation into the growing procapsid (51,57). The portal complex features both charged and hydrophobic patches that are important for specific interactions with other proteins and DNA.
The Crown Domain
The portal C-terminal crown domain, which is present in all portal protein structures except that of Phi29, has been implicated in interactions with the dsDNA (42). In T4, an asymmetric cryoEM reconstruction has been used to resolve the density for the ejection proteins (gp14, gp15, and gp16) that form a proteinaceous core bound to the portal crown domain. The tunnel created by the stack of ejection proteins is proposed to enable DNA spooling inside the capsid (86). In P22, the C-terminal barrel domain forms a 200-Å left-handed coiled-coil alpha helical barrel that extends into the capsid, which is thought to facilitate DNA packaging and ejection to prevent tangling, as well as to provide a binding site for ejection proteins (84, 105).
The Barrel Domain
The barrel domain can be predicted by monitoring the coiled-coil propensity in the C-terminal ~ 130 residues of portal protein. A survey of all sequenced P22-like podoviruses found that most P22-like members of this diverse family harbor a barrel, which, in contrast, is not found in Phi29-like phages and in T7-like phages. The barrel can also be predicted in a few long-tailed sipho- and myovirus bacteriophages, although with less frequency than in Podoviridae (41). Deletion studies found that a complete (or nearly complete) barrel is required to deliver the DNA inside the host, while a naturally occurring deletion of the 48 C-terminal residues (amino acids 677–725) has no apparent deleterious effect on virus infectivity (59). Larger deletions of the barrel between residues 650 and 602 result in virus particles that package DNA efficiently but are defective in delivering DNA into the host, consistent with an ~ 10-fold lower phage titer (41).
Thus, portal proteins across bacteriophages maintain the portal fold, despite low sequence similarity, which is important for the role of the portal protein as a conduit for DNA as well as the docking site for tail machinery.
PORTAL AS A DNA PACKAGING SENSOR
In addition to acting as a conduit for DNA, plasticity in the P22 portal protein allows the protein to transmit structural signals during the packaging process. Point mutations in the P22 portal, including V64M and V303M, cause over-packaging of the genome, demonstrating that portal acts as a switch to indicate when the capsid is full. Mutations to the well-conserved central helices in portal may increase rigidity and retard structural changes that signal the end of packaging (80, 84, 106).
Structural evidence suggests that the unusual barrel domain in P22 facilitates the role in spooling and stabilizing the DNA within the capsid during packaging. Portal compression due to pressure from the packaged DNA likely induces a conformational change in the portal protein that signals when the head is full (41, 45, 107). Crystal structures and modeling of the P22 procapsid and mature virion portal complexes show a portal maturation that increases the central channel diameter and causes the overall structure to become more compact and symmetrical. The disordered C-terminal domain in the procapsid portal forms an alpha helical barrel in mature viron portal, suggesting that DNA plays a role in changing portal structure (45). Overpackaging mutants likely stabilize the procapsid form of portal, preventing the compression of the core region that induces the conformational change to mature viron portal and triggers an end to packaging (107). In an asymmetric cryoEM reconstruction of the P22 virion, density attributed to DNA wraps around the portal protein and is suggested to be involved in the signal for the completion of headful packaging (108).
In P22, the structural rearrangement of portal protein, especially in the clip region, during maturation as the barrel forms and is compressed is essential as it changes which proteins are able to bind to the portal. The P22 large terminase preferentially binds to the procapsid conformation of the portal, whereas the adaptor plug protein gp4 binds the mature virion conformation of the portal (45,109). Induced fit of the portal to the adaptor protein then enables subsequent binding of the plug protein gp10 and the tail only to particles that have encapsulated DNA (110–112). Thus, portal structural changes play a significant role in regulating viral maturation. The P22 portal complex undergoes conformational changes that allow for sequential ordering of the maturation process.
InT4, the portal clip region has been proposed to play an integral role in sensing DNA for both packaging initiation and completion. Mutational analysis of arginine and lysine residues (R338 and K342) in the tunnel entrance in the clip indicates that these residues are essential for genome packaging. These residues are proposed to anchor the beginning of the viral DNA during packaging initiation (82). Mutations in the T4 clip region result in packaging termination defects as well, suggesting that this region is critical for signal transduction between the portal and terminase proteins (94, 113). Positively charged residues in the T4 portal tunnel loop have been shown to stabilize DNA at the end of packaging, likely to prevent spilling of the DNA prior to the addition of the plug proteins (101). Thus, the portal protein plays an active role in all stages of the packaging process.
A recent single-molecule study on T4 DNA packaging in vitro demonstrated that portal acts as a fail-safe clamp to prevent DNA detachment during packaging caused by backsliding when the terminase motor slips. Ordyan et al. (114) demonstrate that terminase gripping on DNA is regulated by the nucleotide (ATP) state of the translocation motor. In conditions with ADP or no nucleotide, the terminase does not grip as strongly as when ATP is present. The portal protein acts as a clamp, however, gripping DNA regardless of the nucleotide state of the terminase. This indicates, perhaps, that residues in portal channel entrance or the tunnel loops clamp DNA, keeping it tethered to the capsid during low-gripping terminase states (101, 114). Similar to the P22 portal complex, electrostatic interactions and conformational changes in the T4 portal protein facilitate DNA packaging from initiation to termination.
The Phi29 portal protein has also been shown to grip DNA through two rings of positively charged lysine residues (K200 and K209) in the clip region. The distance between the lysine rings has been calculated to be 20 Å, approximately the same distance between two phosphate groups in the major groove of the DNA backbone. These residues may help to grip the DNA to enable the pause-and-slip mechanism for DNA packaging, similar to what has been noted in T4 (43).
Single-molecule experiments and structural data have been used to demonstrate that the Phi29 portal not only grips DNA during packaging but also responds structurally. Capsid pressure from internal DNA affects terminase packaging rates and pausing indirectly through allosteric interaction between the portal/pRNA complex and the terminase machinery. CryoEM structures support the hypothesis that DNA contacts the portal wing domain inside the capsid and could induce portal and pRNA structural changes, leading to occlusion of the ATP binding site on the terminase and slowing packaging (115). In addition, DNA binding to the Phi29 C-terminal domain in the portal protein wing domain induces conformational changes in the channel to prevent DNA slippage prior to plug addition at the end of packaging (116). Similar to the processes observed in P22 and T4 portals, the Phi29 portal protein is not a static pore but rather acts as a gauge that responds structurally to transmit the headful signal to other packaging components. Analogous to the Phi29 portal, biochemical data and cryoEM structural analysis suggest that lysine residues (K331 and K342) in the SPP1 portal protein interact with DNA (92, 98). Thus, the gripping function of these residues may be conserved across portals.
Plasticity in the portal structure is also required for DNA packaging in SPP1. Stabilization of adjacent α5 in the stem domain by crosslinking has been shown to inhibit DNA packaging but not DNA ejection, indicating that packaging requires coordinated movement between α5 and the tunnel loops to grip the DNA, as well as to communicate with the terminase protein (117). Mutational evidence supports the hypothesis that portal plays a role in each step of the DNA packaging process (46).
Bacteriophage T7 also exhibits structural rearrangement in the portal complex that facilitates maturation. CryoEM reconstructions of the bacteriophage T7 procapsids and mature virions suggest that internal DNA pressure causes large-scale structural rearrangement of the internal core proteins (gp14, −15, and −16), which are bound to the portal, and compression of the portal toward the pentameric vertex. Further protrusion of the clip region out of the capsid after compression presumably triggers disassociation of the terminase and allows for tail attachment (118). The portal complex of the thermophilic bacteriophage, P23–45, undergoes the opposite transition, from a compressed to an extended conformation in the procapsid and expanded capsid forms. This conformational change also shows coordinated movement of the wing and clip domains (102). DNA packaging has long-reaching conformational effects across the portal protein.
HSV-1 portal protein also features a clip domain that appears to be structurally flexible as well as strongly positively charged. Positively charged residues likely help to grip terminal DNA at the channel entrance. Liu et al. (95) proposed that this region undergoes conformational change allowing for the propagation of the headful signal during packaging as well as the switch in binding partners from the terminase complex to the portal cap protein. Likewise, the P22 cryoEM reconstruction also shows DNA spooled around the wing domain inside the capsid, suggesting that the portal complex facilitates DNA organization within the capsid but could also facilitate transmission of the headful signal across the domains (108).
Thus, the portal complex across phages plays an active role in DNA packaging, from the initial capture of DNA through to the attachment of tail machinery. Portals are dynamic pressure gauges, changing structurally due to increased pressure from DNA packaging and transmitting this signal to facilitate terminase complex dissociation and the subsequent plug and tail protein additions.
GENOME RELEASE
In addition to morphogenic changes that occur during genome packaging, further conformational rearrangements are induced during tail assembly and genome exit. In bacteriophage P22, portal conformational changes to a mature virion form that allow for the terminase machinery detachment and attachment of the plug proteins and tail machinery (45, 109). In mature P22 phages, the ejection proteins localize to the portal barrel domain inside the capsid, priming the phage for DNA ejection (105,119). A recent cryoEM reconstruction of P22 virions with an amber mutation in the tail needle protein, which prevents DNA leakage after packaging, showed that the portal protein maintains its mature virion conformation after genome release from the capsid. In the micrographs, additional density on the tail machinery is hypothesized to be tubes composed of the ejection proteins (gp7, gp16, and gp20) (120). In P22, portal structural rearrangements upon maturation prepare the phage for genome ejection.
In bacteriophage SPP1, portal structural changes due to DNA packaging allow for the adapter and plug proteins to bind sequentially to the portal via electrostatic interactions (99). Binding of the tail needle to the host receptor causes allosteric structural changes throughout the tail machinery that trigger genome release (92). Exposure to the host cellular environment causes the reduction of disulfide bonds within the plug protein, leading to structural changes that open the central channel for DNA exit (92,99). CryoEM reconstructions of tail machinery prior to and after genome release show structural rearrangements that stem from the tail needle and extend through the entire portal protein, such that key DNA binding residues no longer jut into the channel (92). Unlike in P22, the SPP1 portal, therefore, is responsive not only to DNA and protein binding but also to the external environmental stimuli during ejection.
The portal vertex in phage lambda is unique in that it senses temperature as well as internal DNA pressure, ensuring that DNA ejection occurs only when host cells are replicating. At lower temperatures, DNA ejection times lag significantly, even in the presence of the host epitope, as compared to higher temperatures, despite no change in the internal DNApressure (47). This effect has also been noted in herpesviruses where DNA ejection occurs almost exclusively at 37°C (121). As well as temperature, proteolytic cleavage of the portal protein may enable genome uncoating in both phage lambda and herpesviruses. In HSV-1, in vitro DNA ejection is correlated with low levels of portal (UL6) proteolytic cleavage, indicating that cleavage to just one subunit of the dodecameric complex can be destabilizing enough to initiate genome release (121). Conversely, in phage lambda the N terminus of the portal protein (gpB) must be cleaved by the lambda protease, gpC. This cleavage event does not affect portal incorporation or DNA packaging but is necessary for portal structural maturation, indicating that it is important for either the addition of the tail machinery or DNA ejection (15). Thus, the portal protein acts as a sensor of its environment and is able to transmit those signals when appropriate to release the genome into the host cell.
CONCLUSION
Despite decades of genetics work along with biochemistry and biophysics, the mechanism behind portal ring assembly and incorporation into procapsids is not well understood. Further research is required to elucidate the portal’s role in all steps of viral assembly. Notwithstanding the limitations in our mechanistic and structural understanding of the portal protein, promising research is demonstrating the potential of HSV-1 portal as an antiviral drug target in HSV-1 and HSV-2 in vitro using small molecules (100, 122, 123). Studies of bacteriophage assembly have enabled researchers to delve into more complex systems and may help to indicate additional targets for future antiviral therapies.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 GM076661 to C.M.T.andR01 GM100888 to G.C.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Asija K, Teschke CM. 2018. Lessons from bacteriophages part 2: a saga of scientific breakthroughs and prospects for their use in human health. PLOS Pathog. 14:e1006970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asija K, Teschke CM. 2018. Lessons from bacteriophages part 1: deriving utility from protein structure, function, and evolution. PLOS Pathog. 14:e1006971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prevelige PE Jr., Thomas D, King J 1993. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 64:824–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prevelige PE Jr., King J 1993. Assembly of bacteriophage P22: a model for ds-DNA virus assembly. Prog. Med. Virol. 40:206–21 [PubMed] [Google Scholar]
- 5.Prevelige PE, Thomas D, King J. 1988. Scaffolding protein regulates the polymerization of P22 coat subunits into icosahedral shells in vitro. J. Mol. Biol. 202:743–57 [DOI] [PubMed] [Google Scholar]
- 6.Traub F, Maeder M. 1984. Formation of the prohead core of bacteriophage T4 in vivo. J. Virol. 49:892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Driel R, Couture E. 1978. Assembly of the scaffolding core of bacteriophage T4 preheads. J. Mol. Biol. 123:713–19 [DOI] [PubMed] [Google Scholar]
- 8.Newcomb WW, Brown JC. 1991. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol. 65:613–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller MT, King J 1981. Purification of the coat and scaffolding proteins from procapsids of bacteriophage P22. Virology 112:529–47 [DOI] [PubMed] [Google Scholar]
- 10.Fuller MT, King J. 1982. Assembly in vitro of bacteriophage P22 procapsids from purified coat and scaffolding subunits. J. Mol. Biol. 156:633–65 [DOI] [PubMed] [Google Scholar]
- 11.Homa FL, Brown JC. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107–22 [DOI] [PubMed] [Google Scholar]
- 12.Preston VG, Al-Kobaisi MF, McDougall IM, Rixon FJ. 1994. The herpes simplex virus gene UL26 proteinase in the presence of the UL26.5 gene product promotes the formation of scaffold-like structures. J. Gen. Virol. 75:2355–66 [DOI] [PubMed] [Google Scholar]
- 13.Black LW, Rao VB. 2012. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv. Virus Res. 82:119–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kochan J, Murialdo H. 1983. Early intermediates in bacteriophage lambda prohead assembly II. Identification of biologically active intermediates. Virology 131:100–15 [DOI] [PubMed] [Google Scholar]
- 15.Medina E, Wieczorek D, Medina EM, Yang Q, Feiss M, Catalano CE. 2010. Assembly and maturation of the bacteriophage lambda procapsid: gpC is the viral protease.J. Mol. Biol. 401:813–30 [DOI] [PubMed] [Google Scholar]
- 16.Casjens S, Adams MB, Hall C, King J 1985. Assembly-controlled autogenous modulation of bacteriophage P22 scaffolding protein gene expression. J. Virol. 53:174–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee CS, Guo P. 1995. Sequential interactions of structural proteins in phage φ29 procapsid assembly. J. Virol. 69:5024–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64:1007–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, et al. 2007. Measurements of single DNA molecule packaging dynamics in bacteriophage λ reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 373:1113–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. 2001. The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature 413:748–52 [DOI] [PubMed] [Google Scholar]
- 21.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. 2007. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. PNAS 104:16868–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasad BVV, Prevelige PE, Marietta E, Chen RO, Thomas D, et al. 1993. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J. Mol. Biol. 231:65–74 [DOI] [PubMed] [Google Scholar]
- 23.Tavares P 2018. The bacteriophage head-to-tail interface In Virus Protein and Nucleoprotein Complexes, ed. Harris JR, Bhella D, pp. 305–28. Singapore: Springer Singapore; [DOI] [PubMed] [Google Scholar]
- 24.Alam TI, Rao VB. 2008. The ATPase domain of the large terminase protein, gp17, from bacteriophage T4 binds DNA: implications to the DNA packaging mechanism. J. Mol. Biol. 376:1272–81 [DOI] [PubMed] [Google Scholar]
- 25.Rao VB, Feiss M. 2015. Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu. Rev. Virol. 2:351–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh CS, Sippy J, Charbonneau B, Crow Hutchinson J, Mejia-Romero OE, et al. 2016. DNA topology and the initiation of virus DNA packaging. PLOS ONE 11:e0154785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heming JD, Conway JF, Homa FL. 2017. Herpesvirus capsid assembly and DNA packaging. Cell Biol. Herpes Viruses 223:119–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casjens SR, Gilcrease EB. 2009. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Bacteriophages 502:91–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garver K, Guo P. 1997. Boundary of pRNA functional domains and minimum pRNA sequence requirement for specific connector binding and DNA packaging of phage phi29. RNA 3:1068–79 [PMC free article] [PubMed] [Google Scholar]
- 30.Draper B, Rao VB. 2007. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J. Mol. Biol. 369:79–94 [DOI] [PubMed] [Google Scholar]
- 31.Keller N, delToro D, Grimes S, Jardine PJ, Smith DE. 2014. Repulsive DNA-DNA interactions accelerate viral DNA packaging in phage phi29. Phys. Rev Lett. 112:248101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murialdo H, Becker A. 1978. A genetic analysis of bacteriophage lambda prohead assembly in vitro. J. Mol. Biol. 125:57–74 [DOI] [PubMed] [Google Scholar]
- 33.Newcomb WW, Homa FL, Brown JC. 2005. Involvement of the portal at an early step in herpes simplex virus capsid assembly. J. Virol. 79:10540–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yap NL, Rao VB. 1996. Novel mutants in the 5’ upstream region of the portal protein gene 20 overcome a gp40-dependent prohead assembly block in bacteriophage T4. J. Mol. Biol. 263:539–50 [DOI] [PubMed] [Google Scholar]
- 35.Bazinet C, King J. 1988. Initiation of P22 procapsid assembly in vivo. J. Mol. Biol. 202:77–86 [DOI] [PubMed] [Google Scholar]
- 36.Dröge A, Santos MA, Stiege AC, Alonso JC, Lurz R, et al. 2000. Shape and DNA packaging activity of bacteriophage SPP1 procapsid: protein components and interactions during assembly. J. Mol. Biol. 296:117–32 [DOI] [PubMed] [Google Scholar]
- 37.van Driel R, Couture E. 1978. Assembly of bacteriophage T4 head-related structures: II. In vitro assembly of prehead-like structures. J. Mol. Biol. 123:115–28 [DOI] [PubMed] [Google Scholar]
- 38.Guo PX, Erickson S, Xu W, Olson N, Baker TS, Anderson D. 1991. Regulation of the phage cp29 prohead shape and size by the portal vertex. Virology 183:366–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laemmlli UK, Mölbert E, Showe M, Kellenberger E. 1970. Form-determining function of the genes required for the assembly of the head of bacteriophage T4. J. Mol. Biol. 49:99–113 [DOI] [PubMed] [Google Scholar]
- 40.Moore SD, Prevelige PE Jr. 2002. Bacteriophage p22 portal vertex formation in vivo. J. Mol. Biol. 315:975–94 [DOI] [PubMed] [Google Scholar]
- 41.Tang J, Lander GC, Olia A, Li R, Casjens S, et al. 2011. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in P22. Structure 19:496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orlova EV, Gowen B, Dröge A, Stiege A, Weise F, et al. 2003. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 22:1255–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guasch A, Pous J, Ibarra B, Gomis-Rüth FX, Valpuesta JMA, et al. 2002. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage cp29 connector particle. J. Mol. Biol. 315:663–76 [DOI] [PubMed] [Google Scholar]
- 44.Grimes S, Ma S, Gao J, Atz R, Jardine PJ. 2011. Role of cp29 connector channel loops in late-stage DNA packaging. J. Mol. Biol. 410:50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lokareddy RK, Sankhala RS, Roy A, Afonine PV, Motwani T, et al. 2017. Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation. Nat. Commun. 8:14310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isidro A, Henriques AO, Tavares P. 2004. The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology 322:253–63 [DOI] [PubMed] [Google Scholar]
- 47.Freeman KG, Behrens MA, Streletzky KA, Olsson U, Evilevitch A. 2016. Portal stability controls dynamics of DNA ejection from phage. J. Phys. Chem. B 120:6421–29 [DOI] [PubMed] [Google Scholar]
- 48.Nobrega FL, Vlot M, de Jonge PA, Dreesens LL, Beaumont HJE, et al. 2018. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 16:760–73 [DOI] [PubMed] [Google Scholar]
- 49.Earnshaw W, King J. 1978. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J. Mol. Biol. 126:721–47 [DOI] [PubMed] [Google Scholar]
- 50.Fu CY, Prevelige PE Jr. 2009. In vitro incorporation of the phage Phi29 connector complex. Virology 394:149–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu CY, Uetrecht C, Kang S, Morais MC, Heck AJ, et al. 2010. A docking model based on mass spectrometric and biochemical data describes phage packaging motor incorporation. Mol. Cell. Proteom. 9:1764–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newcomb WW, Thomsen DR, Homa FL, Brown JC. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Botstein D, Waddell CH, King J. 1973. Mechanism of head assembly and DNA encapsulation in Salmonella phage p22. I. Genes, proteins, structures and DNA maturation. J. Mol. Biol. 80:669–95 [DOI] [PubMed] [Google Scholar]
- 54.Earnshaw W, Casjens S, Harrison SC. 1976. Assembly of the head of bacteriophage P22: X-ray diffraction from heads, proheads and related structures. J. Mol. Biol. 104:387–410 [DOI] [PubMed] [Google Scholar]
- 55.King J, Lenk EV, Botstein D. 1973. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22: II. Morphogenetic pathway. J. Mol. Biol. 80:697–731 [DOI] [PubMed] [Google Scholar]
- 56.Motwani T, Lokareddy RK, Dunbar CA, Cortines JR, Jarrold MF, et al. 2017. A viral scaffolding protein triggers portal ring oligomerization and incorporation during procapsid assembly. Sci. Adv. 3:e1700423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, et al. 2011. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. PNAS 108:1355–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Traub F, Keller B, Kuhn A, Maeder M. 1984. Isolation of the prohead core of bacteriophage T4 after cross-linking and determination of protein composition. J. Virol. 49:902–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bazinet C, Benbasat J, King J, Carazo JM, Carrascosa JL. 1988. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry 27:1849–56 [DOI] [PubMed] [Google Scholar]
- 60.Weigele PR, Sampson L, Winn-Stapley D, Casjens SR. 2005. Molecular genetics of bacteriophage P22 scaffolding protein’s functional domains. J. Mol. Biol. 348:831–44 [DOI] [PubMed] [Google Scholar]
- 61.Greene B, King J. 1996. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology 225:82–96 [DOI] [PubMed] [Google Scholar]
- 62.Moore SD, Prevelige PE Jr. 2002. A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein. J. Virol. 76:10245–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bazinet C, Villafane R, King J. 1990. Novel second-site suppression of a cold-sensitive defect in phage P22 procapsid assembly. J. Mol. Biol. 216:701–16 [DOI] [PubMed] [Google Scholar]
- 64.Rizzo AA, Suhanovsky MM, Baker ML, Fraser LCR, Jones LM, et al. 2014. Multiple functional roles of the accessory I-domain of bacteriophage P22 coat protein revealed by NMR structure and cryoEM modeling. Structure 22:830–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gordon CL, King J. 1993. Temperature-sensitive mutations in the phage P22 coat protein which interfere with polypeptide chain folding. J. Biol. Chem. 268:9358–68 [PubMed] [Google Scholar]
- 66.Singer GP, Newcomb WW, Thomsen DR, Homa FL, Brown JC. 2005. Identification of a region in the herpes simplex virus scaffolding protein required for interaction with the portal. J. Virol. 79:132–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang K, Baines JD. 2009. Proline and tyrosine residues in scaffold proteins of herpes simplex virus 1 critical to the interaction with portal protein and its incorporation into capsids. J. Virol. 83:8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang K, Baines JD. 2009. Tryptophan residues in the portal protein of herpes simplex virus 1 critical to the interaction with scaffold proteins and incorporation of the portal into capsids. J. Virol. 83:11726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li R, Cherwa JE, Prevelige PE. 2013. cp29 Scaffolding and connector structure-function relationship studied by trans-complementation. Virology 444:355–62 [DOI] [PubMed] [Google Scholar]
- 70.Hsiao CL, Black LW. 1978. Head morphogenesis of bacteriophage T4 II. The role of gene 40 in initiating prehead assembly. Virology 91:15–25 [DOI] [PubMed] [Google Scholar]
- 71.Urbaneja MA, Rivas S, Carrascosa JL, Valpuesta JM. 1994. An intrinsic-tryptophan-fluorescence study of phage cp29 connector/nucleic acid interactions. Eur. J. Biochem. 225:747–53 [DOI] [PubMed] [Google Scholar]
- 72.Parker MH, Casjens S, Prevelige PE. 1998. Functional domains of bacteriophage P22 scaffolding protein. J. Mol. Biol. 281:69–79 [DOI] [PubMed] [Google Scholar]
- 73.Poh SL, el Khadali F, Berrier C, Lurz R, Melki R, Tavares P. 2008. Oligomerization of the SPP1 scaffolding protein. J. Mol. Biol. 378:551–64 [DOI] [PubMed] [Google Scholar]
- 74.Tuma R, Parker MH, Weigele P, Sampson L, Sun Y, et al. 1998. A helical coat protein recognition domain of the bacteriophage P22 scaffolding protein. J. Mol. Biol. 281:81–94 [DOI] [PubMed] [Google Scholar]
- 75.Parent KN, Schrad JR, Cingolani G. 2018. Breaking symmetry in viral icosahedral capsids as seen through the lenses of X-ray crystallography and cryo-electron microscopy. Viruses 10:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poliakov A, Duijn EV, Lander G, Fu C-Y, Johnson JE, et al. 2007. Macromolecular mass spectrometry and electron microscopy as complementary tools for investigation of the heterogeneity of bacteriophage portal assemblies. J. Struct. Biol. 157:371–83 [DOI] [PubMed] [Google Scholar]
- 77.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carrascosa JL, Carazo JM, Ibanez C, Santistebanz A. 1985. Structure of phage cp29 connector protein assembled in vivo. Virology 141:190–200 [DOI] [PubMed] [Google Scholar]
- 79.Lurz R, Orlova EV, Günther D, Dube P, Droge A, et al. 2001. Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol. 310:1027–37 [DOI] [PubMed] [Google Scholar]
- 80.Kochan J, Carrascosa JL, Murialdo H. 1984. Bacteriophage lambda preconnectors: purification and structure. J. Mol. Biol. 174:433–47 [DOI] [PubMed] [Google Scholar]
- 81.Donate LE, Herranz L, Secilla JP, Carazo J, Fujisawa H, Carrascosa J. 1988. Bacteriophage T3 connector: three-dimensional structure and comparison with other viral head-tail connecting regions. J. Mol. Biol. 201:91–100 [DOI] [PubMed] [Google Scholar]
- 82.Sun L, Zhang X, Gao S, Rao PA, Padilla-Sanchez V, et al. 2015. Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution. Nat. Commun. 6:7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Agirrezabala X, Martín-Benito J, Valle M, González JM, Valencia A, et al. 2005. Structure of the connector of bacteriophage T7 at 8 Å resolution: structural homologies of a basic component of a DNA translocating machinery. J. Mol. Biol. 347:895–902 [DOI] [PubMed] [Google Scholar]
- 84.Olia AS, Prevelige PE Jr., Johnson JE, Cingolani G 2011. Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol. 18:597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, et al. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guo F, Liu Z, Vago F, Ren Y, Wu W, et al. 2013. Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. PNAS 110:6811–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jin Y, Sdao SM, Dover JA, Porcek NB, Knobler CM, et al. 2015. Bacteriophage P22 ejects all of its internal proteins before its genome. Virology 485:128–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang C, Tu J, Liu J, Molineux IJ. 2019. Structural dynamics of bacteriophage P22 infection initiation revealed by cryo-electron tomography. Nat. Microbiol. 18:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Molineux IJ. 2001. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40:1–8 [DOI] [PubMed] [Google Scholar]
- 90.Xu J, Xiang Y. 2017. Membrane penetration by bacterial viruses. J. Virol. 91:e00162–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cuervo A, Carrascosa JL. 2012. Viral connectors for DNA encapsulation. Curr. Opin. Biotechnol. 23:529–36 [DOI] [PubMed] [Google Scholar]
- 92.Chaban Y, Lurz R, Brasilès S, Cornilleau C, Karreman M, et al. 2015. Structural rearrangements in the phage head-to-tail interface during assembly and infection. PNAS 112:7009–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Olia AS, Al-Bassam J, Winn-Stapley DA, Joss L, Casjens SR, Cingolani G. 2006. Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. J. Mol. Biol. 363:558–76 [DOI] [PubMed] [Google Scholar]
- 94.Dixit AB, Ray K, Thomas JA, Black LW. 2013. The C-terminal domain of the bacteriophage T4 terminase docks on the prohead portal clip region during DNA packaging. Virology 446:293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Y, Jih J, Dai X, Bi G, Zhou ZH. 2019. Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome. Nature 570:257–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nellissery JK, Szczepaniak R, Lamberti C, Weller SK. 2007. A putative leucine zipper within the herpes simplex virus type 1 UL6 protein is required for portal ring formation. J. Virol. 81:8868–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang K, Wills E, Baines JD. 2009. The putative leucine zipper of the UL6-encoded portal protein of herpes simplex virus 1 is necessary for interaction with pUL15 and pUL28 and their association with capsids. J. Virol. 83:4557–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, et al. 2007. Structural framework for DNA translocation via the viral portal protein. EMBOJ. 26:1984–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lhuillier S, Gallopin M, Gilquin B, Brasilès S, Lancelot N, et al. 2009. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. PNAS 106:8507–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kornfeind EM, Visalli RJ. 2018. Human herpesvirus portal proteins: structure, function, and antiviral prospects. Rev. Med. Virol. 28:e1972 [DOI] [PubMed] [Google Scholar]
- 101.Padilla-Sanchez V, Gao S, Kim HR, Kihara D, Sun L, et al. 2014. Structure-function analysis of the DNA translocating portal of the bacteriophage T4 packaging machine. J. Mol. Biol. 426:1019–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bayfield OW, Klimuk E, Winkler DC, Hesketh EL, Chechik M, et al. 2019. Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids. PNAS 116:3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Albright BS, Nellissery J, Szczepaniak R, Weller SK. 2011. Disulfide bond formation in the herpes simplex virus 1 UL6 protein is required for portal ring formation and genome encapsidation. J. Virol. 85:8616–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rodríguez-Casado A, Thomas GJ. 2003. Structural roles of subunit cysteines in the folding and assembly of the DNA packaging machine (portal) of bacteriophage P22. Biochemistry 42:3437–45 [DOI] [PubMed] [Google Scholar]
- 105.Wu W, Leavitt JC, Cheng N, Gilcrease EB, Motwani T, et al. 2016. Localization of the houdinisome (ejection proteins) inside the bacteriophage P22 virion by bubblegram imaging. mBio 7:e01152–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, et al. 1992. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J. Mol. Biol. 224:1055–74 [DOI] [PubMed] [Google Scholar]
- 107.Bedwell GJ, Prevelige PE. 2017. Targeted mutagenesis of the P22 portal protein reveals the mechanism of signal transmission during DNA packaging. Virology 505:127–38 [DOI] [PubMed] [Google Scholar]
- 108.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, et al. 2006. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312:1791–95 [DOI] [PubMed] [Google Scholar]
- 109.Zheng H, Olia AS, Gonen M, Andrews S, Cingolani G, Gonen T. 2008. A conformational switch in bacteriophage P22 portal protein primes genome injection. Mol. Cell 29:376–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liang L, Zhao H, An B, Tang L. 2017. High-resolution structure of podovirus tail adaptor suggests repositioning of an octad motif that mediates the sequential tail assembly. PNAS 115:313–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tang L, Marion WR, Cingolani G, Prevelige PE, Johnson JE. 2005. Three-dimensional structure ofthe bacteriophage P22 tail machine. EMBO J. 24:2087–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Olia AS, Bhardwaj A, Joss L, Casjens S, Cingolani G. 2007. Role of gene 10 protein in the hierarchical assembly of the bacteriophage P22 portal vertex structure. Biochemistry 46:8776–84 [DOI] [PubMed] [Google Scholar]
- 113.Dixit AB, Ray K, Black LW. 2012. Compression of the DNA substrate by a viral packaging motor is supported by removal ofintercalating dye during translocation. PNAS 109:20419–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ordyan M, Alam I,Mahalingam M, Rao VB, Smith DE. 2018. Nucleotide-dependent DNA gripping and an end-clamp mechanism regulate the bacteriophage T4 viral packaging motor. Nat. Commun. 9:5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berndsen ZT, Keller N, Smith DE. 2015. Continuous allosteric regulation of a viral packaging motor by a sensor that detects the density and conformation ofpackaged DNA. Biophys. J. 108:315–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Geng J, Fang H, Haque F, Zhang L, Guo P. 2011. Three reversible and controllable discrete steps of channel gating of a viral DNA packaging motor. Biomaterials 32:8234–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cuervo A, Vaney MC, Antson AA, Tavares P, Oliveira L. 2007. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J. Biol. Chem. 282:18907–13 [DOI] [PubMed] [Google Scholar]
- 118.Agirrezabala X, Martín-Benito J, Castón JR, Miranda R, Valpuesta JM, Carrascosa JL. 2005. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 24:3820–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Johnson JE, Chiu W 2007. DNA packaging and delivery machines in tailed bacteriophages. Curr. Opin. Struct. Biol. 17:237–43 [DOI] [PubMed] [Google Scholar]
- 120.McNulty R, Cardone G, Gilcrease EB, Baker TS, Casjens SR, Johnson JE. 2018. Cryo-EM elucidation of the structure of bacteriophage P22 virions after genome release. Biophys. J. 114:1295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Newcomb WW, Booy FP, Brown JC. 2007. Uncoating the herpes simplex virus genome. J. Mol. Biol. 370:633–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van Zeijl M, Fairhurst J, Jones TR, Vernon SK, Morin J, et al. 2000. Novel class ofthiourea compounds that inhibit herpes simplex virus type 1 DNA cleavage and encapsidation: resistance maps to the UL6 gene. J. Virol. 74:9054–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Newcomb WW, Brown JC. 2002. Inhibition of herpes simplex virus replication by WAY-150138: assembly of capsids depleted of the portal and terminase proteins involved in DNA encapsidation. J. Virol. 76:10084–88 [DOI] [PMC free article] [PubMed] [Google Scholar]