Abstract
Attention-deficit/hyperactivity disorder (ADHD) is a common and often persistent neurodevelopmental disorder. Beyond gene-finding, neurobiological parameters, such as brain structure, connectivity, and function, have been used to link genetic variation to ADHD symptomatology. We performed a systematic review of brain imaging genetics studies involving 62 ADHD candidate genes in childhood and adult ADHD cohorts. Fifty-one eligible research articles described studies of 13 ADHD candidate genes. Almost exclusively, single genetic variants were studied, mostly focussing on dopamine-related genes. While promising results have been reported, imaging genetics studies are thus far hampered by methodological differences in study design and analysis methodology, as well as limited sample sizes. Beyond reviewing imaging genetics studies, we also discuss the need for complementary approaches at multiple levels of biological complexity and emphasize the importance of combining and integrating findings across levels for a better understanding of biological pathways from gene to disease. These may include multi-modal imaging genetics studies, bioinformatic analyses, and functional analyses of cell and animal models.
Keywords: ADHD, brain imaging genetics, endophenotype, candidate genes, animal models
1. INTRODUCTION
Attention-deficit/hyperactivity disorder (ADHD) is a common neurodevelopmental disorder (Faraone et al., 2015). The world-wide prevalence has been estimated at 5% in children and between 2.5 and 4.9% in adults (Polanczyk and Rohde, 2007; Simon et al., 2009). Approximately 55–75% still carry the diagnosis in adulthood or remit only partially displaying several impairments also in adulthood (Faraone et al., 2006). ADHD is characterized by age-inappropriate levels of inattention and/or hyperactivity and impulsivity (Frances, 2000), but the clinical phenotype is heterogeneous (Frances, 2000; American Psychiatric Association, 2013). Severity level and presentation of ADHD can change during a person’s lifetime, with adult patients displaying less obvious symptoms of hyperactivity and impulsivity (Buitelaar et al., 2011; Haavik et al., 2010). The phenotypic heterogeneity of the disorder is apparent from a large diversity of psychiatric co-morbidities, frequently seen both in children (Biederman and Faraone, 2005; Gillberg et al., 2004; Lycett et al., 2015; Rappley, 2005; Reinhardt and Reinhardt, 2013) and in adults (McGough et al., 2005; Miller et al., 2007; Ollendick et al., 2008; Sobanski et al., 2007; Wilens et al., 2009).
1.1. Identification of ADHD candidate genes
The etiology of ADHD is strongly influenced by genetic factors, as demonstrated by twin and adoption studies (Faraone and Mick, 2010; Kotte et al., 2013; Thapar et al., 2013; Burt, 2009). Heritability estimates range between 70 and 90% (Faraone and Mick, 2010; Larsson et al., 2013b). Despite this substantial heritability, identification of ADHD risk genes has been challenging (Franke et al., 2009; Gizer et al., 2009). One reason for this may be that ADHD has a complex, polygenic genetic background, in which multiple genetic variants (many of them with small effects) contribute to the etiology of the disorder in most patients. Although a substantial fraction of ADHD etiology is due to genes, many environmental risk factors and potential gene-environment interactions are also linked with an increased risk for the disorder (Banerjee et al., 2007; Han et al., 2015). Furthermore, it has been shown that persistent ADHD and its paediatric form are genetically linked, but overlap only partially (Chang et al., 2013).
Multiple molecular genetic studies, employing mostly hypothesis-driven and some hypothesis-free approaches, have been used to identify ADHD risk genes. Because of the high prevalence of ADHD in the population, the search for genetic factors has mainly focused on common genetic variants, which generally have small effect sizes (Li et al., 2014; Neale et al., 2010b).
While many of the individual hypothesis-driven, candidate gene-based association studies have been underpowered, meta-analysis of those studies identified significant associations for common genetic variants in several candidate genes (Gizer et al., 2009; Faraone et al., 2005; Li et al., 2006). Those are the dopamine and serotonin transporter encoding genes, SLC6A3/DAT1 and SLC6A4/5HTT, genes coding for the D4 and D5 dopamine receptors, DRD4 and DRD5, a serotonin receptor, HTR1B, and the gene for the synaptosomal-associated protein 25, SNAP25. Some additional genes (encoding dopamine beta-hydroxylase [DBH], adrenoceptor alpha 2A [ADRA2A], tryptophan hydroxylase 2 [TPH2], and monoamine oxidase A [MAOA]) were found suggestively associated with ADHD in meta-analyses (Gizer et al., 2009; Li et al., 2006; Faraone et al., 2005). In addition, an in depth analysis of 51 genes in a European multisite sample of 674 families with ADHD combined type probands, collected for the International Multisite ADHD Gene project (the IMAGE project), identified associations with ADHD candidate genes, such as ADRAB2, DAT1, DRD4, TPH2, and MAOA (Brookes et al., 2006). For more extensive reviews of ADHD candidate genes see references (Li et al., 2014; Faraone and Mick, 2010; Banaschewski et al., 2010; Franke et al., 2012; Hawi et al., 2015).
Genetic linkage studies provided a first possibility to perform hypothesis-free genetic studies in the early 2000s. However, gene identification through linkage analysis has been limited (Banaschewski et al., 2010). A meta-analysis of seven linkage studies revealed a locus on the short arm of chromosome 16 to be relevant for ADHD etiology (Zhou et al., 2008). An interesting candidate gene in the locus is cadherin 13 (CDH13), a gene also found in the top-ranks of several genome-wide association studies (Rivero et al., 2015). Linkage analysis also identified the latrophilin 3 (LPHN3) gene on chromosome 4, which was subsequently confirmed through association testing (Arcos-Burgos et al., 2010; Ribases et al., 2011). Genome-wide association studies (GWAS) of common single nucleotide polymorphisms (SNPs) have been the main hypothesis-free approach to studying the genetics of ADHD during the last ten years. However, with nine GWASs on ADHD and ADHD-symptoms published to date (Hinney et al., 2011; Lasky-Su et al., 2008; Lesch et al., 2008; Mick et al., 2010; Neale et al., 2008; Neale et al., 2010a; Stergiakouli et al., 2012; Yang et al., 2013; Sanchez-Mora et al., 2014), no locus has yet been identified that meets genome-wide levels of significance (Li et al., 2014; Neale et al., 2010b), nor has meta-analysis of these studies provided one (Van Hulzen et al., 2016). An interesting genome-wide approach to identify ADHD genes has been the analysis of overlap with other psychiatric disorders. A cross-disorder GWAS across five main neuropsychiatric disorders (schizophrenia [SCZ], bipolar disorder [BD], autism spectrum disorder [ASD], major depressive disorder [MDD], and ADHD) identified five genome-wide significant findings, four of which - in/near the genes ITIH3, AS3MT, CACNA1C, and CACNB2 - were shared with ADHD (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013).
Following promising results in other psychiatric disorders (Purcell et al., 2014; Cukier et al., 2014; Cruceanu et al., 2013; Kerner et al., 2013), studies of rare variants were also performed for ADHD and were successful in identifying genetic variants related to the disorder (Elia et al., 2010; Williams et al., 2010; Williams et al., 2012; Ramos-Quiroga et al., 2014; Lesch et al., 2011; Yang et al., 2013). Genome-wide analysis of (rare) copy number variants (CNVs) showed an enrichment of rare CNVs in patients with ADHD (Williams et al., 2010), and implicated the genes CHRNA7 and NPY in ADHD etiology (Williams et al., 2012; Lesch et al., 2011), as well as genes encoding several glutamate receptors (Akutagava-Martins et al., 2014; Elia et al., 2012), and regions on 15q11–15q13 (Valbonesi et al., 2015) and 16p13.11 (Williams et al., 2010). The picture emerging from those initial studies is that the rare variant contribution of ADHD genetics is highly heterogeneous, similar to the common variant contribution. Nevertheless, given the success of CNV studies, exome and whole-genome sequencing are now being used, allowing the identification of rare single nucleotide variants and small insertions/deletions contributing to ADHD etiology. A first study indeed found enrichment of rare variants in a predefined set of 51 candidate genes in adult patients with persistent ADHD (Demontis et al., 2016).
The genetic factors associated with ADHD are distributed across the genome, but tend to be enriched within specific functional categories. By clustering ADHD-related genes within functional networks or pathways, several biological processes have been shown to be involved in the etiology of the disorder. Top-findings of five GWASs in ADHD showed convergence on the biological process of neurite outgrowth (Poelmans et al., 2011). Comparable enrichment analyses revealed that most significantly enriched functions for the ADHD-GWAS association signals were related to nervous system development, neuron projection morphogenesis, oxogenesis, cell-cell communication, glutamatergic synapse/receptor signalling, and multicellular organismal development (Hawi et al., 2015) or neuron projections and synaptic components (Yang et al., 2013), which is consistent with a neurodevelopmental pathophysiology of ADHD. These findings were strengthened by results from a recent study, that used two GWAS datasets to identify pathways associated with ADHD by applying six pathway analysis methods (Mooney et al., 2016). Cross-method convergent results revealed a number of brain-relevant pathways, such as RhoA signaling, glycosaminoglycan biosynthesis, fibroblast growth factor receptor activity, and pathways containing potassium channel genes (Mooney et al., 2016). Another study revealed that CNVs involved in ADHD converge on biologically meaningful gene clusters related to ion channel pathways, organonitrogen compound catabolic processes, and transmembrane transport (Thapar et al., 2015). A combined analysis of ADHD candidate genes, derived both from SNP-based and CNV-based studies, showed that genes involved in biological processes, such as synaptic transmission, catecholamine metabolic processes, G-protein signalling pathways, and cell migration were over-represented among the top-findings of such studies (Cristino et al., 2014). More generally, the genome-wide analysis of five major psychiatric disorders (also including ADHD), supported a role for calcium channel signalling genes for all five disorders, suggesting that genetic variation in calcium-channel activity genes can have pleiotropic effects in the development of psychopathology (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013).
1.2. Brain correlates of ADHD
The effects of ADHD genetic risk factors on (aberrant) behaviour are likely to be mediated through effects on cell biology and brain system development and functioning. Several aspects of brain development, structure, function, and connectivity have been found altered in ADHD (Cortese et al., 2012; Greven et al., 2015; Shaw et al., 2007b; Shaw et al., 2012; van Ewijk et al., 2012; Glahn et al., 2010; Onnink et al., 2015; Mostert et al., 2016; Hoogman et al., submitted).
Two indirect neuroimaging techniques that have been used in imaging (genetics) studies in ADHD are positron emission tomography (PET) and single photon emission computed tomography (SPECT). Both are based on the measurement of a radionuclide’s decay, during which a positron or a γ-ray is emitted, generating photons. The high sensitivity and limitless penetration depth of PET and SPECT enable imaging to examine metabolic activity, cerebral perfusion, neurotransmitter turnover, and receptor binding potentials within examined brain regions or receptor systems (Rahmim and Zaidi, 2008). Until now, the majority of recent PET studies using ADHD samples have focused on examining differences and changes in neurotransmitter binding and receptor density (Zimmer, 2009). In vivo imaging of the dopamine transporter (DAT) is particularly relevant for ADHD, given that DAT is the target of stimulant medications and, subsequently, a target protein for studies of pathophysiology. PET and SPECT studies have been useful in comparing striatal DAT availability between ADHD patients and controls (Jucaite et al., 2005; Spencer et al., 2005; Ludolph et al., 2008). A meta-analysis of nine PET studies revealed that striatal DAT density was 14% higher in patients with ADHD compared to healthy controls (Fusar-Poli et al., 2012). Besides that, striatal density in people with ADHD seems to depend on previous psychostimulant exposure, with lower density in drug-naive subjects and higher density in previously medicated patients (Fusar-Poli et al., 2012). In addition to studies exploring DAT density and binding, Volkow and co-workers examined postsynaptic dopamine receptor availability and found that medication-naïve adults with ADHD showed decreased dopamine D2/D3 receptor availability in the left caudate compared to healthy controls. Following administration of methylphenidate (MPH), the ADHD group demonstrated decreased dopamine activity in the caudate compared with controls (Volkow et al., 2007). One SPECT study investigated D2 receptor availability as a function of MPH therapy in ADHD and concluded that D2 receptor availability is significantly reduced in patients with ADHD in all four regions of the striatum (Ilgin et al., 2001).
Structural magnetic resonance imaging (sMRI) allows to noninvasively characterize the structure of the human brain. With the help of sMRI, the different magnetic properties of brain tissues are used to non-invasively map the spatial distribution of these structural properties of the human brain. Thereby, the different brain tissues (grey and white matter) and cortical and subcortical structures of the brain can be accurately mapped, and different aspects of brain structure can be quantified and compared. In general, sMRI has pointed to total brain volume and total grey matter reductions up to 3–5% in ADHD patients compared to controls (Castellanos et al., 2002; Valera et al., 2007; Greven et al., 2015). To investigate, whether these reductions are global or regional, several brain regions of interest (ROIs) have been studied. A meta-analysis reported significant volume differences in cerebellar regions, total and right cerebral volume, right caudate, and frontal brain areas (Valera et al., 2007). To investigate the most prominent changes in grey matter intensity, detected by using voxel-based morphometry (VBM) analyses, four meta-analyses have been performed to date (Ellison-Wright et al., 2008; Frodl and Skokauskas, 2012; Nakao et al., 2011). Most consistently, grey matter reductions in the ventrolateral prefrontal/insular-striatal regions, such as the right insula, putamen, globus pallidus, and caudate nucleus have been described in ADHD patients (Ellison-Wright et al., 2008; Frodl and Skokauskas, 2012; Nakao et al., 2011; Norman et al., 2016). A recent study could also show that participants with ADHD had significantly smaller grey matter volume in five clusters located in the precentral gyrus, medial and orbital frontal cortex, and (para)cingulate cortices (Bralten et al., 2015). Unaffected siblings of patients with ADHD showed intermediate volumes, significantly different from controls in four of these five clusters (all except the precentral gyrus), suggesting that the volume reductions are unlikely to be a consequence of disease, but may rather contribute to ADHD etiology (Bralten et al., 2015).
Brain differences observed in ADHD have been hypothesized to be partly attributable to a delay in maturational processes (Castellanos et al., 2002). Indeed, the few longitudinal imaging studies of ADHD patients support this hypothesis: for CT measures, Shaw and co-workers investigated growth trajectories of different points of the cortex and reported that cortical thickness maturation in participants with ADHD lagged behind that of healthy controls of approximately three years throughout the cerebrum, but most prominent in the PFC (Shaw et al., 2007a). In addition, also the SA developmental trajectory was found to be delayed in ADHD, especially in the right PFC (Shaw et al., 2012). Support for the developmental delay hypothesis in ADHD also came from cross-sectional meta-analyses of VBM studies, which found increasing age associated with more normal grey matter values in affected brain areas (Frodl and Skokauskas, 2012). The recent large mega-analysis of subcortical regions across 60 years of the lifespan by the Enhancing NeuroImaging Genetics Through Meta Analysis (ENIGMA) ADHD Working Group extended the delayed maturation theory also to the volumes of most subcortical regions (Hoogman et al., submitted). First of all, they observed significant smaller volumes for the nucleus accumbens, amygdala, caudate, hippocampus, putamen, and intracranial volume (ICV) in ADHD cases relative to controls (Hoogman et al., submitted). Age analyses suggested different brain volume trajectories across age for patients and controls. These results from the cross-sectional lifespan analyses were consistent with the early maturation delay hypotheses of ADHD and hint at delays in brain growth and degeneration across the lifespan (Hoogman et al., submitted; Rubia, 2007).
Next to volumetric differences observed in grey matter, white matter structure has also been found to be altered in ADHD, leading to a potential disorganization of the brain’s connectivity. Diffusion tensor imaging (DTI) enables non-invasive investigations of the macrostructural integrity and orientation of white matter fibre bundles. DTI measures the directional diffusion of water molecules along neuronal membranes, allowing to map white matter pathways within the brain. One measure frequently derived from DTI is fractional anisotropy (FA). Anisotropy indicates that diffusion occurs in a directional manner, whereas isotropy indicates diffusion in all directions. Other measures derived from DTI include mean diffusivity (MD), which is an average of axial diffusivity (AD) and the perpendicular diffusivities, and radial diffusivity (RD), which is the average of perpendicular diffusivities, the mode of anisotropy, which is sensitive to crossing fibres, and the apparent diffusion coefficient, which indicates the magnitude of diffusion (Le Bihan, 2003; Le Bihan et al., 2001; Yoncheva et al., 2016). In a meta-analysis comparing DTI findings between patients with ADHD and healthy controls five areas with disturbed microstructural integrity in people with ADHD were highlighted, located in white matter tracts subserving the fronto-striatal-cerebellar neurocircuitry (van Ewijk et al., 2012). Most consistently, studies reported white matter anomalies in the corpus callosum in childhood ADHD (van Ewijk et al., 2014) and adult ADHD (Dramsdahl et al., 2012; Onnink et al., 2015). Although the exact neurobiological meaning is not fully understood, reduced FA in the corpus callosum of adult patients with ADHD was driven by changes in RD rather than AD, suggesting that aberrant myelination is a pathophysiological factor in adult ADHD (Onnink et al., 2015). However, replication from longitudinal studies is still lacking, and the differences between patients and controls seem to be widespread and heterogeneous across studies (van Ewijk et al., 2012).
A method to investigate potential changes in brain activity is functional magnetic resonance imaging (fMRI). FMRI is primarily sensitive to the oxygenation of the blood, the so-called blood-oxygen-level-dependent (BOLD) signal. It measures brain function based on the premise that active cells consume oxygen, thereby causing changes in blood oxygenation and subsequently leading to increased blood flow, although the exact link between cell activation, oxygen saturation, and blood flow is still under debate; for review see (Hillman, 2014). Generally in fMRI, alterations in blood flow after a stimulus (e.g. a certain task) or during a resting state are measured. Comparing anatomical or functional brain measures in individuals with ADHD, their unaffected siblings, and healthy comparison subjects, is one of the best ways to examine the suitability of these neural markers as endophenotypes. With respect to functional brain studies, van Rooij and colleagues (2015c) recently reported a distinction in hemodynamic patters during a stop-signal task between patients with ADHD, their unaffected siblings, and control subjects, suggesting the familial nature of these activation patterns. Thus, inhibition-related neural activation could be considered as a valuable endophenotype for ADHD. Several reviews have provided excellent overviews of cognitive and brain (candidate) endophenotypes for ADHD (del Campo et al., 2012; Gallo and Posner, 2016; Rommelse et al., 2011). In accordance with those reports, dysregulation of structure and function of the fronto-subcortical-cerebellar pathways that control attention, response to reward, salience thresholds, inhibitory control, and motor behaviour are among the most promising endophenotype candidates, and task-based functional MRI studies in ADHD have largely focused on these neurocognitive domains. More specifically, fMRI studies using inhibitory control, working memory, and attentional tasks in patients with ADHD and healthy comparison subjects have shown underactivation of fronto-striatal, frontoparietal, and ventral attention networks in the patients (Cortese et al., 2012). The fronto-parietal network mediates goal-directed executive processes, whereas the ventral attention network facilitates reorientation of attention towards salient and behaviourally relevant external stimuli. Meta-analyses of fMRI studies of inhibition and attention revealed that patients with ADHD have consistent functional abnormalities in two distinct domain-dissociated fronto-basal ganglia networks. These include the inferior frontal cortex, supplementary motor areas, and anterior cingulate cortex (ACC) for inhibition and the dorsolateral prefrontal cortex (PFC), parietal, and cerebellar areas for attention processes (Hart et al., 2013). Studies using reward-processing paradigms reported reduced activation of the ventral striatum of participants with ADHD in the anticipation phase of reward relative to controls (Plichta and Scheres, 2014), and differences between patients and controls have also been observed during reward receipt (von Rhein et al., 2015). Additionally, a meta-analysis of fMRI studies of timing reported consistent reductions in activation in typical areas of timing, such as the left inferior frontal gyrus (IFG)/insula, cerebellum, and the left parietal lobe in ADHD patients relative to controls (Hart et al., 2012).
In resting state MRI (rs-fMRI), the temporal correlations in neural activity across anatomically disparate brain regions are analysed to examine functional connectivity based on spontaneous brain activity, neural organization, and circuit architecture. Rs-fMRI studies of ADHD have mainly focused on neural circuits implicated in the disorder, especially the default-mode network (DMN), cognitive control network, and cortico-striato-thalamo-cortical loops (Posner et al., 2014). It was shown that ADHD is associated with less-pronounced or absent anti-correlations between the DMN and the cognitive control network, lower connectivity within the DMN itself, and lower connectivity within the cognitive and motivational loops of fronto-striatal circuits (Posner et al., 2014). A recent study in a large sample of adult participants with ADHD and healthy controls, showed that functional connectivity in the executive control network, and to a lesser extent also the cerebellum network, was stronger in the ADHD group (Mostert et al., 2016). Additionally, hyperactivity/impulsivity symptoms were positively correlated with the connectivity strength in these networks (Mostert et al., 2016).
Functional near-infrared spectroscopy (fNIRS) measures concentration changes of oxygenated, deoxygenated, and total haemoglobin in brain haemodynamics by measuring the absorption of near-infrared light projected through the scalp (Gervain et al., 2011). Thereby, fNIRS provides an indirect measure of neural activity based on changes in blood oxygenation due to metabolic processes within the cortex. Compared to fMRI, fNIRS is less sensitive to movement artefacts, and since the emitters and detectors can be worn in a cap, functional neural activity can be studied, while the participant is interacting with its environment. This makes fNIRS an ideal tool to study brain development, e.g. in children with ADHD (Vanderwert and Nelson, 2014). FNIRS has greater spatial resolution compared to event-related potential (ERP) or EEG techniques, however, since it is dependent on light penetration and reflection, fNIRS can only examine the cortical surface within 2–3 cm of the scalp (Vanderwert and Nelson, 2014). The majority of fNIRS studies on ADHD investigated children with the disorder. These studies particularly focused on alterations in PFC activity during different experimental paradigms, such as Stroop tasks (Negoro et al., 2010; Xiao et al., 2012), working memory tasks (Schecklmann et al., 2010), the Trail Making Test (Weber et al., 2005), or Go/NoGo paradigms (Xiao et al., 2012; Inoue et al., 2012); they consistently pointed towards an attenuated oxygen metabolism within the frontal lobe (Ehlis et al., 2014). Studies in adult ADHD patients suggest that this hypofunctionality is persistently observed throughout development (Schecklmann et al., 2013; Ehlis et al., 2008).
The functional brain imaging techniques electroencephalography (EEG) and magnetoencephalography (MEG) have also been used for the study of ADHD (genetics). EEG directly measures electrical activity from large populations of cells and therefore offers a very good temporal resolution, far superior to fMRI. However, it has a poor spatial resolution, as electric fields smear as they pass through the skull (Ahmad et al., 2016). Every electric field also has a magnetic field, which can be detected by MEG. The spatial resolution of MEG is slightly better compared to EEG, but MEG only measures information strictly from the sulci, thus it is more limited and misses information (van Diessen et al., 2015). The frequency bands mostly studied in ADHD are theta (θ), alpha (α), and beta (β), either individually, or compared to each other (such as theta/beta power or amplitude ratio). In a resting state, (lower frequency) θ band activity can reflect drowsiness or “cortical slowing”. The α band activity is usually observed during eyes closed conditions at rest, particularly in posterior brain regions, and it is negatively associated with central nervous system arousal. In contrast, β band activity generally accompanies mental activity and concentration. The θ/β power ratio has been proposed to capture the relative contributions of two relevant frequency bands for ADHD; however, the true functional significance of this measure remains unknown (Loo and Makeig, 2012). It has been reported that patients with ADHD exhibit increased fronto-central theta (θ) band activity and increased theta-to-beta (θ/β) power ratio during rest compared to non-ADHD controls (Loo and Makeig, 2012). While (limited) discriminant validity of these EEG measures for ADHD has been suggested, significant EEG heterogeneity also exists across ADHD-diagnosed individuals (Clarke et al., 2011). In addition to differences in frequency bands, event-related potential (ERP) studies explored various aspects of brain functioning in ADHD and identified a substantial number of ERP correlates of ADHD (Johnstone et al., 2013). Robust differences between ADHD patients and healthy controls have been reported in several components related to attention (among others including orienting and vigilance), inhibitory control, and performance monitoring, such as error and reward/punishment processes (Johnstone et al., 2013). MEG studies comparing ADHD patients to healthy controls are scarce and have been geared towards investigating attention-related processes. Alterations in oscillation patterns of brain regions involved in such processes have been observed in patients (ter Huurne et al., 2013; Franzen et al., 2013; Heinrichs-Graham et al., 2014).
Importantly, the brain phenotypes found affected in people with ADHD are often moderately to highly heritable. Findings from twin studies showed that brain structure is under strong genetic control. Additionally, twin studies showed that genetic effects varied regionally within the brain, with high heritability estimates (h2) for frontal lobe volumes ranging from 0.9 to 0.95, for region-based cortical surface areas ranging from 0.48 to 0.77, and moderate estimates for e.g. the hippocampus (h2-range = 0.4 – 0.69) (Peper et al., 2007). Surface area was predominantly more heritable than cortical thickness (h2-range = 0.34 – 0.64) (McKay et al., 2014). Global fractional anisotropy (h2 = 0.55) as well as radial diffusivity (h2 = 0.72) of white matter showed high heritability (Kochunov et al., 2015; McKay et al., 2014). Moreover, basal neural activity during a resting state condition has also been shown to be under genetic control, as functional connectivity within the default-mode network as a whole was significantly heritable (h2 = 0.42) (Glahn et al., 2010). Additional examples for moderate heritabilities of neural activity are e.g. brain activation in the cerebellum and cerebral cortex during working memory tasks (h2-range = 0.5 – 0.65) (Blokland et al., 2014). Strong genetic determination has also been reported for different psychophysiological brain phenotypes measured by EEG, e.g. (Iacono et al., 2014; Smit et al., 2010), and MEG, where different frequency band heritabilities have been described by van Pelt and coworkers (2012).
1.3. Rationale for this review
This review aims to provide a systematic overview of brain imaging genetics studies in ADHD, as brain imaging phenotypes are frequently used as endophenotypes in ADHD research. Endophenotypes (or intermediate phenotypes) have been considered a promising strategy in order to gain more insight into the mechanisms leading from a genetic/biological basis of the disease to the full clinical phenotype (Faraone et al., 2014a). Endophenotypes are (1) those characteristics of a disorder that are linked more closely to its neurobiological substrates than its clinical symptoms (Doyle et al., 2005) and (2) share genetic susceptibility factors with the disorder itself (Gottesman and Gould, 2003). As described above, neuroimaging phenotypes, e.g. derived from sMRI (Hulshoff Pol et al., 2006) and DTI measurements (Jahanshad et al., 2013) are highly heritable. Those brain phenotypes altered in ADHD have therefore been considered key endophenotypes for the disorder, and investigating the genetic influences on these brain measures has been offered as a way for capturing underlying liability for ADHD (Dresler et al., 2014; Durston, 2010; Wu et al., 2014). Compared to existing reviews of brain imaging genetics studies in ADHD (Durston, 2003; Durston, 2010; Durston et al., 2009; Wu et al., 2014; Dresler et al., 2014), this review is more comprehensive by including both childhood and adult ADHD studies, a large spectrum of brain imaging modalities, and by investigating a more complete list of ADHD candidate genes. Beyond the systematic review, we also emphasize the need for additional approaches, describing complementary methods, which provide insight from alternative angles into the biological pathways leading from an ADHD risk gene to disease. Especially, we argue that the integration of methods at different analytical levels (e.g. in silico, cell, brain, cognition, and behaviour) is needed to unravel the function of ADHD candidate genes.
2. METHODS
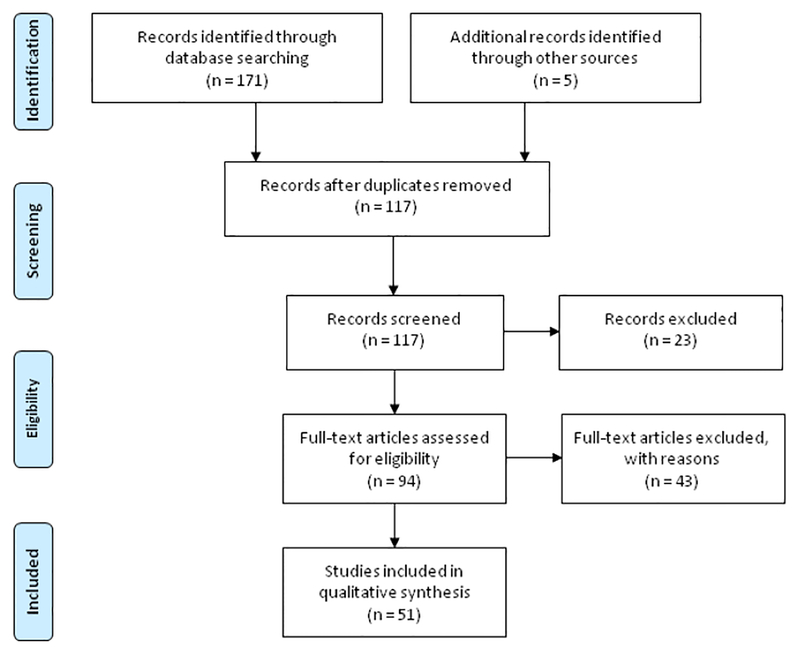
For this review, we selected genes that were previously found associated with ADHD. The selection was based on a recent review of ADHD candidate genes, which described 70 genes that are (with at least some evidence) related to ADHD risk (Li et al., 2014), see Table 1. We discarded eight genes, for which we did not find evidence for association with ADHD based on the analysis of genetic variation: ARVCF, ATP2C2, CPLX4, DNM1, EMP2, IL20RA, MMP7, and TRIO (Table 1). On November 28th 2016, we searched for all remaining 62 genes, all brain imaging modalities, and ADHD using PubMed (www.ncbi.nlm.nih.gov/pubmed) with the following search algorithm (example is shown for the SLC6A3/DAT1 gene): (((SLC6A3 OR solute carrier family 6 neurotransmitter transporter, member 3 protein human OR DAT1 OR dopamine transporter gene OR dopamine transporter [All fields])) AND (gene* OR genetic* OR imaging genetic OR imaging genetics OR genotype OR polymorphism OR SNP OR single nucleotide polymorphism)) AND (structural magnetic resonance imaging OR functional magnetic resonance imaging OR sMRI OR fMRI OR electroencephalography OR diffusion tensor imaging OR DTI OR resting-state functional magnetic resonance imaging OR rsfMRI OR EEG OR magnetoencephalography OR MEG OR single photon emission computed tomography OR SPECT OR positron emission tomography OR PET OR near-infrared spectroscopy OR NIRS OR volume [Title/Abstract])) AND (ADHD OR Attention-deficit/hyperactivity disorder OR [All fields] NOT “review” [Publication Type]). Titles and abstracts of the retrieved records were evaluated for relevant publications. Studies were required to investigate genetic variants in/near the selected ADHD candidate genes, and only studies including patients with ADHD were included. Review articles, medical hypotheses, non-English articles, and studies on animal models were not considered. The preferred reporting items for systematic reviews and meta-analysis (PRISMA) diagram in Figure 1 describes the number of articles identified and their classification.
Table 1:
Candidate genes containing common variants associated with ADHD, adapted from (Li et al., 2014), which served to select genes for this review.
| Gene | Protein | Chr position | References for genes associated with ADHD |
|---|---|---|---|
| ADRA1B | Adrenoceptor alpha1B | 5q33.3 | (Segurado et al., 2011)a; (Hawi et al., 2013)a |
| ADRA2A* | Adrenoceptor alpha 2A | 10q25.2 | (Roman et al., 2003)a; (Shiffrin et al., 2013)b |
| ADRA2C | Adrenoceptor alpha 2C | 4p16.3 | (De Luca et al., 2004)a; (Cho et al., 2008) |
| ADRB1 | Adrenoceptor beta 1 | 10q25.3 | (Pascoli et al., 2005)d |
| ADRB2 | Adrenoceptor beta 2, surface | 5q31-q32 | (Lasky-Su et al., 2008a)a, c; (Brookes et al., 2006a) |
| ASTN2 | Astrotactin2 | 9q33 | (Lesch et al., 2008)a; (Lionel et al., 2011); (Lionel et al., 2014) |
| BCHE | Butyrylcholinesterase | 3q26.1-q26.2 | (Lesch et al., 2011)a; (Jacob et al., 2013) |
| BDNF | Brain-derived neurotrophic factor | 11p14.1 | (Friedel et al., 2005)a; (Lee and Song, 2015)b |
| CALY | Calcyon neuron-specific vesicular protein | 10q26.3 | (Plaisancie et al., 2014)a |
| CCSER1/FAM190A | Coiled-coil serine-rich protein 1 | 4q22.1 | (Lantieri et al., 2010)a,c |
| CDH13 | Cadherin 13 | 16q23.3 | (Lesch et al., 2008)a; (Neale et al., 2010)c |
| CHRNA3 | Cholinergic Receptor, Nicotinic, Alpha 3 | 15q25.1 | (Polina et al., 2014)a |
| CHRNA4 | Cholinergic Receptor, Nicotinic, Alpha 4 | 20q13.33 | (Guan et al., 2009)a; (Wallis et al., 2009) |
| CHRNA7 | Cholinergic Receptor, Nicotinic, Alpha 7 | 5q13.3 spanning CHRNA7 | (Williams et al., 2012)a |
| CNTF | Ciliary neurotrophic factor | 11q12 | (Ribases et al., 2008)a; (Tzang et al., 2014) |
| COMT* | Catechol-O-methyltransferase | 22q11.21 | (Eisenberg et al., 1999)a; (Lee and Song, 2015) |
| CPLX2 | Complexin 2 | 5q35.2 | (Lionel et al., 2011)a |
| DBH | Dopamine beta-hydroxylase (dopamine beta-mono oxygenase) | 9q34 | (Daly et al., 1999)a; (Gizer et al., 2009)b |
| DDC | Dopa decarboxylase (aromatic L-amino acid decarboxylase) | 7p12.1 | (Ribases et al., 2009)a; (Lasky-Su et al., 2008b)c |
| DIRAS2 | DIRAS family, GTP-binding RAS-like 2 | 9q22.32 | (Reif et al., 2011)a |
| DRD1* | Dopamine receptor D1 | 5q34-q35 | (Misener et al., 2004)a; (Ribases et al., 2012) |
| DRD2/ ANNK1 | Dopamine receptor D2/ Ankyrin repeat and kinase domain containing 1 | 11q22-q23 | (Comings et al., 1991)a; (Pan et al., 2015)b |
| DRD3 | Dopamine receptor D3 | 3q13.3 | (Guan et al., 2009)a; (Wu et al., 2012)b |
| DRD4* | Dopamine receptor D4 | 11p15 11p15 |
(LaHoste et al., 1996)a; (Gizer et al., 2009)b; (Wu et al., 2012)b (Barr et al., 2001)a; (Yang et al., 2008)d; (Gizer et al., 2009)b |
| DRD5 | Dopamine receptor D5 | 4p16.1 | (Daly et al., 1999)a; (Gizer et al., 2009)b; (Wu et al., 2012)b |
| FADS2 | Fatty acid desaturase 2 | 11q12.2 | (Brookes et al., 2006b)a |
| FTO | Fat mass and obesity associated | 16q12.2 | (Choudhry et al., 2013)a |
| GDNF | Glial cell derived neurotrophic factor | 5p13.1-p12 | (Simchon-Tenenbaum et al., 2015)d; (Shim et al., 2015) |
| GPRC5B | G protein-coupled receptor, class C, group 5, member B | 16p12 | (Albayrak et al., 2013)a |
| GRIN2A | Glutamate receptor, ionotropic, N-methyl D-aspartate 2A | 16p13.2 | (Turic et al., 2004)a |
| GRM5 | Glutamate receptor, metabotropic 5 | 11q14.3 | (Elia et al., 2010)a; (Hinney et al., 2011)c; (Elia et al., 2012) |
| GRM7 | Glutamate receptor, metabotropic 7 | 3p26-p25 | (Mick et al., 2008)a,c; (Elia et al., 2012); (Park et al., 2014) |
| HES1 | Hes family bHLH transcription factor 1 | 3q28-q29 | (Brookes et al., 2006a)a; (Lasky-Su et al., 2008b)c |
| HTR1A | 5-Hydroxytryptamine (serotonin) receptor 1A, G protein-coupled | 5q11.2-q13 | (Shim et al., 2010)a; (Zuo et al., 2015)d |
| HTR1B | 5-Hydroxytryptamine (serotonin) receptor 1B, G protein-coupled | 6q13 | (Hawi et al., 2002)a; (Gizer et al., 2009)b |
| HTR1E | 5-Hydroxytryptamine (serotonin) receptor 1E, G protein-coupled | 6q14-q15 | (Lasky-Su et al., 2008b)a,c |
| HTR2A | 5-Hydroxytryptamine (serotonin) receptor 2A, G protein-coupled | 13q14-q21 | (Quist et al., 2000)a; (Lasky-Su et al., 2008a)c; (Lasky-Su et al., 2008b)c |
| HTR2C | 5-Hydroxytryptamine (serotonin) receptor 2C, G protein-coupled | Xq23 | (Li et al., 2006)a; (Xu et al., 2009) |
| HTR3A | 5-Hydroxytryptamine (serotonin) receptor 3A, G protein-coupled | 11q23.1-q23.2 | (Hu et al., 2009)d |
| HTR3B | 5-Hydroxytryptamine (serotonin) receptor 3B, G protein-coupled | 11q23.1 | (Oades et al., 2008)a |
| LPHN3* | Latrophilin 3 | 4q13.1 | (Arcos-Burgos et al., 2010)a; (Hwang et al., 2015)d; (Ribases et al., 2011)d; (Labbe et al., 2012)a |
| MAOA* | Monoamine oxidase A | Xp11.4-p11.3 | (Payton et al., 2001)a; (Liu et al., 2011)d |
| MAOB | Monoamine oxidase B | Xp11.4-p11.3 | (Li et al., 2008)a; (Ribases et al., 2009)d |
| NOS1* | Nitric oxide synthase 1 | 12q24.22 | (Reif et al., 2009)a; (Franke et al., 2009)c; (Weber et al., 2015)b |
| PNMT | Phenylethanolamine N-methyltransferase |
17q12 | (Brookes et al., 2006a)a |
| PRKG1 | Protein kinase, cGMP-dependent, type I | 10q11.2 | (Neale et al., 2010)a |
| SLC1A3 | Solute Carrier Family 1 (Glial High Affinity Glutamate Transporter), Member 3 | 5p13 | (Turic et al., 2005)a; (Elia et al., 2009) |
| SLC6A2/NET1* | Solute Carrier Family 6 (Neurotransmitter Transporter), Member 2 | 16q12.2 | (Bobb et al., 2005)a; (Hohmann et al., 2015) |
| SLC6A3/ DAT1* | Solute Carrier Family 6 (Neurotransmitter Transporter), Member 3; Dopamine transporter 1 | 5p15.3 | (Cook et al., 1995)a; (Gizer et al., 2009)b (Galili-Weisstub and Segman, 2003)a; (Gizer et al., 2009)b (Brookes et al., 2006c)a; (Gizer et al., 2009)b |
| SLC6A4/ 5HTT* | Solute Carrier Family 6 (Neurotransmitter Transporter), Member 4; serotonin transporter | 17q11.2 | (Manor et al., 2001)a; (Gizer et al., 2009)b; (Landaas et al., 2010)b |
| SLC9A9/NHE9 | Solute Carrier Family 9, Subfamily A, Member 9 | 3q24 | (de Silva et al., 2003)a; (Stergiakouli et al., 2012)c; (Mick et al., 2010)c |
| SLC18A2/VMAT2 | Solute Carrier Family 18 (Vesicular Monoamine Transporter), Member2 | 10q25 | (Toren et al., 2005)a |
| SNAP25* | Synaptosomal-associated protein, 25kDa | 20p12-p11.2 | (Brophy et al., 2002)a; (Gizer et al., 2009)b |
| SPOCK3 | Sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 | 4q32.3 | (Weber et al., 2014)a; (Gao et al., 2015) |
| STX1A | Syntaxin1A | 7q11.2 | (Sanchez-Mora et al., 2013)d |
| SYP | Synaptophysin | Xp11.23-p11.22 | (Brookes et al., 2006a)a; (Liu et al., 2013)d |
| SYT1 | Synaptotagmin I | 12q21.2 | (Guan et al., 2009)a; (Lasky-Su et al., 2008a)c |
| TCERG1L | Transcription elongation regulator 1-like | 10q26.3 | (Neale et al., 2010)a,c |
| TH | Tyrosine hydroxylase | 11p15.5 | (Segurado et al., 2011)a |
| TPH1 | Tryptophan hydroxylase 1 | 11p15.3-p14 | (Gizer et al., 2009)b |
| TPH2* | Tryptophan hydroxylase 2 | 12q15 | (Walitza et al., 2005)a; (Sheehan et al., 2005)a; (Gizer et al., 2009)b |
| VAMP2 | Vesicle-associated membrane protein 2 (synaptobrevin 2) | 17p13.1 | (Gao et al., 2015)a |
Bold text indicates significant result at P < 0.05 in Gizer et al., 2009.
Association first reported by.
Meta-analysis article.
GWAS finding.
Association in large sample or validation using animal model.
Gene with at least one case-control imaging genetics study; ADHD = Attention-deficit/hyperactivity disorder, chr = chromosome.
Figure 1.
Preferred Reporting Items for Systematic reviews and Meta-Analysis (PRISMA) flowchart of the literature search and study selection for qualitative analysis. Note: see http://www.prismastatement.org for more information in this reporting system.
3. RESULTS
For 25 out of the 62 ADHD candidate genes, we retrieved a total number of 171 reports linking genetic variation in/near the gene to neuroimaging by using the above mentioned search term in PubMed (Figure 1). To those, we added two recent papers from our own group (Sokolova et al., 2015; Van Ewijk et al., in revision) and three additional papers that were missing, from reading the retrieved reports (Dresler et al., 2010; Albrecht et al., 2014; Fernandez-Jaen et al., 2016). After removal of 54 duplicates, we screened 117 records and discarded an additional 66 papers, mostly because they described animal studies, did not include ADHD patients, or did not fulfil our eligibility criteria otherwise. We finally included 51 original research articles on brain imaging genetics studies for 13 ADHD candidate genes (ADRA2A, COMT, DRD1, DRD4, HTR1B, LPHN3, MAOA, NOS1, SLC6A2/NET1, SLC6A3/DAT1, SLC6A4/5HTT, SNAP25, TPH2; Table 2 and 3). Most of the studies investigated a single gene (32 in Caucasians, 6 in Asians), thirteen studies investigated multiple genes (12 in Caucasians, 1 in Asians). The dopamine transporter gene (SLC6A3/DAT1) and the dopamine D4 receptor gene (DRD4) were the most frequently studied ADHD candidate genes (Table 2 and 3).
Table 2:
Overview of imaging genetics studies identified per gene.
Papers were added manually (two recent papers from our own group (1× NOS1, 1× SLC6A3/DAT1) and three additional papers that were missing from the PubMed search (3× SLC6A3/DAT1).
This number still includes duplicate articles, since some studies investigated more than one candidate gene.
No records were retrieved for the following genes: ADRA1B, ADRA2C, ADRB1, ADRB2, ASTN2, CALY, CCSER1, CDH13, CHRNA3, CNTF, CPLX2, DDC, DIRAS2, FADS2, FTO, GDNF, GPRC5B, GRIN2A, GRM7, HES1, HTR1A, HTR1E, HTR2A, HTR2C, HTR3A, HTR3B, PNMT, PRKG1, SLC1A3, SLC18A2, SLC9A9, SPOCK3, STX1A, SYT1, TCERG1L, TPH1, VAMP2.
Table 3:
Imaging genetics studies of ADHD candidate genes in ADHD (case-control) samples (for candidate gene list see Table 1).
| Gene (s) | Variant | Imaging modality |
Imaging/cognitive phenotype or task |
Genotype groups compared |
Samples size (mean age in years) |
Primary results (main effect of genotype) |
Reference |
|---|---|---|---|---|---|---|---|
| ADRA2A | rs1800544, rs553668 | DTI | White matter integrity, FA values | C-allele carriers vs. GG-carriers, T-allele carriers vs. CC-carriers | 53 ADHD (9.1)§ | rs1800544 C-allele carriers: ↓ FA in right postcentral gyrus. rs553668 T-allele carriers: ↓FA in right middle frontal cortex. |
(Park et al., 2013) |
| rs1800544 | SPECT with 99mTc-HMPAO | Regional brain perfusion | C-allele carriers vs. GG-carriers | 21 ADHD (9.9)‡ § | C-allele carriers: ↓ perfusion in bilateral orbitofrontal regions. | (Kim et al., 2010) | |
| COMT | rs4680 | sMRI (VBM) | Striatum, cerebellum, temporal lobe and IFG volume | Met-carriers vs. Val/Val-carriers | 38 ADHD (10.3) 24 HC (10.1) |
ADHD Met-carriers: ↓ GM volume in IFG (whole brain level) compared to HC. ADHD Val/Val: ↑ GM volume in right CN (ROI analysis) compared to ADHD Met-carriers and HC. |
(Villemonteix et al., 2015) |
| GM volume | Met-carriers vs. Val/Val-carriers | 17 ADHD (10.3)§ 15 HC (12.8)§ |
ADHD Met-carriers: ↓ GM volume in left putamen. | (Shimada et al., 2015) | |||
| DTI | FA and RD values | Met-carriers vs. Val/Val-carriers | 71 ADHD (10.9) 24 HC (10.8) |
ADHD Val/Val: ↓ FA and ↑ RD in the right cingulum (cingulated gyrus) compared to ADHD Met-carriers and HC Val/Val. | (Kabukcu Basay et al., 2016) | ||
| DRD4 | exon 3 VNTR | sMRI | Superior frontal, middle frontal, anterior cingulate, and cerebellum cortices volumes | ADHD 7R-carriers vs. non-7R-carriers | 24 ADHD (38.1) 19 ADHD+BPD (35.8) 20 HC (33.2) |
7R- carriers: ↓ volumes of superior frontal cortex and cerebellum cortex compared to non-carriers. No effects in ADHD+BPD or HC. |
(Monuteaux et al., 2008) |
| TBV, PFC, cerebellum, CN, and pallidum volume | 7R-carriers vs. non-7R-carriers | 41 ADHD (9.7) 56 HC (17.6) |
No volumetric differences between 7R and non 7R-carriers. No group × genotype interactions. |
(Castellanos et al., 1998) | |||
| EEG | β and θ frequency bands during a CPT | 7R-carriers vs. non-7R-carriers | 340 children (11.1; 304 ADHD) 191 parents (44.3; 80 ADHD) |
Childhood 7R-carriers: ↑ frontal θ and ↓ global β2 power. Adult 7R-carriers: similar β2 power in ‘eyes closed’ condition, but ↓β2 power in ‘eyes open’ and CPT conditions. |
(Loo et al., 2010) | ||
| LPHN3 | rs2305339, rs734644, rs1397547, rs1397548, haplotype | EEG | Go/No-Go task (CPT) | High risk group (2 copies of risk haplotype = AGCC) vs. low risk group | 114 ADHD high risk group (34.85) 102 ADHD low risk group (34.92) |
High risk group: ↑ anterior Go-centroid of P300, ↓mean NGA. | (Fallgatter et al., 2013) |
| rs6551665, rs1947274, rs2345039 haplotype | Proton magnetic resonance spectroscopy (1H-MRS) | NAA/Cr ratio in striatum, lateral and medial thalamus, cingulate gyrus, and cerebellar vermis | Risk haplotype vs. protective haplotype | Risk haplotype: 13 ADHD, 2 HC Protective haplotype: 1 ADHD, 9 HC Different haplotypes: 8 HC | Risk haplotype carriers: ↓ NAA/Cr in left lateral thalamus, left medial thalamus, right striatum, ↑NAA/Cr in inferior–posterior cerebellar vermis. Carriers of two copies of risk haplotype had lowest levels of NAA/Cr. | (Arcos-Burgos et al., 2010) | |
| MAOA | rs1137070 | fMRI | Working memory task | TT-carriers vs. CC-carriers | 21 ADHD (23.9) ठ19 HC (25.2) ठ|
ADHD TT-carriers: ↑ activation in left inferior frontal lobe, pars opercularis. | (Ko et al., 2015) |
| NOS1 | Exon 1f-VNTR | DTI | WM integrity, FA and MD values | SS-carriers vs. SL/LL-carriers | 178 ADHD (17.6) 122 HC (16.8) |
Female SS-carriers: ↑ MD in right parietal WM tracts. Males: no difference between genotype groups. No genotype × diagnostic group interaction. |
(Van Ewijk et al., in revision) |
| fMRI | Reward anticipation task/modified MID task | SS-carriers vs. SL/LL-carriers | 63 ADHD (38.3) 41 HC (38.0) |
SS-carriers: ↑ activity in VS. No group × genotype interactions. | (Hoogman et al., 2011) | ||
| SLC6A2/NET1 | rs5569, rs28386840 | SPECT with [99mTc] HMPAO | Cerebral perfusion in response to MPH treatment (8 weeks) | rs5569: GG-carriers vs. GA/AA-carriers; rs28386840: AA-carriers vs. AT/TT-carriers | 37 ADHD (8.9) § | No differences in baseline clinical assessments or cerebral perfusion based on genotype. rs5569 GG-carriers: After 8 weeks of treatment hyperperfusion in right inferior temporal gyrus and middle temporal gyrus. |
(Park et al., 2012) |
| rs28386840, rs2242246, rs15534, rs40615 |
PET with (S,S)-[18F]FMeNER-D2 | NET BPND in thalamus, midbrain with pons, putamen, and cerebellum | Minor allele carriers vs. major allele homozygotes | 20 ADHD (30.8) 20 HC (30.4) |
rs28386840 and rs2242446 ADHD major allele carriers (A/T): ↑ NET BPND in the thalamus compared to major allele carrying controls. No difference was detected for the minor allele between groups. rs15534 and rs40615 HC major allele carriers (C/T): ↑ NET BPND in the cerebellum compared to major allele carrying patients. |
(Sigurdardottir et al., 2015) | |
| SLC6A3/DAT1 | 3’ UTR VNTR | SPECT with[99mTc] TRODAT-1 | Striatal DAT availability | 10R/10R-carriers vs. 9R-carriers | 29 ADHD (37.7) | No differences in DAT availability between 10R/10R-carriers and 9R-carriers. | (Krause et al., 2006) |
| SPECT with [123I]IPT in response to MPH treatment | Basal ganglia DAT density | 10R/10R-carriers vs. 9R-carriers | 11 ADHD (9.8)§ | 10R/10R-carriers: ↑ DAT density in basal ganglia. | (Cheon et al., 2005) | ||
| SPECT with [99mTc]-ECD in response to MPH treatment | rCBF during a CPT | 10R/10R-carriers vs. 9R-carriers | 8 ADHD, age range 8–12 ‡ | 10R/10R-carriers: ↑ rCBF in medial frontal and left basal ganglia areas in response to MPH. | (Rohde et al., 2003) | ||
| 3’ UTR, intron 8 VNTR, and haplotype | PET with [11C] altropane | DAT binding in CN | 9R-carriers vs. 10R/10R-carriers; 6R-carriers vs. 5R/5R-carriers |
34 ADHD (32.8) 34 HC (27.7) |
ADHD and HC 9R-carriers: ↑ DAT binding in CN. No association between intron 8 polymorphism or 3’-UTR-intron 8 haplotype with DAT binding. | (Spencer et al., 2013) | |
| rs2652511, rs2937639 haplotype | PET with [11C] cocaine | Ventral striatal DAT expression | CG-haplotype (rs2652511 C-allele and rs2937639 G-allele carriers) vs. rest | 6 ADHD 9 HC | CG-haplotype: No effect on diagnosis. Haplotype was more frequent in individuals with high DAT expression. | (Drgon et al., 2006) | |
| 3’UTR and intron 8 VNTR haplotype | sMRI | Bilateral striatal volumes (nucleus accumbens, CN, and putamen) | Three DAT1 alleles (10/10 genotype, and the haplotypes 10–6 and 9–6) | 118 ADHD (35.9) 111 HC (37) 301 ADHD (17.2) 186 HC (16.6) 1718 HC (26.1) |
Adult ADHD 9–6 haplotype carriers ↑ 5.9 % larger striatum volume relative to participants not carrying this haplotype (in adult ADHD patients only). Effect was not replicated in adolescent case-control and adult population-based cohort. |
(Onnink et al., 2016) | |
| 3’ UTR VNTR | sMRI | Cingulated cortex thickness | 10R/10R-carriers vs. 9R-carriers | 98 ADHD (10.9) | 10R/10R-carriers: ↑ thickness in right cingulated gyrus and right BA 24. | (Fernandez-Jaen et al., 2016) | |
| 3’ UTR VNTR | sMRI | PFC thickness | 10R/10R-carriers vs. 9R-carriers | 63 ADHD (10.9) | 10R/10R-carriers: ↓ cortical thickness in right BA 46 (lateral PFC). No other prefrontal ROI differed significantly. | (Fernandez-Jaen et al., 2015) | |
| 3’ UTR VNTR | sMRI | CN volume | 9R-carriers vs. 10R/10R-carriers | 33 ADHD (10.5) 26 HC (10.6) |
9R-carriers: ↑ volumes of CN. | (Shook et al., 2011) | |
| 3’UTR and intron 8 VNTR haplotype | fMRI | Striatal activity during reward anticipation task | 9–6 haplotype carriers vs. non 9–6 haplotype carriers | 87 ADHD (38.3) 77 HC (38) |
No differences in striatal activity compared with non 9–6 haplotype carriers nor 9R- and 10R/10R-carriers. | (Hoogman et al., 2013) | |
| 87 ADHD (38.3) 77 HC (38); same as above |
Bayesian Constraint-based Causal Discovery (BCCD) algorithm confirmed that there is no direct link between DAT1 genetic variability and brain activation, but suggested an indirect link mediated through inattention symptoms and diagnostic status of ADHD. | (Sokolova et al., 2015) | |||||
| VS and CN activity during reward-predicting cues | SLC6A3 10–6 dosage (2 copies vs. <2 copies) | 29 ADHD (combined type; 15.8) ‡ 30 HC (15.6) ‡ |
ADHD: Activation in CN ↓ as number of copies ↑, but in control group reverse was found. | (Paloyelis et al., 2012) | |||
| 3’ UTR VNTR | fMRI | Working memory task | 9R-carriers vs. 10R/10R-carriers | 53 ADHD (35.7) 38 HC (31.2) |
9R-carriers: ↓ left medial PFC activation compared to 10R/10R-carriers. Group × genotype interaction showed that 10R/10R-ADHD patients had ↑ activity in pre- SMA/dorsal ACC compared to HC. |
(Brown et al., 2011) | |
| Go/No-Go task | 10R/10R-carriers vs. 9R-carriers | 20 ADHD (14.1) 38 HC (13.12) |
10R/10R carriers: ↑ activity in frontal, medial, and parietal regions during response inhibition compared to 9R-carriers; ↓error response in the parahippocampal gyrus. | (Braet et al., 2011) | |||
| 10R/10R-carriers vs. 9R-carriers | 33 ADHD (11.1) | 10R/10R carriers: ↑ activity in left striatum, right dorsal premotor cortex, and temporoparietal cortical junction compared to 9R-carriers. | (Bedard et al., 2010) | ||||
| 9R-carriers vs. 10R/10R-carriers | 10 ADHD (14.6)‡ 10 unaffected siblings (14.8)‡ 9 HC (15.3)‡ |
9R-carriers: ↑ activity in CN and ↓ in cerebellar vermis compared to 10R/10R-carriers. Group × genotype interaction: effect in CN is observed in ADHD and unaffected siblings, but not HC. | (Durston et al., 2008) | ||||
| Multi-source interference task | 10R/10R-carriers vs. 9R-carriers | 42 ADHD (35.4) | 9R-carriers: ↓ activity in dorsal ACC compared to 10R/10R-carriers. | (Brown et al., 2010) | |||
| EEG | Go/No-Go task | 10R/10R-carriers vs. 9R-carriers | 161 ADHD (35.6) 109 HC (35.8) |
ADHD 9R-carriers: ↓ NGA (indicating impaired cognitive response control); No genotype effect in control group | (Dresler et al., 2010) | ||
| Feedback-based learning task; measuring ERPs | 10R/10R-carriers vs. 9R-carriers | 27 ADHD (11.5) 18 PDD (11.3) 20 HC (11.5) |
10R/10R-carriers: ↓ Pe to errors and ↓ SPN in anticipation of negative feedback, particularly with learning. | (Althaus et al., 2010) | |||
| EEG, single dose of 10mg MPH (double-blind, placebo-controlled) | Sustained attention and vigilance task | 10R/10R-carriers vs. 9R-carriers | 27 ADHD (10.1) | 10R/10R-carriers: medication-related EEG changes of ↑ central and parietal p power, ↓ right frontal θ power, ↓ θ/β ratios. 9R-carriers: showed opposite pattern. | (Loo et al., 2003) | ||
| SLC6A4/5HTT | 5-HTTLPR | sMRI (VBM) | GM volume | S-carriers vs. LL-carriers | 291 ADHD 78 subthreshold ADHD 332 HC; Average age: 17 years | S-carriers: stress exposure is associated with ↓ GM volume in precentral gyrus, middle and superior frontal gyri, frontal pole, and cingulated gyrus. Association of GxE interaction with ADHD symptom count was mediated by GM volume in frontal pole and anterior cingulated gyrus only. | (van der Meer et al., 2015) |
| SNAP25 | rs1051312, rs3746544 | fNIRS | MPH treatment-related hemodynamic changes during interference condition of Stroop task | rs1051312: TT- vs. TC/CC-carriers, rs3746544: TT- vs. TG/GG-carriers; 4 groups for interaction analysis |
15 ADHD (26.1) 16 ADHD (9.7) |
rs1051312 TT-carriers: ↓ right [HHb] with treatment rs3746544 TT-carriers: changes in genotype right [HbO2] and [HHb] as well as left [HHb]. Interaction analysis - participants with rs1051312 CC/TC and rs3746544 GG/TG: ↑ right [HHb] with MPH use. Interaction analysis - participants with rs1051312 TT and rs3746544 TT or rs1051312 TT and rs3746544 GG/TG: ↓ right prefrontal [HHb] with MPH use. |
(Oner et al., 2011) |
| TPH2 | rs4570625, rs11178997 | EEG | Go/No-Go task (CPT) | rs4570625: GG- vs. T-carriers, rs11178997: TT- vs. TA-carriers | 124 ADHD (34.7) 84 HC (34.8) |
rs11178997 TT-carriers and rs4570625 GG- carriers: ↓ NGA in both HC and ADHD. No group × genotype interactions. |
(Baehne et al., 2009) |
| SLC6A3/DAT1, DRD4 | 3’ UTR VNTR, exon 3 VNTR | SPECT with [99mTc]TRODAT-1 in response to MPH treatment | Striatal DAT binding potential | 10/10R-carriers vs. 9R-carriers; 7R-carriers vs. non-7R-carriers; 10/10R+7R-carriers vs. rest | 17 ADHD/SUDs, age range 15–21 years ‡ | 10/10R-carriers: no effect on DAT occupancy after MPH treatment. 7R-carriers: no effect on DAT occupancy after MPH treatment. 10/10R+7R-carriers: ↓ DAT occupancy after MPH treatment in right and left CN and putamen. |
(Szobot et al., 2011) |
| SPECT with [99mTc] ECD | rCBF during a CPT | 10/10R-carriers vs. 9R-carriers; 7R-carriers vs. non-7R-carriers; 10/10R+7R-carriers vs. rest | 34 ADHD (11.6) ‡ | 10/10R-carriers: no effect on rCBF. 7R-carriers: no effect on rCBF. 10/10R+7R-carriers: ↑ rCBF in right middle temporal gyrus area. | (Szobot et al., 2005) | ||
| sMRI | PFC GM and CN volume | 9R-carriers vs. 10R/10R-carriers, 4R/4R-carriers vs. rest |
26 ADHD (12.1) 26 unaffected siblings (11.6) 20 HC (10.7); all ‡ |
SLC6A3 ADHD 10/10R-carriers: ↓ CN volumes DRD4 unaffected siblings 7R-carriers: ↑ prefrontal GM volume. No effects on CN, or TBV. No interactions between ADHD status and genotype. |
(Durston et al., 2005) | ||
| 3’UTR and intron 8 VNTR haplotype, exon 3 VNTR | sMRI | Striatum, frontal cortex, and hippocampus volumes | 10–6 haplotype carriers vs. non-10–6 haplotype carriers, 7R-carriers vs. non-7R-carriers | 316 ADHD (17.2) 187 HC (16.5) |
SLC6A3 10–6 haplotype-carriers: ↓ left striatal volume, irrespective of treatment. DRD4 7R-carriers: frontal cortex volume is associated with stimulant treatment at younger age. |
(Schweren et al., 2016) | |
| SLC6A3/DAT1, DRD4, SLC6A4/5HTT | sMRI | Total GM, caudate, and putamen volume | 9R-carriers vs. rest, 7R-carriers vs. rest, S-allele-carriers vs. rest | 368 high ADHD severity (17.3) 374 low ADHD severity (16.8) |
For total GM, differential age effects were found for SLC6A3 9R- and SLC6A4 L/L carriers, depending on the amount of positive peer affiliation. For putamen volume, DRD4 7R-carriers and SLC6A3 10/10 homozygotes showed opposite age relations. Results were independent of ADHD severity. | (Richards et al., 2016) | |
| SLC6A3/DAT1, DRD4, DRD1 | 3’ UTR VNTR, exon 3 VNTR, rs4532 | sMRI; longitudinal study (mean follow-up, 6 years) | Cortical thickness | 9R-carriers vs. 10R/10R-carriers, 7R-carriers vs. non-7R-carriers, C-allele carriers vs. non-C-allele carriers | 105 ADHD (10.1; 13.1; 15.9) 103 HC (10.0; 12.4; 14.4) |
SLC6A3 9R-carriers: No effect on cortical development. DRD4 7R-carriers: thinner right orbitofrontal/inferior prefrontal and posterior parietal cortex. ADHD 7R-carriers: distinct trajectory of cortical development; normalization of right parietal cortical region. DRD1: No effect of genotype on clinical outcome or cortical development. |
(Shaw et al., 2007) |
| SLC6A3/DAT1, DRD4, COMT | 3’ UTR VNTR, exon 3 VNTR, rs4680 | DTI | WM integrity, FA values | 9R-carriers vs. 10R/10R-carriers; 4R/4R-carriers vs. rest; Met-carriers vs. Val/Val | 58 stimulant- and atomoxe-tine-naïve ADHD (8.7) § | SLC6A3 9R-carriers: no effect on WM integrity DRD4 4R/4R-carriers: no effect on WM integrity. Met-carriers: ↓ Network of WM connections linking 18 brain regions | (Hong et al., 2014) |
| SLC6A3/DAT1, COMT | 3’UTR and intron 8 VNTR haplotype, rs37020, rs460000, rs4680 |
fMRI | Stop-signal task | 10–6 haplotype-carriers vs. non-10–6 haplotype-carriers; rs37020: CC vs. rest; rs460000: GG vs. rest; Val/Val vs Met-carriers | 185 ADHD (17.3) 111 unaffected siblings (17.3) 124 HC (16.5) |
No genotype × ADHD interaction effects. SLC6A3 10–6 haplotype-homozygotes: ↑ activity related to successful stop-trials in pre- supplementary motor areas, ↓ activity in superior frontal and temporal pole areas. rs37020 AA-carriers: ↓ activity during failed stop-trials in IFG, pre-supplementary motor areas, and post-central gyrus. rs4680 Val/Val-carriers: ↓ activity during successful stop-trials in thalamus, frontal pole, and left IFG; ↑ activity in hippocampus during failed stop-trials. |
(van Rooij et al., 2015b) |
| SLC6A3/DAT1, DRD4 | 3’UTR and intron 8 VNTR haplotype, exon 3 VNTR | EEG | CPT | 10–6 haplotype-carriers vs. non-10–6 haplotype-carriers; 7R-carriers vs. non-7R-carriers | 94 ADHD; 31 HC;age range 8–16; all ‡ |
SLC6A3 10–6 haplotype-carriers: ↑ activity related to inhibitory response control (Nogo-P3) DRD4 7R-carriers: ↓ activity related to attentional orienting (Cue-P3) and cognitive or response preparation (CNV). No genotype × ADHD interactions. |
(Albrecht et al., 2014) |
| DRD4, COMT | exon 3 VNTR, rs4680 | EEG | Go/No-Go task | 7R-carriers vs. non-7R-carriers; Val/Val vs. Val/Met vs. Met/Met-carriers | 181 ADHD (35.3, age range: 18–60) 114 HC |
Single genes and diagnosis had no effect on neural correlates of prefrontal response control (NGA). DRD4 VNTR and COMTSNP epistatically interacted on NGA. |
(Heinzel et al., 2013) |
| DRD1, SLC6A2/NET1 | rs4532 and rs265981, rs998424 and rs3785157 | sMRI | TCV, volumes of total GM and WM, CN, cerebellum, frontal, temporal, parietal lobes | 2 and 3 genotype groups per SNP | 114 ADHD (9) 79 HC (16) |
DRD1 and NET1 SNPs: No genotype effects on GM or WM volume and no group × genotype interactions. | (Bobb et al., 2005) |
| SLC6A4/5HTT, HTR1B | 5-HTTLPR, rs6296 | fMRI | Stop-signal task | 3 genotype groups per variant | 184 ADHD (17.3) 111 unaffected siblings (17.3) 124 HC (16.5) |
SLC6A4 SS-genotype group: ↓ activation in frontal nodes and ↑ activation in posterior nodes. HTR1B genotype: associated with differential activation in anterior cingulate, occipital, inferior temporal, and cerebellar regions during successful stop trials. No associations between SLC6A4 and HTR1B variants and ADHD or ADHD-related neural activation. |
(van Rooij et al., 2015a) |
| SLC6A4/5HTT, NR3C1 | 5-HTTLPR, rs6189, rs6198 | sMRI | GM volume | S-allele carriers vs. rest; NR3C1 risk haplotype-carriers (rs6189G and rs6198G) vs. rest | 539 ADHD, unaffected siblings, and HC combined (17.2) | NR3C1 risk haplotype-carriers : ↑ positive relation between stress exposure and ADHD severity; which was stronger for SLC6A4 L-allele homozygotes. Interactions were reflected in GM volume of cerebellum, parahippocampal gyrus, intracalcarine cortex, and angular gyrus. | (van der Meer et al., 2016) |
BP = binding potential, BPND = nondisplaceable binding potential BPD = bipolar disorder, CN = caudate nucleus, CPT = continuous performance test, dACC = dorsal anterior cingulated cortex, DAT = dopamine transporter, DTI = Diffusion tensor imaging, ECD = ethyl-cisteinate-dimer, EEG = electroencephalography, ERP = event related potential, FA = fractional anisotropy, fMRI = functional magnetic resonance imaging, fNIRS = functional near-infrared spectroscopy, HC = healthy control, IFG = inferior frontal gyrus, NAA/Cr = ratio of N-acetylaspartate to creatine, NAcc = nucleus accumbens, NGA = NoGo-anteriorization, PDD = Pervasive Developmental Disorder, Pe = error-related positivity, PET = positron emission tomography, PFC = prefrontal cortex, pre-SMA = pre-supplementary motor area, rCBF = regional cerebral blood flow, RD = radial diffusivity, ROI = region of interest, sMRI, structural magnetic resonance imaging, SPECT = single-photon emission tomography, SPN = stimulus-preceding negativity, SUD = substance use disorder, TBV = total brain volume, TCV = total cerebral volume, UTR = untranslated region, VBM = voxel-based morphometry, VNTR = variable number tandem repeat, VS = ventral striatum, 4R = 4 repeat allele, 5R = 5 repeat allele, 7R = 7 repeat allele, 9R = 9 repeat allele, 10R = 10 repeat allele, [HHb] = deoxyhemoglobin, [HbO2] = oxygenated hemoglobin, [99mTc] = technetium-99; 1H-MRS = Proton magnetic resonance spectroscopy, [99m]Tc-HMPAO = technetium-99m hexamethylpropylene amine oxime,
only females,
only males;
Asian sample
3.1. Findings for the dopamine transporter gene (SLC6A3, DAT1)
The gene DAT1 (official name SLC6A3) codes for a solute carrier protein (DAT) responsible for the reuptake of dopamine from the synaptic cleft into the presynaptic neuron, representing a primary mechanism of dopamine regulation in the striatum (Ciliax et al., 1999). The most widely studied polymorphism in SLC6A3/DAT1 is a variable number of tandem repeat (VNTR) sequence in the 3’ untranslated region (3’UTR) that is 40 base pairs (bp) in length. Most common alleles are those with 9 and 10 repeats (9R and 10R). Additionally, a 30 bp VNTR in intron 8 of the gene (most common alleles with 5 and 6 repeats [5R and 6R]), is sometimes studied together with the 3’UTR VNTR as a haplotype. The 10R/10R genotype of the 3’UTR VNTR and the 10–6 haplotype of the two VNTRs are thought to be risk factors for childhood ADHD (Asherson et al., 2007; Brookes et al., 2006). In contrast, the 9R/9R genotype and the 9–6 haplotype are associated with persistent ADHD (Franke et al., 2010). This suggests a differential association of the gene with ADHD depending on age, and a role of DAT1 in modulating the ADHD phenotype across the lifespan. In addition to the VNTRs, several SNPs in DAT1 have been studied for their effect on ADHD and/or brain phenotypes.
Two studies performed PET to study the role of DAT1 genotype on DAT availability, one using 11Altropane as the ligand and one 11Cocaine. In an early study investigating a very small sample of 6 patients with ADHD and 9 controls, Drgon and colleagues studied a haplotype of two SNPs (rs2652511, rs2937639) in the 5’ regulatory region of the SLC6A3/DAT1 gene, and found the CG-allele associated with ventral striatal in vivo DAT availability independent of diagnosis; this finding was confirmed through investigation of striatal DAT expression in post-mortem brain samples (2006). Spencer and coworkers observed that, in adults, the 9R genotype of the 3’UTR VNTR increased DAT binding in caudate nucleus both in patients and healthy controls, whereas the intron 8 VNTR and a haplotype of both variants were not associated with DAT binding (2013); (Table 3).
Four SPECT studies, using different ligands, investigated the effect of the 3’UTR VNTR on DAT availability (Table 3). Two early case-only studies, both in children with ADHD (n=8 and 11, respectively), showed that basal ganglia DAT density was increased (Cheon et al., 2005) and that the regional cerebral blood flow (rCBF) was larger in medial frontal and left basal ganglia during a continuous performance task (CPT) in response to MPH treatment in 10R/10R-homozygotes compared to 9R-carriers (Rohde et al., 2003). A somewhat larger study in adults with ADHD did not identify a difference in striatal DAT availability between 10R/10R-homozygotes and 9R-carriers (Krause et al., 2006). Another SPECT study in boys with ADHD observed a genotypic effect of the 3’UTR VNTR variant increasing rCBF during a CPT only in the presence of risk alleles at both SLC6A3/DAT1 (10R/10R) and DRD4 (7R); this effect was present in the right middle temporal gyrus, an area associated with working memory and selective attention (Szobot et al., 2005); (Table 3). An interaction effect between the two polymorphisms was subsequently also shown by the same group in adolescent patients with ADHD plus substance use disorder (Szobot et al., 2011); (Table 3). In this case, participants homozygous for the SLC6A3/DAT1 10R-allele and carrying the DRD4 7R-allele exhibited decreased DAT occupancy after MPH treatment in the right and left caudate nucleus and putamen (Szobot et al., 2011). A recent meta-analysis, including healthy subjects and patients with different psychiatric disorders including ADHD, assessed the association of the 3’UTR variant with DAT availability (Faraone et al., 2014b). The PET studies provided significant evidence that the 9R-allele was associated with increased DAT availability in human adults, independent of the diagnostic status. The SPECT studies were highly heterogeneous, but when the analysis was limited to the most commonly used ligand, stratification by affection status dramatically reduced heterogeneity and revealed a significant association of the 9R allele with increased DAT availability for healthy subjects. The authors concluded that in humans, the 3’UTR polymorphism of SLC6A3/DAT1 regulates dopamine activity in the striatal brain regions independent of the presence of neuropsychiatric illness.
Eight sMRI studies for SLC6A3/DAT1 were performed thus far (Table 3). Two studies reported a smaller volume of the striatal caudate nucleus in homozygotes for the 10R allele when compared to children with the 9R/10R genotype (Durston et al., 2005; Shook et al., 2011). While Durston and coworkers found this effect to be most pronounced in children with ADHD, rather than their unaffected siblings or healthy comparison subjects, the overall genotype effect was independent of diagnosis. Two recent studies investigated cortical thickness in children and adolescents with ADHD. Fernández-Jaén and colleagues suggested that thickness of the lateral PFC and the cingulated cortex might be influenced by the presence of the 10R-allele (2015; 2016). Interestingly, homozygotes for the 10R allele showed increased thickness in the right cingulated gyrus (Fernandez-Jaen et al., 2016), but decreased cortical thickness in the lateral PFC (Fernandez-Jaen et al., 2015). In addition, a large observational study in an adolescent cohort revealed that irrespective of stimulant treatment, left striatal volume was reduced in participants with ADHD carrying one 10–6 haplotype (Schweren et al., 2016). A recent cross-sectional sMRI study included three cohorts (a childhood/adolescent ADHD case-control sample, an adult ADHD case-control sample, and an adult population-based sample) and showed that only in the adult ADHD case-control cohort, carriers of the DAT1 adult ADHD risk haplotype 9–6 had a 5.9% larger striatum volume relative to participants not carrying this haplotype. The effect varied by diagnostic status, with the risk haplotype affecting striatal volumes only in patients with ADHD (Onnink et al., 2016). A longitudinal study did not reveal any effect of SLC6A3/DAT1 genotype on cortical thickness (Shaw et al., 2007b), consistent with the predominant expression of DAT1 in subcortical (striatal) structures (and cerebellum). A recent gene-environment (GxE) interaction study reported differential age effects for SLC6A3/DAT1 9R-allele carriers for total grey matter volume and for SLC6A3/DAT1 10R-allele homozygotes on putamen volume, depending on positive peer affiliation (Richards et al., 2016) (Table 3). These findings were independent of ADHD severity. The presence of such differential age-dependent GxE effects might explain the diverse and sometimes opposing results of genetic and environmental effects on brain phenotypes (Richards et al., 2016). A single DTI study was performed in a sample of children with ADHD to assess DAT1’s effect on structural connectivity (Table 3). However, the investigated 3’UTR VNTR genotype did not appear to affect white matter integrity (Hong et al., 2014).
In total, nine fMRI studies were performed to investigate the effect of SLC6A3/DAT1 genotype on brain activity related to different tasks, most often examining reward processing and different aspects of executive functioning (Table 3). The studies included childhood, adolescent, and adult ADHD samples. Three fMRI studies investigated the role of the SLC6A3/DAT1 VNTR haplotype using reward-processing paradigms. Reward processing is altered in ADHD, and meta-analysis has shown that activation of the ventral striatum in anticipation of reward is lower in patients with ADHD than in controls (Plichta and Scheres, 2014). In a study in male adolescents, the activation of the caudate nucleus within the ventral striatum was found reduced in the ADHD group as the number of 10–6 haplotype copies increased (Paloyelis et al., 2012). A sizeable study in adult ADHD cases and controls, on the other hand, found no effect of SLC6A3/DAT1 haplotype on striatal activity (Hoogman et al., 2013). The latter dataset was re-analyzed using a Bayesian constraint-based causal discovery algorithm; this analysis suggested that any links between the genetic haplotype in DAT1 and reward anticipatory brain activity may be indirect only, mediated through inattention symptoms (Sokolova et al., 2015).
In studies of response inhibition, tested through a Go/No-Go task in children and adolescents, the 10R/10R genotype was found to be linked to higher (Bedard et al., 2010), but also lower (Durston et al., 2008) striatal activation. Interestingly, Durston and colleagues observed genotypic effects in the caudate nucleus in the patients and their unaffected siblings, but not in healthy controls (2008). Beyond the striatum, SLC6A3/DAT1 genotype effects were also found in additional brain regions, such as during cerebellar activation in children with ADHD (Durston et al., 2008), and in frontal, medial, and parietal regions, where activity was increased during response inhibition in adolescents homozygous for the 10R allele (Braet et al., 2011). Increased activity in the (temporo-)parietal regions in homozygous carriers of the 10R-allele was also observed in a second study, in addition to increased activity in the right dorsal premotor cortex (Bedard et al., 2010). The effects of the SLC6A3/DAT1 haplotype and rs37020 genotype on neural activation during response inhibition have been investigated as well (van Rooij et al., 2015b) (Table 3). Homozygous carriers of the SLC6A3 10–6 haplotype exhibited increased activity related to successful stop-trials in pre-supplementary motor areas and reduced activity in superior frontal and temporal pole areas, whereas homozygous carriers of the rs37020 A-allele showed reduced activity during failed stop-trials in the IFG, pre-supplementary motor areas, and post-central gyrus (van Rooij et al., 2015b). Despite these widespread effects on neural activation changes of the response inhibition network, these changes were independent of ADHD diagnostic status (van Rooij et al., 2015b). As expression of DAT1 is limited outside of striatum and cerebellum, these latter effects are likely due to direct or indirect connections between the regions of gene expression and the rest of the brain.
A working memory task in adult participants elicited increased activity in the dorsal ACC in patients homozygous for the 10R allele, whereas this genotype caused reduced activity in controls (Brown et al., 2011). Additionally, the authors showed a marginal association of the SLC6A3/DAT1 genotype with task-related suppression in the left medial PFC (Brown et al., 2011). Also in a multi-source interference task, the 10R/10R homozygotes had increased activity in the dorsal ACC compared to carriers of the 9R-allele (Brown et al., 2010). The dorsal ACC is thought to play a crucial role in numerous cognitive control functions including attention modulation, competition monitoring, complex motor control, novelty, error detection, working memory, anticipation of cognitively demanding tasks, and the modulation of reward-based decision making (Shenhav et al., 2013). Functional abnormalities associated with the dorsal ACC have been repeatedly reported in ADHD (Cao et al., 2009; Castellanos et al., 2008; Tamm et al., 2004; Zang et al., 2007; Tian et al., 2006), and the results above suggest that these effects might be most pronounced in 10R-allele homozygotes, which constitute approximately 71.9% of the Caucasian population (Doucette-Stamm et al., 1995). No studies in patients have yet investigated effects of DAT1 genotype on functional brain connectivity assessed through resting state fMRI. A single study in healthy participants using seed-based analysis revealed that carriers of the 9R-allele showed stronger connectivity between dorsal caudate nucleus and insula, dorsal ACC, and dorsolateral prefrontal regions, as well as between ventral striatum and centrolateral prefrontal cortex, suggesting wide-spread effects of the SLC6A3/DAT1 genotype on functional connectivity of striatal structures with the rest of the brain (Gordon et al., 2015).
Four EEG studies in ADHD childhood samples investigated the effects of SLC6A3/DAT1 variation (Table 3). One study examined the influence of the common 9R and 10R alleles on prefrontal brain functioning and cognitive response control in participants with adult ADHD and healthy controls (Dresler et al., 2010). By means of a Go-NoGo task (CPT) they inspected a neurophysiological marker of cognitive response control (NoGo-anteriorization, NGA). The NGA is a endophenotypic marker of prefrontal functioning, reflecting neural correlates of both response inhibition and execution in a Go-NoGo test situation (Fallgatter and Strik, 1999). It is a topographic event-related potential parameter quantifying the brain’s electrical field frontalization during motor inhibition (NoGo, when compared with response execution Go). As such, the NGA reflects “NoGo” activation of the medial PFC (anterior cingulate cortex, ACC) (Fallgatter et al., 2002). The NGA and the electrical field frontalization during NoGo trials have been found reduced in patients with ADHD (as well as those with schizophrenia) compared with healthy controls, reflecting diminished activation of the medial PFC in these patient groups (Fallgatter and Muller, 2001; Fallgatter et al., 2005). In the study of SLC6A3/DAT1 genotype carriers of the 9R-allele within the ADHD group showed significantly reduced NGA, whereas no influence of genotype was observed in the control group (Dresler et al., 2010). A second EEG study investigated the effect of the 10–6 haplotype on response control, by using a CPT task in a childhood case-control sample (Albrecht et al., 2014). Independent of ADHD diagnosis, 10–6 haplotype-carriers exhibited elevated brain activity related to inhibitory response control (Albrecht et al., 2014). Another study investigated the effect of the SLC6A3/DAT1 3’UTR genotype on neurophysiological correlates of performance monitoring by measuring event-related potentials (ERPs) during a feedback-based learning task. The authors showed that 10R/10R homozygotes had a smaller error-related positivity (Pe) response to errors and a smaller stimulus-preceding negativity (SPN) in the anticipation of negative feedback, especially with learning (Althaus et al., 2010), suggesting that SLC6A3/DAT1 genotype influences a system that is sensitive to aversive stimuli and their conscious processing (Althaus et al., 2010). The third EEG study investigated MPH medication-related changes on cortical power spectra during a sustained attention and vigilance task in a patient-only sample. ADHD patients have been shown to have increased frontocentral θ band activity and increased θ/β ratio compared to non-ADHD controls during rest (Loo and Makeig, 2012). Generally, the β band activity has been associated with attentional arousal, and it has been suggested that the θ/β ratio may reflect increased impulsivity and difficulty negotiating the speed-accuracy trade-off (faster speed but poorer performance) in ADHD (Loo and Makeig, 2012). In their study of DAT1 genetic variation during MPH treatment, the authors reported that those children with ADHD homozygous for the 10R-allele, exhibited medication-related cortical changes of increased central and parietal β power, decreased right frontal θ power, and lower θ/β ratios; 9R carriers showed the opposite pattern (Loo et al., 2003).
Summarizing, sMRI studies (as well as PET/SPECT by definition) find SLC6A3/DAT1 genotype effects mainly in regions, in which the gene is preferentially is expressed. These local effects can have widespread consequences on brain activity, as fMRI and EEG studies reveal. However, it is still unclear how the PET/SPECT findings, such as high DAT binding and density in nucleus accumbens and dorsal striatum (Kuczenski and Segal, 2002; Segal and Kuczenski, 1997) or the differences observed in brain activity are related to structural volume differences in the specific brain regions of ADHD patients. Therefore, studies could benefit from combining different imaging methodologies. The effects of genotype are often observed independent of diagnostic status, as would be expected for bona fide risk factors. Additionally, credibility of most of the existing studies can be questioned, since many of them are limited by their small sample sizes (Mier et al., 2010; Munafo et al., 2008). The actual neuroanatomical and brain activity-based mechanisms by which SLC6A3/DAT1 genotype increases ADHD risk remain to be clarified, as the results of studies published thus far are too patchy (and based on too small samples) to allow a coherent story to be defined. Moreover, links with cognitive performance and behaviour have often not been investigated. A further complicating matter for the interpretation of studies on SLC6A3/DAT1 is the switch in ADHD risk allele from childhood to adulthood. Longitudinal studies or at least cross-sectional studies including large samples of both children and adult participants (preferably also including persistent and remitted forms of ADHD) are therefore needed to advance the field.
3.2. Findings for the dopamine receptor D4 gene (DRD4)
The dopamine D4 receptor, encoded by DRD4, is a G protein-coupled receptor and belongs to the dopamine D2-like receptor family (Oldenhof et al., 1998). The most widely studied DRD4 polymorphism in ADHD has been the 48 bp VNTR in exon 3 of the gene, with the 2-, 4-, and 7-repeat alleles being the most common alleles. Allele frequencies vary significantly across ethnic groups (Chang et al., 1996; Van Tol et al., 1992), and the ADHD risk allele in the Caucasian population (7R) seems to be a different one from that in Asians (Nikolaidis and Gray, 2010; Wang et al., 2004; Li et al., 2006). The DRD4 48 bp VNTR seems to show differential association with ADHD in children, where it may be one of the strongest risk factors among the common genetic variants (Gizer et al., 2009), and adults, where no association with the disorder could be observed (Sanchez-Mora et al., 2011), though it might occur as part of gene-environment interactions (Sanchez-Mora et al., 2015). DRD4 is abundantly expressed in areas of the brain affected in ADHD, including frontal lobe regions, such as the orbitofrontal cortex and anterior cingulate (Lahti et al., 1995; De La Garza and Madras, 2000; Li et al., 2007b); the risk allele has been found to affect receptor binding and to produce a blunted response to dopamine (Asghari et al., 1995), although the functionality of the DRD4 VNTR has recently been challenged by meta-analysis (Pappa et al., 2015). The exon 3 VNTR has been the sole target of imaging genetic studies involving patients with ADHD, in which different imaging modalities have been used (Table 3).
An early sMRI study in children and adolescents did not report differences between carriers and non-carriers of the 7R-allele in selected regional and global brain volumes (Castellanos et al., 1998). A second study observed smaller prefrontal grey matter volumes in children homozygous for the 4R-allele, an effect that appeared particularly pronounced in unaffected siblings of patients with ADHD (Durston et al., 2005). In contrast, adult patients carrying the 7R-allele were found to have smaller volumes of the superior frontal and cerebellar cortex (Monuteaux et al., 2008). In a longitudinal study, DRD4 genotype also affected cortical development (by measuring cortical thickness), with 7R-carriers showing thinner prefrontal and parietal cortex; patients with ADHD carrying this allele had a distinct trajectory of cortical development characterized by normalization of parietal cortical regions (Shaw et al., 2007b). Additionally, an observational cohort study evaluating the effects of cumulative stimulant treatment, revealed associations between treatment and frontal and hippocampal volume dependent on DRD4 genotype and age (Schweren et al., 2016). More specifically, carriers of the 7R-allele showed decreased frontal cortex volume at younger age and lower treatment levels, whereas left hippocampal volume was increased in those with treatment at younger age (Schweren et al., 2016) (Table 3). By studying GxE interaction effects, carriers of the 7R-allele were found to have larger putamen volumes over age when exposed to high positive peer affiliation (Richards et al., 2016). This result was independent of ADHD severity. Effects of DRD4 genotype on structural brain connectivity were investigated in a single study in Asian children with ADHD, resulting in a report of no effects for 4R-homozygotes (Hong et al., 2014). A very large recent study in healthy participants (n=765) revealed an absence of significant effects of the DRD4 5R-allele on FA as well (Takeuchi et al., 2015). However, widespread changes in another measure of structural connectivity, mean diffusivity (MD), were observed, with increased MD in 5R-carriers in grey and white matter areas of the cerebral cortex, and in subcortical areas including the globus pallidus, amygdala, midbrain areas, and the brain stem (Takeuchi et al., 2015).
An EEG study administering a CPT as a measure of response inhibition and sustained attention, reported that children with the 7R-allele showed increased frontal θ and reduced global β power. Similar effects of DRD4 genotype were also apparent on the β frequency band in the parents (Loo et al., 2010); in both generations, carriers of the 7R-allele had reduced cortical activation upon performing the CPT task compared to participants not carrying this allele (Loo et al., 2010). Similarly, Albrecht and colleagues also applied the CPT to a childhood case-control sample and reported significant effects of the 7R-allele on ERP components. Specifically, they observed reduced activity related to attentional orienting and cognitive or response preparation in 7R-carriers (2014). This effect was found to be independent of an ADHD diagnosis (Albrecht et al., 2014). Extrapolating to behaviour, a recent meta-analysis showed that longer DRD4 variants (including the 7R-allele) were associated with lower levels of executive functioning (Pappa et al., 2015). In contrast to the continuous performance measure, an EEG study of inhibition applying another Go/No-Go task to adults with and without ADHD did not find an effect of DRD4 genotype on the NGA neural correlate of prefrontal response control (Heinzel et al., 2013). However, in this study, an epistatic interaction between the DRD4 VNTR and a SNP in the COMT gene (rs4680; Val/Met) was observed for the NGA; in homozygous carriers of the DRD4 7R-allele, the NGA followed a more pronounced U-relationship based on an increasing number of COMT Met-alleles compared with 7R-heterozygotes. In COMT Val/Met carriers, DRD4 7R-homozygosity was associated with decreased NGA compared with 7R-heterozygosity. This interaction could be localized to the right premotor and supplementary motor areas (Heinzel et al., 2013). These findings were independent of diagnostic status.
Thus, though existing evidence does not support firm conclusions about the pathways from gene to disease, genotypic variability in DRD4 may affect brain structure and/or activity and mark a particular developmental trajectory in cortical brain structure related to adult outcome of ADHD.
3.3. Findings for other ADHD candidate genes
Adrenergic neurotransmitter systems influence attentional processes and certain aspects of executive control (Arnsten, 2006), and the gene encoding the alpha-2A adrenergic receptor (ADRA2A) has been found to be a candidate gene for ADHD. The alpha-2A adrenergic receptor is the most prevalent noradrenergic receptor in the PFC (Park et al., 2005; Arnsten et al., 1996). Two variants appear to be risk factors for ADHD, the rs1800544 SNP in the promoter region, for which the G-allele was considered risk-increasing (Park et al., 2005), and the rs553668 SNP in the promoter region, for which the T-allele was considered the risk allele (Park et al., 2005). However, meta-analyses have not confirmed associations with ADHD risk for either SNP (Gizer et al., 2009). Alpha-2 adrenoreceptors have also been implicated in other major neuropsychiatric diseases that are often comorbid with ADHD, such as major depressive disorder and schizophrenia (Langer, 2015).
Genotypic effects of ADRA2A on the brain were studied in only two studies of Asian children with ADHD. A SPECT study showed that carriers of the non-risk allele for rs1800544 exhibited reduced perfusion in bilateral orbitofrontal regions (Kim et al., 2010). A DTI study investigated the effect of both SNPs (rs1800544 and rs553668) on white matter integrity. Carriers of the rs553668 non-risk-allele (C-allele) showed reduced fractional anisotropy (FA) in the right postcentral gyrus, whereas carriers of the rs553668 T-allele (non-risk group) showed reduced FA in the right middle frontal cortex (Park et al., 2013).
The Catechol-O-methyl-transferase enzyme (encoded by the COMT gene, located at 22q11.21) is involved in the degradation of the catecholamines dopamine and norepinephrine. It is highly expressed in the frontal lobe regions, where it is responsible for the regulation of dopamine levels (Hong et al., 1998). Studies investigating the association between COMT and ADHD have largely focused on a functional SNP (rs4860) in exon 4 that leads to an amino acid substitution (valine → methionine). This polymorphism has been shown to considerably affect COMT enzyme activity, such that homozygous carriers of the valine-allele show 3–4 times higher activity than homozygous carriers of the methionine-allele, resulting in decreased dopamine availability in valine homozygotes (Chen et al., 2004). An initial small study suggested that the valine-allele was associated with increased risk for ADHD (Eisenberg et al., 1999), although a recent meta-analysis failed to confirm this association (Lee and Song, 2015). The functional COMT rs4680 variant also seems relevant to other psychiatric and cognitive phenotypes. A recent study evaluating the association of the COMT genotype with schizophrenia in a systematic review and meta-analysis in 32,816 subjects, revealing a significant association (Gonzalez-Castro et al., 2016). Another recent meta-analysis revealed an association of the COMT genotype and reward learning, suggesting that variability in dopamine signalling associated with COMT rs4680 influences individual differences in reward processing, which may potentially contribute to psychopathology characterized by reward dysfunction, such as ADHD (Corral-Frias et al., 2016).
Two very small sMRI studies, both using VBM, examined the genotypic effect of rs4680 on the brain in children with and without ADHD (Table 3). Villemonteix and colleagues observed that children with ADHD and at least one methionine-allele had reduced grey matter volume in the insula/IFG relative to children without ADHD (2015). In an additional ROI analysis within the small ADHD group only (n=34), the study showed that those children with ADHD who were homozygous for the valine-allele had increased grey matter volume in the caudate nucleus compared to ADHD children carrying the methionine-allele, and also compared to children without ADHD (Villemonteix et al., 2015). A second sMRI study in a Japanese childhood sample also found a relation of COMT genotype with striatal volume pointing in the same direction, as the authors showed that the smaller grey matter volume observed in the left putamen in children with ADHD was moderated by the methionine-allele, whose carriers showed smaller volume than the valine homozygotes (Shimada et al., 2015). Effects of COMT on structural connectivity were investigated by two DTI studies. One study in Asian children with ADHD revealed that carriers of the methionine-allele showed a weakened network of white matter connections linking 18 different brain regions (Hong et al., 2014) (Table 3). This finding is in line with a report on healthy participants, showing that the methionine-allele was associated with impaired structural maturation of brain white matter connectivity (Thomason et al., 2010). Based on these findings, Hong and coworkers formulated the hypothesis that higher dopamine availability may inhibit myelination (2014). Recently, Kabukcu Basay and colleagues reported that children with ADHD, who were homozygous for the Valine-allele, had reduced FA and increased RD values in the right cingulum bundle (2016). This indicates demyelination effects in the white matter connections between the cingulated cortex to the PFC, premotor regions, cortical association areas in the parietal and occipital lobes, thalamus, and hippocampus (Kabukcu Basay et al., 2016). The cingulated cortex is known to be involved in complex cognitive processing (Bush et al., 2000) and functions that are believed to be impaired in ADHD (Bush, 2011; Makris et al., 2008).
A single fMRI study investigated the association of COMT rs4680 genotype on task performance and whole-brain neural activation during response inhibition (van Rooij et al., 2015b). Although the COMT Val158Met variant resulted in differential activation patterns during successful and failed stop-trials in the combined ADHD-control sample, no interactions between genetic effects and ADHD diagnostic status were observed in any of the whole-brain fMRI results (van Rooij et al., 2015b) (Table 3). This indicates that genetic variation in the COMT gene exerts large-scale effects on neural activation changes of the response inhibition network, but these changes are independent of ADHD. The single EEG study of COMT in ADHD, using a Go/No-Go task in adults with and without ADHD, did not reveal an effect of COMT genotype on brain activity individually. However, an epistatic interaction of DRD4 × COMT genotype on neurophysiological correlates of prefrontal function was observed (Heinzel et al., 2013). In homozygous carriers of the DRD4 7R allele, the anteriorization of the NoGo response (NGA, explained above) followed a more pronounced U-relationship with increasing numbers of Met alleles compared with 7R heterozygotes. In COMT Val/Met carriers, DRD4 7R homozygotes showed a significantly decreased NGA compared with 7R heterozygotes. The genotype-dependent effects on the NGA were localized in the right premotor and supplementary motor area. The epistatic interactions were independent of ADHD diagnosis (Heinzel et al., 2013). Given the role of fronto-striatal circuits and dopamine in reward processing (Plichta and Scheres, 2014; von Rhein et al., 2015), an important domain of impairment in ADHD (see above), it is surprising that no studies have yet investigated the role of COMT in brain activity related to reward anticipation and receipt.
The dopamine D1 receptor (encoded by the DRD1 gene) is the most abundant dopamine receptor subtype in the brain; it is highly expressed in the striatum and cerebral cortex (Bergson et al., 1995). Common genetic variation in the DRD1 gene (rs5326) has been associated with schizophrenia risk (Pan et al., 2014) and impaired cognition in patients with bipolar disorder (Zhao et al., 2015), both often found comorbid with ADHD. Several studies explored associations between ADHD and genetic variants of DRD1, such as SNPs rs4532 and rs265981 (Bobb et al., 2005). In the initial study, participants with ADHD were more likely than healthy controls to have the C-allele or rs4532 and the A-allele of rs265981 (Bobb et al., 2005). However, results of a meta-analysis of rs4532 did not support its association with ADHD (Wu et al., 2012), and also replication for rs265981 is still pending. Nevertheless, two sMRI studies investigated the effect of genetic variation in DRD1 on cortical thickness (Shaw et al., 2007b) and on brain volume (Bobb et al., 2005). Neither study found an effect of DRD1 genotype or group × genotype interactions (Table 3).
The 5-Hydroxytryptamine receptor 1B (encoded by the HTR1B gene) is the most widely studied serotonin receptor gene in relation to ADHD. HTR1B is a G protein-coupled receptor that inhibits cyclic AMP formation (Murphy et al., 1998). It is highly expressed in the dorsal raphe nucleus, which is involved in the sleep/wake cycle, and to lesser degrees in the striatum and frontal regions, such as the dorsolateral PFC (Ichikawa et al., 2005). The initial study, investigating 273 nuclear families with ADHD, reported preferential transmission of the rs6296 G-allele to ADHD probands (Hawi et al., 2002). Results of a meta-analysis supported this association between childhood ADHD and rs6296 genotype (Gizer et al., 2009). A single fMRI study investigated the relationship of rs6296 genotype with neural correlates of response inhibition, using a stop-signal task in a childhood sample (van Rooij et al., 2015a). The rs6296 genotype was associated with widespread differential activation during successful and failed stop trials (Table 3). However, the direction of these effects was inconsistent, with both increased and decreased activation for the GG genotype being observed in frontal and posterior nodes, and the differential activation patterns were independent of ADHD diagnosis (van Rooij et al., 2015a).
The latrophilin 3 gene (LPHN3; official name Adhesion G Protein-Coupled Receptor L3 [ADGRL3]) codes for a member of the LPHN subfamily of G-protein-coupled receptors (GPCRs). Subtype 3 is the most brain-specific LPHN (Sugita et al., 1998; Ichtchenko et al., 1998) and is expressed in regions implicated in ADHD, i.e. the caudate nucleus, cerebellum, amygdala, and cerebral cortex (Arcos-Burgos et al., 2010). LPHN3 was identified as an ADHD risk gene downstream of genetic linkage studies in multicase families from a genetic isolate. In multisite association studies, initially, a haplotype of three SNPs (rs6551665, rs1947274, and rs2345039) was shown to be associated with ADHD (Arcos-Burgos et al., 2010). The association of LPHN3 genotypes with ADHD has been replicated in children (rs6551665) (Hwang et al., 2015) and adults with ADHD (multiple markers) (Ribases et al., 2011). By use of proton magnetic resonance spectroscopy (1H-MRS), Arcos-Burgos and colleagues showed that individuals carrying the LPHN3 susceptibility haplotype exhibited a decreased ratio of N-acetylaspartate to creatine (NAA/Cr ratio) in the left lateral and medial thalamus as well as the right striatum, and an increased ratio in inferior–posterior cerebellar vermis; this suggested that the maintenance of neuron viability is altered in those carrying the ADHD risk haplotype (2010). Since the ADHD susceptibility haplotype itself did not cause any significant coding region changes or canonical splice site alterations, it was suggested that non-coding variations may be likely contributors to ADHD (Domene et al., 2011).
Although LPHN3 is among the best-supported candidate genes for ADHD, thus far, imaging genetics studies of the gene are still limited in the literature. No studies on structural or connectivity alterations related to the risk variant of the gene in patients with ADHD have been published yet, nor have any fMRI studies. An EEG study using a Go/No-Go task revealed that adult patients with ADHD carrying a ‘high-risk’ LPHN3 haplotype (comprised out of rs2305339, rs734644, rs1397547, and rs1397548) showed a more anterior Go-centroid of the P300, had a reduced NGA, and had worse behavioural task performance due to more omission errors (Fallgatter et al., 2013).
Monamine oxidase A, encoded by MAOA, is an enzyme, which catalyzes the oxidative deamination of amines, such as dopamine, norepinephrine, and serotonin. For this gene located on the X-chromosome, studies have largely focused on a functional 30 bp VNTR 1.2 kb upstream of the gene, which has been previously associated with impulsivity and aggression (Caspi 2002, Manuck 2000). The polymorphism consists of alleles of 2, 3, 3.5, 4, and 5 repeat copies, and evidence suggests that the 2 and 3 repeat (‘low-activity/MAOA-L’) alleles are less efficiently transcribed than the longer (‘high-activity/MAOA-H’) alleles (Deckert et al., 1999). Although the MAOA gene has received a lot of attention as a candidate gene for ADHD given the prior findings for impulsivity, a meta-analysis did not indicate a significant association between ADHD and the high activity alleles of the VNTR (Gizer et al., 2009). A SNP, rs1137070 (located in exon 14), has also been reported to contribute to impulsivity and the outcome of ADHD (Li et al., 2007a; Liu et al., 2011). Especially, the C-allele of rs1137070 has been associated with high ADHD scores and poor outcomes (Li et al., 2007a).
With evidence for a role of MAOA in aggression being more consistent than for ADHD (e.g. (Brunner et al., 1993; Caspi et al., 2002; Byrd and Manuck, 2014), imaging genetics studies have largely concentrated on population groups other than patients with ADHD, e.g. (Holz et al., 2016; Meyer-Lindenberg et al., 2006). A recent fMRI study forms an exception. Using a phonological working memory task in male Asian adults with and without ADHD, the authors showed that the effect of MAOA (rs1137070 T- versus C-allele) interacted with diagnosis in the left inferior frontal lobe, pars opercularis (Table 3); further analysis demonstrated that the increased brain activation observed in this region in patients was only significantly different to controls among those hemizygous for the T-allele (Ko et al., 2015).
The NOS1 gene codes for nitric oxide synthase 1, an enzyme that synthesizes nitric oxide from L-arginine. Nitric oxide is a reactive free radical, which can act as a biological mediator in several processes, including dopaminergic and serotonergic neurotransmission (Kiss and Vizi, 2001) and neurite outgrowth (Chen et al., 2006). The NOS1 gene has a complex structure, including 12 alternative untranslated first exons (exon 1a-1l). In exon 1f, a VNTR that affects gene expression has been linked to hyperactive and impulsive behaviour in humans (Reif et al., 2009; Weber et al., 2015); the short allele was shown to be the risk factor for ADHD, especially in females.
No studies have yet reported effects of NOS1 on brain structure. A recent case-control DTI study of structural connectivity in adolescents revealed that female homozygous carriers of the ADHD risk allele showed higher MD values in several major white matter tracts of the brain compared with long allele carriers. This effect was present in both female patients and controls (Van Ewijk et al., in revision). The white matter tracts found affected by NOS1 genotype overlap with those earlier found associated with ADHD (Onnink et al., 2015; van Ewijk et al., 2012; Wu et al., 2016). Since higher MD values can be indicative of demyelination, lower axonal density, or axonal degeneration, homozygosity of the short allele might thus be a risk factor for aberrant development of white matter tracts involved in ADHD etiology.
NOS1 exon 1f is particularly highly expressed in striatum. Therefore, the single study of gene effects on brain function investigated the effect of the VNTR on ventral striatal activity during reward anticipation using fMRI (Hoogman et al., 2011) (Table 3). The study revealed that homozygous carriers of short alleles of NOS1 demonstrated higher ventral striatal activity than carriers of the other NOS1 VNTR genotypes (Hoogman et al., 2011). Again, this effect was independent of diagnostic status. Similar effects of the genotype were also observed for behavioural impulsivity, with those carrying the ADHD risk factor acting more impulsive than other participants. As the authors did not perform mediation studies, it remains to be investigated, whether the observed genotype effects on brain connectivity and/or activity directly link to behavioural effects.
The norepinephrine transporter gene (SLC6A2, NET1) codes for a protein responsible for the reuptake of norepinephrine (as well as dopamine) from the synaptic cleft back into the presynaptic neuron (Pacholczyck 1991). The gene is highly expressed in the frontal lobes (Stahl, 2003). Candidate gene studies of SLC6A2/NET1 selected numerous SNPs to test for association, but conflicting results have been reported, with each study yielding evidence of association, but differing in which specific SNPs were associated with ADHD (Gizer et al., 2009).
A SPECT study examined the effects of rs5569 and rs28386840 on cerebral perfusion in response to MPH treatment in Asian children with ADHD (Table 3) (Park et al., 2012). At baseline, no differences were observed, but after eight weeks of MPH treatment increased regional brain perfusion in the right inferior temporal gyrus and middle temporal gyrus in homozygous carriers of the rs5569 G-allele was demonstrated (Park et al., 2012). Given that no previous studies had reported a significant association between this polymorphism and ADHD, no ‘risk’ allele was indicated. A PET study reported the influence of four genetic variants within the transporter gene on in vivo norepinephrine transporter binding in adults with and without ADHD (Table 3) (Sigurdardottir et al., 2015); the authors found differences in cerebellar and thalamic norepinephrine transporter binding depending on genotype between adult patients and controls (Sigurdardottir et al., 2015). For the two SNPs rs28386840 and rs2242446, patients carrying the major alleles (A/T) showed increased norepinephrine transporter binding in the thalamus compared to controls carrying the major alleles. For the SNPs rs15534 and rs40615, controls carrying the major alleles (C/T) showed increased norepinephrine transporter binding in the cerebellum compared to patients carrying the major alleles (Sigurdardottir et al., 2015). In the patients with ADHD, a high correlation between hyperactivity/impulsivity symptoms and norepinephrine transporter binding in the cerebellum was detected, an effect which was strongly moderated by genotype (Sigurdardottir et al., 2015).
With this knowledge on SNP functionality related to ADHD, studies using additional imaging modalities are warranted, but thus far, only a single sMRI study investigated the effect of genetic variation in SLC6A2/NET1 on brain volume (Table 3). No effect of SLC6A2/NET1 genotype (for SNPs rs998424 and rs3785157 – different SNPs from those having been investigated through PET and SPECT) and no group × genotype interactions were reported (Bobb et al., 2005).
The serotonin transporter gene (5HTT, SERT; official name SLC6A4) codes for a solute carrier protein responsible for the reuptake of serotonin from the synaptic cleft back into the presynaptic neuron, which is the primary mechanism for regulating serotonergic activity in the brain (Lesch et al., 1996). A functional polymorphism exists in the promoter region of this gene (5HTTLPR) in the form of a 44-bp insertion/deletion yielding short (S) and long (L) alleles. The long variant is associated with more rapid serotonin reuptake, resulting in lower levels of active serotonin (Lesch et al., 1996). A SNP in the long allele, rs25531, additionally modifies its activity (Lesch et al., 1996). SLC6A4/5HTT has been implicated in emotion regulation, (emotional) memory, and learning processes (Araragi and Lesch, 2013; Barzman et al., 2015; Meneses and Liy-Salmeron, 2012). The serotonin transporter is expressed in regions implicated in attention, memory, and motor activities, such as the amygdala, hippocampus, thalamus, putamen, and ACC (Frankle et al., 2004; Oquendo et al., 2007). The 5HTTLPR has been extensively studied for its role in depression and anxiety – especially in the context of environmental adversity, with the S-allele being the risk allele (Caspi et al., 2003; Oo et al., 2016). For ADHD, the evidence for association is more limited, although an earlier meta-analysis provided significant evidence of an association between ADHD (in children) and the ‘long’ variant of 5HTTLPR (Gizer et al., 2009). However, an international multicentre study reported a slight, non-significant overrepresentation of the S-allele in adult patients with ADHD (Landaas et al., 2010).
Given the evidence for SLC6A4 as a depression gene, imaging genetics research has largely concentrated on non-ADHD samples; in healthy individuals, functional genetic variation in the SLC6A4/5HTT gene has been linked to emotion regulation through effects on brain activation in the amygdala and the wider ‘threat circuit’, while effects on regional brain volumes are inconsistent (Klein et al., under review; Jonassen and Landro, 2014). Three sMRI studies have been performed for 5HTTLPR in ADHD thus far, all investigating GxE interactions. The initial study showed that the interaction between exposure to environmental stress and carriership of the 5HTTLPR S-allele, which was linked to increased ADHD severity in a longitudinal study of ADHD families (van der Meer et al., 2014), was associated with reduced cortical grey matter volume in the precentral gyrus, middle and superior frontal gyri, frontal pole, and cingulate gyrus in S-allele carriers compared with participants homozygous for the L-allele (van der Meer et al., 2015). Importantly, this paper showed that only some of these regions, the frontal pole and the ACC, actually mediated the effects of the gene-environment interaction on ADHD severity. Similarly, van der Meer and colleagues reported that individuals carrying the NR3C1 risk haplotype, who were homozygous for the 5HTTLPR L-allele, showed a negative relation between stress and grey matter volume (2016). However, no mediation effects were found, meaning that the local effects of these interaction on grey matter volume did not significantly explain their association with ADHD severity (van der Meer et al., 2016). Such studies testing the mediation of effects through the observed brain phenotypes (as opposed to those just being epiphenomena of the genetic variation) are largely lacking in the imaging genetics literature thus far, but are critically needed to map gene to disease pathways. However, sample sizes will have to be substantial to allow valid conclusions to be drawn from such studies. Another sMRI study used the same dataset and revealed that, in agreement with age-related reductions of total grey matter volume found in longitudinal studies (Brain Development Cooperative, 2012; Raznahan et al., 2014), participants scoring high on positive peer affiliation and carrying two 5HTTLPR L-alleles had smaller total grey matter volumes with age (Richards et al., 2016). Moreover, participants with the same genotype, but low positive peer affiliation had larger GM volumes with age (Richards et al., 2016). These findings were independent of ADHD severity and were in line with a longitudinal study reporting regional GM reductions with age in adolescents exposed to high positive maternal behavior, but increased putamen volumes when exposed to maternal aggression (Whittle et al., 2014).
In contrast to the sMRI studies, which investigated GxE effects related to stress and social environments, the only fMRI study focused on the relationship between 5HTTLPR genetic variation and neural correlates of response inhibition using a stop-signal task (van Rooij et al., 2015a) (Table 3). Using a childhood sample, van Rooij and colleagues revealed that homozygous carriers of the 5HTTLPR S-allele showed decreased activation in the frontal nodes and increased activation in the posterior nodes in successful stop trials (2015a). However, no significant associations were found between differential neural activation and ADHD diagnosis or ADHD severity (van Rooij et al., 2015a).
The synaptosomal-associated protein 25 (encoded by the SNAP25 gene) is involved in axonal growth and synaptic plasticity, as well as in the docking and fusion of synaptic vesicles in presynaptic neurons necessary for the regulation of neurotransmitter release (Sollner et al., 1993). Several studies have tested for linkage and association between SNAP25 and ADHD, and these studies have consistently genotyped multiple SNPs within the gene rather than focusing on any single polymorphism. An early meta-analysis of SNP rs3746544 suggested SNAP25 as ADHD risk gene (Faraone et al., 2005). A more recent meta-analysis of rs3746544 also provided evidence of a modest but significant association between childhood ADHD and the T-allele, whereas no evidence of association with rs1051312 was found (Gizer et al., 2009).
Again, only a single imaging genetics study is currently available for this gene. In this case, the interaction of MPH treatment-related hemodynamic changes with genetic variation in SNAP25 (rs3746544 and rs1051312) was studied in small samples of children and adults with ADHD on and off single dose MPH using fNIRS (Table 3). Through fNIRS, brain haemodynamics in prefrontal cortices can be measured, since brain activation causes increased cerebral blood flow, but not all of this oxygenized blood is used, therefore oxygenated haemoglobin increases and deoxyhaemoglobin decreases during e.g. sustained attention (Villringer and Chance, 1997). Homozygous carriers of the rs3746544 T-allele exhibited changes in right oxygenated haemoglobin and right as well as left deoxyhaemoglobin levels. Additionally, homozygous carriers of the rs1051312 T-allele showed decreased right prefrontal deoxyhaemoglobin with treatment. Combination of the genotypes also showed interaction effects on right prefrontal deoxyhaemoglobin levels (Oner et al., 2011).
Tryptophan hydroxylase 2 catalyzes the reaction of tryptophan to 5-hydroxytryptophan, which is subsequently decarboxylated to form the neurotransmitter serotonin. Two isoforms of tryptophan hydroxylase have been identified (encoded by the TPH1 and TPH2 genes). The TPH2 gene codes for a rate-limiting enzyme in the production of serotonin in serotonergic neurons in the midbrain raphe nuclei, while TPH1 seems to be involved in synthesizing serotonin in peripheral tissues (Walther et al., 2003). For several SNPs of the TPH2 gene, an association with ADHD was found (Brookes et al., 2006; Sheehan et al., 2005; Walitza et al., 2005). For rs4570625, the G-allele was shown to be transmitted more often to offspring with ADHD (Walitza et al., 2005). For rs11178997, the T-allele was identified as the risk allele for ADHD (Walitza et al., 2005). However, the initial findings could not be replicated in a larger, multicentre study (Brookes et al., 2006). Mouse models in conjunction with approaches focusing on TPH2 variants in humans described a role of serotonin in brain development and in disorders related to negative emotionality, aggression, antisocial behaviour, and bipolar disorder (Waider et al., 2011; Gao et al., 2012; Gao et al., 2016). The TPH2 gene has also been a candidate for the investigation of gene by environment interaction studies, in depression and suicidal behaviour as well as in aggression; for review see (Mandelli and Serretti, 2013; Lesch et al., 2012).
As the evidence for a role of TPH2 in ADHD is limited, only a single imaging genetics study involving patients with this disorder has yet been performed for this gene. This EEG study investigated the genotypic effect of the two SNPs of the TPH2 gene mentioned above (rs4570625 and rs11178997) on the NGA during a Go/NoGo task in adult patients with ADHD and healthy individuals (Table 3). Risk alleles of each of the SNPs were found associated with a reduction in the NGA in both participant groups, indicating an effect on ADHD-relevant prefrontal brain function independent of a specific psychiatric diagnosis (Baehne et al., 2009). These promising first findings may warrant further analysis of TPH2 variants, especially in combination with adverse environmental factors.
4. OUTLOOK
In this review we set out to summarize the current literature on imaging genetics studies of candidate genes involving patients with ADHD. As no genome-wide association studies have yet reported loci/genes with significant evidence for association with ADHD, we used a liberal definition of a candidate (having shown association with the disorder in at least one earlier study) and used an adapted version of the list of ADHD candidate genes recently compiled by Li and coworkers (2014). Of the 62 candidate genes selected for review, we found that only 12 had been studied with any brain imaging technique in an ADHD population. For most of those 12, only a very limited number of studies was available. The two most frequently studied ADHD candidate genes were the SLC6A3/DAT1 and DRD4 genes, and even for those two genes most findings await replication.
Brain imaging phenotypes offer an important level of investigation in mapping the biological pathways from gene to disease. Brain structure, function, and connectivity can provide endophenotypes (or intermediate phenotypes) for a disease (Gottesman and Gould, 2003). As those brain endophenotypes are thought to lie in-between a genetic factor and the clinical phenotype, it has been argued that effect sizes for effects of genes on those brain phenotypes may be larger than the ones for effects on behaviour. However, for structural brain phenotypes (i.e. brain volumes) and for structural connectivity, this has been refuted by recent large-scale studies; see (Franke et al., 2016; Jahanshad et al., in preparation). For brain activity measured by fMRI, comparisons of cognitive and brain imaging studies for several genetic variants (Rose and Donohoe, 2013) and evidence from meta-analyses (Mier et al., 2010; Murphy et al., 2013) suggests that effect sizes might be larger. However, it is still unclear, how large the effects of publication bias on those results are (Murphy et al., 2013). Nevertheless, power estimates indicate that several hundred samples are needed, while the majority of studies discussed in this review had less than 50 participants per group (see Table 3). This has implications for the generalizability/replicability of positive findings (as the sample investigated may not sufficiently well represent the population it was sampled from) as well as harbouring the risk of false-negative findings (Button et al., 2013). Importantly, the term endophenotype has also often been misused in recent years, as for many of the so-called endophenotypes, the required criteria have not all been investigated. As such, many of the endophenotypes may not be intermediates between gene and behavioural phenotype, but rather be simple markers of disease (Kendler and Neale, 2010). Assessing, whether a brain phenotype observed for a risk genotype really mediates between the risk variant and the behavioural/clinical phenotype has been done in hardly any of the papers reviewed here. The paper by van der Meer and colleagues, in which such an analysis has been performed, shows the importance of such mediation analyses, as only a subgroup of the brain substrates of the gene-environment interaction investigated was also linked to ADHD severity (2015).
Shortly summarizing the results of the review, our understanding of the mechanisms underlying the action of genetic risk factors for ADHD is still limited. A promising start has been made. We see, for example, that SLC6A3/DAT1 genotype regulates dopamine activity in striatal regions implicated in ADHD neurobiology (Spencer et al., 2013). Genetic variants also appear to affect brain structure and function beyond the regions of gene expression (Hong et al., 2014; van der Meer et al., 2014; Villemonteix et al., 2015), which is likely due to effects on structural and/or functional connectivity. Some studies reported differential genotype effects for ADHD patients and controls, whereas others did not observe genotype by diagnosis interactions (Table 3). However, such findings have not been replicated yet, and genotype effects may differ in the context of other ADHD-related risk factors.
As there is room for improvement of study designs, and research focusing on new ADHD candidate genes identified through hypothesis-free, genome-wide association studies will soon be needed, we address main challenges and opportunities for imaging genetics studies in the following paragraph. Additionally, since imaging genetics approaches cannot explain the full complexity of a biological system such as the human brain, we also highlight additional levels of investigation and methodologies that can complement the insights provided by imaging genetics studies.
4.1. Main challenges and opportunities for imaging genetics studies
The size of individual studies is potentially the biggest challenge in imaging genetics today, as also discussed above. Given the small (to potentially modest) effect sizes to be expected (Franke et al., 2016; Murphy et al., 2013; Rose and Donohoe, 2013), larger samples are essential in order to gain the necessary statistical power.
Common confounding factors encountered in the published brain imaging genetics studies are mainly related to the study sample itself, and many are aggravated by the limited sample sizes used. False-negative findings may occur due to the large variability of the ADHD phenotype. Additionally, comorbidities occur frequently in ADHD (Kessler et al., 2006; McGough et al., 2005; Wilens et al., 2009), and differences in medication use may further increase phenotypic heterogeneity (Schweren et al., 2013). Together with the low effect sizes of individual common genetic variants and limited sample sizes, phenotypic heterogeneity in a sample challenges the discovery of effects. Meaningful subtyping of this heterogeneous condition to decrease phenotypic heterogeneity and maximize power might therefore be helpful. Besides that, potential differences of effects between age groups, with gender distribution, intelligence levels, and between ethnicities can still make it challenging to replicate and generalize findings.
Although most genetic risk factors investigated here were derived from studies of ADHD in children, our review shows that imaging genetics studies have employed childhood, adolescent, and adult ADHD samples. Knowing about the age-dependence of the genetic contribution to ADHD (Chang et al., 2013; Pingault et al., 2015), which e.g. results in differential association of SLC6A3/DAT1 (Franke et al., 2010) and DRD4 (Sanchez-Mora et al., 2011) with ADHD in children and adults, makes the definition of appropriate research questions highly important. Only one study thus far used a longitudinal approach, with a follow-up period of six years (Shaw et al., 2007b). Longitudinal samples are best suited to investigate potential age-dependent changes in the effects of genetic factors on the brain, and therefore more longitudinal studies of ADHD including imaging and genetic assessments are needed. This is especially important in light of a recent discussion on whether not only the onset of ADHD can occur in adulthood, but also whether childhood-onset and adult-onset ADHD may be distinct syndromes or trajectories (Agnew-Blais et al., 2016; Caye et al., 2016; Faraone and Biederman, 2016; Moffitt et al., 2015). Therefore, longitudinal studies including participants with remitted versus persistent ADHD, can help us in understanding the genetic and neurobiological correlates of this multifactorial disorder.
All of the imaging genetics studies reviewed here employed hypothesis-driven approaches. This means, that candidate genetic variants were only investigated in relation to specified candidate brain phenotypes and by this it severely limits our ability to create new knowledge on genetic effects on the brain. Even main effects of a variant may have been missed, as our ability to define the right hypotheses is hampered by the still so limited understanding of ADHD etiology. Besides that, most studies investigated single genetic variants (SNPs or VNTRs), although we know that frequently more than one risk allele exists in a single gene/locus; see e.g. (Schizophrenia Working Group of the Psychiatric Genomics et al., 2014). Also, only few studies looked for epistatic or gene-environment interaction effects.
In the face of limited sample sizes available for an imaging genetics study, data reduction strategies might help to preserve power. By moving to gene-based mass-univariate and multivariate statistics, it is possible to test the combined effect of multiple genetics variants in a single test statistic. Such models thus reduce the number of statistical tests, e.g. through gene-wide analyses, and - by explaining more phenotypic variance - may enable the discovery of gene effects that would have been otherwise undetectable with single variant methods (Hibar et al., 2011; Bralten et al., 2011; Bralten et al., 2013). Alternatively – or rather in addition - analyses of multiple genetic factors may be performed in the context of international collaborations, like ENIGMA and CHARGE (Psaty et al., 2009; Thompson et al., 2014) or using the publicly available data from large national research efforts (like the UK Biobank; http://www.ukbiobank.ac.uk/). These are mainly population studies, but since ADHD is currently viewed as an extreme on a continuum of population traits (Larsson et al., 2013a; Chen et al., 2008), we can learn a lot from studies performed in healthy individuals. While for this review, we specifically selected studies that had included patients with ADHD, it is indeed apparent also from those studies that genes affect traits rather than disorders (Hoogman et al., 2011). We have referred to some relevant studies in healthy individuals already in the previous sections. An additional example is an fMRI study investigating a genetic variant in SNAP25, which was found associated with altered activation of the posterior cingulate cortex during a working memory task, a finding that was replicated in a second, independent sample (Soderqvist et al., 2010). With working memory being one of the cognitive domains affected in ADHD (Alderson et al., 2013), this finding can contribute to unravelling the biological pathways from gene to disease. However, some studies have also suggested differential genotype effects in subjects with ADHD and healthy participants (Durston et al., 2008; Monuteaux et al., 2008; Onnink et al., 2016). In those cases, imaging genetics studies in healthy participants will not reveal the full spectrum of the genetic effects, and studies in patients are warranted. The ENIGMA-ADHD Working Group, with its sample spanning 60 years of the human lifespan (Hoogman et al., submitted), is an important resource for the investigation of diagnosis-specific effects.
There are a number of additional areas, in which we foresee that imaging genetics studies will profit from technical advances as well as recent insights into disease etiology. Importantly, a new GWAS meta-analysis is now underway by the ADHD Working Group of the Psychiatric Genomics Consortium (PGC) and the Danish iPSYCH consortium, which will provide several bona fide ADHD risk genes identified in hypothesis-free analyses (PGC and iPSYCH ADHD working groups, in preparation). Those will be interesting targets for investigation through imaging genetics approaches. In addition, the introduction of next generation sequencing (NGS) technology in psychiatric research will allow us to go beyond studying SNPs and VNTRs. It is likely that also more rare genetic variants, identified through exome and whole-genome sequencing will find their way into imaging genetics studies, e.g. through mutational load or burden tests (Medland et al., 2014).
Most imaging genetics studies in the field of ADHD have used MRI as their main brain imaging technique. For subcortical brain structures, grey matter volume has been the most frequently investigated characteristic. For the cortex, measurements of surface area (SA) and cortical thickness (CT) were found to be genetically and phenotypically rather independent (Winkler et al., 2010). Volume has been shown to be more closely related to SA than CT (Winkler et al., 2010). Therefore, it has been suggested that SA and CT measurements should be considered separately in imaging genetics studies (Winkler et al., 2010), in addition or instead of cortical volume. Until now, only very few studies have made use of this opportunity, however. Because of its low invasiveness, high spatial resolution, and wide availability of MRI scanners, this technique dominates the brain imaging field. Imaging genetic studies investigating genetic variation in the SLC6A2/NET1, SLC6A3/DAT1, and DRD4 also used PET and SPECT (Table 3). These modalities enable direct localization and quantification of e.g. binding capacities of transporters and receptors, but are quite invasive. An alternative method for investigating the neurochemistry of the brain in vivo might be proton magnetic resonance spectroscopy (1H-MRS), which allows for non-invasive quantification of several neurometabolites, such as N-acetylaspartate, (phospho-) creatine, choline, myo-inositol, glutamate and glutamine, and gamma aminobutyric acid (GABA) (Naaijen et al., 2015). While no significant differences in GABA levels were found in ADHD compared with controls (Schur et al., 2016), a possible increased signal of a combination of glutamate, glutamine, and GABA in the striatum of ADHD patients was observed, as well as an increased signal in the ACC in a paediatric ADHD sample and a reduced signal in an adult ADHD sample; reviewed by (Naaijen et al., 2015). The neurodevelopmental changes in fronto-striatal glutamatergic circuits across the lifespan suggested by this might be interesting targets for future imaging genetics studies. By combining different methods, it should be possible to create a comprehensive picture of how polymorphisms in ADHD-related genes affect the brain at chemical, structural, and functional levels. To date, also no MEG genetics studies has been reported. The high temporal resolution of this modality might be of great additional value in understanding genetic effects on brain function. Resting-state fMRI studies are also still lacking from the imaging genetics literature. Especially the combination and integration of different modalities in the study of individual participants may provide more comprehensive insights into gene effects (Kobiella et al., 2011). With respect to functional brain markers that might serve as a useful endophenotype, it is crucial to use a functional contrast that isolates brain activity specifically associated with the cognitive process of interest. For example, stop/NoGo and Go conditions in traditional stop-signal/Go-NoGo tasks not only differ in the involvement of response inhibition, but also in other processes such as novelty/probability of occurrence and perceptual processing (Boehler et al., 2010; Sanchez-Carmona et al., 2016). Results of a recent study highlight the importance of controlling for the different strategies adopted by participants to perform selective stopping tasks before analyzing brain activation patterns (Sanchez-Carmona et al., 2016). Thus, activity emerging from a functional contrast (e.g., Go versus NoGo) will probably reflect a mixture of different processes. This is an important issue to be considered, when interpreting task-based fMRI studies and designing new studies.
Another opportunity to improve the design of imaging genetics studies is the use of mediation and moderation analyses. In mediation analysis, a causal explanation for the effect of an independent on a dependent variable is statistically modelled. The assumption of causality is important in this type of analysis and thus should be justified by a plausible biological theory or appropriate experimental constraints. In moderation analysis, the influence of a third variable on the association of an independent and dependent variable is modelled. The simple moderation and mediation models can be combined to account for more complex data structures and biological models (Hayes and Scharkow, 2013; van der Meer et al., 2014). Such models have e.g. been employed in psychological (Aram et al., 2010; Graziano et al., 2011) and medical research questions (Nigg et al., 2008), but could be also promising for analyses of the pathway from genes to phenotypes in complex disorders, such as ADHD, which involves different biological and non-biological factors acting synergistically during an individual’s developmental trajectory (Bale et al., 2010; Krain and Castellanos, 2006). Indeed, van der Meer and coworkers recently used such analyses to inspect the moderation of the effect of stress on ADHD severity by the 5HTTLPR genetic variant. The researchers could show that an interaction of 5HTTLPR genotype and stress was associated with ADHD severity (van der Meer et al., 2014), and that this gene-environment interaction had several substrates in the brain. Importantly however, only a subset of those substrates did really mediate the effects of the gene-environment interaction, whereas others were epiphenomena (van der Meer et al., 2015).
While imaging genetics studies with the goal of mapping pathways from gene to disease up to now have always started with the selection of the candidate gene/variant to study, the advances brought about by international consortia now start to allow entirely data-driven approaches to be used for imaging genetics. For example, we have recently developed a comprehensive pipeline for the analysis of genetic overlap of GWAS results for disease risk (in that case schizophrenia risk data from the PGC) and the GWAS results for brain volume (Franke et al., 2016). The latter was based on data from the ENIGMA consortium (Stein et al., 2012; Medland et al., 2014; Thompson et al., 2014), in which MRI scans and genome-wide genotyping data from up to 30,717 individuals were analysed, and several genetic factors contributing to the volumes of specific brain structures were identified (Hibar et al., 2015). Finding such overlap between disease risk variants and those for brain phenotypes would be indicative of etiologic sharing, and would thus directly flag the pathways from gene to disease (Franke et al., 2016). For their previous studies, ENIGMA used mainly regional brain volume measures (Hibar et al., 2015; Stein et al., 2012), but one can also envisage more comprehensive voxel-wise genome-wide scans (vGWAS) to examine evidence for associations across the genome at each voxel in the brain image (Stein et al., 2010). While these approaches still are limited by the fact that a stringent correction for type I errors dramatically increases the threshold for statistical significance (as genomes and images are both highly dimensional), data reduction strategies are being devised that can preserve power in such settings (Medland et al., 2014). With respect to the different imaging modalities, structural MRI and DTI seem to be best suited for larger collaborations and data sharing, but also fMRI imaging genetics meta-analyses have been shown to be feasible (Mier et al., 2010; Murphy et al., 2013). First analyses of overlap between ADHD GWAS findings and results of the ENIGMA volume analyses are currently underway (Klein et al., in preparation).
4.2. Complementary methods for the evaluation of the mechanisms underlying ADHD risk genes
Imaging genetics analyses of the human brain provide information on the effect of ADHD risk genes/variants on brain structure, activity, and connectivity, but other levels of investigation are needed to provide a more complete picture (Figure 2). We also need information about the molecular networks, in which an ADHD gene acts, and the cellular processes that are affected by it. In the following section we highlight different methodologies and approaches that can be used to shed light on these additional levels of complexity contributing to the mechanisms underlying the effects of ADHD genes on behaviour and disease.
Figure 2:
This schematic representation of the endophenotype concept shows the pathway from gene to disease at different levels of complexity in psychiatric genetics. The figure has been modified from a previous publication (Figure 1, (Franke et al., 2009)). Polygenicity (schematically depicted by gene A to I) is involved in causing disease symptoms. A reduced number of genes is involved in disease-related endophenotypes. These can be studied at various biological levels, e.g. biochemical processes and cell function can be assessed by biological assays in cell or animal models by measuring e.g. neuron morphology or synaptic functioning. Neuroimaging methods (structural and functional) can be applied to assess relevant endophenotypes at the level of brain morphology (‘Morphology brain region A-C’). Endophenotypes, related to the ‘function of brain units’, can be e.g. investigated by functional MRI or through performance measurements on neuropsychological tests. Aberrations at this level can result in altered behavior and disease-related behavioral traits, that subsequently lead to disease symptoms. Environmental influences can impact on all levels and need more attention in future studies. Bioinformatic pathway and network analyses can help to integrate data from various sources and to identify molecular networks or cellular processes in which ADHD-related genes are enriched.
4.2.1. Bioinformatics approaches help to integrate findings from different types of molecular studies
Bioinformatics is a broad field of research, which can support our efforts to map pathways from gene to disease in several different ways. A first goal to be pursued through bioinformatics analyses is the clarification of the actual effects of risk variants on gene expression and regulation. Risk variants for a disease are often found in non-protein coding sequences, and therefore the molecular consequences are difficult to evaluate (Paul et al., 2014; Civelek and Lusis, 2014). We will not go into this type of bioinformatics analyses any deeper, but would want to point the interested reader to a recent example for a comprehensive bioinformatic follow-up study demonstrating how to elucidate the mechanisms by which a genetic risk variant (e.g. rs1344706; ZNF408A gene) confers susceptibility to disease, in this case schizophrenia and bipolar disorder (Hess et al., 2015).
Bioinformatics can also help to unravel the molecular networks and cellular processes that an ADHD risk gene is involved in. ADHD-associated genetic factors are distributed throughout the genome; however, they have been found to be enriched within functional categories. This clustering of ADHD-related genes within functional networks or pathways has helped to identify biological processes of importance to ADHD etiology. As also mentioned earlier, through those bioinformatics studies we learned that functions related to nervous system development, cell migration, neuron projection morphogenesis/neurite outgrowth, oxogenesis, cell-cell communication, glutamatergic synapse/receptor signalling, multicellular organismal development, RhoA signalling, glycosaminoglycan biosynthesis, fibroblast growth factor receptor activity, ion (potassium) channel function, transmembrane transport, as well as synaptic transmission, catecholamine metabolic processes, G-protein signalling and organonitrogen compound catabolic processes are enriched in the results of hypothesis-free, genome-wide ADHD-GWAS and CNV studies (Poelmans et al., 2011; Hawi et al., 2015; Yang et al., 2013; Cristino et al., 2014; Mooney et al., 2016; Thapar et al., 2015). Those studies mainly used childhood ADHD data; little is yet known about biological pathways leading to persistence of ADHD. A first, small-scale GWAS of rare and common genetic variants in adults with persistent ADHD showed that the top SNPs implicated biological pathways involved in the regulation of gene expression, cell adhesion, and inflammation (Zayats et al., 2015). The Network and Pathway Analysis Subgroup of the PGC focussed on common pathways underlying additional, ADHD-related psychiatric disorders in adults, i.e. schizophrenia, major depression, and bipolar disorder (The Network Pathway Analysis Subgroup of the Psychiatric Genomics Consortium, 2015). Histone methylation processes (playing important roles in the regulation of gene expression) showed the strongest association, and the researchers also found significant evidence for involvement of immune and neuronal signalling pathways and for processes occurring at the postsynaptic density (The Network Pathway Analysis Subgroup of the Psychiatric Genomics Consortium, 2015).
Comprehensive studies applied to the ADHD-related autism spectrum disorders (ASD) point out, how the combination and integration across several molecular modalities can advance our knowledge about pathways from gene to disease. For example, systematic integration of findings from multiple levels of genomics data and studies of mouse models highlighted the period of fetal development and the processes of chromatin structure, neurite outgrowth, steroidogenesis, synaptic function, and neuron-glial signalling (Poelmans et al., 2013; Chen et al., 2015). Additional studies revealed that ASD genes grouped together in terms of functional annotations, protein-protein interactions and coexpression (Voineagu et al., 2011; Ben-David and Shifman, 2012; O’Roak et al., 2012; Neale et al., 2012), and gene interaction and coevolutionary patterns (Gilman et al., 2011). By integrating gene expression data representing normal human fetal development, developmental timing and cellular specificity of the molecular pathways disrupted in ASD could be clarified (Willsey et al., 2013; Parikshak et al., 2013). Such integrative studies are still lacking for ADHD, but highly warranted in order to shed more light on the onset and neurodevelopmental trajectory of the disorder.
4.2.2. Animal models provide proof of causality for genes and molecular processes found associated with ADHD
The complexity and crucial role of the brain in the human body largely restrict studies in humans to the described non-invasive imaging methods for investigating brain structure and function. However, to fully understand the role of a disease gene in the fine-tuning of this sophisticated organ, we also need to be able to manipulate genes and monitor the effects of such manipulation on molecular and cellular processes. For this, animal models are indispensable. They can be genetically modified to enable determination of causality, have a natural complexity of the nervous system, and allow a tight control of environmental influences, including diet and drug delivery (Lange et al., 2012; van der Voet et al., 2016). During the last decades, studies were performed on several ADHD animal models, including monkey (Ma et al., 2005; Seu et al., 2009), rat (Ruocco et al., 2014; Williams et al., 2009b; Russell et al., 1995), mouse (Wallis et al., 2012; Zhu et al., 2014; Zimmermann et al., 2015), zebrafish (Lange et al., 2012), and fruit fly (van der Voet et al., 2016). The particularity of each model, like the different levels of complexity of the nervous system, provide complementary information to the broad picture of ADHD.
Most studies in monkeys were mainly based on drug administration to stimulate/inhibit certain brain regions or neurotransmitter receptors and to study its effect on a nearly as sophisticated brain structure as the human (Ma et al., 2005; Seu et al., 2009).
The rat and mouse models additionally provided evidence for involvement of genes in ADHD (Gainetdinov et al., 1999; Simchon et al., 2010; Wallis et al., 2012; Zhuang et al., 2001; Zimmermann et al., 2015). This was done by measuring face-valid hyperactivity, inattentiveness, or impulsivity, and also looking for physiological and histological abnormalities in reverse and forward genetic approaches. Reverse genetic approaches are employed for disease candidate genes identified in human studies; the orthologues of the observed human genes are manipulated in the animal model to evoke measurable phenotypes and to study their involvement on cellular and/or behavioural levels (Rivero et al., 2013; Wallis et al., 2012). Forward genetics use the opposite tactic by studying inbred animal strains for ADHD-associated conducts, and analysing their protein expression pattern and behaviour (Dimatelis et al., 2015; Womersley et al., 2015; Li et al., 2007b). In reverse genetics approaches, early studies proved the involvement of candidate genes from association studies in ADHD, e.g. by showing ADHD-like behavioural phenotypes in Dat1 knock-out mice and the coloboma mouse showing a mutation in Snap25 (Heyser et al., 1995; Giros et al., 1996). Most knock-out and transgenic mouse models targeted genes involved in dopamine transmission. These mutant models provide an excellent opportunity to evaluate the contribution of dopamine-related processes to brain pathophysiology, to analyse the neuronal circuits and molecular mechanisms involved in the action of ADHD medication, and to test novel treatments for ADHD; for review see (Leo and Gainetdinov, 2013). More recent studies of a mouse model containing a null mutation of the latrophilin 3 gene (Lphn3), which showed a hyperactive phenotype in an open field test, revealed that this ADHD candidate gene is involved in gene expression regulation of monoamine signalling genes, such as dopamine and serotonin receptors and transporters, neurotransmitter metabolism genes, and neural development genes (Wallis et al., 2012). Additionally, actin depolymerising factor and n-cofillin double mutant mice displayed hyperlocomotion, impulsivity, impaired working memory, and disturbed morphology of striatal excitatory synapses, accompanied by strongly increased glutamate release (Zimmermann et al., 2015). Of note, the hyperlocomotion and impulsivity were reversed by methylphenidate (Zimmermann et al., 2015).
While reverse genetics approaches are most often applied in ADHD research to date, forward genetics has led to new models to study ADHD, like the spontaneous hypertensive rat (SHR), which was created to study hypertension by inbreeding albino Wistar rats showing elevated blood pressure (Okamoto and Aoki, 1963). These rats also showed deficits in attention, and ADHD-associated conducts, such as increased impulsivity and hyperactivity, and are therefore widely employed as a model for ADHD (Li et al., 2007b; Williams et al., 2009a; Williams et al., 2009c; Williams et al., 2009b). Studies of the molecular and cellular characteristics of this forward genetic model have provided evidence that proteins involved in energy metabolism, neurotransmitter function, neural development, and myelination are differently expressed in the striatum and PFC (Dimatelis et al., 2015; Womersley et al., 2015). The observed changes in the striatal energy metabolism support the neuroenergetics hypothesis of Killeen and colleagues, which states that inadequate neuronal energy supply can lead to ADHD symptoms (2013). Moreover, the GABAergic system was shown to be involved in ADHD-like behaviour of SHR rats by investigating the physiological response of GABA on hippocampal slices and finding GABA significantly altered during early-life stress in these animals (Sterley et al., 2013b; Sterley et al., 2013a).
Novel animal models for the study of ADHD include zebrafish (Danio rerio) and the fruit fly (Drosophila melanogaster) (Lange et al., 2012; van der Voet et al., 2016). These models have the great advantage of being relatively inexpensive, having a wide range of genomic tools available, and being highly suitable for fast, high throughput studies of (candidate) genes. The combination of those characteristics, with the availability of valid and quantifiable phenotypic readouts for ADHD-relevant traits makes these animal models a potent addition to non-invasive studies in humans. Even though both zebrafish and fruit fly diverged from the human lineage early in evolution, there is still a high level of conservation of neuronal genes; for example, in a screen for intellectual disability genes, more than 70% of candidate genes had an unambiguous orthologue in the fruit fly (Oortveld et al., 2013; van der Voet et al., 2014). Also genes involved in the phenotypic manifestation of hyperactivity were crucial during early evolution and thus can be found in species with a much simpler nervous system. Several studies show the strength of such models. In zebrafish, e.g. the orthologues of the human ADHD candidate genes LPHN3, PER1, and PER2 have been studied and shown to cause hyperactivity (Huang et al., 2015; Lange et al., 2012; Wang et al., 2015a). The lphn3.1 mutant fish showed, besides the behavioural phenotypes, misplaced dopamineric neurons in the brain (Lange et al., 2012). Functional studies on Per2 null mutants in zebrafish showed differential expression of the circadian clock genes aanat2 and bmal1b and indicated the involvement of Per2 in the circadian regulation (Wang et al., 2015a). In addition, per1b mutant fish display hyperactive and impulsivity-like behaviours and low levels of dopamine, of which hyperactivity could be rescued by ADHD medication (Huang et al., 2015). It could be shown that the circadian clock has direct influence on the structure and abundance of dopaminergic neurons and lower expression of transcription factors directly regulating development, maintenance and differentiation of these neurons (Huang et al., 2015). In the fruit fly, manipulation of the orthologues of the dopamine-related genes DAT1 and LPHN3, caused characteristic darkness-dependent hyperactivity, an ADHD-like behaviour (van der Voet et al., 2016). Also in those models, ADHD medication was able to reverse the behavioural phenotype (van der Voet et al., 2016). Finally, the vacuolar protein sorting-associated protein 4A (VPS4A) in the striatal node was significantly associated with dysfunctional reward, a cognitive (e.g. poor inhibitory control or timing estimation), but also affective (e.g. delay aversion or hyposensitivity to reward) deficit observed in ADHD (Jia et al., 2016). The orthologue was studied in the fruit fly, showing that flies with an overexpression of Vps4 have a reduced overall activity similar to a Drd1 knockout fly, while a knockdown of Vps4 leads to hyperactivity, suggesting the involvement of VPS4A in the regulation of DRD1-mediated activity (Jia et al., 2016).
At the level of animal models, the integration of knowledge from models of different evolutional complexity will optimally support the elucidation of gene to disease pathways, as it is already apparent from the initial study of LPHN3 orthologues across mouse, zebrafish, and fruit fly.
4.2.3. Modelling psychiatric disorders at the cellular level by using human induced pluripotent stem cell (hiPSC)-derived neurons
ADHD is a multifactorial disorder, and in most patients, several to multiple genetic factors are likely to contribute to disease. Animal models are mostly based on highly penetrant single gene mutations, which limits the translation of findings from molecular and cellular levels to the human situation. A human model, especially one derived from a patient him-/herself, might therefore be preferable in certain situations. Until recently, the only way to study cellular processes and molecular pathways in patient brain cells was through post-mortem material. Due to the scarceness of available postmortem brain tissues, only a very limited number of studies was performed to date, concentrated on demonstrating an influence of ADHD-related genetic variants on gene expression (Weber et al., 2015; Hawi et al., 2013; Brookes et al., 2010; Brookes et al., 2007). Fortunately, cell reprogramming technology has been developed throughout the past decade, which provides a powerful tool to simulate neural developmental processes in vitro in a petri dish (Brennand et al., 2015b; Takahashi et al., 2007; Yu et al., 2007). Neurons from both healthy individuals and psychiatric patients can be derived by 1) reprogramming human skin fibroblasts or blood mononuclear cells into induced pluripotent stem cells (iPSCs) and then differentiating these into neurons (Zhang et al., 2013), or by 2) directly reprogramming skin fibroblasts into neural stem cells or neurons (Pang et al., 2011; Vadodaria et al., 2015; Brennand and Gage, 2012). The human induced pluripotent stem cell (hiPSC)-derived neurons from a patient then have a genetic background identical to the disorder state, and thereby provide the possibility to characterize the effects of the cocktail of genetic perturbations leading to disease in this patient (Brennand et al., 2015a; Madison et al., 2015; O’Shea and McInnis, 2015; Wang et al., 2015b; Lim et al., 2015). Such effects may be specific to cell types and/or specific developmental stages (Duan, 2015; Hockemeyer and Jaenisch, 2016). Moreover, a temporal analysis of disease initiation and progression can be performed in the cell type(s) most relevant to a disorder (Brennand et al., 2012). For example, studying hiPSC neurons from patients with disease-associated large CNVs provides the opportunity to perform comprehensive molecular analyses of the effects of these large CNVs in several cell lineages (Urban and Purmann, 2015). Until now, several studies were published using hiPSC-derived neurons from patients with ASD, schizophrenia, or bipolar disorder (Marchetto et al., 2010; Ananiev et al., 2011; Urbach et al., 2010; Sheridan et al., 2011; Chiang et al., 2011; Brennand et al., 2011; Wen et al., 2014). For example, hiPSC-derived neurons from patients with schizophrenia were shown to have synapse deficits and transcriptional dysregulation (Wen et al., 2014). Additionally, by comparing hiPSC-derived neurons from patients with bipolar disorder and controls, alterations in key components of the microRNA processing pathway were identified, potentially altering neuronal cell fate determination (O’Shea and McInnis, 2015). To overcome confounding effects of variable genetic backgrounds, when comparing cells from patients to those from healthy controls, genome-editing technology can be applied (Duan, 2015), such as Zinc-finger nucleases (ZFN) (Reinhardt et al., 2013), transcription activator-like effector nucleases (TALEN) (Wen et al., 2014), and recently developed clustered regularly interspaced short palindromic repeats (CRISPR) (Ran et al., 2013; Wang et al., 2015b; Liu et al., 2016). Such genome-editing enables the generation of isogenic hiPSC-derived neurons that differ only at the genetic site of interest (Ding et al., 2013; Bedell et al., 2012; Choi et al., 2013). In this way, specific mutations can be either introduced in control cells or corrected in patient cells to investigate causality (Duan, 2015; Hendriks et al., 2016).
Beyond mechanistic insights, hiPSC neurons might also serve as a platform for high throughput screening to identify novel therapeutics for psychiatric disorders (Schadt et al., 2014). For example, the ability to test drugs to rescue synaptic deficiency in Rett syndrome neurons has been demonstrated (Brennand et al., 2012). Generally, it is suggested that these cell models are suited for longitudinal observations, studying on-off-medication effects, and investigating the mechanisms of comorbid disorders in individual patients (O’Shea and McInnis, 2015).
A current limitation of hiPSC-based models is the high heterogeneity of the derived differentiated neuronal populations (Gore et al., 2011; Hu et al., 2010; Lister et al., 2011; Osafune et al., 2008). Although almost pure glutamatergic neurons can be differentiated from hiPSCs, the differentiation efficiency for other types of neurons, such as dopaminergic, GABAergic, serotonergic, or cholinergic neurons, is relatively low, which can cause variability in the outcome of in vitro experiments (Swistowski et al., 2010; Vadodaria et al., 2015; Zhang et al., 2013; Liu et al., 2013; Hu et al., 2016). Therefore, ideally a comparison of several hiPSC-derived neuronal lines from multiple patients should be performed (Brennand and Gage, 2012). It seems advantageous to model the psychiatric disorders in a pure, specified type of neuron; however, it is also reasonable to carry out investigations at the network level in different types of neurons interacting, better mimicking the multidimensional integration in the brain through 3D culture systems or brain ‘organoids’, which are currently being developed (Kim et al., 2015; Lancaster et al., 2013).
Based on the above, it is clear that the reprogramming technology will revolutionize our use of model systems. Although still very much under development, and not yet applied to ADHD risk genes, first results for other psychiatric disorders already suggest that valuable findings about molecular and cellular pathways from gene to disease can be derived from iPSC-derived neurons.
5. CONCLUSION
Results from studies described in this review show that imaging genetics approaches are highly suitable to provide more insight into the pathways from gene to behaviour via the brain in ADHD. However, this field is clearly still in its early stages. Inconsistency of findings, due to the use of relatively small sample sizes, clinical and biological heterogeneity of ADHD, methodological differences in study design, and analysis methodology, make it difficult, yet, to draw firm conclusions about effects of genes on brain morphology, function, and connectivity. Individual genes need to be investigated more extensively and in larger samples, and additional genes need to be studied – preferably focussing on those implicated in ADHD through genome-wide, hypothesis-free approaches. We emphasize that a combination and integration of imaging genetics studies with complementary approaches at different levels of biological complexity - including bioinformatics as well as cell and animal models - will be necessary to fully map the biological pathways from gene to disease.
HIGHLIGHTS.
We present a systematic review of brain imaging genetics studies in ADHD.
We found imaging genetics studies for 13 ADHD candidate genes, mostly DAT1 and DRD4.
First promising results are described, however comparability of studies was limited.
Brain imaging genetics can help to map pathways from gene to disease.
We discuss complementary approaches, e.g. integrating findings across levels of organismal complexity and using bioinformatic, cell and animal models.
ACKNOWLEDGEMENTS
Funding: This works was supported by the Netherlands Organization for Scientific Research (NWO), i.e. the NWO Brain & Cognition Excellence Program (grant 433-09-229) and the Vici Innovation Program (grant 016-130-669 to BF). Additional support was received from the European Community’s Seventh Framework Programme (FP7/2007 – 2013) under grant agreements n° 602805 (Aggressotype), n° 602450 (IMAGEMEND), and n° 278948 (TACTICS) as well as from the European Community’s Horizon 2020 Programme (H2020/2014 – 2020) under grant agreements n° 643051 (MiND) and n° 667302 (CoCA). The work was also supported by grants for the ENIGMA Consortium (grant number U54 EB020403) from the BD2K Initiative of a cross-NIH partnership, and by the ECNP Network ADHD across the Lifespan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
None of the authors report conflicts of interest. Barbara Franke discloses having received educational speaking fees from Merz and Shire.
REFERENCES
- AGNEW-BLAIS JC, POLANCZYK GV, DANESE A, WERTZ J, MOFFITT TE & ARSENEAULT L (2016). Evaluation of the Persistence, Remission, and Emergence of Attention-Deficit/Hyperactivity Disorder in Young Adulthood. JAMA Psychiatry, 73, 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AHMAD RF, MALIK AS, KAMEL N, REZA F & ABDULLAH JM (2016). Simultaneous EEG-fMRI for working memory of the human brain. Australas Phys Eng Sci Med, 39, 363–378. [DOI] [PubMed] [Google Scholar]
- AKUTAGAVA-MARTINS GC, SALATINO-OLIVEIRA A, GENRO JP, CONTINI V, POLANCZYK G, ZENI C, CHAZAN R, KIELING C, ANSELMI L, MENEZES AM, GREVET EH, BAU CH, ROHDE LA & HUTZ MH (2014). Glutamatergic copy number variants and their role in attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 165B, 502–509. [DOI] [PubMed] [Google Scholar]
- ALBRECHT B, BRANDEIS D, UEBEL-VON SANDERSLEBEN H, VALKO L, HEINRICH H, XU X, DRECHSLER R, HEISE A, KUNTSI J, MULLER UC, ASHERSON P, STEINHAUSEN HC, ROTHENBERGER A & BANASCHEWSKI T (2014). Genetics of preparation and response control in ADHD: the role of DRD4 and DAT1. J Child Psychol Psychiatry, 55, 914–923. [DOI] [PubMed] [Google Scholar]
- ALDERSON RM, KASPER LJ, HUDEC KL & PATROS CH (2013). Attention-deficit/hyperactivity disorder (ADHD) and working memory in adults: a meta-analytic review. Neuropsychology, 27, 287–302. [DOI] [PubMed] [Google Scholar]
- ALTHAUS M, GROEN Y, WIJERS AA, MINDERAA RB, KEMA IP, DIJCK JD & HOEKSTRA PJ (2010). Variants of the SLC6A3 (DAT1) polymorphism affect performance monitoring-related cortical evoked potentials that are associated with ADHD. Biol Psychol, 85, 19–32. [DOI] [PubMed] [Google Scholar]
- AMERICAN PSYCHIATRIC ASSOCIATION (2013). Diagnostic and Statistical Manual of Mental Disorders: DSM 5: Books4US. [Google Scholar]
- ANANIEV G, WILLIAMS EC, LI H & CHANG Q (2011). Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS One, 6, e25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARAM D, BAZELET I & GOLDMAN H (2010). Early literacy and parental writing mediation in young children with and without ADHD. European Journal of Special Needs Education, 25, 397–412. [Google Scholar]
- ARARAGI N & LESCH KP (2013). Serotonin (5-HT) in the regulation of depression-related emotionality: insight from 5-HT transporter and tryptophan hydroxylase-2 knockout mouse models. Curr Drug Targets, 14, 549–570. [DOI] [PubMed] [Google Scholar]
- ARCOS-BURGOS M, JAIN M, ACOSTA MT, SHIVELY S, STANESCU H, WALLIS D, DOMENE S, VELEZ JI, KARKERA JD, BALOG J, BERG K, KLETA R, GAHL WA, ROESSLER E, LONG R, LIE J, PINEDA D, LONDONO AC, PALACIO JD, ARBELAEZ A, LOPERA F, ELIA J, HAKONARSON H, JOHANSSON S, KNAPPSKOG PM, HAAVIK J, RIBASES M, CORMAND B, BAYES M, CASAS M, RAMOS-QUIROGA JA, HERVAS A, MAHER BS, FARAONE SV, SEITZ C, FREITAG CM, PALMASON H, MEYER J, ROMANOS M, WALITZA S, HEMMINGER U, WARNKE A, ROMANOS J, RENNER T, JACOB C, LESCH KP, SWANSON J, VORTMEYER A, BAILEY-WILSON JE, CASTELLANOS FX & MUENKE M (2010). A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol Psychiatry, 15, 1053–1066. [DOI] [PubMed] [Google Scholar]
- ARNSTEN AF (2006). Fundamentals of attention-deficit/hyperactivity disorder: circuits and pathways. J Clin Psychiatry, 67 Suppl 8, 7–12. [PubMed] [Google Scholar]
- ARNSTEN AF, STEERE JC & HUNT RD (1996). The contribution of alpha 2-noradrenergic mechanisms of prefrontal cortical cognitive function. Potential significance for attention-deficit hyperactivity disorder. Arch Gen Psychiatry, 53, 448–455. [DOI] [PubMed] [Google Scholar]
- ASGHARI V, SANYAL S, BUCHWALDT S, PATERSON A, JOVANOVIC V & VAN TOL HH (1995). Modulation of intracellular cyclic AMP levels by different human dopamine D4 receptor variants. J Neurochem, 65, 1157–1165. [DOI] [PubMed] [Google Scholar]
- ASHERSON P, BROOKES K, FRANKE B, CHEN W, GILL M, EBSTEIN RP, BUITELAAR J, BANASCHEWSKI T, SONUGA-BARKE E, EISENBERG J, MANOR I, MIRANDA A, OADES RD, ROEYERS H, ROTHENBERGER A, SERGEANT J, STEINHAUSEN HC & FARAONE SV (2007). Confirmation that a specific haplotype of the dopamine transporter gene is associated with combined-type ADHD. Am J Psychiatry, 164, 674–677. [DOI] [PubMed] [Google Scholar]
- BAEHNE CG, EHLIS AC, PLICHTA MM, CONZELMANN A, PAULI P, JACOB C, GUTKNECHT L, LESCH KP & FALLGATTER AJ (2009). Tph2 gene variants modulate response control processes in adult ADHD patients and healthy individuals. Mol Psychiatry, 14, 1032–1039. [DOI] [PubMed] [Google Scholar]
- BALE TL, BARAM TZ, BROWN AS, GOLDSTEIN JM, INSEL TR, MCCARTHY MM, NEMEROFF CB, REYES TM, SIMERLY RB, SUSSER ES & NESTLER EJ (2010). Early life programming and neurodevelopmental disorders. Biol Psychiatry, 68, 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANASCHEWSKI T, BECKER K, SCHERAG S, FRANKE B & COGHILL D (2010). Molecular genetics of attention-deficit/hyperactivity disorder: an overview. Eur Child Adolesc Psychiatry, 19, 237–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANERJEE TD, MIDDLETON F & FARAONE SV (2007). Environmental risk factors for attention-deficit hyperactivity disorder. Acta Paediatr, 96, 1269–1274. [DOI] [PubMed] [Google Scholar]
- BARZMAN D, GEISE C & LIN PI (2015). Review of the genetic basis of emotion dysregulation in children and adolescents. World J Psychiatry, 5, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEDARD AC, SCHULZ KP, COOK EH JR., FAN J, CLERKIN SM, IVANOV I, HALPERIN JM & NEWCORN JH (2010). Dopamine transporter gene variation modulates activation of striatum in youth with ADHD. Neuroimage, 53, 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEDELL VM, WANG Y, CAMPBELL JM, POSHUSTA TL, STARKER CG, KRUG RG 2ND, TAN W, PENHEITER SG, MA AC, LEUNG AY, FAHRENKRUG SC, CARLSON DF, VOYTAS DF, CLARK KJ, ESSNER JJ & EKKER SC (2012). In vivo genome editing using a high-efficiency TALEN system. Nature, 491, 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEN-DAVID E & SHIFMAN S (2012). Networks of neuronal genes affected by common and rare variants in autism spectrum disorders. PLoS Genet, 8, e1002556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERGSON C, MRZLJAK L, SMILEY JF, PAPPY M, LEVENSON R & GOLDMAN-RAKIC PS (1995). Regional, cellular, and subcellular variations in the distribution of D1 and D5 dopamine receptors in primate brain. J Neurosci, 15, 7821–7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIEDERMAN J & FARAONE SV (2005). Attention-deficit hyperactivity disorder. Lancet, 366, 237–248. [DOI] [PubMed] [Google Scholar]
- BLOKLAND GA, MCMAHON KL, THOMPSON PM, HICKIE IB, MARTIN NG, DE ZUBICARAY GI & WRIGHT MJ (2014). Genetic effects on the cerebellar role in working memory: same brain, different genes? Neuroimage, 86, 392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOBB AJ, ADDINGTON AM, SIDRANSKY E, GORNICK MC, LERCH JP, GREENSTEIN DK, CLASEN LS, SHARP WS, INOFF-GERMAIN G, WAVRANT-DE VRIEZE F, ARCOS-BURGOS M, STRAUB RE, HARDY JA, CASTELLANOS FX & RAPOPORT JL (2005). Support for association between ADHD and two candidate genes: NET1 and DRD1. Am J Med Genet B Neuropsychiatr Genet, 134B, 67–72. [DOI] [PubMed] [Google Scholar]
- BOEHLER CN, APPELBAUM LG, KREBS RM, HOPF JM & WOLDORFF MG (2010). Pinning down response inhibition in the brain--conjunction analyses of the Stop-signal task. Neuroimage, 52, 1621–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAET W, JOHNSON KA, TOBIN CT, ACHESON R, MCDONNELL C, HAWI Z, BARRY E, MULLIGAN A, GILL M, BELLGROVE MA, ROBERTSON IH & GARAVAN H (2011). fMRI activation during response inhibition and error processing: the role of the DAT1 gene in typically developing adolescents and those diagnosed with ADHD. Neuropsychologia, 49, 1641–1650. [DOI] [PubMed] [Google Scholar]
- BRAIN DEVELOPMENT COOPERATIVE, G. (2012). Total and regional brain volumes in a population-based normative sample from 4 to 18 years: the NIH MRI Study of Normal Brain Development. Cereb Cortex, 22, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRALTEN J, ARIAS-VASQUEZ A, MAKKINJE R, VELTMAN JA, BRUNNER HG, FERNANDEZ G, RIJPKEMA M & FRANKE B (2011). Association of the Alzheimer’s gene SORL1 with hippocampal volume in young, healthy adults. Am J Psychiatry, 168, 1083–1089. [DOI] [PubMed] [Google Scholar]
- BRALTEN J, FRANKE B, WALDMAN I, ROMMELSE N, HARTMAN C, ASHERSON P, BANASCHEWSKI T, EBSTEIN RP, GILL M, MIRANDA A, OADES RD, ROEYERS H, ROTHENBERGER A, SERGEANT JA, OOSTERLAAN J, SONUGA-BARKE E, STEINHAUSEN HC, FARAONE SV, BUITELAAR JK & ARIAS-VASQUEZ A (2013). Candidate genetic pathways for attention-deficit/hyperactivity disorder (ADHD) show association to hyperactive/impulsive symptoms in children with ADHD. J Am Acad Child Adolesc Psychiatry, 52, 1204–1212 e1201. [DOI] [PubMed] [Google Scholar]
- BRALTEN J, GREVEN CU, FRANKE B, MENNES M, ZWIERS MP, ROMMELSE NN, HARTMAN C, VAN DER MEER D, O’DWYER L, OOSTERLAAN J, HOEKSTRA PJ, HESLENFELD D, ARIAS-VASQUEZ A & BUITELAAR JK (2015). Voxel-based morphometry analysis reveals frontal brain differences in participants with ADHD and their unaffected siblings. J Psychiatry Neurosci, 41, 140377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENNAND K, SAVAS JN, KIM Y, TRAN N, SIMONE A, HASHIMOTO-TORII K, BEAUMONT KG, KIM HJ, TOPOL A, LADRAN I, ABDELRAHIM M, MATIKAINEN-ANKNEY B, CHAO SH, MRKSICH M, RAKIC P, FANG G, ZHANG B, YATES JR 3RD & GAGE FH (2015a). Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry, 20, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENNAND KJ & GAGE FH (2012). Modeling psychiatric disorders through reprogramming. Dis Model Mech, 5, 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENNAND KJ, MARCHETTO MC, BENVENISTY N, BRUSTLE O, EBERT A, IZPISUA BELMONTE JC, KAYKAS A, LANCASTER MA, LIVESEY FJ, MCCONNELL MJ, MCKAY RD, MORROW EM, MUOTRI AR, PANCHISION DM, RUBIN LL, SAWA A, SOLDNER F, SONG H, STUDER L, TEMPLE S, VACCARINO FM, WU J, VANDERHAEGHEN P, GAGE FH & JAENISCH R (2015b). Creating Patient-Specific Neural Cells for the In Vitro Study of Brain Disorders. Stem Cell Reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENNAND KJ, SIMONE A, JOU J, GELBOIN-BURKHART C, TRAN N, SANGAR S, LI Y, MU Y, CHEN G, YU D, MCCARTHY S, SEBAT J & GAGE FH (2011). Modelling schizophrenia using human induced pluripotent stem cells. Nature, 473, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENNAND KJ, SIMONE A, TRAN N & GAGE FH (2012). Modeling psychiatric disorders at the cellular and network levels. Mol Psychiatry, 17, 1239–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROOKES K, XU X, CHEN W, ZHOU K, NEALE B, LOWE N, ANNEY R, FRANKE B, GILL M, EBSTEIN R, BUITELAAR J, SHAM P, CAMPBELL D, KNIGHT J, ANDREOU P, ALTINK M, ARNOLD R, BOER F, BUSCHGENS C, BUTLER L, CHRISTIANSEN H, FELDMAN L, FLEISCHMAN K, FLIERS E, HOWE-FORBES R, GOLDFARB A, HEISE A, GABRIELS I, KORNLUBETZKI I, JOHANSSON L, MARCO R, MEDAD S, MINDERAA R, MULAS F, MULLER U, MULLIGAN A, RABIN K, ROMMELSE N, SETHNA V, SOROHAN J, UEBEL H, PSYCHOGIOU L, WEEKS A, BARRETT R, CRAIG I, BANASCHEWSKI T, SONUGA-BARKE E, EISENBERG J, KUNTSI J, MANOR I, MCGUFFIN P, MIRANDA A, OADES RD, PLOMIN R, ROEYERS H, ROTHENBERGER A, SERGEANT J, STEINHAUSEN HC, TAYLOR E, THOMPSON M, FARAONE SV & ASHERSON P (2006). The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry, 11, 934–953. [DOI] [PubMed] [Google Scholar]
- BROOKES KJ, HAWI Z, PARK J, SCOTT S, GILL M & KENT L (2010). Polymorphisms of the steroid sulfatase (STS) gene are associated with attention deficit hyperactivity disorder and influence brain tissue mRNA expression. Am J Med Genet B Neuropsychiatr Genet, 153B, 1417–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROOKES KJ, NEALE BM, SUGDEN K, KHAN N, ASHERSON P & D’SOUZA UM (2007). Relationship between VNTR polymorphisms of the human dopamine transporter gene and expression in post-mortem midbrain tissue. Am J Med Genet B Neuropsychiatr Genet, 144B, 1070–1078. [DOI] [PubMed] [Google Scholar]
- BROWN AB, BIEDERMAN J, VALERA E, MAKRIS N, DOYLE A, WHITFIELD-GABRIELI S, MICK E, SPENCER T, FARAONE S & SEIDMAN L (2011). Relationship of DAT1 and adult ADHD to task-positive and task-negative working memory networks. Psychiatry Res, 193, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROWN AB, BIEDERMAN J, VALERA EM, DOYLE AE, BUSH G, SPENCER T, MONUTEAUX MC, MICK E, WHITFIELD-GABRIELI S, MAKRIS N, LAVIOLETTE PS, OSCAR-BERMAN M, FARAONE SV & SEIDMAN LJ (2010). Effect of dopamine transporter gene (SLC6A3) variation on dorsal anterior cingulate function in attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 153B, 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRUNNER HG, NELEN M, BREAKEFIELD XO, ROPERS HH & VAN OOST BA (1993). Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science, 262, 578–580. [DOI] [PubMed] [Google Scholar]
- BUITELAAR J, KAN C & ASHERSON P (2011). ADHD in adults; Characterization, diagnosis, and treatment: Cambridge University Press. [Google Scholar]
- BURT SA (2009). Rethinking environmental contributions to child and adolescent psychopathology: a meta-analysis of shared environmental influences. Psychol Bull, 135, 608–637. [DOI] [PubMed] [Google Scholar]
- BUSH G (2011). Cingulate, frontal, and parietal cortical dysfunction in attention-deficit/hyperactivity disorder. Biol Psychiatry, 69, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUSH G, LUU P & POSNER MI (2000). Cognitive and emotional influences in anterior cingulate cortex. Trends Cogn Sci, 4, 215–222. [DOI] [PubMed] [Google Scholar]
- BUTTON KS, IOANNIDIS JP, MOKRYSZ C, NOSEK BA, FLINT J, ROBINSON ES & MUNAFO MR (2013). Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci, 14, 365–376. [DOI] [PubMed] [Google Scholar]
- BYRD AL & MANUCK SB (2014). MAOA, Childhood Maltreatment, and Antisocial Behavior: Meta-analysis of a Gene-Environment Interaction. Biological psychiatry, 75, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAO X, CAO Q, LONG X, SUN L, SUI M, ZHU C, ZUO X, ZANG Y & WANG Y (2009). Abnormal resting-state functional connectivity patterns of the putamen in medication-naive children with attention deficit hyperactivity disorder. Brain Res, 1303, 195–206. [DOI] [PubMed] [Google Scholar]
- CASPI A, MCCLAY J, MOFFITT TE, MILL J, MARTIN J, CRAIG IW, TAYLOR A & POULTON R (2002). Role of genotype in the cycle of violence in maltreated children. Science, 297, 851–854. [DOI] [PubMed] [Google Scholar]
- CASPI A, SUGDEN K, MOFFITT TE, TAYLOR A, CRAIG IW, HARRINGTON H, MCCLAY J, MILL J, MARTIN J, BRAITHWAITE A & POULTON R (2003). Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science, 301, 386–389. [DOI] [PubMed] [Google Scholar]
- CASTELLANOS FX, LAU E, TAYEBI N, LEE P, LONG RE, GIEDD JN, SHARP W, MARSH WL, WALTER JM, HAMBURGER SD, GINNS EI, RAPOPORT JL & SIDRANSKY E (1998). Lack of an association between a dopamine-4 receptor polymorphism and attention-deficit/hyperactivity disorder: genetic and brain morphometric analyses. Mol Psychiatry, 3, 431–434. [DOI] [PubMed] [Google Scholar]
- CASTELLANOS FX, LEE PP, SHARP W, JEFFRIES NO, GREENSTEIN DK, CLASEN LS, BLUMENTHAL JD, JAMES RS, EBENS CL, WALTER JM, ZIJDENBOS A, EVANS AC, GIEDD JN & RAPOPORT JL (2002). Developmental trajectories of brain volume abnormalities in children and adolescents with attention-deficit/hyperactivity disorder. JAMA, 288, 1740–1748. [DOI] [PubMed] [Google Scholar]
- CASTELLANOS FX, MARGULIES DS, KELLY C, UDDIN LQ, GHAFFARI M, KIRSCH A, SHAW D, SHEHZAD Z, DI MARTINO A, BISWAL B, SONUGA-BARKE EJ, ROTROSEN J, ADLER LA & MILHAM MP (2008). Cingulate-precuneus interactions: a new locus of dysfunction in adult attention-deficit/hyperactivity disorder. Biol Psychiatry, 63, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAYE A, ROCHA TB, ANSELMI L, MURRAY J, MENEZES AM, BARROS FC, GONCALVES H, WEHRMEISTER F, JENSEN CM, STEINHAUSEN HC, SWANSON JM, KIELING C & ROHDE LA (2016). Attention-Deficit/Hyperactivity Disorder Trajectories From Childhood to Young Adulthood: Evidence From a Birth Cohort Supporting a Late-Onset Syndrome. JAMA Psychiatry, 73, 705–712. [DOI] [PubMed] [Google Scholar]
- CHANG FM, KIDD JR, LIVAK KJ, PAKSTIS AJ & KIDD KK (1996). The world-wide distribution of allele frequencies at the human dopamine D4 receptor locus. Hum Genet, 98, 91–101. [DOI] [PubMed] [Google Scholar]
- CHANG Z, LICHTENSTEIN P, ASHERSON PJ & LARSSON H (2013). Developmental twin study of attention problems: high heritabilities throughout development. JAMA Psychiatry, 70, 311–318. [DOI] [PubMed] [Google Scholar]
- CHEN J, LIPSKA BK, HALIM N, MA QD, MATSUMOTO M, MELHEM S, KOLACHANA BS, HYDE TM, HERMAN MM, APUD J, EGAN MF, KLEINMAN JE & WEINBERGER DR (2004). Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet, 75, 807–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN J, ZACHAREK A, LI Y, LI A, WANG L, KATAKOWSKI M, ROBERTS C, LU M & CHOPP M (2006). N-cadherin mediates nitric oxide-induced neurogenesis in young and retired breeder neurospheres. Neuroscience, 140, 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN JA, PENAGARIKANO O, BELGARD TG, SWARUP V & GESCHWIND DH (2015). The emerging picture of autism spectrum disorder: genetics and pathology. Annu Rev Pathol, 10, 111–144. [DOI] [PubMed] [Google Scholar]
- CHEN W, ZHOU K, SHAM P, FRANKE B, KUNTSI J, CAMPBELL D, FLEISCHMAN K, KNIGHT J, ANDREOU P, ARNOLD R, ALTINK M, BOER F, BOHOLST MJ, BUSCHGENS C, BUTLER L, CHRISTIANSEN H, FLIERS E, HOWE-FORBES R, GABRIELS I, HEISE A, KORN-LUBETZKI I, MARCO R, MEDAD S, MINDERAA R, MULLER UC, MULLIGAN A, PSYCHOGIOU L, ROMMELSE N, SETHNA V, UEBEL H, MCGUFFIN P, PLOMIN R, BANASCHEWSKI T, BUITELAAR J, EBSTEIN R, EISENBERG J, GILL M, MANOR I, MIRANDA A, MULAS F, OADES RD, ROEYERS H, ROTHENBERGER A, SERGEANT J, SONUGA-BARKE E, STEINHAUSEN HC, TAYLOR E, THOMPSON M, FARAONE SV & ASHERSON P (2008). DSM-IV combined type ADHD shows familial association with sibling trait scores: a sampling strategy for QTL linkage. Am J Med Genet B Neuropsychiatr Genet, 147B, 1450–1460. [DOI] [PubMed] [Google Scholar]
- CHEON KA, RYU YH, KIM JW & CHO DY (2005). The homozygosity for 10-repeat allele at dopamine transporter gene and dopamine transporter density in Korean children with attention deficit hyperactivity disorder: relating to treatment response to methylphenidate. Eur Neuropsychopharmacol, 15, 95–101. [DOI] [PubMed] [Google Scholar]
- CHIANG CH, SU Y, WEN Z, YORITOMO N, ROSS CA, MARGOLIS RL, SONG H & MING GL (2011). Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry, 16, 358–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOI SM, KIM Y, SHIM JS, PARK JT, WANG RH, LEACH SD, LIU JO, DENG C, YE Z & JANG YY (2013). Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology, 57, 2458–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CILIAX BJ, DRASH GW, STALEY JK, HABER S, MOBLEY CJ, MILLER GW, MUFSON EJ, MASH DC & LEVEY AI (1999). Immunocytochemical localization of the dopamine transporter in human brain. J Comp Neurol, 409, 38–56. [DOI] [PubMed] [Google Scholar]
- CIVELEK M & LUSIS AJ (2014). Systems genetics approaches to understand complex traits. Nat Rev Genet, 15, 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLARKE AR, BARRY RJ, DUPUY FE, HECKEL LD, MCCARTHY R, SELIKOWITZ M & JOHNSTONE SJ (2011). Behavioural differences between EEG-defined subgroups of children with Attention-Deficit/Hyperactivity Disorder. Clin Neurophysiol, 122, 1333–1341. [DOI] [PubMed] [Google Scholar]
- CORRAL-FRIAS NS, PIZZAGALLI DA, CARRE J, MICHALSKI LJ, NIKOLOVA YS, PERLIS RH, FAGERNESS J, LEE MR, DRABANT CONLEY E, LANCASTER TM, HADDAD S, WOLF A, SMOLLER JW, HARIRI AR & BOGDAN R (2016). COMT Val Met genotype is associated with reward learning: A replication study and meta-analysis. Genes Brain Behav. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CORTESE S, KELLY C, CHABERNAUD C, PROAL E, DI MARTINO A, MILHAM MP & CASTELLANOS FX (2012). Toward systems neuroscience of ADHD: a meta-analysis of 55 fMRI studies. Am J Psychiatry, 169, 1038–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRISTINO AS, WILLIAMS SM, HAWI Z, AN JY, BELLGROVE MA, SCHWARTZ CE, COSTA LDA F & CLAUDIANOS C (2014). Neurodevelopmental and neuropsychiatric disorders represent an interconnected molecular system. Mol Psychiatry, 19, 294–301. [DOI] [PubMed] [Google Scholar]
- CROSS-DISORDER GROUP OF THE PSYCHIATRIC GENOMICS CONSORTIUM (2013). Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet, 381, 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRUCEANU C, AMBALAVANAN A, SPIEGELMAN D, GAUTHIER J, LAFRENIERE RG, DION PA, ALDA M, TURECKI G & ROULEAU GA (2013). Family-based exome-sequencing approach identifies rare susceptibility variants for lithium-responsive bipolar disorder. Genome, 56, 634–640. [DOI] [PubMed] [Google Scholar]
- CUKIER HN, DUEKER ND, SLIFER SH, LEE JM, WHITEHEAD PL, LALANNE E, LEYVA N, KONIDARI I, GENTRY RC, HULME WF, BOOVEN DV, MAYO V, HOFMANN NK, SCHMIDT MA, MARTIN ER, HAINES JL, CUCCARO ML, GILBERT JR & PERICAK-VANCE MA (2014). Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol Autism, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE LA GARZA R 2ND & MADRAS BK (2000). [(3)H]PNU-101958, a D(4) dopamine receptor probe, accumulates in prefrontal cortex and hippocampus of non-human primate brain. Synapse, 37, 232–244. [DOI] [PubMed] [Google Scholar]
- DECKERT J, CATALANO M, SYAGAILO YV, BOSI M, OKLADNOVA O, DI BELLA D, NOTHEN MM, MAFFEI P, FRANKE P, FRITZE J, MAIER W, PROPPING P, BECKMANN H, BELLODI L & LESCH KP (1999). Excess of high activity monoamine oxidase A gene promoter alleles in female patients with panic disorder. Hum Mol Genet, 8, 621–624. [DOI] [PubMed] [Google Scholar]
- DEL CAMPO N, MULLER U & SAHAKIAN BJ (2012). Neural and behavioral endophenotypes in ADHD. Curr Top Behav Neurosci, 11, 65–91. [DOI] [PubMed] [Google Scholar]
- DEMONTIS D, LESCAI F, BORGLUM A, GLERUP S, OSTERGAARD SD, MORS O, LI Q, LIANG J, JIANG H, LI Y, WANG J, LESCH KP, REIF A, BUITELAAR JK & FRANKE B (2016). Whole-Exome Sequencing Reveals Increased Burden of Rare Functional and Disruptive Variants in Candidate Risk Genes in Individuals With Persistent Attention-Deficit/Hyperactivity Disorder. J Am Acad Child Adolesc Psychiatry, 55, 521–523. [DOI] [PubMed] [Google Scholar]
- DIMATELIS JJ, HSIEH JH, STERLEY TL, MARAIS L, WOMERSLEY JS, VLOK M & RUSSELL VA (2015). Impaired Energy Metabolism and Disturbed Dopamine and Glutamate Signalling in the Striatum and Prefrontal Cortex of the Spontaneously Hypertensive Rat Model of Attention-Deficit Hyperactivity Disorder. J Mol Neurosci, 56, 696–707. [DOI] [PubMed] [Google Scholar]
- DING Q, LEE YK, SCHAEFER EA, PETERS DT, VERES A, KIM K, KUPERWASSER N, MOTOLA DL, MEISSNER TB, HENDRIKS WT, TREVISAN M, GUPTA RM, MOISAN A, BANKS E, FRIESEN M, SCHINZEL RT, XIA F, TANG A, XIA Y, FIGUEROA E, WANN A, AHFELDT T, DAHERON L, ZHANG F, RUBIN LL, PENG LF, CHUNG RT, MUSUNURU K & COWAN CA (2013). A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell, 12, 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOMENE S, STANESCU H, WALLIS D, TINLOY B, PINEDA DE, KLETA R, ARCOS-BURGOS M, ROESSLER E & MUENKE M (2011). Screening of human LPHN3 for variants with a potential impact on ADHD susceptibility. Am J Med Genet B Neuropsychiatr Genet, 156B, 11–18. [DOI] [PubMed] [Google Scholar]
- DOUCETTE-STAMM LA, BLAKELY DJ, TIAN J, MOCKUS S & MAO JI (1995). Population genetic study of the human dopamine transporter gene (DAT1). Genet Epidemiol, 12, 303–308. [DOI] [PubMed] [Google Scholar]
- DOYLE AE, FARAONE SV, SEIDMAN LJ, WILLCUTT EG, NIGG JT, WALDMAN ID, PENNINGTON BF, PEART J & BIEDERMAN J (2005). Are endophenotypes based on measures of executive functions useful for molecular genetic studies of ADHD? J Child Psychol Psychiatry, 46, 774–803. [DOI] [PubMed] [Google Scholar]
- DRAMSDAHL M, WESTERHAUSEN R, HAAVIK J, HUGDAHL K & PLESSEN KJ (2012). Adults with attention-deficit/hyperactivity disorder - a diffusion-tensor imaging study of the corpus callosum. Psychiatry Res, 201, 168–173. [DOI] [PubMed] [Google Scholar]
- DRESLER T, BARTH B, ETHOFER T, LESCH K-P, EHLIS A-C & FALLGATTER AJ (2014). Imaging genetics in adult attention-deficit/hyperactivity disorder (ADHD): a way towards pathophysiological understanding? Borderline Personality Disorder and Emotion Dysregulation, 1, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRESLER T, EHLIS AC, HEINZEL S, RENNER TJ, REIF A, BAEHNE CG, HEINE M, BOREATTIHUMMER A, JACOB CP, LESCH KP & FALLGATTER AJ (2010). Dopamine transporter (SLC6A3) genotype impacts neurophysiological correlates of cognitive response control in an adult sample of patients with ADHD. Neuropsychopharmacology, 35, 2193–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRGON T, LIN Z, WANG GJ, FOWLER J, PABLO J, MASH DC, VOLKOW N & UHL GR (2006). Common human 5’ dopamine transporter (SLC6A3) haplotypes yield varying expression levels in vivo. Cell Mol Neurobiol, 26, 875–889. [DOI] [PubMed] [Google Scholar]
- DUAN J (2015). Path from schizophrenia genomics to biology: gene regulation and perturbation in neurons derived from induced pluripotent stem cells and genome editing. Neurosci Bull, 31, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DURSTON S (2003). A review of the biological bases of ADHD: what have we learned from imaging studies? Ment Retard Dev Disabil Res Rev, 9, 184–195. [DOI] [PubMed] [Google Scholar]
- DURSTON S (2010). Imaging genetics in ADHD. Neuroimage, 53, 832–838. [DOI] [PubMed] [Google Scholar]
- DURSTON S, DE ZEEUW P & STAAL WG (2009). Imaging genetics in ADHD: a focus on cognitive control. Neurosci Biobehav Rev, 33, 674–689. [DOI] [PubMed] [Google Scholar]
- DURSTON S, FOSSELLA JA, CASEY BJ, HULSHOFF POL HE, GALVAN A, SCHNACK HG, STEENHUIS MP, MINDERAA RB, BUITELAAR JK, KAHN RS & VAN ENGELAND H (2005). Differential effects of DRD4 and DAT1 genotype on fronto-striatal gray matter volumes in a sample of subjects with attention deficit hyperactivity disorder, their unaffected siblings, and controls. Mol Psychiatry, 10, 678–685. [DOI] [PubMed] [Google Scholar]
- DURSTON S, FOSSELLA JA, MULDER MJ, CASEY BJ, ZIERMANS TB, VESSAZ MN & VAN ENGELAND H (2008). Dopamine transporter genotype conveys familial risk of attention-deficit/hyperactivity disorder through striatal activation. J Am Acad Child Adolesc Psychiatry, 47, 61–67. [DOI] [PubMed] [Google Scholar]
- EHLIS AC, BAHNE CG, JACOB CP, HERRMANN MJ & FALLGATTER AJ (2008). Reduced lateral prefrontal activation in adult patients with attention-deficit/hyperactivity disorder (ADHD) during a working memory task: a functional near-infrared spectroscopy (fNIRS) study. J Psychiatr Res, 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- EHLIS AC, SCHNEIDER S, DRESLER T & FALLGATTER AJ (2014). Application of functional near-infrared spectroscopy in psychiatry. Neuroimage, 85 Pt 1, 478–488. [DOI] [PubMed] [Google Scholar]
- EISENBERG J, MEI-TAL G, STEINBERG A, TARTAKOVSKY E, ZOHAR A, GRITSENKO I, NEMANOV L & EBSTEIN RP (1999). Haplotype relative risk study of catechol-O-methyltransferase (COMT) and attention deficit hyperactivity disorder (ADHD): association of the high-enzyme activity Val allele with ADHD impulsive-hyperactive phenotype. Am J Med Genet, 88, 497–502. [PubMed] [Google Scholar]
- ELIA J, GAI X, XIE HM, PERIN JC, GEIGER E, GLESSNER JT, D’ARCY M, DEBERARDINIS R, FRACKELTON E, KIM C, LANTIERI F, MUGANGA BM, WANG L, TAKEDA T, RAPPAPORT EF, GRANT SF, BERRETTINI W, DEVOTO M, SHAIKH TH, HAKONARSON H & WHITE PS (2010). Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry, 15, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELIA J, GLESSNER JT, WANG K, TAKAHASHI N, SHTIR CJ, HADLEY D, SLEIMAN PM, ZHANG H, KIM CE, ROBISON R, LYON GJ, FLORY JH, BRADFIELD JP, IMIELINSKI M, HOU C, FRACKELTON EC, CHIAVACCI RM, SAKURAI T, RABIN C, MIDDLETON FA, THOMAS KA, GARRIS M, MENTCH F, FREITAG CM, STEINHAUSEN HC, TODOROV AA, REIF A, ROTHENBERGER A, FRANKE B, MICK EO, ROEYERS H, BUITELAAR J, LESCH KP, BANASCHEWSKI T, EBSTEIN RP, MULAS F, OADES RD, SERGEANT J, SONUGABARKE E, RENNER TJ, ROMANOS M, ROMANOS J, WARNKE A, WALITZA S, MEYER J, PALMASON H, SEITZ C, LOO SK, SMALLEY SL, BIEDERMAN J, KENT L, ASHERSON P, ANNEY RJ, GAYNOR JW, SHAW P, DEVOTO M, WHITE PS, GRANT SF, BUXBAUM JD, RAPOPORT JL, WILLIAMS NM, NELSON SF, FARAONE SV & HAKONARSON H (2012). Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet, 44, 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLISON-WRIGHT I, ELLISON-WRIGHT Z & BULLMORE E (2008). Structural brain change in Attention Deficit Hyperactivity Disorder identified by meta-analysis. BMC Psychiatry, 8, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FALLGATTER AJ, BARTSCH AJ & HERRMANN MJ (2002). Electrophysiological measurements of anterior cingulate function. J Neural Transm (Vienna), 109, 977–988. [DOI] [PubMed] [Google Scholar]
- FALLGATTER AJ, EHLIS AC, DRESLER T, REIF A, JACOB CP, ARCOS-BURGOS M, MUENKE M & LESCH KP (2013). Influence of a latrophilin 3 (LPHN3) risk haplotype on event-related potential measures of cognitive response control in attention-deficit hyperactivity disorder (ADHD). Eur Neuropsychopharmacol, 23, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FALLGATTER AJ, EHLIS AC, ROSLER M, STRIK WK, BLOCHER D & HERRMANN MJ (2005). Diminished prefrontal brain function in adults with psychopathology in childhood related to attention deficit hyperactivity disorder. Psychiatry Res, 138, 157–169. [DOI] [PubMed] [Google Scholar]
- FALLGATTER AJ & MULLER TJ (2001). Electrophysiological signs of reduced prefrontal response control in schizophrenic patients. Psychiatry Res, 107, 19–28. [DOI] [PubMed] [Google Scholar]
- FALLGATTER AJ & STRIK WK (1999). The NoGo-anteriorization as a neurophysiological standard-index for cognitive response control. Int J Psychophysiol, 32, 233–238. [DOI] [PubMed] [Google Scholar]
- FARAONE SV, ASHERSON P, BANASCHEWSKI T, BIEDERMAN J, BUITELAAR JK, RAMOSQUIROGA JA, ROHDE LA, SONUGA-BARKE EJS, TANNOCK R & FRANKE B (2015). Attention-deficit/hyperactivity disorder. Nature Reviews Disease Primers, 1, 15020. [DOI] [PubMed] [Google Scholar]
- FARAONE SV & BIEDERMAN J (2016). CAn attention-deficit/hyperactivity disorder onset occur in adulthood? JAMA Psychiatry, 73, 655–656. [DOI] [PubMed] [Google Scholar]
- FARAONE SV, BIEDERMAN J & MICK E (2006). The age-dependent decline of attention deficit hyperactivity disorder: a meta-analysis of follow-up studies. Psychol Med, 36, 159–165. [DOI] [PubMed] [Google Scholar]
- FARAONE SV, BONVICINI C & SCASSELLATI C (2014a). Biomarkers in the diagnosis of ADHD--promising directions. Curr Psychiatry Rep, 16, 497. [DOI] [PubMed] [Google Scholar]
- FARAONE SV & MICK E (2010). Molecular genetics of attention deficit hyperactivity disorder. Psychiatr Clin North Am, 33, 159–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FARAONE SV, PERLIS RH, DOYLE AE, SMOLLER JW, GORALNICK JJ, HOLMGREN MA & SKLAR P (2005). Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry, 57, 1313–1323. [DOI] [PubMed] [Google Scholar]
- FARAONE SV, SPENCER TJ, MADRAS BK, ZHANG-JAMES Y & BIEDERMAN J (2014b). Functional effects of dopamine transporter gene genotypes on in vivo dopamine transporter functioning: a meta-analysis. Mol Psychiatry, 19, 880–889. [DOI] [PubMed] [Google Scholar]
- FERNANDEZ-JAEN A, ALBERT J, FERNANDEZ-MAYORALAS DM, LOPEZ-MARTIN S, FERNANDEZPERRONE AL, JIMENEZ DE LA PENA M, CALLEJA-PEREZ B, RECIO RODRIGUEZ M & LOPEZ ARRIBAS S (2016). Cingulate Cortical Thickness and Dopamine Transporter (DAT1) Genotype in Children and Adolescents With ADHD. J Atten Disord. [DOI] [PubMed] [Google Scholar]
- FERNANDEZ-JAEN A, LOPEZ-MARTIN S, ALBERT J, FERNANDEZ-MAYORALAS DM, FERNANDEZPERRONE AL, DE LA PENA MJ, CALLEJA-PEREZ B, RODRIGUEZ MR, LOPEZ-ARRIBAS S & MUNOZ-JARENO N (2015). Cortical thickness differences in the prefrontal cortex in children and adolescents with ADHD in relation to dopamine transporter (DAT1) genotype. Psychiatry Res, 233, 409–417. [DOI] [PubMed] [Google Scholar]
- FRANCES A (2000). Diagnostic and statistical manual of mental disorders, DSM-IV-TR, Washington DC: American Psychiatric Association. [Google Scholar]
- FRANKE B, FARAONE SV, ASHERSON P, BUITELAAR J, BAU CH, RAMOS-QUIROGA JA, MICK E, GREVET EH, JOHANSSON S, HAAVIK J, LESCH KP, CORMAND B, REIF A & INTERNATIONAL MULTICENTRE PERSISTENT, A. C. (2012). The genetics of attention deficit/hyperactivity disorder in adults, a review. Mol Psychiatry, 17, 960–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKE B, NEALE BM & FARAONE SV (2009). Genome-wide association studies in ADHD. Hum Genet, 126, 13–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKE B, STEIN JL, RIPKE S, ANTTILA V, HIBAR DP, VAN HULZEN KJ, ARIAS-VASQUEZ A, SMOLLER JW, NICHOLS TE, NEALE MC, MCINTOSH AM, LEE P, MCMAHON FJ, MEYER-LINDENBERG A, MATTHEISEN M, ANDREASSEN OA, GRUBER O, SACHDEV PS, ROIZ-SANTIANEZ R, SAYKIN AJ, EHRLICH S, MATHER KA, TURNER JA, SCHWARZ E, THALAMUTHU A, YAO Y, HO YY, MARTIN NG, WRIGHT MJ, SCHIZOPHRENIA WORKING GROUP OF THE PSYCHIATRIC GENOMICS, C., PSYCHOSIS ENDOPHENOTYPES INTERNATIONAL, C., WELLCOME TRUST CASE CONTROL, C., ENIGMA, C., O’DONOVAN MC, THOMPSON PM, NEALE BM, MEDLAND SE & SULLIVAN PF (2016). Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept. Nat Neurosci, 19, 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKE B, VASQUEZ AA, JOHANSSON S, HOOGMAN M, ROMANOS J, BOREATTI-HUMMER A, HEINE M, JACOB CP, LESCH KP, CASAS M, RIBASES M, BOSCH R, SANCHEZ-MORA C, GOMEZ-BARROS N, FERNANDEZ-CASTILLO N, BAYES M, HALMOY A, HALLELAND H, LANDAAS ET, FASMER OB, KNAPPSKOG PM, HEISTER AJ, KIEMENEY LA, KOOIJ JJ, BOONSTRA AM, KAN CC, ASHERSON P, FARAONE SV, BUITELAAR JK, HAAVIK J, CORMAND B, RAMOS-QUIROGA JA & REIF A (2010). Multicenter analysis of the SLC6A3/DAT1 VNTR haplotype in persistent ADHD suggests differential involvement of the gene in childhood and persistent ADHD. Neuropsychopharmacology, 35, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKLE WG, HUANG Y, HWANG DR, TALBOT PS, SLIFSTEIN M, VAN HEERTUM R, ABIDARGHAM A & LARUELLE M (2004). Comparative evaluation of serotonin transporter radioligands 11C-DASB and 11C-McN 5652 in healthy humans. J Nucl Med, 45, 682–694. [PubMed] [Google Scholar]
- FRANZEN JD, HEINRICHS-GRAHAM E, WHITE ML, WETZEL MW, KNOTT NL & WILSON TW (2013). Atypical coupling between posterior regions of the default mode network in attention-deficit/hyperactivity disorder: a pharmaco-magnetoencephalography study. J Psychiatry Neurosci, 38, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRODL T & SKOKAUSKAS N (2012). Meta-analysis of structural MRI studies in children and adults with attention deficit hyperactivity disorder indicates treatment effects. Acta Psychiatr Scand, 125, 114–126. [DOI] [PubMed] [Google Scholar]
- FUSAR-POLI P, RUBIA K, ROSSI G, SARTORI G & BALOTTIN U (2012). Striatal dopamine transporter alterations in ADHD: pathophysiology or adaptation to psychostimulants? A meta-analysis. Am J Psychiatry, 169, 264–272. [DOI] [PubMed] [Google Scholar]
- GAINETDINOV RR, JONES SR & CARON MG (1999). Functional hyperdopaminergia in dopamine transporter knock-out mice. Biol Psychiatry, 46, 303–311. [DOI] [PubMed] [Google Scholar]
- GALLO EF & POSNER J (2016). Moving towards causality in attention-deficit hyperactivity disorder: overview of neural and genetic mechanisms. Lancet Psychiatry, 3, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAO J, JIA M, QIAO D, QIU H, SOKOLOVE J, ZHANG J & PAN Z (2016). TPH2 gene polymorphisms and bipolar disorder: A meta-analysis. Am J Med Genet B Neuropsychiatr Genet, 171, 145–152. [DOI] [PubMed] [Google Scholar]
- GAO J, PAN Z, JIAO Z, LI F, ZHAO G, WEI Q, PAN F & EVANGELOU E (2012). TPH2 gene polymorphisms and major depression--a meta-analysis. PLoS One, 7, e36721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GERVAIN J, MEHLER J, WERKER JF, NELSON CA, CSIBRA G, LLOYD-FOX S, SHUKLA M & ASLIN RN (2011). Near-infrared spectroscopy: a report from the McDonnell infant methodology consortium. Dev Cogn Neurosci, 1, 22–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GILLBERG C, GILLBERG IC, RASMUSSEN P, KADESJO B, SODERSTROM H, RASTAM M, JOHNSON M, ROTHENBERGER A & NIKLASSON L (2004). Co-existing disorders in ADHD -- implications for diagnosis and intervention. Eur Child Adolesc Psychiatry, 13 Suppl 1, I80–92. [DOI] [PubMed] [Google Scholar]
- GILMAN SR, IOSSIFOV I, LEVY D, RONEMUS M, WIGLER M & VITKUP D (2011). Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron, 70, 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIROS B, JABER M, JONES SR, WIGHTMAN RM & CARON MG (1996). Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature, 379, 606–612. [DOI] [PubMed] [Google Scholar]
- GIZER IR, FICKS C & WALDMAN ID (2009). Candidate gene studies of ADHD: a meta-analytic review. Hum Genet, 126, 51–90. [DOI] [PubMed] [Google Scholar]
- GLAHN DC, WINKLER AM, KOCHUNOV P, ALMASY L, DUGGIRALA R, CARLESS MA, CURRAN JC, OLVERA RL, LAIRD AR, SMITH SM, BECKMANN CF, FOX PT & BLANGERO J (2010). Genetic control over the resting brain. Proc Natl Acad Sci U S A, 107, 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONZALEZ-CASTRO TB, HERNANDEZ-DIAZ Y, JUAREZ-ROJOP IE, LOPEZ-NARVAEZ ML, TOVILLA-ZARATE CA & FRESAN A (2016). The Role of a Catechol-O-Methyltransferase (COMT) Val158Met Genetic Polymorphism in Schizophrenia: A Systematic Review and Updated Meta-analysis on 32,816 Subjects. Neuromolecular Med. [DOI] [PubMed] [Google Scholar]
- GORDON EM, DEVANEY JM, BEAN S & VAIDYA CJ (2015). Resting-state striato-frontal functional connectivity is sensitive to DAT1 genotype and predicts executive function. Cereb Cortex, 25, 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORE A, LI Z, FUNG HL, YOUNG JE, AGARWAL S, ANTOSIEWICZ-BOURGET J, CANTO I, GIORGETTI A, ISRAEL MA, KISKINIS E, LEE JH, LOH YH, MANOS PD, MONTSERRAT N, PANOPOULOS AD, RUIZ S, WILBERT ML, YU J, KIRKNESS EF, IZPISUA BELMONTE JC, ROSSI DJ, THOMSON JA, EGGAN K, DALEY GQ, GOLDSTEIN LS & ZHANG K (2011). Somatic coding mutations in human induced pluripotent stem cells. Nature, 471, 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOTTESMAN II & GOULD TD (2003). The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry, 160, 636–645. [DOI] [PubMed] [Google Scholar]
- GRAZIANO PA, MCNAMARA JP, GEFFKEN GR & REID A (2011). Severity of children’s ADHD symptoms and parenting stress: a multiple mediation model of self-regulation. J Abnorm Child Psychol, 39, 1073–1083. [DOI] [PubMed] [Google Scholar]
- GREVEN CU, BRALTEN J, MENNES M, O’DWYER L, VAN HULZEN KJ, ROMMELSE N, SCHWEREN LJ, HOEKSTRA PJ, HARTMAN CA, HESLENFELD D, OOSTERLAAN J, FARAONE SV, FRANKE B, ZWIERS MP, ARIAS-VASQUEZ A & BUITELAAR JK (2015). Developmentally stable whole-brain volume reductions and developmentally sensitive caudate and putamen volume alterations in those with attention-deficit/hyperactivity disorder and their unaffected siblings. JAMA Psychiatry, 72, 490–499. [DOI] [PubMed] [Google Scholar]
- HAAVIK J, HALMOY A, LUNDERVOLD AJ & FASMER OB (2010). Clinical assessment and diagnosis of adults with attention-deficit/hyperactivity disorder. Expert Rev Neurother, 10, 1569–1580. [DOI] [PubMed] [Google Scholar]
- HAN JY, KWON HJ, HA M, PAIK KC, LIM MH, GYU LEE S, YOO SJ & KIM EJ (2015). The effects of prenatal exposure to alcohol and environmental tobacco smoke on risk for ADHD: A large population-based study. Psychiatry Res, 225, 164–168. [DOI] [PubMed] [Google Scholar]
- HART H, RADUA J, MATAIX-COLS D & RUBIA K (2012). Meta-analysis of fMRI studies of timing in attention-deficit hyperactivity disorder (ADHD). Neurosci Biobehav Rev, 36, 2248–2256. [DOI] [PubMed] [Google Scholar]
- HART H, RADUA J, NAKAO T, MATAIX-COLS D & RUBIA K (2013). Meta-analysis of functional magnetic resonance imaging studies of inhibition and attention in attention-deficit/hyperactivity disorder: exploring task-specific, stimulant medication, and age effects. JAMA Psychiatry, 70, 185–198. [DOI] [PubMed] [Google Scholar]
- HAWI Z, CUMMINS TD, TONG J, JOHNSON B, LAU R, SAMARRAI W & BELLGROVE MA (2015). The molecular genetic architecture of attention deficit hyperactivity disorder. Mol Psychiatry. [DOI] [PubMed] [Google Scholar]
- HAWI Z, DRING M, KIRLEY A, FOLEY D, KENT L, CRADDOCK N, ASHERSON P, CURRAN S, GOULD A, RICHARDS S, LAWSON D, PAY H, TURIC D, LANGLEY K, OWEN M, O’DONOVAN M, THAPAR A, FITZGERALD M & GILL M (2002). Serotonergic system and attention deficit hyperactivity disorder (ADHD): a potential susceptibility locus at the 5-HT(1B) receptor gene in 273 nuclear families from a multi-centre sample. Mol Psychiatry, 7, 718–725. [DOI] [PubMed] [Google Scholar]
- HAWI Z, MATTHEWS N, WAGNER J, WALLACE RH, BUTLER TJ, VANCE A, KENT L, GILL M & BELLGROVE MA (2013). DNA variation in the SNAP25 gene confers risk to ADHD and is associated with reduced expression in prefrontal cortex. PLoS One, 8, e60274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAYES AF & SCHARKOW M (2013). The relative trustworthiness of inferential tests of the indirect effect in statistical mediation analysis: does method really matter? Psychol Sci, 24, 1918–1927. [DOI] [PubMed] [Google Scholar]
- HEINRICHS-GRAHAM E, FRANZEN JD, KNOTT NL, WHITE ML, WETZEL MW & WILSON TW (2014). Pharmaco-MEG evidence for attention related hyper-connectivity between auditory and prefrontal cortices in ADHD. Psychiatry Res, 221, 240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEINZEL S, DRESLER T, BAEHNE CG, HEINE M, BOREATTI-HUMMER A, JACOB CP, RENNER TJ, REIF A, LESCH KP, FALLGATTER AJ & EHLIS AC (2013). COMT × DRD4 epistasis impacts prefrontal cortex function underlying response control. Cereb Cortex, 23, 1453–1462. [DOI] [PubMed] [Google Scholar]
- HENDRIKS WT, WARREN CR & COWAN CA (2016). Genome Editing in Human Pluripotent Stem Cells: Approaches, Pitfalls, and Solutions. Cell Stem Cell, 18, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HESS JL, QUINN TP, AKBARIAN S & GLATT SJ (2015). Bioinformatic analyses and conceptual synthesis of evidence linking ZNF804A to risk for schizophrenia and bipolar disorder. Am J Med Genet B Neuropsychiatr Genet, 168B, 14–35. [DOI] [PubMed] [Google Scholar]
- HEYSER CJ, WILSON MC & GOLD LH (1995). Coloboma hyperactive mutant exhibits delayed neurobehavioral developmental milestones. Brain Res Dev Brain Res, 89, 264–269. [DOI] [PubMed] [Google Scholar]
- HIBAR DP, STEIN JL, KOHANNIM O, JAHANSHAD N, SAYKIN AJ, SHEN L, KIM S, PANKRATZ N, FOROUD T, HUENTELMAN MJ, POTKIN SG, JACK CR JR., WEINER MW, TOGA AW, THOMPSON PM & ALZHEIMER’S DISEASE NEUROIMAGING, I. (2011). Voxelwise gene-wide association study (vGeneWAS): multivariate gene-based association testing in 731 elderly subjects. Neuroimage, 56, 1875–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIBAR DP, STEIN JL, RENTERIA ME, ARIAS-VASQUEZ A, DESRIVIERES S, JAHANSHAD N, TORO R, WITTFELD K, ABRAMOVIC L, ANDERSSON M, ARIBISALA BS, ARMSTRONG NJ, BERNARD M, BOHLKEN MM, BOKS MP, BRALTEN J, BROWN AA, MALLAR CHAKRAVARTY M, CHEN Q, CHING CR, CUELLAR-PARTIDA G, DEN BRABER A, GIDDALURU S, GOLDMAN AL, GRIMM O, GUADALUPE T, HASS J, WOLDEHAWARIAT G, HOLMES AJ, HOOGMAN M, JANOWITZ D, JIA T, KIM S, KLEIN M, KRAEMER B, LEE PH, OLDE LOOHUIS LM, LUCIANO M, MACARE C, MATHER KA, MATTHEISEN M, MILANESCHI Y, NHO K, PAPMEYER M, RAMASAMY A, RISACHER SL, ROIZSANTIANEZ R, ROSE EJ, SALAMI A, SAMANN PG, SCHMAAL L, SCHORK AJ, SHIN J, STRIKE LT, TEUMER A, VAN DONKELAAR MM, VAN EIJK KR, WALTERS RK, WESTLYE LT, WHELAN CD, WINKLER AM, ZWIERS MP, ALHUSAINI S, ATHANASIU L, EHRLICH S, HAKOBJAN MM, HARTBERG CB, HAUKVIK UK, HEISTER AJ, HOEHN D, KASPERAVICIUTE D, LIEWALD DC, LOPEZ LM, MAKKINJE RR, MATARIN M, NABER MA, REESE MCKAY D, NEEDHAM M, NUGENT AC, PUTZ B, ROYLE NA, SHEN L, SPROOTEN E, TRABZUNI D, VAN DER MAREL SS, VAN HULZEN KJ, WALTON E, WOLF C, ALMASY L, AMES D, AREPALLI S, ASSAREH AA, BASTIN ME, BRODATY H, BULAYEVA KB, CARLESS MA, CICHON S, CORVIN A, CURRAN JE, CZISCH M, DE ZUBICARAY GI, DILLMAN A, DUGGIRALA R, DYER TD, ERK S, FEDKO IO, FERRUCCI L, FOROUD TM, FOX PT, FUKUNAGA M, RAPHAEL GIBBS J, GORING HH, GREEN RC, GUELFI S, HANSELL NK, HARTMAN CA, HEGENSCHEID K, HEINZ A, HERNANDEZ DG, HESLENFELD DJ, HOEKSTRA PJ, HOLSBOER F, HOMUTH G, HOTTENGA JJ, IKEDA M, JACK CR JR., JENKINSON M, JOHNSON R, KANAI R, KEIL M, KENT JW JR., KOCHUNOV P, KWOK JB, LAWRIE SM, LIU X, LONGO DL, MCMAHON KL, MEISENZAHL E, MELLE I, MOHNKE S, MONTGOMERY GW, MOSTERT JC, MUHLEISEN TW, NALLS MA, NICHOLS TE, NILSSON LG, NOTHEN MM, OHI K, OLVERA RL, PEREZ-IGLESIAS R, BRUCE PIKE G, POTKIN SG, REINVANG I, REPPERMUND S, RIETSCHEL M, ROMANCZUK-SEIFERTH N, ROSEN GD, RUJESCU D, SCHNELL K, SCHOFIELD PR, SMITH C, STEEN VM, SUSSMANN JE, THALAMUTHU A, TOGA AW, TRAYNOR BJ, TRONCOSO J, TURNER JA, VALDES HERNANDEZ MC, VAN ‘T ENT D, VAN DER BRUG M, VAN DER WEE NJ, VAN TOL MJ, VELTMAN DJ, WASSINK TH, WESTMAN E, ZIELKE RH, ZONDERMAN AB, ASHBROOK DG, HAGER R, LU L, MCMAHON FJ, MORRIS DW, WILLIAMS RW, BRUNNER HG, BUCKNER RL, BUITELAAR JK, CAHN W, CALHOUN VD, CAVALLERI GL, CRESPO-FACORRO B, DALE AM, DAVIES GE, DELANTY N, DEPONDT C, DJUROVIC S, DREVETS WC, ESPESETH T, GOLLUB RL, HO BC, HOFFMANN W, HOSTEN N, KAHN RS, LE HELLARD S, MEYER-LINDENBERG A, MULLER-MYHSOK B, NAUCK M, NYBERG L, PANDOLFO M, PENNINX BW, ROFFMAN JL, SISODIYA SM, SMOLLER JW, VAN BOKHOVEN H, VAN HAREN NE, VOLZKE H, WALTER H, WEINER MW, WEN W, WHITE T, AGARTZ I, ANDREASSEN OA, BLANGERO J, BOOMSMA DI, BROUWER RM, CANNON DM, COOKSON MR, DE GEUS EJ, DEARY IJ, DONOHOE G, FERNANDEZ G, FISHER SE, FRANCKS C, GLAHN DC, GRABE HJ, GRUBER O, HARDY J, HASHIMOTO R, HULSHOFF POL HE, JONSSON EG, KLOSZEWSKA I, LOVESTONE S, MATTAY VS, MECOCCI P, MCDONALD C, MCINTOSH AM, OPHOFF RA, PAUS T, PAUSOVA Z, RYTEN M, SACHDEV PS, SAYKIN AJ, SIMMONS A, SINGLETON A, SOININEN H, WARDLAW JM, WEALE ME, WEINBERGER DR, ADAMS HH, LAUNER LJ, SEILER S, SCHMIDT R, CHAUHAN G, SATIZABAL CL, BECKER JT, YANEK L, VAN DER LEE SJ, EBLING M, FISCHL B, LONGSTRETH WT JR., GREVE D, SCHMIDT H, NYQUIST P, VINKE LN, VAN DUIJN CM, XUE L, MAZOYER B, BIS JC, GUDNASON V, SESHADRI S, ARFAN IKRAM M, THE ALZHEIMER’S DISEASE NEUROIMAGING, I., THE, C. C., EPIGEN, IMAGEN, SYS, MARTIN NG, WRIGHT MJ, SCHUMANN G, FRANKE B, THOMPSON PM & MEDLAND SE (2015). Common genetic variants influence human subcortical brain structures. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HILLMAN EM (2014). Coupling mechanism and significance of the BOLD signal: a status report. Annu Rev Neurosci, 37, 161–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HINNEY A, SCHERAG A, JARICK I, ALBAYRAK O, PUTTER C, PECHLIVANIS S, DAUVERMANN MR, BECK S, WEBER H, SCHERAG S, NGUYEN TT, VOLCKMAR AL, KNOLL N, FARAONE SV, NEALE BM, FRANKE B, CICHON S, HOFFMANN P, NOTHEN MM, SCHREIBER S, JOCKEL KH, WICHMANN HE, FREITAG C, LEMPP T, MEYER J, GILSBACH S, HERPERTZDAHLMANN B, SINZIG J, LEHMKUHL G, RENNER TJ, WARNKE A, ROMANOS M, LESCH KP, REIF A, SCHIMMELMANN BG, HEBEBRAND J & PSYCHIATRIC, G. C. A. S. (2011). Genome-wide association study in German patients with attention deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 156B, 888–897. [DOI] [PubMed] [Google Scholar]
- HOCKEMEYER D & JAENISCH R (2016). Induced Pluripotent Stem Cells Meet Genome Editing. Cell Stem Cell, 18, 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLZ N, BOECKER R, BUCHMANN AF, BLOMEYER D, BAUMEISTER S, HOHMANN S, JENNENSTEINMETZ C, WOLF I, RIETSCHEL M, WITT SH, PLICHTA MM, MEYER-LINDENBERG A, SCHMIDT MH, ESSER G, BANASCHEWSKI T, BRANDEIS D & LAUCHT M (2016). Evidence for a Sex-Dependent MAOAx Childhood Stress Interaction in the Neural Circuitry of Aggression. Cereb Cortex, 26, 904–914. [DOI] [PubMed] [Google Scholar]
- HONG J, SHU-LEONG H, TAO X & LAP-PING Y (1998). Distribution of catechol-O-methyltransferase expression in human central nervous system. Neuroreport, 9, 2861–2864. [DOI] [PubMed] [Google Scholar]
- HONG SB, ZALESKY A, PARK S, YANG YH, PARK MH, KIM B, SONG IC, SOHN CH, SHIN MS, KIM BN, CHO SC & KIM JW (2014). COMT genotype affects brain white matter pathways in attention-deficit/hyperactivity disorder. Hum Brain Mapp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOOGMAN M, AARTS E, ZWIERS M, SLAATS-WILLEMSE D, NABER M, ONNINK M, COOLS R, KAN C, BUITELAAR J & FRANKE B (2011). Nitric oxide synthase genotype modulation of impulsivity and ventral striatal activity in adult ADHD patients and healthy comparison subjects. Am J Psychiatry, 168, 1099–1106. [DOI] [PubMed] [Google Scholar]
- HOOGMAN M, BRALTEN J, HIBAR DP, MENNES M, ZWIERS M, SCHWEREN LJ, VAN HULZEN K, MEDLAND S, SHUMSKAYA E, JAHANSHAD N, DE ZEEUW P, SZEKELY E, SUDRE G, WOLFERS T, ONNINK AM, DAMMERS J, MOSTERT JC, VIVES-GILABERT Y, KOHLS G, OBERWELLAND E, SEITZ J, SCHULTE-RÜTHER M, AMBROSIO DI BRUTTOPILO S, DOYLE A, HOVIK MF, DRAMSDAHL M, TAMM L, VAN ERP TG, DALE AM, SCHORK A, CONZELMANN A, ZIERHUT K, BAUR R, MCCARTHY H, YONCHEVA YN, CUBILLO A, CHANTILUKE K, METHA M, PALOYELIS Y, HOHMANN S, BAUMEISTER S, BRAMATI IE, MATTOS P, TOVAR-MOLL F, DOUGLAS P, BANASCHEWSKI T, BRANDEIS D, KUNTSI J, ASHERSON P, RUBIA K, KELLY C, DI MARTINO A, MILHAM MP, CASTELLANOS FX, FRODL T, ZENTIS M, LESCH KP, REIF A, PAULI P, JERNIGAN T, HAAVIK J, PLESSEN KJ, LUNDERVOLD AJ, HUGDAHL K, SEIDMAN L, BIEDERMAN J, ROMMELSE N, HESLENFELD D, HARTMAN C, HOEKSTRA PJ, OOSTERLAAN J, VAN POLIER G, KONRAD K, VIALARROYA O, RAMOS JA, SOLIVA JC, DURSTON S, BUITELAAR J, FARAONE SV, SHAW P, THOMPSON P & FRANKE B (submitted). Subcortical brain volume differences between participants with ADHD and healthy individuals across the lifespan: an ENIGMA collaboration. [Google Scholar]
- HOOGMAN M, ONNINK M, COOLS R, AARTS E, KAN C, ARIAS VASQUEZ A, BUITELAAR J & FRANKE B (2013). The dopamine transporter haplotype and reward-related striatal responses in adult ADHD. Eur Neuropsychopharmacol, 23, 469–478. [DOI] [PubMed] [Google Scholar]
- HU BY, WEICK JP, YU J, MA LX, ZHANG XQ, THOMSON JA & ZHANG SC (2010). Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A, 107, 4335–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU Y, QU ZY, CAO SY, LI Q, MA L, KRENCIK R, XU M & LIU Y (2016). Directed differentiation of basal forebrain cholinergic neurons from human pluripotent stem cells. J Neurosci Methods, 266, 42–49. [DOI] [PubMed] [Google Scholar]
- HUANG J, ZHONG Z, WANG M, CHEN X, TAN Y, ZHANG S, HE W, HE X, HUANG G, LU H, WU P, CHE Y, YAN YL, POSTLETHWAIT JH, CHEN W & WANG H (2015). Circadian modulation of dopamine levels and dopaminergic neuron development contributes to attention deficiency and hyperactive behavior. J Neurosci, 35, 2572–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HULSHOFF POL HE, SCHNACK HG, MANDL RCW, BRANS RGH, VAN HAREN NEM, BAARÉ WFC, VAN OEL CJ, COLLINS DL, EVANS AC & KAHN RS (2006). Gray and white matter density changes in monozygotic and same-sex dizygotic twins discordant for schizophrenia using voxel-based morphometry. Neuroimage, 31, 482–488. [DOI] [PubMed] [Google Scholar]
- HWANG IW, LIM MH, KWON HJ & JIN HJ (2015). Association of LPHN3 rs6551665 A/G polymorphism with attention deficit and hyperactivity disorder in Korean children. Gene, 566, 68–73. [DOI] [PubMed] [Google Scholar]
- JACOB CP, WEBER H, RETZ W, KITTEL-SCHNEIDER S, HEUPEL J, RENNER T, LESCH KP & REIF A (2013). Acetylcholine-metabolizing butyrylcholinesterase (BCHE) copy number and single nucleotide polymorphisms and their role in attention-deficit/hyperactivity syndrome. J Psychiatr Res, 47, 1902–1908. [DOI] [PubMed] [Google Scholar]
- IACONO WG, MALONE SM, VAIDYANATHAN U & VRIEZE SI (2014). Genome-wide scans of genetic variants for psychophysiological endophenotypes: a methodological overview. Psychophysiology, 51, 1207–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICHIKAWA M, OKAMURA-OHO Y, OKUNISHI R, KANAMORI M, SUZUKI H, RITANI A, NITTA H, EGUCHI N, URADE Y & HAYASHIZAKI Y (2005). Expression analysis of genes responsible for serotonin signaling in the brain. Neurobiol Dis, 19, 378–385. [DOI] [PubMed] [Google Scholar]
- ICHTCHENKO K, KHVOTCHEV M, KIYATKIN N, SIMPSON L, SUGITA S & SUDHOF TC (1998). alpha-latrotoxin action probed with recombinant toxin: receptors recruit alpha-latrotoxin but do not transduce an exocytotic signal. EMBO J, 17, 6188–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ILGIN N, SENOL S, GUCUYENER K, GOKCORA N & SENER S (2001). Is increased D2 receptor availability associated with response to stimulant medication in ADHD. Dev Med Child Neurol, 43, 755–760. [DOI] [PubMed] [Google Scholar]
- INOUE Y, SAKIHARA K, GUNJI A, OZAWA H, KIMIYA S, SHINODA H, KAGA M & INAGAKI M (2012). Reduced prefrontal hemodynamic response in children with ADHD during the Go/NoGo task: a NIRS study. Neuroreport, 23, 55–60. [DOI] [PubMed] [Google Scholar]
- JAHANSHAD N, GANJGAHI H, BRALTEN J, DEN BRABER A, FASKOWITZ J, KNODT AR, LEMAITRE H, NIR TM, PATEL B, RITCHIE S, SPROOTEN E, HOOGMAN M, VAN HULZEN K, ZAVALIANGOS-PETROPULU A, ZWIERS M, ALMASY L, BASTIN ME, BERNSTEIN MA, BLANGERO J, CURRAN JC, DEARY IJ, DE ZUBICARAY GI, DUGGIRALA R, FISHER SE, FRANKE B, FOX P, GOLDMAN D, HABERG AK, HARIRI A, HONG LE, HUENTELMAN M, MARTIN NG, MARTINOT JL, MCINTOSH A, MCMAHON KL, MEDLAND SE, MITCHELL BD, MUNOZ MANIEGA S, OLVERA RL, OOSTERLAAN J, PETERSON C, ROYLE N, SAYKIN AJ, SCHUMANN G, STARR J, STEIN EA, SUSSMANN J, VALDES HERNANDEZ MC, VAN’T ENT D, WARDLAW JM, WEINER MW, WILLIAMSON DE, WINKLER AM, WRIGHT MJ, YANG Y, THOMPSON PM, GLAHN DC, NICHOLS TE & KOCHUNOV P (in preparation). Do Candidate SNPs Affect the Brain’s White Matter Microstructure? Large Scale Evaluation of 6165 Diffusion MRI Scans. [Google Scholar]
- JAHANSHAD N, KOCHUNOV PV, SPROOTEN E, MANDL RC, NICHOLS TE, ALMASY L, BLANGERO J, BROUWER RM, CURRAN JE, DE ZUBICARAY GI, DUGGIRALA R, FOX PT, HONG LE, LANDMAN BA, MARTIN NG, MCMAHON KL, MEDLAND SE, MITCHELL BD, OLVERA RL, PETERSON CP, STARR JM, SUSSMANN JE, TOGA AW, WARDLAW JM, WRIGHT MJ, HULSHOFF POL HE, BASTIN ME, MCINTOSH AM, DEARY IJ, THOMPSON PM & GLAHN DC (2013). Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: a pilot project of the ENIGMA-DTI working group. Neuroimage, 81, 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIA T, MACARE C, DESRIVIERES S, GONZALEZ DA, TAO C, JI X, RUGGERI B, NEES F, BANASCHEWSKI T, BARKER GJ, BOKDE AL, BROMBERG U, BUCHEL C, CONROD PJ, DOVE R, FROUIN V, GALLINAT J, GARAVAN H, GOWLAND PA, HEINZ A, ITTERMANN B, LATHROP M, LEMAITRE H, MARTINOT JL, PAUS T, PAUSOVA Z, POLINE JB, RIETSCHEL M, ROBBINS T, SMOLKA MN, MULLER CP, FENG J, ROTHENFLUH A, FLOR H, SCHUMANN G & CONSORTIUM, I. (2016). Neural basis of reward anticipation and its genetic determinants. Proc Natl Acad Sci U S A, 113, 3879–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSTONE SJ, BARRY RJ & CLARKE AR (2013). Ten years on: a follow-up review of ERP research in attention-deficit/hyperactivity disorder. Clin Neurophysiol, 124, 644–657. [DOI] [PubMed] [Google Scholar]
- JONASSEN R & LANDRO NI (2014). Serotonin transporter polymorphisms (5-HTTLPR) in emotion processing: implications from current neurobiology. Prog Neurobiol, 117, 41–53. [DOI] [PubMed] [Google Scholar]
- JUCAITE A, FERNELL E, HALLDIN C, FORSSBERG H & FARDE L (2005). Reduced midbrain dopamine transporter binding in male adolescents with attention-deficit/hyperactivity disorder: association between striatal dopamine markers and motor hyperactivity. Biol Psychiatry, 57, 229–238. [DOI] [PubMed] [Google Scholar]
- KABUKCU BASAY B, BUBER A, BASAY O, ALACAM H, OZTURK O, SUREN S, IZCI AY O, ACIKEL C, AGLADIOGLU K, ERDAL ME, ERCAN ES & HERKEN H (2016). White matter alterations related to attention-deficit hyperactivity disorder and COMT val(158)met polymorphism: children with valine homozygote attention-deficit hyperactivity disorder have altered white matter connectivity in the right cingulum (cingulate gyrus). Neuropsychiatr Dis Treat, 12, 969–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KENDLER KS & NEALE MC (2010). Endophenotype: a conceptual analysis. Mol Psychiatry, 15, 789–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KERNER B, RAO AR, CHRISTENSEN B, DANDEKAR S, YOURSHAW M & NELSON SF (2013). Rare Genomic Variants Link Bipolar Disorder with Anxiety Disorders to CREB-Regulated Intracellular Signaling Pathways. Front Psychiatry, 4, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KESSLER RC, ADLER L, BARKLEY R, BIEDERMAN J, CONNERS CK, DEMLER O, FARAONE SV, GREENHILL LL, HOWES MJ, SECNIK K, SPENCER T, USTUN TB, WALTERS EE & ZASLAVSKY AM (2006). The prevalence and correlates of adult ADHD in the United States: results from the National Comorbidity Survey Replication. Am J Psychiatry, 163, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KILLEEN PR, RUSSELL VA & SERGEANT JA (2013). A behavioral neuroenergetics theory of ADHD. Neurosci Biobehav Rev, 37, 625–657. [DOI] [PubMed] [Google Scholar]
- KIM BN, KIM JW, KANG H, CHO SC, SHIN MS, YOO HJ, HONG SB & LEE DS (2010). Regional differences in cerebral perfusion associated with the alpha-2A-adrenergic receptor genotypes in attention deficit hyperactivity disorder. J Psychiatry Neurosci, 35, 330–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM YH, CHOI SH, D’AVANZO C, HEBISCH M, SLIWINSKI C, BYLYKBASHI E, WASHICOSKY KJ, KLEE JB, BRUSTLE O, TANZI RE & KIM DY (2015). A 3D human neural cell culture system for modeling Alzheimer’s disease. Nature Protocols, 10, 985–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KISS JP & VIZI ES (2001). Nitric oxide: a novel link between synaptic and nonsynaptic transmission. Trends Neurosci, 24, 211–215. [DOI] [PubMed] [Google Scholar]
- KLEIN M, STEIN JL, ENIGMA C, GROUP, P. A. W., IPSYCH, C., BORGLUM A, FARAONE SV, THOMPSON PM, MEDLAND SE, ARIAS VASQUEZ A & FRANKE B (in preparation). Investigating the overlap between common genetic factors for ADHD and subcortical brain volumes [Google Scholar]
- KLEIN M, VAN DONKELAAR MM, VERHOEF E & FRANKE B (under review). Imaging Genetics in Neurodevelopmental Psychopathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KO CH, HSIEH TJ, WANG PW, LIN WC, CHEN CS & YEN JY (2015). The Altered Brain Activation of Phonological Working Memory, Dual Tasking, and Distraction Among Participants With Adult ADHD and the Effect of the MAOA Polymorphism. J Atten Disord. [DOI] [PubMed] [Google Scholar]
- KOBIELLA A, REIMOLD M, ULSHÖFER D, IKONOMIDOU V, VOLLMERT C, VOLLSTÄDT-KLEIN S, RIETSCHEL M, REISCHL G, HEINZ A & SMOLKA M (2011). How the serotonin transporter 5-HTTLPR polymorphism influences amygdala function: the roles of in vivo serotonin transporter expression and amygdala structure. Transl Psychiatry, 1, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOCHUNOV P, FU M, NUGENT K, WRIGHT SN, DU X, MUELLERKLEIN F, MORRISSEY M, ESKANDAR G, SHUKLA DK, JAHANSHAD N, THOMPSON PM, PATEL B, POSTOLACHE TT, STRAUSS KA, SHULDINER AR, MITCHELL BD & HONG LE (2015). Heritability of complex white matter diffusion traits assessed in a population isolate. Hum Brain Mapp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOTTE A, FARAONE SV & BIEDERMAN J (2013). Association of genetic risk severity with ADHD clinical characteristics. Am J Med Genet B Neuropsychiatr Genet, 162B, 718–733. [DOI] [PubMed] [Google Scholar]
- KRAIN AL & CASTELLANOS FX (2006). Brain development and ADHD. Clin Psychol Rev, 26, 433–444. [DOI] [PubMed] [Google Scholar]
- KRAUSE J, DRESEL SH, KRAUSE KH, LA FOUGERE C, ZILL P & ACKENHEIL M (2006). Striatal dopamine transporter availability and DAT-1 gene in adults with ADHD: no higher DAT availability in patients with homozygosity for the 10-repeat allele. World J Biol Psychiatry, 7, 152–157. [DOI] [PubMed] [Google Scholar]
- KUCZENSKI R & SEGAL DS (2002). Exposure of adolescent rats to oral methylphenidate: preferential effects on extracellular norepinephrine and absence of sensitization and cross-sensitization to methamphetamine. J Neurosci, 22, 7264–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAHTI RA, ROBERTS RC & TAMMINGA CA (1995). D2-family receptor distribution in human postmortem tissue: an autoradiographic study. Neuroreport, 6, 2505–2512. [DOI] [PubMed] [Google Scholar]
- LANCASTER MA, RENNER M, MARTIN CA, WENZEL D, BICKNELL LS, HURLES ME, HOMFRAY T, PENNINGER JM, JACKSON AP & KNOBLICH JA (2013). Cerebral organoids model human brain development and microcephaly. Nature, 501, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANDAAS ET, JOHANSSON S, JACOBSEN KK, RIBASES M, BOSCH R, SANCHEZ-MORA C, JACOB CP, BOREATTI-HUMMER A, KREIKER S, LESCH KP, KIEMENEY LA, KOOIJ JJ, KAN C, BUITELAAR JK, FARAONE SV, HALMOY A, RAMOS-QUIROGA JA, CORMAND B, REIF A, FRANKE B, MICK E, KNAPPSKOG PM & HAAVIK J (2010). An international multicenter association study of the serotonin transporter gene in persistent ADHD. Genes Brain Behav, 9, 449–458. [DOI] [PubMed] [Google Scholar]
- LANGE M, NORTON W, COOLEN M, CHAMINADE M, MERKER S, PROFT F, SCHMITT A, VERNIER P, LESCH KP & BALLY-CUIF L (2012). The ADHD-susceptibility gene lphn3.1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development. Mol Psychiatry, 17, 946–954. [DOI] [PubMed] [Google Scholar]
- LANGER SZ (2015). alpha2-Adrenoceptors in the treatment of major neuropsychiatric disorders. Trends Pharmacol Sci, 36, 196–202. [DOI] [PubMed] [Google Scholar]
- LARSSON H, ASHERSON P, CHANG Z, LJUNG T, FRIEDRICHS B, LARSSON JO & LICHTENSTEIN P (2013a). Genetic and environmental influences on adult attention deficit hyperactivity disorder symptoms: a large Swedish population-based study of twins. Psychol Med, 43, 197–207. [DOI] [PubMed] [Google Scholar]
- LARSSON H, CHANG Z, D’ONOFRIO BM & LICHTENSTEIN P (2013b). The heritability of clinically diagnosed attention deficit hyperactivity disorder across the lifespan. Psychol Med, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LASKY-SU J, NEALE BM, FRANKE B, ANNEY RJ, ZHOU K, MALLER JB, VASQUEZ AA, CHEN W, ASHERSON P, BUITELAAR J, BANASCHEWSKI T, EBSTEIN R, GILL M, MIRANDA A, MULAS F, OADES RD, ROEYERS H, ROTHENBERGER A, SERGEANT J, SONUGA-BARKE E, STEINHAUSEN HC, TAYLOR E, DALY M, LAIRD N, LANGE C & FARAONE SV (2008). Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet, 147B, 1345–1354. [DOI] [PubMed] [Google Scholar]
- LE BIHAN D (2003). Looking into the functional architecture of the brain with diffusion MRI. Nat Rev Neurosci, 4, 469–480. [DOI] [PubMed] [Google Scholar]
- LE BIHAN D, MANGIN JF, POUPON C, CLARK CA, PAPPATA S, MOLKO N & CHABRIAT H (2001). Diffusion tensor imaging: concepts and applications. J Magn Reson Imaging, 13, 534–546. [DOI] [PubMed] [Google Scholar]
- LEE YH & SONG GG (2015). BDNF 196 G/A and COMT Val158Met Polymorphisms and Susceptibility to ADHD: A Meta-Analysis. J Atten Disord. [DOI] [PubMed] [Google Scholar]
- LEO D & GAINETDINOV RR (2013). Transgenic mouse models for ADHD. Cell Tissue Res, 354, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LESCH KP, ARARAGI N, WAIDER J, VAN DEN HOVE D & GUTKNECHT L (2012). Targeting brain serotonin synthesis: insights into neurodevelopmental disorders with long-term outcomes related to negative emotionality, aggression and antisocial behaviour. Philos Trans R Soc Lond B Biol Sci, 367, 2426–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LESCH KP, BENGEL D, HEILS A, SABOL SZ, GREENBERG BD, PETRI S, BENJAMIN J, MULLER CR, HAMER DH & MURPHY DL (1996). Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science, 274, 1527–1531. [DOI] [PubMed] [Google Scholar]
- LESCH KP, SELCH S, RENNER TJ, JACOB C, NGUYEN TT, HAHN T, ROMANOS M, WALITZA S, SHOICHET S, DEMPFLE A, HEINE M, BOREATTI-HUMMER A, ROMANOS J, GROSS-LESCH S, ZERLAUT H, WULTSCH T, HEINZEL S, FASSNACHT M, FALLGATTER A, ALLOLIO B, SCHAFER H, WARNKE A, REIF A, ROPERS HH & ULLMANN R (2011). Genome-wide copy number variation analysis in attention-deficit/hyperactivity disorder: association with neuropeptide Y gene dosage in an extended pedigree. Mol Psychiatry, 16, 491–503. [DOI] [PubMed] [Google Scholar]
- LESCH KP, TIMMESFELD N, RENNER TJ, HALPERIN R, ROSER C, NGUYEN TT, CRAIG DW, ROMANOS J, HEINE M, MEYER J, FREITAG C, WARNKE A, ROMANOS M, SCHAFER H, WALITZA S, REIF A, STEPHAN DA & JACOB C (2008). Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm, 115, 1573–1585. [DOI] [PubMed] [Google Scholar]
- LI D, SHAM PC, OWEN MJ & HE L (2006). Meta-analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder (ADHD). Hum Mol Genet, 15, 2276–2284. [DOI] [PubMed] [Google Scholar]
- LI J, KANG C, ZHANG H, WANG Y, ZHOU R, WANG B, GUAN L, YANG L & FARAONE SV (2007a). Monoamine oxidase A gene polymorphism predicts adolescent outcome of attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 144B, 430–433. [DOI] [PubMed] [Google Scholar]
- LI Q, LU G, ANTONIO GE, MAK YT, RUDD JA, FAN M & YEW DT (2007b). The usefulness of the spontaneously hypertensive rat to model attention-deficit/hyperactivity disorder (ADHD) may be explained by the differential expression of dopamine-related genes in the brain. Neurochem Int, 50, 848–857. [DOI] [PubMed] [Google Scholar]
- LI Z, CHANG SH, ZHANG LY, GAO L & WANG J (2014). Molecular genetic studies of ADHD and its candidate genes: A review. Psychiatry Res. [DOI] [PubMed] [Google Scholar]
- LIM CS, YANG JE, LEE YK, LEE K, LEE JA & KAANG BK (2015). Understanding the molecular basis of autism in a dish using hiPSCs-derived neurons from ASD patients. Molecular Brain, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LISTER R, PELIZZOLA M, KIDA YS, HAWKINS RD, NERY JR, HON G, ANTOSIEWICZ-BOURGET J, O’MALLEY R, CASTANON R, KLUGMAN S, DOWNES M, YU R, STEWART R, REN B, THOMSON JA, EVANS RM & ECKER JR (2011). Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature, 471, 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU J, GAO C, CHEN W, MA W, LI X, SHI Y, ZHANG H, ZHANG L, LONG Y, XU H, GUO X, DENG S, YAN X, YU D, PAN G, CHEN Y, LAI L, LIAO W & LI Z (2016). CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl Psychiatry, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU L, GUAN LL, CHEN Y, JI N, LI HM, LI ZH, QIAN QJ, YANG L, GLATT SJ, FARAONE SV & WANG YF (2011). Association analyses of MAOA in Chinese Han subjects with attention-deficit/hyperactivity disorder: family-based association test, case-control study, and quantitative traits of impulsivity. Am J Med Genet B Neuropsychiatr Genet, 156B, 737–748. [DOI] [PubMed] [Google Scholar]
- LIU Y, LIU H, SAUVEY C, YAO L, ZARNOWSKA ED & ZHANG SC (2013). Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nature Protocols, 8, 1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOO SK, HALE ST, HANADA G, MACION J, SHRESTHA A, MCGOUGH JJ, MCCRACKEN JT, NELSON S & SMALLEY SL (2010). Familial clustering and DRD4 effects on electroencephalogram measures in multiplex families with attention deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry, 49, 368–377. [PMC free article] [PubMed] [Google Scholar]
- LOO SK & MAKEIG S (2012). Clinical utility of EEG in attention-deficit/hyperactivity disorder: a research update. Neurotherapeutics, 9, 569–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOO SK, SPECTER E, SMOLEN A, HOPFER C, TEALE PD & REITE ML (2003). Functional effects of the DAT1 polymorphism on EEG measures in ADHD. J Am Acad Child Adolesc Psychiatry, 42, 986–993. [DOI] [PubMed] [Google Scholar]
- LUDOLPH AG, KASSUBEK J, SCHMECK K, GLASER C, WUNDERLICH A, BUCK AK, RESKE SN, FEGERT JM & MOTTAGHY FM (2008). Dopaminergic dysfunction in attention deficit hyperactivity disorder (ADHD), differences between pharmacologically treated and never treated young adults: a 3,4-dihdroxy-6-[18F]fluorophenyl-l-alanine PET study. Neuroimage, 41, 718–727. [DOI] [PubMed] [Google Scholar]
- LYCETT K, SCIBERRAS E, MENSAH FK & HISCOCK H (2015). Behavioral sleep problems and internalizing and externalizing comorbidities in children with attention-deficit/hyperactivity disorder. Eur Child Adolesc Psychiatry, 24, 31–40. [DOI] [PubMed] [Google Scholar]
- MA CL, ARNSTEN AF & LI BM (2005). Locomotor hyperactivity induced by blockade of prefrontal cortical alpha2-adrenoceptors in monkeys. Biol Psychiatry, 57, 192–195. [DOI] [PubMed] [Google Scholar]
- MADISON JM, ZHOU F, NIGAM A, HUSSAIN A, BARKER DD, NEHME R, VAN DER VEN K, HSU J, WOLF P, FLEISHMAN M, O’DUSHLAINE C, ROSE S, CHAMBERT K, LAU FH, AHFELDT T, RUECKERT EH, SHERIDAN SD, FASS DM, NEMESH J, MULLEN TE, DAHERON L, MCCARROLL S, SKLAR P, PERLIS RH & HAGGARTY SJ (2015). Characterization of bipolar disorder patient-specific induced pluripotent stem cells from a family reveals neurodevelopmental and mRNA expression abnormalities. Mol Psychiatry, 20, 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAKRIS N, BUKA SL, BIEDERMAN J, PAPADIMITRIOU GM, HODGE SM, VALERA EM, BROWN AB, BUSH G, MONUTEAUX MC, CAVINESS VS, KENNEDY DN & SEIDMAN LJ (2008). Attention and executive systems abnormalities in adults with childhood ADHD: A DT-MRI study of connections. Cereb Cortex, 18, 1210–1220. [DOI] [PubMed] [Google Scholar]
- MANDELLI L & SERRETTI A (2013). Gene environment interaction studies in depression and suicidal behavior: An update. Neurosci Biobehav Rev, 37, 2375–2397. [DOI] [PubMed] [Google Scholar]
- MARCHETTO MC, CARROMEU C, ACAB A, YU D, YEO GW, MU Y, CHEN G, GAGE FH & MUOTRI AR (2010). A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell, 143, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCGOUGH JJ, SMALLEY SL, MCCRACKEN JT, YANG M, DEL’HOMME M, LYNN DE & LOO S (2005). Psychiatric comorbidity in adult attention deficit hyperactivity disorder: findings from multiplex families. Am J Psychiatry, 162, 1621–1627. [DOI] [PubMed] [Google Scholar]
- MCKAY DR, KNOWLES EE, WINKLER AA, SPROOTEN E, KOCHUNOV P, OLVERA RL, CURRAN JE, KENT JW JR., CARLESS MA, GORING HH, DYER TD, DUGGIRALA R, ALMASY L, FOX PT, BLANGERO J & GLAHN DC (2014). Influence of age, sex and genetic factors on the human brain. Brain Imaging Behav, 8, 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEDLAND SE, JAHANSHAD N, NEALE BM & THOMPSON PM (2014). Whole-genome analyses of whole-brain data: working within an expanded search space. Nat Neurosci, 17, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MENESES A & LIY-SALMERON G (2012). Serotonin and emotion, learning and memory. Rev Neurosci, 23, 543–553. [DOI] [PubMed] [Google Scholar]
- MEYER-LINDENBERG A, BUCKHOLTZ JW, KOLACHANA B, A RH, PEZAWAS L, BLASI G, WABNITZ A, HONEA R, VERCHINSKI B, CALLICOTT JH, EGAN M, MATTAY V & WEINBERGER DR (2006). Neural mechanisms of genetic risk for impulsivity and violence in humans. Proc Natl Acad Sci U S A, 103, 6269–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MICK E, TODOROV A, SMALLEY S, HU X, LOO S, TODD RD, BIEDERMAN J, BYRNE D, DECHAIRO B, GUINEY A, MCCRACKEN J, MCGOUGH J, NELSON SF, REIERSEN AM, WILENS TE, WOZNIAK J, NEALE BM & FARAONE SV (2010). Family-based genome-wide association scan of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry, 49, 898–905 e893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIER D, KIRSCH P & MEYER-LINDENBERG A (2010). Neural substrates of pleiotropic action of genetic variation in COMT: a meta-analysis. Mol Psychiatry, 15, 918–927. [DOI] [PubMed] [Google Scholar]
- MILLER TW, NIGG JT & FARAONE SV (2007). Axis I and II comorbidity in adults with ADHD. J Abnorm Psychol, 116, 519–528. [DOI] [PubMed] [Google Scholar]
- MOFFITT TE, HOUTS R, ASHERSON P, BELSKY DW, CORCORAN DL, HAMMERLE M, HARRINGTON H, HOGAN S, MEIER MH, POLANCZYK GV, POULTON R, RAMRAKHA S, SUGDEN K, WILLIAMS B, ROHDE LA & CASPI A (2015). Is Adult ADHD a Childhood-Onset Neurodevelopmental Disorder? Evidence From a Four-Decade Longitudinal Cohort Study. Am J Psychiatry, 172, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONUTEAUX MC, SEIDMAN LJ, FARAONE SV, MAKRIS N, SPENCER T, VALERA E, BROWN A, BUSH G, DOYLE AE, HUGHES S, HELLIESEN M, MICK E & BIEDERMAN J (2008). A preliminary study of dopamine D4 receptor genotype and structural brain alterations in adults with ADHD. Am J Med Genet B Neuropsychiatr Genet, 147B, 1436–1441. [DOI] [PubMed] [Google Scholar]
- MOONEY MA, MCWEENEY SK, FARAONE SV, HINNEY A, HEBEBRAND J, CONSORTIUM, I., GERMAN, A. G. G., NIGG JT & WILMOT B (2016). Pathway analysis in attention deficit hyperactivity disorder: An ensemble approach. Am J Med Genet B Neuropsychiatr Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOSTERT JC, SHUMSKAYA E, MENNES M, ONNINK AM, HOOGMAN M, KAN CC, ARIAS VASQUEZ A, BUITELAAR J, FRANKE B & NORRIS DG (2016). Characterising resting-state functional connectivity in a large sample of adults with ADHD. Prog Neuropsychopharmacol Biol Psychiatry, 67, 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUNAFO MR, BROWN SM & HARIRI AR (2008). Serotonin transporter (5-HTTLPR) genotype and amygdala activation: a meta-analysis. Biol Psychiatry, 63, 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURPHY DL, ANDREWS AM, WICHEMS CH, LI Q, TOHDA M & GREENBERG B (1998). Brain serotonin neurotransmission: an overview and update with an emphasis on serotonin subsystem heterogeneity, multiple receptors, interactions with other neurotransmitter systems, and consequent implications for understanding the actions of serotonergic drugs. J Clin Psychiatry, 59 Suppl 15, 4–12. [PubMed] [Google Scholar]
- MURPHY SE, NORBURY R, GODLEWSKA BR, COWEN PJ, MANNIE ZM, HARMER CJ & MUNAFO MR (2013). The effect of the serotonin transporter polymorphism (5-HTTLPR) on amygdala function: a meta-analysis. Mol Psychiatry, 18, 512–520. [DOI] [PubMed] [Google Scholar]
- NAAIJEN J, LYTHGOE DJ, AMIRI H, BUITELAAR JK & GLENNON JC (2015). Fronto-striatal glutamatergic compounds in compulsive and impulsive syndromes: a review of magnetic resonance spectroscopy studies. Neurosci Biobehav Rev, 52, 74–88. [DOI] [PubMed] [Google Scholar]
- NAKAO T, RADUA J, RUBIA K & MATAIX-COLS D (2011). Gray matter volume abnormalities in ADHD: voxel-based meta-analysis exploring the effects of age and stimulant medication. Am J Psychiatry, 168, 1154–1163. [DOI] [PubMed] [Google Scholar]
- NEALE BM, KOU Y, LIU L, MA’AYAN A, SAMOCHA KE, SABO A, LIN CF, STEVENS C, WANG LS, MAKAROV V, POLAK P, YOON S, MAGUIRE J, CRAWFORD EL, CAMPBELL NG, GELLER ET, VALLADARES O, SCHAFER C, LIU H, ZHAO T, CAI G, LIHM J, DANNENFELSER R, JABADO O, PERALTA Z, NAGASWAMY U, MUZNY D, REID JG, NEWSHAM I, WU Y, LEWIS L, HAN Y, VOIGHT BF, LIM E, ROSSIN E, KIRBY A, FLANNICK J, FROMER M, SHAKIR K, FENNELL T, GARIMELLA K, BANKS E, POPLIN R, GABRIEL S, DEPRISTO M, WIMBISH JR, BOONE BE, LEVY SE, BETANCUR C, SUNYAEV S, BOERWINKLE E, BUXBAUM JD, COOK EH JR., DEVLIN B, GIBBS RA, ROEDER K, SCHELLENBERG GD, SUTCLIFFE JS & DALY MJ (2012). Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature, 485, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEALE BM, LASKY-SU J, ANNEY R, FRANKE B, ZHOU K, MALLER JB, VASQUEZ AA, ASHERSON P, CHEN W, BANASCHEWSKI T, BUITELAAR J, EBSTEIN R, GILL M, MIRANDA A, OADES RD, ROEYERS H, ROTHENBERGER A, SERGEANT J, STEINHAUSEN HC, SONUGA-BARKE E, MULAS F, TAYLOR E, LAIRD N, LANGE C, DALY M & FARAONE SV (2008). Genome-wide association scan of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 147B, 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEALE BM, MEDLAND S, RIPKE S, ANNEY RJ, ASHERSON P, BUITELAAR J, FRANKE B, GILL M, KENT L, HOLMANS P, MIDDLETON F, THAPAR A, LESCH KP, FARAONE SV, DALY M, NGUYEN TT, SCHAFER H, STEINHAUSEN HC, REIF A, RENNER TJ, ROMANOS M, ROMANOS J, WARNKE A, WALITZA S, FREITAG C, MEYER J, PALMASON H, ROTHENBERGER A, HAWI Z, SERGEANT J, ROEYERS H, MICK E, BIEDERMAN J & GROUP, I. I. C. (2010a). Case-control genome-wide association study of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry, 49, 906–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEALE BM, MEDLAND SE, RIPKE S, ASHERSON P, FRANKE B, LESCH KP, FARAONE SV, NGUYEN TT, SCHAFER H, HOLMANS P, DALY M, STEINHAUSEN HC, FREITAG C, REIF A, RENNER TJ, ROMANOS M, ROMANOS J, WALITZA S, WARNKE A, MEYER J, PALMASON H, BUITELAAR J, VASQUEZ AA, LAMBREGTS-ROMMELSE N, GILL M, ANNEY RJ, LANGELY K, O’DONOVAN M, WILLIAMS N, OWEN M, THAPAR A, KENT L, SERGEANT J, ROEYERS H, MICK E, BIEDERMAN J, DOYLE A, SMALLEY S, LOO S, HAKONARSON H, ELIA J, TODOROV A, MIRANDA A, MULAS F, EBSTEIN RP, ROTHENBERGER A, BANASCHEWSKI T, OADES RD, SONUGA-BARKE E, MCGOUGH J, NISENBAUM L, MIDDLETON F, HU X, NELSON S & PSYCHIATRIC, G. C. A. S. (2010b). Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry, 49, 884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEGORO H, SAWADA M, IIDA J, OTA T, TANAKA S & KISHIMOTO T (2010). Prefrontal dysfunction in attention-deficit/hyperactivity disorder as measured by near-infrared spectroscopy. Child Psychiatry Hum Dev, 41, 193–203. [DOI] [PubMed] [Google Scholar]
- NIGG JT, KNOTTNERUS GM, MARTEL MM, NIKOLAS M, CAVANAGH K, KARMAUS W & RAPPLEY MD (2008). Low blood lead levels associated with clinically diagnosed attention-deficit/hyperactivity disorder and mediated by weak cognitive control. Biol Psychiatry, 63, 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIKOLAIDIS A & GRAY JR (2010). ADHD and the DRD4 exon III 7-repeat polymorphism: an international meta-analysis. Soc Cogn Affect Neurosci, 5, 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORMAN LJ, CARLISI C, LUKITO S, HART H, MATAIX-COLS D, RADUA J & RUBIA K (2016). Structural and Functional Brain Abnormalities in Attention-Deficit/Hyperactivity Disorder and Obsessive-Compulsive Disorder: A Comparative Meta-analysis. JAMA Psychiatry, 73, 815–825. [DOI] [PubMed] [Google Scholar]
- O’ROAK BJ, VIVES L, GIRIRAJAN S, KARAKOC E, KRUMM N, COE BP, LEVY R, KO A, LEE C, SMITH JD, TURNER EH, STANAWAY IB, VERNOT B, MALIG M, BAKER C, REILLY B, AKEY JM, BORENSTEIN E, RIEDER MJ, NICKERSON DA, BERNIER R, SHENDURE J & EICHLER EE (2012). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature, 485, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’SHEA KS & MCINNIS MG (2015). Induced pluripotent stem cell (iPSC) models of bipolar disorder. Neuropsychopharmacology, 40, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKAMOTO K & AOKI K (1963). Development of a strain of spontaneously hypertensive rats. Jpn Circ J, 27, 282–293. [DOI] [PubMed] [Google Scholar]
- OLDENHOF J, VICKERY R, ANAFI M, OAK J, RAY A, SCHOOTS O, PAWSON T, VON ZASTROW M & VAN TOL HH (1998). SH3 binding domains in the dopamine D4 receptor. Biochemistry, 37, 15726–15736. [DOI] [PubMed] [Google Scholar]
- OLLENDICK TH, JARRETT MA, GRILLS-TAQUECHEL AE, HOVEY LD & WOLFF JC (2008). Comorbidity as a predictor and moderator of treatment outcome in youth with anxiety, affective, attention deficit/hyperactivity disorder, and oppositional/conduct disorders. Clin Psychol Rev, 28, 1447–1471. [DOI] [PubMed] [Google Scholar]
- ONER O, AKIN A, HERKEN H, ERDAL ME, CIFTCI K, AY ME, BICER D, ONCU B, BOZKURT OH, MUNIR K & YAZGAN Y (2011). Association among SNAP-25 gene DdeI and MnlI polymorphisms and hemodynamic changes during methylphenidate use: a functional near-infrared spectroscopy study. J Atten Disord, 15, 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ONNINK AM, FRANKE B, VAN HULZEN K, ZWIERS MP, MOSTERT JC, SCHENE AH, HESLENFELD DJ, OOSTERLAAN J, HOEKSTRA PJ, HARTMAN CA, VASQUEZ AA, KAN CC, BUITELAAR J & HOOGMAN M (2016). Enlarged striatal volume in adults with ADHD carrying the 9–6 haplotype of the dopamine transporter gene DAT1. J Neural Transm (Vienna). [DOI] [PMC free article] [PubMed] [Google Scholar]
- ONNINK AM, ZWIERS MP, HOOGMAN M, MOSTERT JC, DAMMERS J, KAN CC, VASQUEZ AA, SCHENE AH, BUITELAAR J & FRANKE B (2015). Deviant white matter structure in adults with attention-deficit/hyperactivity disorder points to aberrant myelination and affects neuropsychological performance. Prog Neuropsychopharmacol Biol Psychiatry, 63, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OO KZ, AUNG YK, JENKINS MA & WIN AK (2016). Associations of 5HTTLPR polymorphism with major depressive disorder and alcohol dependence: A systematic review and meta-analysis. Aust N Z J Psychiatry. [DOI] [PubMed] [Google Scholar]
- OORTVELD MA, KEERTHIKUMAR S, OTI M, NIJHOF B, FERNANDES AC, KOCHINKE K, CASTELLS-NOBAU A, VAN ENGELEN E, ELLENKAMP T, ESHUIS L, GALY A, VAN BOKHOVEN H, HABERMANN B, BRUNNER HG, ZWEIER C, VERSTREKEN P, HUYNEN MA & SCHENCK A (2013). Human intellectual disability genes form conserved functional modules in Drosophila. PLoS Genet, 9, e1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OQUENDO MA, HASTINGS RS, HUANG YY, SIMPSON N, OGDEN RT, HU XZ, GOLDMAN D, ARANGO V, VAN HEERTUM RL, MANN JJ & PARSEY RV (2007). Brain serotonin transporter binding in depressed patients with bipolar disorder using positron emission tomography. Arch Gen Psychiatry, 64, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OSAFUNE K, CARON L, BOROWIAK M, MARTINEZ RJ, FITZ-GERALD CS, SATO Y, COWAN CA, CHIEN KR & MELTON DA (2008). Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol, 26, 313–315. [DOI] [PubMed] [Google Scholar]
- PALOYELIS Y, MEHTA MA, FARAONE SV, ASHERSON P & KUNTSI J (2012). Striatal sensitivity during reward processing in attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry, 51, 722–732 e729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAN Y, YAO J & WANG B (2014). Association of dopamine D1 receptor gene polymorphism with schizophrenia: a meta-analysis. Neuropsychiatr Dis Treat, 10, 1133–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PANG ZP, YANG N, VIERBUCHEN T, OSTERMEIER A, FUENTES DR, YANG TQ, CITRI A, SEBASTIANO V, MARRO S, SUDHOF TC & WERNIG M (2011). Induction of human neuronal cells by defined transcription factors. Nature, 476, 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAPPA I, MILEVA-SEITZ VR, BAKERMANS-KRANENBURG MJ, TIEMEIER H & VAN IJZENDOORN MH (2015). The magnificent seven: A quantitative review of dopamine receptor d4 and its association with child behavior. Neuroscience & Biobehavioral Reviews, 57, 175–186. [DOI] [PubMed] [Google Scholar]
- PARIKSHAK NN, LUO R, ZHANG A, WON H, LOWE JK, CHANDRAN V, HORVATH S & GESCHWIND DH (2013). Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell, 155, 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK L, NIGG JT, WALDMAN ID, NUMMY KA, HUANG-POLLOCK C, RAPPLEY M & FRIDERICI KH (2005). Association and linkage of alpha-2A adrenergic receptor gene polymorphisms with childhood ADHD. Mol Psychiatry, 10, 572–580. [DOI] [PubMed] [Google Scholar]
- PARK MH, KIM JW, YANG YH, HONG SB, PARK S, KANG H, KIM BN, SHIN MS, YOO HJ & CHO SC (2012). Regional brain perfusion before and after treatment with methylphenidate may be associated with the G1287A polymorphism of the norepinephrine transporter gene in children with attention-deficit/hyperactivity disorder. Neurosci Lett, 514, 159–163. [DOI] [PubMed] [Google Scholar]
- PARK S, HONG SB, KIM JW, YANG YH, PARK MH, KIM BN, SHIN MS, YOO HJ & CHO SC (2013). White-matter connectivity and methylphenidate-induced changes in attentional performance according to alpha2A-adrenergic receptor gene polymorphisms in Korean children with attention-deficit hyperactivity disorder. J Neuropsychiatry Clin Neurosci, 25, 222–228. [DOI] [PubMed] [Google Scholar]
- PAUL DS, SORANZO N & BECK S (2014). Functional interpretation of non-coding sequence variation: concepts and challenges. Bioessays, 36, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEPER JS, BROUWER RM, BOOMSMA DI, KAHN RS & HULSHOFF POL HE (2007). Genetic influences on human brain structure: a review of brain imaging studies in twins. Hum Brain Mapp, 28, 464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PGC AND IPSYCH ADHD WORKING GROUPS (in preparation).
- PINGAULT JB, VIDING E, GALERA C, GREVEN CU, ZHENG Y, PLOMIN R & RIJSDIJK F (2015). Genetic and Environmental Influences on the Developmental Course of Attention-Deficit/Hyperactivity Disorder Symptoms From Childhood to Adolescence. JAMA Psychiatry, 72, 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PLICHTA MM & SCHERES A (2014). Ventral-striatal responsiveness during reward anticipation in ADHD and its relation to trait impulsivity in the healthy population: a meta-analytic review of the fMRI literature. Neurosci Biobehav Rev, 38, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POELMANS G, FRANKE B, PAULS DL, GLENNON JC & BUITELAAR JK (2013). AKAPs integrate genetic findings for autism spectrum disorders. Transl Psychiatry, 3, e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POELMANS G, PAULS DL, BUITELAAR JK & FRANKE B (2011). Integrated genome-wide association study findings: identification of a neurodevelopmental network for attention deficit hyperactivity disorder. Am J Psychiatry, 168, 365–377. [DOI] [PubMed] [Google Scholar]
- POLANCZYK G & ROHDE LA (2007). Epidemiology of attention-deficit/hyperactivity disorder across the lifespan. Curr Opin Psychiatry, 20, 386–392. [DOI] [PubMed] [Google Scholar]
- POSNER J, PARK C & WANG Z (2014). Connecting the dots: a review of resting connectivity MRI studies in attention-deficit/hyperactivity disorder. Neuropsychol Rev, 24, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PSATY BM, O’DONNELL CJ, GUDNASON V, LUNETTA KL, FOLSOM AR, ROTTER JI, UITTERLINDEN AG, HARRIS TB, WITTEMAN JC, BOERWINKLE E & CONSORTIUM, C. (2009). Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet, 2, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PURCELL SM, MORAN JL, FROMER M, RUDERFER D, SOLOVIEFF N, ROUSSOS P, O’DUSHLAINE C, CHAMBERT K, BERGEN SE, KAHLER A, DUNCAN L, STAHL E, GENOVESE G, FERNANDEZ E, COLLINS MO, KOMIYAMA NH, CHOUDHARY JS, MAGNUSSON PK, BANKS E, SHAKIR K, GARIMELLA K, FENNELL T, DEPRISTO M, GRANT SG, HAGGARTY SJ, GABRIEL S, SCOLNICK EM, LANDER ES, HULTMAN CM, SULLIVAN PF, MCCARROLL SA & SKLAR P (2014). A polygenic burden of rare disruptive mutations in schizophrenia. Nature, 506, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAHMIM A & ZAIDI H (2008). PET versus SPECT: strengths, limitations and challenges. Nucl Med Commun, 29, 193–207. [DOI] [PubMed] [Google Scholar]
- RAMOS-QUIROGA JA, SANCHEZ-MORA C, CASAS M, GARCIA-MARTINEZ I, BOSCH R, NOGUEIRA M, CORRALES M, PALOMAR G, VIDAL R, COLL-TANE M, BAYES M, CORMAND B & RIBASES M (2014). Genome-wide copy number variation analysis in adult attention-deficit and hyperactivity disorder. J Psychiatr Res, 49, 60–67. [DOI] [PubMed] [Google Scholar]
- RAN FA, HSU PD, WRIGHT J, AGARWALA V, SCOTT DA & ZHANG F (2013). Genome engineering using the CRISPR-Cas9 system. Nature Protocols, 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAPPLEY MD (2005). Clinical practice. Attention deficit-hyperactivity disorder. N Engl J Med, 352, 165–173. [DOI] [PubMed] [Google Scholar]
- RAZNAHAN A, SHAW PW, LERCH JP, CLASEN LS, GREENSTEIN D, BERMAN R, PIPITONE J, CHAKRAVARTY MM & GIEDD JN (2014). Longitudinal four-dimensional mapping of subcortical anatomy in human development. Proc Natl Acad Sci U S A, 111, 1592–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REIF A, JACOB CP, RUJESCU D, HERTERICH S, LANG S, GUTKNECHT L, BAEHNE CG, STROBEL A, FREITAG CM, GIEGLING I, ROMANOS M, HARTMANN A, ROSLER M, RENNER TJ, FALLGATTER AJ, RETZ W, EHLIS AC & LESCH KP (2009). Influence of functional variant of neuronal nitric oxide synthase on impulsive behaviors in humans. Arch Gen Psychiatry, 66, 41–50. [DOI] [PubMed] [Google Scholar]
- REINHARDT MC & REINHARDT CA (2013). Attention deficit-hyperactivity disorder, comorbidities, and risk situations. J Pediatr (Rio J), 89, 124–130. [DOI] [PubMed] [Google Scholar]
- REINHARDT P, SCHMID B, BURBULLA LF, SCHONDORF DC, WAGNER L, GLATZA M, HOING S, HARGUS G, HECK SA, DHINGRA A, WU GM, MULLER S, BROCKMANN K, KLUBA T, MAISEL M, KRUGER R, BERG D, TSYTSYURA Y, THIEL CS, PSATHAKI OE, KLINGAUF J, KUHLMANN T, KLEWIN M, MULLER H, GASSER T, SCHOLER HR & STERNECKERT J (2013). Genetic Correction of a LRRK2 Mutation in Human iPSCs Links Parkinsonian Neurodegeneration to ERK-Dependent Changes in Gene Expression. Cell Stem Cell, 12, 354–367. [DOI] [PubMed] [Google Scholar]
- RIBASES M, RAMOS-QUIROGA JA, SANCHEZ-MORA C, BOSCH R, RICHARTE V, PALOMAR G, GASTAMINZA X, BIELSA A, ARCOS-BURGOS M, MUENKE M, CASTELLANOS FX, CORMAND B, BAYES M & CASAS M (2011). Contribution of LPHN3 to the genetic susceptibility to ADHD in adulthood: a replication study. Genes Brain Behav, 10, 149–157. [DOI] [PubMed] [Google Scholar]
- RICHARDS JS, ARIAS VASQUEZ A, FRANKE B, HOEKSTRA PJ, HESLENFELD DJ, OOSTERLAAN J, FARAONE SV, BUITELAAR JK & HARTMAN CA (2016). Developmentally Sensitive Interaction Effects of Genes and the Social Environment on Total and Subcortical Brain Volumes. PLoS One, 11, e0155755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIVERO O, SELTEN MM, SICH S, POPP S, BACMEISTER L, AMENDOLA E, NEGWER M, SCHUBERT D, PROFT F, KISER D, SCHMITT AG, GROSS C, KOLK SM, STREKALOVA T, VAN DEN HOVE D, RESINK TJ, NADIF KASRI N & LESCH KP (2015). Cadherin-13, a risk gene for ADHD and comorbid disorders, impacts GABAergic function in hippocampus and cognition. Transl Psychiatry, 5, e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIVERO O, SICH S, POPP S, SCHMITT A, FRANKE B & LESCH KP (2013). Impact of the ADHD-susceptibility gene CDH13 on development and function of brain networks. Eur Neuropsychopharmacol, 23, 492–507. [DOI] [PubMed] [Google Scholar]
- ROHDE LA, ROMAN T, SZOBOT C, CUNHA RD, HUTZ MH & BIEDERMAN J (2003). Dopamine transporter gene, response to methylphenidate and cerebral blood flow in attention-deficit/hyperactivity disorder: a pilot study. Synapse, 48, 87–89. [DOI] [PubMed] [Google Scholar]
- ROMMELSE NN, GEURTS HM, FRANKE B, BUITELAAR JK & HARTMAN CA (2011). A review on cognitive and brain endophenotypes that may be common in autism spectrum disorder and attention-deficit/hyperactivity disorder and facilitate the search for pleiotropic genes. Neurosci Biobehav Rev, 35, 1363–1396. [DOI] [PubMed] [Google Scholar]
- ROSE EJ & DONOHOE G (2013). Brain vs behavior: an effect size comparison of neuroimaging and cognitive studies of genetic risk for schizophrenia. Schizophr Bull, 39, 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUBIA K (2007). Neuro-anatomic evidence for the maturational delay hypothesis of ADHD. Proc Natl Acad Sci U S A, 104, 19663–19664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUOCCO LA, TRENO C, GIRONI CARNEVALE UA, ARRA C, BOATTO G, NIEDDU M, PAGANO C, ILLIANO P, BARBATO F, TINO A, CARBONI E, LAVIOLA G, LACIVITA E, LEOPOLDO M, ADRIANI W & SADILE AG (2014). Prepuberal stimulation of 5-HT7-R by LP-211 in a rat model of hyper-activity and attention-deficit: permanent effects on attention, brain amino acids and synaptic markers in the fronto-striatal interface. PLoS One, 9, e83003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUSSELL V, DE VILLIERS A, SAGVOLDEN T, LAMM M & TALJAARD J (1995). Altered dopaminergic function in the prefrontal cortex, nucleus accumbens and caudate-putamen of an animal model of attention-deficit hyperactivity disorder--the spontaneously hypertensive rat. Brain Res, 676, 343–351. [DOI] [PubMed] [Google Scholar]
- SANCHEZ-CARMONA AJ, ALBERT J & HINOJOSA JA (2016). Neural and behavioral correlates of selective stopping: Evidence for a different strategy adoption. Neuroimage, 139, 279–293. [DOI] [PubMed] [Google Scholar]
- SANCHEZ-MORA C, RAMOS-QUIROGA JA, BOSCH R, CORRALES M, GARCIA-MARTINEZ I, NOGUEIRA M, PAGEROLS M, PALOMAR G, RICHARTE V, VIDAL R, ARIAS-VASQUEZ A, BUSTAMANTE M, FORNS J, GROSS-LESCH S, GUXENS M, HINNEY A, HOOGMAN M, JACOB C, JACOBSEN KK, KAN CC, KIEMENEY L, KITTEL-SCHNEIDER S, KLEIN M, ONNINK M, RIVERO O, ZAYATS T, BUITELAAR J, FARAONE SV, FRANKE B, HAAVIK J, JOHANSSON S, LESCH KP, REIF A, SUNYER J, BAYES M, CASAS M, CORMAND B & RIBASES M (2014). Case-Control Genome-Wide Association Study of Persistent Attention-Deficit Hyperactivity Disorder Identifies FBXO33 as a Novel Susceptibility Gene for the Disorder. Neuropsychopharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANCHEZ-MORA C, RIBASES M, CASAS M, BAYES M, BOSCH R, FERNANDEZ-CASTILLO N, BRUNSO L, JACOBSEN KK, LANDAAS ET, LUNDERVOLD AJ, GROSS-LESCH S, KREIKER S, JACOB CP, LESCH KP, BUITELAAR JK, HOOGMAN M, KIEMENEY LA, KOOIJ JJ, MICK E, ASHERSON P, FARAONE SV, FRANKE B, REIF A, JOHANSSON S, HAAVIK J, RAMOS-QUIROGA JA & CORMAND B (2011). Exploring DRD4 and its interaction with SLC6A3 as possible risk factors for adult ADHD: a meta-analysis in four European populations. Am J Med Genet B Neuropsychiatr Genet, 156B, 600–612. [DOI] [PubMed] [Google Scholar]
- SANCHEZ-MORA C, RICHARTE V, GARCIA-MARTINEZ I, PAGEROLS M, CORRALES M, BOSCH R, VIDAL R, VILADEVALL L, CASAS M, CORMAND B, RAMOS-QUIROGA JA & RIBASES M (2015). Dopamine receptor DRD4 gene and stressful life events in persistent attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. [DOI] [PubMed] [Google Scholar]
- SCHADT EE, BUCHANAN S, BRENNAND KJ & MERCHANT KM (2014). Evolving toward a human-cell based and multiscale approach to drug discovery for CNS disorders. Front Pharmacol, 5, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHECKLMANN M, EHLIS AC, PLICHTA MM, DRESLER T, HEINE M, BOREATTI-HUMMER A, ROMANOS M, JACOB C, PAULI P & FALLGATTER AJ (2013). Working memory and response inhibition as one integral phenotype of adult ADHD? A behavioral and imaging correlational investigation. J Atten Disord, 17, 470–482. [DOI] [PubMed] [Google Scholar]
- SCHECKLMANN M, ROMANOS M, BRETSCHER F, PLICHTA MM, WARNKE A & FALLGATTER AJ (2010). Prefrontal oxygenation during working memory in ADHD. J Psychiatr Res, 44, 621–628. [DOI] [PubMed] [Google Scholar]
- SCHIZOPHRENIA WORKING GROUP OF THE PSYCHIATRIC GENOMICS, C., RIPKE S, NEALE BM, CORVIN A, WALTERS JTR, FARH KH, HOLMANS PA, LEE P, BULIK-SULLIVAN B, COLLIER DA, HUANG H, PERS TH, AGARTZ I, AGERBO E, ALBUS M, ALEXANDER M, AMIN F, BACANU SA, BEGEMANN M, BELLIVEAU RA, BENE J, BERGEN SE, BEVILACQUA E, BIGDELI TB, BLACK DW, BRUGGEMAN R, BUCCOLA NG, BUCKNER RL, BYERLEY W, CAHN W, CAI G, CAMPION D, CANTOR RM, CARR VJ, CARRERA N, CATTS SV, CHAMBERT KD, CHAN RCK, CHAN RYL, CHEN EYH, CHENG W, CHEUNG EFC, CHONG SA, CLONINGER CR, COHEN D, COHEN N, CORMICAN P, CRADDOCK N, CROWLEY JJ, CURTIS D, DAVIDSON M, DAVIS KL, DEGENHARDT F, DEL FAVERO J, DEMONTIS D, DIKEOS D, DINAN T, DJUROVIC S, DONOHOE G, DRAPEAU E, DUAN J, DUDBRIDGE F, DURMISHI N, EICHHAMMER P, ERIKSSON J, ESCOTT-PRICE V, ESSIOUX L, FANOUS AH, FARRELL MS, FRANK J, FRANKE L, FREEDMAN R, FREIMER NB, FRIEDL M, FRIEDMAN JI, FROMER M, GENOVESE G, GEORGIEVA L, GIEGLING I, GIUSTI-RODRÍGUEZ P, GODARD S, GOLDSTEIN JI, GOLIMBET V, GOPAL S, GRATTEN J, DE HAAN L, HAMMER C, HAMSHERE ML, HANSEN M, HANSEN T, HAROUTUNIAN V, HARTMANN AM, HENSKENS FA, HERMS S, HIRSCHHORN JN, HOFFMANN P, HOFMAN A, HOLLEGAARD MV, HOUGAARD DM, IKEDA M, JOA I, JULIÀ A, KAHN RS, KALAYDJIEVA L, KARACHANAK-YANKOVA S, KARJALAINEN J, KAVANAGH D, KELLER MC, KENNEDY JL, KHRUNIN A, KIM Y, KLOVINS J, KNOWLES JA, KONTE B, KUCINSKAS V, KUCINSKIENE ZA, KUZELOVAPTACKOVA H, KÄHLER AK, LAURENT C, LEE J, LEE SH, LEGGE SE, LERER B, LI M, LI T, LIANG KY, LIEBERMAN J, LIMBORSKA S, LOUGHLAND CM, LUBINSKI J, LÖNNQVIST J, MACEK M, MAGNUSSON PKE, MAHER BS, MAIER W, MALLET J, MARSAL S, MATTHEISEN M, MATTINGSDAL M, MCCARLEY RW, MCDONALD C, MCINTOSH AM, MEIER S, MEIJER CJ, MELEGH B, MELLE I, MESHOLAM-GATELY RI, METSPALU A, MICHIE PT, MILANI L, MILANOVA V, MOKRAB Y, MORRIS DW, MORS O, MURPHY KC, MURRAY RM, MYIN-GERMEYS I, MÜLLER-MYHSOK B, NELIS M, NENADIC I, NERTNEY DA, NESTADT G, NICODEMUS KK, NIKITINA-ZAKE L, NISENBAUM L, NORDIN A, O’CALLAGHAN E, O’DUSHLAINE C, O’NEILL FA, OH SY, OLINCY A, OLSEN L, VAN OS J, PSYCHOSIS ENDOPHENOTYPES INTERNATIONAL C, PANTELIS C, PAPADIMITRIOU GN, PAPIOL S, PARKHOMENKO E, PATO MT, PAUNIO T, PEJOVIC-MILOVANCEVIC M, PERKINS DO, PIETILÄINEN O, PIMM J, POCKLINGTON AJ, POWELL J, PRICE A, PULVER AE, PURCELL SM, QUESTED D, RASMUSSEN HB, REICHENBERG A, REIMERS MA, RICHARDS AL, ROFFMAN JL, ROUSSOS P, RUDERFER DM, SALOMAA V, SANDERS AR, SCHALL U, SCHUBERT CR, SCHULZE TG, SCHWAB SG, SCOLNICK EM, SCOTT RJ, SEIDMAN LJ, SHI J, SIGURDSSON E, SILAGADZE T, SILVERMAN JM, SIM K, SLOMINSKY P, SMOLLER JW, SO HC, SPENCER CCA, STAHL EA, STEFANSSON H, STEINBERG S, STOGMANN E, STRAUB RE, STRENGMAN E, STROHMAIER J, STROUP TS, SUBRAMANIAM M, SUVISAARI J, SVRAKIC DM, SZATKIEWICZ JP, SÖDERMAN E, THIRUMALAI S, TONCHEVA D, TOSATO S, VEIJOLA J, WADDINGTON J, WALSH D, WANG D, WANG Q, WEBB BT, WEISER M, WILDENAUER DB, WILLIAMS NM, WILLIAMS S, WITT SH, WOLEN AR, WONG EHM, WORMLEY BK, XI HS, ZAI CC, ZHENG X, ZIMPRICH F, WRAY NR, STEFANSSON K, VISSCHER PM, WELLCOME TRUST CASE-CONTROL C, ADOLFSSON R, ANDREASSEN OA, BLACKWOOD DHR, BRAMON E, BUXBAUM JD, BØRGLUM AD, CICHON S, DARVASI A, DOMENICI E, EHRENREICH H, ESKO T, GEJMAN PV, GILL M, GURLING H, HULTMAN CM, IWATA N, JABLENSKY AV, JÖNSSON EG, KENDLER KS, KIROV G, KNIGHT J, LENCZ T, LEVINSON DF, LI QS, LIU J, MALHOTRA AK, MCCARROLL SA, MCQUILLIN A, MORAN JL, MORTENSEN PB, MOWRY BJ, NÖTHEN MM, OPHOFF RA, OWEN MJ, PALOTIE A, PATO CN, PETRYSHEN TL, POSTHUMA D, RIETSCHEL M, RILEY BP, RUJESCU D, SHAM PC, SKLAR P, ST CLAIR D, WEINBERGER DR, WENDLAND JR, WERGE T, DALY MJ, SULLIVAN PF & O’DONOVAN MC (2014). Biological Insights From 108 Schizophrenia-Associated Genetic Loci. Nature, 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHUR RR, DRAISMA LW, WIJNEN JP, BOKS MP, KOEVOETS MG, JOELS M, KLOMP DW, KAHN RS & VINKERS CH (2016). Brain GABA levels across psychiatric disorders: A systematic literature review and meta-analysis of H-MRS studies. Hum Brain Mapp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHWEREN LJ, DE ZEEUW P & DURSTON S (2013). MR imaging of the effects of methylphenidate on brain structure and function in attention-deficit/hyperactivity disorder. Eur Neuropsychopharmacol, 23, 1151–1164. [DOI] [PubMed] [Google Scholar]
- SCHWEREN LJ, HARTMAN CA, HESLENFELD DJ, GROENMAN AP, FRANKE B, OOSTERLAAN J, BUITELAAR JK & HOEKSTRA PJ (2016). Age and DRD4 Genotype Moderate Associations Between Stimulant Treatment History and Cortex Structure in Attention-Deficit/Hyperactivity Disorder. J Am Acad Child Adolesc Psychiatry, 55, 877–885.e873. [DOI] [PubMed] [Google Scholar]
- SEGAL DS & KUCZENSKI R (1997). Repeated binge exposures to amphetamine and methamphetamine: behavioral and neurochemical characterization. J Pharmacol Exp Ther, 282, 561–573. [PubMed] [Google Scholar]
- SEU E, LANG A, RIVERA RJ & JENTSCH JD (2009). Inhibition of the norepinephrine transporter improves behavioral flexibility in rats and monkeys. Psychopharmacology (Berl), 202, 505–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHAW P, ECKSTRAND K, SHARP W, BLUMENTHAL J, LERCH J, GREENSTEIN D, CLASEN L, EVANS A, GIEDD J & RAPOPORT J (2007a). Attention-deficit/hyperactivity disorder is characterized by a delay in cortical maturation. Proceedings of the National Academy of Sciences, 104, 19649–19654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHAW P, GORNICK M, LERCH J, ADDINGTON A, SEAL J, GREENSTEIN D, SHARP W, EVANS A, GIEDD JN, CASTELLANOS FX & RAPOPORT JL (2007b). Polymorphisms of the dopamine D4 receptor, clinical outcome, and cortical structure in attention-deficit/hyperactivity disorder. Arch Gen Psychiatry, 64, 921–931. [DOI] [PubMed] [Google Scholar]
- SHAW P, MALEK M, WATSON B, SHARP W, EVANS A & GREENSTEIN D (2012). Development of cortical surface area and gyrification in attention-deficit/hyperactivity disorder. Biological psychiatry, 72, 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEEHAN K, LOWE N, KIRLEY A, MULLINS C, FITZGERALD M, GILL M & HAWI Z (2005). Tryptophan hydroxylase 2 (TPH2) gene variants associated with ADHD. Mol Psychiatry, 10, 944–949. [DOI] [PubMed] [Google Scholar]
- SHENHAV A, BOTVINICK MM & COHEN JD (2013). The expected value of control: an integrative theory of anterior cingulate cortex function. Neuron, 79, 217–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHERIDAN SD, THERIAULT KM, REIS SA, ZHOU F, MADISON JM, DAHERON L, LORING JF & HAGGARTY SJ (2011). Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One, 6, e26203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIMADA K, FUJISAWA TX, TAKIGUCHI S, NARUSE H, KOSAKA H, OKAZAWA H & TOMODA A (2015). Ethnic differences in COMT genetic effects on striatal grey matter alterations associated with childhood ADHD: A voxel-based morphometry study in a Japanese sample. World J Biol Psychiatry, 1–7. [DOI] [PubMed] [Google Scholar]
- SHOOK D, BRADY C, LEE PS, KENEALY L, MURPHY ER, GAILLARD WD, VANMETER JW, COOK EH JR., STEIN M & VAIDYA CJ (2011). Effect of dopamine transporter genotype on caudate volume in childhood ADHD and controls. Am J Med Genet B Neuropsychiatr Genet, 156B, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIGURDARDOTTIR HL, KRANZ GS, RAMI-MARK C, JAMES GM, VANICEK T, GRYGLEWSKI G, KAUTZKY A, HIENERT M, TRAUB-WEIDINGER T, MITTERHAUSER M, WADSAK W, HACKER M, RUJESCU D, KASPER S & LANZENBERGER R (2015). Effects of norepinephrine transporter gene variants on NET binding in ADHD and healthy controls investigated by PET. Hum Brain Mapp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMCHON Y, WEIZMAN A & REHAVI M (2010). The effect of chronic methylphenidate administration on presynaptic dopaminergic parameters in a rat model for ADHD. Eur Neuropsychopharmacol, 20, 714–720. [DOI] [PubMed] [Google Scholar]
- SIMON V, CZOBOR P, BALINT S, MESZAROS A & BITTER I (2009). Prevalence and correlates of adult attention-deficit hyperactivity disorder: meta-analysis. Br J Psychiatry, 194, 204–211. [DOI] [PubMed] [Google Scholar]
- SMIT DJ, BOERSMA M, VAN BEIJSTERVELDT CE, POSTHUMA D, BOOMSMA DI, STAM CJ & DE GEUS EJ (2010). Endophenotypes in a dynamically connected brain. Behav Genet, 40, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOBANSKI E, BRUGGEMANN D, ALM B, KERN S, DESCHNER M, SCHUBERT T, PHILIPSEN A & RIETSCHEL M (2007). Psychiatric comorbidity and functional impairment in a clinically referred sample of adults with attention-deficit/hyperactivity disorder (ADHD). Eur Arch Psychiatry Clin Neurosci, 257, 371–377. [DOI] [PubMed] [Google Scholar]
- SODERQVIST S, MCNAB F, PEYRARD-JANVID M, MATSSON H, HUMPHREYS K, KERE J & KLINGBERG T (2010). The SNAP25 gene is linked to working memory capacity and maturation of the posterior cingulate cortex during childhood. Biol Psychiatry, 68, 1120–1125. [DOI] [PubMed] [Google Scholar]
- SOKOLOVA E, HOOGMAN M, GROOT P, CLAASSEN T, VASQUEZ AA, BUITELAAR JK, FRANKE B & HESKES T (2015). Causal discovery in an adult ADHD data set suggests indirect link between DAT1 genetic variants and striatal brain activation during reward processing. Am J Med Genet B Neuropsychiatr Genet. [DOI] [PubMed] [Google Scholar]
- SOLLNER T, WHITEHEART SW, BRUNNER M, ERDJUMENT-BROMAGE H, GEROMANOS S, TEMPST P & ROTHMAN JE (1993). SNAP receptors implicated in vesicle targeting and fusion. Nature, 362, 318–324. [DOI] [PubMed] [Google Scholar]
- SPENCER TJ, BIEDERMAN J, FARAONE SV, MADRAS BK, BONAB AA, DOUGHERTY DD, BATCHELDER H, CLARKE A & FISCHMAN AJ (2013). Functional genomics of attention-deficit/hyperactivity disorder (ADHD) risk alleles on dopamine transporter binding in ADHD and healthy control subjects. Biol Psychiatry, 74, 84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPENCER TJ, BIEDERMAN J, MADRAS BK, FARAONE SV, DOUGHERTY DD, BONAB AA & FISCHMAN AJ (2005). In vivo neuroreceptor imaging in attention-deficit/hyperactivity disorder: a focus on the dopamine transporter. Biol Psychiatry, 57, 1293–1300. [DOI] [PubMed] [Google Scholar]
- STAHL SM (2003). Neurotransmission of cognition, part 3. Mechanism of action of selective NRIs: both dopamine and norepinephrine increase in prefrontal cortex. J Clin Psychiatry, 64, 230–231. [PubMed] [Google Scholar]
- STEIN JL, HUA X, LEE S, HO AJ, LEOW AD, TOGA AW, SAYKIN AJ, SHEN L, FOROUD T, PANKRATZ N, HUENTELMAN MJ, CRAIG DW, GERBER JD, ALLEN AN, CORNEVEAUX JJ, DECHAIRO BM, POTKIN SG, WEINER MW, THOMPSON P & ALZHEIMER’S DISEASE NEUROIMAGING, I. (2010). Voxelwise genome-wide association study (vGWAS). Neuroimage, 53, 1160–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEIN JL, MEDLAND SE, VASQUEZ AA, HIBAR DP, SENSTAD RE, WINKLER AM, TORO R, APPEL K, BARTECEK R, BERGMANN O, BERNARD M, BROWN AA, CANNON DM, CHAKRAVARTY MM, CHRISTOFOROU A, DOMIN M, GRIMM O, HOLLINSHEAD M, HOLMES AJ, HOMUTH G, HOTTENGA JJ, LANGAN C, LOPEZ LM, HANSELL NK, HWANG KS, KIM S, LAJE G, LEE PH, LIU X, LOTH E, LOURDUSAMY A, MATTINGSDAL M, MOHNKE S, MANIEGA SM, NHO K, NUGENT AC, O’BRIEN C, PAPMEYER M, PUTZ B, RAMASAMY A, RASMUSSEN J, RIJPKEMA M, RISACHER SL, RODDEY JC, ROSE EJ, RYTEN M, SHEN L, SPROOTEN E, STRENGMAN E, TEUMER A, TRABZUNI D, TURNER J, VAN EIJK K, VAN ERP TG, VAN TOL MJ, WITTFELD K, WOLF C, WOUDSTRA S, ALEMAN A, ALHUSAINI S, ALMASY L, BINDER EB, BROHAWN DG, CANTOR RM, CARLESS MA, CORVIN A, CZISCH M, CURRAN JE, DAVIES G, DE ALMEIDA MA, DELANTY N, DEPONDT C, DUGGIRALA R, DYER TD, ERK S, FAGERNESS J, FOX PT, FREIMER NB, GILL M, GORING HH, HAGLER DJ, HOEHN D, HOLSBOER F, HOOGMAN M, HOSTEN N, JAHANSHAD N, JOHNSON MP, KASPERAVICIUTE D, KENT JW JR., KOCHUNOV P, LANCASTER JL, LAWRIE SM, LIEWALD DC, MANDL R, MATARIN M, MATTHEISEN M, MEISENZAHL E, MELLE I, MOSES EK, MUHLEISEN TW, NAUCK M, NOTHEN MM, OLVERA RL, PANDOLFO M, PIKE GB, PULS R, REINVANG I, RENTERIA ME, RIETSCHEL M, ROFFMAN JL, ROYLE NA, RUJESCU D, SAVITZ J, SCHNACK HG, SCHNELL K, SEIFERTH N, SMITH C, STEEN VM, VALDES HERNANDEZ MC, VAN DEN HEUVEL M, VAN DER WEE NJ, VAN HAREN NE, VELTMAN JA, VOLZKE H, WALKER R, WESTLYE LT, WHELAN CD, AGARTZ I, BOOMSMA DI, CAVALLERI GL, DALE AM, DJUROVIC S, DREVETS WC, HAGOORT P, HALL J, HEINZ A, JACK CR JR., FOROUD TM, LE HELLARD S, MACCIARDI F, MONTGOMERY GW, POLINE JB, PORTEOUS DJ, SISODIYA SM, STARR JM, SUSSMANN J, TOGA AW, VELTMAN DJ, WALTER H, WEINER MW, ALZHEIMER’S DISEASE NEUROIMAGING, I., CONSORTIUM, E., CONSORTIUM, I., SAGUENAY YOUTH STUDY, G., BIS JC, IKRAM MA, SMITH AV, GUDNASON V, TZOURIO C, VERNOOIJ MW, LAUNER LJ, DECARLI C, SESHADRI S, COHORTS FOR, H., AGING RESEARCH IN GENOMIC EPIDEMIOLOGY, C., ANDREASSEN OA, APOSTOLOVA LG, BASTIN ME, BLANGERO J, BRUNNER HG, BUCKNER RL, CICHON S, COPPOLA G, DE ZUBICARAY GI, DEARY IJ, DONOHOE G, DE GEUS EJ, ESPESETH T, FERNANDEZ G, GLAHN DC, GRABE HJ, HARDY J, HULSHOFF POL HE, JENKINSON M, KAHN RS, MCDONALD C, MCINTOSH AM, MCMAHON FJ, MCMAHON KL, MEYER-LINDENBERG A, MORRIS DW, MULLERMYHSOK B, NICHOLS TE, OPHOFF RA, PAUS T, PAUSOVA Z, PENNINX BW, POTKIN SG, SAMANN PG, SAYKIN AJ, SCHUMANN G, SMOLLER JW, WARDLAW JM, WEALE ME, MARTIN NG, FRANKE B, WRIGHT MJ, THOMPSON PM & ENHANCING NEURO IMAGING GENETICS THROUGH META-ANALYSIS, C. (2012). Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet, 44, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STERGIAKOULI E, HAMSHERE M, HOLMANS P, LANGLEY K, ZAHARIEVA I, DE CG, PSYCHIATRIC, G. C., HAWI Z, KENT L, GILL M, WILLIAMS N, OWEN MJ, O’DONOVAN M & THAPAR A (2012). Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am J Psychiatry, 169, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STERLEY TL, HOWELLS FM & RUSSELL VA (2013a). Evidence for reduced tonic levels of GABA in the hippocampus of an animal model of ADHD, the spontaneously hypertensive rat. Brain Res, 1541, 52–60. [DOI] [PubMed] [Google Scholar]
- STERLEY TL, HOWELLS FM & RUSSELL VA (2013b). Maternal separation increases GABA(A) receptor-mediated modulation of norepinephrine release in the hippocampus of a rat model of ADHD, the spontaneously hypertensive rat. Brain Res, 1497, 23–31. [DOI] [PubMed] [Google Scholar]
- SUGITA S, ICHTCHENKO K, KHVOTCHEV M & SUDHOF TC (1998). alpha-Latrotoxin receptor CIRL/latrophilin 1 (CL1) defines an unusual family of ubiquitous G-protein-linked receptors. G-protein coupling not required for triggering exocytosis. J Biol Chem, 273, 32715–32724. [DOI] [PubMed] [Google Scholar]
- SWISTOWSKI A, PENG J, LIU Q, MALI P, RAO MS, CHENG L & ZENG X (2010). Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells, 28, 1893–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SZOBOT C, ROMAN T, CUNHA R, ACTON P, HUTZ M & ROHDE LA (2005). Brain perfusion and dopaminergic genes in boys with attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 132B, 53–58. [DOI] [PubMed] [Google Scholar]
- SZOBOT CM, ROMAN T, HUTZ MH, GENRO JP, SHIH MC, HOEXTER MQ, JUNIOR N, PECHANSKY F, BRESSAN RA & ROHDE LA (2011). Molecular imaging genetics of methylphenidate response in ADHD and substance use comorbidity. Synapse, 65, 154–159. [DOI] [PubMed] [Google Scholar]
- TAKAHASHI K, TANABE K, OHNUKI M, NARITA M, ICHISAKA T, TOMODA K & YAMANAKA S (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131, 861–872. [DOI] [PubMed] [Google Scholar]
- TAKEUCHI H, TOMITA H, TAKI Y, KIKUCHI Y, ONO C, YU Z, SEKIGUCHI A, NOUCHI R, KOTOZAKI Y, NAKAGAWA S, MIYAUCHI CM, IIZUKA K, YOKOYAMA R, SHINADA T, YAMAMOTO Y, HANAWA S, ARAKI T, HASHIZUME H, KUNITOKI K, SASSA Y & KAWASHIMA R (2015). Cognitive and neural correlates of the 5-repeat allele of the dopamine D4 receptor gene in a population lacking the 7-repeat allele. Neuroimage, 110, 124–135. [DOI] [PubMed] [Google Scholar]
- TAMM L, MENON V, RINGEL J & REISS AL (2004). Event-related FMRI evidence of frontotemporal involvement in aberrant response inhibition and task switching in attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry, 43, 1430–1440. [DOI] [PubMed] [Google Scholar]
- TER HUURNE N, ONNINK M, KAN C, FRANKE B, BUITELAAR J & JENSEN O (2013). Behavioral consequences of aberrant alpha lateralization in attention-deficit/hyperactivity disorder. Biol Psychiatry, 74, 227–233. [DOI] [PubMed] [Google Scholar]
- THAPAR A, COOPER M, EYRE O & LANGLEY K (2013). What have we learnt about the causes of ADHD? J Child Psychol Psychiatry, 54, 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THAPAR A, MARTIN J, MICK E, ARIAS VASQUEZ A, LANGLEY K, SCHERER SW, SCHACHAR R, CROSBIE J, WILLIAMS N, FRANKE B, ELIA J, GLESSNER J, HAKONARSON H, OWEN MJ, FARAONE SV, O’DONOVAN MC & HOLMANS P (2015). Psychiatric gene discoveries shape evidence on ADHD’s biology. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THE NETWORK PATHWAY ANALYSIS SUBGROUP OF THE PSYCHIATRIC GENOMICS CONSORTIUM (2015). Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci, 18, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THOMASON ME, DOUGHERTY RF, COLICH NL, PERRY LM, RYKHLEVSKAIA EI, LOURO HM, HALLMAYER JF, WAUGH CE, BAMMER R, GLOVER GH & GOTLIB IH (2010). COMT genotype affects prefrontal white matter pathways in children and adolescents. Neuroimage, 53, 926–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THOMPSON PM, STEIN JL, MEDLAND SE, HIBAR DP, VASQUEZ AA, RENTERIA ME, TORO R, JAHANSHAD N, SCHUMANN G, FRANKE B, WRIGHT MJ, MARTIN NG, AGARTZ I, ALDA M, ALHUSAINI S, ALMASY L, ALMEIDA J, ALPERT K, ANDREASEN NC, ANDREASSEN OA, APOSTOLOVA LG, APPEL K, ARMSTRONG NJ, ARIBISALA B, BASTIN ME, BAUER M, BEARDEN CE, BERGMANN O, BINDER EB, BLANGERO J, BOCKHOLT HJ, BOEN E, BOIS C, BOOMSMA DI, BOOTH T, BOWMAN IJ, BRALTEN J, BROUWER RM, BRUNNER HG, BROHAWN DG, BUCKNER RL, BUITELAAR J, BULAYEVA K, BUSTILLO JR, CALHOUN VD, CANNON DM, CANTOR RM, CARLESS MA, CASERAS X, CAVALLERI GL, CHAKRAVARTY MM, CHANG KD, CHING CR, CHRISTOFOROU A, CICHON S, CLARK VP, CONROD P, COPPOLA G, CRESPO-FACORRO B, CURRAN JE, CZISCH M, DEARY IJ, DE GEUS EJ, DEN BRABER A, DELVECCHIO G, DEPONDT C, DE HAAN L, DE ZUBICARAY GI, DIMA D, DIMITROVA R, DJUROVIC S, DONG H, DONOHOE G, DUGGIRALA R, DYER TD, EHRLICH S, EKMAN CJ, ELVSASHAGEN T, EMSELL L, ERK S, ESPESETH T, FAGERNESS J, FEARS S, FEDKO I, FERNANDEZ G, FISHER SE, FOROUD T, FOX PT, FRANCKS C, FRANGOU S, FREY EM, FRODL T, FROUIN V, GARAVAN H, GIDDALURU S, GLAHN DC, GODLEWSKA B, GOLDSTEIN RZ, GOLLUB RL, GRABE HJ, GRIMM O, GRUBER O, GUADALUPE T, GUR RE, GUR RC, GORING HH, HAGENAARS S, HAJEK T, HALL GB, HALL J, HARDY J, HARTMAN CA, HASS J, HATTON SN, HAUKVIK UK, HEGENSCHEID K, HEINZ A, HICKIE IB, HO BC, HOEHN D, HOEKSTRA PJ, HOLLINSHEAD M, HOLMES AJ, HOMUTH G, HOOGMAN M, HONG LE, HOSTEN N, HOTTENGA JJ, HULSHOFF POL HE, HWANG KS, JACK CR JR., JENKINSON M, JOHNSTON C, JONSSON EG, KAHN RS, KASPERAVICIUTE D, KELLY S, KIM S, KOCHUNOV P, KOENDERS L, KRAMER B, KWOK JB, LAGOPOULOS J, LAJE G, LANDEN M, LANDMAN BA, LAURIELLO J, LAWRIE SM, LEE PH, LE HELLARD S, LEMAITRE H, LEONARDO CD, LI CS, LIBERG B, LIEWALD DC, LIU X, LOPEZ LM, LOTH E, LOURDUSAMY A, LUCIANO M, MACCIARDI F, MACHIELSEN MW, MACQUEEN GM, MALT UF, MANDL R, MANOACH DS, MARTINOT JL, MATARIN M, MATHER KA, MATTHEISEN M, MATTINGSDAL M, MEYER-LINDENBERG A, MCDONALD C, MCINTOSH AM, MCMAHON FJ, MCMAHON KL, MEISENZAHL E, MELLE I, MILANESCHI Y, MOHNKE S, MONTGOMERY GW, MORRIS DW, MOSES EK, MUELLER BA, MUNOZ MANIEGA S, MUHLEISEN TW, MULLER-MYHSOK B, MWANGI B, NAUCK M, NHO K, NICHOLS TE, NILSSON LG, NUGENT AC, NYBERG L, OLVERA RL, OOSTERLAAN J, OPHOFF RA, PANDOLFO M, PAPALAMPROPOULOU-TSIRIDOU M, PAPMEYER M, PAUS T, PAUSOVA Z, PEARLSON GD, PENNINX BW, PETERSON CP, PFENNIG A, PHILLIPS M, PIKE GB, POLINE JB, POTKIN SG, PUTZ B, RAMASAMY A, RASMUSSEN J, RIETSCHEL M, RIJPKEMA M, RISACHER SL, ROFFMAN JL, ROIZ-SANTIANEZ R, ROMANCZUK-SEIFERTH N, ROSE EJ, ROYLE NA, RUJESCU D, RYTEN M, SACHDEV PS, SALAMI A, SATTERTHWAITE TD, SAVITZ J, SAYKIN AJ, SCANLON C, SCHMAAL L, SCHNACK HG, SCHORK AJ, SCHULZ SC, SCHUR R, SEIDMAN L, SHEN L, SHOEMAKER JM, SIMMONS A, SISODIYA SM, SMITH C, SMOLLER JW, SOARES JC, SPONHEIM SR, SPROOTEN E, STARR JM, STEEN VM, STRAKOWSKI S, STRIKE L, SUSSMANN J, SAMANN PG, TEUMER A, TOGA AW, TORDESILLAS-GUTIERREZ D, TRABZUNI D, TROST S, TURNER J, VAN DEN HEUVEL M, VAN DER WEE NJ, VAN EIJK K, VAN ERP TG, VAN HAREN NE, VAN ‘T ENT D, VAN TOL MJ, VALDES HERNANDEZ MC, VELTMAN DJ, VERSACE A, VOLZKE H, WALKER R, WALTER H, WANG L, WARDLAW JM, WEALE ME, WEINER MW, WEN W, WESTLYE LT, WHALLEY HC, WHELAN CD, WHITE T, WINKLER AM, WITTFELD K, WOLDEHAWARIAT G, WOLF C, ZILLES D, ZWIERS MP, THALAMUTHU A, SCHOFIELD PR, FREIMER NB, LAWRENCE NS, DREVETS W & ALZHEIMER’S DISEASE NEUROIMAGING INITIATIVE, E. C. I. C. S. Y. S. G. (2014). The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav, 8, 153–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIAN L, JIANG T, WANG Y, ZANG Y, HE Y, LIANG M, SUI M, CAO Q, HU S, PENG M & ZHUO Y (2006). Altered resting-state functional connectivity patterns of anterior cingulate cortex in adolescents with attention deficit hyperactivity disorder. Neurosci Lett, 400, 39–43. [DOI] [PubMed] [Google Scholar]
- URBACH A, BAR-NUR O, DALEY GQ & BENVENISTY N (2010). Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell, 6, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- URBAN AE & PURMANN C (2015). Using iPSCs and genomics to catch CNVs in the act. Nat Genet, 47, 100–101. [DOI] [PubMed] [Google Scholar]
- VADODARIA KC, MERTENS J, PAQUOLA A, BARDY C, LI X, JAPPELLI R, FUNG L, MARCHETTO MC, HAMM M, GORRIS M, KOCH P & GAGE FH (2015). Generation of functional human serotonergic neurons from fibroblasts. Mol Psychiatry. [DOI] [PubMed] [Google Scholar]
- VALBONESI S, MAGRI C, TRAVERSA M, FARAONE SV, CATTANEO A, MILANESI E, VALENTI V, GENNARELLI M & SCASSELLATI C (2015). Copy number variants in attention-deficit hyperactive disorder: identification of the 15q13 deletion and its functional role. Psychiatr Genet, 25, 59–70. [DOI] [PubMed] [Google Scholar]
- VALERA EM, FARAONE SV, MURRAY KE & SEIDMAN LJ (2007). Meta-analysis of structural imaging findings in attention-deficit/hyperactivity disorder. Biol Psychiatry, 61, 1361–1369. [DOI] [PubMed] [Google Scholar]
- VAN DER MEER D, HARTMAN CA, RICHARDS J, BRALTEN JB, FRANKE B, OOSTERLAAN J, HESLENFELD DJ, FARAONE SV, BUITELAAR JK & HOEKSTRA PJ (2014). The serotonin transporter gene polymorphism 5-HTTLPR moderates the effects of stress on attention-deficit/hyperactivity disorder. J Child Psychol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN DER MEER D, HOEKSTRA PJ, BRALTEN J, VAN DONKELAAR M, HESLENFELD DJ, OOSTERLAAN J, FARAONE SV, FRANKE B, BUITELAAR JK & HARTMAN CA (2016). Interplay between stress response genes associated with attention-deficit hyperactivity disorder and brain volume. Genes Brain Behav, 15, 627–636. [DOI] [PubMed] [Google Scholar]
- VAN DER MEER D, HOEKSTRA PJ, ZWIERS M, MENNES M, SCHWEREN LJ, FRANKE B, HESLENFELD DJ, OOSTERLAAN J, FARAONE SV, BUITELAAR JK & HARTMAN CA (2015). Brain Correlates of the Interaction Between 5-HTTLPR and Psychosocial Stress Mediating Attention Deficit Hyperactivity Disorder Severity. Am J Psychiatry, 172, 768–775. [DOI] [PubMed] [Google Scholar]
- VAN DER VOET M, HARICH B, FRANKE B & SCHENCK A (2016). ADHD-associated dopamine transporter, latrophilin and neurofibromin share a dopamine-related locomotor signature in Drosophila. Mol Psychiatry, 21, 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN DER VOET M, NIJHOF B, OORTVELD MA & SCHENCK A (2014). Drosophila models of early onset cognitive disorders and their clinical applications. Neurosci Biobehav Rev, 46 Pt 2, 326–342. [DOI] [PubMed] [Google Scholar]
- VAN DIESSEN E, NUMAN T, VAN DELLEN E, VAN DER KOOI AW, BOERSMA M, HOFMAN D, VAN LUTTERVELD R, VAN DIJK BW, VAN STRAATEN EC, HILLEBRAND A & STAM CJ (2015). Opportunities and methodological challenges in EEG and MEG resting state functional brain network research. Clin Neurophysiol, 126, 1468–1481. [DOI] [PubMed] [Google Scholar]
- VAN EWIJK H, BRALTEN J, VAN DUIN E, HAKOBJAN M, HOOGMAN M, OOSTERLAAN J & FRANKE B (in revision). Female-specific association of NOS1 genotype with white matter microstructure in ADHD patients and controls. J Child Psychol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN EWIJK H, HESLENFELD DJ, ZWIERS MP, BUITELAAR JK & OOSTERLAAN J (2012). Diffusion tensor imaging in attention deficit/hyperactivity disorder: a systematic review and meta-analysis. Neurosci Biobehav Rev, 36, 1093–1106. [DOI] [PubMed] [Google Scholar]
- VAN EWIJK H, HESLENFELD DJ, ZWIERS MP, FARAONE SV, LUMAN M, HARTMAN CA, HOEKSTRA PJ, FRANKE B, BUITELAAR JK & OOSTERLAAN J (2014). Different mechanisms of white matter abnormalities in attention-deficit/hyperactivity disorder: a diffusion tensor imaging study. J Am Acad Child Adolesc Psychiatry, 53, 790–799 e793. [DOI] [PubMed] [Google Scholar]
- VAN HULZEN KJE, SCHOLZ CJ, FRANKE B, RIPKE S, MCQUILLIN A, SONUGA-BARKE EJ, GROUP, P. A. W., KELSOE JR, LANDEN M, ANDREASSEN OA, GROUP, P. B. D. W., LESCH KP, WEBER H, FARAONE SV, ARIAS VASQUEZ A & REIF A (2016). Genetic overlap between Attention-Deficit/Hyperactivity Disorder and Bipolar Disorder: Evidence from GWAS meta-analysis. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN PELT S, BOOMSMA DI & FRIES P (2012). Magnetoencephalography in twins reveals a strong genetic determination of the peak frequency of visually induced gamma-band synchronization. J Neurosci, 32, 3388–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN ROOIJ D, HARTMAN CA, VAN DONKELAAR MM, BRALTEN J, VON RHEIN D, HAKOBJAN M, FRANKE B, HESLENFELD DJ, OOSTERLAAN J, ROMMELSE N, BUITELAAR JK & HOEKSTRA PJ (2015a). Variation in serotonin neurotransmission genes affects neural activation during response inhibition in adolescents and young adults with ADHD and healthy controls. World J Biol Psychiatry, 16, 625–634. [DOI] [PubMed] [Google Scholar]
- VAN ROOIJ D, HOEKSTRA PJ, BRALTEN J, HAKOBJAN M, OOSTERLAAN J, FRANKE B, ROMMELSE N, BUITELAAR JK & HARTMAN CA (2015b). Influence of DAT1 and COMT variants on neural activation during response inhibition in adolescents with attention-deficit/hyperactivity disorder and healthy controls. Psychol Med, 45, 3159–3170. [DOI] [PubMed] [Google Scholar]
- VAN ROOIJ D, HOEKSTRA PJ, MENNES M, VON RHEIN D, THISSEN AJ, HESLENFELD D, ZWIERS MP, FARAONE SV, OOSTERLAAN J, FRANKE B, ROMMELSE N, BUITELAAR JK & HARTMAN CA (2015c). Distinguishing Adolescents With ADHD From Their Unaffected Siblings and Healthy Comparison Subjects by Neural Activation Patterns During Response Inhibition. Am J Psychiatry, 172, 674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN TOL HH, WU CM, GUAN HC, OHARA K, BUNZOW JR, CIVELLI O, KENNEDY J, SEEMAN P, NIZNIK HB & JOVANOVIC V (1992). Multiple dopamine D4 receptor variants in the human population. Nature, 358, 149–152. [DOI] [PubMed] [Google Scholar]
- VANDERWERT RE & NELSON CA (2014). The use of near-infrared spectroscopy in the study of typical and atypical development. Neuroimage, 85 Pt 1, 264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VILLEMONTEIX T, DE BRITO SA, SLAMA H, KAVEC M, BALERIAUX D, METENS T, BAIJOT S, MARY A, RAMOZ N, SEPTIER M, GORWOOD P, PEIGNEUX P & MASSAT I (2015). Structural correlates of COMT Val158Met polymorphism in childhood ADHD: a voxel-based morphometry study. World J Biol Psychiatry, 16, 190–199. [DOI] [PubMed] [Google Scholar]
- VILLRINGER A & CHANCE B (1997). Non-invasive optical spectroscopy and imaging of human brain function. Trends Neurosci, 20, 435–442. [DOI] [PubMed] [Google Scholar]
- VOINEAGU I, WANG X, JOHNSTON P, LOWE JK, TIAN Y, HORVATH S, MILL J, CANTOR RM, BLENCOWE BJ & GESCHWIND DH (2011). Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature, 474, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOLKOW ND, WANG GJ, NEWCORN J, TELANG F, SOLANTO MV, FOWLER JS, LOGAN J, MA Y, SCHULZ K, PRADHAN K, WONG C & SWANSON JM (2007). Depressed dopamine activity in caudate and preliminary evidence of limbic involvement in adults with attention-deficit/hyperactivity disorder. Arch Gen Psychiatry, 64, 932–940. [DOI] [PubMed] [Google Scholar]
- VON RHEIN D, COOLS R, ZWIERS MP, VAN DER SCHAAF M, FRANKE B, LUMAN M, OOSTERLAAN J, HESLENFELD DJ, HOEKSTRA PJ, HARTMAN CA, FARAONE SV, VAN ROOIJ D, VAN DONGEN EV, LOJOWSKA M, MENNES M & BUITELAAR J (2015). Increased neural responses to reward in adolescents and young adults with attention-deficit/hyperactivity disorder and their unaffected siblings. J Am Acad Child Adolesc Psychiatry, 54, 394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAIDER J, ARARAGI N, GUTKNECHT L & LESCH KP (2011). Tryptophan hydroxylase-2 (TPH2) in disorders of cognitive control and emotion regulation: a perspective. Psychoneuroendocrinology, 36, 393–405. [DOI] [PubMed] [Google Scholar]
- WALITZA S, RENNER TJ, DEMPFLE A, KONRAD K, WEWETZER C, HALBACH A, HERPERTZDAHLMANN B, REMSCHMIDT H, SMIDT J, LINDER M, FLIERL L, KNOLKER U, FRIEDEL S, SCHAFER H, GROSS C, HEBEBRAND J, WARNKE A & LESCH KP (2005). Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase-2 gene in attention-deficit/hyperactivity disorder. Mol Psychiatry, 10, 1126–1132. [DOI] [PubMed] [Google Scholar]
- WALLIS D, HILL DS, MENDEZ IA, ABBOTT LC, FINNELL RH, WELLMAN PJ & SETLOW B (2012). Initial characterization of mice null for Lphn3, a gene implicated in ADHD and addiction. Brain Res, 1463, 85–92. [DOI] [PubMed] [Google Scholar]
- WALTHER DJ, PETER JU, BASHAMMAKH S, HORTNAGL H, VOITS M, FINK H & BADER M (2003). Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science, 299, 76. [DOI] [PubMed] [Google Scholar]
- WANG E, DING YC, FLODMAN P, KIDD JR, KIDD KK, GRADY DL, RYDER OA, SPENCE MA, SWANSON JM & MOYZIS RK (2004). The genetic architecture of selection at the human dopamine receptor D4 (DRD4) gene locus. Am J Hum Genet, 74, 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG M, ZHONG Z, ZHONG Y, ZHANG W & WANG H (2015a). The zebrafish period2 protein positively regulates the circadian clock through mediation of retinoic acid receptor (RAR)-related orphan receptor alpha (Roralpha). J Biol Chem, 290, 4367–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG P, LIN M, PEDROSA E, HRABOVSKY A, ZHANG Z, GUO W, LACHMAN HM & ZHENG D (2015b). CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in neurodevelopment. Mol Autism, 6, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEBER H, KITTEL-SCHNEIDER S, HEUPEL J, WEISSFLOG L, KENT L, FREUDENBERG F, ALTTOA A, POST A, HERTERICH S, HAAVIK J, HALMOY A, FASMER OB, LANDAAS ET, JOHANSSON S, CORMAND B, RIBASES M, SANCHEZ-MORA C, RAMOS-QUIROGA JA, FRANKE B, LESCH KP & REIF A (2015). On the role of NOS1 ex1f-VNTR in ADHD-allelic, subgroup, and meta-analysis. Am J Med Genet B Neuropsychiatr Genet. [DOI] [PubMed] [Google Scholar]
- WEBER P, LUTSCHG J & FAHNENSTICH H (2005). Cerebral hemodynamic changes in response to an executive function task in children with attention-deficit hyperactivity disorder measured by near-infrared spectroscopy. J Dev Behav Pediatr, 26, 105–111. [DOI] [PubMed] [Google Scholar]
- WEN Z, NGUYEN HN, GUO Z, LALLI MA, WANG X, SU Y, KIM NS, YOON KJ, SHIN J, ZHANG C, MAKRI G, NAUEN D, YU H, GUZMAN E, CHIANG CH, YORITOMO N, KAIBUCHI K, ZOU J, CHRISTIAN KM, CHENG L, ROSS CA, MARGOLIS RL, CHEN G, KOSIK KS, SONG H & MING GL (2014). Synaptic dysregulation in a human iPS cell model of mental disorders. Nature, 515, 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITTLE S, SIMMONS JG, DENNISON M, VIJAYAKUMAR N, SCHWARTZ O, YAP MB, SHEEBER L & ALLEN NB (2014). Positive parenting predicts the development of adolescent brain structure: a longitudinal study. Dev Cogn Neurosci, 8, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILENS TE, BIEDERMAN J, FARAONE SV, MARTELON M, WESTERBERG D & SPENCER TJ (2009). Presenting ADHD symptoms, subtypes, and comorbid disorders in clinically referred adults with ADHD. J Clin Psychiatry, 70, 1557–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS J, SAGVOLDEN G, TAYLOR E & SAGVOLDEN T (2009a). Dynamic behavioural changes in the Spontaneously Hyperactive Rat: 1. Control by place, timing, and reinforcement rate. Behav Brain Res, 198, 273–282. [DOI] [PubMed] [Google Scholar]
- WILLIAMS J, SAGVOLDEN G, TAYLOR E & SAGVOLDEN T (2009b). Dynamic behavioural changes in the Spontaneously Hyperactive Rat: 2. Control by novelty. Behav Brain Res, 198, 283–290. [DOI] [PubMed] [Google Scholar]
- WILLIAMS J, SAGVOLDEN G, TAYLOR E & SAGVOLDEN T (2009c). Dynamic behavioural changes in the Spontaneously Hyperactive Rat: 3. Control by reinforcer rate changes and predictability. Behav Brain Res, 198, 291–297. [DOI] [PubMed] [Google Scholar]
- WILLIAMS NM, FRANKE B, MICK E, ANNEY RJ, FREITAG CM, GILL M, THAPAR A, O’DONOVAN MC, OWEN MJ, HOLMANS P, KENT L, MIDDLETON F, ZHANG-JAMES Y, LIU L, MEYER J, NGUYEN TT, ROMANOS J, ROMANOS M, SEITZ C, RENNER TJ, WALITZA S, WARNKE A, PALMASON H, BUITELAAR J, ROMMELSE N, VASQUEZ AA, HAWI Z, LANGLEY K, SERGEANT J, STEINHAUSEN HC, ROEYERS H, BIEDERMAN J, ZAHARIEVA I, HAKONARSON H, ELIA J, LIONEL AC, CROSBIE J, MARSHALL CR, SCHACHAR R, SCHERER SW, TODOROV A, SMALLEY SL, LOO S, NELSON S, SHTIR C, ASHERSON P, REIF A, LESCH KP & FARAONE SV (2012). Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. Am J Psychiatry, 169, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS NM, ZAHARIEVA I, MARTIN A, LANGLEY K, MANTRIPRAGADA K, FOSSDAL R, STEFANSSON H, STEFANSSON K, MAGNUSSON P, GUDMUNDSSON OO, GUSTAFSSON O, HOLMANS P, OWEN MJ, O’DONOVAN M & THAPAR A (2010). Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet, 376, 1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLSEY AJ, SANDERS SJ, LI M, DONG S, TEBBENKAMP AT, MUHLE RA, REILLY SK, LIN L, FERTUZINHOS S, MILLER JA, MURTHA MT, BICHSEL C, NIU W, COTNEY J, ERCANSENCICEK AG, GOCKLEY J, GUPTA AR, HAN W, HE X, HOFFMAN EJ, KLEI L, LEI J, LIU W, LIU L, LU C, XU X, ZHU Y, MANE SM, LEIN ES, WEI L, NOONAN JP, ROEDER K, DEVLIN B, SESTAN N & STATE MW (2013). Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell, 155, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINKLER AM, KOCHUNOV P, BLANGERO J, ALMASY L, ZILLES K, FOX PT, DUGGIRALA R & GLAHN DC (2010). Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage, 53, 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOMERSLEY JS, DIMATELIS JJ & RUSSELL VA (2015). Proteomic analysis of maternal separation-induced striatal changes in a rat model of ADHD: The spontaneously hypertensive rat. J Neurosci Methods, 252, 64–74. [DOI] [PubMed] [Google Scholar]
- WU J, XIAO H, SUN H, ZOU L & ZHU LQ (2012). Role of dopamine receptors in ADHD: a systematic meta-analysis. Mol Neurobiol, 45, 605–620. [DOI] [PubMed] [Google Scholar]
- WU Z, YANG L & WANG Y (2014). Applying Imaging Genetics to ADHD: the Promises and the Challenges. Mol Neurobiol. [DOI] [PubMed] [Google Scholar]
- WU ZM, BRALTEN J, CAO QJ, HOOGMAN M, ZWIERS MP, AN L, SUN L, YANG L, ZANG YF, FRANKE B & WANG YF (2016). White Matter Microstructural Alterations in Children with ADHD: Categorical and Dimensional Perspectives. Neuropsychopharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIAO T, XIAO Z, KE X, HONG S, YANG H, SU Y, CHU K, XIAO X, SHEN J & LIU Y (2012). Response inhibition impairment in high functioning autism and attention deficit hyperactivity disorder: evidence from near-infrared spectroscopy data. PLoS One, 7, e46569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG L, NEALE BM, LIU L, LEE SH, WRAY NR, JI N, LI H, QIAN Q, WANG D, LI J, FARAONE SV, WANG Y, PSYCHIATRIC, G. C. A. S., DOYLE AE, REIF A, ROTHENBERGER A, FRANKE B, SONUGA-BARKE EJ, STEINHAUSEN HC, BUITELAAR JK, KUNTSI J, BIEDERMAN J, LESCH KP, KENT L, ASHERSON P, OADES RD, LOO SK, NELSON SF, FARAONE SV, SMALLEY SL, BANASCHEWSKI T, ARIAS VASQUEZ A, TODOROV A, CHARACH A, MIRANDA A, WARNKE A, THAPAR A, NEALE BM, CORMAND B, FREITAG C, MICK E, MULAS F, MIDDLETON F, HAKONARSONHAKONARSON H, PALMASON H, SCHAFER H, ROEYERS H, MCGOUGH JJ, ROMANOS J, CROSBIE J, MEYER J, RAMOSQUIROGA JA, SERGEANT J, ELIA J, LANGELY K, NISENBAUM L, ROMANOS M, DALY MJ, RIBASES M, GILL M, O’DONOVAN M, OWEN M, CASAS M, BAYES M, LAMBREGTSROMMELSE N, WILLIAMS N, HOLMANS P, ANNEY RJ, EBSTEIN RP, SCHACHAR R, MEDLAND SE, RIPKE S, WALITZA S, NGUYEN TT, RENNER TJ & HU X (2013). Polygenic transmission and complex neuro developmental network for attention deficit hyperactivity disorder: genome-wide association study of both common and rare variants. Am J Med Genet B Neuropsychiatr Genet, 162B, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YONCHEVA YN, SOMANDEPALLI K, REISS PT, KELLY C, DI MARTINO A, LAZAR M, ZHOU J, MILHAM MP & CASTELLANOS FX (2016). Mode of Anisotropy Reveals Global Diffusion Alterations in Attention-Deficit/Hyperactivity Disorder. J Am Acad Child Adolesc Psychiatry, 55, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YU J, VODYANIK MA, SMUGA-OTTO K, ANTOSIEWICZ-BOURGET J, FRANE JL, TIAN S, NIE J, JONSDOTTIR GA, RUOTTI V, STEWART R, SLUKVIN II & THOMSON JA (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science, 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- ZANG YF, HE Y, ZHU CZ, CAO QJ, SUI MQ, LIANG M, TIAN LX, JIANG TZ & WANG YF (2007). Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev, 29, 83–91. [DOI] [PubMed] [Google Scholar]
- ZAYATS T, ATHANASIU L, SONDERBY I, DJUROVIC S, WESTLYE LT, TAMNES CK, FLADBY T, AASE H, ZEINER P, REICHBORN-KJENNERUD T, KNAPPSKOG PM, KNUDSEN GP, ANDREASSEN OA, JOHANSSON S & HAAVIK J (2015). Genome-wide analysis of attention deficit hyperactivity disorder in Norway. PLoS One, 10, e0122501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG YS, PAK C, HAN Y, AHLENIUS H, ZHANG ZJ, CHANDA S, MARRO S, PATZKE C, ACUNA C, COVY J, XU W, YANG N, DANKO T, CHEN L, WERNIG M & SUDHOF TC (2013). Rapid Single-Step Induction of Functional Neurons from Human Pluripotent Stem Cells. Neuron, 78, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO L, LIN Y, LAO G, WANG Y, GUAN L, WEI J, YANG Z, NI P, LI X, JIANG Z, LI T, HAO X, LIN D, CAO L & MA X (2015). Association study of dopamine receptor genes polymorphism with cognitive functions in bipolar I disorder patients. J Affect Disord, 170, 85–90. [DOI] [PubMed] [Google Scholar]
- ZHOU K, DEMPFLE A, ARCOS-BURGOS M, BAKKER SC, BANASCHEWSKI T, BIEDERMAN J, BUITELAAR J, CASTELLANOS FX, DOYLE A, EBSTEIN RP, EKHOLM J, FORABOSCO P, FRANKE B, FREITAG C, FRIEDEL S, GILL M, HEBEBRAND J, HINNEY A, JACOB C, LESCH KP, LOO SK, LOPERA F, MCCRACKEN JT, MCGOUGH JJ, MEYER J, MICK E, MIRANDA A, MUENKE M, MULAS F, NELSON SF, NGUYEN TT, OADES RD, OGDIE MN, PALACIO JD, PINEDA D, REIF A, RENNER TJ, ROEYERS H, ROMANOS M, ROTHENBERGER A, SCHAFER H, SERGEANT J, SINKE RJ, SMALLEY SL, SONUGA-BARKE E, STEINHAUSEN HC, VAN DER MEULEN E, WALITZA S, WARNKE A, LEWIS CM, FARAONE SV & ASHERSON P (2008). Meta-analysis of genome-wide linkage scans of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 147B, 1392–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU J, LEE KP, SPENCER TJ, BIEDERMAN J & BHIDE PG (2014). Transgenerational transmission of hyperactivity in a mouse model of ADHD. J Neurosci, 34, 2768–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHUANG X, OOSTING RS, JONES SR, GAINETDINOV RR, MILLER GW, CARON MG & HEN R (2001). Hyperactivity and impaired response habituation in hyperdopaminergic mice. Proc Natl Acad Sci U S A, 98, 1982–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZIMMER L (2009). Positron emission tomography neuroimaging for a better understanding of the biology of ADHD. Neuropharmacology, 57, 601–607. [DOI] [PubMed] [Google Scholar]
- ZIMMERMANN AM, JENE T, WOLF M, GORLICH A, GURNIAK CB, SASSOE-POGNETTO M, WITKE W, FRIAUF E & RUST MB (2015). Attention-Deficit/Hyperactivity Disorder-like Phenotype in a Mouse Model with Impaired Actin Dynamics. Biol Psychiatry, 78, 95–106. [DOI] [PubMed] [Google Scholar]
- ALBAYRAK O, PUTTER C, VOLCKMAR AL, CICHON S, HOFFMANN P, NOTHEN MM, JOCKEL KH, SCHREIBER S, WICHMANN HE, FARAONE SV, NEALE BM, HERPERTZ-DAHLMANN B, LEHMKUHL G, SINZIG J, RENNER TJ, ROMANOS M, WARNKE A, LESCH KP, REIF A, SCHIMMELMANN BG, SCHERAG A, HEBEBRAND J, HINNEY A & PSYCHIATRIC, G. C. A. S. (2013). Common obesity risk alleles in childhood attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 162B, 295–305. [DOI] [PubMed] [Google Scholar]
- BARR CL, FENG Y, WIGG KG, SCHACHAR R, TANNOCK R, ROBERTS W, MALONE M & KENNEDY JL (2001). 5’-untranslated region of the dopamine D4 receptor gene and attention-deficit hyperactivity disorder. Am J Med Genet, 105, 84–90. [PubMed] [Google Scholar]
- BROOKES KJ, CHEN W, XU X, TAYLOR E & ASHERSON P (2006b). Association of fatty acid desaturase genes with attention-deficit/hyperactivity disorder. Biol Psychiatry, 60, 1053–1061. [DOI] [PubMed] [Google Scholar]
- BROOKES KJ, MILL J, GUINDALINI C, CURRAN S, XU X, KNIGHT J, CHEN CK, HUANG YS, SETHNA V, TAYLOR E, CHEN W, BREEN G & ASHERSON P (2006c). A common haplotype of the dopamine transporter gene associated with attention-deficit/hyperactivity disorder and interacting with maternal use of alcohol during pregnancy. Arch Gen Psychiatry, 63, 74–81. [DOI] [PubMed] [Google Scholar]
- BROPHY K, HAWI Z, KIRLEY A, FITZGERALD M & GILL M (2002). Synaptosomal-associated protein 25 (SNAP-25) and attention deficit hyperactivity disorder (ADHD): evidence of linkage and association in the Irish population. Mol Psychiatry, 7, 913–917. [DOI] [PubMed] [Google Scholar]
- CHO SC, KIM JW, KIM BN, HWANG JW, SHIN MS, PARK M, KIM SA, CHO DY, YOO HJ, CHUNG US, SON JW & PARK TW (2008). Association between the alpha-2C-adrenergic receptor gene and attention deficit hyperactivity disorder in a Korean sample. Neurosci Lett, 446, 108–111. [DOI] [PubMed] [Google Scholar]
- CHOUDHRY Z, SENGUPTA SM, GRIZENKO N, THAKUR GA, FORTIER ME, SCHMITZ N & JOOBER R (2013). Association between obesity-related gene FTO and ADHD. Obesity (Silver Spring), 21, E738–744. [DOI] [PubMed] [Google Scholar]
- COMINGS DE, COMINGS BG, MUHLEMAN D, DIETZ G, SHAHBAHRAMI B, TAST D, KNELL E, KOCSIS P, BAUMGARTEN R, KOVACS BW & et al. (1991). The dopamine D2 receptor locus as a modifying gene in neuropsychiatric disorders. JAMA, 266, 1793–1800. [PubMed] [Google Scholar]
- COOK EH JR., STEIN MA, KRASOWSKI MD, COX NJ, OLKON DM, KIEFFER JE & LEVENTHAL BL (1995). Association of attention-deficit disorder and the dopamine transporter gene. Am J Hum Genet, 56, 993–998. [PMC free article] [PubMed] [Google Scholar]
- DALY G, HAWI Z, FITZGERALD M & GILL M (1999). Mapping susceptibility loci in attention deficit hyperactivity disorder: preferential transmission of parental alleles at DAT1, DBH and DRD5 to affected children. Mol Psychiatry, 4, 192–196. [DOI] [PubMed] [Google Scholar]
- DE LUCA V, MUGLIA P, VINCENT JB, LANKTREE M, JAIN U & KENNEDY JL (2004). Adrenergic alpha 2C receptor genomic organization: association study in adult ADHD. Am J Med Genet B Neuropsychiatr Genet, 127B, 65–67. [DOI] [PubMed] [Google Scholar]
- DE SILVA MG, ELLIOTT K, DAHL HH, FITZPATRICK E, WILCOX S, DELATYCKI M, WILLIAMSON R, EFRON D, LYNCH M & FORREST S (2003). Disruption of a novel member of a sodium/hydrogen exchanger family and DOCK3 is associated with an attention deficit hyperactivity disorder-like phenotype. J Med Genet, 40, 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELIA J, CAPASSO M, ZAHEER Z, LANTIERI F, AMBROSINI P, BERRETTINI W, DEVOTO M & HAKONARSON H (2009). Candidate gene analysis in an ongoing genome-wide association study of attention-deficit hyperactivity disorder: suggestive association signals in ADRA1A. Psychiatr Genet, 19, 134–141. [DOI] [PubMed] [Google Scholar]
- FRIEDEL S, HORRO FF, WERMTER AK, GELLER F, DEMPFLE A, REICHWALD K, SMIDT J, BRONNER G, KONRAD K, HERPERTZ-DAHLMANN B, WARNKE A, HEMMINGER U, LINDER M, KIEFL H, GOLDSCHMIDT HP, SIEGFRIED W, REMSCHMIDT H, HINNEY A & HEBEBRAND J (2005). Mutation screen of the brain derived neurotrophic factor gene (BDNF): identification of several genetic variants and association studies in patients with obesity, eating disorders, and attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 132B, 96–99. [DOI] [PubMed] [Google Scholar]
- GALILI-WEISSTUB E & SEGMAN RH (2003). Attention deficit and hyperactivity disorder: review of genetic association studies. Isr J Psychiatry Relat Sci, 40, 57–66. [PubMed] [Google Scholar]
- GAO Q, LIU L, CHEN Y, LI H, YANG L, WANG Y & QIAN Q (2015). Synaptosome-related (SNARE) genes and their interactions contribute to the susceptibility and working memory of attention-deficit/hyperactivity disorder in males. Prog Neuropsychopharmacol Biol Psychiatry, 57, 132–139. [DOI] [PubMed] [Google Scholar]
- GUAN L, WANG B, CHEN Y, YANG L, LI J, QIAN Q, WANG Z, FARAONE SV & WANG Y (2009). A high-density single-nucleotide polymorphism screen of 23 candidate genes in attention deficit hyperactivity disorder: suggesting multiple susceptibility genes among Chinese Han population. Mol Psychiatry, 14, 546–554. [DOI] [PubMed] [Google Scholar]
- HAWI Z, MATTHEWS N, BARRY E, KIRLEY A, WAGNER J, WALLACE RH, HEUSSLER HS, VANCE A, GILL M & BELLGROVE MA (2013). A high density linkage disequilibrium mapping in 14 noradrenergic genes: evidence of association between SLC6A2, ADRA1B and ADHD. Psychopharmacology (Berl), 225, 895–902. [DOI] [PubMed] [Google Scholar]
- HOHMANN S, HOHM E, TREUTLEIN J, BLOMEYER D, JENNEN-STEINMETZ C, SCHMIDT MH, ESSER G, BANASCHEWSKI T, BRANDEIS D & LAUCHT M (2015). Association of norepinephrine transporter (NET, SLC6A2) genotype with ADHD-related phenotypes: findings of a longitudinal study from birth to adolescence. Psychiatry Res, 226, 425–433. [DOI] [PubMed] [Google Scholar]
- HU YF, CARON MG & SIEBER-BLUM M (2009). Norepinephrine transport-mediated gene expression in noradrenergic neurogenesis. BMC Genomics, 10, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LABBE A, LIU A, ATHERTON J, GIZENKO N, FORTIER ME, SENGUPTA SM & RIDHA J (2012). Refining psychiatric phenotypes for response to treatment: contribution of LPHN3 in ADHD. Am J Med Genet B Neuropsychiatr Genet, 159B, 776–785. [DOI] [PubMed] [Google Scholar]
- LAHOSTE GJ, SWANSON JM, WIGAL SB, GLABE C, WIGAL T, KING N & KENNEDY JL (1996). Dopamine D4 receptor gene polymorphism is associated with attention deficit hyperactivity disorder. Mol Psychiatry, 1, 121–124. [PubMed] [Google Scholar]
- LANTIERI F, GLESSNER JT, HAKONARSON H, ELIA J & DEVOTO M (2010). Analysis of GWAS top hits in ADHD suggests association to two polymorphisms located in genes expressed in the cerebellum. Am J Med Genet B Neuropsychiatr Genet, 153B, 1127–1133. [DOI] [PubMed] [Google Scholar]
- LASKY-SU J, ANNEY RJ, NEALE BM, FRANKE B, ZHOU K, MALLER JB, VASQUEZ AA, CHEN W, ASHERSON P, BUITELAAR J, BANASCHEWSKI T, EBSTEIN R, GILL M, MIRANDA A, MULAS F, OADES RD, ROEYERS H, ROTHENBERGER A, SERGEANT J, SONUGA-BARKE E, STEINHAUSEN HC, TAYLOR E, DALY M, LAIRD N, LANGE C & FARAONE SV (2008a). Genome-wide association scan of the time to onset of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 147B, 1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI J, WANG Y, HU S, ZHOU R, YU X, WANG B, GUAN L, YANG L, ZHANG F & FARAONE SV (2008). The monoamine oxidase B gene exhibits significant association to ADHD. Am J Med Genet B Neuropsychiatr Genet, 147, 370–374. [DOI] [PubMed] [Google Scholar]
- LI J, WANG Y, ZHOU R, ZHANG H, YANG L, WANG B & FARAONE SV (2006). Association between polymorphisms in serotonin 2C receptor gene and attention-deficit/hyperactivity disorder in Han Chinese subjects. Neurosci Lett, 407, 107–111. [DOI] [PubMed] [Google Scholar]
- LIONEL AC, CROSBIE J, BARBOSA N, GOODALE T, THIRUVAHINDRAPURAM B, RICKABY J, GAZZELLONE M, CARSON AR, HOWE JL, WANG Z, WEI J, STEWART AF, ROBERTS R, MCPHERSON R, FIEBIG A, FRANKE A, SCHREIBER S, ZWAIGENBAUM L, FERNANDEZ BA, ROBERTS W, ARNOLD PD, SZATMARI P, MARSHALL CR, SCHACHAR R & SCHERER SW (2011). Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med, 3, 95ra75. [DOI] [PubMed] [Google Scholar]
- LIONEL AC, TAMMIMIES K, VAAGS AK, ROSENFELD JA, AHN JW, MERICO D, NOOR A, RUNKE CK, PILLALAMARRI VK, CARTER MT, GAZZELLONE MJ, THIRUVAHINDRAPURAM B, FAGERBERG C, LAULUND LW, PELLECCHIA G, LAMOUREUX S, DESHPANDE C, CLAYTON-SMITH J, WHITE AC, LEATHER S, TROUNCE J, MELANIE BEDFORD H, HATCHWELL E, EIS PS, YUEN RK, WALKER S, UDDIN M, GERAGHTY MT, NIKKEL SM, TOMIAK EM, FERNANDEZ BA, SORENI N, CROSBIE J, ARNOLD PD, SCHACHAR RJ, ROBERTS W, PATERSON AD, SO J, SZATMARI P, CHRYSLER C, WOODBURY-SMITH M, BRIAN LOWRY R, ZWAIGENBAUM L, MANDYAM D, WEI J, MACDONALD JR, HOWE JL, NALPATHAMKALAM T, WANG Z, TOLSON D, COBB DS, WILKS TM, SORENSEN MJ, BADER PI, AN Y, WU BL, MUSUMECI SA, ROMANO C, POSTORIVO D, NARDONE AM, MONICA MD, SCARANO G, ZOCCANTE L, NOVARA F, ZUFFARDI O, CICCONE R, ANTONA V, CARELLA M, ZELANTE L, CAVALLI P, POGGIANI C, CAVALLARI U, ARGIROPOULOS B, CHERNOS J, BRASCH-ANDERSEN C, SPEEVAK M, FICHERA M, OGILVIE CM, SHEN Y, HODGE JC, TALKOWSKI ME, STAVROPOULOS DJ, MARSHALL CR & SCHERER SW (2014). Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes. Hum Mol Genet, 23, 2752–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU L, CHEN Y, LI H, QIAN Q, YANG L, GLATT SJ, FARAONE SV & WANG Y (2013). Association between SYP with attention-deficit/hyperactivity disorder in Chinese Han subjects: differences among subtypes and genders. Psychiatry Res, 210, 308–314. [DOI] [PubMed] [Google Scholar]
- MANOR I, EISENBERG J, TYANO S, SEVER Y, COHEN H, EBSTEIN RP & KOTLER M (2001). Family-based association study of the serotonin transporter promoter region polymorphism (5-HTTLPR) in attention deficit hyperactivity disorder. Am J Med Genet, 105, 91–95. [PubMed] [Google Scholar]
- MICK E, NEALE B, MIDDLETON FA, MCGOUGH JJ & FARAONE SV (2008). Genome-wide association study of response to methylphenidate in 187 children with attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet, 147B, 1412–1418. [DOI] [PubMed] [Google Scholar]
- MISENER VL, LUCA P, AZEKE O, CROSBIE J, WALDMAN I, TANNOCK R, ROBERTS W, MALONE M, SCHACHAR R, ICKOWICZ A, KENNEDY JL & BARR CL (2004). Linkage of the dopamine receptor D1 gene to attention-deficit/hyperactivity disorder. Mol Psychiatry, 9, 500–509. [DOI] [PubMed] [Google Scholar]
- OADES RD, LASKY-SU J, CHRISTIANSEN H, FARAONE SV, SONUGA-BARKE EJ, BANASCHEWSKI T, CHEN W, ANNEY RJ, BUITELAAR JK, EBSTEIN RP, FRANKE B, GILL M, MIRANDA A, ROEYERS H, ROTHENBERGER A, SERGEANT JA, STEINHAUSEN HC, TAYLOR EA, THOMPSON M & ASHERSON P (2008). The influence of serotonin- and other genes on impulsive behavioral aggression and cognitive impulsivity in children with attention-deficit/hyperactivity disorder (ADHD): Findings from a family-based association test (FBAT) analysis. Behav Brain Funct, 4, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAN YQ, QIAO L, XUE XD & FU JH (2015). Association between ANKK1 (rs1800497) polymorphism of DRD2 gene and attention deficit hyperactivity disorder: a meta-analysis. Neurosci Lett, 590, 101–105. [DOI] [PubMed] [Google Scholar]
- PARK S, KIM BN, CHO SC, KIM JW, KIM JI, SHIN MS, YOO HJ, HAN DH & CHEONG JH (2014). The metabotropic glutamate receptor subtype 7 rs3792452 polymorphism is associated with the response to methylphenidate in children with attention-deficit/hyperactivity disorder. J Child Adolesc Psychopharmacol, 24, 223–227. [DOI] [PubMed] [Google Scholar]
- PASCOLI V, VALJENT E, CORBILLE AG, CORVOL JC, TASSIN JP, GIRAULT JA & HERVE D (2005). cAMP and extracellular signal-regulated kinase signaling in response to d-amphetamine and methylphenidate in the prefrontal cortex in vivo: role of beta 1-adrenoceptors. Mol Pharmacol, 68, 421–429. [DOI] [PubMed] [Google Scholar]
- PAYTON A, HOLMES J, BARRETT JH, HEVER T, FITZPATRICK H, TRUMPER AL, HARRINGTON R, MCGUFFIN P, O’DONOVAN M, OWEN M, OLLIER W, WORTHINGTON J & THAPAR A (2001). Examining for association between candidate gene polymorphisms in the dopamine pathway and attention-deficit hyperactivity disorder: a family-based study. Am J Med Genet, 105, 464–470. [DOI] [PubMed] [Google Scholar]
- PLAISANCIE J, BOUNEAU L, CANCES C, GARNIER C, BENESTEAU J, LEONARD S, BOURROUILLOU G, CALVAS P, VIGOUROUX A, JULIA S & BIETH E (2014). Distal 10q monosomy: new evidence for a neurobehavioral condition? Eur J Med Genet, 57, 47–53. [DOI] [PubMed] [Google Scholar]
- POLINA ER, ROVARIS DL, DE AZEREDO LA, MOTA NR, VITOLA ES, SILVA KL, GUIMARAES-DA-SILVA PO, PICON FA, BELMONTE-DE-ABREU P, ROHDE LA, GREVET EH & BAU CH (2014). ADHD diagnosis may influence the association between polymorphisms in nicotinic acetylcholine receptor genes and tobacco smoking. Neuromolecular Med, 16, 389–397. [DOI] [PubMed] [Google Scholar]
- QUIST JF, BARR CL, SCHACHAR R, ROBERTS W, MALONE M, TANNOCK R, BASILE VS, BEITCHMAN J & KENNEDY JL (2000). Evidence for the serotonin HTR2A receptor gene as a susceptibility factor in attention deficit hyperactivity disorder (ADHD). Mol Psychiatry, 5, 537–541. [DOI] [PubMed] [Google Scholar]
- REIF A, NGUYEN TT, WEISSFLOG L, JACOB CP, ROMANOS M, RENNER TJ, BUTTENSCHON HN, KITTEL-SCHNEIDER S, GESSNER A, WEBER H, NEUNER M, GROSS-LESCH S, ZAMZOW K, KREIKER S, WALITZA S, MEYER J, FREITAG CM, BOSCH R, CASAS M, GOMEZ N, RIBASES M, BAYES M, BUITELAAR JK, KIEMENEY LA, KOOIJ JJ, KAN CC, HOOGMAN M, JOHANSSON S, JACOBSEN KK, KNAPPSKOG PM, FASMER OB, ASHERSON P, WARNKE A, GRABE HJ, MAHLER J, TEUMER A, VOLZKE H, MORS ON, SCHAFER H, RAMOS-QUIROGA JA, CORMAND B, HAAVIK J, FRANKE B & LESCH KP (2011). DIRAS2 is associated with adult ADHD, related traits, and co-morbid disorders. Neuropsychopharmacology, 36, 2318–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIBASES M, HERVAS A, RAMOS-QUIROGA JA, BOSCH R, BIELSA A, GASTAMINZA X, FERNANDEZ-ANGUIANO M, NOGUEIRA M, GOMEZ-BARROS N, VALERO S, GRATACOS M, ESTIVILL X, CASAS M, CORMAND B & BAYES M (2008). Association study of 10 genes encoding neurotrophic factors and their receptors in adult and child attention-deficit/hyperactivity disorder. Biol Psychiatry, 63, 935–945. [DOI] [PubMed] [Google Scholar]
- RIBASES M, RAMOS-QUIROGA JA, HERVAS A, BOSCH R, BIELSA A, GASTAMINZA X, ARTIGAS J, RODRIGUEZ-BEN S, ESTIVILL X, CASAS M, CORMAND B & BAYES M (2009). Exploration of 19 serotoninergic candidate genes in adults and children with attention-deficit/hyperactivity disorder identifies association for 5HT2A, DDC and MAOB. Mol Psychiatry, 14, 71–85. [DOI] [PubMed] [Google Scholar]
- RIBASES M, RAMOS-QUIROGA JA, HERVAS A, SANCHEZ-MORA C, BOSCH R, BIELSA A, GASTAMINZA X, LESCH KP, REIF A, RENNER TJ, ROMANOS M, WARNKE A, WALITZA S, FREITAG C, MEYER J, PALMASON H, CASAS M, BAYES M & CORMAND B (2012). Candidate system analysis in ADHD: evaluation of nine genes involved in dopaminergic neurotransmission identifies association with DRD1. World J Biol Psychiatry, 13, 281–292. [DOI] [PubMed] [Google Scholar]
- ROMAN T, SCHMITZ M, POLANCZYK GV, EIZIRIK M, ROHDE LA & HUTZ MH (2003). Is the alpha-2A adrenergic receptor gene (ADRA2A) associated with attention-deficit/hyperactivity disorder? Am J Med Genet B Neuropsychiatr Genet, 120B, 116–120. [DOI] [PubMed] [Google Scholar]
- SANCHEZ-MORA C, CORMAND B, RAMOS-QUIROGA JA, HERVAS A, BOSCH R, PALOMAR G, NOGUEIRA M, GOMEZ-BARROS N, RICHARTE V, CORRALES M, GARCIA-MARTINEZ I, COROMINAS R, GUIJARRO S, BIGORRA A, BAYES M, CASAS M & RIBASES M (2013). Evaluation of common variants in 16 genes involved in the regulation of neurotransmitter release in ADHD. Eur Neuropsychopharmacol, 23, 426–435. [DOI] [PubMed] [Google Scholar]
- SEGURADO R, BELLGROVE MA, MANCONI F, GILL M & HAWI Z (2011). Epistasis between neurochemical gene polymorphisms and risk for ADHD. Eur J Hum Genet, 19, 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIFFRIN ND, GRUBER J, GLATT SJ & FARAONE SV (2013). No association between MspI allele of the ADRA2A polymorphism and ADHD: meta-analysis of family-based studies. Psychiatr Genet, 23, 174–175. [DOI] [PubMed] [Google Scholar]
- SHIM SH, HWANGBO Y, KWON YJ, JEONG HY, LEE BH, HWANG JA & KIM YK (2010). A case-control association study of serotonin 1A receptor gene and tryptophan hydroxylase 2 gene in attention deficit hyperactivity disorder. Prog Neuropsychopharmacol Biol Psychiatry, 34, 974–979. [DOI] [PubMed] [Google Scholar]
- SHIM SH, HWANGBO Y, YOON HJ, KWON YJ, LEE HY, HWANG JA & KIM YK (2015). Increased levels of plasma glial-derived neurotrophic factor in children with attention deficit hyperactivity disorder. Nord J Psychiatry, 69, 546–551. [DOI] [PubMed] [Google Scholar]
- SIMCHON-TENENBAUM Y, WEIZMAN A & REHAVI M (2015). Alterations in brain neurotrophic and glial factors following early age chronic methylphenidate and cocaine administration. Behav Brain Res, 282, 125–132. [DOI] [PubMed] [Google Scholar]
- TOREN P, REHAVI M, LUSKI A, ROZ N, LAOR N, LASK M & WEIZMAN A (2005). Decreased platelet vesicular monoamine transporter density in children and adolescents with attention deficit/hyperactivity disorder. Eur Neuropsychopharmacol, 15, 159–162. [DOI] [PubMed] [Google Scholar]
- TURIC D, LANGLEY K, MILLS S, STEPHENS M, LAWSON D, GOVAN C, WILLIAMS N, VAN DEN BREE M, CRADDOCK N, KENT L, OWEN M, O’DONOVAN M & THAPAR A (2004). Follow-up of genetic linkage findings on chromosome 16p13: evidence of association of N-methyl-D aspartate glutamate receptor 2A gene polymorphism with ADHD. Mol Psychiatry, 9, 169–173. [DOI] [PubMed] [Google Scholar]
- TURIC D, LANGLEY K, WILLIAMS H, NORTON N, WILLIAMS NM, MOSKVINA V, VAN DEN BREE MB, OWEN MJ, THAPAR A & O’DONOVAN MC (2005). A family based study implicates solute carrier family 1-member 3 (SLC1A3) gene in attention-deficit/hyperactivity disorder. Biol Psychiatry, 57, 1461–1466. [DOI] [PubMed] [Google Scholar]
- TZANG RF, HSU CD, LIOU YJ, HONG CJ & TSAI SJ (2014). Family-based association study of ciliary neurotrophic factor receptor and norepinephrine transporter genes in attention-deficit hyperactivity disorder. Psychiatr Genet, 24, 118–119. [DOI] [PubMed] [Google Scholar]
- WALLIS D, ARCOS-BURGOS M, JAIN M, CASTELLANOS FX, PALACIO JD, PINEDA D, LOPERA F, STANESCU H, PINEDA D, BERG K, PALACIO LG, BAILEY-WILSON JE & MUENKE M (2009). Polymorphisms in the neural nicotinic acetylcholine receptor alpha4 subunit (CHRNA4) are associated with ADHD in a genetic isolate. Atten Defic Hyperact Disord, 1, 19–24. [DOI] [PubMed] [Google Scholar]
- WEBER H, SCHOLZ CJ, JACOB CP, HEUPEL J, KITTEL-SCHNEIDER S, ERHARDT A, HEMPEL S, SCHMIDT B, KIEL T, GESSNER A, LESCH KP & REIF A (2014). SPOCK3, a risk gene for adult ADHD and personality disorders. Eur Arch Psychiatry Clin Neurosci, 264, 409–421. [DOI] [PubMed] [Google Scholar]
- XU X, BROOKES K, SUN B, ILOTT N & ASHERSON P (2009). Investigation of the serotonin 2C receptor gene in attention deficit hyperactivity disorder in UK samples. BMC Res Notes, 2, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG J-W, JANG W-S, HONG SD, JI YI, KIM DH, PARK J, KIM SW & JOUNG YS (2008). A case-control association study of the polymorphism at the promoter region of the DRD4 gene in Korean boys with attention deficit-hyperactivity disorder: Evidence of association with the − 521 C/T SNP. Progress in Neuro-Psychopharmacology and Biological Psychiatry, 32, 243–248. [DOI] [PubMed] [Google Scholar]
- ZUO L, SABA L, LIN X, TAN Y, WANG K, KRYSTAL JH, TABAKOFF B & LUO X (2015). Significant association between rare IPO11-HTR1A variants and attention deficit hyperactivity disorder in Caucasians. Am J Med Genet B Neuropsychiatr Genet, 168, 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]