Abstract
Clinical narratives are a valuable source of information for both patient care and biomedical research. Given the unstructured nature of medical reports, specific automatic techniques are required to extract relevant entities from such texts. In the natural language processing (NLP) community, this task is often addressed by using supervised methods. To develop such methods, both reliably-annotated corpora and elaborately designed features are needed. Despite the recent advances on corpora collection and annotation, research on multiple domains and languages is still limited. In addition, to compute the features required for supervised classification, suitable language- and domain-specific tools are needed.
In this work, we propose a novel application of recurrent neural networks (RNNs) for event extraction from medical reports written in Italian. To train and evaluate the proposed approach, we annotated a corpus of 75 cardiology reports for a total of 4,365 mentions of relevant events and their attributes (e.g., the polarity). For the annotation task, we developed specific annotation guidelines, which are provided together with this paper.
The RNN-based classifier was trained on a training set including 3,335 events (60 documents). The resulting model was integrated into an NLP pipeline that uses a dictionary lookup approach to search for relevant concepts inside the text. A test set of 1,030 events (15 documents) was used to evaluate and compare different pipeline configurations. As a main result, using the RNN-based classifier instead of the dictionary lookup approach allowed increasing recall from 52.4% to 88.9%, and precision from 81.1% to 88.2%. Further, using the two methods in combination, we obtained final recall, precision, and F1 score of 91.7%, 88.6%, and 90.1%, respectively. These experiments indicate that integrating a well-performing RNN-based classifier with a standard knowledge-based approach can be a good strategy to extract information from clinical text in non-English languages.
Keywords: Information Extraction, Natural Language Processing, Neural Networks
1. Introduction
The increasing amount of data available in electronic health records represents a valuable source to both improve the monitoring of patients and to facilitate biomedical research. As clinical data are often available only in the form of unstructured texts, the task of extracting and accessing information from them has become an important topic in biomedical research. Given the considerable effort required to manually review multiple textual reports, developing techniques for the automatic extraction of relevant information represents a necessary step to enhance searching and summarization [1].
Clinical natural language processing (NLP) has been an active area of research for many years [2,3]. Several efforts have been put into identifying and addressing critical challenges, such as the lack of shared standards and annotated datasets [4]. For the English language, significant advances have been promoted by the annotation of clinical corpora, and supported by the organization of specific competitions [5–9]. The 2012 Informatics for Integrating Biology and the Bedside (i2b2) NLP challenge [8] focused on the extraction of events (e.g., “magnetic resonance imaging”), temporal expressions (e.g., “May 18th 2010”), and temporal relations (e.g., “A before B”) from clinical narratives related to intensive care. The SemEval 2016 Clinical TempEval Task [9] required participants to identify event expressions as well as temporal expressions and relations in a corpus of clinical notes related to oncology (the THYME corpus [10]). For event extraction, all state-of-the-art solutions that have been proposed involve supervised approaches, mainly exploiting conditional random fields (CRFs), support vector machines (SVMs), and – more recently – neural network (NN) models. Although these supervised approaches have been proven to perform well on resources in English, the substantial lack of shared annotated corpora still represents a significant barrier for non-English languages [11].
While approaches based on CRFs and SVMs often rely on complex linguistic features (e.g., lexical, morphological, syntactic features), NN models are able to learn representations starting from tokens or characters, without the need for language-specific resources and manually engineered features [12]. Thanks to these characteristics, NNs have been applied to several entity recognition tasks [13–16] on different domains and languages [17–19]. Among existing NN architectures, recurrent neural networks (RNNs) are specialized for processing sequences of input data: they make use of sequential information through cyclical connections [20]. As NER involves the identification of entities represented by multiple words, this task can be addressed as a sequence labelling problem, in which a sequence of tokens has to be transformed into a sequence of labels indicating the inclusion in the named entities. For this reason, RNNs are particularly suitable to solve NER tasks, as it has been shown in many different works on this topic [14–16,18].
Long Short Term Memory (LSTM) networks are a class of RNNs that are particularly suitable to store and access information over long contexts [21]. These models have been investigated for a long time in the area of general domain NER, especially for English texts [15]. As regards other languages, Lample et al. have used NN architectures based on LSTMs to identify named entities in Spanish, German, and Dutch [16]. More specifically, the authors proposed two models: (i) a bidirectional LSTM with a sequential conditional random layer above it, and (ii) a model that builds and labels chunks using an algorithm inspired by transition-based parsing (with states represented by stack LSTMs). The proposed models obtained a state-of-the-art performance for all the analyzed languages. As for the clinical domain, LSTM networks have been applied to several entity extraction problems. Jagannatha and Yu used these neural models to detect medications in the textual portions of oncology EHRs, showing that they significantly outperformed CRFs [22]. Wu et al. explored the use of LSTM networks to identify clinical concepts from different types of clinical notes, demonstrating their superior performance with respect to traditional supervised approaches [23]. Liu et al. investigated the performance of LSTM networks for both clinical entity recognition and protected health information recognition in clinical notes [24]: experiments showed that character-level word representations could be used to further improve the performance of LSTM architectures.
In the Italian NLP community, few works have dealt with supervised clinical information extraction. Esuli et al. explored different approaches based on CRFs to extract information from radiology reports [25]. To evaluate the proposed approaches, they annotated 500 mammography reports with segments belonging to one of 9 classes (e.g., mammography standardized code, technical info). The authors found that combining a two-stage method with standard linear-chain CRFs outperformed a traditional single-stage CRFs system. Attardi et al. performed recognition of relevant entities such as symptoms, diseases, and treatments, from clinical records written in Italian [26]. They created an annotated corpus of 10,000 sentences, and performed experiments with a customizable statistical sequence labeller [27]. In this case, the best result was obtained in a configuration using a L2-regularized L2-loss support vector classifier. Gerevini et al. worked on the automatic classification of Italian radiological reports following a multilevel schema [28]. In particular, they annotated a corpus of 346 reports using five levels of classification (exam type, test result, lesion neoplastic nature, lesion site, and lesion type). The annotated corpus was used to run experiments with different learning algorithms (Naïve Bayes, SVMs, decision trees, random forests, and NNs), leading to encouraging results. As for the application of neural networks to entity recognition in the Italian language, Bonadiman et al. proposed a NN model that uses a sliding window of word contexts to predict tags for NER in the general domain [18]. To the best of our knowledge, no work has explored NN models to extract clinical entities from texts in the Italian language.
In this paper, we focus on the problem of identifying clinical information in free text written in Italian. Our main goal is to present a methodology for event annotation and extraction, running experiments with neural network models. As a main difference from typical named entities, which are not related to temporality and are generally expressed through nouns, events are characterized by temporal features and can be expressed with various parts of speech (POS), such as nouns and verbs. In this work, we aim at extracting all the events that can be relevant for a patient’s history, with a focus on clinical concepts (e.g., diseases, medications) and other generic episodes (e.g., hospital admissions). To this end, we annotated 75 medical reports in the cardiology domain with 4,365 number of mentions of relevant events, and used this annotated corpus to develop an RNN-based extraction method. Also, we integrated the resulting classifier into a pipeline that previously relied on a rule-based dictionary lookup approach to extract clinical information [29]. This research extends previous work on information extraction from medical reports written in Italian, where we had used 50 annotated documents to explore different RNN classifiers that extract relevant events [30].
As pointed out by Velupillai et al. [11], annotating a corpus with the information of interest is one of the most important steps to enable the development and the evaluation of a supervised system. In light of this consideration, in this paper we report our efforts to manually annotate relevant information in the considered corpus, and then we describe the approaches used for event extraction. To identify events’ boundaries and semantic types, we compared different pipeline configurations, using the dictionary lookup approach and a supervised RNN-based model, both separately and in combination. Besides identifying event boundaries, we were also interested in extracting some attributes of the events, such as the polarity (i.e., positive or negative mention). In the next sections, the developed NLP modules will be presented in detail.
2. Materials and methods
2.1. Dataset
The corpus considered in this work includes 75 annotated medical reports written in Italian and related to the cardiology domain. These documents were randomly chosen from a larger corpus of about 11,000 reports made available by the Molecular Cardiology Laboratories of the Istituti Clinici Scientifici Maugeri (ICSM) hospital in Pavia, Italy. The documents contain the patient’s clinical and family history, the results of performed tests, possible drug prescriptions, and a diagnostic conclusion. In the considered corpus, the patient’s clinical history represents the richest textual portion, including the majority of the events to be extracted.
2.2. Annotation procedure
To annotate documents in an effective way, we first needed to formalize the extraction problem, by (i) identifying the target information for the extraction, and (ii) defining guidelines for the annotation process. In this section, we report the steps we followed to annotate relevant events. The developed annotation guidelines are available as a supplementary file (S1).
2.2.1. Available annotation guidelines and their adaptation
In our project, we aim at ultimately building a system that retrieves and summarizes the events included in multiple unstructured reports, taking into account the available temporal details. To this end, we developed an annotation strategy based on previous work, leveraging the guidelines used in the Clinical TempEval Task [9]. In particular, we analyzed and adapted the THYME annotation guidelines, which were created to annotate clinical events, temporal expressions, and temporal relations in clinical notes [10]. To define the event semantic types to be considered, we also relied on the 2012 i2b2 guidelines for event annotation [8].
As a first step, we selected a set of 20 reports to be manually reviewed and discussed with clinicians. In this phase, we identified the relevant events to be annotated. Following the definition given in the THYME guidelines, “an EVENT is anything that’s relevant on the clinical timeline i.e., anything that would show up on a detailed timeline of the patient’s care or life”. This definition includes both clinical concepts, such as diagnoses or diagnostic procedures, and general events conveying a legal meaning, such as discussions with patients (e.g., “we discussed the risks of this procedure”). In the THYME project, events can be represented with noun phrases, adjective phrases, or verbs; although each event could span multiple words, only the syntactic head of each concept is annotated. As a main difference, in this work we mostly focus on the annotation of clinical concepts expressed through noun phrases, without considering events related to legal issues. Moreover, given the difficulty to automatically reconstruct the whole span of an event starting from its syntactic head, we decided to produce manual (or gold) annotations that correspond to multi-token entities. As another difference, to guide the annotation task, we identified four types of relevant events that are frequently mentioned in the cardiology reports: Problems (e.g., syncopal episode, Brugada Syndrome), Tests (e.g., electrocardiogram, effort stress test), Treatments (mostly drug prescriptions), and Occurrences (generic events that are not included in the first three groups, e.g., admission, medical visit). These semantic types represent a subset of those used in the 2012 i2b2 guidelines.
2.2.2. The annotation process
To annotate Problems, Tests, and Treatments, we decided to use the UMLS Metathesaurus as a guide, considering the entries belonging to the matching semantic types as the main events to be annotated [31]. For example, concepts that could be traced back to the UMLS “Pharmacologic Substance” or “Therapeutic or Preventive Procedure” semantic types were annotated as Treatments. As for Occurrences, we defined a list of relevant events to be searched for. For this group only, we also considered verbs as candidates for annotation. We thus annotated verbs/events that (i) happen to the patient and (ii) are relevant to his/her clinical history (e.g., “admitted”, “discharged”). As an important remark, we decided to annotate also concepts with overlapping text spans, such as “Test with Flecainide” (Test) and “Flecainide” (Treatment), whenever we thought it was important to maintain information about both events.
In the annotation process, we annotated both the concept boundaries and its semantic type. In addition, we reused three Event’s attributes from the THYME project, which we considered particularly relevant in our extraction task:
The DocTimeRel, which is the relation between an event and the document creation time. This relation (which can be one among OVERLAP, BEFORE, BEFORE/OVERLAP, or AFTER) captures the temporal aspects of the event.
The polarity of the event, which can be either POSITIVE or NEGATIVE. Events are usually regarded as negative when they did not happen, or were found not to be true.
The contextual modality of the event, which can be ACTUAL, HEDGED, HYPOTHETICAL, or GENERIC. Actual events are those having already happened or being scheduled. Hedged events are concepts mentioned with any sort of hedging (e.g., “Suspicious for X”). Hypothetical events are those that might happen in the future, without certainty (e.g., “If X happens, then…”). Generic events are concepts mentioned in a general sense that should not appear on the patient’s clinical timeline (e.g., “In all patients with disease X…”).
As the events in our corpus often refer to family members (arrhythmias can be inherited diseases), we decided to consider an additional attribute, i.e., the experiencer of each event, which represents the subject the event itself refers to (the patient or another person). For example, in a sentence like “the patient’s mother showed prolonged QT interval”, the experiencer attribute of the event QT interval would take the value “other”. This definition of experiencer is similar to the one proposed by Harkema et al. for the development of the ConText algorithm [32], which will be illustrated in Section 2.3.3.
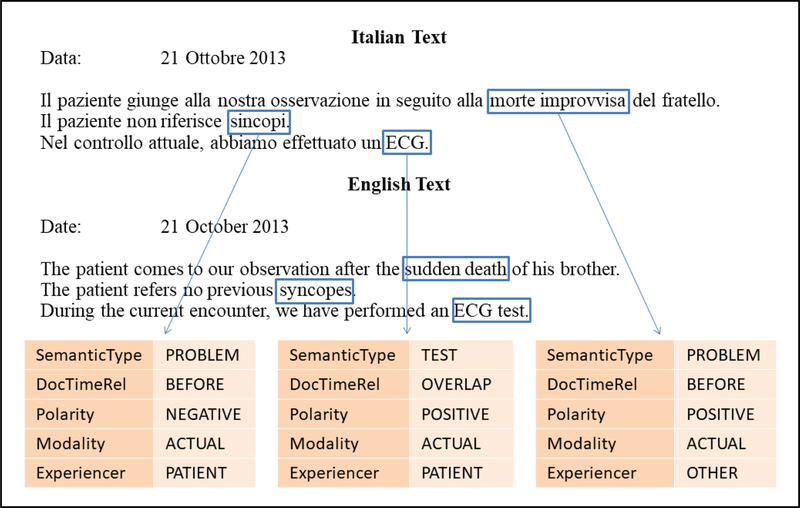
Figure 1 shows an example of the performed annotations on a portion of text taken from one of our reports (sentences have been paraphrased). The English translation is reported for convenience. In this case, three events are highlighted: sudden death, which happened to the patient’s brother in the past (DocTimeRel is “Before” and experiencer is “Other”), syncopes, referring to the patient’s history in a negated way since he did not suffer from this problem (DocTimeRel is “Before” and polarity is “Negative”), and ECG test, that was performed on the patient during the current visit (DocTimeRel is “Actual”). To perform annotations, we used the Anafora Tool [33].
Figure 1. Annotations example.
In the reported textual portion, three annotated events are highlighted (sudden death, syncopes, and ECG). For each event, five attributes of interest are specified (semantic type, DocTimeRel, polarity, modality, and experiencer).
2.2.3. Annotated dataset creation
To create the annotated dataset, a subset of 75 documents were randomly chosen in the cardiology corpus (without including the initial 20 documents used for events’ definition). This corpus size was selected to reach a trade-off between the considerable time required to annotate each document (around 1 hour per annotator) and the need to use a large corpus for supervised training.
To carry out the annotations, two researchers with Biomedical Informatics background were involved (one PhD student and one Assistant Professor). For the first 7 documents, the two annotators discussed the events to be extracted, and relevant observations were included into the guidelines. The remaining documents were then annotated in a “blind” way, without any other consultation. To build the final dataset to be used for methods development, an adjudication phase was conducted, in which the two annotators jointly selected the annotations to be kept (in terms of event spans and attributes). See Section 3.1 for total number of events and their distributions.
2.3. Event extraction methods
For processing our medical reports, we developed an NLP pipeline using the UIMA framework [34]. The pipeline uses the Italian TextPro tool to identify sentences and tokens [35], and a configuration file containing regular expressions to identify the document sections. For extracting the text spans denoting events, we compared two different approaches: a knowledge-based approach that uses a dictionary lookup of a controlled vocabulary, which does not require annotated data, and a supervised machine learning approach based on RNNs.
To run experiments and evaluate results on event extraction, we randomly split our annotated dataset into training and test sets, made up of 60 (80%) and 15 (20%) documents, respectively. We trained our previously developed supervised algorithm on the training set, and used a randomly selected development set (20% of training) to experiment with the state-of-the-art architecture proposed by Lample et al. [16]. The test set was set aside to compare the performance of all selected configurations. First, we considered the dictionary lookup approach and the RNN supervised classifiers separately. Then, we merged the output of the two approaches (using the best performing supervised method).
2.3.1. Dictionary lookup
The dictionary lookup approach is a simple, yet useful solution for clinical event extraction. In this approach, concepts included in specific dictionaries are searched for in the texts. In an effort to leverage on the availability of shared resources, we used the Italian version of UMLS, and the FederFarma Italian dictionary of drugs [36]. To better account for all the domain-specific concepts mentioned in the reports, we developed two additional vocabularies (available as a supplementary file, S2), containing 38 domain-specific procedures (Tests), which were not represented in the Italian version of the UMLS, and 30 general events of interest (Occurrences). To expand the list of concepts to be searched for, we also created a dictionary of 29 acronyms used in the cardiology reports.
To identify UMLS concepts, we used the cTAKES UMLS Lookup Annotator [37], targeting the search to the semantic types corresponding to our events of interest. To use this annotator, we extracted the relevant concepts from the Italian UMLS Metathesaurus and created a dictionary resource in the format required by the cTAKES UMLS Lookup Annotator.
In the proposed dictionary lookup approach, the search is performed on TextPro normalized tokens, allowing the identification of both singular and plural entries. As we were interested in presenting the feasibility of an approach based on RNN models, rather than performing a comparison with a fully developed rule-based linguistic approach, we did not generate other morpho-syntactic and orthographic variants of the concepts.
2.3.2. RNN classifier
To develop a supervised algorithm for event extraction, we converted our annotated corpus to the format known as IOB (inside, outside, beginning). This format requires to classify each token as belonging to the span of one event (B or I) or not (O). As we mentioned in Section 2.2, we were interested in extracting events that fall into one of four semantic types. To take this requirement into account, we included the information on the semantic type in the token labels, using the format “IOB label – semantic type”. In this framework, the sentence “we performed an ECG test” would be translated to the sequence “O O O B-TEST I-TEST”. Since the standard IOB format does not allow classifying a single token into multiple entities (though it is possible with the Anafora tool [33]), for overlapping events such as “Test with Flecainide” and “Flecainide” (i.e., the token “Flecainide” would be both I-TEST and B-TREATMENT) we decided to keep only the longest event (“Test with Flecainide”) with its semantic type (“B-TEST I-TEST I-TEST”). It is important to highlight that, when computing the overall pipeline performance on the test set, we maintained overlapping entries as separate concepts to be extracted.
2.3.2.1. GRU model with POS features
In a preliminary work, we used a set of 50 reports annotated by a single subject to develop an RNN classifier for event extraction [30]. Among existing recurrent architectures, we explored LSTM networks and GRU networks, which represent a simpler variation of the LSTM model [38]. We experimented with both the standard unidirectional model and its bidirectional configuration, which scans the inputs forward and backward. Moreover, we tested the contribution of Part-of-Speech (POS) features as additional inputs to the network. These experiments resulted in the model using the forward GRU network with both token embeddings and POS embeddings performing the best. In the present work, we re-trained this model on the enlarged training set (60 documents) and integrated the resulting classifier into the event extraction pipeline.
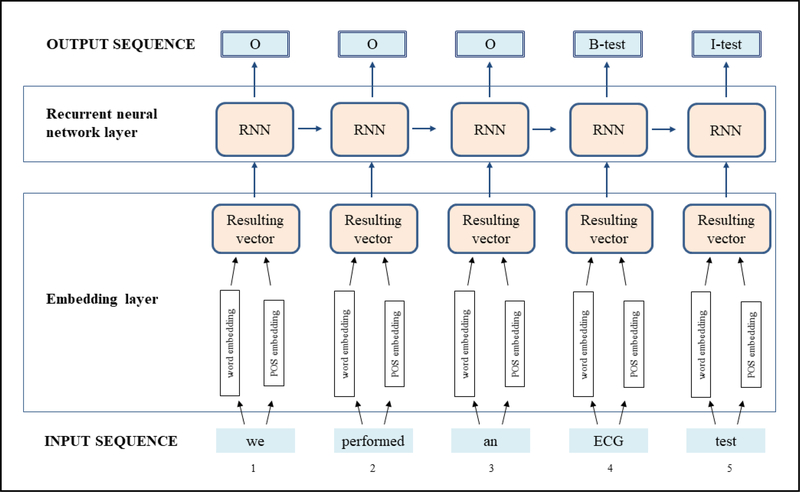
Figure 2 shows the methodological approach used in the GRU-POS architecture. The classification model takes as input the sentences of N tokens identified in the preprocessing phase. For each sentence, the output is a sequence of B, I, O labels including semantic types. As shown in Figure 2, the model includes an embedding layer and an RNN layer. The embedding layer is used to convert tokens and their POS tags to numerical vector representations, i.e., the embeddings, which serve as inputs to the neural network layer.
Figure 2. GRU-POS architecture.
The neural network architecture includes an embedding layer and an RNN layer. The network takes as input the sentences of N tokens identified in the preprocessing phase. For each sentence, the output is a sequence of B, I, O labels including semantic types.
In NN language models, words embeddings are usually learned from a corpus of documents using a variety of unsupervised techniques [39]. Most of these techniques rely on the distributional hypothesis, which states that words that occur in the same context tend to have similar meanings. In this framework, given a target word, its context is defined as the set of surrounding words (e.g., the 2 preceding and the 2 following words). Among the models used for learning embeddings, one of the most used is the continuous bag-of-words (CBOW), which predicts target words starting from their context [40]. The embeddings learned in this way were shown to effectively capture the syntactic and semantic features of words, and can be used to support many NLP tasks [41]. In our model, to account for the fact that the identified BI sequences also correspond to specific POS sequences (e.g., noun-adjective, noun-preposition-noun), both the tokens and their POS tags are converted into real-valued vectors of fixed size, which are then concatenated. We used 200-dimensional vectors for token embeddings, and 40-dimensional vectors for POS embeddings. To choose these dimensions we considered that the number of possible POS tags is smaller than the vocabulary size, and we assumed that POS tags could be represented by using a smaller number of features with respect to those used for the tokens.
In the GRU-POS architecture, both token and POS embeddings are learned during the training of the network, i.e., to solve the specific B, I, O classification task. However, it has been shown that initializing embeddings with vectors pre-trained in an unsupervised way can be helpful, as it provides a good starting point that guides the network training [42]. For this reason, we pre-trained word embeddings on a large corpus obtained by merging our clinical corpus (4,860,575 tokens) with 3000 general domain documents (2,550,769 tokens) gathered from the PAISA corpus, a collection of web texts in Italian [43]. Combining these two sources was useful to extend the amount of data available for computing the embeddings, i.e., the coverage.
2.3.2.2. Bi-LSTM with CRF layer
To compare our previously developed architecture to state-of-the-art work in other languages, we tested the model proposed by Lample et al., where a CRF layer is used on top of a bidirectional LSTM layer [16]. This model relies on two sources of information about words: pre-trained word embeddings (in a similar way to our GRU-POS approach), and additional character-based word representations (CHAR embeddings) learned in a supervised way.
As a first step, we tested the contribution of word embeddings alone by considering the following pre-training configurations: (i) no pre-training, (ii) pre-training with a variable number of PAISA documents (3000, 6000, 9000), (iii) pre-training with no update of the weights during the network training. We evaluated all these configurations on the development set and selected the one that performed best. Then, we investigated the additional contribution of POS features and CHAR embeddings. For both word and POS embeddings, we used the same dimensions we considered for our GRU-POS model. For CHAR embeddings, we used 25-dimensional vectors. To implement all configurations, we relied on the YASET tool [44].
2.3.3. Attribute extraction methods
As mentioned in Section 2.2, in addition to text spans and semantic types, we were interested in extracting four attributes for each event.
For identifying the DocTimeRel attribute, we used an SVM classifier that takes as input a set of features related to the event, and gives as output its relation to the document creation time (Overlap, Before, Before/overlap, After). In particular, the classifier uses 8 features: the first token of the event, its POS tag, the section in which the event is found, the temporal tense of the first verb in the sentence, and four features representing the event’s context (the 2 preceding and the 2 following tokens). To compute the POS tag and the verb temporal tense, we relied on TextPro annotations. To identify the section of the document in which the event mention occurs, we used a rule-based section annotator, which exploits an external configuration file containing the name of the different sections. The DocTimeRel classifier was trained on the same set used to train the RNN classifier (60 documents).
For extracting the polarity, the modality, and the experiencer attributes, we used the ConText algorithm, a rule-based approach that was proven to perform well on several languages [32]. Since the original ConText lexicon is available in English, we translated it to Italian, and we used the algorithm to identify negations, hedged conditions, and the event experiencer. For identifying generic and hypothetical events, we defined suitable rules by manually looking at a few reports in the training set.
2.4. Evaluation metrics
The extraction of event text spans was evaluated against manual annotations (gold events) using precision, recall, and F1 score. We defined as true positives (TPs) the gold events whose boundaries are correctly identified by the system, in other words there is an exact match between offsets. The gold events that are not detected by the system are considered as false negatives (FNs), while events extracted by the system but not found in the gold annotations are considered as false positives (FPs). Precision (P), recall (R), and F1 score (F1) are computed as follows:
For events marked as TPs, the performance of attribute extraction was computed as well. For all attributes (semantic type, DocTimeRel, polarity, modality, and experiencer), we computed the F1 score of each possible value, which allows highlighting the effect of the misclassifications performed on minority classes.
3. Results
3.1. Annotated corpus
The corpus used to run experiments was annotated by two annotators. The inter-annotator agreement (IAA) was computed using the F1 score, which is particularly suitable to measure the classification agreement in the case of entity recognition tasks [45]. To compute the F1 score, we considered the annotations of the first annotator as the gold standard, and the annotations of the second annotator as the system’s output (swapping the roles of the two annotators would result in the same final measure). First, we considered as TPs only the annotations which exactly overlapped each other (in terms of event text spans), obtaining an F1 score of 85.5% (strict F1). Then, we relaxed this assumption by including also partially matching annotations (e.g., “aritmia ventricolare” and “aritmia ventricolare maligna”). In this second case, we obtained an F1 score of 92.2% (relaxed F1).
In the adjudication phase, we reviewed the main reasons for disagreement, which can be summarized as follows. First, one annotator systematically marked a higher number of test results (e.g., “ST elevation”), which were not considered by the second annotator. Moreover, a specific group of Treatment interventions (e.g. “cardioverter defibrillator implant”) was always annotated by one annotator, while the other focused on the identification of drug prescriptions only. Finally, a few disagreements were mostly due to oversights, especially for the set of documents which were analyzed first.
After carefully adjudicating all documents, the final set of annotations was defined. In Table 1 we report the total number of events annotated in our corpus (after the adjudication phase), divided into training and test sets. To give a sense of the documents average size, we also include the number of sentences and tokens detected in the preprocessing phase.
Table 1.
Number of gold events, documents, sentences, and tokens in the training and test sets.
| Training set | Test set | All corpus | |
|---|---|---|---|
| Gold Events | 3335 | 1030 | 4365 |
| Documents | 60 | 15 | 75 |
| Sentences | 3347 | 941 | 4288 |
| Tokens | 44115 | 13148 | 57263 |
In Table 2 we report the distribution of attribute values for the 4,365 annotated events, grouped into training set (3,335 events) and test set (1,030 events). As mentioned, we focused on clinical events represented by noun phrases, with the only exception of Occurrence entities. By analyzing gold annotations, we found that only 12 out of 705 mentions (1.7%) were represented by verbs.
Table 2.
Distribution of attribute values in the complete annotated dataset.
| Attribute | Value | Training set | Test set | All corpus | |||
|---|---|---|---|---|---|---|---|
| Semantic type | Problem | 1504 | (45.1%) | 463 | (45.0%) | 1967 | (45.1%) |
| Test | 880 | (26.4%) | 295 | (28.6%) | 1175 | (26.9%) | |
| Occurrence | 542 | (16.3%) | 163 | (15.8%) | 705 | (16.2%) | |
| Treatment | 409 | (12.3%) | 109 | (10.6%) | 518 | (11.9%) | |
| DocTimeRel | Overlap | 1565 | (46.9%) | 400 | (38.8%) | 1965 | (45.0%) |
| Before | 1222 | (36.6%) | 437 | (42.4%) | 1659 | (38.0%) | |
| After | 517 | (15.5%) | 185 | (18.0%) | 702 | (16.1%) | |
| Before/overlap | 31 | (0.9%) | 8 | (0.8%) | 39 | (0.9%) | |
| Polarity | Positive | 2882 | (86.4%) | 915 | (88.8%) | 3797 | (87.0%) |
| Negative | 453 | (13.6%) | 115 | (11.2%) | 568 | (13.0%) | |
| Modality | Actual | 3020 | (90.6%) | 918 | (89.1%) | 3938 | (90.2%) |
| Hypothetical | 116 | (3.5%) | 54 | (5.2%) | 170 | (3.9%) | |
| Hedged | 107 | (3.2%) | 35 | (3.4%) | 142 | (3.3%) | |
| Generic | 92 | (2.8%) | 23 | (2.2%) | 115 | (2.6%) | |
| Experiencer | Patient | 3070 | (92.1%) | 961 | (93.3%) | 4031 | (92.3%) |
| Other | 265 | (7.9%) | 69 | (6.7%) | 334 | (7.7%) | |
3.2. Event extraction results
In this section, we report the results obtained for the event extraction task. As mentioned, we performed experiments with the dictionary lookup approach, the GRU-POS classifier, and different BI-LSTM-CRF configurations. To compare the RNN-based classifiers to a reference supervised algorithm, we also evaluated an SVM model based on embedding-related features. Specifically, the SVM classifies each token using as features its embedding, and the embeddings and the B, I, O labels for the two previous tokens.
The BI-LSTM-CRF configurations were tested on the development set (supplementary file S3, Table S3.1). Among pre-training configurations, the best performing one used word vectors pre-trained on the corpus resulting from merging our cardiology corpus with 3000 PAISA documents (the same corpus that we used for the GRU-POS architecture). Increasing the number of PAISA documents did not improve the results, which is probably due to the fact that these documents are too different (in terms of content) from the cardiology corpus. As expected, removing the pre-training procedure or preventing weights from being updated by the network resulted in a decrease in performance. The selected BI-LSTM-CRF model (i.e., pre-training on cardiology notes merged to 3000 PAISA documents) was then used to test the contribution of POS features (BI-LSTM-CRF-POS) and of CHAR embeddings (BI-LSTM-CRF-CHAR), with the best result obtained by the latter on the development set. We also explored the combination of POS and CHAR features which, however, resulted in a slight drop in performance. This might be due to the bias-variance effect, where increasing the model complexity can lead to worse generalizability results. Detailed analyses are reported in the supplementary file S3 (“Neural Model Selection and Training Details” section).
In Table 3 we report the results obtained on the test set. As the events annotated in our corpus are not necessarily included in the external dictionaries, the dictionary lookup approach did not perform well, resulting in an F1 score of 63.7% (row 1). The supervised SVM and the RNN-based classifiers obtained better extraction results, mostly thanks to an improvement in recall. Comparing the SVM and the GRU-POS models, we found that the GRU-POS network (row 3) outperformed the SVM classifier (row 2), with a considerable increase in recall (from 75.3% to 86.8%). As for the BI-LSTM-CRF architecture, the POS configuration resulted in a slightly better performance (87.9% versus 87.4% F1 score), with a drop in precision and an improvement in recall (row 4). The BI-LSTM-CRF-CHAR architecture obtained the best result across all RNN-based configuration, with a final F1-score of 88.6% (row 5).
Table 3.
Comparison among different configurations for the extraction of event text spans (test set).
| Row | Extraction method | TP | FP | FN | P | R | F1 |
|---|---|---|---|---|---|---|---|
| 1 | Dictionary lookup | 540 | 126 | 490 | 81.1% | 52.4% | 63.7% |
| 2 | SVM | 776 | 102 | 254 | 88.4% | 75.3% | 81.3% |
| 3 | GRU-POS | 894 | 121 | 136 | 88.1% | 86.8% | 87.4% |
| 4 | BI-LSTM-CRF-POS | 916 | 139 | 114 | 86.8% | 88.9% | 87.9% |
| 5 | BI-LSTM-CRF-CHAR | 916 | 122 | 114 | 88.2% | 88.9% | 88.6% |
| 6 | Dictionary lookup + BEST | 944 | 122 | 86 | 88.6% | 91.7% | 90.1% |
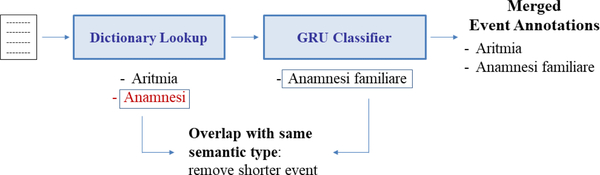
To investigate whether the combination of supervised and knowledge-based approaches could lead to an improved performance, we merged the outputs of the BI-LSTM-CRF-CHAR classifier and the dictionary lookup approach, and evaluated the results on this new output (Table 3, row 6. We refer to this configuration with the name “Dictionary lookup + BEST”). Both methods identify events in the text, creating corresponding annotations according to the identified boundaries and semantic types. In the merging phase, we added the annotations found by the supervised classifier to the list of annotations already performed by the dictionary lookup approach. To deal with overlapping annotations, we considered two different cases: (i) if the overlapping events are characterized by the same semantic type (e.g., “genetic defect responsible for Brugada Syndrome” and “Brugada Syndrome”, both with type Problem) we keep only the longest annotations (which could lead to an error, in the case both entities are relevant); (ii) if the overlapping events are characterized by different semantic types (e.g., “test with Flecainide” with type Test and “Flecainide” with type Treatment), we keep both events. In Figure 3, an example of the first type of overlap is shown: as “Anamnesi” (History) and “Anamnesi familiare” (Family history) are characterized by the same semantic type (i.e., Test), only the longest event is annotated by the pipeline.
Figure 3. Merge of event annotations when using both dictionary lookup and RNN-based classifier.
The annotations found by the RNN-based classifier are added to the list of annotations obtained by the dictionary lookup approach, managing possible overlapping annotations.
Comparing the results obtained in row 5 and row 6 of Table 3, we can notice that combining dictionary entries with the BI-LSTM-CRF-CHAR classifier output allowed a further improvement in recall, with a final F1 score of 90.1%. As expected, by integrating the annotations produced by both approaches, the system was able to extract the highest number of gold events (944 TPs), thus allowing decreasing FNs from 114 to 86.
3.3. Event attributes extraction results
In this section, we report the results obtained for the identification of event attributes. To perform the evaluation, we selected the best performing event extraction configuration (“Dictionary lookup + BEST”), and considered the events extracted with correct boundaries (944). For each of these events, we compared the attribute values computed by the pipeline to the corresponding gold values.
In Table 4 the performance of the different attribute extraction systems are shown. As some attributes are characterized by a strongly unbalanced distribution among classes, we computed the F1 score for all different values. For Polarity, Modality and Experiencer, which are particularly unbalanced, we also computed 95% confidence intervals for precision and recall (supplementary file S3, Table S3.2).
Table 4.
Attribute extraction performance (test set).
| Attribute | Value | P | R | F1 |
|---|---|---|---|---|
| Semantic Type | Problem Test Treatment Occurrence |
99.0% 98.6% 96.2% 100% |
98.7% 99.7% 97.1% 98.1% |
98.9% 99.1% 96.6% 99.1% |
| DocTimeRel | Overlap Before After Before/overlap |
97.0% 71.9% 99.2% 0% |
76.8% 99.2% 66.1% 0% |
85.7% 83.4% 79.3% 0% |
| Polarity | Positive Negative |
98.1% 88.7% |
98.6% 85.5% |
98.3% 87.0% |
| Modality | Actual Hypothetical Hedged Generic |
94.6% 77.3% 76.7% 100% |
99.0% 32.7% 74.2% 63.6% |
96.8% 45.9% 75.4% 77.8% |
| Experiencer | Patient Other |
95.8% 82.4% |
99.3% 42.4% |
97.6% 56.0% |
4. Discussion
Recognizing relevant entities in clinical free text is a challenging task, often addressed by using supervised approaches that require elaborately designed features. To meet this need, we presented a methodology for event extraction based on RNN models, which are able to automatically compute the features required for supervised learning. This approach was developed on a set of cardiology reports written in Italian.
This work presents two main novelties. First, to the best of our knowledge, no other work has exploited recurrent neural networks to perform event recognition on clinical narratives written in Italian. Second, we have proposed an annotation guideline for clinical narratives in the Italian language that integrates and adapts advices taken from state of the art guidelines [8,10,46]. Although our guidelines largely rely on related work on English clinical text, we believe that describing their application to a different language could be relevant for the clinical NLP community.
4.1. Inter-annotator agreement
To evaluate the quality of the developed guidelines and the manually annotated corpus, we computed the IAA, obtaining a strict F1 of 85.5% and a relaxed F1 of 92.2%. Despite the differences in the task definition, these scores can be compared to those computed on the THYME corpus (strict F1 of 80.4% for the Event tag), showing that the agreement achieved in this paper can be considered a good starting point. In future work, the performed disagreement analysis will be exploited to further improve the guidelines and the annotation task.
4.2. Extraction performance
To assess the system performance on event identification, we compared the proposed RNN-based model to a strategy based on dictionary lookup and to an SVM classifier, using an independent test set. Although dictionary lookup can be an effective method to identify standardized concepts included in external dictionaries, it fails in extracting the non-standard, domain specific terms frequently used in our reports. In our case, this approach misses a high number of relevant events, resulting in low system recall (52.4%) and an overall F1 score of 63.7%. These results are comparable to the ones obtained by Chiaramello et al., who used the MetaMap tool to extract UMLS medical concepts from Italian clinical notes [47]. In their case, the MetaMap annotation showed recall, precision and F-measure equal to 53%, 98% and 69%, respectively. As the main reason for annotation failures, the authors identified the impossibility of generating concepts variants for the Italian language. As regards the presented SVM baseline, which relies on embedding-related features, we obtained an F1 score of 81.3%, mostly given by a considerable increase in recall with respect to dictionary lookup (from 52.4% to 75.3%). Specifically, FNs decreased from 490 to 254, and TPs increased from 540 to 776. Compared to the SVM, the proposed RNN-based classifier resulted in an even better performance, allowing a further improvement in recall up to 88.9% (FNs decreased to 114), with an F1 score of 88.6%.
To investigate whether it was possible to improve the overall performance by exploiting the availability of external dictionaries, we integrated the developed classifier into a dictionary-based NLP pipeline for clinical information extraction. Although the considered dictionaries only partially include the relevant terms found in reports, merging the outputs of the two proposed approaches allowed an improvement in results (F1 score up to 90.1%), with an increase in recall, and a slight increase in precision. Given the event extraction problem, an improvement in recall could be expected when using an external dictionary, as this approach is supposed to find additional entries with respect to those extracted by the supervised classifier (this is particularly true if we consider the presence of overlapping entities). However, the impact of the proposed merging technique on precision could be variable, depending on the quality of the dictionary (which could contain ambiguous entries) and the text spans extracted by the classifier. This is an important aspect to be aware of, depending on the considered clinical use-case, i.e., whether it is more desirable to improve precision or recall.
To further discuss the results obtained in this paper, we carefully reviewed the errors related to each event extraction configuration. In particular, we were interested in comparing the FNs and the FPs given by the supervised BI-LSTM-CRF-CHAR model and the combined approach. Integrating the output given by the supervised classifier and the dictionary lookup allowed an increase in the number of extracted events, leading to a reduction of FNs. After an error analysis, we found out that, thanks to the use of external lexicons, the integrated system was able to find very specific and infrequent medical concepts, such as “Hashimoto’s Thyroiditis” and the “Tareg” drug, that could not be extracted by the RNN-based classifier (neither these terms nor similar ones were ever seen in the training set). Among false negatives that were common to all configurations, we noticed particularly complex events such as “durata prolungata della fase terminale del QRS” (“Prolonged duration of the terminal phase of QRS”) and “FOP con shunt sx-dx” (where FOP is an abbreviation for “patent foramen ovale” and sx-dx stand for left and right, respectively). As for entities that were found by RNN-based classifiers, but not by the dictionary or the SVM, we noticed word compounds that were not particularly common in the training set, such as “impianto del defibrillatore” (“defibrillator implant”) and “Ecocolordopplergrafia cardiaca” (“cardiac colorized Doppler echocardiography”). When looking at false positives, we realized that some word compounds made of noun+adjective were often mistaken for events by all neural classifiers. For example, the word compound “anamnesi numerosi” (anamnesis numerous) in the sentence “In anamnesi numerosi episodi sincopali” (in anamnesis numerous syncopal episodes) was extracted as an event, while only the word “anamnesis” was marked in the gold annotations. As another source of false positives, extracted text spans often included only a portion of the gold annotation. For instance, the string “Miocardioscintigrafia” (myocardial scintigraphy) was extracted instead of the longer portion “Miocardioscintigrafia da sforzo” (effort myocardial scintigraphy). As an interesting observation, the number of FPs extracted by the dictionary lookup approach and the BI-LSTM-CRF-CHAR model was similar (126 vs. 122). By comparing these two sets of errors, we found 15 overlapping entries, 8 of which (53%) represented a “partial” gold entity (e.g., “FOP” instead of “FOP con shunt sx-dx”). The remaining 111 FPs from the dictionary lookup approach were corrected or superseded by the supervised model, which – conversely – resulted in 107 additional errors. Most of the corrected entities corresponded to incomplete strings (e.g., “Blocco di branca destra”, right bundle branch block) that were completed (e.g., “Blocco di branca destra incompleto”, incomplete right bundle branch block), while additional errors were due to either wrong entity boundaries or incorrectly extracted terms.
Besides extracting event spans and semantic types, we also developed ad-hoc methods to extract other event attributes. Correctly extracting these details is a crucial task, as events that are mentioned with a negative or hypothetical meaning should not be represented as part of a patient’s history. To the best of our knowledge, this is the first time this problem is addressed for Italian clinical narratives. Overall, the resulting system achieved good accuracies, indicating that rule-based approaches such as ConText represent an effective method to extract details on the polarity, modality, and experiencer of event mentions. For these attributes, slightly lower F1 scores were noticed for those values that were less represented in the dataset, as shown in Table 4. As regards the SVM-based extraction of the DocTimeRel attribute, a good overall performance was obtained as well (F1 scores above 80%), with the only exception of the Before/overlap value. For this value, the limited number of examples available in the training set did not allow a correct classification on the test set, resulting in an F1 score of 0%.
4.3. Comparison to related work
Although the challenges related to clinical information extraction depend on the task complexity (e.g., corpus heterogeneity and size, event definition) we can compare our results to previous work on clinical texts in Italian. The system proposed by Esuli et al., based on a two-stage CRF method, resulted in a F1 score of 85.9% for a single-annotator experiment [25]. Comparing our work to the paper by Esuli et al., we can point out two main differences in the information extraction task definition. First, the authors aim to extract longer segments compared to ours (the average segment length is 17.33 words), which might explain the lower results they obtained. On the other hand, their approach is evaluated at the token level (each token counts as a TP, FP, TN, or FN for a given tag), which credits a system also for partial success. As regards the system presented by Attardi et al., which relies on a statistical sequence labeler, the authors obtained higher F1 scores with respect to ours (e.g., 98.26% for the annotation of body-parts and treatments) [26]. In this case, though, the reference annotations were mostly generated automatically, which is likely to have biased the creation of the corpus used for evaluation. As another difference in the considered task, the authors developed different classifiers to account for different semantic types (e.g., body-parts and treatments, and other mentions were classified with two different models). Targeting the classification model to each semantic type may have increased the performance by reducing the variability of the features. Finally, in the work by Gerevini et al., the authors propose a supervised method for automatic report classification, which represents a different task with respect to event extraction [28]. While their work is focused on labelling each whole report according to five levels of classification (exam type, test result, lesion neoplastic nature, lesion site, and lesion type), we aim to identify events and their attributes in the text.
As regards general-domain applications using LSTM approaches, the BI-LSTM-CRF model proposed by Lample et al., which we explored in this work too, achieved an F1 score of 90.94% on the English dataset [16]. Using the same approach on the Spanish dataset (the most similar language to Italian) led to an F1 score of 85.75%, which is in line with our results. This finding shows the great potential for extending LSTM approaches to different domains and languages.
With respect to the English language, Italian presents some specific challenges for NLP. First, it has a higher degree of inflection: nouns are characterized by a gender (e.g., “house” is feminine while “table” is masculine), and adjectives are inflected depending on the noun they refer to (e.g., bella/bello/belle/belli represent feminine/masculine and singular/plural forms of “beautiful”). Verbs follow three patterns of conjugation (amARE, credERE, dormIRE), and their base form is modified according to the subject’s person, number, and possibly gender (Io amO, tu amI, noi amiAMO, essi amANO, standing for “I/you/we/they love”). Moreover, in Italian it is not mandatory to specify personal pronouns (Amo is equivalent to Io amo) and the word order is more flexible than in English (e.g., adjectives can precede or follow nouns). Given these characteristics, “Naïve” dictionary lookup is more challenging in the Italian language, as there are more forms to be considered (moreover, the coverage of available medical dictionaries is lower). On the other hand, it might be easier to automatically generate lexical variants for synonym search in this language; this, however, is a non-trivial undertaking and so we considered it outside the scope of this work. As regards the RNN-based extraction step, we might expect an embedding vocabulary based on word tokens to have more sparsity in Italian as compared to English, given the higher degree of inflection. To partially address this, pre-training on larger datasets can be helpful: the results obtained in our experiments were promising even when using embeddings at the word level. To the best of our knowledge, only one work has applied recurrent architectures to extract named entities from Italian news [18]. Compared to general domain text, clinical notes are characterized by an additional layer of complexity, as sentences do not always respect syntactic rules, and domain-specific jargon (abbreviations, acronyms) is commonly used. Despite the overall complexity of the considered task, the RNN-based approach that we proposed performs well. In particular, the use of LSTM-based architectures allows improving the system recall with respect to other approaches. Limiting false negatives is a challenge, as relevant events are usually mentioned in the reports by using terms that are often institution-specific (or even expert-specific), and are thus not likely to be found in standardized terminologies. Building an effective classifier that is able to correctly extract this information is an important step to support patient history review and decision making. Our experiments indicate that integrating a well-performing supervised approach with a dictionary lookup strategy can lead to a further improvement in results.
4.4. Limitations and future work
The methods presented in this work suffer from some limitations. First, although we put a considerable effort into the corpus annotation, the size of the annotated dataset is still small in comparison to other works [7]. This work shows promise that the task is learnable with high accuracy even with small datasets, but, because of the limited scope, questions remain about how general this conclusion is. In the future, more documents will be annotated with the information of interest. In addition, to assess the generalizability of our approach, we plan to extend our annotations to documents coming from a different domain. Specifically, we are already working on an application involving anatomic pathology reports in the oncology domain. Another limitation related to the annotation process concerns the definition of the event tags: in this work, we mostly considered clinical events expressed through noun phrases, without taking into account verbs representing actions (e.g., “the patient complained...”). In future work, it would be interesting to extend annotations to verbs denoting events. Finally, with respect to the proposed extraction methodology, we did not consider variants of concepts other than singular/plural forms for the dictionary lookup approach. In the future, we will develop a more complex linguistic rule-based approach, which is likely to further improve the extraction results. As regards the RNN-based extraction step, it would be interesting to explore models that operate over sub-word units, which would help better represent the morphological aspects of words.
5. Conclusions
Developing classifiers that do not require complex NLP preprocessing and feature engineering is an important step to analyze documents across different domains and languages. This is particularly true for the clinical domain, as narratives are often available only in the institution’s local language. In this work, we presented a corpus of Italian medical reports annotated for event mentions, and explored RNN architectures for automatically extracting these events. The developed classifier achieved good results, indicating that LSTM models are a good choice to learn useful data representations.
Supplementary Material
Acknowledgements
We would like to acknowledge the Centre for Health Technologies (CHT) at the University of Pavia for making this work possible. We would also like to thank Dr. Sumithra Velupillai at the Institute of Psychiatry, Psychology and Neuroscience (IoPPN), King’s College London, for providing helpful insight. Research reported in this publication was supported by the National Library of Medicine and National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers R01LM012973 and R01GM114355. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the anonymous reviewers for critically reading the manuscript and suggesting improvements.
Bibliography
- 1.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inf. 2008;128–44. [PubMed] [Google Scholar]
- 2.Friedman C A broad-coverage natural language processing system. Proc AMIA Symp. 2000;270–4. [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman C, Hripcsak G. Natural language processing and its future in medicine. Acad Med J Assoc Am Med Coll. 1999. August;74(8):890–5. [DOI] [PubMed] [Google Scholar]
- 4.Chapman WW, Nadkarni PM, Hirschman L, D’Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc JAMIA. 2011. October;18(5):540–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc JAMIA. 2011;18(5):552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pradhan S, Elhadad N, South BR, Martinez D, Christensen L, Vogel A, et al. Evaluating the state of the art in disorder recognition and normalization of the clinical narrative. J Am Med Inform Assoc JAMIA. 2015. January;22(1):143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elhadad N, Pradhan S, Lipsky Gorman S, Chapman WW, Manandhar S, Savova GK. SemEval-2015 task 14: Analysis of clinical text. In: Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015) 2015. p. 303–10. [Google Scholar]
- 8.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc JAMIA. 2013. September;20(5):806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bethard S, Savova G, Chen W-T, Derczynski L, Pustejovsky J, Verhagen M. Semeval-2016 task 12: Clinical TempEval. Proc 10th Int Workshop Semantic Eval SemEval-2016 2016;1052–1062. [Google Scholar]
- 10.Styler IV WF, Bethard S, Finan S, Palmer M, Pradhan S, de Groen PC, et al. Temporal annotation in the clinical domain. Trans Assoc Comput Linguist. 2014;2:143–154. [PMC free article] [PubMed] [Google Scholar]
- 11.Velupillai S, Mowery D, South BR, Kvist M, Dalianis H. Recent Advances in Clinical Natural Language Processing in Support of Semantic Analysis. Yearb Med Inform. 2015. August 13;10(1):183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg Y A Primer on Neural Network Models for Natural Language Processing. arXiv:151000726. 2015. October 2; [Google Scholar]
- 13.Collobert R, Weston J, Bottou L, Karlen M, Kavukcuoglu K, Kuksa P. Natural language processing (almost) from scratch. J Mach Learn Res. 2011;12(Aug):2493–2537. [Google Scholar]
- 14.Mesnil G, He X, Deng L, Bengio Y. Investigation of recurrent-neural-network architectures and learning methods for spoken language understanding. In: Proceedings of INTERSPEECH 2013 2013. [Google Scholar]
- 15.Hammerton J Named entity recognition with long short-term memory. In: Proceedings of the seventh conference on Natural language learning at HLT-NAACL 2003 2003. p. 172–175. [Google Scholar]
- 16.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural Architectures for Named Entity Recognition. In: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies San Diego, California: Association for Computational Linguistics; 2016. p. 260–270. [Google Scholar]
- 17.Li P, Huang H. Clinical Information Extraction via Convolutional Neural Network. arXiv:160309381. 2016. March 30; [Google Scholar]
- 18.Bonadiman D, Severyn A, Moschitti A. Deep Neural Networks for Named Entity Recognition in Italian. In: Proceedings of CLiC-IT 2015 2015. [Google Scholar]
- 19.Santos CN dos, Guimarães V. Boosting Named Entity Recognition with Neural Character Embeddings. ArXiv150505008 Cs. 2015. May 19; [Google Scholar]
- 20.Goodfellow I, Bengio Y, Courville A. Chapter 10: Sequence Modeling: Recurrent and Recursive Nets. In: Deep Learning; p. 321–65. [Google Scholar]
- 21.Hochreiter S, Schmidhuber J. Long Short-Term Memory. Neural Comput. 1997. November;9(8):1735–80. [DOI] [PubMed] [Google Scholar]
- 22.Jagannatha AN, Yu H. Bidirectional RNN for Medical Event Detection in Electronic Health Records. In: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; 2016. p. 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Jiang M, Xu J, Zhi D, Xu H. Clinical Named Entity Recognition Using Deep Learning Models. AMIA Annu Symp Proc. 2018. April 16;2017:1812–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Yang M, Wang X, Chen Q, Tang B, Wang Z, et al. Entity recognition from clinical texts via recurrent neural network. BMC Med Inform Decis Mak. 2017. July 5;17(Suppl 2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esuli A, Marcheggiani D, Sebastiani F. An enhanced CRFs-based system for information extraction from radiology reports. J Biomed Inform. 2013. June;46(3):425–35. [DOI] [PubMed] [Google Scholar]
- 26.Attardi G, Cozza V, Sartiano D. Annotation and extraction of relations from Italian medical records. In: Proceedings of the 6th Italian Information Retrieval Workshop 2015. [Google Scholar]
- 27.Attardi G, Berardi G, Rossi SD, Simi M. The Tanl Tagger for Named Entity Recognition on Transcribed Broadcast News at Evalita 2011 In: Evaluation of Natural Language and Speech Tools for Italian. Springer, Berlin, Heidelberg; 2013. p. 116–25. [Google Scholar]
- 28.Gerevini AE, Lavelli A, Maffi A, Maroldi R, Minard A-L, Serina I, et al. Automatic Classification of Radiological Reports for Clinical Care. In: Proceedings of AIME 2017, 16th Conference on Artificial Intelligence in Medicine Springer, Cham; 2017. p. 149–59. [DOI] [PubMed] [Google Scholar]
- 29.Viani N, Larizza C, Tibollo V, Napolitano C, Priori SG, Bellazzi R, et al. Information extraction from Italian medical reports: An ontology-driven approach. Int J Med Inf. 2018. March;111:140–8. [DOI] [PubMed] [Google Scholar]
- 30.Viani N, Miller TA, Dligach D, Bethard S, Napolitano C, Priori SG, et al. Recurrent Neural Network Architectures for Event Extraction from Italian Medical Reports In: Artificial Intelligence in Medicine. Springer, Cham; 2017. p. 198–202. [Google Scholar]
- 31.Unified Medical Language System (UMLS) [Internet]. [cited 2017 Jan 7]. Available from: https://www.nlm.nih.gov/research/umls/
- 32.Harkema H, Dowling JN, Thornblade T, Chapman WW. Context: An Algorithm for Determining Negation, Experiencer, and Temporal Status from Clinical Reports. J Biomed Inform. 2009. October;42(5):839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen W-T, Styler W. Anafora: A Web-based General Purpose Annotation Tool. In: Proceedings of the NAACL HLT 2013 2013. p. 14–19. [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10(3–4):327–348. [Google Scholar]
- 35.Pianta E, Girardi C, Zanoli R. The TextPro Tool Suite. In: Proceedings of the 6th edition of the Language Resources and Evaluation Conference 2008. [Google Scholar]
- 36.FederFarma [Internet]. [cited 2017 Jan 7]. Available from: https://www.federfarma.it/
- 37.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc JAMIA. 2010;17(5):507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho K, van Merrienboer B, Bahdanau D, Bengio Y. On the Properties of Neural Machine Translation: Encoder-Decoder Approaches. arXiv:14091259. 2014. September 3; [Google Scholar]
- 39.Mikolov T, Karafiát M, Burget L, Cernockỳ J, Khudanpur S. Recurrent neural network based language model. In: Proceedings of INTERSPEECH 2010 2010. p. 1045–8. [Google Scholar]
- 40.Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. arXiv:13013781. 2013; [Google Scholar]
- 41.Mikolov T, Yih W, Zweig G. Linguistic Regularities in Continuous Space Word Representations. In: Proceedings of NAACL-HLT 2013 2013. p. 746–751. [Google Scholar]
- 42.Erhan D, Bengio Y, Courville A, Manzagol P-A, Vincent P, Bengio S. Why does unsupervised pre-training help deep learning? J Mach Learn Res. 2010;11(Feb):625–660. [Google Scholar]
- 43.Lyding V, Stemle E, Borghetti C, Brunello M, Castagnoli S, Dell’Orletta F, et al. The PAISA corpus of italian web texts. In: Proceedings of the WaC-9 Workshop 2014. p. 36–43. [Google Scholar]
- 44.Tourille J, Doutreligne M, Ferret O, Névéol A, Paris N, Tannier X. Evaluation of a Sequence Tagging Tool for Biomedical Texts. In: Proceedings of the Ninth International Workshop on Health Text Mining and Information Analysis Brussels, Belgium: Association for Computational Linguistics; 2018. p. 193–203. [Google Scholar]
- 45.Hripcsak G, Rothschild AS. Agreement, the F-Measure, and Reliability in Information Retrieval. J Am Med Inform Assoc JAMIA. 2005;12(3):296–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caselli T, Lenzi VB, Sprugnoli R, Pianta E, Prodanof I. Annotating events, temporal expressions and relations in Italian: the It-TimeML experience for the Ita-TimeBank. In: Proceedings of the 5th Linguistic Annotation Workshop Association for Computational Linguistics; 2011. p. 143–151. [Google Scholar]
- 47.Chiaramello E, Pinciroli F, Bonalumi A, Caroli A, Tognola G. Use of “off-the-shelf” information extraction algorithms in clinical informatics: A feasibility study of MetaMap annotation of Italian medical notes. J Biomed Inform. 2016. October;63:22–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.