Abstract
Introduction:
Telomere Length is critically important in normal cells and telomere shortening in combination with other oncogenic changes— promotes genome instability, potentially stimulating initiation of the early stages of cancer.
Aim:
The present study was carried out to detect human telomerase reverse transcriptase expression in oral cancer and pre-cancerous lesions by immunohistochemistry.
Materials and methods:
An observational study was planned in which a total of 45 biopsy specimen of oral mucosa was obtained. Of these, 15 (33.3%) belonged to normal subjects, 15 (33.3%) to subjects found to have Oral submucousal fibrosis and 15 (33.3%) subjects with Oral squamous cell carcinoma.
Results:
Among cases of OSCC, majority was of well differentiated grade (80.0%), only 1 (6.7%) case was poorly differentiated and rest was of moderately differentiated (13.3%) Labelling intensity of OSCC (78.07 ± 22.31) was maximum followed by that of Normal (44.47 ± 6.32) and minimum of OSMF (26.67 ± 15.05) and intergroup difference and between group differences were also found to be significant. Labelling score of OSCC (154.47 ± 94.74) was maximum followed by that of Normal (84.73 ± 51.51) and minimum of OSMF (46.73 ± 44.25) and intergroup difference and between groups differences (Normal vs OSCC, and OSMF and OSCC) were found to be statistically significant.
Conclusion:
The present study highlights only the discriminating ability of hTERT for differentiating the malignant condition from premalignant and normal mucosa. Hence, further studies on a larger sample size, with inclusion of other premalignant conditions too are recommended in order to understand the pattern of hTERT expression changes.
Keywords: Immunohistochemistry, oral squamous cell carcinoma, telomerase, telomere
INTRODUCTION
Cancer is the second most common cause of death in the Western world, after cardiovascular diseases.[1] Oral cancer is one of the most common cancer forms globally. As per an estimate in 2010, worldwide, about 300,000 people were projected to be diagnosed with oral cancer in 2010.[2] Of these, around 126,000 were predicted to die from the disease.[2] In the year 2012, cancers of the oral cavity and pharynx collectively contributed to substantial morbidity and mortality worldwide, with an estimated 526,481 annual incident cases.[3] Among different types of cancers, it ranks sixth in terms of number.[4]
Oral cancer mainly affects the poor owing to a higher exposure to risk factors such as the use of tobacco.[5] Although tobacco use and smoking has been recognized as the most common risk factor for oral cancer, there are a number of other factors too that have been shown to play a role in the causation of cancer in general and oral cancer in particular. Other factors frequently cited are ultraviolet light, nutritional and dietary factors, precancerous lesions, immunosuppression, genetic and dental factors. Oral cancer is a multifactorial disease. Exposure to one of the three broad groups of carcinogenic stimuli, namely chemical, physical and viral, is known to induce cancer in genetically and systemically conditioned oral mucosa.
In recent years, attempts to weigh the risk of cancer at the molecular level are being made and substantial progress in the knowledge has been made. Recent evidence has shown that telomere biology is central to the maintenance of genomic stability and telomeric dysfunction is thought to be an early stage in carcinogenesis.[6]
Telomeres are repeat TTAGGG sequences at the end of linear chromosomes, which guard against loss of genetic material during cellular replication. Due to an inherent end-replication problem, chromosomes are exposed to a potential loss of genetic material, with telomeres acting as a buffer against loss of chromatin. Repeated cell cycles eventually lead to a critically shortened telomere length, signaling cellular senescence and triggering apoptosis. This arrest in proliferation is thought to protect against malignant transformation, and a failure to do so results in catastrophic genomic instability and carcinogenesis.[6]
Telomerase is a ribonucleoprotein enzyme that adds a species dependent telomere, which are specialized structures containing unique simple repetitive sequences (TTAGGG in vertebrate) at the end of chromosomes.[7,8] The enzyme compensates for the end-replication problem and allows cells to proliferate indefinitely.[9] It has been shown that telomerase is activated in most human cancer tissues but not in most normal tissues and tissues adjacent to malignant or benign tumors.[10] In addition, the previous studies have shown that the lack of telomerase activity correlates with critically shortened telomeres and frequent spontaneous cancer remission.[11] Thus, the expression of telomerase is important and may be a rate-limiting step for tumor progression.[10]
In recent years, apart from polymerase chain reaction-based telomeric repeat amplification protocol, which is a complicated assessment requiring highly sophisticated equipment and skilled workforce, several immunohistochemical protocols have emerged that have shown to be efficient tools for the detection of telomerase activity in normal tissue and different premalignant and malignant conditions[12,13,14] including oral cancerous and precancerous lesions.[15,16,17]
With this background, the present study was carried out with an aim to assess the telomerase expression by the immunohistochemistry in oral precancer and cancer.
MATERIALS AND METHODS
The present case–control study included histopathologically confirmed cases of oral squamous cell carcinoma (OSCC) and oral submucous fibrosis (OSMF) after obtaining the Institutional Ethical Committee clearance (ELMC/R_Cell/EC/2016/54), showing dense fibrous connective tissue with epithelial atrophy. Site-matched normal control tissues were obtained from the noninflamed buccal mucosa during the removal of an impacted third molar of patients attending the hospital. All incisional biopsies were performed under local anesthesia (2% lignocaine). Tissues were fixed in 10% buffered formalin and paraffin embedded.
Study population
After obtaining informed consent, the study was done on patients presenting with complaints of different oral mucosal lesions at the Outpatient Department of Dental and Surgery of Era's Lucknow Medical College and Hospital, Lucknow, as well as King George's Medical University, Lucknow. The biopsy obtained was divided into three groups of oral cancer, precancer and normal healthy oral mucosa, each containing 15 samples histopathologically on the basis of hematoxylin and eosin staining.
Immunohistochemical detection of human telomerase reverse transcriptase (hTERT) protein telomerase expression was measured immunohistochemically by evaluating the expression of hTERT protein using TERT polyclonal antibody (Elabscience) following the manufacturer's protocol. The sample was brought down to water and dewaxed by three changes of xylene, absolute alcohol, 90%, 70% and 50%, each for 3 min. The slide was kept in antigen retrieval in a Coplin jar. The jar was placed into the pressure cooker on a hot plate. After one whistle, the plate was switched off and waited till all the pressure was released. The pressure cooker was flooded. The slides were not removed till it cooled down. The slides were washed with three changes of distilled water (3 min) and then were washed with three changes of tris buffer. The primary antibody was applied in a moist chamber for 1 h at room temperature. The slides were washed with –three changes of tris buffer. The secondary antibody was applied for 30 min at room temperature. DAB chromogen solution was applied for 5–10 min. The color was observed under a microscope. When the color developed (khaki color), the slide was placed in running tap water for 5 min. Counterstaining with hematoxylin was done for 1 min. Bluing was done with Scott mixture. The slides were dehydrated, cleared and mounted.
Scoring
Cellular localization of the stain was defined as being either nuclear/cytoplasmic/both. The staining intensity (SI) of the samples was graded and assigned numerical scores: 0 – no stain, 1 – mild, 2 – moderate and 3 – intense stain. Three investigators performed the assessment independently and each investigator used the positive control as a standard bench mark. Nuclear labeling indices (LI) were calculated as the percentage of hTERT-stained cells per thousand cells counted. The counting was done using eyepiece graticule under high-power objective (×40). The nuclear labeling scores (LSs) of the samples were determined using the formula LI × SI92. To eliminate the bias in LS, five different sites were counted by two different examiners.
Statistical analysis
The statistical analysis was done using SPSS (Statistical Package for the Social Sciences, version 15; IBM, USA) statistical analysis software. The values were represented in number (%) and mean ± standard deviation. The results with P < 0.05 were considered to be statistically significant.
RESULTS
The age group of patients ranged from 36 to 65 years. Overall mean age was 51.44 ± 6.03 years. The mean age of patients in normal, OSMF and OSCC groups was 52.73 ± 3.59, 49.60 ± 7.39 and 52.00 ± 6.40 years, respectively. On evaluating the data statistically, the difference among the groups was not found to be significant (P = 0.338) [Table 1].
Table 1.
Group-wise and age-wise distribution
| Group | Number of cases (%) | Mean age (years) |
|---|---|---|
| Normal healthy oral mucosa | 15 (33.33) | 52.73 |
| OSMF | 15 (33.33) | 49.60 |
| OSCC | 15 (33.33) | 52.00 |
OSMF: Oral submucous fibrosis, OSCC: Oral squamous cell carcinoma
Staining intensity
In 15 cases of OSMF, the SI scores were 0, 1, 2 and 3 in 2 (13.3%), 7 (46.7%), 5 (33.3%) and 1 (16.7%) cases. The median score was 1, and the interquartile range spanned from 1 to 2. Of 15 cases of OSCC, none had score 0, 4 (26.7%) had score 1, 8 (53.3%) had score 2 and 3 (20%) had score 3. The median score was 2, and the interquartile range spanned from 1 to 2. On evaluating the intergroup difference in SI using the Kruskal–Wallis test, they were not found to be significant (P = 0.131) [Tables 2 and 3].
Table 2.
Comparison of staining intensity in the three study groups
| Group | Number of cases | Score (%) | Median score (IQR) | |||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |||
| Normal | 15 | 0 (0.0) | 7 (46.7) | 4 (26.7) | 4 (26.7) | 2 (1-3) |
| OSMF | 15 | 2 (13.3) | 7 (46.7) | 5 (33.3) | 1 (16.7) | 1 (1-2) |
| OSCC | 15 | 0 (0.0) | 4 (26.7) | 8 (53.3) | 3 (20.0) | 2 (1-2) |
OSMF: Oral submucous fibrosis, OSCC: Oral squamous cell carcinoma, IQR: Interquartile range
Table 3.
Comparison of staining intensity between malignant and nonmalignant groups
| Group | Number of cases | Score (%) | Median score (IQR) | |||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |||
| Malignant | 15 | 0 (0) | 4 (26.7) | 8 (53.3) | 3 (20.0) | 2 (1-2) |
| Nonmalignant | 30 | 2 (6.7) | 14 (46.7) | 9 (30.0) | 5 (16.7) | 1 (1-2) |
IQR: Interquartile range
Labeling intensity
Labeling index values ranged from 0 to 150. Overall mean labeling index was 49.73 ± 26.60. The mean labeling index was minimum for OSMF (26.67 ± 15.05) followed by normal (44.47 ± 6.32) and maximum for OSCC (78.07 ± 22.31) group. On evaluating the data statistically using ANOVA, the intergroup differences were found to be significant statistically (P < 0.001) [Table 4].
Table 4.
Comparison of labeling intensity among different groups
| Serial number | Group | Number of cases | Labeling intensity | SD | Minimum | Maximum |
|---|---|---|---|---|---|---|
| 1 | Normal | 15 | 44.47 | 6.32 | 36 | 55 |
| 2 | OSMF | 15 | 26.67 | 15.05 | 0 | 55 |
| 3 | OSCC | 15 | 78.07 | 22.31 | 50 | 150 |
| Total | 45 | 49.73 | 26.60 | 0 | 150 |
OSMF: Oral submucous fibrosis, OSCC: Oral squamous cell carcinoma, SD: Standard deviation
Statistical evaluation of between-group differences was done using Tukey's honestly significant difference test. The mean difference ± standard error between normal and OSMF, normal and OSCC and OSMF and OSCC groups was −17.80 ± 5.83, −33.60 ± 5.83, and −51.40 ± 5.83, respectively. All the between-group differences were significant statistically (P < 0.05). Thus, the order of labeling index scores in different groups was OSMF <normal <OSCC [Tables 5 and 6].
Table 5.
Comparison of labeling score among different groups
| Serial number | Group | Number of cases | Labeling score | SD | Minimum | Maximum |
|---|---|---|---|---|---|---|
| 1 | Normal | 15 | 84.73 | 51.51 | 36 | 165 |
| 2 | OSMF | 15 | 46.73 | 44.25 | 0 | 165 |
| 3 | OSCC | 15 | 154.47 | 94.74 | 75 | 450 |
| Total | 45 | 95.31 | 79.74 | 0 | 450 |
OSMF: Oral submucous fibrosis, OSCC: Oral squamous cell carcinoma, SD: Standard deviation, LS: Labeling score
Table 6.
Labeling Intensity among different grades of oral squamous cell carcinoma
| Serial number | Group | Number of cases | Labeling intensity | SD | Minimum | Maximum |
|---|---|---|---|---|---|---|
| 1 | Well differentiated | 12 | 80.92 | 24.24 | 50 | 150 |
| 2 | Moderately differentiated | 2 | 67.50 | 3.54 | 65 | 70 |
| 3 | Poorly differentiated | 1 | 65.00 | |||
| Total | 15 | 78.07 | 22.31 | 50 | 150 |
SD: Standard deviation
Labeling score
Labeling score values ranged from 0 to 450. Overall mean labeling score was 95.31 ± 79.74. The mean labeling score was minimum for OSMF (46.73 ± 44.25) followed by normal (84.73 ± 51.51) and maximum for OSCC (154.47 ± 94.74) group. On evaluating the data statistically using ANOVA, the intergroup differences were found to be significant statistically (P < 0.001). Thus, the order of labeling scores in different groups was OSMF ~ normal < OSCC [Table 7].
Table 7.
Labeling score among different grades of oral squamous cell carcinoma
| Serial number | Group | Number of cases | Labeling score | SD | Minimum | Maximum |
|---|---|---|---|---|---|---|
| 1 | Well differentiated | 12 | 154.33 | 105.83 | 75 | 450 |
| 2 | Moderately differentiated | 2 | 167.50 | 38.89 | 140 | 195 |
| 3 | Poorly differentiated | 1 | 130.00 | |||
| Total | 15 | 154.47 | 94.74 | 75 | 450 |
SD: Standard deviation
The order of labeling intensity was well-differentiated, moderately differentiated and poorly differentiated respectively, and for labeling scores, the order was poorly differentiated, well-differentiated and moderately differentiated grade [Figure 1].
Figure 1.
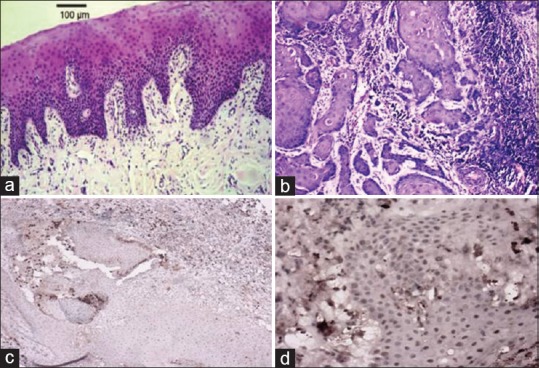
(a) Normal oral mucosa (H&E, ×100). (b) Well-differentiated oral squamous cell carcinoma (H&E, ×100). (c and d) Immunohistochemical expression of human telomerase reverse transcriptase in well-differentiated oral squamous cell carcinoma ×100, ×400, respectively
DISCUSSION
The transformation from normal mucosa to cancerous condition is quite eventful involving a number of cytological and molecular changes. These changes are associated with genetic changes that affect cell cycle, apoptosis, angiogenesis and telomere length. Telomeres are the extreme ends of double-stranded eukaryotic chromosomes comprising a tandem array of TTAGGG repeats and DNA-binding proteins. In humans, it consists of repeats of TTAGGG with a 3’ end overhang that helps in the formation of D-loop and T-loop structures. Telomeres protect the chromosomal ends from degradation by exonucleases and prevent recognition as double-stranded DNA breaks, end-to-end fusions and ring chromosome formation. Thus, telomeres play a vital role in the regulation of gene expression, functional organization of the chromosome and in controlling the replicative life of cells and entry into senescence.[18]
At the chromosomal level, the telomere activity is controlled and stabilized by a ribonucleoprotein complex called telomerase. The telomerase complex consists of RNA template (hTR), a catalytic subunit called hTERT and associated protein (hTP-1).[19] hTERT expression (molecular weight ~130 kDa) is one of the critical determinants of telomerase activity.[20,21,22]
Thus, the study of hTERT can be considered as a surrogate marker of telomerase activity and can help in understanding the transitional pattern from normal tissue to a malignant condition.
Considering the weak cytoplasmic expression and taking clues from previous studies, we focused only on the nuclear stains. Nuclear staining was also studied using two parameters – first one was the percentage of stained nuclei in per thousand cells which was termed as labeling intensity and second was a more objective parameter giving due weight to both span (labeling intensity) and intensity (stain intensity) and deriving a score as a product of labeling intensity and stain intensity and was termed as LS. This scoring system was also used by Palani et al. in their study. The use of multiplication product of two semiquantitative parameters helps to convert them into a quantitative parameter and thus provides a greater discriminating efficiency. However, Raghunandan et al. in their study did not use a scoring system based on multiple parameters and banked on measuring differences in different groups on the basis of independent parameters only. However, other workers like Haraguchi et al. and Luzar et al. similar to the present study preferred to use an integrated scoring system for differentiation among different groups.[23,24,25]
In the present study, all the cases were males. The reason for this could be the fact that the risk of oral cancer is multifolds in males as compared to females. In Northern India, the incidence of oral cancer is related with the habit of tobacco and gutkha use, both the habits being predominated by males. However, despite this predominance of males, the high proportion of males in the present study is the only incidence and does not reflect any epidemiological risk. Consecutively, in premalignant and control groups, matching was done leading to a study in an all male population. The assessment of hTERT activity was done in terms of SI scores, labeling intensity scores and LS, respectively, as per the criteria described by Palani et al.[15] The location of expression was also noted in terms of nuclear and cytoplasmic localization.
Incidentally, the present study found the coexpression of hTERT in both nucleus and cytoplasm in all the cases of all the three groups studied. As far as SI was concerned, we did not find a significant difference among the study groups. On collective assessment between malignant and nonmalignant states too, the difference was not found to be significant statistically.
In the present study, the hTERT LSs also failed to discriminate between different grades of OSCC, although previous studies have shown a possible discriminatory role of hTERT expression for differentiation of different grades or stages of OSCC,[16,26,27] primarily owing to a high dominance of well-differentiated OSCC grade (n = 12/15; 80%) as compared to moderately differentiated (n = 2; 13.3%) and poorly differentiated (n = 1/15; 6.7%) cases, thus leaving the scope for incidental findings. Despite certain limitations such as the absence of females, disproportionate distribution of grades and inclusion of only one premalignant condition (OSMF only), the present study made a point that telomerase activity is enhanced in the OSCC cases.
In the present study, the labeling intensity which was comparable to the intensity of hTERT staining used by Raghunandan et al.,[23] was minimum in OSMF cases (26.67 ± 15.05) followed by normal oral mucosa (44.47 ± 6.32) and OSCC (78.07 ± 22.31), respectively, and showed a significant intergroup difference.
CONCLUSION
However, owing to these limitations, the present study was able to highlight only the discriminating ability of hTERT for differentiating the malignant condition from premalignant and normal mucosa. Hence, further studies on a larger sample size with the inclusion of other premalignant conditions too are recommended to understand the pattern of hTERT expression changes in different types of premalignant and malignant oral lesions and to understand the probable physiology behind the progression from normal to malignant status.
Financial support and sponsorship
This study was funded by the Intramural Research Grant (Era's Lucknow Medical College and Hospital, Lucknow).
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Johnson N. Tobacco use and oral cancer: A global perspective. J Dent Educ. 2001;65:328–39. [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Cheng YS, Rees T, Wright J. A review of research on salivary biomarkers for oral cancer detection. Clin Transl Med. 2014;3:3. doi: 10.1186/2001-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway DI, Petticrew M, Marlborough H, Berthiller J, Hashibe M, Macpherson LM. Socioeconomic inequalities and oral cancer risk: A systematic review and meta-analysis of case-control studies. Int J Cancer. 2008;122:2811–9. doi: 10.1002/ijc.23430. [DOI] [PubMed] [Google Scholar]
- 6.Baichoo E, Boardman LA. Toward a molecular classification of colorectal cancer: The role of telomere length. Front Oncol. 2014;4:158. doi: 10.3389/fonc.2014.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–73. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 8.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, et al. The RNA component of human telomerase. Science. 1995;269:1236–41. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 9.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–9. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–5. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 11.Hiyama E, Hiyama K, Yokoyama T, Matsuura Y, Piatyszek MA, Shay JW. Correlating telomerase activity levels with human neuroblastoma outcomes. Nat Med. 1995;1:249–55. doi: 10.1038/nm0395-249. [DOI] [PubMed] [Google Scholar]
- 12.Hiyama E, Hiyama K, Yokoyama T, Shay JW. Immunohistochemical detection of telomerase (hTERT) protein in human cancer tissues and a subset of cells in normal tissues. Neoplasia. 2001;3:17–26. doi: 10.1038/sj.neo.7900134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bravaccini S, Sanchini MA, Amadori A, Medri L, Saragoni L, Calistri D, et al. Potential of telomerase expression and activity in cervical specimens as a diagnostic tool. J Clin Pathol. 2005;58:911–4. doi: 10.1136/jcp.2004.024158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo J, Park SY, Kang SJ, Kim BK, Shim SI, Kang CS. Expression of telomerase activity, human telomerase RNA, and telomerase reverse transcriptase in gastric adenocarcinomas. Mod Pathol. 2003;16:700–7. doi: 10.1097/01.MP.0000077517.44687.B6. [DOI] [PubMed] [Google Scholar]
- 15.Palani J, Lakshminarayanan V, Kannan R. Immunohistochemical detection of human telomerase reverse transcriptase in oral cancer and pre-cancer. Indian J Dent Res. 2011;22:362. doi: 10.4103/0970-9290.84281. [DOI] [PubMed] [Google Scholar]
- 16.Kumar SK, Zain RB, Ismail SM, Cheong SC. Human telomerase reverse transcriptase expression in oral carcinogenesis – A preliminary report. J Exp Clin Cancer Res. 2005;24:639–46. [PubMed] [Google Scholar]
- 17.Abrahao AC, Bonelli BV, Nunes FD, Dias EP, Cabral MG. Immunohistochemical expression of p53, p16 and hTERT in oral squamous cell carcinoma and potentially malignant disorders. Braz Oral Res. 2011;25:34–41. doi: 10.1590/s1806-83242011000100007. [DOI] [PubMed] [Google Scholar]
- 18.Popli DB, Sircar K, Chowdhry A. Telomerase: An exploration toward the end of cancer. Indian J Dent Res. 2017;28:574–84. doi: 10.4103/ijdr.IJDR_690_16. [DOI] [PubMed] [Google Scholar]
- 19.Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66:407–25. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J, Southworth LK, Sarin KY, Venteicher AS, Ma W, Chang W. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4:e10. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryce LA, Morrison N, Hoare SF, Muir S, Keith WN. Mapping of the gene for the human telomerase reverse transcriptase, hTERT, to chromosome 5p15.33 by fluorescence in situ hybridization. Neoplasia. 2000;2:197–201. doi: 10.1038/sj.neo.7900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–5. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raghunandan BN, Sanjai K, Kumaraswamy J, Papaiah L, Pandey B, Jyothi BM. Expression of human telomerase reverse transcriptase protein in oral epithelial dysplasia and oral squamous cell carcinoma: An immunohistochemical study. J Oral Maxillofac Pathol. 2016;20:96–101. doi: 10.4103/0973-029X.180953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haraguchi K, Yada N, Sato S, Habu M, Hayakawa M, Takahashi O, et al. The methylation status and expression of human telomerase reverse transcriptase is significantly high in oral carcinogenesis. APMIS. 2017;125:797–807. doi: 10.1111/apm.12723. [DOI] [PubMed] [Google Scholar]
- 25.Luzar B, Poljak M, Marin IJ, Eberlinc A, Klopcic U, Gale N. Human telomerase catalytic subunit gene re-expression is an early event in oral carcinogenesis. Histopathology. 2004;45:13–9. doi: 10.1111/j.1365-2559.2004.01892.x. [DOI] [PubMed] [Google Scholar]
- 26.Muller HJ. The remaking of chromosomes. Collecting Net. 1938;13:15. [Google Scholar]
- 27.New York: American Federation for Aging Research; 2011. American Federation for Aging Research. Telomeres and Telomerase. An Introduction to Aging Science Brought to you by the American Federation for Aging Research. [Google Scholar]


