Abstract
Objective:
MR-perfusion post-processing still lacks standardization. This study evaluates the results of perfusion analysis with two established software solutions in a large series of patients with different diseases when a highly standardized processing workflow is ensured.
Methods:
Multicenter data of 260 patients (80 with brain tumors, 124 with cerebrovascular disease and 56 with dementia examined with the same MR protocol) were analyzed. Raw data sets were processed with two software suites: Olea sphere and NordicICE. Group differences were analyzed with paired t-tests and one-way ANOVA.
Results:
Perfusion metrics were significantly different for all examined diseases in the unaffected brain for both software suites [ratio cortex/white matter left hemisphere: mean transit time (MTT) 0.991 vs 0.847, p < 0.05; relative cerebral bloodflow (rBF) 3.23 vs 4.418, p < 0.001; relative cerebral bloodvolume (rBVc) 2.813 vs 3.884, p < 0.001; right hemisphere: MTT 1.079 vs 0.854, p < 0.05; rBF 3.262 vs 4.378, p < 0.001; rBVc 2.762 vs 3.935, p < 0.001)]. Perfusion results were also significantly different in patients with stroke (ratio cortex/white matter affected hemisphere: MTT 1.058 vs 0.784; p < 0.001), dementia (ratio cortex/white matter left hemisphere: rBVc 1.152 vs 1.795, p < 0.001; right hemisphere: rBVc 1.396 vs 1.662, p < 0.05) and brain tumors (ratio cortex/whole tumor rBVc: 0.778 vs 0.919, p < 0.001 and ratio cortex/tumor hotspot rBVc: 0.529 vs 0.512, p < 0.05).
Conclusion:
Despite a highly standardized workflow, parametric perfusion maps are depended on the chosen software. Radiologists should consider software related variances when using dynamic susceptibility contrast perfusion for clinical imaging and research.
Advances in knowledge:
This multicenter study compared perfusion parameters calculated by two commercial dynamic susceptibility contrast perfusion post-processing software solutions in different central nervous system disorders with a large sample size and a highly standardized processing workflow. Despite, parametric perfusion maps are depended on the chosen software which impacts clinical imaging and research.
Introduction
Dynamic susceptibility contrast (DSC) perfusion is used in various neurological conditions such as cerebrovascular diseases, psychiatric disorders and brain tumors. In acute stroke, DSC helps to identify tissue at risk1,2 and thus is an important diagnostic tool to initiate therapy and to estimate prognosis. In case of vessel stenosis, DSC provides measures of the resulting brain tissue perfusion deficit and an estimation of the cerebrovascular reserve.3
In patients with suspected Alzheimer’s disease, it has been shown that DSC perfusion can reveal impaired cerebral microcirculation.4 Additionally, in patients with first episode major psychosis, DSC has been successfully used to show hemodynamic changes and to reliably distinguish patients from healthy controls.5
In patients with brain tumors, DSC perfusion is an indispensable tool for initial tumor grading, for differentiation of recurrent glioma from similar appearing treatment effects following radiochemotherapy in follow-up imaging, in treatment response assessment and it also plays an important role for identification of genomic mutations (eglomerular filtration rate mutation) to identify patients with a potentially poor clinical outcome.6,7
Despite the widespread application of DSC perfusion for clinical diagnosis and research, perfusion post-processing still lacks standardization.8,9 Repeatability and comparability of DSC perfusion are affected by image acquisition and post-processing. Differing acquisition parameters including temporal and spatial resolution, acquisition time, employed contrast agent and also the post-processing, which includes the accuracy of the arterial input function detection, the chosen calibration and normalization techniques as well as the employed deconvolution, have an impact on the estimated parametric perfusion maps.10 Small studies in patients with brain tumors have shown that intra- and interobserver reproducibility cannot be ensured.11–13
In this multicenter study, we tried to ensure maximal standardization regarding the post-processing of the obtained raw perfusion data sets from three different sites including subgroups of patients with high-grade brain tumors, cerebrovascular disease and dementia to compare the results of the estimated perfusion maps of two widely used, FDA-approved software packages regarding reproducibility and comparability.
Material and methods
Patients
This multicenter study comprised a total of 260 patients in three disease groups from three sites. The first subgroup consists of 44 brain tumor (intra-axial) patients measured at Site 1. The second group includes another 34 brain tumor (intra-axial) patients measured at Site 2. Group 3 consist of 124 patients with intra- or extracranial stenosis of the anterior or posterior circulation and Group 4 comprises 56 patients with suspected dementia, both groups were measured at Site 3.
Informed consent was obtained from all subjects. The Clinical Investigation Ethics Committee of the respective sites approved the study protocol and the research was conducted in accordance with the Declaration of Helsinki.
Imaging protocol
DSC-perfusion was performed with clinical scanners at a magnetic field strength of 1.5 or 3 T using a single-shot EPI gradient echo sequence (imaging parameters for the different sites/scanners are given in Table 1). Contrast agent and automatic injection parameters were standardized. We used 1 mmol ml−1 Gadubutrol (Gadovist, Bayer Healthcare, Leverkusen, Germany) as contrast agent. First, an intravenous bolus of 5 ml Gadovist was injected in the left or right cubital vein using an MR compatible injector as a pre-bolus to diminish the effects of contrast agent extravasation. Anatomical T2W and T1W post-contrast data sets were acquired. This was followed by a second standard amount of 5 ml Gadovist for DSC perfusion, which was injected 8–12 min after the first bolus. Injection rate was 2 ml s−1 for the first injection and 5 ml s−1 for the second injection, each followed by a saline flush of 30 ml (0.9% NaCl). Thus, all patients received a total amount of 10 ml Gadovist.
Table 1. .
Imaging parameters for DSC (EPI-Gradient echo)
| Patient Group 1 | Patient Group 2 | Patient Group 3 | Patient Group 4 | |
|---|---|---|---|---|
| Scanner | 3 T (Trio Tim, Siemens) | 1.5 T (Avanto, Siemens) | 1.5 T (Aera, Siemens) | 3 T (Trio Tim, Siemens) |
| TE/TR/FOV | 54/2380/230 x 230 | 30/1340/230 x 230 | 45/1960/230 x 230 | 32/1840/230 x 230 |
| Slice thickness | 5 mm | 5 mm | 5 mm | 6 mm |
| Matrix | 128 × 86 | 128 × 128 | 128 × 128 | 128 × 128 |
| Temporal resolution | 2.5 s | 2.5 s | 2.5 s | 2.5 s |
| n | 44 | 34 | 124 | 56 |
DSC, dynamic susceptibility contrast; FOV, field of view; TE, echo time; TR, repetition time.
Image processing and analysis
DSC data were transferred to an external workstation for image processing with two different, FDA-approved software packages (Olea sphere v. 2.3 SP2, Olea medical, La Ciotat, France and NordicICE v. 2.3.14, Nordic NeuroLabs, Bergen, Norway).
Data sets were pre-processed in a standardized manner using the pre-defined procedure of Olea sphere and we set the same options in NordicICE. Background segmentation was adjusted to remove extracranial tissue using an automatically detected noise threshold. To maintain data integrity and limit confounding factors, we did not apply spatial or temporal smoothing, due to different technical implementations of these functions, that we could not fully control. We excluded initial images of the series if transient signal intensity effects were present and adjusted the prebolus range accordingly. The raw signal was converted “SI to delR2”, i.e. into relative change in R2* (reciprocal of T2*) vs time.
Automatic arterial pixel selection was chosen for computing an arterial input function (AIF). The AIF was generated automatically by the software for each individual dataset using a global clustering method which examines the time series for all voxels and identifies a suitable AIF.14 AIF was chosen due to the following criteria of quality: early take off, peak height, Full Width at HalfMaximum (FWHM), low noise and time to peak. AIF parameters were exported. With those presets, hemodynamic parameter maps of relative cerebral blood volume (rBV), relative cerebral blood flow (rBF) and mean transit time (MTT) were calculated. rBV parametric maps were calculated with correction for contrast agent leakage (rBVc).15 The workflow is depicted in Figure 1.
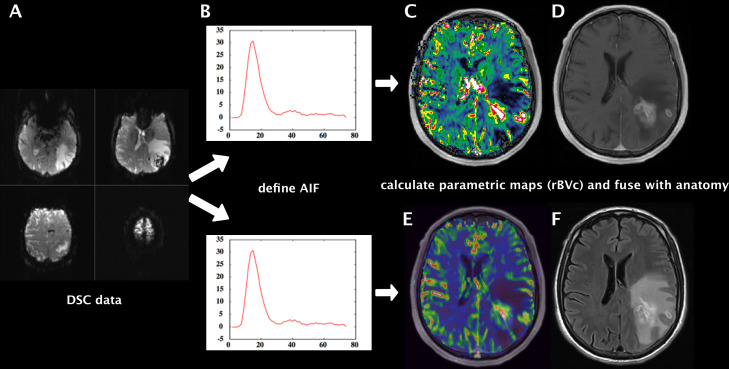
Figure 1. .
Workflow of highly standardized preprocessing of raw DSC-perfusion data sets (A). After background segmentation, removal of extracranial tissue and noise threshold detection, the raw signal was converted into relative change in R2* vs time and the AIF was generated automatically (B). We used the same presets for both software suites and calculated hemodynamic parameter maps of rBV, rBF and MTT. Example shows rBVc map fused with T1W + Gd produced with NordicICE (C) and with Olea sphere (E). Anatomical images T1W + Gd (D) and T2W FLAIR (F) of a representative brain tumor case (left parietooccipital glioblastoma). AIF, arterial inputfunction; DSC, dynamic susceptibility contrast; FLAIR, fluidattenuated inversion recovery; MTT, mean transit time; rBF, relative cerebralblood flow; rBV, relative cerebral blood volume.
Circular regions of interest (ROIs) were placed in the cortex and in deep white matter of the frontal lobe in all patients bilaterally. ROI area was defined as three for cortex and five for deep white matter in a software inherent arbitrary unit (mean surface for cortex ROI = 25.9 mm2, for deep white matter ROI = 80.7 mm2). Disease specific ROIs were placed for subgroups 1 and 2 in the tumor: (1) covering the whole lesion (size depended on tumor size) and (2) covering only the tumor hotspot (highest rBVc, size 3). For subgroup 3, cortical and deep white matter ROIs were placed in the MTT map in the affected vascular territory (prolonged MTT due to vessel stenosis). For subgroup 4, two ROIs (size 3) were placed in the hippocampus of each hemisphere.
For exact anatomical placement of ROIs, the parametric perfusion maps were co-registered and fused with an anatomical data set. ROI placement was performed by two neuroradiologists well trained in perfusion MRI (Figure 2). ROI data was saved in an Excel file and ROI positioning data were exported in matrix co-ordinates. An in-house developed software (PI-Viewer) was used to transform ROI matrix co-ordinates of the respective slices in a format that can be imported into NordicICE. ROIs were then imported into NordicICE and ROI data were saved.
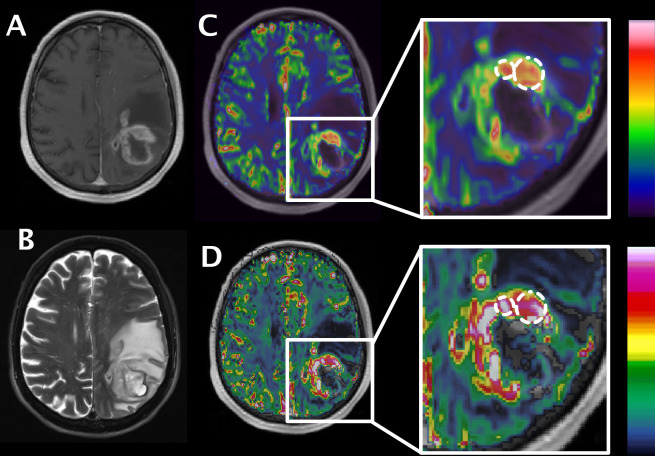
Figure 2. .
ROI positioning in a representative brain tumor case. Anatomical images T1W + Gd (A) and T2W fat-sat (B) of a left parietooccipital glioblastoma. rBVc map fused with T1W + Gd produced with Olea sphere (C) and with NordicICE (D). Larger ROI represents “whole tumor” with sparing of necrotic/cystic component; smaller ROI represents the tumor hotspot. We placed ROIs in Olea sphere and exported the ROI matrix coordinates. These coordinates were transformed into a format that can be imported into NordicICE with a dedicated, in-house developed software. ROI, region of interest.
We used two iterative deconvolution models, oscillation-index standard truncated singular value deconvolution (oSVD16) in Olea sphere and an iterative SVD method with no free parameters using Tikhonov regularization in NordicICE,17 with 100 iterations. Additionally, (only in Olea sphere) parametric maps were calculated using Bayesian hemodynamic parameter estimation (BAY), which is considered to lead to a more reliable and accurate estimation of perfusion indices.18–20
Statistical analysis
For each perfusion parameter measured, i.e. MTT, rBF and rBVc, we calculated the ratio with respect to cortical perfusion: cortex/deep white matter, cortex/whole tumor, cortex/tumor hotspot and cortex/hippocampus to take into account arbitrary units used by each perfusion software. The general values of cortex/deep white matter were evaluated for both hemispheres separately for every subgroup. Disease specific values of cortex/whole tumor and cortex/tumor hotspot for subgroups 1 and 2 (tumor), of cortex/white matter for subgroup 3 (cerebrovascular disease) and of cortex/hippocampus for subgroup 4 (dementia) were evaluated for the affected hemisphere.
Differences in perfusion ratios of all patients and of all subgroups for each software/deconvolution were assessed separately by paired student’s t-tests and one-way ANOVA after verification of normal distribution of the data. We also calculated mean and median values as well as standard deviation and the range (min, max) for each ratio. The value p < 0.05 was considered statistically significant. Statistical analysis was performed using SPSS v. 19 (IBM, Ehningen, Germany).
Results
Comparing both software suites, perfusion metrics of cortex/white matter were significantly different for all measured perfusion parameters. For the left hemisphere, the ratio of MTT was 0.991 (NordicICE, NI) vs 0.847 (Olea sphere, OS); rBF 3.230 vs 4.418 and rBVc 2.813 vs 3.884; all p < 0.05. For the right hemisphere, the ratio of MTT was 1.079 vs 0.854, rBF 3.262 vs 4.378 and rBVc 2.762 vs 3.935; all p < 0.05 (Table 2). The results of the detailed analysis of all subgroups are given in Tables 3–6. Regarding disease specific perfusion values of the four subgroups, both software solutions yielded different hemodynamic parameter values. For both tumor subgroups, rBVc was different regarding the ratio cortex/whole tumor with 0.778 (NI) vs 0.919 (OS); p < 0.001 for subgroup 1 and 0.768 vs 0.738; p < 0.05 for subgroup 2. Regarding the ratio cortex/tumor hotspot, rBVc was also different with 0.529 (NI) vs 0.512 (OS); p < 0.05 for subgroup 1 and 0.611 vs 0.581; p < 0.001 for subgroup 2 (Tables 7 and 8). For subgroup 3 (cerebrovascular disease) the ratio cortex/white matter was significantly different regarding MTT for the affected hemisphere: 1.058 (NI) vs 0.784 (OS); p < 0.001 but not for the unaffected hemisphere 0.819 vs 0.782: p = 0.7 (Table 5). For subgroup 4 (dementia), the ratios cortex/hippocampus were different for both hemispheres regarding MTT: left 0.877 (NI) vs 1.462 (OS); p < 0.05, right 1.165 vs 1.575; p < 0.05 and rBVc: left 1.152 vs 1.795; p < 0.001, right 1.396 vs 1.662; p < 0.05. There was no difference regarding rBF in this subgroup (Table 9). Detailed results of disease-specific ROIs of all subgroups including the comparison of SVD with the results of Bayesian deconvolution are given in Table 5 and Tables 7–9.
Table 2. .
Ratio cortex/white matter all subjects
| Left hemisphere | Right hemisphere | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
||
| MTT | N | 260 | 260 | 260 | MTT | N | 260 | 260 | 260 |
| Mean | 0.991 | 0.847 | 0.629 | Mean | 1.079 | 0.854 | 0.636 | ||
| SD | 1.1374 | 0.4193 | 0.4205 | SD | 1.2125 | 0.3906 | 0.3879 | ||
| Median | 0.769 | 0.796 | 0.590 | Median | 0.812 | 0.801 | 0.557 | ||
| Range (Min, Max) | (0.00,10.72) | (0.03,3.96) | (0.01,4.47) | Range (Min, Max) | (0.05,13.00) | (0.00,4.31) | (0.01,3.62) | ||
| p-value | 0.03001 | < 0.0001 | p-value | 0.0315 | < 0.0001 | ||||
| rBF | N | 260 | 260 | 260 | rBF | N | 260 | 260 | 260 |
| Mean | 3.230 | 4.418 | 6.520 | Mean | 3.262 | 4.378 | 6.575 | ||
| SD | 2.1272 | 1.6958 | 3.8639 | SD | 2.0701 | 1.5207 | 3.9608 | ||
| Median | 2.784 | 4.182 | 5.546 | Median | 2.845 | 4.046 | 5.448 | ||
| Range (Min, Max) | (0.32,13.34) | (1.07,13.02) | (1.65,27.22) | Range (Min, Max) | (0.31,11.83) | (1.46,9.07) | (1.64,28.33) | ||
| p-value | < 0.0001 | < 0.0001 | p-value | < 0.0001 | < 0.0001 | ||||
| rBVc | N | 260 | 260 | 260 | rBVc | N | 260 | 260 | 260 |
| Mean | 2.813 | 3.884 | 3.888 | Mean | 2.762 | 3.935 | 3.935 | ||
| SD | 1.7993 | 1.4015 | 1.4147 | SD | 1.8129 | 1.2853 | 1.2937 | ||
| Median | 2.503 | 3.583 | 3.583 | Median | 2.661 | 3.682 | 3.682 | ||
| Range (Min, Max) | (0.24,15.54) | (1.22,12.67) | (1.22,12.67) | Range (Min, Max) | (0.21,22.76) | (1.66,10.00) | (1.66,10.00) | ||
| p-value | < 0.0001 | < 0.0001 | p-value | < 0.0001 | < 0.0001 | ||||
MTT, mean transit time; SD, standard deviation; rBF, relative cerebral blood flow; rBV, relative cerebral blood volume.
Table 3. .
Ratio cortex/white matter, subgroup 1 (tumor)
| Left hemisphere | Right hemisphere | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
||
| MTT | N | 46 | 46 | 46 | MTT | N | 46 | 46 | 46 |
| Mean | 0.805 | 0.909 | 0.826 | Mean | 0.869 | 0.924 | 0.821 | ||
| SD | 0.2230 | 0.2230 | 0.2149 | SD | 0.5336 | 0.2639 | 0.3114 | ||
| Median | 0.797 | 0.902 | 0.817 | Median | 0.753 | 0.905 | 0.737 | ||
| Range (Min, Max) | (0.35,1.32) | (0.51,1.68) | (0.45,1.42) | Range (Min, Max) | (0.33,3.69) | (0.47,2.24) | (0.43,2.08) | ||
| p-value | 0.0027 | 0.4217 | p-value | 0.0041 | 0.9786 | ||||
| rBF | N | 46 | 46 | 46 | rBF | N | 46 | 46 | 46 |
| Mean | 3.556 | 3.107 | 3.612 | Mean | 3.699 | 3.481 | 4.317 | ||
| SD | 1.5879 | 0.9616 | 1.3104 | SD | 1.5966 | 1.1861 | 1.7372 | ||
| Median | 3.330 | 3.024 | 3.234 | Median | 3.271 | 3.320 | 3.974 | ||
| Range (Min, Max) | (1.35,9.47) | (1.07,5.42) | (1.65,7.15) | Range (Min, Max) | (1.49,9.13) | (1.46,7.85) | (1.64,8.99) | ||
| p-value | 0.0022 | 0.2674 | p-value | 0.2917 | 0.0134 | ||||
| rBVc | N | 46 | 46 | 46 | rBVc | N | 46 | 46 | 46 |
| Mean | 2.729 | 2.970 | 2.970 | Mean | 3.181 | 3.415 | 3.418 | ||
| SD | 0.8689 | 0.8426 | 0.8431 | SD | 1.6610 | 0.9981 | 0.9997 | ||
| Median | 2.498 | 2.861 | 2.861 | Median | 2.795 | 3.324 | 3.324 | ||
| Range (Min, Max) | (1.21,4.90) | (1.22,5.97) | (1.22,5.97) | Range (Min, Max) | (1.34,9.29) | (1.66,6.19) | (1.66,6.19) | ||
| p-value | 0.0022 | 0.0021 | p-value | 0.0002 | 0.0002 | ||||
MTT, mean transit time; SD, standard deviation; SVD, singular value deconvolution; rBF, relative cerebral blood flow; rBV, relative cerebral blood volume.
Table 4. .
Ratio cortex/white matter, subgroup 2 (tumor)
| Left hemisphere | Right hemisphere | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
||
| MTT | N | 34 | 34 | 34 | MTT | N | 34 | 34 | 34 |
| Mean | 2.126 | 0.955 | 0.688 | Mean | 1.827 | 0.963 | 0.737 | ||
| SD | 2.7539 | 0.8290 | 0.8517 | SD | 2.8683 | 0.8497 | 0.7459 | ||
| Median | 0.851 | 0.702 | 0.414 | Median | 0.779 | 0.706 | 0.544 | ||
| Range (Min, Max) | (0.35,10.72) | (0.03,3.96) | (0.01,4.47) | Range (Min, Max) | (0.32,13.00) | (0.00,4.31) | (0.01,3.62) | ||
| p-value | 0.0553 | < 0.0001 | p-value | 0.0357 | < 0.0001 | ||||
| rBF | N | 34 | 34 | 34 | rBF | N | 34 | 34 | 34 |
| Mean | 3.266 | 5.080 | 7.388 | Mean | 3.637 | 4.259 | 5.744 | ||
| SD | 2.8729 | 2.1733 | 3.7865 | SD | 2.9061 | 1.1969 | 2.0676 | ||
| Median | 2.041 | 4.548 | 6.564 | Median | 2.866 | 4.018 | 5.068 | ||
| Range (Min, Max) | (0.38,10.20) | (2.80,13.02) | (3.49,23.51) | Range (Min, Max) | (0.42,11.83) | (2.39,7.61) | (3.32,11.72) | ||
| p-value | 0.0147 | < 0.0001 | p-value | 0.01415 | 0.0008 | ||||
| rBVc | N | 34 | 34 | 34 | rBVc | N | 34 | 34 | 34 |
| Mean | 4.002 | 4.203 | 4.203 | Mean | 4.620 | 3.564 | 3.564 | ||
| SD | 2.3050 | 1.9043 | 1.9043 | SD | 3.2907 | 0.8409 | 0.8409 | ||
| Median | 3.253 | 3.619 | 3.619 | Median | 3.325 | 3.479 | 3.479 | ||
| Range (Min, Max) | (1.63,13.27) | (2.36,12.67) | (2.36,12.67) | Range (Min, Max) | (1.47,14.54) | (2.29,5.53) | (2.29,5.53) | ||
| p-value | 0.0890 | 0.0890 | p-value | 0.04449 | 0.04449 | ||||
Table 5. .
Ratio cortex/white matter, subgroup 3 (stroke)
| Affected hemisphere | Unaffected hemisphere | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
||
| MTT | N | 124 | 124 | 124 | MTT | N | 124 | 124 | 124 |
| Mean | 1.058 | 0.784 | 0.480 | Mean | 0.819 | 0.782 | 0.491 | ||
| SD | 0.7295 | 0.2520 | 0.2147 | SD | 0.4598 | 0.2754 | 0.2350 | ||
| Median | 0.906 | 0.762 | 0.459 | Median | 0.745 | 0.756 | 0.479 | ||
| Range (Min, Max) | (0.05,4.60) | (0.08,1.68) | (0.04,1.28) | Range (Min, Max) | (0.00,3.38) | (0.05,2.38) | (0.07,1.39) | ||
| p-value | 0.0001 | < 0.0001 | p-value | 0.7927 | < 0.0001 | ||||
| rBF | N | 124 | 124 | 124 | rBF | N | 124 | 124 | 124 |
| Mean | 2.645 | 4.953 | 8.376 | Mean | 2.689 | 4.928 | 7.991 | ||
| SD | 1.5693 | 1.5943 | 4.6975 | SD | 1.9928 | 1.5970 | 4.2485 | ||
| Median | 2.314 | 4.700 | 7.094 | Median | 2.243 | 4.668 | 6.922 | ||
| Range (Min, Max) | (0.31,8.22) | (1.96,9.07) | (2.06,28.33) | Range (Min, Max) | (0.32,13.34) | (2.17,10.67) | (2.46,27.22) | ||
| p-value | < 0.0001 | < 0.0001 | p-value | < 0.0001 | < 0.0001 | ||||
| rBVc | N | 124 | 124 | 124 | rBVc | N | 124 | 124 | 124 |
| Mean | 3.235 | 4.376 | 4.385 | Mean | 2.310 | 4.259 | 4.266 | ||
| SD | 3.3508 | 1.3764 | 1.3909 | SD | 1.8318 | 1.3106 | 1.3379 | ||
| Median | 2.120 | 4.165 | 4.165 | Median | 1.918 | 3.933 | 3.933 | ||
| Range (Min, Max) | (0.21,22.76) | (2.30,10.00) | (2.29,10.00) | Range (Min, Max) | (0.24,15.54) | (2.16,9.67) | (2.16,10.46) | ||
| p-value | < 0.0001 | < 0.0001 | p-value | < 0.0001 | < 0.0001 | ||||
MTT, mean transit time; SD, standard deviation; SVD, singular value deconvolution; rBF, relative cerebral blood flow; rBV, relative cerebral blood volume.
Table 6. .
Ratio cortex/white matter, subgroup 4 (dementia)
| Left hemisphere | Right hemisphere | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
||
| MTT | N | 56 | 56 | 56 | MTT | N | 56 | 56 | 56 |
| Mean | 0.834 | 0.873 | 0.738 | Mean | 0.843 | 0.886 | 0.768 | ||
| SD | 0.3346 | 0.4293 | 0.3823 | SD | 0.3511 | 0.2508 | 0.2914 | ||
| Median | 0.750 | 0.796 | 0.644 | Median | 0.760 | 0.842 | 0.674 | ||
| Range (Min, Max) | (0.39,2.01) | (0.43,3.57) | (0.33,3.15) | Range (Min, Max) | (0.15,2.36) | (0.33,1.64) | (0.27,1.60) | ||
| p-value | 0.1390 | 0.0069 | p-value | 0.3571 | 0.0465 | ||||
| rBF | N | 56 | 56 | 56 | rBF | N | 56 | 56 | 56 |
| Mean | 4.138 | 3.964 | 5.126 | Mean | 4.044 | 3.912 | 4.947 | ||
| SD | 1.9557 | 1.3210 | 2.3522 | SD | 2.4063 | 1.2439 | 2.1891 | ||
| Median | 3.660 | 3.878 | 4.690 | Median | 3.223 | 3.795 | 4.290 | ||
| Range (Min, Max) | (0.80,10.03) | (1.32,8.55) | (1.72,13.39) | Range (Min, Max) | (0.69,11.28) | (1.96,7.69) | (2.21,11.90) | ||
| p-value | 0.4057 | 0.0002 | p-value | 0.9807 | 0.0003 | ||||
| rBVc | N | 56 | 56 | 56 | rBVc | N | 56 | 56 | 56 |
| Mean | 3.275 | 3.614 | 3.614 | Mean | 3.184 | 3.613 | 3.588 | ||
| SD | 1.5061 | 1.2243 | 1.2243 | SD | 1.5261 | 1.2031 | 1.1935 | ||
| Median | 3.066 | 3.532 | 3.532 | Median | 2.903 | 3.358 | 3.332 | ||
| Range (Min, Max) | (0.97,7.61) | (1.75,8.20) | (1.75,8.20) | Range (Min, Max) | (0.41,7.43) | (2.11,8.46) | (2.11,8.46) | ||
| p-value | 0.0306 | 0.0313 | p-value | 0.0089 | 0.0130 | ||||
MTT, mean transit time; SD, standard deviation; SVD, singular value deconvolution; rBF, relative cerebral blood flow; rBV, relative cerebral blood volume.
Table 7. .
Ratio cortex/whole tumor affected hemisphere, subgroups 1 and 2 (tumor)
| Subgroup 1 | Subgroup 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
||
| MTT | N | 44 | 44 | 44 | MTT | N | 34 | 34 | 34 |
| Mean | 0.782 | 0.819 | 0.675 | Mean | 1.057 | 1.203 | 1.084 | ||
| SD | 0.4099 | 0.4148 | 0.4011 | SD | 1.0413 | 0.9664 | 0.9256 | ||
| Median | 0.676 | 0.731 | 0.576 | Median | 0.766 | 1.049 | 0.928 | ||
| Range (Min, Max) | (0.18,2.35) | (0.23,2.08) | (0.11,1.70) | Range (Min, Max) | (0.43,6.17) | (0.06,5.69) | (0.06,5.35) | ||
| p-value | 0.4270 | 0.2624 | p-value | 0.1952 | 0.4151 | ||||
| rBF | N | 44 | 44 | 44 | rBF | N | 34 | 34 | 34 |
| Mean | 1.279 | 1.016 | 1.488 | Mean | 1.050 | 0.749 | 0.893 | ||
| SD | 1.1872 | 0.8622 | 1.7827 | SD | 1.1898 | 0.5694 | 0.7287 | ||
| Median | 0.854 | 0.642 | 0.924 | Median | 0.653 | 0.574 | 0.634 | ||
| Range (Min, Max) | (0.19,4.87) | (0.22,3.93) | (0.23,10.95) | Range (Min, Max) | (0.10,6.19) | (0.24,3.02) | (0.21,3.65) | ||
| p-value | 0.0031 | 0.5972 | p-value | 0.0706 | 0.5977 | ||||
| rBVc | N | 44 | 44 | 44 | rBVc | N | 34 | 34 | 34 |
| Mean | 0.778 | 0.919 | 0.919 | Mean | 0.768 | 0.738 | 0.737 | ||
| SD | 0.5816 | 0.7702 | 0.7701 | SD | 0.9136 | 0.5144 | 0.5146 | ||
| Median | 0.569 | 0.590 | 0.590 | Median | 0.479 | 0.584 | 0.584 | ||
| Range (Min, Max) | (0.20,2.53) | (0.24,3.82) | (0.24,3.82) | Range (Min, Max) | (0.21,5.14) | (0.23,2.67) | (0.23,2.67) | ||
| p-value | 0.0002 | 0.0002 | p-value | 0.0446 | 0.0446 | ||||
MTT, mean transit time; SD, standard deviation; SVD, singular value deconvolution; rBF, relative cerebral blood flow; rBV, relative cerebral blood volume.
Table 8. .
Ratio cortex/tumor hotspot affected hemisphere, subgroups 1 and 2 (tumor)
| Subgroup 1 | Subgroup 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
||
| MTT | N | 44 | 44 | 44 | MTT | N | 33 | 33 | 33 |
| Mean | 1.013 | 0.884 | 0.834 | Mean | 1.102 | 0.955 | 0.858 | ||
| SD | 1.7954 | 0.5398 | 0.5256 | SD | 1.1751 | 0.5229 | 0.5558 | ||
| Median | 0.668 | 0.868 | 0.826 | Median | 0.794 | 0.877 | 0.784 | ||
| Range (Min, Max) | (0.12,12.32) | (0.18,3.34) | (0.09,2.07) | Range (Min, Max) | (0.36,5.66) | (0.06,2.16) | (0.11,2.43) | ||
| p-value | 0.2881 | 0.7654 | p-value | 0.5112 | 0.5936 | ||||
| rBF | N | 44 | 44 | 44 | rBF | N | 33 | 33 | 33 |
| Mean | 0.886 | 0.623 | 0.815 | Mean | 0.779 | 0.571 | 0.713 | ||
| SD | 0.8930 | 0.5228 | 0.8081 | SD | 0.9191 | 0.4287 | 0.5989 | ||
| Median | 0.529 | 0.389 | 0.477 | Median | 0.485 | 0.453 | 0.540 | ||
| Range (Min, Max) | (0.09,3.82) | (0.16,2.69) | (0.13,4.37) | Range (Min, Max) | (0.08,5.15) | (0.16,2.15) | (0.15,2.88) | ||
| p-value | 0.0149 | 0.7048 | p-value | 0.0202 | 0.6557 | ||||
| rBVc | N | 44 | 44 | 44 | rBVc | N | 33 | 33 | 33 |
| Mean | 0.529 | 0.512 | 0.512 | Mean | 0.611 | 0.581 | 0.581 | ||
| SD | 0.5563 | 0.3634 | 0.3633 | SD | 0.8431 | 0.3896 | 0.3896 | ||
| Median | 0.355 | 0.365 | 0.365 | Median | 0.365 | 0.493 | 0.493 | ||
| Range (Min, Max) | (0.09,3.33) | (0.17,1.69) | (0.17,1.69) | Range (Min, Max) | (0.16,4.68) | (0.20,1.91) | (0.20,1.91) | ||
| p-value | 0.0446 | 0.0433 | p-value | 0.0005 | 0.0005 | ||||
MTT, mean transit time; SD, standard deviation; SVD, singular value deconvolution; rBF, relative cerebral blood flow; rBV, relative cerebral blood volume.
Table 9. .
Ratio cortex/hippocampus, subgroup 4 (dementia)
| Left hemisphere | Right hemisphere | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
Statistics | NI (SVD) |
Olea (SVD) |
Olea (Bayesian) |
||
| MTT | N | 54 | 54 | 54 | MTT | N | 54 | 54 | 54 |
| Mean | 0.877 | 1.462 | 1.194 | Mean | 1.165 | 1.575 | 0.951 | ||
| SD | 0.6183 | 1.6592 | 1.4783 | SD | 2.7305 | 2.6800 | 1.2180 | ||
| Median | 0.728 | 0.955 | 0.661 | Median | 0.554 | 0.762 | 0.553 | ||
| Range (Min, Max) | (0.20,2.97) | (0.22,7.52) | (0.10,6.44) | Range (Min, Max) | (0.11,19.88) | (0.27,14.78) | (0.13,6.43) | ||
| p-value | 0.0034 | 0.8618 | p-value | 0.0018 | 0.3288 | ||||
| rBF | N | 54 | 54 | 54 | rBF | N | 54 | 54 | 54 |
| Mean | 1.405 | 1.538 | 2.009 | Mean | 1.722 | 1.539 | 1.867 | ||
| SD | 1.3507 | 1.0249 | 1.6327 | SD | 1.4836 | 0.8743 | 1.2297 | ||
| Median | 0.963 | 1.238 | 1.482 | Median | 1.528 | 1.332 | 1.536 | ||
| Range (Min, Max) | (0.13,5.78) | (0.18,4.83) | (0.34,6.47) | Range (Min, Max) | (0.13,9.04) | (0.36,4.11) | (0.43,6.99) | ||
| p-value | 0.3919 | 0.0013 | p-value | 0.6740 | 0.2958 | ||||
| rBVc | N | 54 | 54 | 54 | rBVc | N | 54 | 54 | 54 |
| Mean | 1.152 | 1.795 | 1.795 | Mean | 1.396 | 1.662 | 1.662 | ||
| SD | 1.6852 | 2.3977 | 2.3978 | SD | 1.9554 | 1.5576 | 1.5576 | ||
| Median | 0.807 | 1.214 | 1.214 | Median | 0.813 | 1.262 | 1.262 | ||
| Range (Min, Max) | (0.08,11.63) | (0.19,17.36) | (0.19,17.36) | Range (Min, Max) | (0.07,9.88) | (0.27,10.41) | (0.27,10.41) | ||
| p-value | < 0.0001 | < 0.0001 | p-value | 0.0035 | 0.0035 | ||||
MTT, mean transit time; SD, standard deviation; SVD, singular value deconvolution; rBF, relative cerebral blood flow; rBV, relative cerebral blood volume.
Discussion
Our results show that perfusion estimates differ between both software packages for all measured parameters regarding cortex and white matter in three different groups of neurological diseases: brain tumors, cerebrovascular disease and dementia.
Previous studies that evaluated the influence of different software algorithms on perfusion results have indicated a certain “software-dependency”.11–13,21 However, these studies only comprised rather small groups of patients, the largest study comprised 53 patients,12 and only patients with brain tumors. There are only two studies that analyzed DSC processing in cerebrovascular disease, in both cases acute stroke; one study including 18 data sets1 and one simulation study.2 These studies mainly focused on deconvolution techniques.
Regarding the importance of perfusion metrics on diagnosis and therapy decisions, reliable results of the calculated parameters and comparability are most important. Previous studies have shown a wide range of perfusion metrics, for instance rBV estimates from 3.84 ± 1.40 to 8.79 ± 5.01 in 24 patients with glioblastoma, indicating a great variability, which may be potentially caused by the operator.11 Therefore, we strongly focused on eliminating interoperator variability be establishing a highly-standardized workflow of processing perfusion raw data. To ensure exactly the same ROI positions in both computed parameter maps, we used an elaborated workflow with dedicated software for correct conversion and transfer of ROI matrix coordinates between both software packages.
Perfusion imaging in brain tumors is important for initial tumor grading and for differentiation of recurrent glioma from similar appearing treatment effects following radiochemotherapy in follow-up imaging. Leakage corrected rBV (rBVc) has been proposed to correlate with glioma tumor grade15 and is widely used for clinical imaging and research. However, it has been suggested that software-specific cut-off values for discrimination of low- and high-grade gliomas should be used.21 In brain tumor subgroups, the calculated rBVc values were significantly different when comparing the two software packages (Tables 7 and 8). There was no difference in the estimated rBVc when comparing oSVD and BAY values of Olea sphere. This is not surprising because the calculated blood volume is only dependent on the area under the R2* curve18,22 and is not influenced by the deconvolution model. However, given the standardization of the pre-processing, this means that factors other than the pre-processing and the deconvolution must account for the observed difference of the results between Olea sphere and NordicICE. This may include modeling implementation of T1 leakage correction as well as differences caused by AIF determination. It confirms the necessity for software-specific cut-off values when using rBV for diagnosis or grading of brain tumors and that clinicians and researches need to be cautious when comparing results obtained with different software. To overcome these issues with comparability, a framework to standardize the software algorithms between different vendors would be highly beneficial. Maybe the use of open-source software like it is widely used in diffusion imaging for scientific purposes (http://fsl.fmrib.ox.ac.uk/fsl)23 will help standardize perfusion imaging.
In addition to the two subgroups with brain tumors, we also analyzed patients with intra- or extracranial stenosis. MTT values were significantly different when comparing the two iterative deconvolution techniques of Olea sphere and NordicICE (Table 5). Non-adaptive singular value deconvolution is widely used for deconvolving the tissue signal from the arterial input function in stroke perfusion imaging.1 This technique has earlier been described to be sensitive to delay and dispersion of the arriving contrast agent bolus leading to over- or underestimation of cerebral blood flow in tissues where the bolus arrives earlier than in the chosen AIF.16 oSVD, as used in our setting, in contrast is far less sensitive for differences in tracer arriving time16 leading to a more robust estimation of cerebral blood flow. The oSVD approach is to define an oscillation index and repeat the deconvolution interactively until the oscillations in the resulting residue function R(t) are below the defined limit. Nevertheless, the iterative deconvolution algorithms are different between Olea sphere and NordicICE. Bayesian parameter estimation is a probabilistic method that is considered to deliver even more accurate and robust hemodynamic parameters.18,20 It has been reported to outperform oSVD especially in cases of high cerebral blood flow.19 Thus, this finding is probably of less relevance in patients with cerebrovascular diseases.
In the dementia subgroup, MTT and rBVc values were significantly different between Olea sphere and NordicICE for the disease specific perfusion ratio cortex/hippocampus. rBF, however, did not differ. Since rBF is typically used in the diagnostic work-up of dementia to differentiate subtypes, from a technical perspective, this DSC-parameter may complement imaging-based diagnosis in dementia in the future.
Conclusion
Parametric perfusion maps are depended on the chosen software, even when a highly-standardized processing workflow is maintained. This applies to healthy brain tissue as well as to affected brain tissue in specific diseases in patients with stroke, dementia and brain tumors. Radiologists should be aware of software related variances when using DSC perfusion for clinical imaging and research.
Footnotes
Acknowledgment: Investigator initiated trial supported by Bayer Healthcare. At site two the study was funded by the Swedish Cancer Society.
Disclosure: The scientific guarantor of this publication is Prof. Dr. Arnd Doerfler. The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article. Investigator initiated trial was supported by Bayer Healthcare. One of the authors has significant statistical expertise. No complex statistical methods were necessary for this paper. Institutional Review Board approval was obtained. Written informed consent was obtained from all subjects (patients) in this study. Methodology: case-control, observational, and performed at three institutions.
Contributor Information
Manuel Alexander Schmidt, Email: manuel.schmidt@uk-erlangen.de.
Michael Knott, Email: michaelknott@uk-erlangen.de.
Philip Hoelter, Email: philiphoelter@uk-erlangen.de.
Tobias Engelhorn, Email: tobiasengelhorn@uk-erlangen.de.
Elna Marie Larsson, Email: elnamarielarsson@radiol.uu.se.
Than Nguyen, Email: Thnguyen@ottawahospital.ca.
Marco Essig, Email: messig@exchange.ca.
Arnd Doerfler, Email: arnddoerfler@uk-erlangen.de.
REFERENCES
- 1.Zaro-Weber O, Livne M, Martin SZ, von Samson-Himmelstjerna FC, Moeller-Hartmann W, Schuster A, et al. Comparison of the 2 most popular deconvolution techniques for the detection of Penumbral flow in acute stroke. Stroke 2015; 46: 2795–9. doi: 10.1161/STROKEAHA.115.010246 [DOI] [PubMed] [Google Scholar]
- 2.Meijs M, Christensen S, Lansberg MG, Albers GW, Calamante F. Analysis of perfusion MRI in stroke: to deconvolve, or not to deconvolve. Magn Reson Med 2016; 76: 1282–90. doi: 10.1002/mrm.26024 [DOI] [PubMed] [Google Scholar]
- 3.Kawano T, Ohmori Y, Kaku Y, Muta D, Uekawa K, Nakagawa T, et al. Prolonged mean transit time detected by dynamic susceptibility contrast magnetic resonance imaging predicts cerebrovascular reserve impairment in patients with moyamoya disease. Cerebrovasc Dis 2016; 42(1-2): 131–8. doi: 10.1159/000445696 [DOI] [PubMed] [Google Scholar]
- 4.Eskildsen SF, Gyldensted L, Nagenthiraja K, Nielsen RB, Hansen MB, Dalby RB, et al. Increased cortical capillary transit time heterogeneity in Alzheimer's disease: a DSC-MRI perfusion study. Neurobiol Aging 2017; 50: 107–18. doi: 10.1016/j.neurobiolaging.2016.11.004 [DOI] [PubMed] [Google Scholar]
- 5.Squarcina L, Perlini C, Peruzzo D, Castellani U, Marinelli V, Bellani M, et al. The use of dynamic susceptibility contrast (DSC) MRI to automatically classify patients with first episode psychosis. Schizophr Res 2015; 165: 38–44. doi: 10.1016/j.schres.2015.03.017 [DOI] [PubMed] [Google Scholar]
- 6.Barajas RF, Cha S. Benefits of dynamic susceptibility-weighted contrast-enhanced perfusion MRI for glioma diagnosis and therapy. CNS Oncol 2014; 3: 407–19. doi: 10.2217/cns.14.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Dijken BRJ, van Laar PJ, Holtman GA, van der Hoorn A. Diagnostic accuracy of magnetic resonance imaging techniques for treatment response evaluation in patients with high-grade glioma, a systematic review and meta-analysis. Eur Radiol 2017; 27: 4129–44. doi: 10.1007/s00330-017-4789-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boxerman JL, Shiroishi MS, Ellingson BM, Pope WB. Dynamic susceptibility contrast MR imaging in glioma: review of current clinical practice. Magn Reson Imaging Clin N Am 2016; 24: 649–70. doi: 10.1016/j.mric.2016.06.005 [DOI] [PubMed] [Google Scholar]
- 9.Wintermark M, Albers GW, Alexandrov AV, Alger JR, Bammer R, Baron J-C, et al. Acute stroke imaging research roadmap. AJNR Am J Neuroradiol 2008; 29: 1621–8. doi: 10.1161/STROKEAHA.107.512319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jafari-Khouzani K, Emblem KE, Kalpathy-Cramer J, Bjørnerud A, Vangel MG, Gerstner ER, et al. Repeatability of cerebral perfusion using dynamic susceptibility contrast MRI in glioblastoma patients. Transl Oncol 2015; 8: 137–46. doi: 10.1016/j.tranon.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orsingher L, Piccinini S, Crisi G. Differences in dynamic susceptibility contrast Mr perfusion maps generated by different methods implemented in commercial software. J Comput Assist Tomogr 2014; 38: 647–54. doi: 10.1097/RCT.0000000000000115 [DOI] [PubMed] [Google Scholar]
- 12.Hu LS, Kelm Z, Korfiatis P, Dueck AC, Elrod C, Ellingson BM, et al. Impact of software modeling on the accuracy of perfusion MRI in glioma. AJNR Am J Neuroradiol 2015; 36: 2242–9. doi: 10.3174/ajnr.A4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conte GM, Castellano A, Altabella L, Iadanza A, Cadioli M, Falini A, et al. Reproducibility of dynamic contrast-enhanced MRI and dynamic susceptibility contrast MRI in the study of brain gliomas: a comparison of data obtained using different commercial software. Radiol Med 2017; 122: 294–302. doi: 10.1007/s11547-016-0720-8 [DOI] [PubMed] [Google Scholar]
- 14.Mouridsen K, Christensen S, Gyldensted L, Østergaard L. Automatic selection of arterial input function using cluster analysis. Magn. Reson. Med. 2006; 55: 524–31. doi: 10.1002/mrm.20759 [DOI] [PubMed] [Google Scholar]
- 15.Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol 2006; 27: 859–67. [PMC free article] [PubMed] [Google Scholar]
- 16.Wu O, Østergaard L, Weisskoff RM, Benner T, Rosen BR, Sorensen AG. Tracer arrival timing-insensitive technique for estimating flow in Mr perfusion-weighted imaging using singular value decomposition with a block-circulant deconvolution matrix. Magn Reson Med 2003; 50: 164–74. doi: 10.1002/mrm.10522 [DOI] [PubMed] [Google Scholar]
- 17.Calamante F, Gadian DG, Connelly A. Quantification of bolus-tracking MRI: improved characterization of the tissue residue function using Tikhonov regularization. Magn Reson Med 2003; 50: 1237–47. doi: 10.1002/mrm.10643 [DOI] [PubMed] [Google Scholar]
- 18.Sasaki M, Kudo K, Boutelier T, Pautot F, Christensen S, Uwano I, et al. Assessment of the accuracy of a Bayesian estimation algorithm for perfusion CT by using a digital phantom. Neuroradiology 2013; 55: 1197–203. doi: 10.1007/s00234-013-1237-7 [DOI] [PubMed] [Google Scholar]
- 19.Boutelier T, Kudo K, Pautot F, Sasaki M. Bayesian hemodynamic parameter estimation by bolus tracking perfusion weighted imaging. IEEE Trans Med Imaging 2012; 31: 1381–95. doi: 10.1109/TMI.2012.2189890 [DOI] [PubMed] [Google Scholar]
- 20.Kudo K, Boutelier T, Pautot F, Honjo K, Hu J-Q, Wang H-B, et al. Bayesian analysis of perfusion-weighted imaging to predict infarct volume: comparison with singular value decomposition. Magn Reson Med Sci 2014; 13: 45–50. doi: 10.2463/mrms.2013-0085 [DOI] [PubMed] [Google Scholar]
- 21.Kudo K, Uwano I, Hirai T, Murakami R, Nakamura H, Fujima N, et al. Comparison of different post-processing algorithms for dynamic susceptibility contrast perfusion imaging of cerebral gliomas. MRMS 2017; 16: 129–36. doi: 10.2463/mrms.mp.2016-0036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris RJ, Cloughesy TF, Hardy AJ, Liau LM, Pope WB, Nghiemphu PL, et al. Mri perfusion measurements calculated using advanced deconvolution techniques predict survival in recurrent glioblastoma treated with bevacizumab. J Neurooncol 2015; 122: 497–505. doi: 10.1007/s11060-015-1755-8 [DOI] [PubMed] [Google Scholar]
- 23.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. Fsl.. Neuroimage 2012; 62: 782–90. doi: 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]