Abstract
The presence of disseminated tumour cells (DTCs) in BM predicts poorer metastasis-free survival of breast cancer patients with localized disease. DTCs persist in distant tissues despite systemic administration of adjuvant chemotherapy. Many assume this is because the majority of DTCs are quiescent. Here, we challenge this notion and provide evidence that the microenvironment of DTCs protects them from chemotherapy, independent of cell cycle status. We show that chemoresistant DTCs occupy the perivascular niche (PVN) of distant tissues, where they are protected from therapy by vascular endothelium. Inhibiting integrin-mediated interactions between DTCs and the PVN, driven partly by endothelial-derived von Willebrand Factor and vascular cell adhesion molecule-1, sensitizes DTCs to chemotherapy. Importantly, chemosensitization is achieved without inducing DTC proliferation or exacerbating chemotherapy-associated toxicities, and ultimately results in prevention of bone metastasis. This suggests that prefacing adjuvant therapy with integrin inhibitors is a viable clinical strategy to eradicate DTCs and prevent metastasis.
Despite chemotherapeutic regimens and endocrine therapies that substantially improve patient survival, late, distant recurrence of breast cancer remains a problem. Nearly 10% of all patients with invasive breast carcinoma1, and up to 17% of patients with estrogen receptor positive (ER+) disease2 relapse five or more years after adjuvant treatment. Cells that disseminate from the primary tumour prior to its detection, and persist at distant sites despite systemic therapy are thought to be the source of these distant recurrences3–7. Indeed, elimination of disseminated tumour cells (DTCs) enhances metastasis-free survival of breast cancer patients8, motivating a targeted and selective approach to eradicate DTCs before they emerge.
Currently, no such therapy exists. Instead, patients with invasive breast cancer are treated with regimens that include dose-dense Adriamycin/doxorubicin and cyclophosphamide (AC), and/or paclitaxel9. Non-proportional statistical modeling of patient survival shows that such regimens do not prevent late recurrence10, implying that chemotherapies do not effectively eradicate DTCs. This assertion has been confirmed in clinical specimens3, 5, 11, where the continued presence of DTCs is associated with poorer metastasis-free survival12, 13, and in animal models14, where single DTCs persist despite application of cytotoxic therapy.
It is commonly assumed that DTCs resist chemotherapy because the vast majority are quiescent (i.e., Ki67-negative)15. This assumption ignores a growing body of literature showing that the microenvironment mediates resistance of solid primary tumours and of hematopoietic malignancies16–21. In particular, a number of recent studies identified factors deposited within the perivascular niche (PVN) that protect tumour cells from radiotherapy22 and chemotherapy17, 18. In light of our prior demonstration that quiescent disseminated breast tumour cells reside within a PVN23, we hypothesized that this niche may also confer resistance to therapy. If so, and if the mechanisms are distinct from those that regulate quiescence, it would open the door for new strategies to prevent metastasis4.
Here, we provide experimental support for this hypothesis. Namely, we show that chemoresistant DTCs associate with the PVN, where they are protected from chemotherapy by vascular endothelium— irrespective of their cell cycle status. We show further that inhibiting key integrin-mediated interactions between DTCs and the PVN sensitizes DTCs to chemotherapy, and results in metastasis prevention in a mouse model of ER+ breast cancer bone metastasis. Importantly, chemosensitization is achieved without inducing quiescent DTCs to enter the cell cycle, and without exacerbating chemotherapy-associated toxicities. These data suggest that prefacing adjuvant therapy with integrin inhibitors is a viable strategy to eradicate DTCs and prevent metastasis.
Results.
Chemotherapy selects for perivascular DTCs.
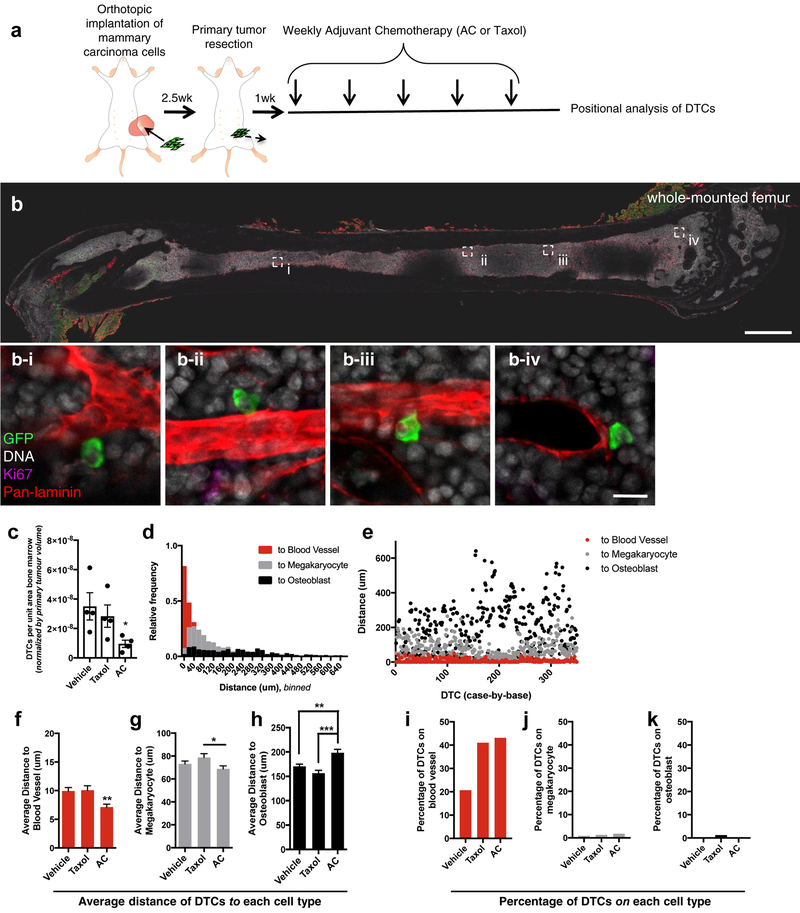
To determine whether DTCs that persist beyond the application of dose-dense chemotherapy occupy a specific niche, we implanted 4T07 cells expressing firefly luciferase and enhanced green fluorescent protein (ffluc-eGFP) into syngeneic (Balb/c) mice, and treated these mice after primary tumour resection with dose-dense AC or paclitaxel for five weeks. We scaled down human dosing9 to account for the differences in body surface area between a human and a mouse (Fig. 1a)24. Femurs from treated mice were stained, whole-mounted and imaged to readily identify eGFP+ tumour cells (Fig. 1b), quantify their number (Fig. 1c), and measure their distance to sites of interest (fig. 1d–k). Given that both DTCs and hematopoietic stem cells are characterized by long-term quiescence and therapeutic resistance25, 26, we measured the distance from eGFP+ DTCs to the three major niches shown to harbour hematopoietic stem cells: osteoblastic27-, megakaryocytic28- and perivascular29–33- niches. Analysis of 350 DTCs revealed that DTCs were concentrated in the PVN of bone marrow (BM) following AC treatment (Fig. 1d–e); the average distance to the nearest blood vessel was 7.13 μm (Fig. 1f). Such intimate localization to megakaryocytes or osteoblasts was not observed; on average, DTCs were 68.8 μm and 198 μm from these cell types (Fig. 1 g–h). Similar data were obtained following treatment with dose-dense paclitaxel (Fig. 1f–h). Comparison to vehicle-treated animals revealed that the average distance of DTCs to the vasculature decreased by 28.1% in mice treated with the AC regimen, whereas the average distance to the osteoblastic niche increased. Given the substantial reduction in DTC number observed in mice treated with AC (Fig. 1c), these data suggest that DTCs localized on vasculature, away from the bone marrow/bone interface, are resistant to AC (Fig. 1f–h). Analysis of DTCs residing directly on vasculature confirmed this assertion. AC- and paclitaxel-based regimens doubled the percentage of DTCs residing on BM vasculature (Fig. 1i). Neither enriched for DTCs neighboring megakaryocytes or osteoblasts (Fig. 1j–k).
Figure 1: Dose-dense chemotherapeutic regimens select for perivascular disseminated tumour cells.
a) Schematic of the experiment to identify if DTCs localize to a specific nice following dose-dense chemotherapy. After resection of the ffluc-eGFP+ primary tumour, mice were treated with five cycles of weekly adriamycin (2mg/kg) and cyclophosphamide (AC; 60mg/kg), paclitaxel (Taxol; 20 mg/kg), or vehicle. DTC position within BM was analyzed by staining and whole-mounting femurs one week after final treatment. b) Tile scans encompassing the entire exposed face of the BM and the full Z-depth (~100 μm) that could be imaged using this method were acquired. Scale bar, 1 mm. b i-iv) Four examples of perivascular, GFP+, Ki67− DTCs are shown in BM following AC treatment (scale bar, 20 μm). c) Quantification of DTCs in femoral BM of vehicle-, Taxol- and AC-treated animals, normalized by BM area analyzed and volume of the primary tumour at resection. For each treatment, n=4 femurs from 4 different animals were analyzed. * P= 0.022 vs. vehicle-treated mice by Kruskal-Wallis and Dunn’s test to correct for multiple comparisons. 350 DTCs were analyzed across n=3 femurs from three different animals, and d) binned in histogram format or e) dot-plotted as individual distances to the nearest blood vessel (red), megakaryocyte (gray) or osteoblast (black). f-h) The average of these distances to the nearest f) blood vessel, g) megakaryocyte and h) osteoblast for mice that underwent treatment with vehicle (n=488 DTCs analyzed across 3 animals), AC (n=350) or Taxol (n=327). ** P= 0.0039 (vs. vehicle) or P=0.0061 (vs. Taxol) for (f); * P=0.047 for (g); **P=0.0017 for (h) and *** P<0.0001 for (h); all by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test. i-k) The percentage of DTCs quantified across three mice in direct contact with i) blood vessels, j) megakaryocytes and k) osteoblasts for mice that underwent treatment with vehicle, Taxol or AC. For c, f-h, centre line represents the mean, and error bars the s.e.m. Statistical tests and P values are indicated. Source data are provided in Supplementary Table 1.
To ensure that the effect observed was not specific to femur, we performed cytometry on DTCs harboured within tibia of the same mice (Supplementary Fig. 1a–b). Here, AC treatment caused a 45.7% reduction in average distance of DTCs to the BM vasculature (Supplementary Fig. 1c). Selection for DTCs residing directly upon vascular basement membrane was a contributor, as the percentage of DTCs occupying this compartment increased 1.22-fold (Supplementary Fig. 1f). On the other hand, the spatial relationship between DTCs and megakaryocytic- or osteoblastic- niches was essentially unchanged (Supplementary Fig. 1 d–e, g–h).
Organotypic culture reveals that microvascular endothelium protects breast tumour cells from chemotherapy.
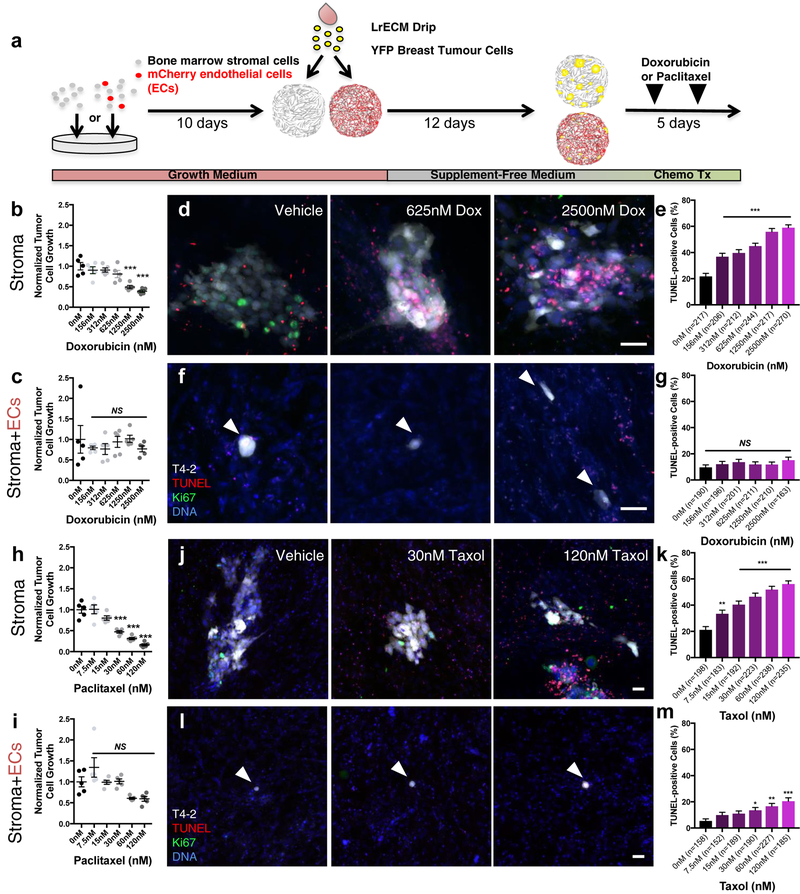
Endothelial-derived (i.e., angiocrine34) factors regulate a host of biological processes spanning development and disorder34. Previously, we developed organotypic models of the BM’s microvasculature, called BM microvascular niches (MVNs), to study whether localization of dormant DTCs to the PVN reflected a functional consequence of DTC-endothelial cell interactions23. Here, we applied these same models to determine whether endothelial cells protect breast tumour cells from chemotherapy. BM mesenchymal stem cells (MSCs, which do not drift towards an osteogenic or adipogenic phenotype over the course of these experiments, Supplementary Fig. 2) and BM MVNs were seeded sparsely with breast tumour cells to approach the scarcity of DTCs in BM35, 36. After 12 additional days, cultures were treated with increasing doses of doxorubicin or paclitaxel, and ‘DTC’ burden was analyzed five days later (Fig. 2a). Basal (HMT-3522-T4–2, T4–2) breast tumour cells exhibited a dose dependent response to doxorubicin (Fig. 2b) and paclitaxel (Fig. 2h) on BM stroma. In contrast, neither doxorubicin (Fig. 2c) nor paclitaxel (Fig. 2i) significantly reduced DTC burden on BM MVNs at any dose tested. The disparity observed in DTC response between these two niches stemmed from a lack of DTC death in MVNs. Whereas doxorubicin induced DTC apoptosis up to 59.0% on BM stroma (Fig. 2d–e), DTC death in BM MVNs increased only 5.45% between vehicle and the highest dose of doxorubicin tested (2500 nM; Fig. 2f–g). Response to paclitaxel was similar, topping out at a rate of cell killing on MVNs (20.5%) that was only one-third the rate observed on BM stroma (Fig. 2j–k, l-m). Five-fluoruracil (5-FU), a chemotherapeutic used commonly as an adjuvant in breast cancer treatment37, and lapatinib, an EGF receptor-targeted therapy38, also yielded dose-dependent responses on BM stroma that were obviated on MVNs (Supplementary Fig. 3a–h). MVN-mediated protection from doxorubicin-induced cell death was observed with a luminal, ER+ breast tumour cell line (MCF-7), also (Supplementary Fig. 3i–l) Combined with our observations in murine BM (Fig. 1), these data demonstrate that the PVN promotes chemotherapeutic resistance of DTCs, and that endothelium is the principal cellular component of the PVN protecting DTCs from chemotherapy. However, these data do not delineate whether this effect is due simply to growth restraints imposed on DTCs by the PVN23, or because of unique chemoprotective factors supplied by endothelium.
Figure 2: Microvascular endothelium protects breast tumour cells from chemotherapy.
a) Schematic of experiment to determine whether endothelium protects DTCs from chemotherapy. Dose response of HMT-3522-T4–2 (T4–2) breast cancer cells on BM b) stroma and c) MVNs to doxorubicin (n=5 independent experiments). NS, no significance, *** P<0.0001 compared to vehicleby one-way ANOVA and Dunnett’s multiple comparison test.. Following treatment, cultures were TUNEL-stained to assess apoptosis. d) Representative images of TUNEL-stained T4–2 ‘DTCs’ treated with increasing doses of doxorubicin (Dox) on BM stroma. Scale bar, 50 μm e) Percentage of TUNEL-positive DTCs per DTC cluster as a function of dose. *** P<0.0001 for all conditions vs. vehicle by one-way ANOVA and Dunnett’s multiple comparisons test. f) Representative images of TUNEL-stained T4–2 DTCs treated with increasing doses of Dox on BM MVNs. Scale bar, 50 μm. g) Percentage of TUNEL-positive DTCs per DTC cluster as a function of dose. NS denotes no significance compared to vehicle by one-way ANOVA and Dunnett’s multiple comparisons test. h, i) Dose response of T4–2 DTCs on BM h) stroma and i) MVNs to paclitaxel/Taxol (n=5 independent experiments). NS denotes no significance, *** P<0.0001 by one-way ANOVA and Dunnett’s multiple comparison test, compared to vehicle. j) Representative images of TUNEL-stained T4–2 cells treated with increasing doses of Taxol on BM stroma. Scale bar, 50 μm. k) Percentage of TUNEL-positive DTCs per DTC cluster as a function of dose. *** P<0.0001 compared to vehicle by one-way ANOVA and Dunnett’s multiple comparisons test. l) Representative images of TUNEL-stained T4–2 cells treated with increasing doses of Taxol on BM MVNs. Scale bar, 50 μm. m) Percentage of TUNEL-positive DTCs per DTC cluster as a function of dose.*P=0.0437, **P=0.0012, *** P<0.0001 compared to vehicle by one-way ANOVA and Dunnett’s multiple comparisons test. For b-c, e, g-I, k, m, centre line represents the mean, and error bars the s.e.m. Statistical tests and P values are indicated. For TUNEL analyses, the number of cells (predominantly single cells and clusters of 2–4 cells) analyzed per condition across n=3 independent experiments is stated within each panel. Source data are provided in Supplementary Table 1.
Cell cycle status and therapeutic resistance are decoupled in the perivascular niche.
To look for evidence of a niche-specific, cell-cycle independent response to chemotherapy, we took three independent approaches. First, we examined only single DTCs that persisted on BM stroma or BM MVNs over the course of 17 days, after application of the highest dose of doxorubicin (Fig. 3a) or paclitaxel (Fig. 3b). In each case, death of even single, ostensibly non-proliferative DTCs was cut in half on MVNs (Fig. 3a–b).
Figure 3: Perivascular niche-mediated chemoprotection does not depend on DTC cell cycle status.
Analysis of single DTCs on stroma (MSC) or BM MVNs (MSC+EC) following treatment with a) 2500 nM doxorubicin or b) 120 nM paclitaxel. Data depict the percentage of TUNEL-positive single cells over n=3 independent experiments. In a) *** P=0.0004 and in b) P=0.0713 by unpaired, two-tailed t test. c) Schematic for experiment to determine how cell cycle status influences response to doxorubicin on BM stroma and BM MVNs. Here, cultures were seeded with T4–2 cells expressing tdTomato and a mVenus-p27K− reporter, treated with 2500nM doxorubicin and NucView405 to label apoptotic cells, and imaged for 72 hours. d) Stills from time-lapse videos on BM stroma (top) or BM MVNs (bottom). Q indicates a quiescent cell that subsequently undergoes apoptosis (white asterisk); P denotes a proliferative cell that subsequently undergoes apoptosis (white asterisk); both on stroma. Scale bar, 20 μm. e) Quantification of relative percentages of p27+ and p27− T4–2 cells that apoptose within 72h of doxorubicin treatment on BM stroma (MSC) and on BM MVNs (MSC+EC) from n=6 independent videos per condition. ** P=0.0027 comparing MSC to MSC+EC by Two-way ANOVA. f) Schematic for experiment to determine whether ‘priming’ MVNs with IGF-1 to trigger DTC proliferation sensitizes DTCs to doxorubicin. g) Representative images of quiescent, microvascular-associated DTCs at Day 10 (left), and after priming with vehicle or with 300 ng/ml IGF-1 at Day 17 (middle). Cultures were fixed and stained at this time point to analyze the proliferative status and viability of tumour cells (right). Scale bars, 50 μm. h) Percentage of TUNEL-positive DTCs per DTC cluster as a function of priming with vehicle or 300 ng/mL IGF-1, and treating with 2500 nM doxorubicin. n=30 and 32 cells from single cells or clusters of mostly 2–4 cells were analyzed for the vehicle- and IGF-1- primed conditions, respectively, over n =3 independent experiments. P=0.793 by unpaired, two-tailed t test. For a-b, e, h centre line represents the mean, and error bars the s.e.m. Statistical tests and P values are indicated. Source data are provided in Supplementary Table 1.
Second, we engineered T4–2 cells to simultaneously express tdTomato and a mutant reporter of p27 (mVenus-p27K−), which delineates quiescent cells39. TdTomato-mVenus-p27K− T4–2 cells were seeded on BM stroma or MVNs, and treated simultaneously after 12 days with 2500nM doxorubicin and a DEVD substrate (NucView405) that fluoresces upon caspase-3-mediated cleavage40. This approach allowed us to monitor DTC apoptosis on BM stroma and MVNs as a function of cell cycle status in real-time (Fig. 3c). In both niches, there was clear evidence of p27-positive and p27-negative tumour cells undergoing apoptosis in response to doxorubicin (Fig. 3d, Supp. Movies 1–3). As expected, substantially fewer events were observed on BM MVNs than BM stroma (Fig. 3d–e). However, the distribution of affected populations skewed in a manner that suggested MVNs protect tumour cells from apoptosis irrespective of their proliferation status (Fig. 3e). Namely, apoptosis on BM stroma occurred predominantly in p27-negative tumour cells, whereas death on MVNs occurred evenly between p27-positive and -negative populations (Fig. 3e). This result suggested that stimulating tumour cell proliferation on MVNs would not sensitize tumour cells to apoptosis. We developed an approach to test this directly.
Our prior work implicated insulin-like growth factor (IGF) binding proteins as negative regulators of breast tumour cell outgrowth23, suggesting that IGF-1 supplementation would stimulate proliferation of quiescent breast tumour cells on MVNs. Administration of IGF-1 after 10 days of co-culture (Supplementary Fig. 4a) stimulated proliferation of quiescent DTCs, judged by a three-fold increase in DTC outgrowth (Supplementary Fig. 4b), and a nearly five-fold increase in Ki67-positive cells (Supplementary Fig. 4c). However, priming co-cultures with IGF-1 prior to treatment with doxorubicin (Fig. 3f) did not sensitize re-emerging tumour cells to chemotherapy (Fig. 3g–h). Thus, all three lines of evidence presented here suggest that perivascular DTCs resist chemotherapy not because they are quiescent, but because components within the PVN protect them from chemotherapy. Therefore, identifying and interfering with this signaling became our priority.
Integrin-mediated interactions between DTCs and microvascular endothelium protect DTCs from chemotherapy.
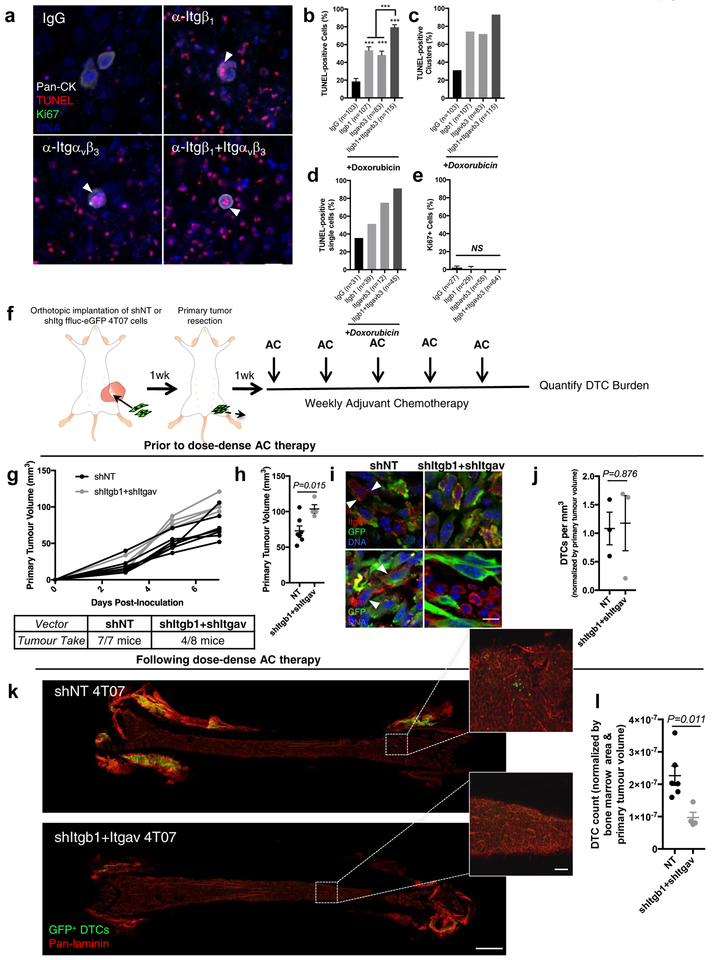
To identify putative, endothelial-derived factors that protect DTCs from chemotherapy, we sequenced the whole transcriptome of BM stroma and BM MVNs by RNAseq. Pathway analysis revealed that ‘integrin binding’ constituted one of the most significantly enriched molecular functions in MVNs (Fig. 4a). A number of the most significantly enriched integrin-cell surface interaction genes in this dataset (Fig. 4b–c) were confirmed by quantitative proteomic profiling of extracellular matrix (ECM; Fig. 4d). This— along with a growing body of evidence linking therapeutic resistance to integrin-mediated adhesion to ECM16, 19, 21, 41, 42 — led us to interrogate a series of integrin receptors that could mediate protective interactions between DTCs and molecules within the PVN. We used an array of high-quality integrin-targeted function blocking antibodies to prime MVNs prior to doxorubicin treatment (Fig. 4e). Given the abundance of single DTCs and the typical size of DTC clusters reported in patient BM43, we focused on single breast tumour cells and clusters ≤ 10 cells. Priming with isotype control antibody did not sensitize DTCs to apoptosis (compare Fig. 4f–g to Fig. 2f–g). Blocking adhesion of a number of integrin subunits led to a substantial increase in sensitivity to doxorubicin (Fig. 4g–i), and antibodies targeting integrin β1 and integrin αvβ3 yielded the best response, killing 56.0–64.6% of cells overall (a ~six-fold increase above baseline), and sensitizing up to 78.6% of single DTCs to doxorubicin (Fig. 4h). Further, these antibodies were the only two to markedly sensitize DTC clusters to doxorubicin (Fig. 4i).
Figure 4: Integrin β1 and integrin αvβ3 protect perivascular disseminated tumour cells from chemotherapy.
a) Functional enrichment analysis conducted on whole transcriptome sequencing data derived from BM stroma and BM MVNs (n=2 biologically independent samples; 3-fold differential expression cutoff). Enrichment was tested by Fisher’s exact test and controlled for multiple testing by false discovery rate (FDR). FDR (-log10) of significantly enriched Gene Ontology molecular function terms are plotted. Integrin binding was amongst the most enriched molecular functions. b) Gene set enrichment analysis of Reactome “Integrin cell surface interactions” pathway. The leading edge subset of genes defining the enrichment are outlined. c) Heatmap of relative expression values in stroma and MVNs of the 27 target genes that define the leading edge GSEA analysis in (b). d) Quantitative, ECM-targeted proteomic data displayed as a volcano plot, showing significantly enriched genes in BM stroma and BM MVNs. Data are derived from n=3 biologically independent samples; P values generated by unpaired, nonparametric, two-sided t test (Mann-Whitney). e) Schematic of experiment to test whether priming DTCs with inhibitory antibodies targeting integrin subunits sensitizes DTCs to doxorubicin. f) Representative images of fixed and TUNEL-stained T4–2 ‘DTCs’ on BM MVNs following treatment with the array of function blocking antibodies listed (or isotype/IgG control) and 2500nM doxorubicin. White arrowheads denote apoptotic DTCs; scale bar, 20 μm. Quantification of TUNEL-positive DTCs and DTC clusters following treatment with the array of function blocking antibodies listed (or isotype/IgG control) and 2500nM doxorubicin on BM MVNs yielded the total percentage of g) TUNEL-positive cells across 1–10 DTC/DTC clusters, h) TUNEL-positive single DTCs and i) TUNEL-positive DTCs within clusters of 2–4 DTCs. The number of cells/cell clusters analyzed per condition is stated within each panel. For (g) and (i), * P<0.05, ** P<0.01 and *** P<0.001 when compared to control (IgG) by one-way ANOVA and Dunnett’s multiple comparisons test. Centre line represents the mean, error bars the s.e.m. Source data are provided in Supplementary Table 1.
To ensure that this response was specific to the DTC compartment, we employed short hairpin RNA (shRNA)-mediated targeting of integrin β1 in T4–2 cells, and achieved a robust 76% knockdown of integrin β1 protein (Supplementary Fig. 5a–b). The number of DTCs or DTC clusters present after 12 days of co-culture was unchanged between shITGB1 and non-targeting (NT) control (Supplementary Fig. 5c, d), suggesting that depletion of integrin β1 does not compromise DTC survival on its own. However, integrin β1 knockdown substantially reduced DTC proliferation (Supplementary Fig. 5c, e). Upon treatment with doxorubicin, DTC burden was further compromised in the shITGB1 condition (Supplementary Fig. 5g), and the percentage of TUNEL-positive DTCs increased drastically (Supplementary Fig. 5f, h–j), on par with levels of TUNEL-labeling achieved with function blocking antibody (Fig. 4f–h). These results confirm that targeting DTC integrin β1 compromises survival; but only upon application of doxorubicin. These data also lend additional support to the concept presented in Figure 3 that chemotherapeutic response of perivascular DTCs is not linked to proliferation state.
Integrin αvβ3 has a restricted binding repertoire, which narrowed considerably when we examined transcriptomic (Fig. 4c) and proteomic data (Fig. 4d) for integrin αvβ3 ligands expressed abundantly in MVNs. Staining BM for von Willebrand Factor (VWF), osteopontin (SPP1), and tenascin-C (TNC) revealed that only VWF was expressed on the basal surface of BM microvascular endothelium (Fig. 5a), consistent with prior reports44–46. To determine whether endothelial VWF protects breast tumour cells from chemotherapy, a loss-of-function approach was employed. We identified two shRNA clones that knocked down EC expression of VWF by 48% and 87%, (Fig. 5b–c) and confirmed, via immunofluorescence, that knockdown sustained over the 27-day course of our experiments (Fig. 5d). Loss of VWF did not impact breast tumour cell survival (Fig. 5e) or outgrowth (Fig. 5f) in the absence of chemotherapy. However, when depletion of endothelial VWF was combined with doxorubicin, up to 70% of single T4–2 cells underwent apoptosis in a manner that correlated with the level of VWF knockdown (Fig. 5g–i). These levels nearly mirrored those obtained with integrin αvβ3 function-blocking antibody, suggesting that integrin αvβ3 protects DTCs from chemotherapy through signaling triggered by endothelial VWF.
Figure 5: Endothelial von Willebrand Factor and VCAM-1 confer DTC resistance to doxorubicin.
a) IF staining for VWF, SPP1, TNC and VCAM-1 to assess localization to BM microvessels. Scale bar, 20 μm (VWF); 10 μm (SPP1, TNC, and VCAM1). b) Representative immunoblot of ECs infected with non-targeting (NT) shRNA or two different shRNA clones targeting VWF. Blot was co-stained for VWF and the nuclear envelope proteins Lamin A/C (loading control). c) Densitometry from n=3 biologically independent samples, normalized to intensity of shNT band on each blot. **P=0.008, ***P=0.0002 by one-way ANOVA and Dunnett’s multiple comparisons test. d) Representative immunostains (n=3 independent experiments) for VWF in BM MVNs composed of shNT ECs or shVWF ECs after 27-days in selection-free media. Scale bar, 100 μm e) Number of clusters and f) tumor cell outgrowth (n=5 independent experiments) in the absence of doxorubicin (n=5 independent experiments). NS, no significance between shNT and shVWF conditions by one-way ANOVA and Dunnett’s multiple comparisons test. Quantification of g) TUNEL-positive DTCs (overall), h) single DTCs and i) 2–4 cell DTC clusters following treatment with 2500nM doxorubicin as a function of endothelial VWF expression. In (g) *P=0.0266 and *** P<0.0001 vs. shNT by one-way ANOVA and Dunnett’s multiple comparisons test. j) TUNEL-positive DTCs/DTC clusters and k) tumour cell outgrowth on BM MVNs following treatment with only VCAM1 blocking antibody or mouse IgG control (n=5 independent experiments). Stated P-values by unpaired, two-tailed t test. l) The percentage of TUNEL-positive DTCs (overall), m) single DTCs and n) clusters of 2–4 DTCs from BM MVNs primed with VCAM1 function blocking antibody or isotype control prior to treatment with 2500nM doxorubicin (as in Fig. 4e). ** P=0.0054 by unpaired, two-tailed t test. For TUNEL analyses, the number of cells (predominantly single cells and clusters of 2–4 cells) analyzed per condition across n=3 independent experiments is stated within each panel. For c, e-g, I, j-l, n centre line represents the mean, and error bars the s.e.m. Source data are provided in Supplementary Table 1.
The potent effect of inhibiting the integrin α4 and β1 subunits on breast tumour cell survival (Fig. 4f–i) suggested that an endothelial ligand for DTC integrin α4β1 mediates chemotherapeutic resistance. Vascular cell adhesion molecule-1 (VCAM-1)— an integrin α4β1 ligand shown previously to mediate chemoresistance of leukemia cells47 — was enriched in BM MVNs (Fig. 4c) and expressed by BM endothelium (Fig. 5a). Therefore, we used a well-characterized antibody that blocks adhesion to VCAM-1 (ref.48) in order to assess whether endothelial VCAM-1 protects DTCs from doxorubicin. Consistent with our observations on VWF depleted endothelium, inhibiting DTC binding to VCAM-1 in the absence of doxorubicin did not alter tumour cell survival or proliferation (Fig. 5j–k). However, upon doxorubicin administration, a near doubling of TUNEL positive cells was observed overall (Fig. 5l), driven primarily by a substantial induction of apoptosis in single breast tumour cells (Fig. 5m–n). These levels of apoptosis induction did not fully approach those obtained by blocking integrin α4 adhesion, suggesting that VCAM-1 is one of multiple endothelial-derived integrin α4β1 ligands that confer chemotherapeutic resistance to DTCs.
Blocking integrin-mediated signalling synergizes with chemotherapy to deplete DTCs from BM and prevent metastasis.
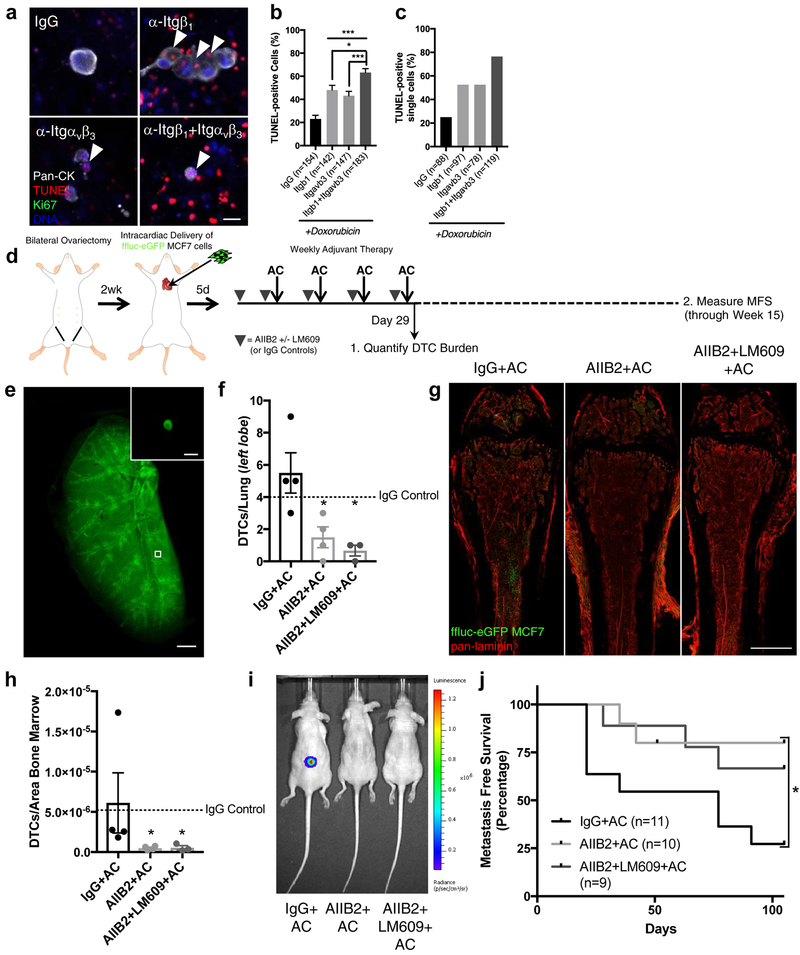
Given our goal of eradicating DTCs from BM, we sought to determine whether combined inhibition of integrin β1 and integrin αvβ3 would result in an additive effect. Combined treatment with function blocking antibodies targeting integrin-β1 and -αvβ3 led to at least one TUNEL-positive tumour cell in 93% of all DTC/clusters analyzed, sensitization of 79.6% of DTCs overall (vs. 48.3–53.6% with either antibody on its own), and sensitization of 91% of all single DTCs to doxorubicin (vs. 35.5% for IgG-primed BM MVNs, 51.3% for MVNs primed with antibody targeting integrin β1, and 75% for MVNs primed with antibody targeting integrin αvβ3) (Fig. 6a–d). Importantly, priming with these integrin inhibitors did not sensitize DTCs to chemotherapy by inducing cell cycle re-entry; treatment with antibodies alone followed by analysis of Ki67 status revealed that both antibodies essentially pushed all DTCs into G0/early G1 (Fig. 6e).
Figure 6: Targeting integrin-β1 and integrin-αvβ3 sensitizes BM DTCs to chemotherapy.
a) TUNEL-stained T4–2 ‘DTCs’ on BM MVNs following treatment with integrin β1 and/or integrin αvβ3 function blocking antibodies and 2500nM doxorubicin. White arrowheads: apoptotic DTCs. Scale bar, 20 μm. Percentage of b) TUNEL-positive DTCs (from predominantly single cells and clusters of 2–4 cells), c) DTC clusters with at least one TUNEL-positive DTC; and d) TUNEL-positive single cells. ***P<0.0001 comparing primed conditions to isotype control, or comparing dual-priming to single agent priming, by one-way ANOVA and Tukey’s multiple comparison test. e) Percentage of Ki67+ DTCs after priming. NS, no significance by one-way ANOVA and Tukey’s multiple comparison test. f) Schematic of experiment to test the combined effect of blocking integrin β1 and integrin αvβ3 on DTC survival in response to chemotherapy in vivo. g) Kinetics of primary tumour growth and take rates for shNT and shItgb1+Itgav 4T07 cells. h) Average primary tumour volume of shNT (n=7 animals) and shItgb1+Itgav (n=4 animals) cohorts at resection. *P=0.0148 by unpaired, two-tailed t test. i) Representative images of n=3 biologically independent shNT and shItgb1+Itgav 4T07 primary tumours, stained for activated integrin-β1 (top row) and integrin αv (bottom row). White arrowheads, integrin expression on GFP+ tumour cells. Scale bar, 10 μm. j) DTCs/mm3 of BM in an untreated cohort of mice (n=3 femurs from three distinct animals). P=0.876 by unpaired, two-tailed t test. k) Representative images of whole-mounted femurs derived from animals treated with dose-dense AC. Scale bar, 1 mm; inset, 100 μm. l) DTCs/femur, normalized by the total area of BM analyzed and by primary tumour volume. n=7 femurs from seven different animals were analyzed for the shNT condition; n=4 femurs from four different animals were analyzed for the shItgb1+shItgav condition. P=0.011 by unpaired, two-tailed t test. For TUNEL and Ki67 analyses, the number of cells (predominantly single cells and clusters of 2–4 cells) analyzed per condition across n=3 independent experiments is stated within each panel. For b, e, h, j, l centre line represents the mean, and error bars the s.e.m. Source data are provided in Supplementary Tables 1 and 2.
These results motivated an experiment to determine whether inhibiting the function of DTC integrin-β1 and -αvβ3 would result in more effective AC-mediated elimination of DTCs in vivo. We used syngeneic, immune competent mice tolerized to ffluc-eGFP49 for this experiment, and due to the lack of suitable function blocking antibodies targeting murine integrin αvβ3, employed shRNA-mediated knockdown of integrin αv and integrin β1. Ffluc-eGFP 4T07 cells incorporating either NT shRNA or shRNA targeting Itgb1 and Itgav (resulting in a downregulation of 81% and 55% at the protein level, respectively) were inoculated in the inguinal fat pads of syngeneic mice (Fig. 6f). Although take rate of the double knockdown line was diminished (Fig. 6g), tumours that did take grew more rapidly than their NT counterparts (Fig. 6g–h), despite sustained reduction of integrin β1 and integrin αv within the primary tumour (Fig. 6i). The number of DTCs harboured in BM did not vary between the two groups in the absence of chemotherapy (Fig. 6j). However, after five rounds of dose dense AC, analysis of whole-mounted femurs revealed that inhibition of integrin β1 and integrin αv resulted in a 57.2% reduction in DTC burden within BM (Fig. 6k–I, Supplementary Table 2).
Despite this promising result, we were unable to address the critical question of whether successful eradication of BM DTCs prevents metastasis in this or any other immune-competent model. We believe this is due to persistent immunogenicity of ffluc and eGFP, even in animals engineered to express these xenogens at an embryonic stage. Consequently, we evaluated an immune compromised model of breast cancer metastasis to bone employing intracardiac delivery of MCF-7, ER+ cells that cluster with the luminal A breast cancer subtype50. Luminal breast cancers are the most bone metastatic breast cancer subtype51, and comprise the majority of late breast cancer recurrences52. In BM MVNs, inhibition of integrin-β1 and -αvβ3 sensitized MCF-7 to doxorubicin in an additive fashion (Fig. 7a–c).
Figure 7: Targeting integrin-β1 sensitizes DTCs to chemotherapy and prevents bone metastasis.
a) Representative images of TUNEL-stained MCF-7 cells on BM MVNs following priming with integrin-β1 and/or -αvβ3 function-blocking antibodies, and 2500 nM doxorubicin. White arrowheads, apoptotic DTCs. Scale bar, 20 μm. Percentage of b) TUNEL-positive cells from predominantly single cells and clusters of 2–4 cells and c) TUNEL-positive single cells. The number of cells/clusters analyzed per condition across n=3 independent experiments is stated within each panel.***P≤ 0.0007 for IgG primed vs. single agent priming; *P= 0.0149 for integrin β1 primed vs. dual agent priming; ***P=0.0004 for integrin αvβ3 primed vs. dual agent priming, all by one-way ANOVA and Tukey’s post-test. d) Schematic of experiment to test the effect of blocking integrin β1 and integrin αvβ3 function on DTC survival and bone metastasis in vivo. e) Representative image of the left lobe of a Ce3D55 cleared lung, stained for eGFP to quantify lung DTCs. Scale bar, 1mm. Inset: representative DTC. Scale, 10 μm. f) DTCs/lung following treatment (Day 29). *P=0.022 for AIIB2+AC (n=4 lungs from four animals) vs. IgG+AC (n=4 lungs from four animals), P=0.013 for AIIB2+LM609+AC (n=3 lungs from three animals) vs. IgG+AC; by one-way ANOVA and Dunnett’s post-test. Dashed line: mean value for mice treated with IgG only. g) Representative images of whole-mounted left femoral heads from animals treated with IgG+AC, AIIB2+AC and AIIB2+LM609+AC. Scale bar, 1 mm. h) DTCs/area BM following treatment (Day 29). *P=0.050 for AIIB2+AC (n=4 femurs from four animals) vs. IgG+AC (n=4 femurs from four animals); P=0.043 for AIIB2+LM609+AC (n=3 femurs from three animals) vs. IgG+AC, by Kruskall-Wallis test and Dunn’s post-test. Dashed line: mean value for mice treated with IgG only. i) Bioluminescence of mice from each treatment group at week 15. j) Kaplan-Meier analysis of metastasis-free survival. *P=0.039 by Log-rank Mantel-Cox test; P=0.034 by Gehan-Breslow-Wilcoxon method. Tick mark represents one censored mouse that died of unknown causes. Cohort size: IgG+AC: 11 animals; AIIB2+AC: 10 animals; AIIB2+LM709+AC: 9 animals. For b, f, h centre line represents the mean, error bars: s.e.m. Source data for b, f, h, j provided in Supplementary Table 1.
Intracardiac delivery of MCF-7 cells in ovariectomized, athymic nude mice allowed the study of luminal breast cancer pathogenesis in the background of estrogen suppression. Female mice were inoculated with 1×105 ffluc-eGFP MCF-7 cells 14–16 days after bilateral ovariectomy, primed with antibody 5 days later, then treated weekly with antibody and AC therapy for four cycles (Fig. 7d). Mice were divided into two cohorts: one sacrificed the day after treatment in order to quantify DTC burden; the other monitored weekly for bioluminescence to track metastasis-free survival (Fig. 7d). In contrast to mice primed with isotype control, mice primed with integrin-β1 inhibitory antibody AIIB253 or a combination of AIIB2 and the integrin-αvβ3 inhibitory antibody LM60954 exhibited a marked and significant reduction of DTC burden in Ce3D-cleared55 and wholemounted lung (Fig. 7e–f), and in wholemounted femur (Fig. 7g–h). Of note, priming with AIIB2 or AIIB2+LM609 with AC treatment led to a 93.9% and a 94.2% reduction in DTC burden within femoral BM (Fig. 7h).
Because mice inoculated with MCF-7 eventually experience bone metastasis at one of a number of sites including the skull, mandible, vertebrae, and femur, we could finally address the question of whether a reduction of DTC burden equates to metastasis prevention in this model. Over a prescribed 15-week period (Fig. 7d), 73% of mice primed with IgG and treated with AC succumbed to bone metastasis, whereas only 22% of mice primed with AIIB2 and 33% of mice primed with AIIB2 and LM609 relapsed (Fig. 7i–j). Of note, and in agreement with previous work56, 57, priming with AIIB2 (or AIIB2 and LM609) did not manifest in overt toxicities affecting body weight (Supplementary Fig. 6a), nor did this regimen alter the density of BM vasculature (Supplementary Fig. 6b). Splenic weight (Supplementary Fig. 6c) was also unchanged over the course of treatment, indicating gross changes of immune cell content did not occur58. This prompted a more thorough examination of viable immune cell populations in BM of uninoculated, immune-competent mice. Priming with AIIB2 did not skew AC-driven effects on the number of viable CD45+ leukocytes, CD11b+ myeloid cells, B220+ B cells and CD3+ T cells (Supplementary Fig. 6d).
These data confirm that augmenting standard-of-care therapy with an antibody blocking integrin β1 function results in a drastic reduction in DTC burden and prevention of bone metastasis— all without enhancing chemotherapy-associated toxicity. These data also illustrate the predictive power of organotypic MVNs to accurately reveal the biology and vulnerabilities of disseminated breast tumour cells.
Discussion.
Here, we present data showing that the BM microvascular niche protects disseminated breast tumour cells from chemotherapy in a cell cycle-independent fashion. We show that integrin-mediated interactions between DTCs and molecules within the MVN, including VWF and VCAM-1, protect DTCs from chemotherapy. Disrupting these interactions sensitizes DTCs to chemotherapy, and prevents bone metastasis. Importantly, integrin inhibition does not trigger quiescent DTCs to enter the cell cycle, nor does it exacerbate chemotherapy-associated toxicities. Thus, we anticipate this finding could ultimately inspire modifications to adjuvant therapy that eradicate DTC burden up front, and prevent future metastasis.
A key finding of this work is that sensitization to cytotoxic therapy can be achieved in quiescent cells. Inhibiting integrin β1 and/or integrin αvβ3 sustains cell cycle exit, and enhances chemotherapeutic response. This finding is reminiscent of prior work in normal and malignant breast epithelial cells demonstrating that protection from apoptosis-inducing agents is achieved not by quiescence-induction, but by laminin-induced polarity via integrin α6β4-mediated hemidesmosome formation21. The present work provides several lines of evidence to support the hypothesis4 that DTC quiescence and chemoresistance are decoupled. Polarity at the level of single DTCs may be a crucial discriminator.
Translating our findings from organotypic culture to preclinical models led to one surprise. Inhibition of integrin β1 and integrin αvβ3 behaved additively in organotypic culture. However, inhibition of integrin β1 alone was sufficient to sensitize the vast majority of BM DTCs to AC therapy, and prevent bone metastasis of MCF-7 cells in immune compromised mice. This result suggests that compromising integrin β1 function is particularly detrimental to metastasis-initiating DTCs, and emphasizes the need to compliment tractable culture models with animal models that allow measurement of metastasis-free survival as a preclinical endpoint.
Despite the marked improvement in metastasis prevention gained from coupling integrin β1 inhibition with adjuvant therapy, 22% of mice still succumbed to bone metastases. This indicates that some metastasis-initiating cells are left behind despite targeting integrin β1. One variable we have not accounted for here is evolution of the DTC microenvironment in response to chemotherapy. Chemotherapy-induced DNA damage of even quiescent cells causes an array of factors to be secreted, including drivers of chemotherapeutic resistance59, 60. Characterizing the chemotherapy-induced secretome of BM vascular endothelium may reveal factors that stimulate DTC survival further. We are working currently to catalogue this secretome, and postulate that blocking molecules that stimulate parallel (rather than convergent) signaling pathways will enhance elimination of metastasis-initiating DTCs.
Our work provides critical insight about how therapeutic resistance of BM DTCs can be overcome. Still, we cannot conclude definitively that elimination of DTCs from BM indicates DTC eradication and metastasis prevention at the systemic level. Addressing this point will determine whether therapies that diminish BM DTC burden prevent metastasis in other cancers, including other breast cancer subtypes, that are more prone to relapse outside of the bone. For now, we can conclude that adjuvant chemotherapy can be modified to target the PVN, resulting in depletion of the BM DTC reservoir, and prevention of bone metastasis.
Methods
Animal studies.
All animal work was performed in accordance with institutional, IACUC (specifically, Fred Hutchinson Cancer Research Center (Fred Hutch) protocols 50865 and 50928) and AAALAS guidelines and ethical regulations.
Modeling spontaneous dissemination and chemoresistance in immune-competent mice.
To quantify the location of DTCs following spontaneous dissemination from orthotopic tumours, 1 × 106 ffluc-eGFP 4T07 cells61 (Karmonos Cancer Institute, Wayne State University) were injected into the inguinal mammary gland of 6–8-week-old female C.FVB-Tg(GNrhr-luc/EGFP)L8Mrln/LmwJ (aka “Glowing Head (GH)”)49 Balb/c mice in 30 μl of a 1:1 solution of LrECM (Growth-factor reduced Cultrex; Trevigen): PBS. Tumours were resected at a final volume of ~250 mm3 approximately 2.5 weeks later, fixed in 1.6% paraformaldehyde (PFA)/30% sucrose/PBS overnight (4 °C), equilibrated in 1:1 Optimal Cutting Temperature (OCT; Fisher Scientific) embedding compound:30% sucrose/PBS for 2 hours (4 °C) and embedded in OCT compound. Bioluminescent imaging (BLI) was performed on a IVIS Spectrum (Perkin Elmer) one-week post-resection following intraperitoneal delivery of 100 μl D-Luciferin (10 mg/mL, BioVision, Inc.) to confirm successful resection; any mice with residual BLI signal were excluded from future study.
To mimic dose-dense chemotherapy in human patients, mice were treated one week post-resection with 5 weekly cycles of either Adriamycin/doxorubicin (2 mg/kg) and cyclophosphamide (60 mg/kg; AC therapy), paclitaxel (20 mg/kg), or vehicle control24. Chemotherapeutics were purchased from SelleckChem. BLI imaging was performed once more after the cessation of therapy to ensure that relapse at the primary site had not occurred.
Animals were euthanized six weeks post-resection and perfused with 25 mL ice cold PBS via the left ventricle (with simultaneous ligation of the inferior vena cava). Femurs and tibia were denuded of connective tissue, fixed in 4% PFA (Electron Microscopy Sciences)/PBS for 2 hours at room temperature, transferred to 30% sucrose/PBS overnight (4 °C), equilibrated in 1:1 OCT:30% sucrose/PBS for 2 hours (4 °C) and finally embedded flat (i.e., with the femoral head and the condyles parallel to the surface) in 24×24×5 mm embedding molds in OCT compound.
For experiments to study the effect of integrin β1 and integrin αvβ3 on the survival of bone marrow DTCs, shNT or shItgb1+shItgav ffluc-eGFP 4T07 cells were implanted into the inguinal mammary gland of GH-Balb/c mice as described above, in a randomized and blinded fashion. Orthotopic tumours were surgically resected after 7 days. One-week post-resection, animals underwent BLI to confirm absence of local recurrence. Mice were subsequently treated with five cycles of AC, and euthanized six weeks post-resection. Femurs were processed as described in the previous paragraph.
Modeling bone metastasis of luminal breast cancer in the setting of estrogen suppression.
Ovariectomy Surgery:
5–6 week old female, athymic nude mice (Charles River; Strain 490) were ovariectomized by dual dorsal incisions to expose and dissect the ovaries without damaging the uterine horn. Ovaries were embedded and H&E and stained by the Fred Hutch Experimental Histopathology Core. Successful ovariectomies were confirmed for every mouse included in preclinical experiments by a veterinary histopathologist.
Intracardiac injection:
Fourteen-16 days after ovariectomy, anesthetized mice were positioned in dorsal recumbency on a VisualSonics Vevo 2100 Ultrasound Imaging System to guide injection of 1×105 viable ffluc-eGFP MCF-7 cells in 100 μl PBS. MCF-7 were triturated upon harvest by passing 5-times through a 25 G needle to obtain a single cell solution, and drawn into a 1 ml syringe mounted with a 22 G needle for injections. Needle height and angle was adjusted on a stereotactic rig, and the needle was guided via ultrasound imaging into the left ventricle. The cell mixture was injected slowly only upon visualization of blood refluxing into the syringe. Mice were monitored over the subsequent week via BLI. Mice with residual signal in their heart one-week post-injection were excluded from further study.
Treatment:
Five days post-injection, mice were primed with antibody, then treated weekly (days 7, 14, 21 and 28 post-injection) with antibody +/− AC. Antibodies consisted of isotype control (250 μg mouse IgG and 5 mg/kg rat IgG), rat anti-integrin β1 (AIIB2, 2.7 mg/ml, 5 mg/kg56, Fred Hutch Antibody Technology Core, 1.23 EU/mL), or AIIB2 and mouse anti-integrin αvβ3 (LM609, 3.5mg/ml, 250 μg/mouse57, UCSD/Cheresh Laboratory, 1.1 EU/mg). Antibodies were delivered intraperitoneally. Two hours following antibody treatment, AC (2 mg/kg Adriamycin/doxorubicin and 60 mg/kg cyclophosphamide) or vehicle control were delivered intraperitoneally. To quantify DTCs remaining after treatment, one cohort of mice was euthanized on Day 29. A second cohort of mice was used to measure metastasis-free survival over a prescribed course of 15 weeks.
Quantification of metastasis-free survival:
Mice were imaged weekly via BLI for a total of 15 weeks using the same methodology described above. Mice with a recurrent bioluminescent signal ≥104 p/sec/cm2/sr that increased over consecutive weeks were considered to have metastases.
Bone marrow whole-mounting and immunofluorescent staining.
Bone marrow wholemounts were generated as detailed by Nombela-Arrieta et al62. OCT-embedded femurs or tibia were shaved at 50 μm intervals on a Leica Cryostat CM3050 S (Leica Microsystems) to expose bone marrow on one face, reversed, and sectioned to expose bone marrow on the opposite face. Sliced femurs were thawed from OCT, placed in a 1.7 mL Eppendorf tube, washed twice with PBS, and blocked overnight (4 °C) in a solution containing sterile-filtered (0.2 μm) 20% goat serum+0.5% Triton X-100 in PBS under constant, gentle agitation by circumferential rotation. Following blocking/permeabilization, bones were incubated for ~72 hours in block containing a combination of chicken anti-GFP, rabbit anti-pan-laminin, Rat anti-VE-Cadherin, Rabbit anti-VWF, Goat anti-Osteopontin, Rabbit anti-Tenascin-C, Mouse anti-VCAM-1, and/or Mouse anti-Ki67-A647 (details regarding these antibodies is provided in Supplementary Table 3). Bones were washed the following day a minimum of 6 times in PBS before washing overnight at 4 °C. Femurs were then counterstained in blocking solution containing secondary antibodies (Supplementary Table 3) and 4′,6-Diamidine-2′-phenylindole dihydrochloride (DAPI; 2 μg/mL). Femurs were washed again the following day in PBS and finally placed into a solution of PBS + 0.02% sodium azide to preserve the bone prior to imaging.
For imaging, whole-mounts were generated by gently placing femurs in aqueous mounting medium (Fluoromount-G, Southern Biotech) in a 3.5 cm plate containing a No 1.5 coverslip insert (MatTek Corporation P35G-1.5–20-C), and covering the femur with a 18×18 mm coverslip in order to press the marrow flush against the imaging window. Tile scans were generated by placing this culture dish on a Zeiss LSM700 and scanning the entire bone using four laser lines and either a 0.17 NA 2.5× Fluar objective to quantify DTC number, or a 0.3 NA 10× air objective to measure distances between DTCs and specific niches over the entire imageable depth of the marrow (≤100 μm). Images of individual DTCs were acquired using a 0.55 NA 20× air objective.
Immunofluorescent staining primary tumour sections.
Twenty-five micron sections of fixed and OCT-embedded primary tumours were cut and mounted on Superfrost Plus glass slides. Sections were washed twice with PBS to rehydrate, and incubated in 0.5 M glycine/PBS for one hour at room temperature (RT). Tissues were simultaneously permeabilized and blocked in a solution containing 0.1% TritonX-100, 5% BSA and 10% Goat Serum in PBS, before staining with primary antibodies overnight (4 °C). Chicken anti-GFP, mouse anti-integrin β1, and mouse anti-integrin αv antibodies (Supplementary Table 3) were diluted in a blocking solution of 5% BSA/PBS and applied overnight. The following day, slides were washed repeatedly in PBS and counter stained as described above for femoral wholemounts, before imaging on a Zeiss LSM700.
Lung clearing via the Ce3D method and staining GFP+ DTCs.
Mice were euthanized and perfused with 25 mL ice cold PBS through the left ventricle. The left lobe of the lung was harvested and fixed in 1% PFA/PBS overnight, rocking at 4°C. Lungs were blocked in 1.5 mL of 1% normal mouse serum (MP Biomedicals), 1% bovine serum albumin (Sigma), 0.3% Triton X-100 (Sigma) in PBS for eight hours at 37 °C in a shaking incubator. Chicken anti-GFP antibody was added to blocking solution and lungs were incubated in primary antibody solution for 72 hours while shaking at 37 °C. Lungs were washed for 24 hours in 0.2% Triton X-100 and 0.5% 1-thioglycerol (Sigma) in PBS while shaking at 37 °C. Lungs were submerged in 1.5 mL block solution containing goat anti-chicken 488 (1:250) and shaken for another 48 hours at 37 °C. Wash was repeated for another 24 hours.
After staining, lungs were cleared using Ce3D55 clearing solution (40% N-methylacetamide [Sigma] v/v, 86% w/v Histodenz [Sigma], 0.1% Triton X-100and 0.5% 1-thioglycerol in PBS). Lungs were covered and incubated in 1 mL Ce3D clearing solution on a circumferential rotor for 72 hours (RT). Ce3D clearing solution was replaced and lungs were incubated for another 72 hours. Cleared lungs were stored in Ce3D clearing solution at room temperature until imaged. Imaging to quantify DTCs was performed manually in single blinded fashion on a Zeiss AXIO Zoom.V16 with the aperture set to 65%. Lungs were scanned at 37.5X, and structures of interest were confirmed to be cellular in nature at 112X. A cleared lung from an uninoculated mouse was used as a control; no DTCs or DTC-like structures were observed in this tissue.
Quantification of primary tumours and DTCs in mice.
Mammary tumour measurements.
Mammary tumours were measured in vivo and ex vivo using digital calipers to measure the long (L) and short (w) axes of the tumour. Volume was calculated using the equation V= 0.5Lw2.
DTC localization to specific niches.
ZEN Black (Zeiss) was used to load Z-stacked tile scans of femurs and tibia. Distances between GFP-positive DTCs to the nearest pan-laminin- or VE-Cadherin-positive blood vessel, the nearest megakaryocyte (based on their unique nuclear morphology) and the nearest osteoblast (the interface between bone marrow and bone) were measured by hand using this software, and tabulated for each identified DTC.
DTC concentration.
To gauge the effect of integrin inhibition on DTC burden and response to AC, a custom macro was applied to tile-scanned images in order to count DTCs in NIH Image J software. Bone marrow was traced manually, and the channel containing GFP-positive DTCs was thresholded to remove all background and create a binary image. DTCs were masked, and “Analyze Particles” was run to count all particles with a size ≥ 50 μm2, and circularity between 0.00 and 1.00. The obtained value was normalized by the total area of bone marrow imaged. For models involving tumour cell inoculation in the mammary fat pad, this value was normalized further by the volume of the primary tumour measured ex vivo upon surgical resection to account for any differences in the pool of tumour cells available to disseminate.
Two femurs from two uninoculated GH-Balb/c mice that had undergone the entire staining protocol (including primary antibodies) were used as a reference. These returned a zero value. Initial measurements were confirmed with manual counts to ensure accuracy.
Assessing vessel density in femoral bone marrow.
Vessel density within the femoral bone marrow was approximated using the macro for DTC concentration, applied instead to the pan-laminin channel. Here, however, area fraction measurements were conducted within the traced region of the thresholded image to provide a percent area occupied by vasculature within the marrow.
Flow-based analysis of immune populations in response to AC therapy +/− AIIB2.
Wild type, 6-week old female FVB mice were either left untreated, or treated over the course of five weeks with weekly cycles of IgG, AIIB2, IgG+AC or AIIB2+AC (n=3 mice per group); all at the same doses described above. Mice were euthanized one week post-treatment.
Bone marrow from 1 femur/mouse was isolated at the time of euthanasia by cutting each end of the femur and flushing marrow out with 1× HBSS and a 27.5-gauge insulin syringe. Freshly isolated bone marrow from each mouse was filtered over a 40 μm filter, treated with ACK lysis buffer for 4 min at RT, and resuspended in PBS + 2% FBS. 1×106 bone marrow cells/femur were stained with CD16/32 Fc block (BioLegend, clone 93, 1:100) and LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit (Invitrogen, 1:1000) in PBS for 30 min at RT, followed by washing in PBS + 5% BSA (FACS buffer). Subsequently, samples were stained with antibodies against CD45, CD3, CD11b, and B220 (see Supplementary Table 3 for details) all at 1:100 in FACS buffer for 30 min at 4 °C. Samples were again washed in FACS buffer followed by fixation in BD Cytofix™ (BD Biosciences) for 30 min at 4 °C, washed again in FACS buffer and resuspended in 325 μl FACS buffer. 5 μl of each sample was mixed with AccuCheck counting beads (Invitrogen) at 200,000 beads/mL in a total volume of 200 μl. All samples were analyzed on a BD FACSymphony™ flow cytometer (BD Biosciences) and analysed with FlowJo™ v10 software.
Analyses were gated off of forward by side scatter and single cells. Dead cells were quantified by positive staining for the viability stain. Live cells were further gated off of CD45+ and analyzed as B220+ (B cells) or as CD11b+CD3- (myeloid cells) and CD11b-CD3+ (T cells). Absolute cell counts were quantified by first identifying the total cell number from samples that had AccuCheck beads added to them and dividing the cell events by the number of bead events, multiplied by the concentration of beads, the bead volume/cell volume, and the total volume of the cell sample to be analyzed. The total cell number calculated was multiplied by the frequency of single cells and subsequent gated populations of interest, yielding absolute cell counts.
Cell culture and reagents.
Freshly isolated human umbilical vein endothelial cells (ECs) were provided kindly by Dr. Andrew Putnam (University of Michigan) and propagated in fully supplemented EGM-2 growth medium (Lonza). ECs were processed from umbilical cords obtained by a process considered exempt by the University of Michigan’s institutional review board (notice of determination dated August 21, 2014) because the tissue is normally discarded, and no identifying information is provided to the researchers who receive the cords. Bone marrow-derived human MSCs were obtained commercially (ScienCell) and propagated in low glucose DMEM supplemented with 10% fetal bovine serum (ThermoFisher). All primary human cells were used in experiments by passage 11. Malignant HMT-3522-T4–2 (T4–2) cells were grown in H14 medium on collagen-coated tissue culture flasks63. MCF-7 cells were grown in high glucose DMEM supplemented with 10% FBS.
mCherry-E4orf1-ECs were generated as described previously23. YFP-T4–2 and –MCF-7 were generated by infection of tumour cells with pLentiCMV/YFP lentivirus followed by selection for 96 h with 1 μg/ml puromycin. tdTomato-mVenus-p27K− T4–2 cells were generated by sequential infection with pLentiCMV/tdTomato lentivirus, selection for 96 h with 1 μg/ml puromycin, and retrovirally infected with pMXs-IRES-puro/mVenus-p27K− (ref. 48) generously provided by the Paddison Laboratory (Fred Hutchinson Cancer Research Center). As reported previously, this vector encodes for a mutant form of p27 that lacks cyclin-dependent kinase inhibitory activity39. Cells were starved to drive them into a quiescent state and then mVenus-positive cells were flow sorted to establish a stable population of tdTomato-mVenus-p27K− T4–2 cells.
Generation of bone marrow microvascular niches.
Bone marrow microvascular niches (MVNs) were generated as previously detailed23. MSCs were seeded alone at a density of 5×104 cells/well in 96-well culture plates or with mCherry-E4-ECs at a 5:1 ratio to generate bone marrow MVNs. Cells were suspended in EGM-2 at a concentration 5×104 cells/100 μl (stroma only) or 6×104 cells/100 μl (stroma+ECs). After depositing 100 μl of cellular suspension per well of a 96-well plate, plates were left undisturbed on a flat surface for 20 min to allow even cell seeding prior to incubation.
After 10 days, YFP tumour cells were suspended in unsupplemented DMEM/F12 (500 cells/ml). YFP tumour cells were seeded (100 μl/well) after washing cultures thrice with PBS. Cells were allowed to settle for 15–25 min at room temperature, then a drip of LrECM in DMEM/F12 was slowly added to each well (final concentration=20%). The LrECM drip condensed for 10–15min at room temperature before polymerizing fully at 37°C. If required, cultures were imaged immediately after seeding on a Zeiss LSM700 confocal microscope using a 0.3 NA 10× air objective. The objective was centered to each well before acquisition of 512×512 pixel, 6×6 tiles (zoom =0.7) that captured the near-entirety of each well. Cultures were maintained with media changes every 72 h and imaged again at day 10, 12 and 17 (depending on the experiment).
Treatment with doxorubicin and paclitaxel (Tocris BioScience) were conducted by generating 1000× stocks of each concentration tested and diluting said stock into a minimum of 1 mL of DMEM/F12. Cultures were treated at Day −12 and −15 post-seeding of tumour cells. Treatments were controlled by treating with the volumetric equivalent of vehicles (ddH2O for doxorubicin, DMSO for paclitaxel).
Function blocking experiments were conducted by first priming MVNs at Day 10 post-seeding of tumour cells with 100 μg/mL of antibody or the appropriate isotype control in DMEM/F12. Between 46–48 hours later, medium was removed and 2X solutions of antibody were added to the appropriate wells. A 2X solution of doxorubicin was added exactly two hours later. This was repeated at Day 15 post-seeding of tumour cells, and cultures were analyzed at Day 17. Supplementary Table 3 lists all function blocking antibodies (and isotype controls) employed— all purchased/produced in preservative-free format.
Immunofluorescent/TUNEL staining of microvascular niches.
MVNs were fixed in 4% PFA/PBS for 15 min (4 °C), washed 3-times with ice cold PBS, and left in 0.5M Glycine/PBS until staining (4 °C) in order to neutralize any unreacted PFA.
For staining, cultures were washed once with PBS prior to permeabilization in 0.5% Triton-X100/PBS for 15 min (RT), then washed three more times with PBS. If TUNEL staining was to be conducted, a two-component In Situ Cell Death Detection Kit (Roche/Sigma 12156792910) was thawed and mixed on ice according to manufacturer’s instructions. 30 μl of staining solution was added to each well prior to incubation for 1 hour (37 °C). Cultures were washed three-times in PBS prior to proceeding with our standard staining protocol.
Cultures were blocked in 5% BSA/PBS (0.2 μm filtered) for 1 hour (RT) prior to staining in primary antibody diluted in blocking solution overnight with gentle tilt-shaking (4 °C). Primary antibodies used were: chicken anti-GFP or rabbit anti-pan-cytokeratin (AbCam ab9377, 1:500) to visualize breast tumour cells, rabbit anti-VWF, and mouse anti-Ki67-A647 (see Supplemetary Table 3 for details). Cultures were washed the following morning in PBS (6 washes over one hour) prior to staining with secondary antibodies (Supplementary Table 3) diluted in blocking solution for 4–6 hours (RT). DAPI was added to this solution (2 μg/mL) to visualize nuclei. Finally, cultures were washed extensively with PBS, culminating in an overnight wash prior to storage in PBS+0.02% sodium azide. Plates were stored in hydrated glass containers (4 °C) before and after imaging.
Von Kossa and Oil Red O staining of microvascular niches.
Bone marrow microvascular niche cultures were generated as described above. Cultures were maintained in EGM-2 for 10 days then fixed with 10% neutral buffered formalin solution (Sigma) at room temperature for 40 min. Cultures were washed three times with ddH20 and stored at 4°C in ddH20 overnight.
As a positive control, human MSCs were differentiated into adipocytes or osteoblasts using established protocols64. Briefly, 1×104 MSCs were seeded into 96 well plates in low glucose DMEM containing 10% FBS. The next day, differentiation media were applied. Adipogenic media consisted of 1 μM dexamethasone (Sigma), 50 μM indomethacin (Sigma), 500 nM 3-isobutyl-1-methylxanthine (Sigma), 5 μg/mL insulin, and 2% FBS in low glucose DMEM. Osteogenic media contained 50 μM ascorbic acid (Sigma), 10 nM dexamethasone (Sigma), 10 μM β-glycerophosphate (EMD Millipore), and 2% FBS in low glucose DMEM. MSCs were cultured in differentiation media for 14- (osteogenic) or 21- days (adipogenic), fixed, and stained as follows:
Von Kossa staining.
All reagents for Von Kossa staining were acquired from American MasterTech Scientific. Formalin fixed cultures were washed five times with ddH20 then incubated in 5% sodium nitrate solution and exposed to UV light at 345 nm for 40 min (RT). Following UV exposure, cultures were washed five times in ddH20, then incubated in 5% sodium thiosulfate solution for 3 min (RT). Washes in ddH20 were repeated another five times. Cultures were then incubated in nuclear Fast Red Stain for 5 min (RT), washed five times with ddH20, and stored at 4 °C in 0.02% sodium azide/ddH20. Cultures were imaged on a Zeiss AXIO Zoom.V16 with the aperture set to 37% to acquire maximal depth in the Z plane, and magnification set to 22X to capture each well of a 96 well plate in its entirety.
Oil Red O staining.
All reagents used in Oil Red O staining were obtained from American MasterTech Scientific. Cultures were washed five times in ddH20 then incubated in 100% propylene glycol for two min (RT). Oil Red O Stain was preheated to 60°C and added to MVNs immediately following propylene glycol incubation. Cultures were incubated in warm Oil Red O Stain for 6 min at 60 °C, washed once in 80% propylene glycol/ddH20, then washed five times in ddH20. Cultures were incubated in Modified Mayer’s Hematoxylin for one min (RT), washed again in ddH20 five times, and stored at 4 °C in 0.02% sodium azide/ ddH20 until imaged. Images were acquired on a Nikon Eclipse 80i equipped with a DS-Vi1 high-speed color camera, using a 10X/0.3 NA objective.
Time-lapse acquisition.
Time-lapse sequences were acquired with a Zeiss LSM 700 confocal microscope fitted with an environmental chamber to maintain temperature (37 °C), humidity and CO2 (5%). Twelve days after seeding tdTomato-mVenus-p27K− T4–2 cells, cultures were treated with 2500 nM Doxorubicin and 1 μM of caspase-3 and −7 substrate NucView 405 (Biotium)40 in unsupplemented DMEM/F12. Images (5×5 tiles, 512×512 resolution, 8-bit) were acquired at minimal laser power every 20 min for up to 72 h. Six distinct timelapses were acquired for each condition and used to quantify the number of apoptotic events that took place after a single doxorubicin treatment, and to record whether these events involved p27+ or p27− cells. For the purposes of this analysis, cells that switched p27 status over the course of acquisition were omitted.
Quantification of tumour cell growth, proliferative status and apoptosis in microvascular niches.
Quantification of normalized tumour cell area fraction:
A macro was written using NIH ImageJ software to remove bias from data quantification. For the YFP channel only, day 10, 12 and 17 images (i.e., prior to priming, prior to chemotherapeutic treatment, and post-treatment) were subjected to the following: contrast was enhanced such that 0.5% of pixels were saturated. The image was then sharpened and a constant threshold was applied to all samples within a given experiment to eliminate variability. The total area fraction of the 6×6 tiled image occupied by YFP+ cells was then calculated. For each image, the measured area fraction at day 17 was normalized by the corresponding pre-treatment value in order to account for any variations in seeding density or outgrowth from well-to-well. Values obtained for each condition were averaged, and these averages were subsequently normalized to the vehicle-treated control where indicated.
Quantification of TUNEL staining:
After TUNEL staining, 12-bit, 2048×2048 pixel images were acquired on a Zeiss LSM700 using a 0.3 NA 10X air objective. For each pan-cytokeratin-positive tumour cell/cluster, the number of cells composing that cluster was counted manually (based on DAPI signal), and the Ki67 status of each cell in that cluster was tabulated, as was the TUNEL status. Cells were counted as TUNEL-positive if they had strong (intensity >1×103), non-punctate nuclear signal. Values reported are the percentage of TUNEL-positive cells for each cluster analyzed, averaged for each condition over the total number of cells/clusters analyzed.
Quantification of Ki67-positivity:
Tumour cells/clusters containing at least one cell with a strong nuclear Ki67 signal were counted manually. The Ki67 positive fraction was obtained by dividing this number by the total number of YFP+ clusters per well.
Whole transcriptome sequencing and analysis.
RNA was isolated from two biologically independent MSC and MSC+EC cultures that were cultured 12 days in EGM-2 medium using a Qiagen RNeasy Mini Kit according to manufacturer’s instructions. RNA concentration, purity, and integrity was assessed by NanoDrop (Thermo Fisher Scientific Inc) and Agilent Bioanalyzer. RNA-seq libraries were constructed from 1 ug total RNA using the Illumina TruSeq Stranded mRNA LT Sample Prep Kit according to the manufacturer’s protocol. Barcoded libraries were pooled and sequenced on the Illumina HiSeq 2500 generating 50 bp paired end reads. Sequencing reads were mapped to the hg19 human genome using TopHat v2.1.0 (ref.65). Gene level abundance was quantified from the filtered alignments in R using the Genomic Alignments Bioconductor package. Differential expression was assessed using transcript abundances as inputs to the edgeR Bioconductor package in R66. For edgeR analysis, genes filtered for a minimum expression level of at least 1 count per million reads (CPM) in at least two samples were used to calculate expression differences using an exact test with a negative binomial distribution, applying a significance level of 0.05 with Benjamin-Hochberg false discovery rate (FDR) adjustment. Gene expression results were ranked by their edgeR statistics and used to conduct Gene Set Enrichment Analysis (GSEA)67 to determine patterns of pathway activity in different groups. We utilized the curated pathways from within the MSigDBv5.1. Over-representation enrichment analysis was performed using the String database68 by Fisher’s exact test with correction for multiple testing. RNA sequencing data are deposited in the Gene Expression Omnibus database under the accession number GSE119153.
shRNA-mediated knockdown of ITGB1, VWF, Itgb1 and Itgav.
To target human ITGB1 and VWF, lentivirus was produced from pGIPZ constructs containing non-targeting shRNA, shRNA targeting ITGB1, or shRNA targeting VWF. HMT-3422-T4–2 cells or ECs were infected at 5 MOI and selected in 1 μg/mL puromycin. To target murine Itgb1 and Itgav, lentivirus was produced from pLKO.1 constructs, and ffluc-eGFP 4T07 cells were infected at 5 MOI. Stable knockdowns of both genes were established by sequential infection and subsequent culture in 5 μg/mL puromycin. Non-targeting vector was used as a control.
Sequences for all shRNA clones targeting human ITGB1, human VWF, murine Itgb1, and murine Itgav are provided in Supplementary Table 4.
Western blotting.
Tumour cells were lysed in 2% SDS/PBS containing cOmplete protease inhibitor cocktail (Roche) and PhosSTOP phosphatase inhibitor cocktail (Roche). Lysate (20–50 μg) was then separated on a Tris-Glycine 4–20% gel. Human and murine Integrin β1 subunit were probed with a mouse monoclonal antibody (BD Biosciences 610467, Clone 18, 1:500). The murine Integrin αv subunit was probed with a mouse monoclonal antibody (BD Biosciences 611012, Clone 21, 1:500). Human VWF was probed with a rabbit polyclonal antibody (Dako, A0082, 1:500). Blots were simultaneously probed with a rabbit polyclonal antibody to the nuclear membrane protein Lamin A/C as a loading control (Santa Cruz Biotechnology sc-20681, 1:4000), and developed on a Li-Cor Odyssey. Densitometry was performed on Odyssey software v3.0. See Supplementary Table 3 for further details on antibodies.
Analysis of bone marrow stroma and MVNs via LC-MS/MS.
Reagents.
Reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. Sodium chloride was from Acros Organics (part of Thermo Fisher). Microcentrifuge tubes and other consumables were from Axygen Inc. (Union City, CA) and RINO Screw Camp Tubes from Next Advance (Averill Park, NY). Formic acid (FA), and hydroxylamine (NH2OH) hydrochloride were from Fluka (Buchs, Switzerland). Anhydrous potassium carbonate, guanidine hydrochloride, sodium hydroxide, and acetonitrile (LC-MS grade) were from Fisher Scientific (Pittsburgh, PA). Trypsin (sequencing grade, TPCK treated) was from Promega (Madison, WI).
Sample Preparation.
Cultures were established for 12 days in EGM-2, washed extensively with PBS to remove medium, collected and pooled into Eppendorf tubes and flash frozen on dry ice prior to storage at −80 °C until analysis.
Prior to analysis, samples were thawed and homogenized in freshly prepared high-salt buffer (50 mM Tris-HCl, 3 M NaCl, 25 mM EDTA, 0.25% w/v CHAPS, pH 7.5) containing 1× protease inhibitor (Halt Protease Inhibitor, Thermo Scientific) at a concentration of 10 mg/mL. Homogenization took place in a bead beater (Bullet Blender Storm 24, Next Advance, 1 mm glass beads) for 3 min at 4 °C. Samples were then spun for 20 min @18,000g at 4 °C, and the supernatant removed and stored as Fraction 1. A fresh aliquot of high-salt buffer was added to the remaining pellet at 10 mg/mL of the starting weight, vortexed at 4 °C for 15 min, and spun for 15 min. The supernatant was removed and stored as Fraction 2. This high-salt extraction was repeated once more to generate Fraction 3, after which freshly prepared guanidine extraction buffer (6 M guanidinium chloride adjusted to pH 9.0 with NaOH) was added at 10 mg/mL and vortexed for 1 hour at room temperature. The samples were then spun for 15 min, the supernatant removed, and stored as Fraction 4 (sECM). Fractions 1, 2, & 3 (Cellular) were combined and all fractions were stored at −80 °C until further analysis.
Hydroxylamine (NH2OH) Digestion.
The remaining pellets from each tissue, representing insoluble ECM proteins, were digested with hydroxylamine as previously described69. Briefly, after chaotrope extraction pellets were treated with freshly prepared hydroxylamine buffer (1 M NH2OH-HCl, 4.5 M guanidine-HCl, 0.2 M K2CO3, pH adjusted to 9.0 with NaOH) at 10 mg/mL of the starting tissue weight. The samples were briefly vortexed, then incubated at 45 °C with vortexing for 16 hours. Following incubation, the samples were spun for 15 min at 18,000 × g, the supernatant removed, and stored as Fraction 5 (iECM) at −80 °C until further proteolytic digestion with trypsin. The final pellet was stored at −80 °C until further analysis.
Trypsin Digestion.
For each sample, 200 μL of the Cellular fraction, and 100 μL of the sECM and iECM fractions, respectively, were subsequently subjected to reduction, alkylation, and enzymatic digestion with trypsin. A filter-aided sample preparation (FASP) approach, as well as C18 cleanup, was performed as previously described70.
LC-MS/MS Analysis.
Samples were analyzed on an Q Exactive HF mass spectrometer (Thermo Fisher Scientific) coupled to an EASY-nanoLC 1000 system through a nanoelectrospray source. The analytical column (100 μm i.d. × 150 mm fused silica capillary) was packed in house with 2.7 μm 80 Å Cortex C18 resin (Phenomenex; Torrance, CA)). The flow rate was adjusted to 400 nL/min, and peptides were separated over a 120-min linear gradient of 4–28% ACN with 0.1% FA. Data acquisition was performed using the instrument supplied Xcalibur™ (version 2.1) software. The mass spectrometer was operated in positive ion mode. Full MS scans were acquired in the Orbitrap mass analyzer over the 300–2000 m/z range with 60,000 resolution. Automatic gain control (AGC) was set at 1.00E+06 and the fifteen most intense peaks from each full scan were fragmented via HCD with normalized collision energy of 28. MS2 spectra were acquired in the Orbitrap mass analyzer with 15,000 resolution with AGC set at 1.00E+05. All replicates of each tissue were run sequentially and pre-digested yeast alcohol dehydrogenase standard (nanoLCMS Solutions LLC, Rancho Cordova, CA) was run between sample groups to monitor drift in analytical performance.
Database Searching and Protein Identification.
MS/MS spectra were extracted from raw data files and converted into .mgf files using Pava (UCSF). Peptide spectral matching was performed with Mascot (Ver. 2.5) against the Uniprot mouse database (release 201701). Mass tolerances were +/− 10 ppm for parent ions, and +/− 0.2 Da for fragment ions. Trypsin specificity was used for cellular and sECM fractions, allowing for 1 missed cleavage. For iECM fraction, C-terminal N and trypsin were used, allowing for 1 missed cleavage. Met oxidation, Pro hydroxylation, protein N-terminal acetylation, and peptide N-terminal pyroglutamic acid formation were set as variable modifications with Cys carbamidomethylation set as a fixed modification. Scaffold (version 4.4.6, Proteome Software, Portland, OR, USA) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Mascot scoring algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified unique peptides. Metaboanalyst was used for multivariate analysis of proteomic data.
Code Availability.
Code used in this study (e.g., ImageJ macros for image analysis) are freely available from the corresponding author upon reasonable request.
Statistics and Reproducibility.
Statistical analyses were conducted with GraphPad Prism 7 software. Data that were distributed normally were compared via unpaired, two-tailed t test (if only two conditions were tested) or via one- or two-way ANOVA (for experiments containing three or more conditions). Post-testing to correct for multiple comparisons was chosen based on whether a decision was made a priori to compare conditions to vehicle (Dunnett’s), or to compare all treatment groups with each other (Tukey’s). Data that were not distributed normally were tested via Kruskal-Wallis and Dunn’s post-test to correct for multiple comparisons. Please refer to figure legends for individual n- and P- values, and the specific statistical test(s) employed. Unless noted otherwise, data are reported such that the centre line represents the mean, and error bars represent the s.e.m. Experiments were repeated independently with similar results.
Supplementary Material
Acknowledgements.
We thank Drs. Glenn Merlino and Chi-Ping Day for generously providing C.FVB-Tg(GNrhr-luc/EGFP)L8Mrln/LmwJ mice, Dr. Andrew Putnam for providing human umbilical vein endothelial cells, Dr. Slobodan Beronja for providing murine targeted shRNA constructs, Dr. Patrick Paddison for providing the mVenus-p27K- vector, Dr. Christopher Morse for instruction on bilateral ovariectomy, Dr. Sunil Hingorani for in-depth discussions about preclinical trial design, and Dr. Paul Lampe for helpful instruction regarding antibody purification. We are also thankful to Dr. Beronja for his critical feedback on this work. This study was catalyzed by start-up funds provided by the Fred Hutchinson Cancer Research Center (Fred Hutch) and a grant from the Cuyamaca Foundation (to CMG), and supported to its completion by an Era of Hope Award from the Department of Defense (DoD) Breast Cancer Research Program (BCRP; W841XWH-15-1-0201; CMG, KCH), a grant from the Breast Cancer Research Foundation (IIDRP-17-001; CMG), a grant from the National Breast Cancer Coalition’s Artemis Project for Metastasis Prevention, a Physical Sciences Oncology Project Grant from the NIH/NCI (U54CA193461-01; CMG), and by the Comparative Medicine, Antibody Technology, Experimental Histopathology and Genomics Shared Resources of the Fred Hutch/University of Washington Cancer Consortium (P30 CA015704). PSN’s laboratory is supported by the Prostate Cancer Foundation and NIH/NCI grants P50CA097186, U54CA224079, and R01CA165573 and DOD Awards PC170431 and PC160662. KCH’s laboratory is supported further by a grant from the NIH/NCI (R33CA183685). AD is supported by a fellowship from the Terri Brodeur Breast Cancer Foundation, CAG is supported by a postdoctoral fellowship from the Susan G. Komen Foundation, JD is supported by a Postdoctoral Breakthrough award by the DoD BCRP, ARL was supported by a Cardiovascular Pathology Training Grant from the NIH (5T32HL007213-38) and is currently supported by a fellowship from the NIH/NCI 1F31CA228424-01, and SBC is supported by a Cellular and Molecular Biology Training Grant from the NIH (T32GM007270).
Footnotes
Data Availability
RNAseq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE119153.
Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012002.
Source data for Figs. 1c, 1f–h, 2b–c, 2e, 2g, 2h–i, 2k, 2m, 3a–b, 3e, 3h, 4g, 4i, 5c, 5e–g, 5i–m, 6b, 6e, 6h, 6j, 6l, 7b, 7f, 7h, 7j; and Supplementary Fig. 1c–e, 3a–l, 4b–c, 5b, 5d–e, 5g–h, 5j, and 6a–d have been provided as Supplementary Table 1.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Competing Interests.
The authors declare no competing interests.
References.
- 1.Karrison TG, Ferguson DJ & Meier P Dormancy of mammary carcinoma after mastectomy. Journal of the National Cancer Institute 91, 80–85 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Pan H et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. The New England journal of medicine 377, 1836–1846 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun S et al. Lack of effect of adjuvant chemotherapy on the elimination of single dormant tumor cells in bone marrow of high-risk breast cancer patients. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 18, 80–86 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Ghajar CM Metastasis prevention by targeting the dormant niche. Nature reviews. Cancer 15, 238–247 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janni W et al. Persistence of disseminated tumor cells in the bone marrow of breast cancer patients predicts increased risk for relapse--a European pooled analysis. Clinical cancer research : an official journal of the American Association for Cancer Research 17, 2967–2976 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Sosa MS, Bragado P & Aguirre-Ghiso JA Mechanisms of disseminated cancer cell dormancy: an awakening field. Nature reviews. Cancer 14, 611–622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein CA Parallel progression of primary tumours and metastases. Nature reviews. Cancer 9, 302–312 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Naume B et al. Clinical outcome with correlation to disseminated tumor cell (DTC) status after DTC-guided secondary adjuvant treatment with docetaxel in early breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 32, 3848–3857 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Gradishar WJ et al. NCCN Guidelines Insights: Breast Cancer, Version 1.2017. Journal of the National Comprehensive Cancer Network : JNCCN 15, 433–451 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Jatoi I et al. Time-Varying Effects of Breast Cancer Adjuvant Systemic Therapy. Journal of the National Cancer Institute 108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thurm H et al. Rare expression of epithelial cell adhesion molecule on residual micrometastatic breast cancer cells after adjuvant chemotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research 9, 2598–2604 (2003). [PubMed] [Google Scholar]
- 12.Mansi JL et al. Bone marrow micrometastases in primary breast cancer: prognostic significance after 6 years’ follow-up. European journal of cancer 27, 1552–1555 (1991). [DOI] [PubMed] [Google Scholar]
- 13.Braun S et al. A pooled analysis of bone marrow micrometastasis in breast cancer. The New England journal of medicine 353, 793–802 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Naumov GN et al. Ineffectiveness of doxorubicin treatment on solitary dormant mammary carcinoma cells or late-developing metastases. Breast cancer research and treatment 82, 199–206 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Pantel K et al. Differential expression of proliferation-associated molecules in individual micrometastatic carcinoma cells. Journal of the National Cancer Institute 85, 1419–1424 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Boyerinas B et al. Adhesion to osteopontin in the bone marrow niche regulates lymphoblastic leukemia cell dormancy. Blood 121, 4821–4831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao Z et al. Angiocrine factors deployed by tumor vascular niche induce B cell lymphoma invasiveness and chemoresistance. Cancer cell 25, 350–365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Z et al. Molecular Checkpoint Decisions Made by Subverted Vascular Niche Transform Indolent Tumor Cells into Chemoresistant Cancer Stem Cells. Cancer cell 31, 110–126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damiano JS, Cress AE, Hazlehurst LA, Shtil AA & Dalton WS Cell adhesion mediated drug resistance (CAM-DR): role of integrins and resistance to apoptosis in human myeloma cell lines. Blood 93, 1658–1667 (1999). [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nature medicine 18, 1359–1368 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weaver VM et al. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer cell 2, 205–216 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hambardzumyan D et al. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes & development 22, 436–448 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghajar CM et al. The perivascular niche regulates breast tumour dormancy. Nature cell biology 15, 807–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acharyya S et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 150, 165–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trumpp A, Essers M & Wilson A Awakening dormant haematopoietic stem cells. Nature reviews. Immunology 10, 201–209 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Winkler IG et al. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nature medicine 18, 1651–1657 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Calvi LM et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425, 841–846 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Bruns I et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nature medicine 20, 1315–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler JM et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell stem cell 6, 251–264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi H et al. Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nature cell biology 12, 1046–1056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding L, Saunders TL, Enikolopov G & Morrison SJ Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiel MJ et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Chen JY et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 530, 223–227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butler JM, Kobayashi H & Rafii S Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nature reviews. Cancer 10, 138–146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantel K, Braun S, Passlick B & Schlimok G Minimal residual epithelial cancer: diagnostic approaches and prognostic relevance. Progress in histochemistry and cytochemistry 30, 1–60 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Schmidt-Kittler O et al. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proceedings of the National Academy of Sciences of the United States of America 100, 7737–7742 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gradishar WJ et al. Breast cancer version 3.2014. Journal of the National Comprehensive Cancer Network : JNCCN 12, 542–590 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Xia W et al. Anti-tumor activity of GW572016: a dual tyrosine kinase inhibitor blocks EGF activation of EGFR/erbB2 and downstream Erk1/2 and AKT pathways. Oncogene 21, 6255–6263 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Oki T et al. A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0–G1 transition. Scientific reports 4, 4012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosch M & Franklin-Tong VE Temporal and spatial activation of caspase-like enzymes induced by self-incompatibility in Papaver pollen. Proceedings of the National Academy of Sciences of the United States of America 104, 18327–18332 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirata E et al. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin beta1/FAK signaling. Cancer cell 27, 574–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seguin L et al. An integrin beta(3)-KRAS-RalB complex drives tumour stemness and resistance to EGFR inhibition. Nature cell biology 16, 457–468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woelfle U et al. Bi-specific immunomagnetic enrichment of micrometastatic tumour cell clusters from bone marrow of cancer patients. Journal of immunological methods 300, 136–145 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Kusumbe AP, Ramasamy SK, Starsichova A & Adams RH Sample preparation for high-resolution 3D confocal imaging of mouse skeletal tissue. Nature protocols 10, 1904–1914 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Lopes da Silva M & Cutler DF von Willebrand factor multimerization and the polarity of secretory pathways in endothelial cells. Blood 128, 277–285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura-Ishizu A et al. Extracellular matrix protein tenascin-C is required in the bone marrow microenvironment primed for hematopoietic regeneration. Blood 119, 5429–5437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weekes CD, Kuszynski CA & Sharp JG VLA-4 mediated adhesion to bone marrow stromal cells confers chemoresistance to adherent lymphoma cells. Leukemia & lymphoma 40, 631–645 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Dittel BN, McCarthy JB, Wayner EA & LeBien TW Regulation of human B-cell precursor adhesion to bone marrow stromal cells by cytokines that exert opposing effects on the expression of vascular cell adhesion molecule-1 (VCAM-1). Blood 81, 2272–2282 (1993). [PubMed] [Google Scholar]
- 49.Day CP et al. “Glowing head” mice: a genetic tool enabling reliable preclinical image-based evaluation of cancers in immunocompetent allografts. PloS one 9, e109956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holliday DL & Speirs V Choosing the right cell line for breast cancer research. Breast cancer research : BCR 13, 215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrell JC et al. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast cancer research and treatment 132, 523–535 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Copson E et al. Prospective observational study of breast cancer treatment outcomes for UK women aged 18–40 years at diagnosis: the POSH study. Journal of the National Cancer Institute 105, 978–988 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Hall DE et al. The alpha 1/beta 1 and alpha 6/beta 1 integrin heterodimers mediate cell attachment to distinct sites on laminin. The Journal of cell biology 110, 2175–2184 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheresh DA Human endothelial cells synthesize and express an Arg-Gly-Asp-directed adhesion receptor involved in attachment to fibrinogen and von Willebrand factor. Proceedings of the National Academy of Sciences of the United States of America 84, 6471–6475 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W, Germain RN & Gerner MY Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proceedings of the National Academy of Sciences of the United States of America 114, E7321–E7330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park CC et al. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer research 66, 1526–1535 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brooks PC et al. Antiintegrin alpha v beta 3 blocks human breast cancer growth and angiogenesis in human skin. The Journal of clinical investigation 96, 1815–1822 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hawkins ED et al. Regulation of asymmetric cell division and polarity by Scribble is not required for humoral immunity. Nature communications 4, 1801 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Gilbert LA & Hemann MT DNA damage-mediated induction of a chemoresistant niche. Cell 143, 355–366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gomez-Sarosi L, Sun Y, Coleman I, Bianchi-Frias D & Nelson PS DNA Damage Induces a Secretory Program in the Quiescent TME that Fosters Adverse Cancer Phenotypes. Molecular cancer research : MCR 15, 842–851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller BE, Miller FR, Wilburn DJ & Heppner GH Analysis of tumour cell composition in tumours composed of paired mixtures of mammary tumour cell lines. British journal of cancer 56, 561–569 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nombela-Arrieta C et al. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nature cell biology 15, 533–543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Briand P, Nielsen KV, Madsen MW & Petersen OW Trisomy 7p and malignant transformation of human breast epithelial cells following epidermal growth factor withdrawal. Cancer research 56, 2039–2044 (1996). [PubMed] [Google Scholar]
- 64.Tang Y et al. MT1-MMP-dependent control of skeletal stem cell commitment via a beta1-integrin/YAP/TAZ signaling axis. Developmental cell 25, 402–416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim D et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szklarczyk D et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic acids research 45, D362–D368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrett AS et al. Hydroxylamine Chemical Digestion for Insoluble Extracellular Matrix Characterization. Journal of Proteome Research (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goddard ET et al. Quantitative extracellular matrix proteomics to study mammary and liver tissue microenvironments. The International Journal of Biochemistry & Cell Biology 81, 223–232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.