Abstract
Background
V-ATPases are hetero-oligomeric enzymes consisting of 13 subunits and playing key roles in ion homeostasis and signaling. Differential expression of these proton pumps has been implicated in carcinogenesis and metastasis. To elucidate putative molecular signatures underlying these phenomena, we evaluated the expression of V-ATPase genes in esophageal squamous cell carcinoma (ESCC) and extended the analysis to other cancers.
Methods
Expression of all V-ATPase genes were analyzed in ESCC by a microarray data and in different types of tumors available from public databases. Expression of C isoforms was validated by qRT-PCR in paired ESCC samples.
Findings
A differential expression pattern of V-ATPase genes was found in different tumors, with combinations in up- and down-regulation leading to an imbalance in the expression ratios of their isoforms. Particularly, a high C1 and low C2 expression pattern accurately discriminated ESCC from normal tissues. Structural modeling of C2a isoform uncovered motifs for oncogenic kinases in an additional peptide stretch, and an actin-biding domain downstream to this sequence.
Interpretation
Altogether these data revealed that the expression ratios of subunits/isoforms could form a conformational code that controls the H+ pump regulation and interactions related to tumorigenesis. This study establishes a paradigm change by uncovering multi-cancer molecular signatures present in the V-ATPase structure, from which future studies must address the complexity of the onco-related V-ATPase assemblies as a whole, rather than targeting changes in specific subunit isoforms.
Funding
This work was supported by grants from CNPq and FAPERJ-Brazil.
Keywords: ATP6V1C1, ATP6V1C2, Isoform a, Cancer biomarker, Esophageal cancer, V-ATPase, H+ signatures
Research in context.
Evidence before this study
V-ATPases play essential roles in bioenergetics, ion homeostasis and signaling, influencing a variety of cellular processes and have also been associated to distinct hallmarks of cancer. This proton pump is a multimeric complex composed of 13 different subunits (and two more accessory subunits), some with distinct isoforms encoded by different genes, and previous studies have usually focused only on differential expression of single isoforms, generating controversial data on different subunits/isoforms pointed as putative molecular markers in different cancers or tumor types. Recently, a molecular signature of V-ATPase has been identified, in which the downregulation of three subunits isoforms was necessary to predict glioma aggressiveness.
Added value of this study
We found the existence of a multicancer specific assembly for the V-ATPase and described that rather the upregulation of only a specific subunit isoform, the carcinogenesis is characterized by an imbalance of the expression ratio of several subunits and their isoforms. Further exploring the imbalance of C isoforms, we determined that the C1high/C2low-expression patterns can accurately segregate esophageal tumors and normal tissues, and revealed structural differences among the C isoforms, suggesting distinct posttranslational regulations that could affect the pump coupling and regulatory protein-binding sites accessibility.
Implications of all the available evidence
These findings represent a paradigm change by which future studies must address complex cancer V-ATPase molecular signatures rather than focusing only in particular isoforms that usually has been proposed as putative oncogenic markers. The conformational code depicted here possibly reflects regulatory specificities for the H+-pump coupling and membrane targeting, therefore, opening a new avenue to explore the major role of these enzymes in different signaling pathways and carcinogenic process. In such a new framework, integrative analyzes of the holoenzyme encompassing all their multiples subunits and considering the interconnectivity among the isoforms will revolutionize the field of translational/clinic studies by defining complex V-ATPase expression patterns more effective as diagnosis and prognosis biomarkers and/or useful for development of new target therapies.
Alt-text: Unlabelled box
1. Introduction
Esophageal cancer (EC), the sixth most common cause of death from cancer worldwide [1,2], encompass two histological subtypes, esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC), the last one responsible for about 90% of all diagnosed cases [3]. Very poor overall survival of ESCC patients is mainly due to its late stage detection and poor response to treatments [4]. Therefore, it is essential to understand the molecular mechanisms involved in ESCC towards identifying new therapeutic targets and molecular biomarkers that can improve the detection and clinical managements.
The V-type H+-ATPase (V-ATPase) has attracted increasing attention in the field of molecular oncology [5] due to its differential overexpression in tumor cell membranes [6,7] and the consequent intra- and extracellular pH dysregulation [8], both associated with key tumorigenic and metastatic processes, such as, migration and invasion [6,7,9], apoptosis [10,11], regulation of signaling pathways frequently altered in cancer [5,12], immunomodulation [13,14] and therapy resistance [15,16]. Furthermore, proton pumps inhibitors that directly or indirectly modulate the activity of this enzyme have proved to be potential therapeutic drugs to control cancer progression (e.g., Martins et al., 2019; and refences therein) [10].
V-ATPases are ATP-dependent proton pumps that translocate H+ ions across endomembranes of all eukaryotic cells and also through plasma membranes of some specialized cells, acidifying intracellular compartments and the extracellular matrix, respectively [12,17]. Recently, V-ATPase was found to generate proton gradients in outer as well as inner nuclear membranes in prostate cells [18]. V-ATPase is complex enzyme composed of 13 subunits organized into a cytoplasmic domain (V1) responsible for ATP hydrolysis and a proton translocation transmembrane domain (V0). The V1 domain consists of eight (A-H) subunits, while V0 sector, in human cells, is formed by the subunits c, c", a, d and e, as well as two accessory subunits, Ac45 and M8-9 [12,19]. The V-ATPase subunits B, C, E, G, a, d, and e have multiple isoforms encoded by different genes, which have initially been postulated to exhibit tissue specific expression patterns [20], [21], [22]. However, until recently, no clear functional roles have been attributed to the distinct isoforms, except for the four a isoforms described as responsible for the enzyme targeting to different cellular membranes [23]. The isoforms a1 and a2 are found in the H+ pumps of intracellular membranes, while pumps assembled with a3 and a4 are usually directed to plasma membrane [7,22]. In addition, it has also been demonstrated that yeast V-ATPase complexes containing two different a isoforms show distinct cellular localization, coupling efficiency and the main regulation of the pump by association/dissociation of the V1 and V0 domains [24].
Since V-ATPase plays essential roles in a variety of cellular processes, the existence of multiple subunit isoforms might reflect multifunctional conformations, including specific compositions for specific cell types and pathophysiological conditions. However, in different tumor cells lines, functional assays related to tumor growth, migration, invasiveness and metastasis provided similar results regardless of the individual isoform for which gene expression changes were found [7,9,[25], [26], [27], [28]]. Therefore, the classic functional assessments seem to be ineffective in selectively targeting particular V-ATPase subunits/isoforms assemble into different oligomeric states involved in diseases processes [22], including the different tumorigenic molecular phenotypes.
Here, we investigated the differential expression patterns for all V-ATPase subunits/isoforms in human ESCC and performed a comparative analysis with other types of tumors, revealing the existence of complexes containing unique combinations of V-ATPase isoforms, that could explain the oncogenic activations of this proton pump. In perspective, this conformational code of V-ATPase isoforms has potential as cancer biomarker for early diagnosis and more effective prognosis and could help in development of drugs highly specific for tumorigenic/metastatic combinatorial assemblies.
2. Materials and methods
2.1. V-ATPase expression in ESCC transcriptome
The expression profile of the V-ATPase genes in ESCC was analyzed using transcriptome data developed by the Brazilian National Institute of Cancer – INCA [29]. Transcriptome data are available in Gene Expression Omnibus (GEO) under accession number GSE75241. Microarrays were performed using a genechip Human Exon 1.0 ST array (Affymetrix, Inc.) and compared 15 ESCC paired samples (ESCC and tumors and respective surrounding tissues, collected at least 5 cm far from the tumor border) and 5 healthy esophageal tissues of individuals without cancer (collected from patients submitted to endoscopic routine examination not related to cancer or esophageal disorders, performed at the University Hospital Pedro Ernesto-UERJ). The raw data were normalized in Expression Console software (Affymetrix) using robust multi-array average (RMA) method. The differential expression was analyzed using the Affymetrix Transcriptome Analysis Console (TAC), following the software guidelines. Multiple Testing Corrections and False Discovery Rate Prediction were performed by Benjamini-Hochberg procedure.
2.2. V-ATPase expression in Oncomine database
The expression levels of V-ATPase genes in 20 cancer types were obtained from the Oncomine database (https://www.oncomine.org/resource/login.html) [30]. The fold-change of mRNA expression in cancer tissues compared to in their normal tissues was acquired using parameters of a threshold p-value of 1E-4; fold-change of 2; and gene ranking in the top 10%.
2.3. V-ATPase gene expression in the cancer genome atlas (TCGA) database
Gene expression data from EC samples (n = 182), both esophageal adenocarcinoma (n = 87) and squamous cell carcinoma (n = 95), were downloaded from the public database cBioPortal for Cancer Genomics [31,32], which provides visualization, analysis, and download of largescale data sets deposited in The Cancer Genome Atlas (TCGA).
2.4. Patients and samples
Thirty-eight patients enrolled from 2008 to 2015 at the Brazilian National Cancer Institute (INCA) who had confirmed ESCC diagnosis and had not undergone chemotherapy and/or radiotherapy were recruited. Paired biopsies (tumors and normal-appearing surrounding mucosa, collected at least 5 cm far from the tumor border) were included in real-time quantitative PCR analysis. Epidemiological and clinicopathological data were obtained through interviews by using a standardized questionnaire and from patient's medical records, including whenever possible sex, age at diagnosis, tobacco smoking, alcohol intake, tumor stage and differentiation, esophageal localization and overall survival time after diagnosis. Individuals were classified regarding tobacco smoking as smokers (for ever- or ex-smoking) or never-smoking, and regarding alcohol intake as drinkers (for ever- or ex-drinkers) or never-drinkers. The use of the human samples was approved by the Ethic Committee of the institution (INCA - 116/11). All patients, who kindly agreed to participate in the study, signed an informed consent form prior to study enrollment
2.5. Clinicopathological features
The clinicopathological characteristics of the 38 ESCC patients evaluated in the study are presented in Table S1. The median age of patients was 57 years, ranging from 48 to 79 years, male patients represented 84.2% of cases and near 90% of all patients were alcohol consumers and/or smokers. Tumors were most often located in the middle third of the esophagus (47.4%), with a high prevalence of advanced stage (III or IV) of the disease (48%) and poorly or moderately differentiated (97.4%) culminating in a high mortality rate (81.6%) with median overall survival of 7.73 months.
2.6. Real-time PCR
Total RNA was extracted from the fresh tumor and normal surrounding esophageal tissues using RNeasy mini kit (Qiagen), according to the manufacturer's protocol. RNA concentration and purity were determined by spectrophotometrically, and 500 ng of total RNA was used for reverse transcription in a final volume of 20 µL with SuperScript II Reverse Transcriptase (Thermo Fisher Scientific), following the manufacturer's protocol. Expression levels were detected by StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific) and each reaction (12 µL) contained the corresponding cDNA (1:10), 900 nM of each primer and 6 µL of Power SYBR Master Mix PCR (Thermo Fisher Scientific). The reaction conditions were as follows: 95 °C for 10 min; 95 °C for 15 s, 60 °C for 1 min, for 40 cycles; at the end a melting curve analysis was included, and fluorescence was measured from 60 to 99 °C (each sample was analyzed in duplicate). Relative mRNA levels were quantified using the comparative cycle threshold method (ΔΔCt) and normalized by the GAPDH expression and using the normal surrounding tissue as the reference (2−ΔΔCt formula). The expression ratio between ATP6V1C1 and ATP6V1C2 isoform C transcripts was calculated for each patient.
The expression levels of ATP6V1C1, ATP6V1C2 variant 1 (v1) and ATP6V1C2 variants 1 and 2 (v1,2) were analyzed using the oligonucleotides shown in Table S2. The cDNA samples availability determined the respective number of assays, 38 paired samples for the ATP6V1C1, 37 for ATP6V1C2-v1 and 36 for ATP6V1C2-v1,2.
2.7. In silico 3D models of C subunit isoforms, specific peptide domains, and protein-protein interaction sites and docking assays
Conserved polypeptide sequences were estimated using the NCBi database and the program BLASTP 2.8.0, while the analysis of conserved domains was verified by using the CDD/SPARCLE. The identification of sites for phosphorylation/dephosphorylation and protein-protein interactions was performed using the Human Protein Reference Database. The prediction of 3D models for hC1 (NP_001686.1), hC2b (NP_653184.2) and hC2a (NP_001034451.1) isoforms was performed by homology and ab initio modeling by four distinct programs: Modeller 9.19, Swiss-Model, RaptorX and I-Tasser. Analysis of global and local stereochemical quality for all predicted models was also performed by using different programs: Rampage, Verify3D, ProSA, VoroMQA, ProQ3D, Qprob, DeepQA and SVMQA. The best models were refined using the ModRefiner program. Protein-protein docking assays were carried out using the programs ClusProV2 and HADDOCK. In order to select the best interactions and to discard false positives, extensive analysis was carried out with the programs DockScore, PPCheck and CCharPPI. After selection of the protein complexes with native interaction characteristics, the prediction of binding free energy (∆G - kcal/mol) and the dissociation constant (Kd) of the complexes were calculated using the program PRODIGY. Native complexes and their interactions were modeled by the program UCSF Chimera 1.11.2 (Sao Francisco, CA, USA). Multiple sequence alignments were generated using ClustalW (http://www.genome.jp/tools-bin/clustalw). The references of each computational tools used in this section can be accessed in the Supplementary materials.
2.8. Statistical analysis
The data were statistically analyzed using GraphPad Prism 5 Software (San Diego, CA, USA) and differences were considered significant at p value of < 0.05. A Kolmogorov- Smirnov test was used for normality analysis. Paired t-test or Wilcoxon matched pair test was applied when comparing two groups. When comparing three or more groups, one-way ANOVA or Kruskal-Wallis test and Tukey's or Dunn's posttest were used. A receiver operating characteristic (ROC) curve was plotted to investigate whether C isoforms expression could be used as a marker to discriminate ESCC from normal-appearing surrounding mucosa.
3. Results
3.1. Expression profile of V-ATPase subunits in ESCC
The expression profiles of 25 genes encoding V-ATPase subunits were obtained from an ESCC transcriptome developed by the Brazilian National Institute of Cancer – INCA [29]. An unsupervised hierarchical clustering analysis showed that ESCC exhibits striking changes in expression of most isoforms of the V-ATPase subunits in comparison with normal esophageal tissues excised surrounding the tumors, in a profile which clearly segregates these two groups and suggests a pattern of tumor-specific V-ATPase expression (Fig. 1a; the fold change and p-value of each gene is shown in Table S3). The genes were considered differentially expressed when their p-value was less than 0.005. It is of note that the A and B subunits, which constitute the evolutionary conserved catalytic domain of the V-ATPase, presented no significant expression changes (ATP6V1A: 1-fold change and p = 0.78; ATP6V1B1: 1-fold change and p = 0.79; ATP6V1B2: −1.08-fold change and p = 0.45) (Table S3, Figure S1).
Fig. 1.
V-ATPase expression profile in ESCC by microarray database. (a) Unsupervised hierarchical clustering analysis of 15 esophageal samples (tumors and respective surrounding tissues) showing the expression of the 25 genes of V-ATPase. Green indicates decreased gene expression and red increased genes expression. N and gray bar correspond to normal surrounding tissues; T and black bar correspond to tumor. (b) ATP6V1C1 and ATP6V1C2 expression levels in tumor, normal-appearing surrounding tissues and healthy esophageal mucosa from individuals without cancer. *p = 0.0146; ***p = 0.0003; ****p < 0.0001 [ATP6V1C1 - one-way ANOVA with Tukey's post-test. ATP6V1C2 - Kruskal-Wallis test with Dunn's post-test].
The subunit c” (ATP6V0B), which compose the proteolipid ring of the V-ATPase, exhibited higher expression in ESCC samples than in normal tissues. Even more interesting were the changes found among the isoforms expression of the same subunit, as observed for d and G subunits, where d2 (ATP6V0D2) and G2 (ATP6V1G2) isoforms were more expressed in tumor, while d1, G1 and G3 isoforms (ATP6V0D1, ATP6V1G1 and ATP6V1G3, respectively) were less expressed in tumor compared with normal surrounding tissues (Figure S2).
However, among all differentially expressed V-ATPase genes, those encoding the subunits C and a presented the highest differences among their respective isoforms. In ESCC compared with normal surrounding tissue, ATP6V1C1 and TCIRG1 mRNAs were upregulated (2.79 and 1.53-fold change, respectively), while those of ATP6V1C2 and ATP6V0A4 were downregulated (−1.28 and −2.76-fold change, respectively). In addition, significant difference in expression for these four genes were also observed between ESCC and healthy esophageal tissues from individuals without cancer (Fig. 1b and c; Figure S3). The TCIRG1 and ATP6V0A4 genes encode two of the four distinct a isoforms, a3 and a4 (V0 sector), whereas in the V1 domain, ATP6V1C1 and ATP6V1C2 encode the different C subunit isoforms; ATP6V1C1 encodes the C1 isoform (NCBI gene ID: 528) and ATP6V1C2 encodes two alternative transcriptional splice variants, namely C2a and C2b isoforms (NCBI gene ID: 245973).
3.2. ATP6V1C1 and ATP6V1C2 transcripts are differentially expressed in ESCC
Since the C subunit plays crucial regulatory role for V-ATPase structural and functional coupling [22,33,34], as well as taking into account that ESCC exhibited more remarkable expression ratio of the C isoforms comparing to other subunits isoforms, we validated the results of the microarray analysis evaluating ATP6V1C1 and ATP6V1C2 mRNA levels by RT-PCR in 38 ESCC paired samples (tumors and respective normal surrounding tissues). The ATP6V1C1 expression in ESCC relative to respective normal tissue ranged from 0.43 to 7.37-fold change, with the median value of 2.33-fold change. Assuming a fold change cut-off of ≥ 2, about half (54%) of the ESCC samples presented ATP6V1C1 mRNA levels increased when compared to their respective normal surrounding tissues (Fig. 2a), while the expression of the ATP6V1C2 gene was downregulated in most of the tumors analyzed, considering a cut-off ≤ −2 (67.6% for v1; 72.2% for v1,2). For ATP6V1C2 fold change values ranged from −0.46 to −48.9 with the median value of −3.96 for v1 and of −0.51 to −52.9 with the median value of −6.53 for v1,2 (Fig. 2b and c).
Fig. 2.
The expression pattern of V-ATPase C isoforms in ESCC. Left images: RT-PCR analysis of (a) ATP6V1C1, (b) ATP6V1C2-v1 and (c) ATP6V1C2-v1,2 expression in paired ESCC samples organized by tumor stage. Values are expressed as relative to those obtained in tumors respective normal surrounding tissue (= 1 or −1), represented by the red dashed line. Right images: qRT-PCR evaluation of (d) ATP6V1C1, (e) ATP6V1C2-v1 and (f) ATP6V1C2-v1, 2 mRNA levels distribution in the groups of normal surrounding and their paired ESCC tissues. Values are shown in relative units. mRNA levels were normalized by those of GAPDH, used as the housekeeping gene. NA – no information available about tumor stages. ****p < 0.0001 [Wilcoxon matched pair test].
Next, a comparative analysis of mRNA levels of subunit C isoforms in tumors and normal surrounding tissues was performed. Expression levels of ATP6V1C1 in ESCC samples group was approximately 2.5-fold higher than those detected in the normal surrounding tissues group (median values of 0.008196 and 0.003301, respectively) (Fig. 2d). For ATP6V1C2-v1 and ATP6V1C2-v1,2 the mRNA levels were significantly lower in the ESCC group than those in the normal surrounding tissues group, with the median values of −4.3 and −9.6-fold, respectively (v1: median of 0.000489 in ESCC and 0.00241074 normal surrounding; v1,2: median of in 0.000629 ESCC and 0.006044 normal surrounding) (Fig. 2e and f). These findings are in agreement with the data obtained from microarray analysis (Fig. 1), confirming that the V-ATPase C isoforms present a remarkable differential expression in ESCC, when compared to normal surrounding tissues. The protein expression of C1 and C2 isoforms was investigated by immunohistochemistry. For both isoforms the protein expression was diffuse in the tumor area, with C1 exhibiting intense staining, the indicative of over-expression. In the normal surrounding tissues, isoforms C1 and C2 were detected predominantly in the lower layers of the epithelium (Figure S4).
We further examined the expression ratio of ATP6V1C1 and ATP6V1C2 isoforms (C1/C2-v1 and C1/C2-v1,2 ratios). The median value of C1/C2-v1 ratio was of 1.4-fold in normal surrounding tissues and 13.4-fold in the ESCC samples, while the median value of C1/C2-v1,2 ratio was of 0.5-fold and 10.1-fold in the normal surrounding and ESCC tissues, respectively (Figure S5). These results suggest that the expression levels of the different C isoforms are carefully guarded to achieve a proportional balance in normal tissues, which dysregulation towards the C1 prevalence over the C2 is an intrinsic characteristic of ESCC.
3.3. Subunit C isoforms as potential diagnostic biomarker for ESCC
In order to evaluate whether C isoforms mRNA expression could be used to discriminate between tumor and non-tumor esophageal tissues, we performed the Receiver Operating Characteristic (ROC) curve analysis using the gene expression values from ESCC and normal surrounding tissues. The differential expression of ATP6V1C1 and ATP6V1C2 was able to accurately discriminate ESCC samples from normal surrounding tissues (p < 0.0001), with sensitivity and specificity of 79.49% and 84.62% for ATP6V1C1; 83.78% and 83.78% for ATP6V1C2-v1 and 80.56% and 86.11% for ATP6V1C2-v1,2, respectively. The ROC curve analysis for the C1/C2 expression ratios generated an area under curve greater than 0.94, reflecting the high accuracy of the test to discriminate between tumors and normal surrounding tissues. The C1/C2-v1 and C1/C2-v1,2 ratios showed respectively a sensitivity of 86.49% and 88.89% and a specificity of 91.89% and 91.67% (Fig. 3). These results indicate that mRNA expression ratio of the subunit C isoforms can be useful as a diagnostic biomarker for ESCC.
Fig. 3.
ROC curves of C isoforms mRNA expression for the discrimination of ESCC from normal surrounding mucosa samples. ROC curves are relative to mRNA expression of: (a) ATP6V1C1, (b) ATP6V1C2-v1, (c) ATP6V1C2-v1,2, (d) C1/C2-v1 ratio and (e) C1/C2-v1,2 ratio. The area under curve (AUC) indicates the accuracy of the test in discriminating tumors from normal surrounding tissues. Numbers in a box correspond to a cut-off used for the determination of the sensitivity and specificity. ROC, receiver operating characteristic. p < 0.0001 [ROC curve].
3.4. Structural modeling of the subunit Cisoforms
In addition, we performed the analysis of conserved domains among the human V-ATPase subunits C1 (NP_001686.1) (hC1), C2a (NP_001034451.1) (hC2a) and C2b (NP_653184.2) (hC2b) using the program BLASTP 2.8.0 in the mammalian taxon of the database NCBI Protein Reference Sequences. The three amino acid (aa) sequences were found to be highly conserved, and hC1 (382 aa), hC2a (427 aa) and hC2b (381 aa) share at least ~55.3% identity and ~74.2% similarity (Table S4). When compared to isoform hC2b, the isoform hC2a has an additional exon that encodes for 46 aa at the position 276–321 aa. The BlastP analysis suggested that the isoform hC2a is present in all mammals, but absent in other taxa (yeast, nematode, mollusk, arthropod, birds, reptiles, amphibia and higher plants). Next, were performed analysis for protein-protein binding and for phosphorylation sites, exclusively on hC2a additional exon. These analyses pointed to the presence of interesting motifs for serine/threonine kinases and phosphatases, such as ERK1 and 2, Casein Kinase II, PKA, PKC; and three potential binding motifs for proteins which recognize phosphorylated serine‑based motifs with binding domains to WW, MDC1 BRCT and Plk1 PBD (Tables S5 and S6). No tyrosine kinase/phosphatase binding sites were identified in this specific region. Predictions of hC1, hC2a and hC2b three-dimensional structures were performed by homology modeling and ab initio approaches (Fig. 4). The RMSD assessment also indicated that hC1 and hC2b are structurally very similar, while hC2a has an additional structural motif (Table S4). Analysis of global and local stereochemical quality using validation programs revealed that the most stable structure predicted for the additional exon of hC2a would be an alpha-helix conformation flanked by unstructured regions (Fig. 4c). However, since the subunit C binds to, and serves as a substrate for PKA and PKC [35], these proteins were also used to refine the validation of the hC2a structural model. Following a series of protein-protein dockings between proposed hC2a models with PKA and PKC structures (Figure S6), the conformational model that best established PKA/PKC-hC2a interaction complexes was the hC2a predicted model for which the domain corresponding to the additional exon is in a beta-sheet flanked by unstructured regions (Fig. 4d, Figure S6). In addition, based on the actin-binding domain described for the B subunit [36], we found a similar actin-binding domain in each of the C subunit isoforms (Fig. 4e and f).
Fig. 4.
Prediction of three-dimensional structure of C subunit isoforms and putative actin-binding sites. (a) hC1 isoform, (b) hC2b isoform, (c) most promising model of hC2a isoform and (d) model of hC2a isoform validated by protein docking. C1 and C2b were modeled by homology using as template the resolved structure from yeast V-ATPase (3J9V.pdb) and C2a was obtained by ab initio approach. e) Multiple sequence alignment comparing “profilin-like” domain in B subunit (blue box) and similar sequence in C subunit isoforms. (f) 3D structure of hC2a isoform highlighting in green the putative "profilin-like" domain for actin-binding. The additional peptide stretch of 46 aa present in the hC2a isoform is shown in red.
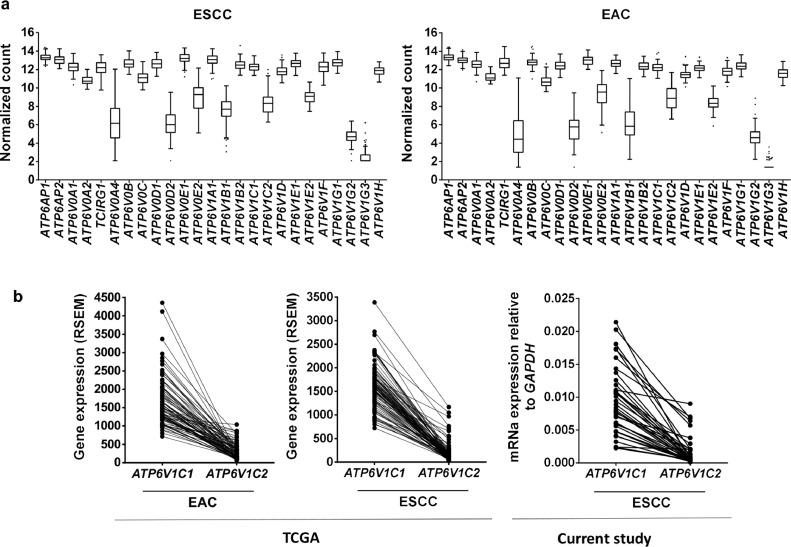
3.5. Expression profile of V-ATPase genes in EC histological subtypes
The expression profile of two genes encoding the V-ATPase subunit C in ESCC prompted us to investigate if such dysregulation is involved in esophageal carcinogenesis in general, independently of histological subtype. To this end, we used TCGA database to analyze the expression levels of V-ATPase genes in ESCC and EAC. The expression profile of all V-ATPase genes presented a very similar pattern in both histological subtypes (Fig. 5a). Regarding to C isoforms, both EAC and ESCC tumors exhibited higher expression levels of ATP6V1C1 than ATP6V1C2. The ATP6V1C1 expression was increased 6.6-fold when compared to ATP6V1C2 expression in EAC tissues. In ESCC samples, ATP6V1C1 was 11.6-fold more expressed than ATP6V1C2, corroborating with the data obtained by qRT-PCR, where ATP6V1C1 was 12.7-fold more expressed in tumors than ATP6V1C2 (Fig. 5b). Finally, the ATP6V1C1 expression levels were similar between the two-histological subtypes (expression median of 1578 for ESCC and 1479 for EAC), however, the average expression of ATP6V1C2 was lower in ESCC than in EAC (expression median of 136 for ESCC and 225 for EAC) (Figure S7).
Fig. 5.
V-ATPase mRNA expression in esophageal cancer histological subtypes. (a) Expression profile of all V-ATPase genes in esophageal adenocarcinoma (EAC) and ESCC. (b) Respective ATP6V1C1 and ATP6V1C2 mRNA levels in EAC and ESCC assessed by RNA-sequencing data from TCGA and by qRT-PCR in the current study.
3.6. Multi-cancer V-ATPase structural signatures
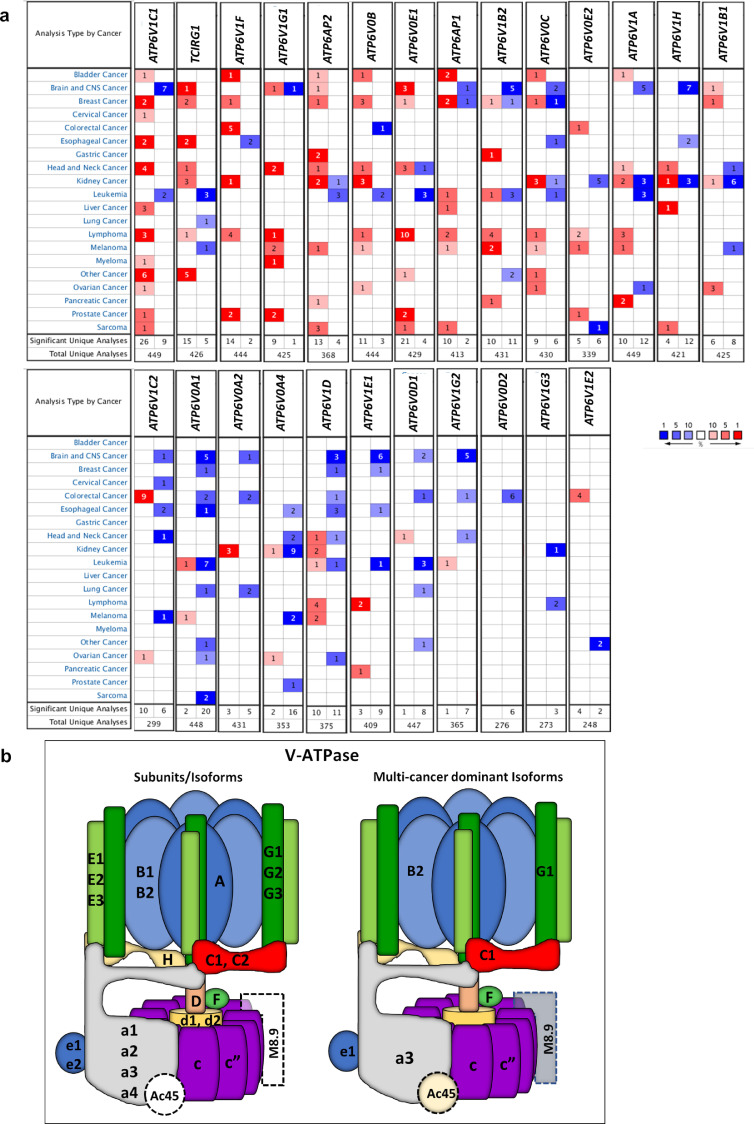
To investigate the expression pattern of V-ATPase genes in other cancer types, we systematically analyzed mRNA transcriptome between normal tissues and tumor tissues of 20 sites comprising different histological types using the Oncomine database. The threshold was designated according to the following values: p-value 1E-4, fold-change 2, and top gene ranks 10%. Although the expression of some subunits such as H, c and B1 (ATP6V1H, ATP6V0C and ATP6V1B1) seems to vary with the tumor type, it was possible to identify a common expression pattern for several key subunits/isoforms in different tumor tissues. Remarkably, upregulation of subunits F and c” (ATP6V1F and ATP6V0B) and of the accessory subunits (ATP6AP1 and ATP6AP2) as well as the imbalance of the subunit isoforms C, G and a, particularly the upregulation of C1, G1 and a3 (ATP6V1C1, ATP6V1G1 and TCIRG1), and downregulation of C2, G2, a1 and a4 (ATP6V1C2, ATP6V1G2, ATP6V0A1 and ATP6V0A4) were identified in most cancer types compared to normal tissues (Fig. 6). These results may imply a tumorigenic coordinated expression of the multimeric V-ATPase, resulting in a quite specific multicancer balance for most of the subunits isoforms.
Fig. 6.
mRNA expression pattern of V-ATPase genes in multiple cancer types. (a) Comparative table indicating the number of the analyses that meet the threshold with mRNA over-expression (left column, red) and under-expression (right column, blue) in cancer versus corresponding normal tissues. Each column refers to a V-ATPase gene and each line refers to a cancer site (some tumor sites include different histological types). The threshold was designed with the following parameters: p-value of 1E-4, fold-change of 2, and gene ranking of 10%. (b) Proposed combinatorial assemblies of V-ATPase subunits/isoforms with dominantly expressed isoforms (on the right) in multiple tumor types based on Oncomine analysis. Accessory subunits are shown in dashed lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Despite the large amount of data accumulated on differential expression of V-ATPase subunits/isoforms in various tumors, up to now no clear molecular signature could be related to the carcinogenesis process. Pharmacological inhibitions [6,10,37,38] as well as the silencing of specific isoforms (a2, a3, c, E1, G1 or C1) resulted in inhibition of migration and invasiveness in different cancer cells and reduction of tumor growth and metastasis [9,[25], [26], [27], [28],[39], [40], [41]]. If by one hand, these studies have confirmed the remarkable role of the V-ATPase in molecular oncology, on the other hand, such a functional assessment has been insufficient to propose any mechanistic insight on the subunits/isoforms differentially expressed in different tumors. Here, using a broader analysis encompassing all V-ATPase subunits/isoforms, it was possible to demonstrate a complementary interdependence between the cancer-related expression changes, mainly in isoforms of subunits that regulate the targeting and coupling of the pump. These combinatorial assemblies of isoforms might reflect conformational codes related to the control of swirling motions of the pump and the coupled electrochemical gradients, implying in the different functions of this housekeeping enzyme that energizes most of the cell membranes and thus must be precisely regulated in the different compartments in any kind of functional cell. In this way, our work can also help to clarify the functional and evolutionary rational for the existence of such isoforms, opening a new structural perspective on the variety of differential conformations of this proton pump in normal as well as in tumor cells.
Some subtle differences in the expression pattern of V-ATPase genes were found in distinct types of tumors. Microarrays of a specific histological type of esophageal cancer and Oncomine multi-cancer analyses (where, for example, C1 gene was upregulated in 12 different tumors and downregulated only in brain cancer and leukemia) indicated that the cancer V-ATPase signatures are not stochastic nor deterministic, but rather represent a hybrid phenomenon that vary among the cancer histological types. It is likely that distinct combinatorial assemblies of V-ATPase isoforms can be found in different tumor stages, and specific expression patterns could be useful towards early diagnostics and more effective prognostics. Indeed, a combination of downregulation of ATP6V1G2, V0A1, and V1C1 isoforms has been proposed by Terrasi and colleagues to predict glioma aggressiveness [42].
The subunits a and C presented the highest expression differences among their respective isoforms not only in ESCC (Fig. 1, Figure S3) but also in other cancers, as revealed by Oncomine analysis (Fig. 6). The isoforms a3 (TCIRG1) and a4 (ATP6V0A4) were upregulated and downregulated, respectively, in tumors as compared to normal tissues. Overexpression of a3 targets the V-ATPase to the plasma membrane and confers higher invasiveness to breast cancer and melanoma cells [7,43]. Moreover, a knockdown of a4 did not affect invasion of breast cancer cells, but resulted in a 2-fold increase in the a3 mRNA levels [7], which is in line with our results suggesting a tightly regulated balance among the isoforms of these V-ATPase subunits. Another multi-cancer V-ATPase signature revealed was the upregulation of the accessory subunit Ac45 (ATP6AP1), reinforcing that Ac45 in conjunction with isoforms a can play a role in invasiveness [44,45] probably by influencing the localization, activity and/or efficiency of the enzyme [23,24,44].
Furthermore, we demonstrated that C isoforms expression profiles are potential diagnostic markers, since their mRNA levels markedly discriminate ESCC from normal surrounding esophageal tissues, with the best results obtained using the C1/C2 ratio (Fig. 2 and 3) rather than any individual expression pattern. This underscores an interconnectivity and fine tuning of C isoforms expression. Indeed, similar levels of expression for both C1 and C2 isoforms were found in normal tissues, contrasting with a wider range of ATP6V1C1 and ATP6V1C2 expression found among the different tumors, invariably involving an expression imbalance characterized by the dominance of C1 isoform (Fig. 2 and Figure S4).
In silico modeling revealed an additional stretch of 46 amino acids with binding sites for different kinases and phosphatases present only in C2a (Fig. 4, Tables S5 and S6), suggesting specific post-translational regulations. Two structures were predicted for the additional peptide stretch of C2a isoform, a beta sheet fold or an alpha-helix bend flanked by unstructured regions. Both could represent functional conformations since C subunits can be found in a membrane-bound holoenzyme and also as a free protein in the cytosol [34]. Based on the actin-binding domain described for the B subunit [36], we detected similar domains in the C subunit isoforms (Fig. 4e and f). The putative actin binding site in the C2a isoform was located immediately downstream the additional peptides stretch, exclusive of this isoform and predicted to be highly mobile. Thus, it appears mechanistically sound that the additional peptide stretch in the C2a isoform may integrate post-translational kinases-dependent regulations and actin binding mechanisms in normal cells, which are disrupted as the C1 isoform becomes prevalent in cancer cells.
This model provides new insights on previous reports, where V-ATPase containing C2a or C2b isoforms exhibited lower coupling efficiency between ATP hydrolysis and proton transport than V-ATPase containing C1 or Vma5p (yeast C subunit) [33]. The interaction of actin and C subunit also contribute to the disassembly and re-assembly regulatory mechanism of the pump [34,46,47]. The single C subunit of insect V-ATPases was also demonstrated to interact and modulate the actin polymerization [48]. It seems likely that human cells that express multiple C isoforms might exhibit a wider array of actin-proton pump interactions with a role in the regulation of normal as well as pathological metabolism. Thus, it is not surprising that cytoskeleton rearrangements somehow interconnected with the V-ATPase activity has been implicated in cancer-related processes, such as migration and invasion [37,39], release of exosomes [49], membrane fusion [50] and receptors recycling [17].
In conclusion, our data originally unveiled multi-cancer molecular signatures for the V-ATPase holoenzyme, indicating a key role for the differential expression of the subunits/isoforms in establishment of a conformational code, which ultimately would lead to specific proton signals and energy transductions related to carcinogenesis. Compelling evidence has been provided for a clear distinctive imbalance of the C subunit isoforms pointing to ATP6V1C1 and ATP6V1C2 as promising targets for development of new cancer biomarkers. Future studies taking advantage of new structural databases and powerful platforms for integrative system biology [51] should promote further in-depth characterization of V-ATPase multi-cancer molecular signatures, illuminating and exploring their mechanistic roles in carcinogenesis, early diagnosis and therapeutic potential.
Funding sources
This work was supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ, Brazil). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001 (PhD fellowship to JCVCS and Post-doctoral fellowship to EPC). JCVCS received Post-doctoral fellowship from Universidade Estadual do Norte Fluminense Darcy Ribeiro (UENF) and FFF received a PhD fellowship from FAPERJ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceptualization: ARF, LFRP, JCV, PNN; Data curation: JCV, PNN, TAS, ARF, LFRP; Formal analysis: JCV, PNN, EPC, ARF, LFRP, TAS; Funding acquisition: ARF, LFRP; Investigation: JCV, PNN, EPC, ARF, LFRP; Methodology: LFRP, ARF, PNN, EPC, TAS, JCV; Project administration: ARF, ALOF, LFRP; Resources: ARF, LFRP; Software: JCV, PNN, EPC, FFF; Supervision: ARF, LFRP, ALOF; Validation: JCV, PNN; Visualization: JCVCS, ARF, FFF; Writing - original draft: JCV, ARF; Writing - review & editing: JCV, ALOF, ARF, LFRP, PNN. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors have no conflicts of interest.
Acknowledgements
We thank all patients involved in this study, the staffs of Endoscopy Service and National Tumor Bank of the Brazilian National Cancer Institute for their contribution in sample collection.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.042.
Contributor Information
Luis Felipe Ribeiro Pinto, Email: lfpinto@inca.gov.br.
Arnoldo Rocha Façanha, Email: arnoldo@uenf.br.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Colombet M., Soerjomataram I. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 3.Rustgi A.K., El-Serag H.B. Esophageal carcinoma. N Engl J Med. 2014;371:2499–2509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- 4.Bird-Lieberman E.L., Fitzgerald R.C. Early diagnosis of oesophageal cancer. Br J Cancer. 2009;101:1–6. doi: 10.1038/sj.bjc.6605126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stransky L., Cotter K., Forgac M. The function of V-ATPases in cancer. Physiol Rev. 2016;96:1071–1091. doi: 10.1152/physrev.00035.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sennoune S.R., Bakunts K., Martinez G.M. Vacuolar H+-ATPase in human breast cancer cells with distinct metastatic potential: distribution and functional activity. Am J Physiol Cell Physiol. 2004;286:C1443–C1452. doi: 10.1152/ajpcell.00407.2003. [DOI] [PubMed] [Google Scholar]
- 7.Capecci J., Forgac M. The function of vacuolar ATPase (V-ATPase) a subunit isoforms in invasiveness of MCF10a and MCF10CA1a human breast cancer cells. J Biol Chem. 2013;288:32731–32741. doi: 10.1074/jbc.M113.503771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb B.A., Chimenti M., Jacobson M.P., Barber D.L. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 9.Cotter K., Capecci J., Sennoune S. Activity of plasma membrane V-ATPases is critical for the invasion of MDA-MB231 breast cancer cells. J Biol Chem. 2015;290:3680–3692. doi: 10.1074/jbc.M114.611210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martins B.X., Arruda R.F., Costa G.A. Myrtenal-induced V-ATPase inhibition - A toxicity mechanism behind tumor cell death and suppressed migration and invasion in melanoma. Biochim Biophys Acta Gen Subj. 2019;1863:1–12. doi: 10.1016/j.bbagen.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Von Schwarzenberg K., Wiedmann R.M., Oak P. Mode of cell death induction by pharmacological vacuolar H +-ATPase (V-ATPase) inhibition. J Biol Chem. 2013;288:1385–1396. doi: 10.1074/jbc.M112.412007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshansky V., Rubinstein J.L., Gruber G. Eukaryotic V-ATPase: novel structural findings and functional insights. Biochim Biophys Acta. 2014;1837:857–879. doi: 10.1016/j.bbabio.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Katara G.K., Jaiswal M.K., Kulshrestha A., Kolli B., Gilman-Sachs A., Beaman K.D. Tumor-associated vacuolar ATPase subunit promotes tumorigenic characteristics in macrophages. Oncogene. 2014;33:5649–5654. doi: 10.1038/onc.2013.532. [DOI] [PubMed] [Google Scholar]
- 14.Ibrahim S.A., Kulshrestha A., Katara G.K., Amin M.A., Beaman K.D. Cancer derived peptide of vacuolar ATPase ‘a2’ isoform promotes neutrophil migration by autocrine secretion of IL-8. Sci Rep. 2016;6:36865. doi: 10.1038/srep36865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Zaguilan R., Raghunand N., Lynch R.M. pH and drug resistance. I. functional expression of plasmalemmal V-type H+-ATPase in drug-resistant human breast carcinoma cell lines. Biochem Pharmacol. 1999;57:1037–1046. doi: 10.1016/s0006-2952(99)00022-2. [DOI] [PubMed] [Google Scholar]
- 16.Liao C., Hu B., Arno M.J., Panaretou B. Genomic screening in vivo reveals the role played by vacuolar H+ ATPase and cytosolic acidification in sensitivity to DNA-damaging agents such as cisplatin. Mol Pharmacol. 2007;71:416–425. doi: 10.1124/mol.106.030494. [DOI] [PubMed] [Google Scholar]
- 17.Nishi T., Forgac M. The vacuolar (H+)-ATPases–nature's most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 18.Santos J.M., Martínez-Zaguilán R., Facanha A.R., Hussain F., Sennoune S.R. Vacuolar H+-ATPase in the nuclear membranes regulates nucleo-cytosolic proton gradients. Am J Physiol - Cell Physiol. 2016;311:C547–C558. doi: 10.1152/ajpcell.00019.2016. [DOI] [PubMed] [Google Scholar]
- 19.Cotter K., Stransky L., McGuire C., Forgac M. Recent insights into the structure, regulation, and function of the V-ATPases. Trends Biochem Sci. 2015;40:611–622. doi: 10.1016/j.tibs.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith A.N., Borthwick K.J., Karet F.E. Molecular cloning and characterization of novel tissue-specific isoforms of the human vacuolar H(+)-ATPase C, G and d subunits, and their evaluation in autosomal recessive distal renal tubular acidosis. Gene. 2002;297:169–177. doi: 10.1016/s0378-1119(02)00884-3. [DOI] [PubMed] [Google Scholar]
- 21.Sun-Wada G.H., Yoshimizu T., Imai-Senga Y., Wada Y., Futai M. Diversity of mouse proton-translocating ATPase: presence of multiple isoforms of the C, d and G subunits. Gene. 2003;302:147–153. doi: 10.1016/s0378-1119(02)01099-5. [DOI] [PubMed] [Google Scholar]
- 22.Toei M., Saum R., Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49:4715–4723. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawasaki-Nishi S., Bowers K., Nishi T., Forgac M., Stevens T.H. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J Biol Chem. 2001;276:47411–47420. doi: 10.1074/jbc.M108310200. [DOI] [PubMed] [Google Scholar]
- 24.Kawasaki-Nishi S., Nishi T., Forgac M. Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J Biol Chem. 2001;276:17941–17948. doi: 10.1074/jbc.M010790200. [DOI] [PubMed] [Google Scholar]
- 25.Feng S., Zhu G., McConnell M. Silencing of atp6v1c1 prevents breast cancer growth and bone metastasis. Int J Biol Sci. 2013;9:853–862. doi: 10.7150/ijbs.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulshrestha A., Katara G.K., Ibrahim S. Vacuolar ATPase ‘a2’ isoform exhibits distinct cell surface accumulation and modulates matrix metalloproteinase activity in ovarian cancer. Oncotarget. 2015;6:3797–3810. doi: 10.18632/oncotarget.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cotter K., Liberman R., Sun-Wada G.H. The a3 isoform of subunit a of the vacuolar ATPase localizes to the plasma membrane of invasive breast tumor cells and is overexpressed in human breast cancer. Oncotarget. 2016;7:46142–46157. doi: 10.18632/oncotarget.10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Son S.W., Kim S.H., Moon E.Y., Kim D.H., Pyo S., Um S.H. Prognostic significance and function of the vacuolar H+-ATPase subunit V1E1 in esophageal squamous cell carcinoma. Oncotarget. 2016;7:49334–49348. doi: 10.18632/oncotarget.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolau-Neto P., Da Costa N.M., de Souza Santos P.T. Esophageal squamous cell carcinoma transcriptome reveals the effect of FOXM1 on patient outcome through novel PIK3R3 mediated activation of PI3K signaling pathway. Oncotarget. 2018;9:16634–16647. doi: 10.18632/oncotarget.24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhodes D.R., Yu J., Shanker K. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J., Aksoy B.A., Dogrusoz U. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerami E., Gao J., Dogrusoz U. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun-Wada G.H., Murata Y., Namba M., Yamamoto A., Wada Y., Futai M. Mouse proton pump ATPase C subunit isoforms (C2-a and C2-b) specifically expressed in kidney and lung. J Biol Chem. 2003;278:44843–44851. doi: 10.1074/jbc.M307197200. [DOI] [PubMed] [Google Scholar]
- 34.Kane P.M., Parra K.J. Assembly and regulation of the yeast vacuolar H(+)-ATPase. J Exp Biol. 2000;203:81–87. doi: 10.1242/jeb.203.1.81. [DOI] [PubMed] [Google Scholar]
- 35.Voss M., Vitavska O., Walz B., Wieczorek H., Baumann O. Stimulus-induced phosphorylation of vacuolar H(+)-ATPase by protein kinase A. J Biol Chem. 2007;282:33735–33742. doi: 10.1074/jbc.M703368200. [DOI] [PubMed] [Google Scholar]
- 36.Chen S.H., Bubb M.R., Yarmola E.G. Vacuolar H+-ATPase binding to microfilaments: regulation in response to phosphatidylinositol 3-kinase activity and detailed characterization of the actin-binding site in subunit B. J Biol Chem. 2004;279:7988–7998. doi: 10.1074/jbc.M305351200. [DOI] [PubMed] [Google Scholar]
- 37.Licon-Munoz Y., Michel V., Fordyce C.A., Parra K.J. F-actin reorganization by V-ATPase inhibition in prostate cancer. Biol Open. 2017;6:1734–1744. doi: 10.1242/bio.028837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costa G.A., de Souza S.B., da Silva Teixeira L.R. Tumor cell cholesterol depletion and V-ATPase inhibition as an inhibitory mechanism to prevent cell migration and invasiveness in melanoma. Biochim Biophys Acta Gen Subj. 2018;1862:684–691. doi: 10.1016/j.bbagen.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 39.Cai M., Liu P., Wei L. Atp6v1c1 may regulate filament actin arrangement in breast cancer cells. PLoS ONE. 2014;9:e84833. doi: 10.1371/journal.pone.0084833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Cristofori A., Ferrero S., Bertolini I. The vacuolar H+ ATPase is a novel therapeutic target for glioblastoma. Oncotarget. 2015;6:17514–17531. doi: 10.18632/oncotarget.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McConnell M., Feng S., Chen W. Osteoclast proton pump regulator Atp6v1c1 enhances breast cancer growth by activating the mTORC1 pathway and bone metastasis by increasing V-ATPase activity. Oncotarget. 2017;8:47675–47690. doi: 10.18632/oncotarget.17544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terrasi A., Bertolini I., Martelli C. Specific V-ATPase expression sub-classifies IDHwt lower-grade gliomas and impacts glioma growth in vivo. EBioMedicine. 2019;41:214–224. doi: 10.1016/j.ebiom.2019.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishisho T., Hata K., Nakanishi M. The a3 isoform vacuolar type H(+)-ATPase promotes distant metastasis in the mouse B16 melanoma cells. Mol Cancer Res. 2011;9:845–855. doi: 10.1158/1541-7786.MCR-10-0449. [DOI] [PubMed] [Google Scholar]
- 44.Jansen E.J., Scheenen W.J., Hafmans T.G., Martens G.J. Accessory subunit Ac45 controls the V-ATPase in the regulated secretory pathway. Biochim Biophys Acta. 2008;1783:2301–2310. doi: 10.1016/j.bbamcr.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 45.Smith G.A., Howell G.J., Phillips C., Muench S.P., Ponnambalam S., Harrison M.A. Extracellular and luminal pH regulation by vacuolar H+-ATPase isoform expression and targeting to the plasma membrane and endosomes. J Biol Chem. 2016;291:8500–8515. doi: 10.1074/jbc.M116.723395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vitavska O., Wieczorek H., Merzendorfer H. A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton. J Biol Chem. 2003;278:18499–18505. doi: 10.1074/jbc.M212844200. [DOI] [PubMed] [Google Scholar]
- 47.Serra-Peinado C., Sicart A., Llopis J., Egea G. Actin filaments are involved in the coupling of V0-V1 domains of vacuolar H+-ATPase at the golgi complex. J Biol Chem. 2016;291:7286–7299. doi: 10.1074/jbc.M115.675272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vitavska O., Merzendorfer H., Wieczorek H. The V-ATPase subunit C binds to polymeric F-actin as well as to monomeric G-actin and induces cross-linking of actin filaments. J Biol Chem. 2005;280:1070–1076. doi: 10.1074/jbc.M406797200. [DOI] [PubMed] [Google Scholar]
- 49.Liegeois S., Benedetto A., Garnier J.M., Schwab Y., Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in caenorhabditis elegans. J Cell Biol. 2006;173:949–961. doi: 10.1083/jcb.200511072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strasser B., Iwaszkiewicz J., Michielin O., Mayer A. The V-ATPase proteolipid cylinder promotes the lipid-mixing stage of SNARE-dependent fusion of yeast vacuoles. EMBO J. 2011;30:4126–4141. doi: 10.1038/emboj.2011.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey M.H., Tokheim C., Porta-Pardo E. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385. doi: 10.1016/j.cell.2018.02.060. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.