Abstract
The transcription factor Bach1 impairs angiogenesis after ischemic injury by suppressing Wnt/β-catenin signaling; however, the specific domains responsible for the anti-angiogenic effects of Bach1 remain unclear. This study determined the role of the BTB domain of Bach1 in ischemic angiogenesis. Bach1 is highly expressed in circulating endothelial cells from acute myocardial infarction patients and is the early induction gene after ischemia. Mice were treated with adenoviruses coding for GFP (AdGFP), Bach1 (AdBach1), or a Bach1 mutant lacking the BTB domain (AdBach1-ΔBTB) after surgically induced hind-limb ischemia. Measures of blood-flow recovery, capillary density, and the expression of vascular endothelial growth factor (VEGF) and heme oxygenase-1 (HO-1) were significantly lower and ROS levels were higher in the AdBach1 group, but not in AdBach1-ΔBTB animals. Furthermore, transfection with AdBach1, but not AdBach1-ΔBTB, in human endothelial cells was associated with significant declines in 1) capillary density and hemoglobin content in the Matrigel-plug assay, 2) proliferation, migration, tube formation, and VEGF and HO-1 expression in endothelial cells. Bach1 binds directly with TCF4, and this interaction is mediated by residues 81–89 of the Bach1 BTB domain and the N-terminal domain of TCF4. Bach1, but not Bach1-ΔBTB, also co-precipitated with histone deacetylase 1 (HDAC1), while the full-length HDAC1 proteins, but not HDAC1 mutants lacking the protein-interaction domain, co-precipitated with Bach1. Collectively, these results demonstrate that the anti-angiogenic activity of Bach1 is crucially dependent on molecular interactions that are mediated by the protein's BTB domain, and this domain could be a drug target for angiogenic therapy.
Keywords: Angiogenesis, Bach1, The BTB domain, VEGF, TCF4
Research in context.
Evidence before this study
We have shown that the transcription factor Bach1 suppressed angiogenesis after ischemic injury by impeding Wnt/β-catenin signaling. Bach1 functions as a competitive inhibitor of β-catenin/TCF4 binding, recruits HDAC1 to the promoter of TCF4-targeted genes. However, the specific domains and residues responsible for the anti-angiogenic effects of Bach1 had yet to be identified.
Added value of this study
We found that Bach1 gene was upregulated in human acute myocardial infarction (AMI) patients in samples enriched for circulating endothelial cells and ischemic myocardium of AMI mouse. Lacking of the BTB domain, Bach1 did not impede angiogenesis in ischemic hind limbs of mice and in vivo Matrigel plug, and failed to reduce proliferation, migration, and tube formation in human endothelial cells. Mechanically, Bach1 bound directly with TCF4, and this interaction was mediated by residues 81–89 of the Bach1 BTB domain. The BTB domain was essential for the Bach1-induced blockade of Wnt-targeted promoter activity and VEGF gene expression, for the binding of Bach1 to TCF4 and HDAC1, and for HDAC activation. The Bach1 BTB domain was also responsible for the Bach1-induced oxidative stress response.
Implications of all the available evidence
Our study suggests that the anti-angiogenic activity of Bach1 is crucially dependent on molecular interactions that are mediated by the protein's BTB domain. Peptides or small molecules that target the Bach1 BTB domain may improve recovery from ischemic injury or disease.
Alt-text: Unlabelled box
1. Introduction
Angiogenesis is essential for prolonging survival of the injured myocardium or muscles following myocardial or peripheral ischemia. Recently, it has been reported that new cardiac blood vessels are formed from pre-existing endothelial cells (EC) [1,2]. Therefore, the cellular mechanisms that promote the regenerative capacity of endogenous EC for enhancing angiogenesis need to be resolved. BTB and CNC homology 1 (Bach1) is a member of the basic region leucine zipper (bZip) family of transcription factors. Bach1 is widely expressed in mammalian tissues, where it functions as a crucial regulator of the cell cycle and differentiation, as well as the oxidative-stress response and heme homeostasis [3]. We have shown that Bach1 impairs angiogenesis in both developing zebrafish [4] and the ischemic hindlimbs of mice [5], and that Bach1 exert an anti-angiogenic effect, at least in part, via the repression of Wnt/β-catenin signaling. In the canonical Wnt-signaling pathway, β-catenin responds to Wnt stimulation through translocating from the cytoplasm into the nucleus, where β-catenin can form a complex with transcription factor 4 (TCF4) to active gene expression [6]; however, Bach1 competitively inhibits β-catenin/TCF4 binding and recruits histone deacetylase (HDAC) 1 to the promoter of TCF4-targeted genes, which suppresses the expression of angiogenic factors including vascular endothelial growth factor (VEGF) and interleukin-8 (IL-8). Wnt/β-catenin signaling also promotes the angiogenic activity of endothelial cells [7], including the proliferation and migration of vascular endothelial cells after myocardial infarction [8]. Thus, a more thorough understanding of the mechanisms by which Bach1 inhibits of Wnt/β-catenin signaling could lead to the development of novel treatments for promoting angiogenesis in ischemic disease.
The bZip domain of Bach1 is located near the protein's C terminus, and the N-terminal region contains the BTB domain. The bZip domain binds DNA [9], while the BTB domain, which has been identified in as many as 40 mammalian transcription factors, interacts with other molecules that regulate gene expression [10,11]; thus, the experiments described in this report were designed to test our hypothesis that the anti-angiogenic activity of Bach1 is mediated by the BTB domain, and to more fully characterize how interactions among Bach1, TCF4, and HDAC1 regulate Wnt/β-catenin signaling.
2. Materials and methods
2.1. Reagents
Trichostatin A (TSA, T1952), Drabkin's reagent (D5941), and an antibody against Flag (F1804) were purchased from Sigma-Aldrich (St. Louis, MO). Human vascular endothelial growth factor (VEGF165, 100–20) was purchased from PeproTech Inc. (Rocky Hill, NJ). Dihydroethidium (DHE, D1168) was obtained from Invitrogen (Carlsbad, CA). Antibodies against Bach1 (Santa Cruz #sc-271211, RRID:AB_10608972), NQO1 antibody (Santa Cruz #sc-32793, RRID:AB_ 628036), normal mouse IgG (sc-2025, RRID:AB_737182), normal rabbit IgG (sc-66931, RRID:AB_1125055), normal goat IgG (sc-2028, RRID:AB_737167), β-actin (sc-47778, RRID:AB_626632), HA (sc-7392, RRID:AB_627809) and Protein A/G PLUS-Agarose (sc-2003, RRID:AB_10201400) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody against CD31 (ab28364, RRID:AB_726362) was purchased from Abcam (Cambridge, MA). GST antibody (#66001, RRID:AB_10951482), GFP-tag antibody (Proteintech #50430–2-AP, RRID:AB_11042881) and HO-1 antibody (Proteintech #10701–1-AP, RRID:AB_2118685) was purchased from Proteintech (Rosemont, IL).
2.2. Isolation and culture of human umbilical vein endothelial cells (HUVECS)
HUVECS were isolated [12] from the fresh umbilical cord of normal parturients with informed consent. The procedure was approved by the Ethics Committee of Experimental Research at Fudan University Shanghai Medical College. HUVECS were identified by the morphology, expression of CD31, and uptake of acetyl-LDL; HUVECS were passaged one to six times before use, and cultured at 37 °C with 5% CO2 in endothelial cell medium (Sciencell, Carlsbad, CA) supplemented with 5% fetal bovine serum, endothelial cell growth supplement (Sciencell, Carlsbad, CA).
2.3. Viral transfection
Recombinant adenoviruses encoding GFP (AdGFP), human Bach1 (AdBach1), or a mutated version of human Bach1 that lacked the BTB domain (AdBach1-ΔBTB) were purchased from GenePharma (Shanghai, China); the AdBach1 and AdBach1-ΔBTB vectors also coded for GFP expression. HUVECS were transfected at a multiplicity of infection (MOI) of 25, and transfection was verified via the presence of GFP expression. No evidence of cellular toxicity was detected.
2.4. Murine hind limb ischemia (HLI) model
All experimental protocols were approved by the Ethics Committee of Experimental Research at Fudan University Shanghai Medical College and performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH publication No 85–23). Hindlimb ischemia (HLI) was conducted in 8-week-old male C57BL/6 mice as described previously. The mice were anesthetized with intraperitoneal sodium pentobarbital (50 mg/kg), the proximal femoral artery and the distal saphenous artery was ligated and removed in one hindlimb; sham surgery was performed in the contralateral hindlimb of each mouse. Mice in the NS group were treated with normal saline (40 μL), and mice in the AdGFP, AdBach1, and AdBach1-ΔBTB groups were treated with the corresponding adenoviruses (total dose: 2 × 108 plaque-forming units); treatments were administered to three sites in the grastrocnemius muscle and three sites in the adductor muscle. The recovery of blood flow in the ligated limbs of mice was performed with a laser Doppler perfusion imaging (LDPI) (Moor Instruments) system. Perfusion was expressed as the ratio of measurements in the ligated and non-ligated (sham-operated) contralateral limb.
2.5. In-vivo matrigel plug assay
The in-vivo Matrigel plug assay was conducted as described previously [13] with modifications. Cells were suspended in 100 μL phosphate-buffered saline (PBS) and mixed with VEGF (100 ng/mL) in 400 μL Matrigel; then, the cell-Matrigel mixture was injected subcutaneously along the abdominal midline of six-week-old female BALB/c nude mice. Seven days later, the Matrigel plugs were collected, photographed, embedded, sectioned, and stained with fluorescent anti-CD31 antibodies. Capillary density was calculated by counting CD31-expressing vascular structures in a blinded fashion on five sections per plug (two fields per section) under a fluorescent microscope. For hemoglobin analysis, the excised Matrigel plugs were homogenized in 100 μL phosphate-buffered saline (PBS) and centrifuged; then, the supernatant was collected and hemoglobin content was measured via the Drabkin assay with a standard curve determined from stock solutions containing known quantities of hemoglobin. Hemoglobin content (mg) was normalized to the weight of the plug (g).
2.6. Construction of vectors
Human Bach1, TCF-4, and HDAC1 genes were generated and cloned into pcDNA3.1 as described previously [5]. Mutants of Flag-tagged versions of the Bach1-ΔBTB sequence, or mutant versions of the sequence lacking portions of the BTB domain were amplified from the full-length cDNA of human Bach1 with appropriate sets of primers. MAREs reporter containing three MAREs elements (GCTGACTCAT) were amplified and cloned into the luciferase vector pGL3-basic. Glutathione S-transferase (GST)-TCF4 full, GST-TCF4-N terminal, GST-TCF4-C terminal, or GST-TCF4 lacking both N terminal and C terminal (TCF4-M) fusion protein was constructed by inserting PCR-generated DNA fragments into PGEX-6P-1 vector. Mutants of HA-tagged versions of the sequence lacking N terminal or C terminal of HDAC1 were amplified from the full-length cDNA of human HDAC1 with appropriate sets of primers. The TOPflash and FOPflash reporter vector were purchased from Millipore (Billerica, MA). A 2.65 kb human VEGF gene promoter in pGL3-basic luciferase reporter was kindly provided by Dr. Fang (Chinese Academy of Sciences, China). All constructs were verified by sequencing.
2.7. Luciferase assay
Cells were transfected with a β-galactosidase plasmid and the pGL3 luciferase reporter or VEGF reporter or MAREs reporter, or FOPflash or TOPflash reporter; transfection was performed with Lipofectamine 2000. Luciferase activity was measured with a luciferase assay kit (Promega, Madison, WI) and expressed as the ratio of VEGF or MAREs reporter/pGL3-basic or TOPflash/FOPflash. Each experiment was conducted three times.
2.8. Quantitative real-time reverse transcription-polymerase chain reaction
The expression level of VEGF gene was analyzed by real-time reverse transcription-polymerase chain reaction (RT-PCR) as described previously [14]. Total RNA was extracted using the TRIzol reagent and cDNA was synthesized. Real-time quantitative PCR was carried out using the qPCR SYBR® Green Master Mix (Yeason, Shanghai, China #11201ES03). Primers are shown in Online Table I. All samples were analyzed using a Bio-Rad real-time analyzer (Bio-Rad Laboratories, Hercules, CA). Each experiment was repeated three independent times.
2.9. Immunoblotting and immunoprecipitation
Immunoblotting and Immunoprecipitation were performed as described previously [5]. Briefly, cells or tissue homogenates were lysed with SDS sample buffer on ice. Samples were heated and subjected for 10 min to centrifugation at 10,000 g. SDS-PAGE was performed and proteins were detected using their respective antibodies. Chemiluminescence was detected on a Tanon-5500 Imaging System (Tanon Science & Technology Ltd, Shanghai, China). The intensity of the bands was measured via densitometric analysis with ImageJ software, and normalized to the control. For immunoprecipitation, 1 mg of total protein was incubated with the 5 μg antibody at 4 °C overnight and then with Protein A/G PLUS-Agarose (Santa Cruz, Santa Cruz, CA) at 4 °C for an additional 4 h; then, the precipitates were washed with RIPA and PBS, resuspended in SDS loading buffer, and immunoblotting was performed via standard protocols. For the in-vitro binding experiment, GST-fused proteins were expressed in Escherichia coli and purified by glutathione-Sepharose 4B beads. The proteins were incubated with glutathione-Sepharose 4B beads for four hours at 4 °C. The beads were washed and incubated with target proteins for 12 h at 4 °C. The beads were washed and resuspended in SDS loading buffer. SDS-PAGE was performed and proteins were detected using their respective antibodies. Each experiment was repeated three independent times. Each experiment was repeated three independent times.
2.10. In vitro pull-down assay
The GST-tagged Bach1 and GST-tagged TCF4 recombinant proteins were expressed and purified with glutathione-Sepharose 4B beads. Purified GST-tagged recombinant proteins Bach1 or TCF4 were then incubated with target proteins for 12 h at 4 °C followed by three times of wash. Conjugated proteins were then eluted by SDS loading buffer, and examined by western blot analysis.
2.11. Cell proliferation
Proliferation was measured by using a counting chamber to determine the number of cells present after 0, 24, 48, and 72 h of culture. Each experiment was conducted three times.
2.12. Cell migration
HUVECS (3 × 105 cells/ml) were plated onto 8-μm-pore Transwell filters in a 12-well plate according to the manufacturer's instructions (Corning, New York, NY) [15]. The number of cells migrating over ten hours was determined using the 0.1% crystal violet staining. Results were compiled as the mean of five randomly fields containing at least two platings. Each experiment was conducted three times.
2.13. Tube formation assay
Cells were seeded into 48-well plates (2 × 104 cells/well) that had been coated with 250 μL growth factor-reduced Matrigel. Twelve hours later, the cells were viewed under an inverted phase-contrast microscope, and tube formation was quantified as the total summed length of all tubes in five randomly selected fields per well. Analyses were performed with ImageJ software, and each experiment was conducted three times.
2.14. Immunofluorescence
Paraffin-embedded sections were deparaffinized, rehydrated, and analyzed via immunofluorescence. Briefly, the sections were incubated with rabbit anti-CD31 antibody in blocking solution overnight and then with Cy3-conjugated donkey anti-rabbit IgG secondary antibodies. Nuclei were counter-stained with DAPI, and the sections were examined under a fluorescence microscope.
2.15. HDAC activity assay
HDAC activity was measured with an HDAC activity assay kit (Millipore, Billerica, MA). Nuclear extracts (15 μg) were incubated with the HDAC assay substrate at 37 °C for 45 min; then, diluted activator reagent was added, samples were incubated at 37 °C for another 20 min, and fluorescence was measured with a microplate recorder (excitation: 390 nm, emission: 460 nm). Procedures were conducted and standard curves were calculated as directed by the manufacturer's protocol.
2.16. Statistical analysis
Data are expressed as mean ± SEM. Differences among three or more groups were evaluated for significance via one-way analysis of variance (ANOVA) and the Tukey post-hoc test. Analyses were conducted with Prism software, and a p-value of less than 0.05 was considered significant.
3. Results
3.1. Bach1 is highly expressed in circulating endothelial cells from acute myocardial infarction patients
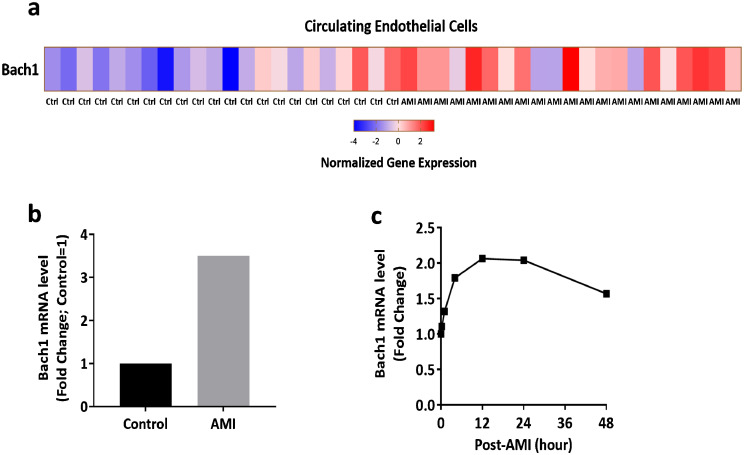
We have shown that Bach1 expression was upregulated in the endothelial cells exposed to hypoxia and in the ischemic hindlimb muscles of mice [5]. To determine whether the Bach1 expression is related to human ischemic diseases, we analyzed the published microarray data of circulating endothelial cells (CECs) from healthy control volunteers (n = 22) and acute myocardial infarction (AMI) patients (n = 21) [16]. The expression of Bach1 in CECs was higher in AMI patients than healthy controls (Fig. 1a), and a 3.5-fold increase in Bach1 expression was observed in AMI compared with controls (Fig. 1b). The publicly available microarray data analyses in AMI mice also indicated that mRNA levels of Bach1 in ischemic/infarcted zone of the left ventricular myocardium were upregulated during the first 48 h following occlusion of the coronary artery (Fig. 1c) [17].
Fig. 1.
Bach1 is highly expressed in circulating endothelial cells from acute myocardial infarction patients and ischemic myocardium of mice. (a) Heat map for the Bach1 gene in the microarray of gene expression analysis of enriched CECs from healthy controls (n = 22) and AMI patients (n = 21), expression levels were represented from low (blue) to high (red). (b) Analysis of the relative Bach1 gene expression from microarray in AMI (n = 21) and controls (n = 22), normalized with controls. (c) Temporal expression profiles for the Bach1 gene expressed in ischemic/infarcted zone of the left ventricular myocardium in mice at the indicated time points (0, 0.25, 1, 4, 12, 24, and 48 h post-occlusion), results were normalized to the control at 0 h.
3.2. The BTB domain of Bach1 is required for the anti-angiogenic activity of Bach1 in the ischemic hind limbs of mice
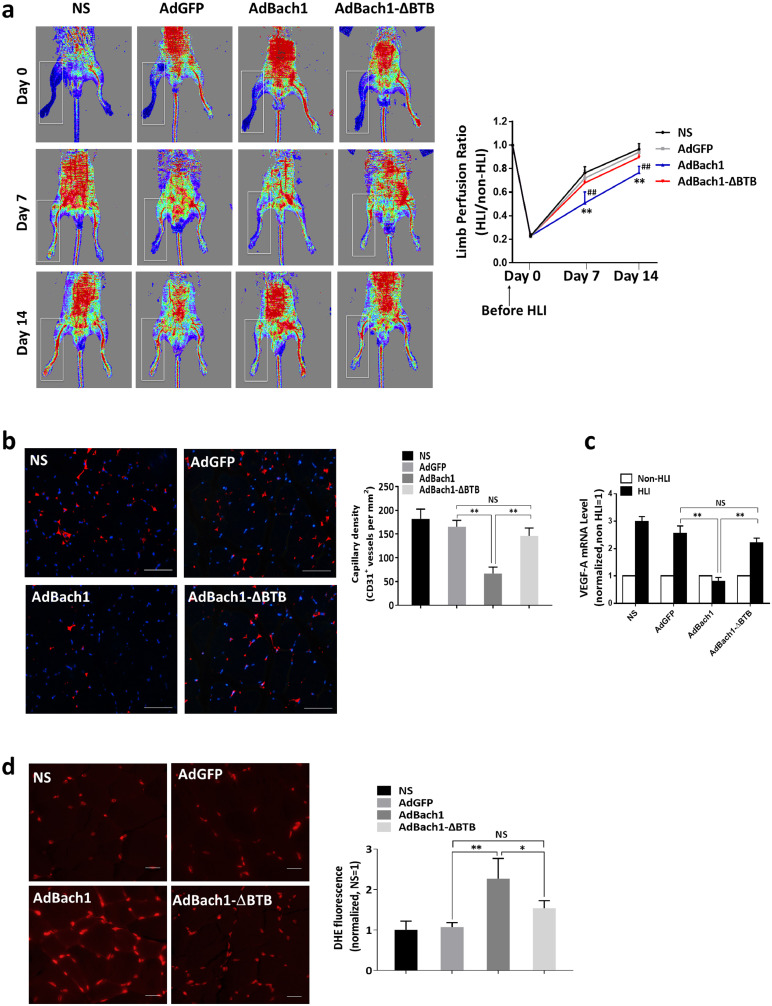
Bach1 was shown to impair ischemia-induced angiogenesis [5]. To determine whether the anti-angiogenic effect of Bach1 during recovery from peripheral ischemic injury is mediated by the BTB domain, hind-limb ischemia (HLI) was surgically induced in C57BL/6 J mice, and the injured limbs were treated with saline (the NS group) or with adenoviruses coding for GFP (the AdGFP group), for Bach1 (the AdBach1 group), or for a mutated version of Bach1 that lacked the BTB domain (the AdBach1-ΔBTB group). Seven and 14 days later, the ratios of perfusion measurements in the ischemic and non-ischemic hind limbs of NS, AdGFP, and AdBach1-ΔBTB animals were similar and significantly greater than the ratio in AdBach1 animals (Fig. 2a). Treatment with AdBach1, but not AdBach1-ΔBTB, also significantly reduced measures of capillary density (Fig. 2b), the expression of VEGF-A (Fig. 2c) and heme oxygenase-1 (HO-1) (Supplemental Fig. 1a) and increased ROS levels (Fig. 2d) in the injured limbs of mice, and differences among the NS, AdGFP, and AdBach1-ΔBTB groups were not significant. Evaluations of GFP fluorescence indicated that the vectors were expressed in ischemic hindlimb muscles (Supplemental Fig. 1a).
Fig. 2.
The BTB domain of Bach1 is required for the Bach1-induced suppression of angiogenesis in a murine model of hind-limb ischemia (HLI). HLI was surgically induced in 8-week-old C57BL/6 J mice; then, the injured limbs were injected with normal saline (the NS group) or with adenoviruses coding for GFP (the AdGFP group), Bach1 (the AdBach1 group), or a Bach1 mutant that lacked the BTB domain (the AdBach1-ΔBTB group). (a) Blood flow was measured in the ischemic (HLI) and nonischemic (non-HLI) limbs 0, 7, and 14 days after HLI induction via laser Doppler imaging and then quantified as the ratio of measurements in the injured and uninjured limbs for each animal (n = 6, **P < 0.01 AdBach1 vs. AdGFP, ##P < 0.01 AdBach1 vs. AdBach1-ΔBTB, one-way analysis of variance). (b) Fourteen days after HLI induction, sections from the injured limbs were immunofluorescently stained for expression of the endothelial cell marker CD31; scale bar, 50 μm. Capillary density was quantified as the number of CD31+ vessels per mm2 (n = 6, **P < 0.01, one-way analysis of variance). (c) VEGF-A mRNA levels were quantified via real-time PCR in thigh adductor and gastrocnemius muscles harvested from the ischemic and nonischemic limbs of mice 14 days after HLI induction (n = 6, **P < 0.01, one-way analysis of variance). (d) ROS production in ischemic muscle tissue on day 14 was determined by an in situ detection of dihydroethedium (DHE) fluorescence (n = 4, *P < 0.05, **P < 0.01, one-way analysis of variance). Scale bar, 20 μm.
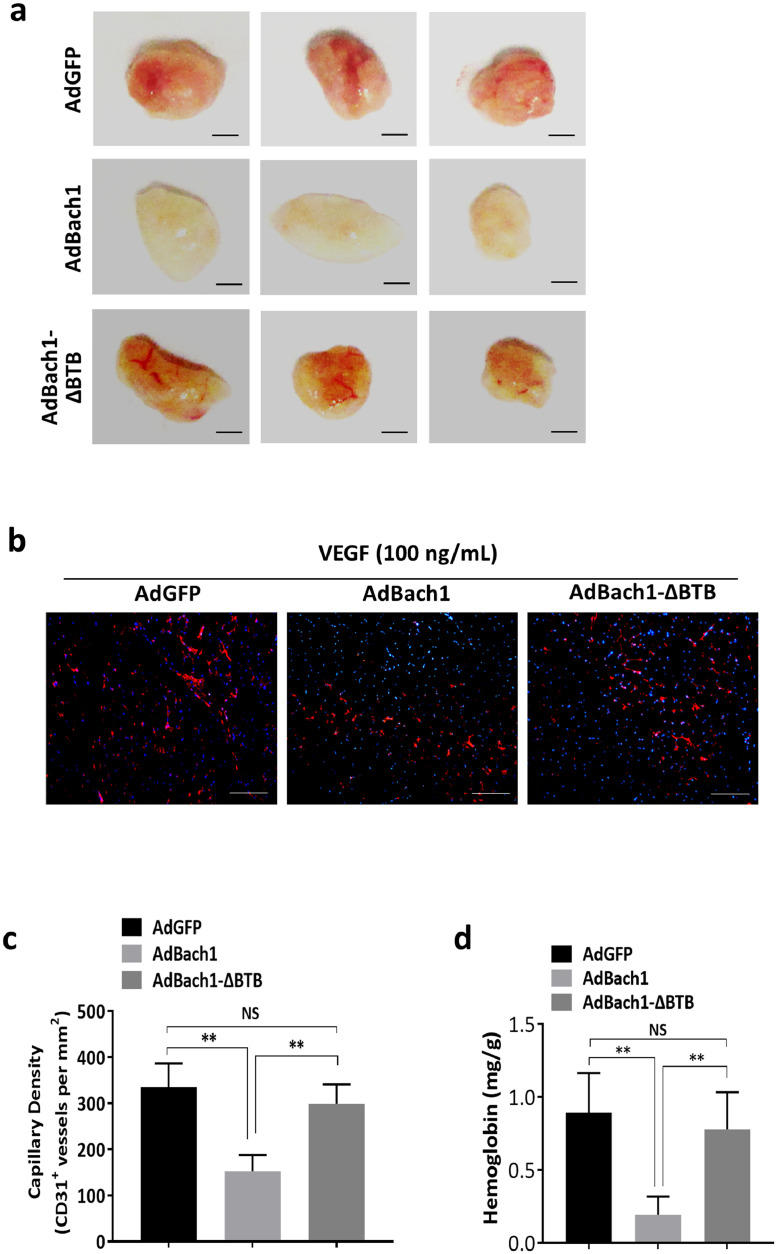
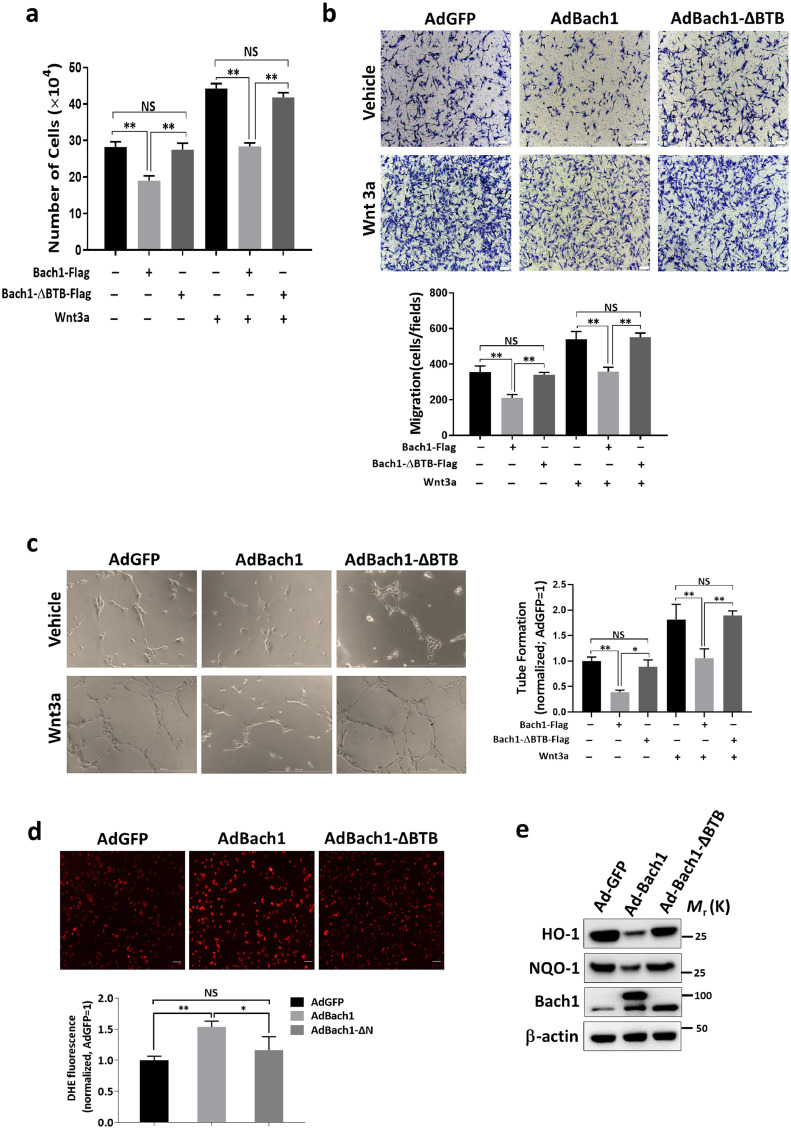
Our observations in the HLI model were supported by monitoring the angiogenic activity of HUVECS that had been transfected with AdGFP (AdGFP HUVECS), AdBach1 (AdBach1 HUVECS), or AdBach1-ΔBTB (AdBach1-ΔBTB HUVECS). When the cells were subcutaneously injected with VEGF and Matrigel into BALB/c nude mice, and the plugs were harvested 7 days later (Fig. 3a), measures of capillary density (Fig. 3B-C) and hemoglobin content (Fig. 3d) were similar in plugs formed with AdGFP or AdBach1-ΔBTB HUVECS but significantly lower in the plugs that contained AdBach1 HUVECS. Transfection with AdBach1, but not AdBach1-ΔBTB, significantly reduced proliferation (Fig. 4a), migration (Fig. 4b), and tube formation (Fig. 4c) in cultured HUVECS. When Wnt signaling was stimulated by culturing the cells in Wnt3a, assessments of tube formation, cell migration, and proliferation were significantly lower in AdBach1-HUVECS than AdBach1-ΔBTB-HUVECS (Fig. 4a–c). Treatment with AdBach1, but not AdBach1-ΔBTB, also significantly increased intracellular ROS levels (Fig. 4d) and induced cell apoptosis (Supplemental Fig. 1b), and decreased the expression of antioxidant genes such as HO-1 and NAD(P)H quinone oxidoreductase 1 (NQO1) (Fig. 4e) in cultured HUVECS. Collectively these observations indicate that the BTB domain of Bach1 is essential for the Bach1-mediated suppression of both blood-vessel growths in ischemic limbs and angiogenic activity in endothelial cells.
Fig. 3.
The BTB domain of Bach1 is required for the Bach1-induced suppression of angiogenesis in implanted, endothelial-cell–containing Matrigel plugs. Human umbilical vein endothelial cells (HUVECs) were transfected with AdGFP, AdBach1, or AdBach1-ΔBTB and mixed with VEGF (100 ng/mL) and Matrigel; then, the mixture was subcutaneously injected into nude mice. Seven days later, (a) the Matrigel plugs were explanted (bar = 2 mm), fixed, sectioned, and (b) immunofluorescently stained for CD31 expression (bar = 200 μm). (c) Capillary density was quantified as the number of CD31+ vessels per mm2 (n = 6, **P < 0.01, one-way analysis of variance). (d) Perfusion of the explanted plugs was quantified via measurements of hemoglobin content (n = 6, **P < 0.01, one-way analysis of variance).
Fig. 4.
The Bach1-induced suppression of angiogenic activity in cultured endothelial cells is mediated by the BTB domain. HUVECs were transfected with AdGFP, AdBach1, or AdBach1-ΔBTB and incubated with or without Wnt3a. (a) Transfected HUVECs were cultured for 72 h, and proliferation was evaluated by using a cell-counting chamber to determine the number of cells (n = 3, **P < 0.01, one-way analysis of variance). (b) Transfected HUVECs were seeded over transwell culture-plate inserts and cultured for 8 h; then, migration was quantified by counting the cells that were present on the lower side of the membrane (n = 3, **P < 0.01, one-way analysis of variance). (c) HUVECs were seeded into plates that had been coated with growth-factor reduced Matrigel. Twelve hours later, tube formation was quantified by viewing the cells with an inverted phase-contrast microscope and measuring the cumulative length of all tubes in each group; results were normalized to measurements in the AdGFP-transfected cells (n = 3, *P < 0.05, **P < 0.01, one-way analysis of variance). (d) ROS production was determined by the detection of dihydroethedium (DHE) fluorescence in transfected HUVECs. Fluorescent levels were expressed as percent increase over the AdGFP group (n = 4, *P < 0.05, **P < 0.01, one-way analysis of variance). (e) HO-1 and NQO-1 protein levels in AdGFP, AdBach1, and AdBach1-ΔBTB HUVECs were measured via Western blot.
3.3. The BTB domain of Bach1 is required for the Bach1-induced inhibition of Wnt/β-catenin signaling and Bach1-TCF4 binding
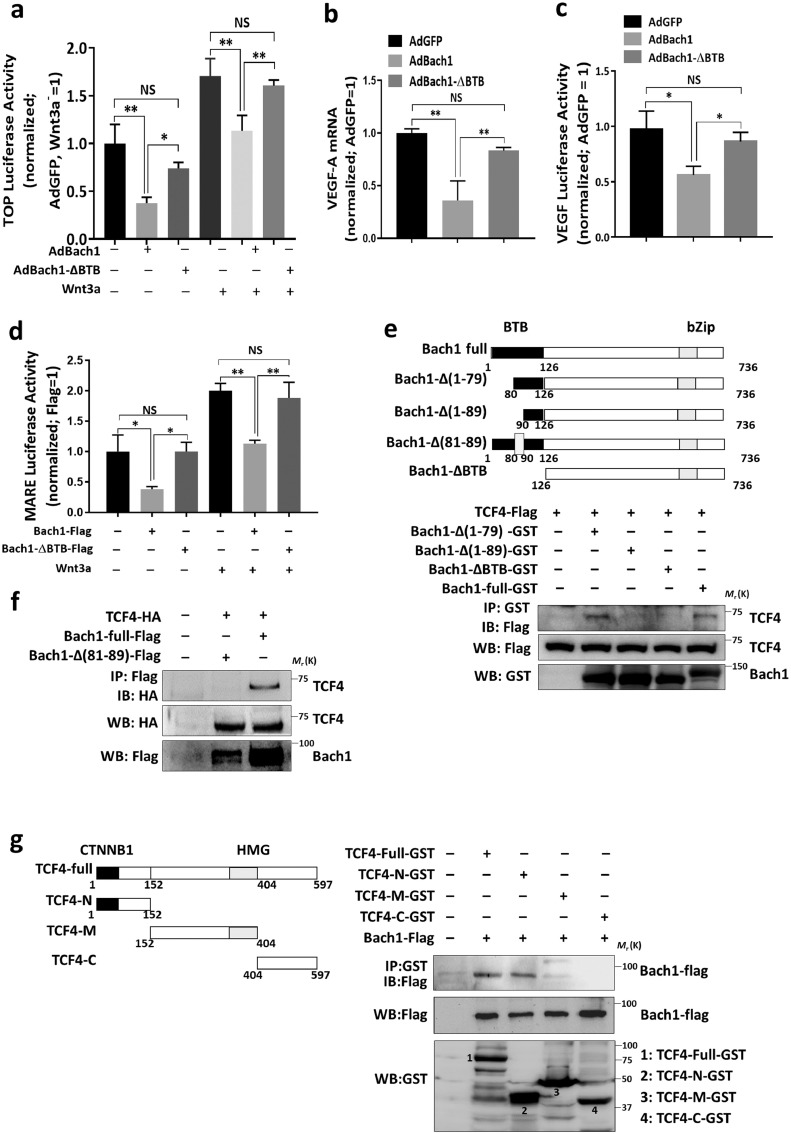
Both physiological and pathological angiogenesis, as well as the proliferation and migration of endothelial cells, are regulated in part by Wnt/β-catenin signaling [7]. Thus, since the results from our earlier publications indicate that Bach1 represses Wnt/β-catenin signaling by interacting with TCF4 and recruiting HDAC1 to the promoter of TCF4-targeted genes [5], we used a Wnt/β-catenin–specific reporter plasmid (TOPflash, which contains a luciferase reporter under the control of four copies of the TCF/LEF-binding element) to investigate whether the role of the BTB domain in the Bach1-induced suppression of angiogenic activity in HUVECS was mediated by Wnt/β-catenin signaling. After transfection with the TOPflash reporter, luciferase activity was significantly lower in AdBach1 HUVECS, but not AdBach1-ΔBTB HUVECS, than in AdGFP HUVECS under both normal culture conditions and when Wnt signaling was stimulated via treatment with Wnt3a (Fig. 5a). Furthermore, the mRNA levels of VEGF-A (Fig. 5b), which is a downstream target of Wnt signaling, as well as the activity of a luciferase reporter containing the VEGF promoter (Fig. 5c), were downregulated in AdBach1 HUVECS but not AdBach1-ΔBTB HUVECS. Bach1 was shown to bind to Maf recognition elements (MAREs) in the gene promoters [18]. We then determined whether Bach1′s BTB domain could affect MAREs-dependent reporter activity. MAREs luciferase activity was significantly lower in AdBach1-HUVECS, but not AdBach1-ΔBTB-HUVECS under both normal conditions and Wnt3a treatment (Fig. 5d).
Fig. 5.
The BTB domain of Bach1 is required for the Bach1-induced suppression of Wnt/β-catenin signaling and Bach1-TCF4 binding. (a) AdGFP, AdBach1, and AdBach1-ΔBTB HUVECs were transfected with a TOPflash or FOPflash reporter, and luciferase activity was quantified in the presence (+) and absence (–) of Wnt3a (200 ng/mL) stimulation; results were normalized to measurements in AdGFP-transfected cells cultured without Wnt3a (n = 3, *P < 0.05, **P < 0.01, one-way analysis of variance). (b) VEGF-A mRNA levels in AdGFP, AdBach1, and AdBach1-ΔBTB HUVECs were measured via real-time PCR and normalized to measurements in AdGFP-transfected cells (n = 3, **P < 0.01, one-way analysis of variance). (c) AdGFP, AdBach1, and AdBach1-ΔBTB HUVECs were transfected with a reporter construct coding for luciferase production from the VEGF promoter; then, luciferase activity was quantified and normalized to measurements in AdGFP-transfected cells (n = 3, *P < 0.05, one-way analysis of variance). (d) HEK293T cells that had been transfected with an empty vector or with a Bach1-Flag or Bach1-ΔBTB-Flag and MARES reporter for 24 h and incubated with or without Wnt3a for 12 h, luciferase activity was quantified and normalized to measurements in control vector-transfected cells (n = 3, *P < 0.05, **P < 0.01, one-way analysis of variance). (e) Bacterially expressed GST-tagged versions of the full Bach1 protein (Bach1-Full-GST) or with mutated versions of Bach1 that lacked either the N-terminal BTB domain or the other Bach1 deletion mutants was incubated with Flag-tagged TCF4; then, the reaction products were precipitated with glutathione-Sepharose 4B beads, and TCF4 was detected in the precipitate via Western blot with anti-Flag antibodies. (f) HEK293T cells were transfected with 2 vectors, one coding for TCF4-HA, and the other coding for Flag-tagged versions of the full Bach1 sequence and Bach1 sequences lacking amino acid residues 81–89 (Bach1-Δ(81–89)), the cells were lysed; then, Bach1 and Bach1-Δ(81–89) were immunoprecipitated from the lysate with an anti-Flag antibody, and TCF4 was detected in the precipitate with an anti-HA antibody. (g) HEK293T cells were transfected with Flag-tagged Bach1 and lysed; then, the lysate was incubated with GST or GST-tagged versions of TCF4 or the indicated TCF4-deletion mutants, the GST-bound proteins were eluted, and the presence of Bach1 was evaluated via Western blot with an anti-Flag antibody.
We have also shown that Bach1 bound TCF4 [5], so we conducted GST-pulldown experiments and immunoprecipitation assays in HEK293T cells to determine the binding site of the Bach1-TCF4 interaction. Our results shows that TCF4 co-immunoprecipitated with both the full-length Bach1 protein and a mutant that lacked residues 1–79 of the BTB domain, but not with the Bach1-ΔBTB protein or with mutants lacking BTB-domain residues 1–89 or 81–89 (Fig. 5e–f). Similarly, the results from pulldown assays in HEK293K cells that had been cotransfected with vectors coding for Flag-tagged Bach1 and a panel of Glutathione S-transferase (GST)-tagged TCF4 deletion mutants indicated that Bach1 co-immunoprecipitated with the full TCF4 protein and with a mutant that lacked all but the N-terminal CTNNB1 domain, but not with either of two mutants in which the CTNNB1 domain was missing (Fig. 5g). Thus, TCF4 binds directly with Bach1, and this interaction is mediated by residues 81–89 of the Bach1 BTB domain and the N-terminal CTNNB1 domain of TCF4.
3.4. The BTB domain of Bach1 interacts with HDAC1 and is required for Bach1-induced HDAC activity
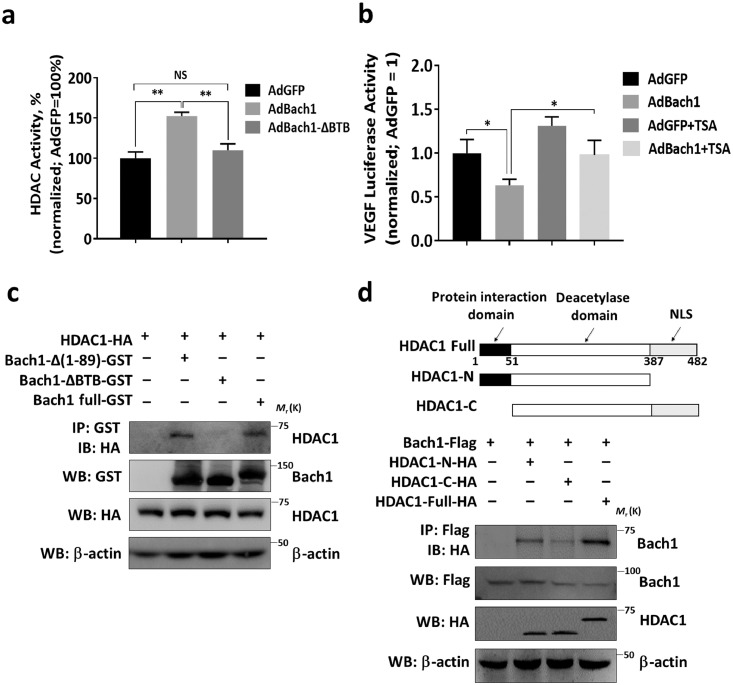
HDAC activity was also significantly greater in AdBach1 HUVECS, but not AdBach1-ΔBTB HUVECS, than in HUVECS transfected with AdGFP (Fig. 6a), and the decline in VEGF-luciferase reporter activity associated with Bach1 overexpression was attenuated by culturing the cells with the HDAC inhibitor Trichostatin A (TSA) (Fig. 6b). To determine whether the Bach1-mediated recruitment of HDAC1 to TCF4-targeted genes occurs via direct interactions between HDAC1 and the Bach1 BTB domain, in vitro pull-down assays were performed. When glutathione-S-transferase–tagged Bach1 constructs containing the full Bach1 sequence (Bach1-Full-GST) or sequences lacking residues 1–89 of the BTB domain (Bach1-Δ(1–89)-GST) or the N-terminal BTB domain (Bach1-ΔBTB-GST) were incubated with HA-tagged HDAC1, HDAC1 co-precipitated with both the full-length version of Bach1 and the mutant lacking BTB residues 1–89, but not with Bach1-ΔBTB (Fig. 6c). Furthermore, when HEK293T cells were transfected to express the full-length Flag-tagged Bach1 protein and HA-tagged versions of HDAC1 or HDAC1 deletion mutants, Bach1 co-immunoprecipitated with full-length HDAC1 and a C-terminal deletion mutant, but not with an HDAC1 mutant that lacked the N-terminal protein-interaction sequence (Fig. 6d). Collectively, these observations demonstrate that the Bach1-induced increase in HDAC1 activity is mediated by direct interactions between HDAC1 and the BTB domain of Bach1.
Fig. 6.
The BTB domain of Bach1 is required for Bach1-induced HDAC1 activity and mediates the Bach1-HDAC1 interaction. (a) HDAC activity was measured in nuclear extracts from HUVECs that had been transfected with AdGFP, AdBach1 or AdBach1-ΔBTB (n = 4, **P < 0.01, one-way analysis of variance). (b) AdGFP and AdBach1 HUVECs were transfected with a reporter construct coding for luciferase production from the VEGF promoter and cultured in the presence or absence of the HDAC inhibitor Trichostatin A (TSA, 1 μmol/L); then, luciferase activity was quantified and normalized to measurements in AdGFP-transfected cells without TSA (n = 3, *P < 0.05, one-way analysis of variance). (c) HA-tagged HDAC1 was incubated with GST-tagged versions of the full Bach1 sequence, the Bach1-ΔBTB sequence, or a Bach1 sequence lacking amino acid residues 1–89; then, the reaction products were precipitated with glutathione-Sepharose 4B beads, and HDAC1 was detected in the precipitate via Western blot with anti-HA antibodies. (d) HEK293T cells were transfected with 2 vectors, one coding for Flag-tagged Bach1 and the other for HA-tagged versions of the full HDAC1 sequence or the indicated HDAC1 deletion mutant; then, the cells were lysed, Bach1 was immunoprecipitated from the lysate with an anti-Flag antibody, and HDAC1 or the HDAC1 deletion mutants were detected in the precipitate with an anti-HA antibody.
4. Discussion
Cellular stress (such as ischemia and proinflammatory factor) may lead to endothelial cell dysfunction and detachment during the acute phase process, and elevated numbers of circulating endothelial cells (CECs) have been implicated in the pathophysiology leading to AMI [16,19,20]. We found that Bach1 gene was upregulated in human AMI in samples enriched for CECs and ischemic myocardium of AMI mouse, indicating that Bach1 is the early induction gene as a pivotal regulator of early post-ischemic molecular events, and is associated with the ischemic induced injury. We have shown that Bach1 induced endothelial cell apoptosis and suppressed ischemic angiogenesis, suggesting Bach1 is a major regulator of endothelial function and angiogenesis [5,21]. Bach1 functions as a competitive inhibitor of β-catenin/TCF4 binding, recruits HDAC1 to the promoter of TCF4-targeted genes, and prevents β-catenin from being acetylated by p300/CREB-binding protein (CBP). Collectively, these three interrelated mechanisms impede Wnt/β-catenin signaling and downregulate the expression of proteins (e.g., VEGF and IL-8) that promote angiogenic activity in human endothelial cells and vessel growth in ischemic limbs [5]; however, the specific domains and residues responsible for the anti-angiogenic effects of Bach1 had yet to be identified. Here, we show that unlike the full-length Bach1 protein, our Bach1-ΔBTB mutant, which lacks the BTB domain, does not impede angiogenesis when administered to the ischemic hind limbs of mice and fails to reduce proliferation, migration, and tube formation when overexpressed in human endothelial cells. The BTB domain was also essential for the Bach1-induced blockade of Wnt-targeted promoter activity (TCF/LEF) and gene expression (VEGF), for the binding of Bach1 to TCF4 and HDAC1, and for HDAC activation. Thus, the BTB domain appears to be directly involved in a number of molecular interactions that facilitate the anti-angiogenic activity of Bach1 [5].
Bach1 is a key factor in the regulation of oxidative stress, and acts as a molecular sensor of intracellular heme levels [22]. Recently, two companion papers demonstrate that Bach1 stabilization is essential for lung cancer metastasis by inhibiting HO-1 or glycolysis [23,24]. We have recently demonstrated that Bach1 is an essential factor in stem cell pluripotency, self-renewal, and lineage specification in human embryonic stem cells (hESCs), and the BTB domain of Bach1 is important for the interaction with polycomb repressive complex 2 subunit EZH2 [25]. Many BTB domain-containing proteins regulate gene transcription by interacting with non-BTB proteins and manipulating chromatin structure [26]. For example, our results are consistent with previous reports that Bach2 suppresses transcription via the recruitment of class II HDACs [27], and BCL6 regulates transcription by interacting with an SMRT/SIN3A/HDAC-containing complex [28,29]. BTB-containing proteins must form dimers or oligomers before binding the recognition sequences of target genes [30], perhaps because the isolated monomers remain unstructured [31], and the BTB domains of human and mouse Bach1 contain N-hook motifs that are essential for dimerization [32]. Furthermore, the results presented here indicate that the regions of Bach1 that mediate the Bach1-TCF4 and Bach1-HDAC1 interactions are located in a well-conserved region within the domain's helical core [33]: the residues involved in TCF4-Bach1 binding are located at the beginning of the α4 helix (residues 81–89), while the site of the HDAC1-Bach1 interaction extends from the end of helix α4 to the beginning of helix α6 (residues 90–126). Bach1, TCF4 and HDAC1 can form a complex to repress the TCF4-targeted genes [5]. Our findings demonstrate that Bach1 cannot bind to TCF4 and HDAC1 and had no inhibiting effect on angiogenesis after ischemic injury when lacking the BTB domain. In addition, Bach1, but not Bach1-ΔBTB, increased ROS generation and apoptosis and suppressed the expression of antioxidant genes (HO-1 and NQO1) in HUVECS, indicating that the BTB domain of Bach1 was also essential for the Bach1-induced oxidative stress response.
In summary, the results presented here show that the anti-angiogenic activity of Bach1 is crucially dependent on a number of molecular interactions that are mediated by the protein's BTB domain. Thus, peptides or small molecules that target the Bach1 BTB domain may improve recovery from ischemic injury or disease.
Author contributions
All authors contributed and critically reviewed and approved the manuscript. M.D. and L.S. conceived and designed the project. J.L., J.M., W.X., and G.J. performed experiments and analyzed the data. H.S., M.A., L.Q., and J.J. performed construction of vectors and statistical calculations. Z.X., W.X., Z.Y., and L.S. helped design experiments and interpreted the results. M.D. supervised the whole study and wrote the manuscript.
Declaration of competing interest
None.
Acknowledgments
Acknowledgments
We thank Dr. W. Kevin Cukier–Meisner for editorial assistance.
Funding sources
This work was supported by the Great Program (91639103 to D. Meng), and General Programs (81670450 and 81873469 to D. Meng, 81570081 and 81770083 to S. Li, 81800234 to L. Jiang, 81572713 to X. Zhi, 81873536 to X. Wang) of the National Natural Science Foundation of China, the National Key Research and Development Program of China (2018YFC1313600 to S. Li), and China Postdoctoral Science Foundation (2019M651371 and 2019T120303 to X. Wei). The funders have no roles in the study design, data collection, data analysis, interpretation, or writing of the report.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.102617.
Contributor Information
Shanqun Li, Email: li.shanqun@zs-hospital.sh.cn.
Dan Meng, Email: dmeng@fudan.edu.cn.
Appendix. Supplementary materials
References
- 1.He L., Huang X., Kanisicak O., Li Y., Wang Y., Li Y. Preexisting endothelial cells mediate cardiac neovascularization after injury. J Clin Invest. 2017;127(8):2968–2981. doi: 10.1172/JCI93868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z., Solomonidis E.G., Meloni M., Taylor R.S., Duffin R., Dobie R. Single-cell transcriptome analyses reveal novel targets modulating cardiac neovascularization by resident endothelial cells following myocardial infarction. Eur Heart J. 2019 doi: 10.1093/eurheartj/ehz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X., Guo J., Wei X., Niu C., Jia M., Li Q. Bach1: function, regulation, and involvement in disease. Oxid Med Cell Longev. 2018;2018 doi: 10.1155/2018/1347969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang L., Yin M., Xu J., Jia M., Sun S., Wang X. The transcription factor bach1 suppresses the developmental angiogenesis of zebrafish. Oxid Med Cell Longev. 2017;2017 doi: 10.1155/2017/2143875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang L., Yin M., Wei X., Liu J., Wang X., Niu C. Bach1 represses Wnt/beta-catenin signaling and angiogenesis. Circ Res. 2015;117(4):364–375. doi: 10.1161/CIRCRESAHA.115.306829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behrens J., von Kries J.P., Kuhl M., Bruhn L., Wedlich D., Grosschedl R. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382(6592):638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 7.Dejana E. The role of wnt signaling in physiological and pathological angiogenesis. Circ Res. 2010;107(8):943–952. doi: 10.1161/CIRCRESAHA.110.223750. [DOI] [PubMed] [Google Scholar]
- 8.Blankesteijn W.M., van Gijn M.E., Essers-Janssen Y.P., Daemen M.J., Smits J.F. Beta-catenin, an inducer of uncontrolled cell proliferation and migration in malignancies, is localized in the cytoplasm of vascular endothelium during neovascularization after myocardial infarction. Am J Pathol. 2000;157(3):877–883. doi: 10.1016/s0002-9440(10)64601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oyake T., Itoh K., Motohashi H., Hayashi N., Hoshino H., Nishizawa M. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16(11):6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerasimova T.I., Gdula D.A., Gerasimov D.V., Simonova O., Corces V.G. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82(4):587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 11.Stogios P.J., Downs G.S., Jauhal J.J., Nandra S.K., Prive G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6(10):R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Y., Qi Y., Kang J., Li N., Tian X., Yan C. Nerve growth factor promotes formation of lumen-like structures in vitro through inducing apoptosis in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2008;366(3):685–691. doi: 10.1016/j.bbrc.2007.11.160. [DOI] [PubMed] [Google Scholar]
- 13.Bonauer A., Carmona G., Iwasaki M., Mione M., Koyanagi M., Fischer A. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science (New York, NY) 2009;324(5935):1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 14.Meng D., Lv D.D., Fang J. Insulin-like growth factor-I induces reactive oxygen species production and cell migration through Nox4 and Rac1 in vascular smooth muscle cells. Cardiovasc Res. 2008;80(2):299–308. doi: 10.1093/cvr/cvn173. [DOI] [PubMed] [Google Scholar]
- 15.Meng D., Mei A., Liu J., Kang X., Shi X., Qian R. NADPH oxidase 4 mediates insulin-stimulated HIF-1alpha and VEGF expression, and angiogenesis in vitro. PLoS One. 2012;7(10):e48393. doi: 10.1371/journal.pone.0048393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muse E.D., Kramer E.R., Wang H., Barrett P., Parviz F., Novotny M.A. A whole blood molecular signature for acute myocardial infarction. Sci Rep. 2017;7(1):12268. doi: 10.1038/s41598-017-12166-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harpster M.H., Bandyopadhyay S., Thomas D.P., Ivanov P.S., Keele J.A., Pineguina N. Earliest changes in the left ventricular transcriptome postmyocardial infarction. Mamm Genome. 2006;17(7):701–715. doi: 10.1007/s00335-005-0120-1. [DOI] [PubMed] [Google Scholar]
- 18.Dhakshinamoorthy S., Jain A.K., Bloom D.A., Jaiswal A.K. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J Biol Chem. 2005;280(17):16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 19.Damani S., Bacconi A., Libiger O., Chourasia A.H., Serry R., Gollapudi R. Characterization of circulating endothelial cells in acute myocardial infarction. Sci Transl Med. 2012;4(126):126ra33. doi: 10.1126/scitranslmed.3003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quilici J., Banzet N., Paule P., Meynard J.B., Mutin M., Bonnet J.L. Circulating endothelial cell count as a diagnostic marker for non-ST-elevation acute coronary syndromes. Circulation. 2004;110(12):1586–1591. doi: 10.1161/01.CIR.0000142295.85740.98. [DOI] [PubMed] [Google Scholar]
- 21.Wang X., Liu J., Jiang L., Wei X., Niu C., Wang R. Bach1 induces endothelial cell apoptosis and cell-cycle arrest through ROS generation. Oxid Med Cell Longev. 2016;2016 doi: 10.1155/2016/6234043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davudian S., Mansoori B., Shajari N., Mohammadi A., Baradaran B. BACH1, the master regulator gene: a novel candidate target for cancer therapy. Gene. 2016;588(1):30–37. doi: 10.1016/j.gene.2016.04.040. [DOI] [PubMed] [Google Scholar]
- 23.Wiel C., Le Gal K., Ibrahim M.X., Jahangir C.A., Kashif M., Yao H. BACH1 stabilization by antioxidants stimulates lung cancer metastasis. Cell. 2019;178(2) doi: 10.1016/j.cell.2019.06.005. 330-45 e22. [DOI] [PubMed] [Google Scholar]
- 24.Lignitto L., LeBoeuf S.E., Homer H., Jiang S., Askenazi M., Karakousi T.R. Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of bach1. Cell. 2019;178(2) doi: 10.1016/j.cell.2019.06.003. 316-29 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei X., Guo J., Li Q., Jia Q., Jing Q., Li Y. Bach1 regulates self-renewal and impedes mesendodermal differentiation of human embryonic stem cells. Sci Adv. 2019;5(3):eaau7887. doi: 10.1126/sciadv.aau7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albagli O., Dhordain P., Deweindt C., Lecocq G., Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6(9):1193–1198. [PubMed] [Google Scholar]
- 27.Hoshino H., Nishino T.G., Tashiro S., Miyazaki M., Ohmiya Y., Igarashi K. Co-repressor SMRT and class II histone deacetylases promote Bach2 nuclear retention and formation of nuclear foci that are responsible for local transcriptional repression. J Biochem. 2007;141(5):719–727. doi: 10.1093/jb/mvm073. [DOI] [PubMed] [Google Scholar]
- 28.Ahmad K.F., Melnick A., Lax S., Bouchard D., Liu J., Kiang C.L. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12(6):1551–1564. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 29.Dhordain P., Lin R.J., Quief S., Lantoine D., Kerckaert J.P., Evans R.M. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26(20):4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katsani K.R., Hajibagheri M.A., Verrijzer C.P. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18(3):698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Igarashi K., Hoshino H., Muto A., Suwabe N., Nishikawa S., Nakauchi H. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for beta-globin locus control region complex. J Biol Chem. 1998;273(19):11783–11790. doi: 10.1074/jbc.273.19.11783. [DOI] [PubMed] [Google Scholar]
- 32.Ito N., Watanabe-Matsui M., Igarashi K., Murayama K. Crystal structure of the Bach1 BTB domain and its regulation of homodimerization. Genes Cells. 2009;14(2):167–178. doi: 10.1111/j.1365-2443.2008.01259.x. [DOI] [PubMed] [Google Scholar]
- 33.Rosbrook G.O., Stead M.A., Carr S.B., Wright S.C. The structure of the Bach2 POZ-domain dimer reveals an intersubunit disulfide bond. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 1):26–34. doi: 10.1107/S0907444911048335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.