Abstract
Background
Lipid accumulation has been highlighted in cancer development and progression, but the exact mechanism remains unclear in renal cell carcinoma (RCC). MicroRNAs (miRNAs) have been confirmed to participate in the pathological processes of cancers, including tumour occurrence and inhibition. However, the role and mechanism of miR-765 have not been elucidated in clear cell renal cell carcinoma (ccRCC).
Methods
Using The Cancer Genome Atlas (TCGA) database and qRT-PCR, we investigated differences in miR-765 and proteolipid protein 2 (PLP2) expression, as well as their clinical relevance. To investigate the function of miR-765 and PLP2 in ccRCC, we performed in vitro and in vivo experiments to explore their biological functions in ccRCC.
Findings
In this study, we showed that miR-765 was upregulated in the plasma of ccRCC patients after tumour resection. Consistently, ccRCC tissues had low expression of miR-765 when compared with corresponding non-cancerous tissues. Overexpression of miR-765 suppressed cell proliferation and metastasis in vitro and in vivo. Mechanistic studies demonstrated that PLP2 was a direct target gene of miR-765. PLP2 was highly expressed in ccRCC tissues, and high PLP2 levels were positively correlated with higher tumour stage and grade and poor prognosis. PLP2 expression was negatively correlated with the miR-765 level in patient samples. We further showed that PLP2 restrained the cell metastasis and proliferation induced by miR-765 and reduced the lipid-eliminating effects of miR-765 in renal cancer cells.
Interpretation
Our findings suggest that miR-765 may function as a tumour suppressor and eliminate lipids in clear cell renal cell carcinoma by targeting PLP2.
Funding
This work was funded the grants from the National Natural Scientific Foundation of China (Grant No. 81672528, 81672524, 81602218, 31741032, 81902588).
Keywords: miR-765, ccRCC, PLP2, Lipid eliminator
Research in context.
Evidence before this study
Clear cell renal cell carcinoma (ccRCC) is one of the most common types of RCC and is named because of the lipid deposition in the cytoplasm. However, the exact mechanism remains unclear. Therefore, it is urgent to explore biomarkers for diagnosis, the causes of lipid accumulation, and whether eliminating lipid accumulation is conducive to inhibiting tumour growth.
Added value of this study
Here, we found that miR-765 was upregulated in the plasma of ccRCC patients after tumour resection and that ccRCC tissues had low expression of miR-765. Overexpression of miR-765 suppressed cell proliferation and metastasis in vitro and in vivo. Furthermore, we identified proteolipid protein 2 (PLP2) as a direct target gene of miR-765. PLP2 was highly expressed in ccRCC tissues, and high PLP2 indicated an advanced tumour stage and grade and poor prognosis. PLP2 inhibited cell metastasis and proliferation and the lipid eliminating abilities of miR-765 in renal cancer cells.
Implications of all the available evidence
This study identified the novel plasma biomarker miR-765 and the miR-765-PLP2 axis that regulates lipid metabolism in ccRCC. The results suggest that the miR-765-PLP2 axis serves as a clinical biomarker and a therapeutic target in ccRCC.
Alt-text: Unlabelled box
1. Introduction
Metabolic abnormalities are a characteristic of tumours [1], and dysfunction of lipid metabolism is one of the most important features of renal cell carcinoma (RCC), especially the clear cell type (ccRCC). The latest research shows that 73,820 new cases, and 14,770 deaths as a result of kidney and renal pelvis cancer are estimated to have occurred in the United States [2]. ccRCC is responsible for the most cancer-related deaths class accounts for 70–85% of RCC [3]. RCC is considered a metabolic disease [4], [5], [6], and obesity and atherosclerosis have been reported to be risk factors for RCC [7,8]. ccRCC is known for abnormal lipid accumulation [4] of cholesterol, cholesterol esters, and neutral lipids (triglycerides) [9]. However, the molecular mechanisms and significance of eliminating this lipid accumulation remain unclear. miRNAs are non-coding RNAs that contribute to multiple cellular processes, such as cancer cell proliferation [10,11], apoptosis [12,13], antiangiogenic mechanisms [14] and metastasis [15,16]. They may act as tumour suppressors or oncogenes by regulating gene transcription [17,18]. It has been shown that circulating miRNAs may act as biomarkers of gait speed in obese adults [19] or have a prognostic role in cancer [20]. More recently, several articles confirmed that plasma miRNAs could be biomarkers of RCC, such as miR-144–3p and miR-210 [21,22]. Previous studies have demonstrated that miR-765 inhibits osteosarcoma [23] and tongue squamous cell carcinoma [24]. Nevertheless, to the best of our knowledge, there is still no relevant research about the role of miR-765 in ccRCC. We found that the plasma level of miR-765 was upregulated after tumour resection in ccRCC patients and that ccRCC tissues had a low level of miR-765 compared with corresponding non-ccerous tissues. The present study systematically describes the function and role of miR-765 in ccRCC. Functionally, miR-765 inhibited RCC cell proliferation, migration and invasion in vitro and in vivo. Mechanistically, miR-765 serves as a tumour suppressor and eliminates lipids by downregulating PLP2 in ccRCC. Our findings reveal that the miR-765-PLP2 axis may be a biomarker and a therapeutic target for ccRCC.
2. Materials and methods
2.1. Patient samples and cells
Thirty-six clear cell renal cell carcinoma (ccRCC) patient surgical specimens, 18 non-ccRCC (nccRCC) patient surgical specimens and 18 blood plasma samples (preoperative and operational day 7 without chemotherapy or radiotherapy for ccRCC) were obtained from the Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology between 2016 and 2018. Informed consent was provided and obtained from patients, and the experimental and study procedures were approved by the Institutional Review Board of Huazhong University of Science and Technology. The Cancer Genome Atlas (TCGA) data were obtained from http://cancergenome.nih.gov/, and Oncomine data were obtained from https://www.oncomine.org. The microarray platform used was the Agilent Human miRNA Microarray 18.0 (Agilent Technologies, Santa Clara, USA), which included 1523 human miRNAs from plasma obtained from 5 patients with ccRCC preoperatively and at day 7 postoperatively. The miRNA Microarray cohort was upload to the Gene Expression Omnibus (GEO) dataset and had been assigned accession number: GSE140835. HK2, 786-O, A498 and Caki-1 cells were cultivated in high-glucose DMEM (Wuhan Boster Biological Technology, Ltd., Wuhan, China) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), as previously described [15].
2.2. RNA extraction and qRT-PCR
RNA extraction and qRT-PCR were performed with the TRizol reagent (Thermo, Massachusetts, USA) and SYBR Green mix (Thermo, Massachusetts, USA) according to the instructions of the manufacturers. The experimental procedure was performed as previously described [10]. The relative expression of miR-765 and PLP2 was calculated by the 2−ΔCt method (ΔCt = Ctgene–Ctnormalizer). The primers for miR-765 and U6 were purchased from RiboBio (RiboBio, Guangzhou, China). The primers for PLP2 and GAPDH were purchased from GENEWIZ (GENEWIZ, Suzhou, China):
PLP2:
Forward: 5′- ATTCATCAACTGGCCCTGGA-3′,
Reverse: 5′- AACGGGGAAGGTGACATAGG-3′,
GAPDH:
Forward: 5′-GAGTCAACGGATTTGGTCGT-3′,
Reverse: 5′-GACAAGCTTCCCGTTCTCAG-3′.
2.3. Cell transfection and reagents
Lentiviruses containing human miR-765 or negative control (NC) and plasmids expressing PLP2 or NC were synthesized by GeneChem (Shanghai, China). MiR-765 mimics, inhibitor and NC were synthesized from RiboBio (Guangzhou, China). Lentiviruses, miR-765 mimics, inhibitor, PLP2 plasmid and NC were transfected in 6-well plates according to the recommendations of the manufacturer as previously described [21]. Oil Red O (ORO) was provided by Wuhan Servicebio Technology. A triglyceride assay kit was purchased from Nanjing Jiancheng Bioengineering Institute [25].
2.4. Cell proliferation analysis
A498 and Caki-1 cells were transfected with miR-765 lentiviruses, mimics, PLP2 plasmid and NC, and then the cells were added to a 96-well plate. The cell proliferation rate (OD value) was detected by Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc., Rockville, MD, USA) according to the manufacturer's protocol as previously described [10].
2.5. Migratory and invasion assays
Cells were withdrawn from plasma for 24 h, and the experimental procedure was performed with polycarbonate membrane inserts (Corning, New York, USA) with or without Matrigel (Thermo Fisher Scientific, Waltham, USA) as previously described [26].
2.6. Luciferase assays
Wild-type and mutant PLP2 3′UTR reporters were purchased from RiboBio (RiboBio, Guangzhou, China). Tumour cells transfected with miR-765 mimics or NC in 24-well plates were cotransfected with 500 ng luciferase reporter. All dual luciferase assays were measured using the Dual Luciferase Assay (Promega, Madison, USA). The experimental procedure was performed as previously described.
2.7. Western blotting
Tissues and cells were obtained by protein lysis with RIPA buffer (Wuhan Boster Biological Technology, Ltd., Wuhan, China), PMSF (Wuhan Boster Biological Technology, Ltd., Wuhan, China) and a protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA). Protein concentrations were measured with a bicinchoninic acid kit (Beyotime Institute of Biotechnology). Western blotting was performed according to the manufacturer's instructions with polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Bedford, MA, USA); proteins were blocked in PBS with 5% non-fat milk and incubated with antibodies against PLP2 (1:1000; A06255; Boster Biological Technology, Ltd., USA) and GAPDH (1:2000; BM3876; Wuhan Boster Biological Technology, Ltd., Wuhan, China) overnight. After 14 h, membranes were incubated with secondary antibodies (1:5000; BA1020; Wuhan Boster Biological Technology, Ltd., Wuhan, China) for 2 h, and the signal was detected by the ChemiDoc XRS+ system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
2.8. Immunohistochemistry (IHC)
Adjacent normal, ccRCC tissues and xenograft tumour were fixed in formalin, dehydrated, and embedded, and then the tissue sections were incubated with primary mouse PLP2 (1:250, ab122792, Abcam) or POU2F2 (1:100, A2776, Abclonal), overnight at 4 °C. The sections were washed three times with PBS, and then the sections were incubated with the secondary antibody for 2 h at room temperature.
2.9. Tumour formation in nude mice
All animal experiments were performed in accordance with the animal protocols approved by the Institutional Animal Use and Care Committee of Huazhong University of Science and Technology. 16 male mice (Beijing HFK Bioscience, Beijing, China) were randomly divided into four groups. Caki cells (1 × 106) with transfected lentiviruses containing human miR-765 and/or PLP2 were injected subcutaneously into the flanks of mice or 2 × 106 Caki-1 intravenously injection through tail vein. The tumour size was measured every 3 days. After 30 days, the mice were euthanized, and the tumours were harvested. Tumour volumes were calculated as follows: volume = (D × d2)/2.
2.10. Bioinformatics analysis
Gene set enrichment analysis (GSEA) was applied to evaluate the role of PLP2 in the pathogenesis of KIRC (http://www.broadinstitute.org/gsea) [27]. A false discovery rate (FDR) <25% and p<0.05 were considered as statistically significant, as previously described [26]
2.11. Statistical analysis
The data of paired samples were analysed with a paired t-test. The samples from four groups were analysed with one-way ANOVA (#), and unpaired samples from two groups were analysed with a t-test (*). Kaplan–Meier (KM) curves were generated to assess the expression level of PLP2, and the survival rate was assessed with a log-rank test. Univariate and multivariate Cox proportional hazard regression models were used to analyse the prognostic significance of PLP2. Error bars represent the mean ± SEM of data; the cut-off for a statistically significant p value and the statistical tools were described previously [10].
3. Results
3.1. MiR-765 is upregulated after tumour resection in plasma and has a low level in the cancer tissues of ccRCC patients
A microarray platform was used to determine the specific plasma miRNAs associated with ccRCC. We compared 5 paired preoperative plasma samples with postoperative-day-7 plasma samples. As shown in Fig. 1(a) and (b), 5 miRNAs were significantly upregulated in postoperative plasma samples compared to preoperative plasmas, and miR-765 was upregulated most significantly. Then, we extended this analysis to 18 patients with ccRCC (Fig. 1(c)). Similarly, the expression of miR-765 was significantly higher in non-cancerous normal tissues than in ccRCC tissues (Fig. 1(d)). Furthermore, the expression of miR-765 was significantly lower in ccRCC tissues from patients from The Cancer Genome Atlas ccRCC dataset (TCGA-KIRC) than in non-cancerous normal tissues (Fig. 1(e)). There was no difference in the expression of miR-765 in The Cancer Genome Atlas kidney chromophobe dataset (TCGA-KICH) or kidney renal papillary cell carcinoma dataset (TCGA-KIRP) (Fig. 1(f) and (g)). We found no difference in miR-765 expression between 18 non-ccRCC (nccRCC) patient cancer tissues and non-cancerous normal tissues. Unfortunately, there was no difference in overall survival (OS) and disease–free survival (DFS) in ccRCC patients or TCGA-KIRC with different miR-765 expression (Supplementary Fig. 1(a)–(d)).
Fig. 1.
Integrated analysis of ccRCC identifies miR-765. (A) and (B) Volcano plot and heat map of plasma miRNA array analysis of preoperative and postoperative plasma samples from patients with ccRCC. The level of miR-765 in plasma were upregulated most significantly after tumour resection. (C) The levels of miR-765 in plasma were upregulated in postoperative samples were compared with preoperative samples in 18 patients with ccRCC. (D) and (E) MiR-765 was significantly higher in normal tissues than in ccRCC tissues in clinical samples and in data from The Cancer Genome Atlas kidney renal clear cell carcinoma (TCGA-KIRC). (F) and (G) There was no difference in miR-765 expression in The Cancer Genome Atlas kidney chromophobe (TCGA-KICH) and kidney renal papillary cell carcinoma (TCGA-KIRP). (H) There was no difference in miR-765 expression in 18 patients with non-ccRCC (nccRCC). Data indicate the means ± SEM. *** P < 0.001 (t-test).
3.2. MiR-765 inhibits the malignant potential of renal cancer cells in vitro
As miR-765 was significantly downregulated in ccRCC tissues, we tested miR-765 expression in the human kidney 2 (HK-2) cell line, a proximal tubular cell line derived from normal kidney and three renal cancer cell lines (786-O, A498 and Caki-1). A498 and Caki-1 cells had lower expression levels of miR-765 than 786-O and HK-2 cells (Fig. 2(a)). Then, we assessed the possible role of miR-765 in cancer cell malignancy in A498 and Caki-1 cells. We overexpressed miR-765 in A498 and Caki-1 cells with lentiviruses carrying miR-765 or a control miRNA (NC) (Fig. 2(b)). MiR-765 overexpression restrained the proliferation and clonogenicity of renal cancer cells (Fig. 2(c) and (d)). Overexpression of miR-765 also significantly repressed migration and invasion abilities as assessed by transwell assays (Fig. 2(e) and (f)).
Fig. 2.
MiR-765 inhibits the malignant potential of renal cancer cells in vitro and in vivo. (A) MiR-765 was downregulated in renal cancer cell lines. (B) Lentiviruses carrying miR-765 or control miRNA (NC) were transfected into renal cancer cell lines. (C) and (D) MiR-765 inhibited cell proliferation in A498 and Caki-1 cells. (E) and (F) Representative pictures showing the effect of miR-765 on the migration and invasion capabilities A498 and Caki-1 cells. Original magnification ×200. Data indicate the means ± SEM. * P < 0.05; ** P < 0.01; *** P < 0.001 (t-test).
3.3. Target genes of miR-765 in ccRCC as predicted with bioinformatics analysis
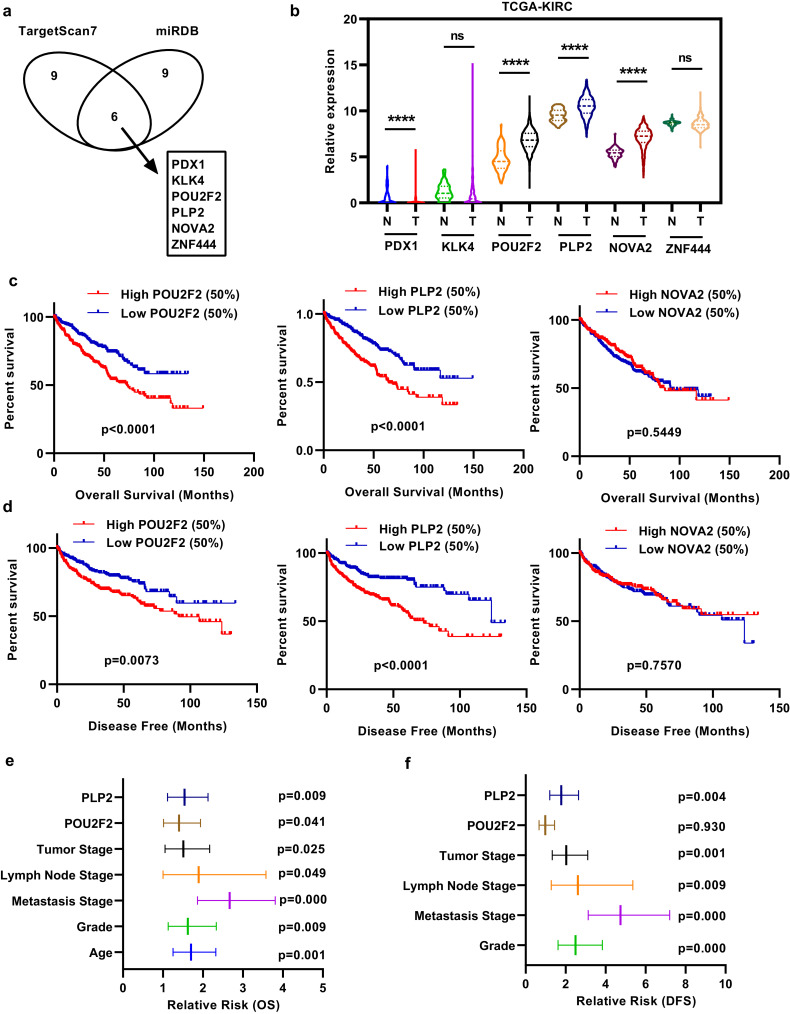
Prediction software (miRDB and TargetScan) was used to predict potential target molecules of miR-765. The top 15 scores of each prediction were used for analysis, and 6 molecules were identified (Fig. 3(a)). We merged this result with correlation analysis in the TCGA-KIRC database to narrow down prospective candidates. The column chart indicates the expression of the 6 target molecules (Fig. 3(b)). As shown in Fig. 3(b), pancreatic and duodenal homeobox 1 (PDX1), POU class 2 homeobox 2 (POU2F2), proteolipid protein 2 (PLP2), and NOVA alternative splicing regulator 2 (NOVA2) had significantly higher expression in ccRCC tissues than in corresponding non-cancerous normal tissues, and PDX1 expression was very low. Kaplan–Meier analysis determined the association between the expression of POU2F2, PLP2 and NOVA2 and OS and DFS in TCGA-KIRC. Patients with high POU2F2 or PLP2 expression exhibited shorter OS and DFS times than those with low POU2F2 or PLP2 expression (Fig. 3(c) and (d)). Univariate and multivariate survival analysis of OS and DFS indicated that high PLP2 expression but not high POU2F2 expression was a potential independent prognostic factor in ccRCC patients (Fig. 3(e) and (f), Tables 1 and 2). IHC showed that both POU2F2 and PLP2 were upregulated in ccRCC (Supplementary Fig. 2(a) and (b)).
Fig. 3.
Target genes of miR-765 predicted with bioinformatics analysis in ccRCC. (A) Bioinformatic prediction of the top 15 candidate mRNAs targeting miR-765 via the TargetScan and miRDB platforms. (B) Column chart depicting the expression of the 6 target genes in TCGA-KIRC. (C) and (D) Overall survival (OS) and disease-free survival (DFS) analysis showed 3 target genes in TCGA-KIRC. (E) and (F) Univariate and multivariate survival analysis of OS and DFS indicated that high PLP2 expression was a potential independent prognostic factor in ccRCC patients. Data indicate the means ± SEM.****P < 0.0001 (t-test).
Table 1.
Univariate and multivariate analyses of POUF2 and PLP2 mRNA level and patient overall survival.
| Univariate analysis |
Multivariate analysisc |
|||||
|---|---|---|---|---|---|---|
| Variable | HRa | 95%CIb | P | HR | 95% CI | P |
| Overall survival | ||||||
| Age (years) | ||||||
| ≤60 vs. >60 | 1.758 | 1.294–2.388 | 0.000 | 1.704 | 1.251–2.321 | 0.001 |
| Gender | ||||||
| Female vs. Male | 0.943 | 0.693–1.284 | 0.825 | |||
| T stage | ||||||
| T3 or T4 vs. T1 or T2 | 3.120 | 2.306–4.220 | 0.000 | 1.511 | 1.053–2.169 | 0.025 |
| N stage | ||||||
| N1 vs. N0 or NX | 3.917 | 2.121–7.234 | 0.000 | 1.894 | 1.002–3.579 | 0.049 |
| M stage | ||||||
| M1 vs. M0 or MX | 4.303 | 3.155–5.868 | 0.000 | 2.667 | 1.865–3.81 | 0.000 |
| Grade | ||||||
| G3 or G4 vs. G1 or G2 | 2.613 | 1.865–3.661 | 0.000 | 1.624 | 1.130–2.335 | 0.009 |
| POU2F2 | ||||||
| High vs. Low | 1.883 | 1.378–2.575 | 0.000 | 1.402 | 1.014–1.939 | 0.041 |
| PLP2 | ||||||
| High vs. Low | 2.053 | 1.506–2.798 | 0.000 | 1.540 | 1.114–2.128 | 0.009 |
Hazard ratio, estimated from Cox proportional hazard regression model.
Confidence interval of the estimated HR.
Multivariate models were adjusted for T, N, M classification and age.
Table 2.
Univariate and multivariate analyses of POUF2 and PLP2 mRNA level and patient disease–free survival.
| Univariate analysis |
Multivariate analysisc |
|||||
|---|---|---|---|---|---|---|
| Variable | HRa | 95%CIb | P | HR | 95% CI | P |
| Disease–free survival | ||||||
| Age (years) | ||||||
| ≤60 vs. >60 | 1.358 | 0.954–1.935 | 0.090 | |||
| Gender | ||||||
| Female vs. Male | 1.425 | 0.959–2.118 | 0.079 | |||
| T stage | ||||||
| T3 or T4 vs. T1 or T2 | 4.601 | 3.185–6.646 | 0.000 | 1.969 | 1.287–3.010 | 0.002 |
| N stage | ||||||
| N1 vs. N0 or NX | 5.991 | 3.008–11.933 | 0.000 | 3.080 | 1.465–6.478 | 0.003 |
| M stage | ||||||
| M1 vs. M0 or MX | 8.627 | 5.943–12.522 | 0.000 | 4.876 | 3.210–7.408 | 0.000 |
| Grade | ||||||
| G3 or G4 vs. G1 or G2 | 3.453 | 2.287–5.214 | 0.000 | 2.418 | 1.570–3.723 | 0.000 |
| POU2F2 | ||||||
| High vs. Low | 1.684 | 1.172–2.420 | 0.005 | 0.966 | 0.654–1.427 | 0.862 |
| PLP2 | ||||||
| High vs. Low | 2.551 | 1.751–3.717 | 0.000 | 1.816 | 1.217–2.710 | 0.004 |
Hazard ratio, estimated from Cox proportional hazard regression model.
Confidence interval of the estimated HR.
Multivariate models were adjusted for T, N and M classification.
3.4. PLP2 is significantly upregulated and predicts poor prognosis in TCGA-KIRC
We investigated PLP2 expression and its ability to predict disease progression in TCGA-KIRC. The expression of PLP2 was significantly higher in ccRCC tissues than in non-cancerous normal tissues (Fig. 4(a) and (b)). The outcome was further confirmed in five additional data sets from the Oncomine database (https://www.oncomine.org) (Fig. 4(c)). High PLP2 expression was found in advanced renal cancer compared with early renal cancer (Fig. 4(d)). High PLP2 expression was also confirmed in patients with lymphatic metastasis or distant metastasis (Fig. 4(e) and (f)). A high level of PLP2 had a positive correlation with advanced grade and pathological TNM stage (T stage, lymphatic metastasis, and distant metastasis grouping) in ccRCC patients (Fig. 4(g) and (h)). Patients with high PLP2 expression exhibited a higher risk of death and recurrence (Fig. 4(i)).
Fig. 4.
PLP2 is significantly upregulated and predicts poor prognosis in the TCGA-KIRC dataset containing 72 normal tissues and 533 ccRCC tissues. (A) and (B) PLP2 was upregulated in cancer tissues compared with para-cancerous tissues. (C) PLP2 expression levels in five additional ccRCC data sets. The levels of PLP2 were compared across different clinicopathological parameters: (D) T stage, (E) lymphatic metastasis, (F) distant metastasis, (G) TNM stage (H) grade and (I) cancer-related death. Data indicate the means ± SEM. *P<0.05; **P<0.01, *** P < 0.001, **** P < 0.0001 (t-test), ### P<0.001(one-way ANOVA).
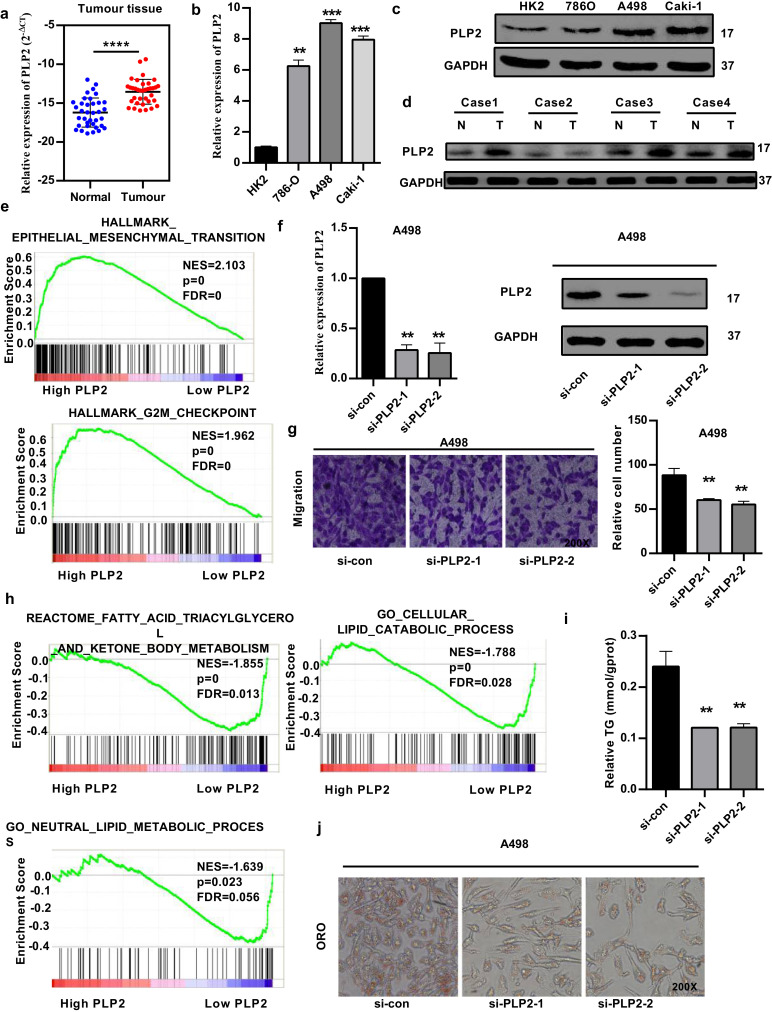
3.5. PLP2 is involved in the biological pathogenesis of ccRCC
As PLP2 was upregulated significantly and predicted poor prognosis in TCGA-KIRC, we tested PLP2 expression with quantitative real-time polymerase chain reaction (qRT-PCR) and western blotting in ccRCC patient samples and renal cancer cells (Fig. 5(a)–(d)). Gene set enrichment analysis (GSEA) revealed that high PLP2 expression was associated with epithelial mesenchymal transition (EMT) and the G2M checkpoint (Fig. 5(e)). Then, we evaluated the role of PLP2 in RCC cell lines. Using small RNA interference technology, we constructed two siRNAs (si-PLP2-1 and si-PLP2-2) and si-con, as shown in Fig. 5(f). Transfection with the siRNAs significantly repressed the migration of cancer cells (A498) (Fig. 5(g)). RCC is considered a metabolic disease, and GSEA demonstrated that high PLP2 expression was associated with fatty acid triacylglycerol metabolism, lipid catabolic processes and neutral lipid metabolic processes (Fig. 5(h)). Silencing of PLP2 significantly promoted neutral lipid catabolic processes (involving triacylglycerol) (Fig. 5(i)) and eliminated abnormal lipid accumulation (Fig. 5(j)) in A498 cancer cells.
Fig. 5.
PLP2 is involved in the biological pathogenesis of ccRCC. (A-D) PLP2 is upregulated in ccRCC tissues and renal cancer cells, as assessed by qRT-PCR and western blotting analysis. (E) Gene set enrichment analysis (GSEA) was used to compare high and low PLP2 expression groups in the TCGA database. Enrichment curves are shown for activated gene sets related to EMT and the G2M checkpoint. (F) Silencing of PLP2 with small RNA interference technology (si-PLP2-1 and si-PLP2-2). (G) si-PLP2 significantly repressed the migration of A498 cells. (H) GSEA enrichment curves showed that low PLP2 expression was associated with fatty acid triacylglycerol metabolism, lipid catabolic processes and neutral lipid metabolic processes. (H) and (I) Silencing of PLP2 significantly promoted neutral lipid catabolic processes and eliminated abnormal lipid accumulation in A498 cancer cells. Original magnification 200x. Data indicate the means ± SEM. **P<0.01, *** P < 0.001 (t-test).
3.6. PLP2 is a direct target of miR-765
To further confirm that PLP2 was a direct target of miR-765, we first determined that PLP2 was negatively correlated with miR-765 in TCGA-KIRC and clinical ccRCC samples (Fig. 6(a) and (b)). qRT-PCR and western blotting were used to verify the protein and mRNA level changes seen with the miR-765 mimics and inhibitor. The results revealed that miR-765 affected both the mRNA and protein levels of PLP2 in renal cancer cells (Fig. 6(c)–(f)). A luciferase reporter assay using wild-type (WT) or mutated (MUT) PLP2 in A498 and Caki-1 cells confirmed that miR-765 binds to the 3′-untranslated region (UTR) of PLP2 (Fig. 6(g) and (h)). These results suggest that PLP2 is a direct target of miR-765.
Fig. 6.
PLP2 is a direct target of miR-765. (A) and (B) PLP2 was negatively correlated with miR-765 in TCGA-KIRC and clinical ccRCC samples. (C-F) qRT-PCR and western blotting analysis of PLP2 expression in miR-765 mimic-, inhibitor- and NC-transfected A498 and Caki-1 cells. (G) and (H) Luciferase reporter assays showing approximately 50% decreased reporter activity after transfection of the wild-type PLP2 3′UTR reporter construct into A498 and Caki-1 cells in combination with miR-765 mimics. Data indicate the means ± SEM. *P<0.05; **P<0.01 (t-test).
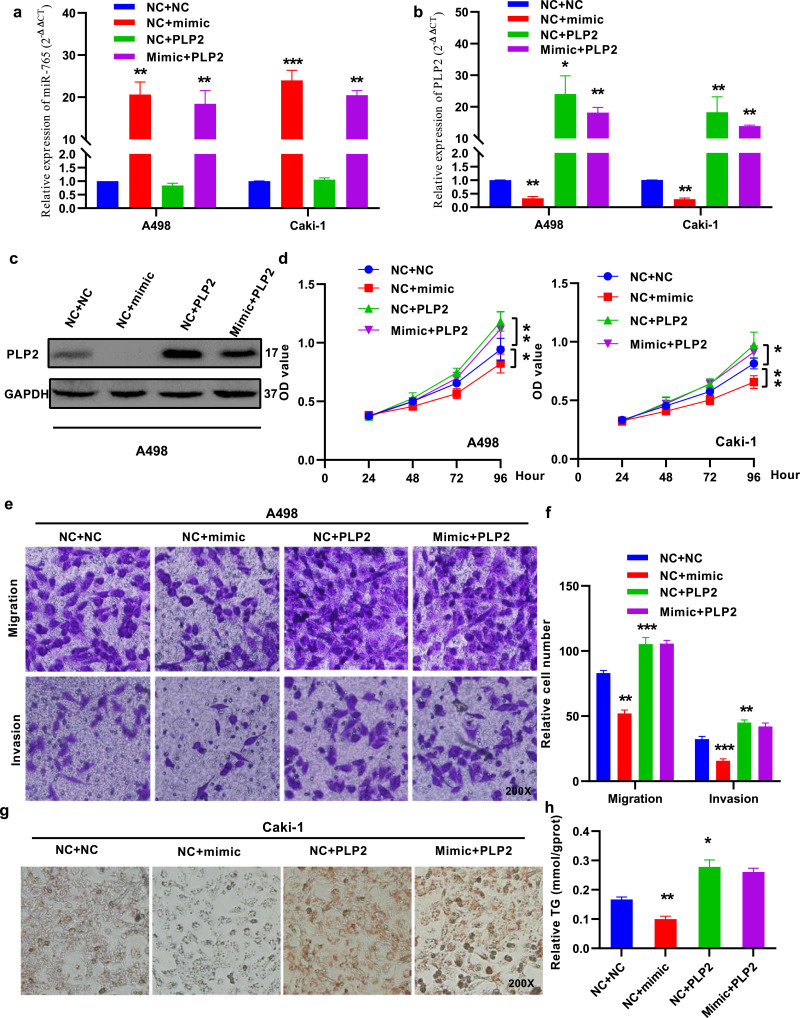
3.7. PLP2 reverses the function of miR-765 in renal cancer
To determine whether PLP2 reversed the suppressive effect of miR-765, we co-transfected miR-765 mimics and PLP2 into renal cancer cells (Fig. 7(a) and (b)). Overexpression of PLP2 reversed the miR-765-induced inhibition of proliferation in A498 and Caki-1 cells (Fig. 7(c) and (d)) and the migration and invasion of A498 cells (Fig. 7(e) and (f)). In addition, the lipid-eliminating role of miR-765 was also reversed by PLP2 in Caki-1 cells (Fig. 7(g) and (h)). Taken together, these data demonstrated that PLP2 reversed the tumour suppression of miR-765 in RCC.
Fig. 7.
PLP2 reverses the function of miR-765 in renal cancer. (A) and (B) The expression of miR-765 and PLP2 was measured by qRT-PCR in A498 and Caki-1 cells. (C) The expression of PLP2 was measured by western blotting in A498 cells. (D) PLP2 overexpression impaired the effects of the miR-765 mimic on cell proliferation in A498 and Caki-1 cells. (E) and (F) PLP2 overexpression impaired the effects of the miR-765 mimic on migration and invasion in A498 cells. (G) and (H) PLP2 overexpression impaired the effects of the miR-765 mimic on lipid elimination in A498 cells. Original magnification 200x. Data indicate the means ± SEM. *P<0.05; **P<0.01, *** P < 0.001 (t-test).
3.8. MiR-765 inhibits malignancy and eliminates lipids in renal cancer cells by PLP2 in vivo
Subcutaneous and intravenously injection of Caki-1 cells stably expressing miR-765 or/and PLP2 into male nude mice was used to demonstrate that miR-765 can inhibit the ccRCC tumour growth induced by PLP2 in vivo. The sizes of subcutaneous tumours were reduced by miR-765, and PLP2 reversed the effect of miR-765 on tumour growth (Fig. 8(a)) and liver metastasis (Fig. 8(d)). The average weight and volume of miR-765-overexpressing tumours were decreased significantly compared to those in the NC group, and overexpression of PLP2 reversed the effect of miR-765 on tumour weight and tumour volume (Fig. 8(b) and (c)). MiR-765 reduced the expression of PLP2 and eliminated lipids, and overexpression of PLP2 increased lipid accumulation and reversed the effect of miR-765, as indicated by ORO staining (Fig. 8(e)). In addition, PLP2 can reverse the angiogenesis-inhibiting effect with vascular endothelial growth factor A (VEGFA) and tumour metastasis ability by Ki67 of miR-765 (Fig. 8(f)). In summary, these data indicated that miR-765 inhibited cell malignancy and eliminated lipids by targeting PLP2 in renal cancer cells.
Fig. 8.
MiR-765 inhibits the malignant potential of renal cancer cells and eliminates lipids in renal cancer cells via PLP2 in vivo. (A), (B) and (C) Tumour size, weight and volume curves from the xenograft formation assay in which Caki-1 cells were subcutaneously injected into the flanks of mice and grown for 30 days. Tumour size was measured every 3 days. (D) PLP2 reversed the effect of miR-765 on tumour liver metastasis. (E) Oil Red O (ORO) staining showed PLP2 reversed the effect of miR-765 on tumour lipid accumulation. (F) PLP2 reversed the angiogenesis-inhibiting effect with vascular endothelial growth factor A (VEGFA) and tumour metastasis ability by Ki67 of miR-765. Original magnification x200. Data indicate the means ± SEM. * P < 0.05; ** P < 0.01; *** P < 0.001 (t-test).
4. Discussion
Currently, many studies have confirmed that circulating miRNAs are dysregulated in patient plasma and can serve as tumour biomarkers [21,22]. Here, for the first time, we demonstrated that miR-765 was upregulated in the plasma of ccRCC patients after tumour resection and that ccRCC tissues had a lower expression of miR-765 than non-cancerous control tissues. MiR-765 was shown to be a tumour suppressor in osteosarcoma [23] and tongue squamous cell carcinoma [24]. Other studies indicated that miR-765 was upregulated in hepatocellular carcinoma and melanoma [28,29]. However, the level and function of miR-765 in ccRCC remain unknown. In this study, miR-765 was significantly downregulated in the plasma and cancer tissues of ccRCC patients and in renal cancer cells. Overexpression of miR-765 inhibited the proliferation and motility of RCC cells in vitro and in vivo. Thus, we identified miR-765 as a tumour suppressor in renal cancer. miRDB (http://mirdb.org/miRDB) and TargetScan (http://www.targetscan.org) were used to determine the candidate genes of miR-765, and proteolipid protein 2 (PLP2) was verified to be a potential functional downstream target. Clinical data analysis found that miR-765 had a negative association with PLP2 in human ccRCC samples. PLP2 was shown to function as an oncogene in hepatocellular carcinoma [30], breast cancer [31] and glioma [32]. However, the function of PLP2 and miRNAs in regulating PLP2 expression in ccRCC remains unknown.
We analysed PLP2 expression and its prognostic role in TCGA-KIRC. PLP2 was significantly upregulated and predicted poor prognosis in ccRCC patients. GSEA demonstrated that high PLP2 expression was significantly associated with EMT, the G2M checkpoint, fatty acid triacylglycerol metabolism, lipid catabolic processes and neutral lipid metabolic processes in ccRCC. Silencing of PLP2 impaired cell proliferation, migration and invasion, promoted neutral lipid catabolic processes and eliminated abnormal lipid accumulation in RCC cells. Overexpression of PLP2 reversed the effects of miR-765 on cell growth, malignant potential and lipid accumulation in RCC cells. Our findings reveal that miR-765 could be a tumour suppressor and eliminate lipids by downregulating PLP2 in ccRCC.
In summary, miR-765 can inhibit cell proliferation and malignant potential and promote lipid catabolic processes in RCC by directly downregulating PLP2. This is the first study to identify PLP2 as a potential target gene of miR-765 in RCC. Low plasma levels of miR-765 may be a novel biomarker, and PLP2 could be a novel predictor and therapeutic target in human ccRCC. However, our research may be limited, and further work is needed.
Declaration of Competing Interest
The authors declare no conflicts of this manuscript.
Acknowledgments
Funding sources
This work was funded the grants from the National Natural Scientific Foundation of China (Grant no. 81672528, 81672524, 81602218, 31741032, 81902588). The funders have no roles in study design, data collection, data analysis, interpretation, or writing of the report.
Ethics statement
This study was approved by the Ethics Committees of Huazhong University of Science and Technology, and all aspects of the study comply with the criteria established by the Declaration of Helsinki.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.102622.
Contributor Information
Xiaoping Zhang, Email: xzhang@hust.edu.cn.
Xuegang Wang, Email: wangxuegang@xmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Finley L.W., Zhang J., Ye J., Ward P.S., Thompson C.B. SnapShot: cancer metabolism pathways. Cell Metab. 2013;17(3):466–4e2. doi: 10.1016/j.cmet.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh J.J., Purdue M.P., Signoretti S., Swanton C., Albiges L., Schmidinger M. Renal cell carcinoma. Nat Rev Dis Primer. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wettersten H.I., Aboud O.A., Lara P.N., Jr., Weiss R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol. 2017;13(7):410–419. doi: 10.1038/nrneph.2017.59. [DOI] [PubMed] [Google Scholar]
- 5.Hakimi A.A., Reznik E., Lee C.H., Creighton C.J., Brannon A.R., Luna A. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29(1):104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinnaird A., Dromparis P., Saleme B., Gurtu V., Watson K., Paulin R. Metabolic modulation of clear-cell renal cell carcinoma with dichloroacetate, an inhibitor of pyruvate dehydrogenase kinase. Eur Urol. 2016;69(4):734–744. doi: 10.1016/j.eururo.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Hager M., Mikuz G., Kolbitsch C., Moser P.L. Association between local atherosclerosis and renal cell carcinomas. Nutr Cancer. 2008;60(3):364–367. doi: 10.1080/01635580802067241. [DOI] [PubMed] [Google Scholar]
- 8.Psutka S.P., Stewart S.B., Boorjian S.A., Lohse C.M., Tollefson M.K., Cheville J.C. Diabetes mellitus is independently associated with an increased risk of mortality in patients with clear cell renal cell carcinoma. J Urol. 2014;192(6):1620–1627. doi: 10.1016/j.juro.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Saito K., Arai E., Maekawa K., Ishikawa M., Fujimoto H., Taguchi R. Lipidomic signatures and associated transcriptomic profiles of clear cell renal cell carcinoma. Sci Rep. 2016;6:28932. doi: 10.1038/srep28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao W., Wang X., Wang T., Xing J. MiR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma. Aging. 2019;11(2):615–633. doi: 10.18632/aging.101763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun X., Cui M., Tong L., Zhang A., Wang K. Upregulation of microRNA-3129 suppresses epithelial ovarian cancer through CD44. Cancer Gene Ther. 2018;25(11–12):317–325. doi: 10.1038/s41417-018-0026-1. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X., Zhang X., Hu S., Zheng M., Zhang J., Zhao J. Identification of miRNA-7 by genome-wide analysis as a critical sensitizer for TRAIL-induced apoptosis in glioblastoma cells. Nucl Acids Res. 2017;45(10):5930–5944. doi: 10.1093/nar/gkx317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharifi M., Moridnia A. Apoptosis-inducing and antiproliferative effect by inhibition of miR-182-5p through the regulation of CASP9 expression in human breast cancer. Cancer Gene Ther. 2017;24(2):75–82. doi: 10.1038/cgt.2016.79. [DOI] [PubMed] [Google Scholar]
- 14.Chen X., Xu X., Pan B., Zeng K., Xu M., Liu X. miR-150-5p suppresses tumor progression by targeting VEGFA in colorectal cancer. Aging. 2018;10(11):3421–3437. doi: 10.18632/aging.101656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao W., Lou N., Ruan H., Bao L., Xiong Z., Yuan C. Mir-144-3p promotes cell proliferation, metastasis, sunitinib resistance in clear cell renal cell carcinoma by downregulating ARID1A. Cel. Physiol Biochem: Int J Exp Cell Phys Biochem Pharmacol. 2017;43(6):2420–2433. doi: 10.1159/000484395. [DOI] [PubMed] [Google Scholar]
- 16.Zare M., Bastami M., Solali S., Alivand M.R. Aberrant miRNA promoter methylation and EMT-involving miRNAs in breast cancer metastasis: diagnosis and therapeutic implications. J Cell Physiol. 2018;233(5):3729–3744. doi: 10.1002/jcp.26116. [DOI] [PubMed] [Google Scholar]
- 17.Lee S.E., Kim S.J., Youn J.P., Hwang S.Y., Park C.S., Park Y.S. MicroRNA and gene expression analysis of melatonin-exposed human breast cancer cell lines indicating involvement of the anticancer effect. J Pineal Res. 2011;51(3):345–352. doi: 10.1111/j.1600-079X.2011.00896.x. [DOI] [PubMed] [Google Scholar]
- 18.Yang S., Sun Z., Zhou Q., Wang W., Wang G., Song J. MicroRNAs, long noncoding RNAs, and circular RNAs: potential tumor biomarkers and targets for colorectal cancer. Cancer Manag Res. 2018;10:2249–2257. doi: 10.2147/CMAR.S166308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T., Brinkley T.E., Liu K., Feng X., Marsh A.P., Kritchevsky S. Circulating mirnas as biomarkers of gait speed responses to aerobic exercise training in obese older adults. Aging. 2017;9(3):900–913. doi: 10.18632/aging.101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulivi P., Petracci E., Marisi G., Baglivo S., Chiari R., Billi M. Prognostic role of circulating miRNAs in early-stage non-small cell lung cancer. J Clin Med. 2019;8(2) doi: 10.3390/jcm8020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Wang T., Chen C., Wu Z., Bai P., Li S. Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. J Cell Biochem. 2018 doi: 10.1002/jcb.27347. [DOI] [PubMed] [Google Scholar]
- 22.Lou N., Ruan A.M., Qiu B., Bao L., Xu Y.C., Zhao Y. miR-144-3p as a novel plasma diagnostic biomarker for clear cell renal cell carcinoma. Urol Oncol. 2017;35(1) doi: 10.1016/j.urolonc.2016.07.012. 36.e7–36.e14. [DOI] [PubMed] [Google Scholar]
- 23.Liang W., Wei X., Li Q., Dai N., Li C.Y., Deng Y. MicroRNA-765 enhances the anti-angiogenic effect of CDDP via APE1 in osteosarcoma. J Cancer. 2017;8(9):1542–1551. doi: 10.7150/jca.18680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding J., Yang C., Yang S. LINC00511 interacts with miR-765 and modulates tongue squamous cell carcinoma progression by targeting LAMC2. J Oral Pathol Med: Off Publ Int Assoc Oral Pathol Am Acad Oral Pathol. 2018;47(5):468–476. doi: 10.1111/jop.12677. [DOI] [PubMed] [Google Scholar]
- 25.Xiong Z., Xiao W., Bao L., Xiong W., Xiao H., Qu Y. Tumor cell “Slimming” regulates tumor progression through PLCL1/UCP1‐Mediated lipid browning. Adv Sci. 2019 doi: 10.1002/advs.201801862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao W., Xiong Z., Xiong W., Yuan C., Xiao H., Ruan H. Melatonin/PGC1A/ UCP1 promotes tumor slimming and represses tumor progression by initiating autophagy and lipid browning. J Pineal Res. 2019 doi: 10.1111/jpi.12607. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie B.H., He X., Hua R.X., Zhang B., Tan G.S., Xiong S.Q. Mir-765 promotes cell proliferation by downregulating INPP4B expression in human hepatocellular carcinoma. Cancer Biomark: Sect A Dis Mark. 2016;16(3):405–413. doi: 10.3233/CBM-160579. [DOI] [PubMed] [Google Scholar]
- 29.Lin J., Zhang D., Fan Y., Chao Y., Chang J., Li N. Regulation of cancer stem cell self-renewal by HOXB9 antagonizes endoplasmic reticulum stress-induced melanoma cell apoptosis via the miR-765-FOXA2 axis. J Investig Dermatol. 2018;138(7):1609–1619. doi: 10.1016/j.jid.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Zimmerman J.W., Pennison M.J., Brezovich I., Yi N., Yang C.T., Ramaker R. Cancer cell proliferation is inhibited by specific modulation frequencies. Br J Cancer. 2012;106(2):307–313. doi: 10.1038/bjc.2011.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou Y., Chen Y., Yao S., Deng G., Liu D., Yuan X. MiR-422a weakened breast cancer stem cells properties by targeting PLP2. Cancer Biol Ther. 2018;19(5):436–444. doi: 10.1080/15384047.2018.1433497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y.H., Hueng D.Y., Tsai W.C. Proteolipid protein 2 overexpression indicates aggressive tumor behavior and adverse prognosis in human gliomas. Int J Mol Sci. 2018;19(11) doi: 10.3390/ijms19113353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.