Abstract
Background
Heterogeneous nuclear ribonucleoprotein (hnRNP) A2/B1 is an important RNA-binding protein that affects the RNA processing, splicing, transport and stability of many genes. hnRNPA2/B1 is expressed during proliferation and metastasis of various cancer types and promotes such processes. However, the precise role and mechanism of hnRNPA2/B1 in breast cancer remain unclear.
Methods
The association of hnRNPA2/B1 with breast cancer metastasis was assessed using tissue chips, mouse models and publicly available data. The role and mechanism of hnRNPA2/B1 in breast cancer metastasis were studied in cell lines and mouse models.
Findings
In contrast to other cancer research findings, hnRNPA2/B1 expression was negatively correlated with breast cancer metastasis. hnRNPA2/B1 inhibited MDA-MB-231 triple-negative breast cancer (TNBC) cell metastasis in vitro and in vivo. hnRNPA2/B1 knockout activated ERK-MAPK/Twist and GR-beta/TCF4 pathways but inhibited STAT3 and WNT/TCF4 signalling pathways. Profilin 2 (PFN2) promoted breast cancer cell migration and invasion, whereas hnRNPA2/B1 bound directly to the UAGGG locus in the 3′-untranslated region of PFN2 mRNA and reduced the stability of PFN2 mRNA.
Interpretation
Our data supported the role of hnRNPA2/B1 in tumour metastasis risk and survival prediction in patients with breast cancer. The inhibitory role of hnRNPA2/B1 in metastasis was a balance of downstream multiple genes and signalling pathways. PFN2 downregulation by hnRNPA2/B1 might partly explain the inhibitory mechanism of hnRNPA2/B1 in breast cancer metastasis. Therefore, hnRNPA2/B1 might be used as a new prognostic biomarker and valuable molecular target for breast cancer treatments.
Keywords: Heterogeneous nuclear ribonucleoprotein A2/B1, Breast cancer metastasis, Epithelial–mesenchymal transition, Profilin 2
Research in context.
Evidence before this study
Heterogeneous nuclear ribonucleoprotein (hnRNP) A2/B1 is an important RNA-binding protein that is expressed in the proliferation and metastasis of various cancer types and promotes such processes.
Added value of this study
In contrast to other cancer research, we report that hnRNPA2/B1 expression was negatively correlated with breast cancer metastasis. hnRNPA2/B1 knockout promoted breast cancer cell metastasis in vitro and in vivo by activating ERK-MAPK/Twist and GR-beta/TCF4 pathways but inhibited STAT3 and WNT/TCF4 signalling pathways, suggesting that the phenotype of inhibiting metastasis might be caused by the balance of multiple genes and the signalling pathways located downstream of hnRNPA2/B1. In addition, PFN2 downregulation by hnRNPA2/B1 might partly explain the inhibitory mechanism of hnRNPA2/B1 in breast cancer metastasis.
Implications of all available evidence
Our data supported the role of hnRNPA2/B1 in tumour metastasis risk and survival prediction in patients with breast cancer. The inhibitory role of hnRNPA2/B1 in metastasis was a balance of downstream multiple genes and signalling pathways. Therefore, hnRNPA2/B1 might be used as a new prognostic biomarker and valuable molecular target for breast cancer treatments.
Alt-text: Unlabelled box
1. Introduction
Metastasis is the main feature of cancer cells and the leading cause of death in clinical patients with cancer. Most patients with cancer die from metastases rather than from their primary tumours [1]. Breast cancer is the most commonly diagnosed malignant tumour and the leading cause of cancer deaths in women worldwide. In 2018, approximately 2.09 million women were diagnosed with breast cancer (11.6% of all cancer sites) worldwide, from which 0.63 million women died [2]. Distal metastasis is also the leading cause of high mortality in breast cancer [3]. Despite advances in therapy, the five-year survival rate of advanced or metastasised breast cancer patients remains as low as 26%, reflecting the need for further insights into the metastatic process and development of new therapies [4]. Understanding the metastasis mechanism of breast cancer and its difference from other tumour metastases is important for treatment and search for therapeutic targets.
Heterogeneous nuclear ribonucleoprotein (hnRNP) A2/B1 has two isoforms, namely, A2 and B1, which are the products of the alternative splicing of the precursor mRNA of the same gene. A2 is 12 amino acids shorter than B1 at the N-terminus and is mainly expressed in the cells at more than 95% [5]. Previous research found that the binding preference of RNA motifs is slightly different between A2 and B1 [6], suggesting that they may have different functions. As an RNA-binding protein, hnRNPA2/B1 is involved in carcinogenesis through its interaction with other proteins [7] and participates in various cellular processes, such as cancer cell metabolism [8,9], migration [10], invasion [11], proliferation [12], survival and apoptosis through RNA processing [13], splicing, transportation [14] and stability of a number of downstream target genes [15]. hnRNPA2/B1 is highly expressed in many cancers, such as pancreatic [16], liver [17], lung [18], breast [19] and prostate cancer [20] as well as in malignant glioma [21]. As an alternative splicing factor, hnRNPA2/B1 alters the alternative splicing of pyruvate kinase isozyme M2 in cancer cells and activates the switching of metabolism to aerobic glycolysis [9]. In KRAS-dependant human pancreatic ductal adenocarcinoma cells, hnRNPA2/B1 knockout significantly reduces the viability, anchorage-independent proliferation and formation of xenograft tumours, increases the apoptosis of cells and inactivates AKT signalling [22]. hnRNPA2/B1 knockout reduces cell viability, migration and invasion and decreases P-STAT3 and MMP-2 in glioblastoma cells [11]. Silencing hnRNPA2/B1 in lung cancer cells increases E-cadherin and inhibits lung cancer metastasis and EMT progression [23]. The above studies indicate the important role of hnRNPA2/B1 in carcinogenesis, invasion and metastasis. However, the precise function of hnRNPA2/B1 and its molecular mechanism in breast cancer have not been comprehensively investigated. In the present study, our results demonstrate that hnRNPA2/B1 has a distinct role and molecular mechanism in breast cancer compared with other tissue-derived cancer cells.
2. Materials and methods
2.1. Cell culture
MDA-MB-231 and MCF-7 human breast cancer cell lines and human embryonic kidney 293T cell line were purchased from the Cell Bank of Shanghai Institutes for Biological Sciences of China. MCF-7 and MDA-MB-231 cell lines were characterised by Genetic Testing Biotechnology Corporation (Suzhou, China) by using short tandem repeat markers. The cells were cultured in complete DMEM (Gibco,Cat#12800-017) containing 10% foetal bovine serum (FBS) (PAN,Cat#ST30-3302) and 100 U/mL each of penicillin and streptomycin at 37 °C and 5% CO2.
2.2. hnRNPA2/B1 knockout cell lines
The hnRNPA2/B1 gene was knocked out in MDA-MB-231 and MCF-7 cells by using the CRISPR-Cas9 system. Two small guide RNAs against hnRNP A2/B1 (Supplementary Table S4) were inserted into the pLX-based vector. The pLX-sgRNA (RRID:Addgene_50662) vectors were co-transduced with pCW-Cas9 (RRID:Addgene_50661)to knock out hnRNP A2/B1. The MCF-7 cell cultures were selected using 0.6 µg/mL puromycin and 3 µg/mL blasticidin (for MDA-MB-231, 0.2 µg/mL puromycin and 20 µg/mL blasticidin). The expression of hnRNP A2/B1 was examined by Western blot and quantitative real-time reverse transcription (qRT)-PCR analyses.
2.3. Overexpression and knockdown of genes
The coding regions of hnRNP A2, hnRNP B1 and PFN2 were cloned by PCR and inserted between the EcoRI and KpnI endonuclease sites of p3 × Flag-CMV-10 expression construct. The primers are listed in Supplementary Table S2. The PFN2 expression vector pLV-PFN2 (Vector ID:VB180625-1016sme) and control vector were purchased from VectorBuilder. For the knockdown of PFN2, siRNAs or negative control siRNA (Genepharma, Shanghai, China) were transfected into the cells by using Lipofectamine 3000 (Invitrogen, CA, USA,Cat# L3000015).
2.4. Cell proliferation assays
Cell proliferation was measured using growth curve assay. The cells were seeded into 96-well plates at a concentration of 5000 cells per well and incubated for different times. CCK-8 was added, and absorbance (OD) at 490 nm was recorded. The cell numbers were converted using OD values.
2.5. Stable isotope labelling with amino acids (SILAC) in cell culture and proteomic analysis
SILAC assay was performed according to a published protocol [24]. The cells were cultured in DMEM containing 10% dialysed FBS and heavy-isotope amino acid media or natural-isotope amino acid media (Thermo, USACat#89983). After six times of passage, the cells labelled with light or heavy amino acid were lysed. Equal amounts of light- or heavy-isotope-labelled proteins were mixed. Liquid chromatography tandem mass spectrometry (LC–MS/MS) analysis was performed (AB Sciex Triple TOF 5600).
2.6. Tissue microarray
Tissue microarray (US Biomax, BR10010d) of 47 cases of infiltrative ductal carcinoma and lymph node metastasis and three cases of infiltrative lobular carcinoma was evaluated through immunohistochemical staining according to the protocol provided by US Biomax Inc. Images were captured by Precipoint M8 Digital Scanning Microscopy System. Optical density analysis of protein expression was conducted with Image Pro Plus. The protein expression level in immunohistochemical images was also determined by a pathologist according to the intensity and area of staining.
2.7. Western blot analysis
The protein samples were separated using sodium dodecyl sulphate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5% bovine serum albumin for 1 h at room temperature and incubated with first antibody overnight at 4 °C. The membranes were subsequently incubated with secondary antibody for 1 h at room temperature. Protein signals were detected using chemiluminescent reagents (Millipore, USA,Cat# 190113-13) and visualised after exposure to the X-ray film. Relative optical density ratio was calculated using Image J software. The antibodies used in this study are listed in Supplementary Table S5.
2.8. Quantitative PCR analysis
Total RNA was extracted using TRIzol (Invitrogen™) and reverse transcribed using PrimeScript™ RT Kit (TaKaRa, Cat#:RR047A). qRT-PCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme,Cat#
Q331-03). mRNA level was quantified using the 2−ΔΔCt algorithm with GAPDH or β-actin as the normaliser gene. The CT plots for GAPDH or β-actin gene are found in Supplementary Fig. S15. All the primers used are found in Supplementary Table S3.
2.9. Confocal immunofluorescence
Cells were seeded on sterile coverslips at 2 × 105 cells/well, fixed with 3.7% paraformaldehyde and permeabilised in 0.1% Triton X-100/PBS. Coverslips were blocked for 30 min by using 1% BSA, washed and exposed to a 1:250 dilution of ActinGreen™ 488 Stain (GeneCopoeia™, Cat#C052T) for 30 min in the dark at room temperature. After washing, the cells were incubated with 1 µg/mL diluted DAPI for 5 min at room temperature. The cells were photographed using FV1000 IX81 laser scanning confocal microscope.
2.10. F-actin analysis
The cells were harvested and incubated with phalloidin F-actin probe (ActinGreen™ 488 Stain, GeneCopoeia™, Cat No.C052T) for 30 min. F-actin was assessed by fluorescence-activated cell sorting analysis on a Gallios Beckman Coulter.
2.11. Migration and Matrigel invasion assay
Migration and Matrigel invasion assays were performed using a Transwell plate. In brief, 1 × 105 cells in 200 µL of serum-free DMEM with 0.1% sterile BSA were added into the upper chamber, and 600 µL of the complete medium was placed into the lower chamber (Corning, Cat#3422). The cells were incubated at 37 °C for 24–48 h. After fixation in 95% ethanol and staining with 0.1% crystal violet, the migrated cells were counted in six randomly selected microscope fields.
For the Transwell experiment induced by EGF, the medium of the lower chamber was DMEM containing 0.1% BSA and 100 ng/mL EGF. For the Transwell experiment treated with Trametinib (MCE, Cat #1187431-43-1), the Trametinib concentration was 100 nM.
2.12. Migratory cell sorting experiment
Migratory cell sorting in vitro and in vivo were performed using Transwell plate and immunomagnetic beads, respectively. The expression of hnRNPA 2/B1 was analysed by Western blot and qPCR.
2.12.1. Migratory cell sorting experiment in vitro
7 × 106 cells (cell invasion in Matrigel-covered membrane) or 5 × 106 cells (cell migration in membrane without Matrigel cover) in 1.5 mL of serum-free DMEM containing 0.1% BSA were added into the upper chamber, and 2.6 mL of the complete medium was added into the lower chamber (Corning, Cat# 3428). The cells were incubated at 37 °C for 24–48 h. The cells on both surfaces of the filter were digested with 0.01 M EDTA–Na2 in PBS and harvested separately.
2.12.2. Migratory cell sorting experiment in vivo
The model of human breast cancer xenografts was established. After 40 days, eyeball blood samples were collected from the mice. Erythrocytes were lysed and centrifuged to obtain a mixture of human and mouse cells. A mixture of human and murine cells from the xenograft was obtained from the same mouse. The eyeball blood samples from the mice injected with PBS were treated as negative controls to evaluate the sorting process. The human breast cancer cell line MDA-MB-231 in the cell mixture were sorted and purified with antihuman HLA-ABC (Biolegend, Cat# 311434) and anti-mouse CD45 (Ebioscience, Cat#13-9457-82) antibodies by using Dynabeads® FlowComp™ Flexi Kit (Invitrogen, Cat#11061D) according to the manufacturer's instructions. The purified human breast cancer cells were collected and counted, and cDNA was obtained from 1000 cells by using Whole Transcriptome Amplification Kit (CellAmp ™, Ver.2).
2.13. RNA immunoprecipitation (RIP) analysis
RIP analysis was performed using hnRNPA2/B1 antibodies and Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA,Cat#17-700) following the manufacturer's protocol.
2.14. Animal studies
Six-week-old BALB/c female nude mice were used for the models of spontaneous lung metastasis, pulmonary metastasis by tail vein injection and cancer blood burden. All animal handling procedures were approved by the Institutional Review Board of Xiamen University.
2.14.1. Model of spontaneous lung metastasis
Tumour cells were injected subcutaneously in the right axilla of 10 mice (3 × 106 cells/100 µL). Tumour volume (in mm3) was measured every 4 days and calculated using the following formula: volume = length × width2 × 0.5. After 48 days, the mice were sacrificed to collect tumour, liver and lung samples. The tissues were fixed with formalin and embedded in paraffin. The slides were stained with H and E. Basic morphology was observed under light microscopy (Olympus IX51).
2.14.2. Model of pulmonary metastasis by tail vein injection
Tumour cells were injected into the lateral tail vein of eight nude mice (3 × 105 cells/100 µL). The mice were sacrificed 6 weeks after injection. The lungs were harvested, fixed and stained with Bouin's solution. The metastatic colonies on the lung surface were counted. The lung and liver samples were fixed with formalin and paraffin embedded. The slides were stained with hematoxylin-eosin and observed by light microscopy (Olympus IX51).
2.14.3. Model of cancer blood burden
Tumour cells were injected subcutaneously in the fat pad under the breast of five mice (3 × 106 cells/100 µL). At 3 or 6 days after transplantation, 100 µL of blood was obtained from the mouse's eye socket to test for metastatic cancer cells. Considering the low abundance of cancer cells in the blood, genomic DNA was abstracted, and qRT-PCR analysis was performed to measure circulating cancer cells with human-specific GAPDH or Alu primers and mouse-specific GAPDH primers.
2.15. Dual-luciferase reporter gene assay
The 3′-UTR of hnRNPA2/B1 mRNA and the mutant were cloned and inserted into pGL3 plasmid. Dual-Luciferase Reporter Assay was performed following the manufacturer's instruction. The full length of the 3′-UTR of mRNA, the first half of 3′-UTR (3′A) and the latter half of the 3′-UTR (3′B) were cloned and inserted into the Xbal-Fsel restriction site downstream to firefly luciferase (pGL3 promoter plasmid). 239T cells were co-transfected with the constructed recombinant (0.2 µg/well) or control plasmids (0.2 µg/well) and Renilla luciferase reporter (4 ng/well) or hnRNP A2 overexpression plasmids. After 24 h, luciferase and Renilla luciferase activities were separately measured using a single-tube luminometer (Promega) following the manufacturer's instruction for the Dual-Luciferase Reporter Assay Kit (Promega, USA,Cat# E1910). The primers are listed in Supplementary Table S2.
2.16. Statistical analysis
Statistical analyses were performed using Graphpad Prism 6.0. Data were presented as mean ± SEM, and the statistical significance of the mean differences were determined using Student's t-test (t-test) and two-way ANOVA as indicated in the figure legend. Sample size (n) is also reported in the figure legend for each experiment, with n as the number of identically treated replicates. All tests were two-tailed, and P values < 0.05 were considered statistically significant.
3. Results
3.1. Downregulated hnRNPA2/B1 is associated with metastasis and prognosis in breast cancer
The expression data of breast cancer in public databases were analysed using the Breast Cancer Integrative Platform [25]. The expression of hnRNPA2/B1 decreased in breast cancer in contrast to that in other types of tumours (Fig. 1A and Supplementary Fig. S1). Patients with high hnRNPA2/B1 expression had longer median survival time than those in the low expression group (Fig. 1B and Supplementary Fig.S1). Moreover, hnRNPA2/B1 was significantly high in breast cancer patients, and good therapeutic responses and prognosis were obtained (Supplementary Fig. S3).
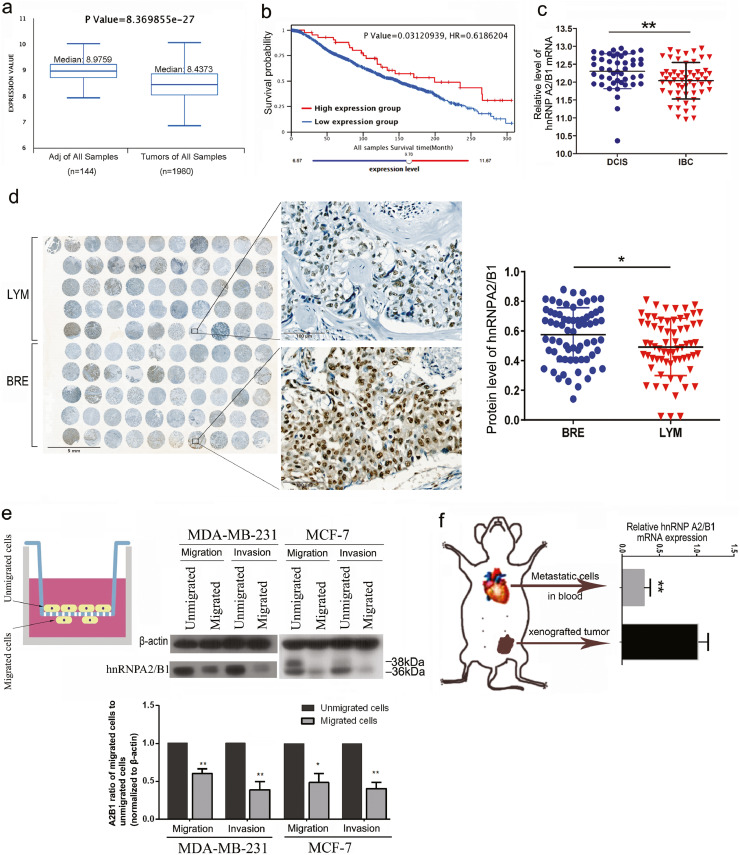
Fig. 1.
hnRNPA2/B1 expression decreases and is correlated with tumour invasion and metastasis in breast cancer. (A) The hnRNPA2/B1 expression in breast tumour was significantly reduced compared with that in adjacent normal samples. Adj: adjacent normal. The expression data of METABRIC from EMBL-EBI were analysed using the Breast Cancer Integrative Platform (http://www.omicsnet.org/bcancer/). (B) Overall survival (OS) analysis indicates that the median survival time of the patients in the high-hnRNPA2B1-expression group (n = 43, median survival time: 199.27 months) was longer than that of the patients in the low-hnRNPA2B1-expression group (n = 1916, median survival time: 148.8 months). (C) The expression data in the gene chip (GSE59246) from the GEO database indicated that hnRNPA2/B1 decreased significantly in invasive breast cancer (IBC, n = 85) compared with ductal carcinoma in situ (DCIS, n = 59). (D) The expression of hnRNPA2/B1 in the corresponding lymph node metastatic carcinoma tissues was significantly reduced compared with that in breast cancer tissues. A tissue microarray with 50 pairs of breast cancer and corresponding lymph node metastasis tissues was analysed by IHC staining and Image Pro-Plus 6.0. BRE: breast cancer tissues; LYM: matched metastatic tissues in lymph nodes. (E) Western blot analysis showed that the expression level of hnRNPA2B1 in highly aggressive cells was significantly lower than that in non-metastatic cells in the cell population derived from the same cell line. Highly invasive cells that penetrated Matrigel were selected using an in vitro invasion chamber. The representative image of three replicate experiments is shown. The quantitative expression of A2B1 was normalised by β-actin. *p < 0.05.**p < 0.01 (Student's t-test). (F) Quantitative PCR showed that the mRNA level of hnRNPA2B1 in metastatic cells was significantly lower than that in the non-metastatic cells of the transplanted tumours in vivo. MDA-MB-231 cells were subcutaneously transplanted in nude mice (n = 3), and the metastatic tumour cells were collected from the blood by using immunomagnetic beads. Human MDA-MB-231 cells in the cell mixture were sorted using anti-human HLA-ABC biotin and anti-mouse CD45 biotin antibodies. All data are expressed as mean ± SEM (n = 3). *p < 0.05.**p < 0.01 (Student's t-test).
The data from another source (http://kmplot.com/analysis/) also show that high hnRNPA2B1 in breast and lung cancer indicates significantly longer overall survival (OS) but shorter OS in gastric, ovarian and liver cancer. The high expression of hnRNPA2B1 indicates longer OS in breast cancer patients with positive lymph node metastasis but shorter OS in patients with negative lymph node metastasis (Supplementary Fig. S4).
The expression data in breast cancer shows that in contrast to its effects in most tumours, high hnRNPA2B1 indicates good prognosis and survival rate. Considering that metastasis is a key factor in tumour-related death, we determined whether hnRNPA2B1 inhibits tumour metastasis. We evaluated hnRNPA2/B1 expression in Ductal Carcinoma in situ and Invasion Breast Cancer (IBC) samples in the gene chip data (GSE59246) from the GEO database. The mRNA expression of hnRNPA2/B1 significantly decreased in IBC (Fig. 1C). We further analysed the tissue sample microarray containing breast cancer and matched the lymph node metastasis from 50 breast cancer patients. The immunohistochemical analysis shows that the positive and strong positive rates of hnRNPA2/B1 are as high as 80% in tumour tissues and 40.8% in lymph nodes. The expression of hnRNPA2/B1 was significantly reduced in the corresponding lymph node metastatic cancer tissues (Fig. 1D and Supplementary Fig. S2A), suggesting the negative correlation of hnRNPA2/B1 expression with tumour cell metastasis.
The comparison between the migrated and non-migrated MDA-MB-231 and MCF-7 cells revealed a significant decrease in the hnRNPA2/B1 expression in invasive cells (Fig. 1E). The hnRNPA2/B1 expression significantly decreased in the metastatic tumour cells in the blood of mice compared with the non-migrated tumour cells in the xenograft (Fig. 1F). These results further confirm that the low expression of hnRNPA2/B1 contributes to the invasive phenotype of breast cancer.
3.2. HnRNPA2/B1 knockout promotes migration and invasion but reduces cell proliferation in vitro
We knocked out hnRNPA2/B1 by CRISPR/CAS9 method (Fig. 2A, Supplementary Fig. S5). The Transwell chamber assays indicated that the migration (Fig. 2B) and invasion (Fig. 2C) of hnRNPA2/B1 knockout MDA-MB-231 and MCF-7 cells significantly increased compared with the control cells. After restoring the expression of the A2 protein, the main product of the hnRNPA2/B1 gene, in two hnRNPA2/B1 KO cell lines (KO20 and KO28) (Fig. 2D), the migration and invasion of MCF-7 cells was inhibited (Fig. 2E and F). Restoring A2 expression in MDA-MB-231 KO cells also inhibited migration and invasion (Supplementary Fig. S7). These results strongly confirm that hnRNPA2/B1 KO can significantly promote the migration and invasion of breast cancer cells in vitro.
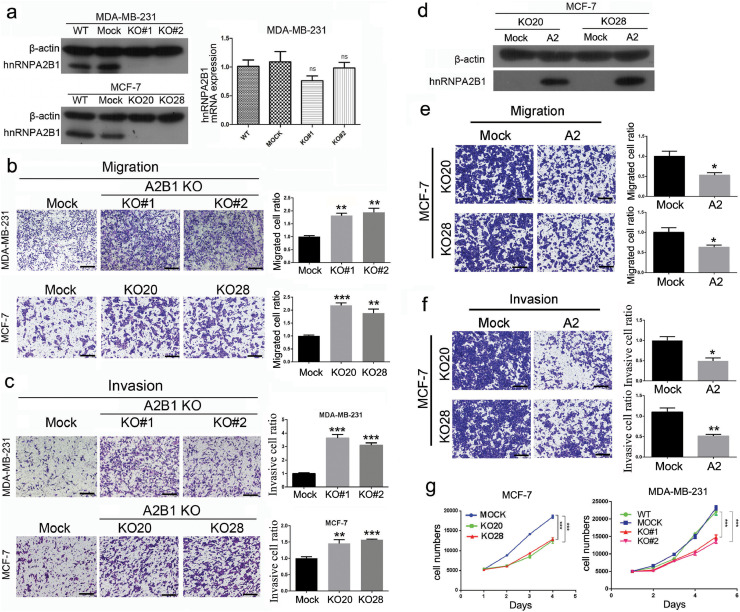
Fig. 2.
hnRNPA2/B1 KO increases breast cancer cell metastasis but reduces cell proliferation in vitro. The representative images and quantification of three replicate experiments (n = 3) are shown. Data are presented as mean ± SEM (n = 3). All the scale bars are 250 µm. (A) hnRNPA2/B1 was knocked out through the CRISPR/CAS9 system. The hnRNPA2/B1 expression levels in MCF-7 and MDA-MB-231 cells were measured by Western blot analysis and real-time PCR. hnRNPA2B1 protein expression was completely knocked out, but the mRNA level of hnRNPA2/B1 did not change significantly. KO#1 and KO#2: two knockout cell clones of MDA-MB-231 developed from different sgRNA treatments. KO20 and KO28: two knockout cell clones of MCF-7 developed from the same sgRNA treatment. ns: not significant (two-way ANOVA) (B) The migratory capacity of MDA-MB-231 and MCF-7 cells with or without hnRNPA2/B1 was measured by Transwell migration assays. **p < 0.01, ***p < 0.001 (Student's t-test). (C) The invasive capacity of MDA-MB-231 and MCF-7 cells with or without hnRNPA2/B1 was determined through Transwell assays. **p < 0.01, ***p < 0.001 (Student's t-test). (D) hnRNPA2/B1 protein was restored in two MCF-7 KO cells. After the hnRNPA2 expression was restored, the migration (E) and invasion (F) abilities of two hnRNPA2/B1 KO cells were significantly reduced as measured by Transwell assays. *p < 0.05, **p < 0.01 (Student's t-test). (G) Cell proliferation of MDA-MB-231 and MCF-7 cells were significantly inhibited by hnRNPA2B1 KO measured through the MTT proliferation assay. *p < 0.05, **p < 0.01, ***p < 0.001 (two-way ANOVA).
The proliferation assay showed that hnRNPA2/B1 KO significantly reduced the proliferation of MCF-7 and MDA-MB-231 cells (Fig. 2G). The restored B1 and A2 in MCF-7 KO cells promoted the proliferation (Supplementary Fig. S7).
3.3. HnRNPA2/B1 knockout promotes breast cancer metastasis but inhibits tumour growth in vivo
The metastasis of tumour in vivo is a multistage process, where cancer cells enter the lymphatic or blood circulation from the primary site (early stage), move into the distal site from the circulatory system, anchor and grow (late stage). The lung metastasis (spontaneous lung metastasis) of the nude mouse xenograft mimics the entire cancer cell metastasis, and the tail vein injection lung metastasis animal model (experimental lung metastasis) mimics the latter stage of metastasis. hnRNPA2/B1 knockout and control cells were implanted subcutaneously into nude mice to explore the potential function of hnRNPA2/B1 throughout the breast cancer metastasis. The histological study of the pathological sections indicates that hnRNPA2/B1 knockout significantly promoted the spontaneous lung metastasis of MDA-MB-231 cells (Fig. 3A and B).
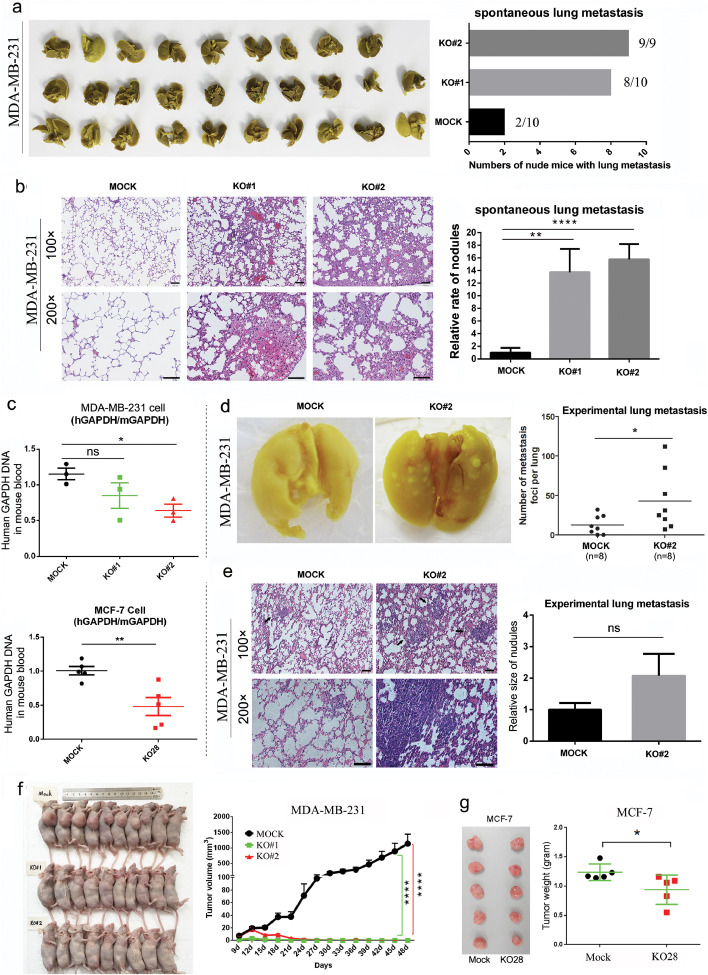
Fig. 3.
hnRNPA2/B1 KO increases breast cancer tumour metastasis but inhibits tumour growth in vivo. (A) hnRNPA2/B1 KO exacerbated the lung metastasis of breast cancer cells. MDA-MB-231 cells were subcutaneously injected into nude mice. After 48 days, lung metastasis occurred in two, eight and nine nude mice in the control (n = 10), KO#1 (n = 10), and KO#2 (n = 9) groups, respectively. (B) The lungs of the tumour-bearing mice were assessed in terms of metastatic foci by histologically studying the pathological sections. Results indicated that the knocked out groups (KO#1 and KO#2) had more lung metastatic nodules. Scale bar:100 µm. mean ± SEM. **p < 0.01, ****p < 0.0001 (Student's t-test). (C) Cancer blood burden assay showed that hnRNPA2/B1 KO significantly decreased the number of human breast cancer cells in the mouse blood system. The DNA level of GAPDH of the human cancer cell in mouse blood was measured through qRT-PCR by using human-specific GAPDH (hGAPDH) and mouse-specific GAPDH (mGAPDH) primers. n = 3 and n = 5 for MDA-MB-231 and MCF-7 cells, respectively. mean ± SEM. ns: nonsignificant difference, P value = 0.1977. *p < 0.05, **p <0.01 (Student's t-test). (D) Experimental lung metastasis assay showed that hnRNPA2/B1 KO promoted the late stage of the lung metastasis of breast cancer cells. MDA-MB-231 cells were injected intravenously into nude mice. Statistical analysis showed that the number of lung metastatic nodules was significantly higher in the KO#2 group (n = 8) than in the mock group (n = 8). *p < 0.05 (Student's t-test), (E) Histological examination of pulmonary nodules. No significant difference was observed in the size of the nodules between the mock and KO#2 groups at p = 0.1135 (t-test). Scale bar: 100 µm. (F) hnRNPA2/B1 KO significantly inhibited the formation and growth of MDA-MB-231 cell xenografts. These data are from the same mice in Fig. 3A and the experiment was repeated twice. mean ± SEM. ****p < 0.0001 (two-way ANOVA). (G) hnRNPA2/B1 KO significantly inhibited the growth of MCF-7 cell xenografts (n = 5/group). Data are presented as mean ± SEM. *p < 0.05 (Student's t-test).
Cancer blood burden test was carried out to further validate the ability of breast cancer cells to metastasise to the blood stream from the primary site. Six days after the subcutaneous fat pad transplantation of mammary gland in nude mice, the metastatic cells in the blood were analysed by quantitative PCR. The results indicate that hnRNPA2/B1 knockout significantly decreased the number of human breast cancer cells in the mouse blood system (Fig. 3C). This occurrence may be the result of decreased cell viability, consistent with previous data on cell proliferation (Fig. 2G). hnRNPA2/B1 knockout MDA-MB-231 cells (KO#2) and control cells (MOCK) were injected into the tail vein of nude mice to investigate the effect of hnRNPA2/B1 on the latter stage of breast cancer cell metastasis. The nodules that metastasised to the lung surface were counted. The average number of pulmonary metastases resulting from MOCK cells was 12.63±4.25, whereas those induced by hnRNPA2/B1 knockout cells was 42.88±13.31 (Fig. 3D). The histological examination of the lung sections confirmed that the pulmonary nodules were invasive cancer. No significant difference was observed in the size of the nodules between the MOCK and KO#2 groups (p = 0.1135) (Fig. 3E). These data indicate that hnRNPA2/B1 knockout has a profound effect on the metastasis of breast cancer cells from blood to distal anchorage growth.
We further examined the effects of hnRNPA2/B1 knockout on the formation and growth of transplanted tumours. hnRNPA2/B1 knockout completely inhibited the formation of transplanted tumours in MDA-MB-231 cells (Fig. 3F). In MCF-7 cells, hnRNPA2/B1 knockout also significantly inhibited the growth of xenograft tumours (Fig. 3G).
In contrast to findings reported in other cancers, the results show that hnRNPA2/B1 KO inhibits the growth of xenograft tumours but promotes spontaneous lung metastasis. Hence, hnRNPA2/B1 may have different effects on tumour growth and metastasis.
3.4. Breast cancer patients with high hnRNPA2/B1 expression have longer survival
We analysed the relationship between hnRNAP2/B1 expression and survival time of breast cancer patients by using the public databases of the Breast Cancer Integrative Platform (BCIP) and KM plotter. Patients with high hnRNAP2/B1 expression had high survival time (Figs. S1C and S4A) (n = 1958 in BCIP, n = 1764 in KM plotter), consistent with the speculation that hnRNAP2/B1 inhibits tumour metastasis, thereby prolonging patient survival. Further analysis of patients with positive or negative lymph node metastasis showed interesting results. Among patients without lymph node metastasis (Figs. S1G and S4G), those with high hnRNAP2/B1 expression had a shorter survival time, consistent with the expectation that hnRNAP2/B1 expression promotes tumour growth and lead to death. In patients with lymph node metastasis (Figs. S1H and S4F), high hnRNAP2/B1 expression could prolong the survival time. The inhibition of hnRNAP2/B1 on tumour metastasis has become a major factor conducive to the survival of patients.
We further investigated whether hnRNAP2/B1 inhibits metastasis in different subtypes of breast cancer. In the BCIP database, patients with triple-negative breast cancer (TNBC) (n = 30) and high A2/B1 expression had higher survival time than the patients (n = 288) with low A2/B1 expression (Fig. S1D). In the KM plotter database, the TNBC patients (n = 101) with high A2/B1 expression had less survival time than those (n = 60) with low expression (Fig. S4Q). The contradicting results from the two databases show that the analysis is no longer reliable due to the decrease in the number of samples, although both results showed a statistical P value of < 0.05. However, the data from the BCIP database (Fig. S1D) supported the results of the TNBC cell line MDA-MB-231.
The relationship between hnRNAP2/B1 expression and survival time was analysed in TNBC patients with lymph node metastasis. In the KM plotter database, no significant correlation was found between hnRNAP2/B1 expression and survival time in TNBC patients, regardless of positive (Fig. S4R) (n = 72) or negative (Fig. S4S) (n = 87) lymph node status. This occurrence indicates that the sample number in existing public databases is extremely small to obtain reliable results of TNBC subtype.
In the BCIP database, for ER-negative patients, high hnRNAP2/B1 expression was associated with high survival time (Fig. S1I). For ER-positive patients, high hnRNAP2/B1 expression was associated with short survival time (Fig. S1J). Although the difference is statistically significant, a small difference in the survival time was observed among ER-positive patients. In the KM plotter database, high hnRNAP2/B1 expression was associated with longer survival time in ER-positive (Fig. S1I) and ER-negative (Fig. S1J) patients. These results suggest that hnRNAP2/B1 can at least inhibit metastasis in ER-negative patients.
3.5. HnRNPA2/B1 knockout promotes F-actin cytoskeleton formation and EMT in human breast cancer cells
EMT plays an important role in the metastasis of tumour cells. Increasing cellular contractility and actin stress fibre formation are the hallmarks of EMT [26]. In the present study, hnRNPA2/B1 knockout reorganised the F-actin cytoskeleton, increased the width of lamellipodium (the best-characterised motile organelle) and increased the number of migrated spindle cells (Supplementary Fig. S6). The percentage of migratory cells in hnRNPA2/B1 knockout cells significantly increased after EGF induction (Fig. 4A). The induction of EGF on cell migration and invasion was confirmed by previous studies [27,28]. We also evaluated the role of EGF in our research system. The Transwell assay showed that EGF promoted the migration and invasion of MDA-MB-231 cells (Supplementary Fig. S8). The flow cytometry results also show an improved F-actin formation in hnRNPA2/B1 knockout cells (Fig. 4B).
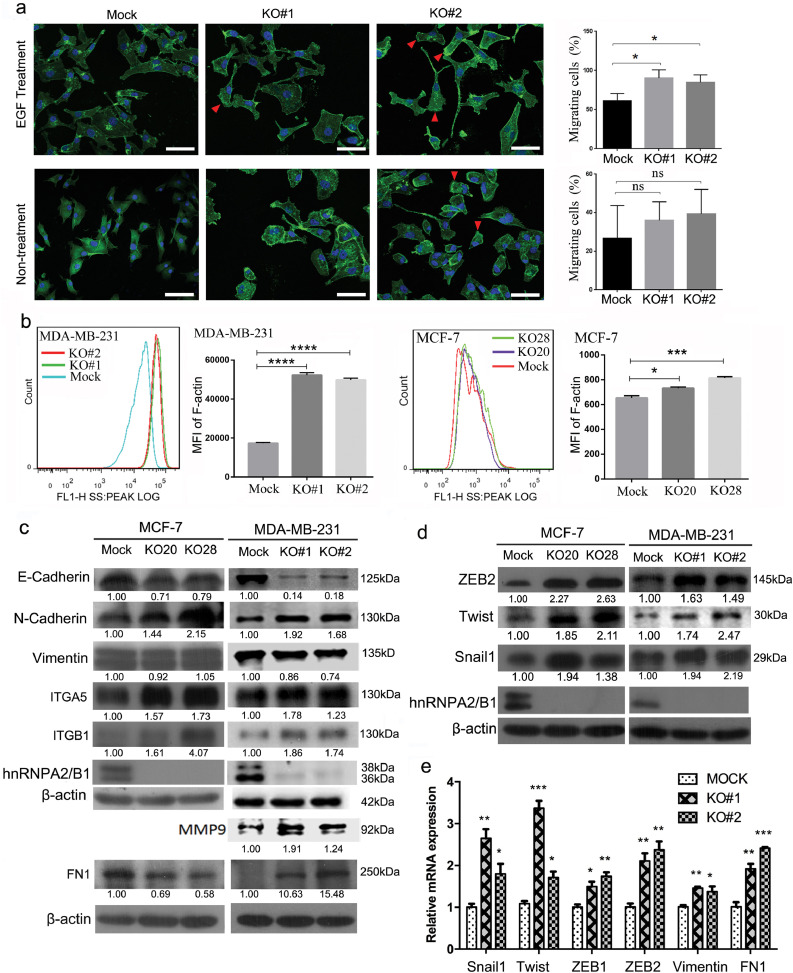
Fig. 4.
hnRNPA2/B1 knockout (KO) promotes the F-actin formation and the EMT progression of human breast cancer cells. (A) The migratory cells in the hnRNPA2B1 KO group increased significantly after induction by EGF tropism. Agarose gel particles containing 200 µg/mL EGF were placed on the edge of the cell culture dish to form a concentration gradient of EGF which was used to induce cell migration. The cells with prominent lamellipodium and nuclei on the lateral side of the cell membrane are defined as migratory cells. MDA-MB-231 cells were stained with FITC-phalloidin for F-actin and Hoechst 33,342 for the nuclei. Scale bar: 100 µm. *p < 0.05 (Student's t-test). (B) hnRNPA2/B1 KO promoted F-actin formation. F-actin in MCF-7 and MDA-MB-231 cells were stained with ActinGreen™ 488 phalloidin and assessed through flow cytometry. The mean fluorescence intensity significantly increased in hnRNPA2B1 KO cells. *p < 0.05, ***p < 0.001 (Student's t-test) (C) Western blot analysis results showed that hnRNPA2/B1 KO significantly altered the expression of some EMT markers, including the decreased E-cadherin expression and the increased N-cadherin, ITGA5, ITGB1 and MMP9 expression. Fibronectin 1 (FN1) was upregulated in hnRNPA2/B1 knockout MDA-MB-231 cells but was downregulated in hnRNPA2/B1 knockout MCF-7 cells. The protein bands were quantified with Image J software and β-actin used for normalisation. The representative images of the three independent experiments are shown. (D) The major EMT transcript factors ZEB2, Twist1 and Snail1 were upregulated by hnRNPA2/B1 KO. All the full panels of each Western blot can be found in Supplementary Fig. S12. (E) Quantitative PCR showed that hnRNPA2/B1 KO in MDA-MB-231 cells increased the mRNA level of the main EMT transcription factors, including Snail1, Twist, ZEB1 and ZEB2. Each mRNA was normalised by GAPDH. Values are relative expression normalised to Mock cells from three biological replicate experiments with three Mock and KO wells in each experiment. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 (Student's t-test).
The detection of EMT markers shows that hnRNPA2/B1 knockout significantly downregulated E-cadherin and upregulated N-cadherin, ITGA5, ITGB1 and MMP9 (Fig. 4C). At the protein and mRNA levels, hnRNPA2/B1 knockout significantly upregulated the expression of Snail1, Twist and ZEB2, which are the main EMT-related transcription factors (Fig. 4D and E). In general, these data show that hnRNPA2/B1 knockout promotes the EMT process in breast cancer cells.
3.6. HnRNPA2/B1 knockout has contradictory effects on different EMT-promoting genes and signalling pathways
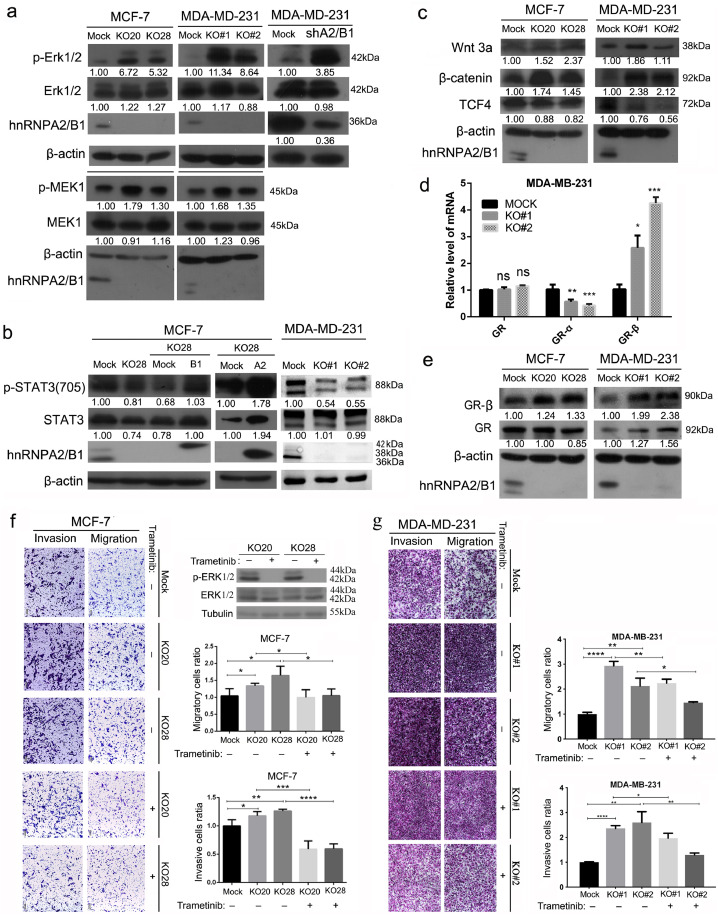
Multiple signalling pathways promote cancer-associated EMT progression and metastasis. The Western blot analysis results show that the phosphorylated ERK1/2 was upregulated in hnRNPA2/B1 knockout or knockdown cells (Fig. 5A), and its downstream transcription factor Twist [29] increased (Fig. 4D). Twist downregulated E-cadherin and upregulated N-cadherin, fibronectin and integrin alpha5 [26,30], as also confirmed by our data (Fig. 4C). The ERK inhibitor Trametinib was used to treat breast cancer cells to further evaluate the effect of ERK phosphorylation on migration and invasion. With the reduction in ERK phosphorylation activated by A2B1 KO, the migration and invasion of MCF-7 KO and MDA-MB-231 KO cells were significantly reduced (Fig. 5F and G). Hence, hnRNPA2/B1 knockout promotes migration and invasion by activating the ERK MAPK/Twist pathway. hnRNPA2/B1 knockout inhibited several EMT-promoting genes and signalling pathways. The treatment also significantly downregulated the phosphorylated STAT3 level in MCF-7 and MDA-MB-231 cells (Fig. 5B). Restoring the expression of hnRNPA2/B1 in knockout cells increased the level of total and phosphorylated protein of STAT3 (Fig. 5B), suggesting the activation effect of hnRNPA2/B1 on the STAT3 pathway. hnRNPA2/B1 knockout has contradictory effects on the core components of the canonical Wnt pathway. hnRNPA2/B1 knockout significantly increased the expression of Wnt3 and beta-catenin, inhibiting its downstream transcription factor TCF4 (also known as TCF7L2) in MCF-7 and MDA-MB-231 cells (Fig. 5C). hnRNPA2/B1 knockout may activate the downstream genes of the Wnt pathway. The glucocorticoid receptor (GR) gene generated two isoforms, namely, GRα and GRβ, by alternative splicing. GRβ directly interacted with TCF4 independent of beta-catenin and enhanced TCF4 activity at high levels [31]. Our transcriptome sequencing data indicated that the transcription factor GR was upregulated in hnRNPA2/B1 knockout cells (data not shown). The qRT-PCR results indicated that in hnRNPA2/B1 knockout cells, GRα mRNA was downregulated, whereas GRβ mRNA was upregulated (Fig. 5D), suggesting the altered splicing of GR isoforms. The Western blot results confirmed that the GRβ protein was upregulated by hnRNPA2/B1 knockout in MCF-7 and MDA-MB-231 cells (Fig. 5E), indicating that TCF4 targeting genes may be activated. hnRNPA2/B1 knockout has contradictory effects on different EMT-promoting genes and signalling pathways, suggesting that the final phenotypic result may be a balance of the effects of different downstream genes of hnRNPA2/B1.
Fig. 5.
Effects of hnRNPA2/B1 KO on EMT-related signalling pathways are complex and even contradictory. Representative images of Western blot analysis and quantitative PCR are shown. The protein bands were quantified with Image J software, and β-actin was used for normalisation. (A) hnRNPA2/B1 KO activated the ERK MAPK pathway. (B) hnRNPA2/B1 KO inhibited the STAT3 pathway in the two breast cancer cell lines, and the restored expression of A2 or B1 protein in MCF-7 cells increased the level of the total STAT3 and phosphorylated STAT3. (C) hnRNPA2/B1 KO showed contradictory effects on the WNT/β-catenin/TCF4 pathway. hnRNPA2/B1 KO increased Wnt 3a and beta-catenin but decreased the transcript factor TCF4. (D) Quantitative PCR showed that hnRNPA2/B1 KO changed the splicing of the two isoforms of the glucocorticoid receptors GRβ and GRα. Data are mean ± SEM (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001 (Student's t-test), ns: not significant. (E) Western blot analysis results confirmed the upregulation of GRβ in hnRNPA2/B1 KO cells. All the full panels of each Western blot can be seen in Supplementary Fig. S13. (F) Transwell results showed that after treatment of MCF-7 KO cells by Trametinib (Tra), ERK phosphorylation inhibitor, the migration and invasion ability of KO20 and KO28 cells decreased significantly. WB experiments showed that phosphorylation of ERK protein was completely inhibited. (G) ERK phosphorylation inhibitor also decreased the migration and invasion in MDA-MB-231 KO cells. All the data are expressed mean ± SEM (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student's t-test).
3.7. HnRNPA2/B1 knockout upregulates PFN2 expression in breast cancer cells and PFN2 promotes the invasive phenotype of breast cancer cells
SILAC technology combined with LC-MS/MS technology was used for quantitative proteomic analysis in MDA-MB-231 and hnRNP A2/B1 KO cells (KO#1, KO#2) to further understand the effect of hnRNPA2/B1 on breast cancer cells. The regulatory pathway of actin cytoskeleton related to tumour metastasis was significantly altered by hnRNPA2/B1 knockout (Supplementary Table S1). These pathways altered by hnRNPA2/B1 knockout suggest that hnRNPA2/B1 mainly affects the assembly of actin cytoskeleton, the interaction between cells and the extracellular matrix microenvironment of breast cancer cells.
We further investigated PFN2, which is associated with actin cytoskeleton assembly and is upregulated by hnRNPA2B1 knockout. The KEGG analysis indicated that PFN2 promotes actin polymerisation, stress fibres and lamellipodia formation, thereby promoting cell mobility (Supplementary Fig. S11).
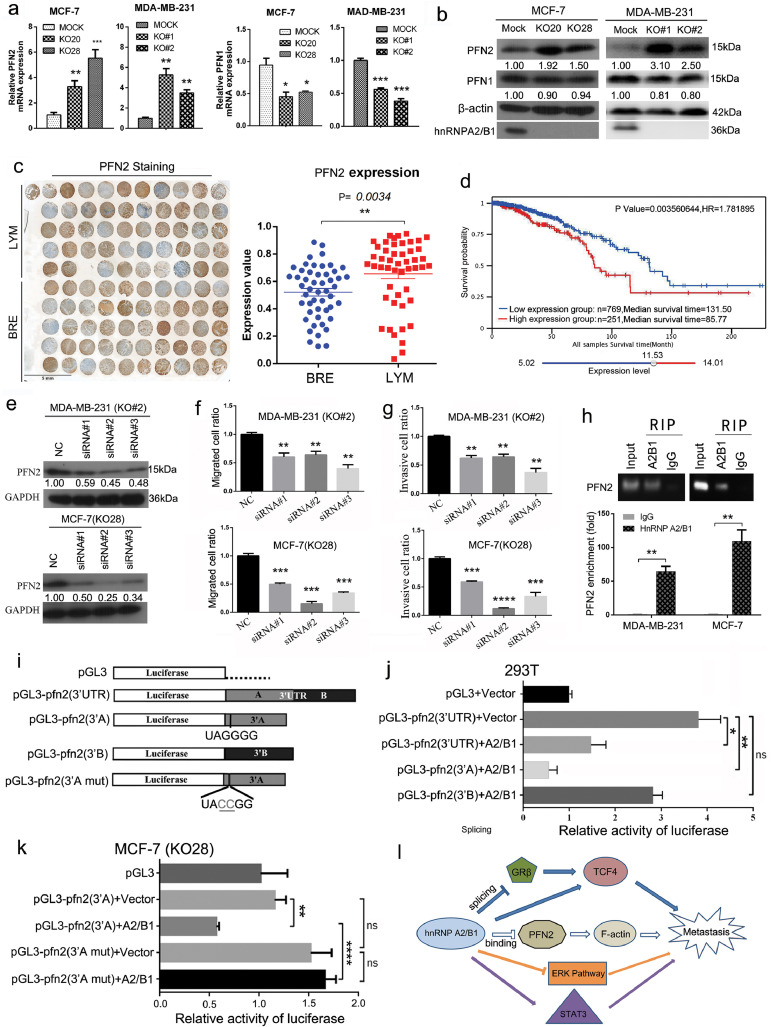
The qRT-PCR results showed that hnRNPA2/B1 knockout downregulated the mRNA expression of PFN1 and upregulated the mRNA expression of PFN2 in MDA-MB-231 and MCF-7 cells (Fig. 6A). The upregulation of PFN2 was confirmed by the Western blot analysis, whereas PFN1 did not dramatically change at the protein level (Fig. 6B).
Fig. 6.
hnRNPA2/B1 decreases the profilin 2 (PFN2) expression by binding PFN2 mRNA 3ʹ-UTR sequence UAGGG and PFN2 promotes the migration and invasion of human breast cancer cells in vitro. (A) Quantitative PCR showed the increased PFN2 expression and the decreased PFN1 expression in hnRNPA2/B1 KO cells of MCF-7 and MDA-MB-231. (B) Western blot analysis results confirmed the significantly upregulated PFN2 and the downregulated PFN1 in hnRNPA2/B1 KO cells. However, the change in PFN1 protein levels was not as drastic. Full panels of each Western blot can be found in Supplementary Fig. S14. (C) Quantitative results showed that the PFN2 expression significantly increased in the metastatic cancer tissues of the lymph nodes. The tissue sample microarray of 50 cases of invasive ductal breast cancer and matched lymph node metastatic tissues was analysed by IHC staining. BRE: breast cancer tissues; LYM: matched metastatic tissues in lymph node. (D) The analysis of the OS of the dataset of TCGA_RNA_Seq (http://www.omicsnet.org/bcancer/) indicated that the patients with a high PFN2 expression showed low survival probability. (E) Small interference RNAs were used to knock down (KD) the expression level of PFN2 in MDA-MB-231 and MCF-7 cells. hnRNPA2/B1 was previously knocked out from the two cells. The KD of PFN2 was tested by Western blot analysis. (F) Transwell assay results indicated that PFN2 KD significantly downregulated the migration of MDA-MB-231 and MCF-7 cells. (G) PFN2 KD significantly decreased the invasion of MDA-MB-231 and MCF-7 cells. (H) RNA immunoprecipitation assay indicated that hnRNPA2/B1 protein was bound to the mRNA of PFN2. (I) The full length of 3ʹ-UTR, the first half of 3ʹ-UTR (3ʹA), and the latter half of the 3ʹ-UTR (3ʹB) were cloned into the Xbal-Fsel restriction site downstream the firefly luciferase reporter plasmids (pGL3 promoter). The hnRNPA2/B1 binding sequence UAGGG was mutated to UACCG. (J) Dual-luciferase reporter assay results indicated that the full length 3ʹ-UTR and the first half of 3ʹ-UTR (3ʹA) in PFN2 mRNA significantly decreased the luciferase activity when hnRNPA2/B1 was co-expressed in 293T cells. (K) Dual-luciferase reporter assay was performed in hnRNPA2/B1 KO cells (MCF-7, KO28).The restored hnRNPA2/B1 protein significantly decreased the luciferase activity in KO28 MCF-7 cells, whereas the mutant of the binding site eliminated the inhibitory effect of hnRNPA2B1 protein on the luciferase activity. Vector: control (empty) vector, A2/B1: the expression vector of hnRNPA2/B1. All the data were expressed as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (Student's t-test). (L) Working model of hnRNPA2/B1 as a negative regulator of human breast cancer metastasis. The effect of hnRNPA2/B1 on breast cancer metastasis was a balance of multiple genes and signalling pathways. hnRNPA2/B1 KO promoted cell mobility by stabilising the mRNA of PFN2 and upregulating its expression. hnRNPA2/B1 KO promoted the activation of ERK MAPK pathway but inhibited the STAT3 pathway. The effects of hnRNPA2/B1 on the WNT/β-catenin/TCF4 pathway were complex and contradictory. On the one hand, hnRNPA2/B1 KO inhibited the TCF4 expression. On the other hand, hnRNPA2/B1 KO promoted the upregulation of GRβ through alternative splicing, thereby maintaining or promoting the TCF4 activity.
The PFN2 expression in lymph node metastases increased significantly by using the tissue microarray of invasive ductal carcinoma (Fig. 6C, Supplementary Fig. S2B), suggesting that PFN2 may promote breast cancer metastasis. Differential and OS analyses were conducted on the PFN2 gene in patients with breast cancer by using the data from KM-plotter (http://kmplot.com/analysis/) and the datasets of the TCGA_RNA_Seq and the METABRIC (http://www.omicsnet.org/bcancer/). The results indicate a significant increase in PFN2 in breast cancer tissues compared with adjacent normal tissue, a significant increase in PFN2 in triple-negative breast cancer compared with non-TNBC and a lower survival probability in patients with high PFN2 expression (Fig. 6D, Supplementary Fig. S9).
Small interference RNAs targeting PFN2 were constructed and transfected into breast cancer cells to study the role of PFN2 on breast cancer metastasis (Fig. 6E). The PFN2 knockdown was found to significantly decrease the migration (Fig. 6F) and invasion (Fig. 6G) of MDA-MB-231 and MCF-7 cells (Supplementary Fig. S10A and B), respectively. Overexpression of PFN2 in MDA-MB-231 cells promoted cell migration and invasion (Supplementary Fig. S10C and D). These data prove that PFN2 promotes breast cancer invasion.
3.8. HnRNPA2/B1 binds to the 3′-UTR region of PFN2 mRNA and reduces its stability
Considering that hnRNPA2/B1 is an RNA-binding protein, we performed an RIP experiment to determine whether hnRNPA2/B1 binds to the mRNA of PFN2. The RIP results showed that hnRNPA2/B1 bound to the mRNAs of PFN1 and PFN2 (Fig. 6H and Supplementary Fig. S10E).
We hypothesised that hnRNPA2/B1 affects the stability of PFN2 by binding to its mRNA 3′-UTR. To verify the hypothesis, we cloned the 3′-UTR region and the fragments (3′A and 3′B) of PFN2 mRNA and inserted them behind the coding region of the luciferase reporter (Fig. 6I). The results of the dual-luciferase reporter gene assay showed that the 3′-UTR and fragment 3′A of PFN2 significantly reduced luciferase activity, whereas fragment 3′B did not affect luciferase activity (Fig. 6J). This finding suggests that hnRNPA2/B1 binds to the fragment A of the 3′-UTR of PFN2 mRNA and reduces the stability of PFN2 mRNA.
We found an RNA sequence UAGGG that can be bound by the hnRNPA2B1 protein [32], which located on fragment 3′A. We mutated the sequence to UACCG. The dual-luciferase reporter gene assay in 293T cells showed that the mutated fragment A significantly increased the luciferase activity (Supplementary Fig. S10F). Considering that the endogenous expression of hnRNPA2/B1 in 293T cells may affect the experimental results, we repeated the dual-luciferase reporter experiment in hnRNPA2/B1 KO MCF-7 cells (KO28). The restored hnRNPA2/B1 expression significantly decreased the luciferase activity, whereas the mutant eliminated the inhibitory effect of A2B1 protein on luciferase activity (Fig. 6K). These results confirm that the UAGGG sequence of the PFN2 3′-UTR is the binding site of the hnRNPA2/B1 protein and mediates the downregulation of PFN2 mRNA.
4. Discussion
In this study, we proved that hnRNPA2/B1 is a negative regulator of breast cancer metastasis. The role of hnRNPA2/B1 in breast cancer metastasis is quite different from that in other tumours. In hepatoma cells, hnRNPA2/B1 induces EMT and promotes the metastasis by activating the transcription factor SNAIL [33]. In prostate cancer [20], cytoplasmic hnRNP A2 may activate the migration-promoting WNT pathway by promoting the expression of beta-catenin. hnRNPA2/B1 knockdown could reduce the adhesion, migration and invasion of glioblastoma by decreasing the expression of phosphor-STAT3 and MMP-2 [11]. hnRNPA2/B1 silencing increased the expression of E-cadherin and downregulated Twist1 and Snail, which are E-cadherin inhibitors in non-epithelial lung cancer cell lines A549 and H1703 [23]. These data suggest that hnRNPA2/B1 promotes metastasis in these tumours. Nevertheless, our cellular phenotype and animal model studies have shown that hnRNPA2/B1 may inhibit metastasis in breast cancer cells. The analyses of clinical samples and databases in support this conclusion.
Importantly, a limitation of our study is that only two cell lines were used: MCF-7 luminal A and MDA-MB-231 TNBC cells. Further, the in vivo studies did not include mammary gland implantation studies of MDA-MB-231 cells. Therefore, the application of these results to other breast cancer cells and subtypes remains to be determined. According to our analysis of breast cancer data in publically available databases, most subtypes with high hnRNPA2B1 expression had significantly longer survival time. Hence, the conclusion of this work may not only apply to luminal A and triple-negative breast cancer.
The different roles of hnRNPA2/B1 in different tumours may be attributed to the fact that hnRNPA2/B1 has many downstream genes [15] and even influence the processing and splicing of microRNA [34,35]. These downstream genes and microRNAs may have different roles or different expression patterns in different tumours. The ultimate effect of hnRNPA2/B1 on metastasis may be a balance of the roles of downstream molecules. In addition, a previous study found that A2 and B1 proteins may have different preferences for different RNA molecules [6]; the proportion of A2 and B1 may change in different tissues and pathological stages [5,36,37], which further increases the complexity of the role of hnRNPA2/B1 in different tumours. These phenomena may have caused the contradictory roles of hnRNPA2/B1 in different cancers.
Our breast cancer tissue chip analysis showed that although 78% of breast cancer was hnRNPA2/B1 positive, its expression level was significantly reduced in the metastatic cancer tissues in the lymph node of the corresponding patients. Data analysis from public databases also showed that the low expression of hnRNPA2/B1 was accompanied by low survival. In this study, the knockout of hnRNPA2/B1 significantly promoted the migration and invasion in vitro and facilitated the metastasis of breast cancer cells in vivo. Our study confirmed for the first time that hnRNPA2/B1 plays a different role in breast cancer and is a negative regulator of breast cancer cell metastasis.
Our results found that hnRNAP2/B1 promoted tumour proliferation. The in vitro cell proliferation experiments and in vivo growth curves of the transplanted tumour showed that A2B1 KO significantly inhibited the proliferation of breast cancer MDA-MB-231 and MCF-7 cells. Significant differences were observed between the tumour sizes of the two types of A2B1 KO cells in vivo. This finding could be attributed to the fact that the two cell lines are different subtypes of breast cancer, and the roles of A2B1 in the two cell lines are subtly different. Proliferation and metastasis are two major causes of cancer death, and hnRNPA2/B1 showed opposite effects on these lethal factors in breast cancer. Clinically, metastasis is the main cause of tumour recurrence and death due to difficulty in controlling metastasis. Therefore, we speculate that high hnRNPA2/B1 expression should be clinically effective in prolonging the survival of breast cancer patients by inhibiting metastasis.
Our data not only demonstrate that hnRNPA2/B1 is a negative regulator of metastasis in breast cancer but also confirm that hnRNPA2/B1 can bind to the UAGGG sequence in the 3′-UTR region of PFN2 mRNA to reduce its stability, whereas PFN2 promotes the metastasis of breast cancer cells. As an actin-binding protein, PNF2 regulates actin dynamics [38] and endocytosis [39]. PFN2 has been reported to promote invasion, metastasis and EMT in lung cancer [40,41] and colorectal cancer [42]. Although other molecular mechanisms may exist, this finding at least partially explains the mechanisms by which hnRNPA2/B1 knockout promotes migration and invasion.
The transcriptome analysis of miRNAs regulated by hnRNPA2B1 indicates that hnRNPA2B1 is involved in the regulation of cell substrate adhesion [35]. In addition to the upregulated PFN2, alpha5/beta1 integrin and its ligand fibronectin were significantly upregulated by hnRNPA2/B1 knockout. Alpha5/beta1integrin can directly or indirectly modulate MMP-2 collagenase activity and then regulate human breast cancer cell invasion [43]. Opposite forces produced by cell–cell adhesion and extracellular matrix-mediated cell migration regulate cell dispersion from multicellular aggregates [44,45]. Alpha5-mediated dispersion from the tumour mass overrides alpha5-mediated tumour cell adhesion in fibronectin-rich microenvironment [46]. Both previous and present findings may explain why Transwell assay with Matrigel could isolate cell subsets with low hnRNPA2/B1 expression. The cells with low hnRNPA2/B1 expression upregulated alpha-5 integrin, and the fibronectin in Matrigel resulted in more migratory cells with high alpha-5 integrin.
We also found that A2/B1 knockout activated ERK MAPK signal pathway, improved the expression of transcription factor Twist downstream of ERK signalling pathway and altered some important cell adhesion molecules, such as the downregulated E-cadherin and the upregulated N-cadherin. Activation of the ERK MAPK/Twist1/ E-cadherin action axis also helps explain the promoting effect of A2/B1 KO on metastasis.
Interestingly, although hnRNPA2/B1 knockout appeared to promote metastasis in vivo and in vitro, STAT3 and TCF4 [31] which are found to promote metastasis [47], [48], [49], [50], are inhibited in hnRNPA2/B1 knockout cells. More interestingly, hnRNPA2/B1 knockout promotes expression of GR beta through alternative splicing in our study, whereas GR beta maintains or facilitates TCF4 activity [31]. These findings further suggest that the downstream genes of hnRNPA2/B1 may play opposite roles in cancer metastasis, and the metastasis regulated by hnRNPA2/B1 should be a balance of these genes in breast cancer. hnRNPA2/B1 has a complex and even contradictory effect on metastasis, as reflected in the fact that A2B1 has opposite effects on the early and late stages of the metastasis of breast cancer cells. Although hnRNPA2/B1 knockout has been shown to promote metastasis in vivo, the animal model studies suggest that hnRNPA2/B1 knockout reduces the number of invasive cancer cells in the blood but promotes metastatic colonisation process. The decreased cell viability by A2B1 KO and the harsh blood environment for cells may explain the decrease in hnRNPA2/B1 knockout cells in the blood system. Similar phenomena were found in previous studies. For example, the loss of PFN-1 promotes vascular dissemination but causes a major impairment in metastatic outgrowth of MDA-231 cells, suggesting that PFN-1 downregulation has contrasting effects on early versus late steps of breast cancer metastasis [51].
Some problems must be further clarified in our study. First, the effect of PFN2 on migration may require further validation in animal models. Second, are there other downstream genes of hnRNPA2/B1 involved in the regulation of metastasis? Third, hnRNPA2/B1 is reduced in metastatic cancer cells; then, who regulates the expression of hnRNPA2/B1 upstream? The answers will shed more light on the mechanisms of hnRNPA2/B1 in tumours.
Overall, our data first demonstrate that hnRNPA2/B1 is a negative regulator of breast cancer metastasis, unlike its roles in other cancers. This negative regulation may be due to the balance of multiple hnRNPA2/B1 downstream genes and signalling pathways. The hnRNPA2/B1 protein binds to PFN2 mRNA and downregulates its stability, thereby affecting the metastasis of breast cancer which at least partly explains the role of hnRNPA2/B1 knockout in promoting migration. hnRNPA2/B1 may be used as a new prognostic biomarker and a valuable molecular target for treatment of breast cancer.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81272921, 81670542, 81871679).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.044.
Appendix. Supplementary materials
References
- 1.Tse J.C., Kalluri R. Mechanisms of metastasis: epithelial-to-mesenchymal transition and contribution of tumor microenvironment. J Cell Biochem. 2007;101(4):816–829. doi: 10.1002/jcb.21215. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Bendre M, Gaddy D, Nicholas RW, Suva LJ. Breast cancer metastasis to bone: it is not all about PTHrP. Clin Orthop Relat Res. 2003;415(Suppl):S39–S45. doi: 10.1097/01.blo.0000093844.72468.f4. [DOI] [PubMed] [Google Scholar]
- 4.Li Z., Kang Y. Emerging therapeutic targets in metastatic progression: a focus on breast cancer. Pharmacol Ther. 2016;161:79–96. doi: 10.1016/j.pharmthera.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozu T., Henrich B., Schafer K.P. Structure and expression of the gene (HNRPA2B1) encoding the human hnRNP protein A2/B1. Genomics. 1995;25(2):365–371. doi: 10.1016/0888-7543(95)80035-k. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen E.D., Balas M.M., Griffin A.M., Roberts J.T., Johnson A.M. Global profiling of hnRNP A2/B1-RNA binding on chromatin highlights LncRNA interactions. RNA Biol. 2018;15(7):901–913. doi: 10.1080/15476286.2018.1474072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vera J., Jaumot M., Estanyol J.M., Brun S., Agell N., Bachs O. Heterogeneous nuclear ribonucleoprotein A2 is a SET-binding protein and a PP2A inhibitor. Oncogene. 2006;25(2):260–270. doi: 10.1038/sj.onc.1209050. [DOI] [PubMed] [Google Scholar]
- 8.Chen M., Zhang J., Manley J.L. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010;70(22):8977–8980. doi: 10.1158/0008-5472.CAN-10-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clower C.V., Chatterjee D., Wang Z., Cantley L.C., Vander Heiden M.G., Krainer A.R. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci USA. 2010;107(5):1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moran-Jones K., Grindlay J., Jones M., Smith R., Norman J.C. hnRNP A2 regulates alternative mRNA splicing of TP53INP2 to control invasive cell migration. Cancer Res. 2009;69(24):9219–9227. doi: 10.1158/0008-5472.CAN-09-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng J., Chen S., Wang F., Zhao H., Xie Z., Xu Z. Effects of hnRNP A2/B1 knockdown on inhibition of glioblastoma cell invasion, growth and survival. Mol Neurobiol. 2016;53(2):1132–1144. doi: 10.1007/s12035-014-9080-3. [DOI] [PubMed] [Google Scholar]
- 12.He Y., Rothnagel J.A., Epis M.R., Leedman P.J., Smith R. Downstream targets of heterogeneous nuclear ribonucleoprotein A2 mediate cell proliferation. Mol Carcinog. 2009;48(2):167–179. doi: 10.1002/mc.20467. [DOI] [PubMed] [Google Scholar]
- 13.Villarroya-Beltri C., Gutierrez-Vazquez C., Sanchez-Cabo F., Perez-Hernandez D., Vazquez J., Martin-Cofreces N. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Percipalle P., Jonsson A., Nashchekin D., Karlsson C., Bergman T., Guialis A. Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res. 2002;30(8):1725–1734. doi: 10.1093/nar/30.8.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geissler R., Simkin A., Floss D., Patel R., Fogarty E.A., Scheller J. A widespread sequence-specific mRNA decay pathway mediated by hnRNPs A1 and A2/B1. Genes Dev. 2016;30(9):1070–1085. doi: 10.1101/gad.277392.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan-Sanders Y., Hammons G.J., Lyn-Cook B.D. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett. 2002;183(2):215–220. doi: 10.1016/s0304-3835(02)00168-4. [DOI] [PubMed] [Google Scholar]
- 17.Mizuno H., Honda M., Shirasaki T., Yamashita T., Yamashita T., Mizukoshi E. Heterogeneous nuclear ribonucleoprotein A2/B1 in association with hTERT is a potential biomarker for hepatocellular carcinoma. Liver Int. 2012;32(7):1146–1155. doi: 10.1111/j.1478-3231.2012.02778.x. [DOI] [PubMed] [Google Scholar]
- 18.Fielding P., Turnbull L., Prime W., Walshaw M., Field J.K. Heterogeneous nuclear ribonucleoprotein A2/B1 up-regulation in bronchial lavage specimens: a clinical marker of early lung cancer detection. Clin Cancer Res. 1999;5(12):4048–4052. [PubMed] [Google Scholar]
- 19.Zhou J., Allred D.C., Avis I., Martinez A., Vos M.D., Smith L. Differential expression of the early lung cancer detection marker, heterogeneous nuclear ribonucleoprotein-A2/B1 (hnRNP-A2/B1) in normal breast and neoplastic breast cancer. Breast Cancer Res Treat. 2001;66(3):217–224. doi: 10.1023/a:1010631915831. [DOI] [PubMed] [Google Scholar]
- 20.Stockley J., Villasevil M.E., Nixon C., Ahmad I., Leung H.Y., Rajan P. The RNA-binding protein hnRNPA2 regulates beta-catenin protein expression and is overexpressed in prostate cancer. RNA Biol. 2014;11(6):755–765. doi: 10.4161/rna.28800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regina GG, Michal C, Asaf S, Sung-Suk S, Arianna B, Luigi C. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 2011;71(13):4464–4472. doi: 10.1158/0008-5472.CAN-10-4410. [DOI] [PubMed] [Google Scholar]
- 22.Barcelo C., Etchin J., Mansour M.R., Sanda T., Ginesta M.M., Sanchez-Arevalo Lobo V.J. Ribonucleoprotein HNRNPA2B1 interacts with and regulates oncogenic KRAS in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147(4):882–892.e8. doi: 10.1053/j.gastro.2014.06.041. [DOI] [PubMed] [Google Scholar]
- 23.Tauler J., Zudaire E., Liu H., Shih J., Mulshine J.L. hnRNP A2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res. 2010;70(18):7137–7147. doi: 10.1158/0008-5472.CAN-10-0860. [DOI] [PubMed] [Google Scholar]
- 24.Zheng L.C., Wang X.Q., Lu K., Deng X.L., Zhang C.W., Luo H. Ephrin-B2/Fc promotes proliferation and migration, and suppresses apoptosis in human umbilical vein endothelial cells. Oncotarget. 2017;8(25):41348–41363. doi: 10.18632/oncotarget.17298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J., Hu S., Chen Y., Li Z., Zhang J., Yuan H. BCIP: a gene-centered platform for identifying potential regulatory genes in breast cancer. Sci Rep. 2017;7:45235. doi: 10.1038/srep45235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Islam T., Resat H. Quantitative investigation of MDA-MB-231 breast cancer cell motility: dependence on epidermal growth factor concentration and its gradient. Mol Biosyst. 2017;13(10):2069–2082. doi: 10.1039/c7mb00390k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S.J., Saadi W., Lin F., Minh-Canh Nguyen C., Li Jeon N. Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis. Exp Cell Res. 2004;300(1):180–189. doi: 10.1016/j.yexcr.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 29.Hong J., Zhou J., Fu J., He T., Qin J., Wang L. Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness. Cancer Res. 2011;71(11):3980–3990. doi: 10.1158/0008-5472.CAN-10-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang M.H., Hsu D.S., Wang H.W., Wang H.J., Lan H.Y., Yang W.H. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12(10):982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q., Lu P.H., Shi Z.F., Xu Y.J., Xiang J., Wang Y.X. Glucocorticoid receptor beta acts as a co-activator of T-Cell factor 4 and enhances glioma cell proliferation. Mol Neurobiol. 2015;52(3):1106–1118. doi: 10.1007/s12035-014-8900-9. [DOI] [PubMed] [Google Scholar]
- 32.Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z.J., Dai Z., Zhou S.L., Hu Z.Q., Chen Q., Zhao Y.M. HNRNPAB induces epithelial-mesenchymal transition and promotes metastasis of hepatocellular carcinoma by transcriptionally activating SNAIL. Cancer Res. 2014;74(10):2750–2762. doi: 10.1158/0008-5472.CAN-13-2509. [DOI] [PubMed] [Google Scholar]
- 34.Alarcon C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 is a mediator of m (6)A-Dependent nuclear RNA processing events. Cell. 2015;162(6):1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klinge CM, Piell KM, Tooley CS, Rouchka EC. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci Rep. 2019;9(1):9430–9451. doi: 10.1038/s41598-019-45636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zech V.F.E., Dlaska M., Tzankov A., Hilbe W. Prognostic and diagnostic relevance of hnRNP A2/B1, hnRNP B1 and S100 A2 in non-small cell lung cancer. Cancer Detect Prev. 2006;30(5):395–402. doi: 10.1016/j.cdp.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Satoh H., Ishikawa H., Kamma H., Fujiwara M., Homma S., Kagohashi K. Expression of hnRNP A2/B1 proteins in small airway epithelial cells. Int J Mol Med. 2004;14(4):605–608. [PubMed] [Google Scholar]
- 38.Schlüter K., Jockusch B.M., Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359(2):97–109. doi: 10.1016/s0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- 39.Gareus R., Di Nardo A., Rybin V., Witke W. Mouse profilin 2 regulates endocytosis and competes with SH3 ligand binding to dynamin 1. J Biol Chem. 2006;281(5):2803–2811. doi: 10.1074/jbc.M503528200. [DOI] [PubMed] [Google Scholar]
- 40.Tang Y.N., Ding W.Q., Guo X.J., Yuan X.W., Wang D.M., Song J.G. Epigenetic regulation of Smad2 and Smad3 by profilin-2 promotes lung cancer growth and metastasis. Nat Commun. 2015;6:8230. doi: 10.1038/ncomms9230. [DOI] [PubMed] [Google Scholar]
- 41.Yan J., Ma C., Gao Y. MicroRNA-30a-5p suppresses epithelial-mesenchymal transition by targeting profilin-2 in high invasive non-small cell lung cancer cell lines. Oncol Rep. 2017;37(5):3146–3154. doi: 10.3892/or.2017.5566. [DOI] [PubMed] [Google Scholar]
- 42.Kim M.J., Lee Y.S., Han G.Y., Lee H.N., Ahn C., Kim C.W. Profilin 2 promotes migration, invasion, and stemness of HT29 human colorectal cancer stem cells. Biosci Biotechnol Biochem. 2015;79(9):1438–1446. doi: 10.1080/09168451.2015.1043118. [DOI] [PubMed] [Google Scholar]
- 43.Galina M, Nadezda K, Ivan C, Natalia U, Albert B.J.C.C. Integrin alpha5beta1 controls invasion of human breast carcinoma cells by direct and indirect modulation of MMP-2 collagenase activity. Cell Cycle. 2009;8(14):2219–2225. doi: 10.4161/cc.8.14.8980. [DOI] [PubMed] [Google Scholar]
- 44.da Rocha-Azevedo B., Grinnell F. Fibroblast morphogenesis on 3D collagen matrices: the balance between cell clustering and cell migration. Exp Cell Res. 2013;319(16):2440–2446. doi: 10.1016/j.yexcr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryan P.L., Foty R.A., Kohn J., Steinberg M.S. Tissue spreading on implantable substrates is a competitive outcome of cell-cell vs. cell-substratum adhesivity. Proc Natl Acad Sci USA. 2001;98(8):4323–4327. doi: 10.1073/pnas.071615398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blandin A.F., Noulet F., Renner G., Mercier M.C., Choulier L., Vauchelles R. Glioma cell dispersion is driven by alpha5 integrin-mediated cell-matrix and cell-cell interactions. Cancer Lett. 2016;376(2):328–338. doi: 10.1016/j.canlet.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 47.Dey N., Barwick B.G., Moreno C.S., Ordanic-Kodani M., Chen Z., Oprea-Ilies G. Wnt signaling in triple negative breast cancer is associated with metastasis. BMC Cancer. 2013;13:537. doi: 10.1186/1471-2407-13-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Fu D., Chen Y., Su J., Wang Y., Li X. G3BP1 promotes tumor progression and metastasis through IL-6/G3BP1/STAT3 signaling axis in renal cell carcinomas. Cell Death Dis. 2018;9(5):501. doi: 10.1038/s41419-018-0504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang T., Ma Z., Liu L., Sun J., Tang H., Zhang B. DDX39 promotes hepatocellular carcinoma growth and metastasis through activating Wnt/beta-catenin pathway. Cell Death Dis. 2018;9(6):675. doi: 10.1038/s41419-018-0591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu H., Chang L.L., Yan F.J., Hu Y., Zeng C.M., Zhou T.Y. AKR1C1 activates STAT3 to promote the metastasis of non-small cell lung cancer. Theranostics. 2018;8(3):676–692. doi: 10.7150/thno.21463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding Z., Joy M., Bhargava R., Gunsaulus M., Lakshman N., Miron-Mendoza M. Profilin-1 downregulation has contrasting effects on early vs. late steps of breast cancer metastasis. Oncogene. 2014;33(16):2065–2074. doi: 10.1038/onc.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.