Abstract
Background
Vascular endothelial zinc finger 1 (Vezf1) is a transcription factor previously shown to regulate vasculogenesis and angiogenesis. We aimed to investigate the role of Vezf1 in the postnatal heart.
Methods
The role of Vezf1 in regulating cardiac growth and contractile function was studied in zebrafish and in primary cardiomyocytes.
Findings
We find that expression of Vezf1 is decreased in diseased human myocardium and mouse hearts. Our experimental data shows that knockdown of zebrafish Vezf1 reduces cardiac growth and results in impaired ventricular contractile response to β-adrenergic stimuli. However, Vezf1 knockdown is not associated with dysregulation of cardiomyocyte Ca2+ transient kinetics. Gene ontology enrichment analysis indicates that Vezf1 regulates cardiac muscle contraction and dilated cardiomyopathy related genes and we identify cardiomyocyte Myh7/β-MHC as key target for Vezf1. We further identify a key role for an MCAT binding site in the Myh7 promoter regulating the response to Vezf1 knockdown and show that TEAD-1 is a binding partner of Vezf1.
Interpretation
We demonstrate a role for Vezf1 in regulation of compensatory cardiac growth and cardiomyocyte contractile function, which may be relevant in human cardiac disease.
Keywords: Vezf1, Cardiac hypertrophy, Cardiac contractile function, TEAD-1
Research in context.
Evidence before this study
Transcription factor Vezf1 has been shown to play an important role in angiogenesis and lymphangiogenesis especially during embryonic development.
Added value of this study
In our study, we find that Vezf1 is necessary for hemodynamic stress-induced increase in cardiac growth and contractile function. Vezf1 is expressed in adult cardiomyocytes, binds transcription factor TEAD-1 and directly regulates expression of cardiomyopathy related genes.
Implications of the available evidence
Our data indicates a novel role for Vezf1 in regulating cardiac structure and function and provides a potential therapeutic target for alleviating development of heart failure.
Alt-text: Unlabelled box
1. Introduction
Cardiovascular diseases are a global health problem and the leading cause of death in the Western world [1]. Sustained increase in cardiac workload, due to cardiovascular pathologies such as systemic hypertension, myocardial infarction (MI) and valvular heart disease, induces cardiac hypertrophy as an adaptive response allowing the heart to maintain cardiac output. Prolonged cardiac hypertrophy, however, predisposes to heart failure, which is characterized by diverse cellular abnormalities in the myocardium, including suppressed angiogenesis, vascular rarefaction, and aberrant Ca2+ cycling, eventually culminating in decreased left ventricular (LV) function [2,3].
Transcription factors, such as GATA-4, NKX2.5 and myocyte enhancer factor 2 (MEF2) have been shown to be nodal for cardiovascular development and later identified to play a role in the postnatal heart [4,5]. GATA-4, for example, is highly expressed during development and reactivated during cardiac pathologies regulating both endothelial cell and cardiomyocyte function [6,7]. A gene trap screen for early cardiovascular genes has previously identified a zinc finger transcription factor, Vezf1 [8]. Vezf1 encodes a nuclear protein containing six zinc finger motifs of the C2H2-type (krüppel-like) and a proline-rich transcriptional transactivation domain and it is highly conserved among vertebrates. In embryos, according to its name, Vezf1 has been shown to be expressed in endothelial cells as well as in mesodermal and neuronal tissues [9,10]. In one study, Vezf1 expression has also been detected in various adult human tissues, including the heart [8]. Human Vezf1 (also known as ZNF161 or DB1) functions as a bona fide transcription factor activating the endothelin-1 promoter [11]. Upstream of Vezf1, RhoB-GTP has been shown to regulate VEZF1-mediated transcription [12]. A role for Vezf1 in epigenetic gene regulation has also been suggested [13]. Vezf1-null mice are embryonic lethal, the main defects being in the vascular endothelium [9]. In a genome-wide quantitative trait loci mapping in zebrafish, Vezf1 was identified as a candidate gene causing genetic susceptibility to cardiotoxicity induced by dioxin-like compounds, suggesting a role for Vezf1 in the vertebrate heart [14]. The Vezf1 gene is located in the long arm of chromosome 17. Deletions in this area (17q21–17q24) have been reported to result in growth retardation, global developmental delay and specific musculoskeletal defects including congenital heart disease [15]. Overall, Vezf1 is poorly characterized and its possible role in the postnatal heart remains to be determined.
Zebrafish (Danio rerio) is a powerful model for studying the molecular and cellular mechanisms of cardiovascular diseases, as these are highly similar between zebrafish and other vertebrates [16]. Here we set out to investigate the role of Vezf1 in healthy and stressed myocardium. We find that Vezf1 gene expression is decreased in diseased human myocardium and in mouse hearts subjected to MI. Our experimental data shows that Vezf1 is required for normal and β-adrenergic stimuli-induced cardiac growth. Vezf1 knockdown in zebrafish blunts the β-adrenergic stress-induced increase in cardiac contractile function, which is not due to impaired Ca2+ cycling. Instead, we find that Vezf1 is expressed in adult cardiomyocytes and directly regulates genes related to cardiac muscle contraction and dilated cardiomyopathy. We further demonstrate that Vezf1 interacts with transcriptional enhancer factor-1 (TEF-1/TEAD-1), and regulates β-myosin heavy chain (β-MHC/Myh7) expression at least partially via an MCAT binding site in the β-MHC promoter. Our data thus identifies Vezf1 as a novel transcription factor necessary for β-adrenergic stress-induced increase in cardiac growth and contractile function.
2. Materials and methods
2.1. Human cardiac samples
The human MI samples were obtained from the Fingesture study, which has stored both clinical and autopsy data from sudden cardiac death (SCD) victims between 1998 and 2018 in Northern Finland (n = 4031) [17]. The study complies with the Declaration of Helsinki and was approved by the Ethics Committee of the Northern Ostrobothnia Hospital District. The National Authority for Medicolegal Affairs (Valvira) approved the review of postmortem data by the investigators. All victims were autopsied at the Department of Forensic Medicine, University of Oulu, Oulu, Finland and subjects with a history and findings of diseased coronary arteries at autopsy were considered cases with coronary artery disease. Samples of healthy cardiac tissue were collected from autopsies of traffic accident victims with no evidence of cardiovascular disease. Heart failure -related datasets GSE5406 and GSE1145 were obtained from the Gene Expression Omnibus database.
2.2. Zebrafish
Larvae of the wild-type Turku strain of zebrafish [18,19] were maintained and bred according to Westerfield [20] at the Biomedicum Helsinki Zebrafish Unit. 2–4 days post fertilization (dpf) zebrafish were used for the experiments. Zebrafish larvae were maintained in E3 medium (5.0 mM NaCl, 0.4 mM CaCl2, 0.3 mM MgSO4 and 0.2 mM KCl) and treated with isoprenaline (Iso) (Sigma-Aldrich, St. Louis, MO) when desired. For heart resection, zebrafish were anesthetized in 0.03% tricaine. Zebrafish without heart resection were euthanized in 0.03% tricaine followed by addition of ice. Experiments on zebrafish were performed in accordance with protocols approved by the National Animal Experiment Board of Finland (ESAVI/4131/04.10.07/2017). Vezf1 splice-blocking morpholino antisense oligonucleotides (SBMO) 5´-CATTGGCCTGCTGGATGGAGAAAGA-3´, Vezf1 translation-blocking MO (TBMO) 5´-ATGAACTCCAGCTCGGCTCCATTGC-3´ and random control oligo (control) 25-N were from Gene Tools, LLC (Philomath, OR). Dosage was determined by titration; 3–4 ng for SBMO and 5–7 ng for TBMO. After confirming similar findings with both morpholinos in phenotype as well as in cardiac physiology, experiments were done with SBMOs, as they better allowed simultaneous qPCR experiments. For the rescue experiments 250 pg of capped Vezf1 mRNA was co-injected with SBMO.
2.3. TUNEL
Zebrafish hearts at 4 dpf were dissected, fixed in 4% paraformaldehyde (PFA) and washed two times in PBS. The samples were stained with ApopTag Red In Situ Apoptosis Detection Kit (Millipore, Burlington, MA), Mef-2 antibody (SC-313, RRID: AB_631,920, Santa Cruz Biotechnology, Dallas, TX) and Dapi (Thermo Fisher Scientific, Waltham, MA). Anti-rabbit Alexa Fluor 488 antibody (A-11,008, RRID: AB_143,165, Thermo Fisher Scientific) was used as secondary antibody. Images were obtained with a Zeiss Apotome (Carl Zeiss Microscopy GmbH, Jena, Germany) equipped with a Hamamatsu ORCA-R2 camera (Hamamatsu Photonics, Shizuoka, Japan) and quantified with ImageJ 1.43 u software (RRID: SCR_003070, NIH, Bethesda, MD).
2.4. Ventricular cardiomyocyte size and number
Hearts were dissected from 4 dpf zebrafish larvae and fixed in 4% PFA and washed in PBS + 0,1% Triton. Hearts were stained with Mef-2 (SC-313, RRID: AB_631,920) and ZN-5 (RRID: AB_10,013,770, Zebrafish International Resource Center) antibodies overnight at +4 °C. Anti-rabbit Alexa Fluor 594 (A-11,012, RRID: AB_141,359) and anti-mouse Alexa Fluor 488 (A-11,001, RRID: AB_2,534,069, Thermo Fisher Scientific) were used as secondary antibodies. Images were captured with a Zeiss AxioImager Z1 microscope by using a Hamamatsu ORCA-R2 camera. Images were analyzed using ImageJ as follows: both anterior and posterior walls were divided to four roughly equal size sectors and the size of four randomly selected cardiomyocytes from each sector was measured and the average cardiomyocyte area of the 32 cardiomyocytes was measured. The total number of cardiomyocytes was counted using the ImageJ Plugin Extended Depth of Field.
2.5. Transmission electron microscopy
Whole zebrafish were fixed in 1% glutaraldehyde and 4% formaldehyde in 0.1 M phosphate buffer, postfixed in 1% osmium tetroxide, dehydrated in acetone and embedded in Epon LX 112 (Ladd Research Industries). Thin sections were cut with a Leica Ultracut UCT ultramicrotome, stained in uranyl acetate and lead citrate and examined in a Philips CM100 transmission electron microscope. The images were captured using a Morada CCD camera (Olympus Soft Imaging Solutions).
2.6. Cardiac physiology and vasculature
The heart function of 2 dpf and 4 dpf larvae was recorded and analyzed from the recorded videos as described earlier [21]. Briefly, larvae were anesthetized in 0.008% tricaine solution and mounted on 3% methylcellulose. Heart and vascular videos were recorded with an Olympus IX70 microscope (Olympus, Tokyo, Japan), a Hamamatsu ORCA-Flash 4.0 camera and HCImage software (RRID:SCR_015041, Hamamatsu Photonics, Shizuoka, Japan) and analyzed with ImageJ. Diameters and distances of vascular structures were measured from the zebrafish trunk from the area where the dorsal aorta (DA) and the posterior cardinal vein (PCV) are parallel. The distance between intersegmental vessels (ISVs) was measured from 3–5 locations of the zebrafish trunk, and the average calculated.
2.7. Ca2+ cycling in zebrafish hearts
For Ca2+ imaging, hearts from 3 dpf larvae were loaded with 55 μM Fura2 (Thermo Fisher Scientific) for 30 min and then de-esterified for 20 min as described [21]. The hearts were perfused with Tyrode's solution and paced with 10 ms bipolar pulses at 1.25 Hz by using a SIU-102 Stimulus Isolation Unit (Warner Instruments, Hamden, CT). Images were recorded with a Hamamatsu ORCA-Flash 4.0 camera at 10–24 frames per second. Polychrome IV (Till Photonics, Gräfelfing, Germany) was used as the light source. The ventricle was selected as region of interest and background fluorescence was subtracted before quantifying the fluorescence in relation to baseline fluorescence (F/F0). Resting Ca2+ concentration and Ca2+ transient amplitude are presented as Fura2 340/380 ratio units. Data were analyzed with HCImage and Axon Clampfit 9.2 (Molecular Devices, San Jose, CA) softwares. Only hearts that appeared unharmed after isolation and showed spontaneous beating with normal atrioventricular conduction were included in the analyses.
2.8. Neonatal rat ventricular cardiomyocytes
Neonatal rat ventricular cardiomyocytes (NRVMs) were isolated from 2 to 4-day old Sprague-Dawley (SD) rats. Rats were euthanized by quick decapitation and ventricles were collected into 2 mg/ml collagenase type 2 solution (Worthington, Lakewood, NJ) prepared in 1xPBS with 50 µM CaCl2. The heart fragments were incubated for 5 min at +37 °C with stirring, and thereafter the collagenase solution was discarded. 5 ml of collagenase solution was added and incubated for 20 min at +37 °C with stirring and the collagenase solution was collected through 100 µm nylon cell strainer (BD, Franklin Lakes, NJ) to DMEM/F-12 (Sigma-Aldrich) containing 30% fetal bovine serum (Thermo Fisher Scientific) and 1% penicillin-streptomycin (PS). This process was repeated 5 times. The cells were next pelleted (200 g, 5 min) and washed with DMEM/F-12 supplemented with 10% FBS and 1% PS. To reduce the number of contaminating non-muscle cells, the cells were pre-plated onto culture dishes (BD) for 2 h at 37 °C in humidified atmosphere with 5% CO2. Isolated NRVMs were plated and incubated in DMEM/F-12 supplemented with 10% FBS and 1% PS overnight. siRNA transfections were performed one day after cell isolation and experiments were started 2–4 days after transfection. For hypoxia experiments, four days after siRNA transfections cells were incubated in DMEM supplemented with 10 mM deoxyglucose and 1 mM sodium dithionite (Honeywell-Fluka, Morris Plains, NJ), and kept in a C-Chamber (BioSpherix, Parish, NY) and oxygen levels controlled at 0.1% with a ProOx C21 controller (BioSpherix). Control (normoxia) cells were incubated in DMEM (Thermo Fisher Scientific) supplemented with 10 mM glucose.
2.9. Adult ventricular cardiomyocytes
Mouse ventricular cardiomyocytes, cardiac endothelial cells and fibroblasts were isolated from 8-week old c57/BL6 mice as described previously [22]. Adult rat ventricular cardiomyocytes (ARVMs) were isolated from 8–12 weeks old male SD rats by retrograde perfusion and enzymatic digestion using collagenase type 2 (Worthington) as previously described [23]. After 2 h incubation, non-attached cells were gently removed and cells were transfected with desired siRNAs. Experiments were started 24–48 h after transfection. Ca2+ transients from cultured ARVMs were measured as previously described [24]. For measurement of adult rat cardiomyocyte size, isolated ARVMs were plated on laminin coated glass chamber slides and treated with isoprenaline (1 µM) when appropriate. For measurement of cardiomyocyte size, cells were fixed with 4% paraformaldehyde, blocked with 10% FBS in PBS and stained with Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific). The cells were viewed with a Nikon Eclipse 80i microscope (Nikon, Tokyo, Japan), and captured with an ORCA-Flash 4.0 camera and NIS-Elements AR 4.30.01 software (RRID: SCR_014329, Nikon). Only rod-shaped cardiomyocytes were measured.
2.10. RNA interference
Specific Vezf1 siRNAs (Vezf1A 5′-CCCAAAACUUCGUUUGUGUtt-3′ and Vezf1B 5′-GGUAUCAUGUAACAUCUGUtt-3′), Tead1 siRNAs (Tead1A 5′-AGACGGAGUAUGCGAGGUUUU-3′ and Tead1B 5′-GAGUAUGCGAGGUUCGAGAUU-3′) and negative control siRNA (SIC001, Sigma-Aldrich) were transfected into the NRVMs and ARVMs at 100 nM concentration by using Lipofectamine 2000 (Thermo Fisher Scientific) as transfection reagent. During transfections NRVMs were incubated in Opti-MEM® I (Thermo Fisher Scientific) for 24 h and thereafter the cells were incubated in serum free medium. ARVMs were transfected for 18 h and thereafter incubated in αMEM (Thermo Fisher Scientific) supplemented with Earle's salt containing 0.01% BSA, 10 mM HEPES, 1 × insulin-transferrin-selenium, 10 mM BDM, 2 mM l-glutamine and 1% PS.
2.11. Plasmids
β-MHC promoter luciferase reporter constructs were a kind gift from Fadia Haddad and are previously described [5]. The activities of β-MHC promoter luciferase reporter and Renilla luciferase pRL-TK were analyzed using Dual-luciferase Reporter Assay System (Promega, Madison, WI) 48 h after transfection. For Vezf1 expression plasmid, coding sequence of mouse Vezf1 (NM_016686.4) was cloned into pcDNA3.1 vector using BamHI cloning site. An empty pcDNA3.1 vector was used as control. Plasmids were transfected into the NRVMs and ARVMs using Lipofectamine 2000 as transfection reagent.
2.12. RNA isolation, cDNA synthesis and qRT-PCR analysis
Trizol (Thermo Fisher Scientific) was used for isolation of RNA from human cardiac tissue samples and an EZNA Total RNA kit I (Omega Bio-tek, Norcross, GA) from isolated cardiomyocytes and from crushed zebrafish larvae. Nuclease-Free water (Thermo Fisher Scientific) was used to dissolve the RNA pellets or eluate the RNA from isolation columns when using the EZNA kit. For cDNA synthesis Transcriptor first strand cDNA synthesis kit (Roche, Basel, Switzerland) was used according to manufacturer's instructions. The relative mRNA levels were analyzed with quantitative RT-PCR (qRT-PCR). When using fluorogenic probes, the qRT-PCR reaction mixtures contained 1 µl of the fluorogenic probe (Sigma-Aldrich), 2.5 µl of each forward and reverse primer (5 µM stock, Sigma-Aldrich), 2 µl of nuclease-free water (Thermo Fisher Scientific) and 12.5 µl of Fast Start Universal Probe Master (Roche). When using SYBR reagent, the qRT-PCR reaction mixtures contained 2.5 µl of each forward and reverse primer, 2.5 µl of nuclease-free water (2 µM stock, Thermo Fisher Scientific) and 12.5 µl of SYBR Green PCR Master Mix (Thermo Fisher Scientific). For the qRT-PCR-plate, 20 µl of the qRT-PCR reaction mix was added together with 5 µl of template cDNA. The sequences of the forward and reverse primers and the fluorogenic probes used for gene-specific cDNA detection are listed in Table S1. The qRT-PCR analyses were done with a 7300 Real Time PCR System (Thermo Fisher Scientific).
2.13. Protein isolation, immunoprecipitation and immunoblotting
Rat cardiac cells and rat left ventricular tissue samples were lysed into 20 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton-X100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate and 1 mM Na3VO4 (pH 7.5) supplemented with 1 mM 1,4-dithiothreitol, 50 mM sodium fluoride, 1:100 protease inhibitor cocktail 1 (Sigma-Aldrich) and 1:100 phosphatase inhibitor cocktail 3 (Sigma-Aldrich). For immunoprecipitation a Pierce Crosslink IP Kit was used according to manufacturer's instructions (Thermo Fisher Scientific). 250 µg of total protein from rat left ventricular samples was immunoprecipitated with 5 µg of Vef1 antibody (RRID: AB_10,606,448, SAB2102675, Sigma-Aldrich). Cell lysates and immunoprecipitation eluates were loaded on SDS‐PAGE (12%) and transferred to nitrocellulose membranes using Trans-Blot® Turbo™ RTA Transfer Kit (BIO-RAD, Hercules, CA) together with Trans-Blot® Turbo™ Transfer System (BIO-RAD) according manufacturer's instructions. The membranes were then blocked with 1:1 ratio of Odyssey® Blocking Buffer (LI-COR Biosciences, Lincoln, NE) and TBS-Tween buffer (50 mM Tris base, 200 mM NaCl, 0.05% Tween 20, pH 7.4) for 1 h at room temperature (RT). Primary antibody incubations were performed in 1:1 ratio of Odyssey® Blocking and TBS-Tween buffer, overnight at +4 °C. Primary antibodies used were 1:1000 β-MHC (MAB1628, RRID: AB_2,282,287, Merck, Darmstadt, Germany), 1:1000 skeletal alpha actin (Ska, ab28052, RRID: AB_867,491, Abcam, Cambridge, United Kingdom), 1:1000 atrial natriuretic peptide (ANP, custom made), 1:2000 Vinculin (ab18058, RRID: AB_444,215, Abcam), 1:500 TEAD-1 (610,922, RRID: AB_398,237, BD) and 1:1,000,000 glyceraldehyde 3‐phosphate dehydrogenase (GAPDH, MAB374, RRID: AB_2,107,445, Millipore). The following day, membranes were washed three times and incubated with appropriate secondary antibody in 1:1 ratio of Odyssey® Blocking and TBS-Tween buffer for 1 h at RT. The secondary antibodies used were 1:5000 Alexa Fluor 680 (A21076, RRID: AB_2,535,736, Thermo Fisher Scientific) and 1:5000 Alexa Fluor 790 (A11371, RRID: AB_2,534,144, Thermo Fisher Scientific). Odyssey Fc Imaging System (LI-COR Biosciences) and Image Studio 4.0 software were used to visualize the blots.
2.14. RNA sequencing analysis
RNA sequencing of RNA samples from neonatal rat ventricular cardiomyocytes was performed by the Finnish Microarray and Sequencing Centre's (Turku centre for Biotechnology, Turku, Finland) analysis service. All the analyses were performed with R language and environment for statistical computing version 3.2 (2015–04–16) and the bioinformatics related Bioconductor module version 3.2.
2.15. Statistical analysis
Data presentations were drawn and statistical analyses were performed using GraphPad Prism6 (California, United States). Data in the graphs are presented as mean ± SD. Normally distributed data was analyzed with Student's T-test or in case of multiple groups, with one-way ANOVA followed by Tukey´s post hoc test. If normality was not possible to verify, data was analyzed with Mann Whitney U test in case of two groups and with Kruskal-Wallis followed by Dunn's post hoc test in case of three or more groups. P < 0.05 was considered significant.
3. Results
3.1. Vezf1 expression is decreased in diseased human myocardium
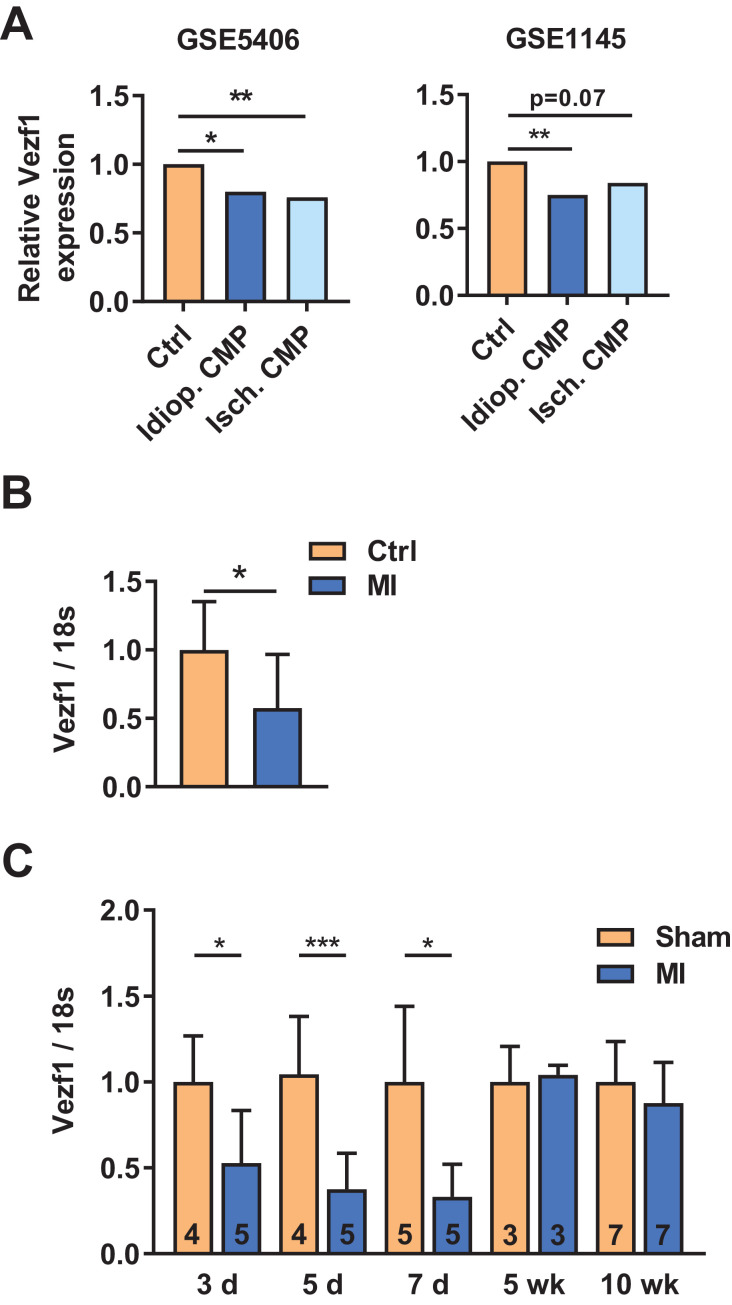
To determine the potential role for Vezf1 in human cardiac disease, we analyzed two different microarray data sets of human heart failure samples (accession numbers GSE5406 and GSE1145) comparing healthy donor hearts to ischemic and idiopathic cardiomyopathy transplantation hearts. We found that Vezf1 expression is decreased by 20% (P < 0.05) and by 25% (P < 0.01) in idiopathic cardiomyopathy, and by 25% (P < 0.01) and 16% (P = 0.07) in ischemic cardiomyopathy compared to the control hearts in GSE5406 and GSE1145, respectively (Fig. 1A). To confirm the findings from the microarray data sets, we analyzed for Vezf1 gene expression in hearts of SCD victims with ischemic heart disease. Heart samples of age-matched victims of traffic accidents without a history or post mortem evidence of cardiovascular disease served as controls. We found that Vezf1 expression is decreased by 43% in hearts of SCD cases with ischemic heart disease compared to control hearts (Fig. 1B, P < 0.05). We then analyzed for Vezf1 expression in hearts of mice subjected to experimental heart failure models and found that LV Vezf1 expression is decreased at 3, 5 and 7 days after MI, but no difference is observed at 5 or 10 weeks after MI (Fig. 1C).
Fig. 1.
Vezf1 expression is decreased in diseased human myocardium. (A) Vezf1 expression in two independent microarray data sets (accession numbers GSE5406 and GSE1145) comparing RNA samples from healthy human donor hearts (Ctrl) to idiopathic and ischemic cardiomyopathy transplantation hearts (Idiop. CMP and Isch. CMP, respectively). * indicates FDR adjusted P-value <0.05, ** indicates FDR adjusted P-value <0.01. (B) qRT-PCR analysis of Vezf1 mRNA levels in healthy control hearts (n = 7) and hearts of sudden cardiac death victims with ischemic heart disease (MI, n = 20). The results are shown as relative to Vezf1 mRNA levels in healthy human hearts (Ctrl). (C) Wild type mice were subjected to myocardial infarction (MI) and RNA was isolated from left ventricular tissue samples 3, 5, 7 days and 5 and 10 weeks later. Shown is qRT-PCR analysis for expression of Vezf1. Results are normalized to expression of 18S (18S ribosomal RNA). *P < 0.05, ***P < 0.001 by Student's T-test. Data are presented as mean ±SD.
3.2. Vezf1 regulates vasculogenesis and angiogenesis
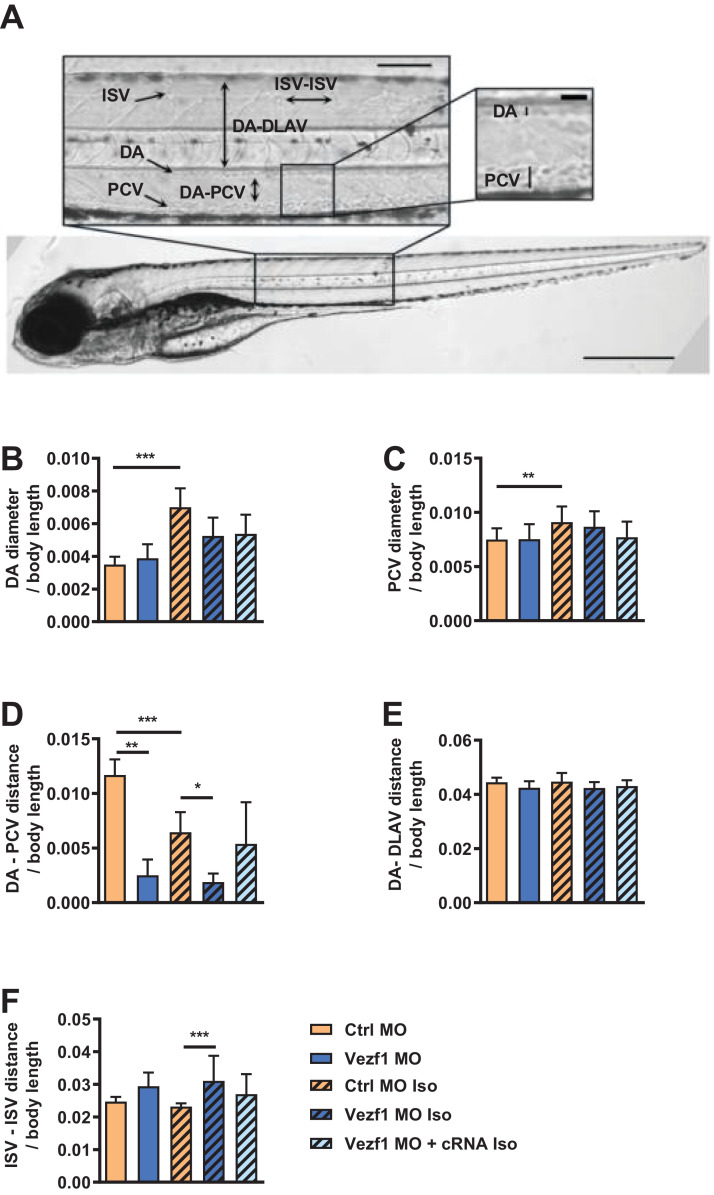
To investigate the role of Vezf1 in cardiovascular biology, we used morpholino (MO) antisense oligonucleotides to deplete Vezf1 in zebrafish. Microinjection of zebrafish embryos with SBMO antisense oligonucleotides resulted in 95% decrease in Vezf1 expression at 1 dpf (p < 0.0001). Molecular mechanisms regulating vessel formation in zebrafish are highly similar to those in humans and optical transparency of developing zebrafish allows high-resolution optical imaging of vascular structures [26]. Analysis of vascular structures in zebrafish at 4 dpf shows that Vezf1 knockdown has no effect on DA and PCV diameter, but reduces the distance between DA and PCV (Fig. 2A–D). Vezf1 knockdown also reduces the DA-PCV distance in zebrafish treated with 300 µM isoprenaline for 48 h (Fig. 2D). Co-injection of zebrafish with capped Vezf1 mRNA (cRNA) was used in parallel experiments to rescue Vezf1 expression upon Vezf1 MO-induced Vezf1 knockdown. As shown in Fig. 2D, Vezf1 rescue with capped Vezf1 mRNA (cRNA) normalizes the DA-PCV distance in Vezf1 knockdown (KD) zebrafish. Vezf1 knockdown results in a non-significant increase in the distance between two consecutive ISVs at baseline (Fig. 2F). However, Vezf1 knockdown significantly increases ISV – ISV distance in zebrafish subjected to 48 h treatment with isoprenaline, indicating that Vezf1 regulates angiogenesis during β-adrenergic stress. Co-injection of Vezf1 cRNA abrogates the antiangiogenic effect of Vezf1 knockdown (Fig. 2F), indicating specificity of the knockdown.
Fig. 2.
Vezf1 regulates vasculogenesis and angiogenesis. Microscopy analysis of vascular structures in zebrafish treated with Ctrl morpholino (MO), Vezf1 MO or Vezf1 MO together with capped Vezf1 mRNA (cRNA) and where indicated, subjected to 300 µM isoprenaline (Iso) for 48 h from 2 dpf to 4 dpf. (A) Whole zebrafish image of Ctrl MO at 4 dpf showing vascular structures. Diameters of dorsal aorta (DA) and posterior cardinal vein (PCV) and distances of two consecutive intersegmental vessels (ISV) from each other, DA-dorsal longitudinal anastomotic vessel (DLAV) and DA-PCV were measured from zebrafish trunk. Scale bar: 200 μm for whole zebrafish and 100 μm and 50 μm for insets. Shown are measurements for (B) DA diameter, (C) PCV diameter, (D) DA-PCV distance, (E) DA-DLAV distance, and (F) ISV-ISV distance. All measurements were made from the area where DA and PCV are parallel. Ctrl MO n = 20, Vezf1 MO n = 6, Ctrl MO + Iso n = 18, Vezf1 MO + Iso n = 12, Vezf1 MO + cRNA + Iso n = 10. *P < 0.05, **P < 0.01, ***P < 0.001. Data are presented as mean ± SD.
3.3. Vezf1 knockdown reduces cardiac growth
Microscopy analysis of zebrafish at 2 dpf and 4 dpf shows that Vezf1 knockdown increases pericardial cavity size in non-stressed hearts (Fig. S1). 48 h isoprenaline treatment from 2 dpf to 4 dpf increases pericardial cavity size in control zebrafish, and Vezf1 knockdown results in a significant further increase of the pericardial cavity size (Fig. S1). Pericardiac edema is commonly associated with mutations affecting cardiac function or vascular integrity in zebrafish [16].
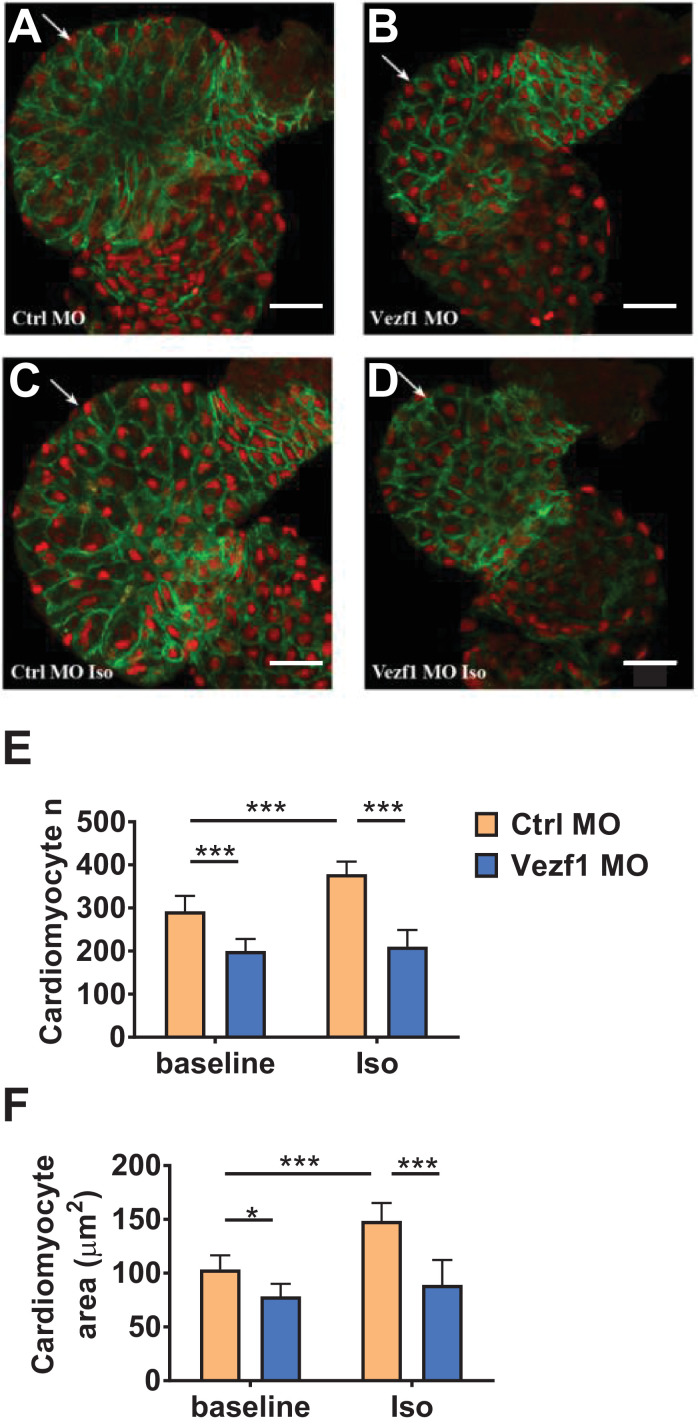
Analysis of zebrafish tissue sections shows that Vezf1 knockdown results in a decrease in the number of ventricular cardiomyocytes at 4 dpf and inhibits the β-adrenergic stress-induced increase in the amount of cardiomyocytes (Fig. 3E). Vezf1 knockdown also results in reduced cardiomyocyte size, or atrophy, at baseline and suppresses isoprenaline-induced increase in the size of cardiomyocytes (Fig. 3F). Vezf1 knockdown has no significant effect on the amount of apoptotic cardiomyocytes at baseline or following 48 h Iso treatment (Fig. S2). Ultrastructural analysis of Vezf1 KD hearts at 2 dpf does not show gross abnormalities in myofilament structure. (Fig. S3).
Fig. 3.
Vezf1 knockdown reduces cardiac growth. Zebrafish were treated with Ctrl morpholino (MO) or Vezf1 MO and where indicated, subjected to 300 µM isoprenaline (Iso) for 48 h from 2 dpf to 4 dpf. Zebrafish hearts were stained with Mef-2 antibody (red) to identify cardiomyocyte nuclei and with ZN-5 antibody (green) to delineate cardiomyocyte cell borders. (A–D) Representative images of hearts of zebrafish treated with Ctrl MO, Vezf1 MO, Ctrl MO + Iso and Vezf1 MO + Iso. The ventricle is marked with an arrow. Scale bar: 25 μm. (E) Analysis for cardiomyocyte number and (F) The area of ventricular cardiomyocytes. n = 9 for each group. *P < 0.05, ***P < 0.001. Data are presented as mean ± SD.
3.4. Loss of Vezf1 impairs cardiac contractile response to β-adrenergic stimuli
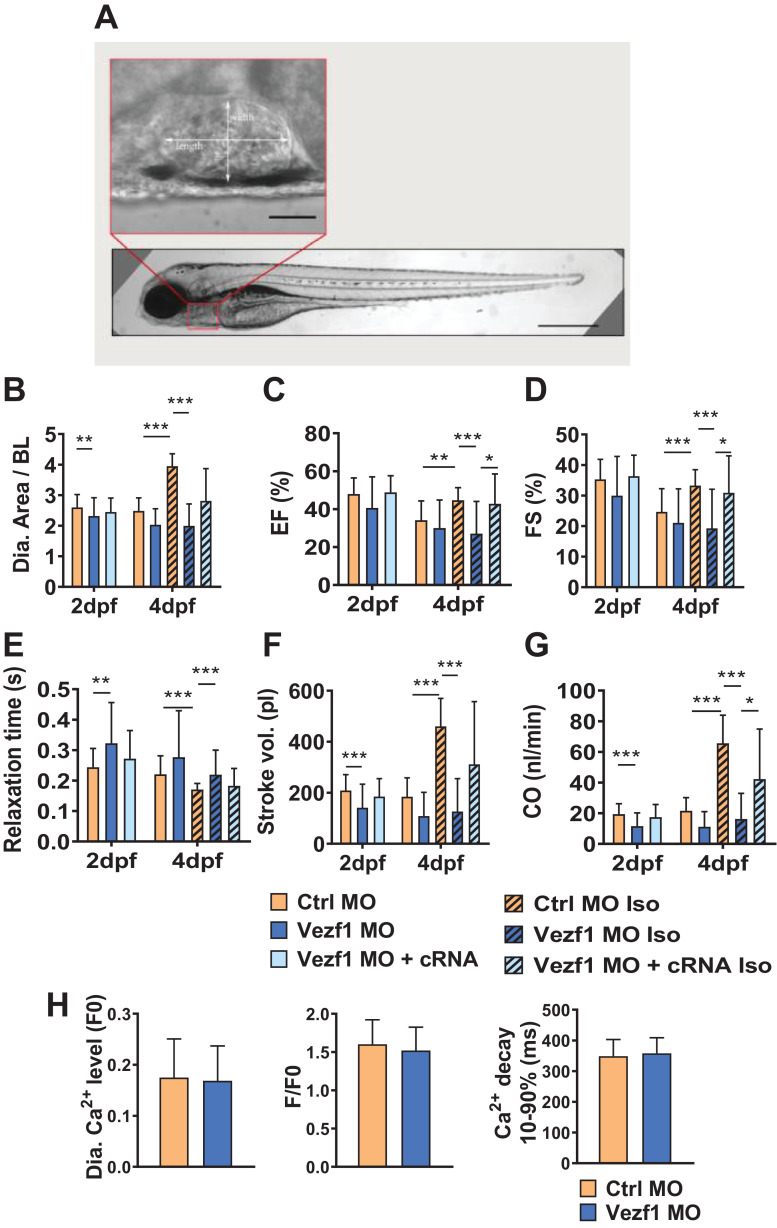
Cardiac function in zebrafish was analyzed from in vivo video recordings (Fig. 4A). We found that Vezf1 knockdown decreases ventricular size at 2 dpf and completely abolishes the isoprenaline-induced increase in ventricular diastolic area at 4 dpf (Fig. 4B). Vezf1 knockdown does not affect cardiac systolic function analyzed at 2 dpf and 4 dpf, but abrogates the isoprenaline-induced enhancement of cardiac contractile function at 4 dpf (Fig. 4C and D). Ventricular relaxation is prolonged in Vezf1 KD hearts at 2 dpf and Vezf1 knockdown also inhibits the isoprenaline-induced shortening of the relaxation time at 4 dpf (Fig. 4E). The defects in isoprenaline-induced ventricular growth and contractility in Vezf1 KD zebrafish culminate in marked decrease in both stroke volume and cardiac output compared to control hearts (Fig. 4F and G). Injection of capped Vezf1 RNA results in full or partial rescue of the phenotype induced by Vezf1 knockdown (Fig. 4B–G). We then performed Ca2+ imaging on 3 dpf isolated zebrafish hearts using Fura2 as calcium indicator. We found Vezf1 KD and control MO ventricles to show similar resting Ca2+ concentration, Ca2+ transient amplitude, and Ca2+ transient decay time (Fig. 4H). No significant arrhythmias were observed.
Fig. 4.
Loss of Vezf1 impairs cardiac contractile response to β-adrenergic stimuli. Zebrafish were treated with Ctrl morpholino (MO), Vezf1 MO or Vezf1 MO together with capped Vezf1 mRNA (cRNA), and where indicated, subjected to 300 µM isoprenaline (Iso) for 48 h from 2 dpf to 4 dpf. (A) Ctrl MO injected zebrafish at 4 dpf with a zoom-in of the ventricle. Scale bar = 500 μm for whole zebrafish and 50 μm for zoom-in of the ventricle. (B–G) Analysis of cardiac structure and function in zebrafish. All cardiac parameters are measured from width and length of the ventricle. Shown are analyses for ventricular diastolic area (B), ejection fraction (C), fractional shortening (D), ventricular relaxation time (E), stroke volume (F), and cardiac output (G). 2 dpf: Ctrl MO n = 36, Vezf1 MO n = 23, Vezf1 MO + cRNA n = 12; 4 dpf: Ctrl MO n = 35, Vezf1 MO n = 22, Ctrl MO Iso n = 36, Vezf1 MO Iso n = 32, Vezf1 MO + cRNA Iso n = 12. (H) Analysis of cardiac Ca2+ cycling at 3 dpf. Shown are analyses for resting cytosolic Ca2+ levels, Ca2+ transient amplitude, and Ca2+ transient decay times. Pacing frequency 1.25 Hz. Ctrl MO n = 15, Vezf1 MO n = 13. *P < 0.05, **P < 0.01, ***P < 0.001. Data are presented as mean ± SD.
3.5. Vezf1 is expressed in adult cardiomyocytes and regulates cardiomyocyte growth and shortening
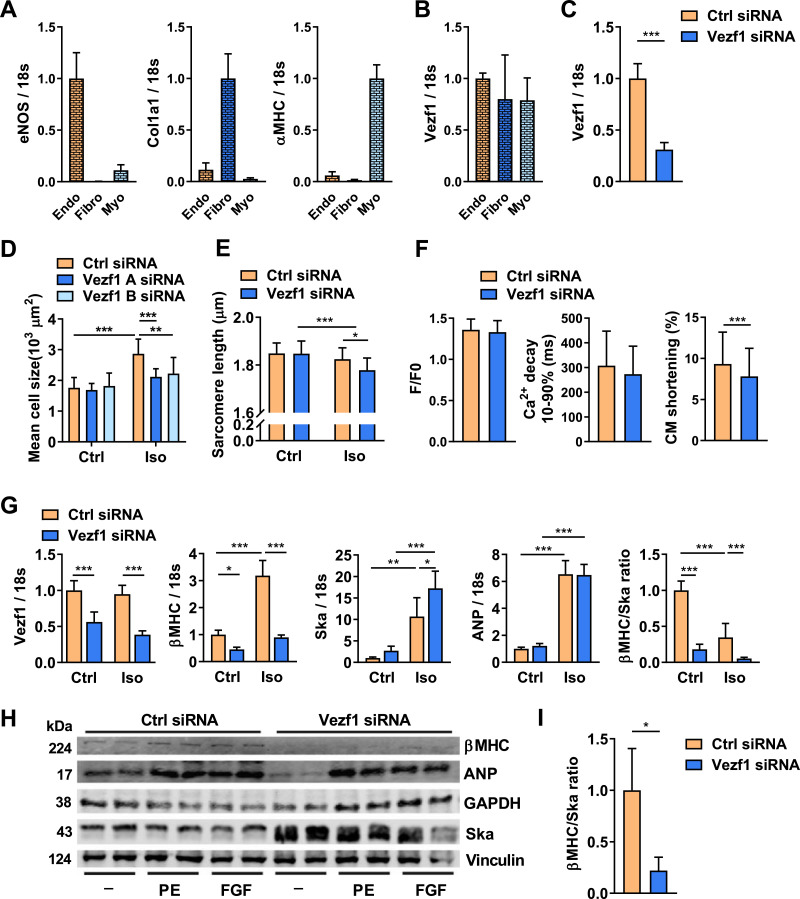
We thus observed that Vezf1 knockdown in zebrafish resulted in reduced cardiac growth and lack of contractile response to β-adrenergic stress, whereas no difference was observed in Ca2+ kinetics. This prompted us to investigate if Vezf1 could serve a direct role in cardiomyocytes. Data derived from studies of embryonic mouse tissues suggest that Vezf1 is primarily expressed in endothelial cells [8]. To investigate if Vezf1 is expressed in adult cardiomyocytes, we fractioned resident cardiac cells from adult mouse myocardium to obtain endothelial cell, fibroblast and cardiomyocyte fractions. qRT-PCR analysis for expression of eNOS, Col1A1 and α-MHC indicated high purity of fractioned endothelial cells, fibroblasts and cardiomyocytes, respectively (Fig. 5A). qRT-PCR analysis for Vezf1 in resident cardiac cells shows that Vezf1 is expressed equally in all three resident cardiac cell types (Fig. 5B).
Fig. 5.
Vezf1 is expressed in adult cardiomyocytes and regulates cardiomyocyte growth and cardiomyopathy related genes. (A) qRT-PCR analysis of the expression of endothelial nitric oxide synthase (eNOS), collagen type I alpha 1 chain (Col1a1), and α myosin heavy chain 6 (α-MHC) mRNAs in pools of fractionated resident mouse cardiac cells. eNOS, Col1a1 and α-MHC were used as markers for endothelial cells (Endo), fibroblasts (Fibro), and cardiomyocytes (Myo), respectively. n = 5. (B) qPCR analysis of Vezf1 mRNA levels in cardiomyocytes and fibroblasts relative to that in the endothelial cells. n = 5. (C) Adult rat ventricular cardiomyocytes were transfected with Vezf1 siRNA (100 nM) or Control siRNA (100 nM) and 3 days later RNA samples were collected. Shown is qRT-PCR analysis for expression of Vezf1. Results are normalized to expression of 18S (18S ribosomal RNA). n = 6. (D and E) Adult rat ventricular cardiomyocytes were transfected with two distinct Vezf1 siRNAs (100 nM) or Control siRNA (100 nM) and 1 day later cells were stimulated with isoprenaline (Iso, 1 µM) for 48 h where indicated. (D) Shown is microscopy analysis for cardiomyocyte size. Ctrl siRNA n = 8, Vezf1 A siRNA n = 9, Vezf1 B siRNA n = 10; Ctrl siRNA + Iso n = 10, Vezf1 A siRNA + Iso n = 15, Vezf1 B siRNA + Iso n = 13. (E) Shown is microscopy analysis for sarcomere length (µm). Ctrl siRNA n = 14, Vezf1 B siRNA n = 18; Ctrl siRNA + Iso n = 22, Vezf1 B siRNA + Iso n = 25. (F) Adult rat ventricular cardiomyocytes were transfected with Vezf1 siRNA (100 nM) or control siRNA and 3 days later Ca2+ cycling and cardiomyocyte (CM) shortening were analyzed. Ctrl n = 44, Vezf1 siRNA n = 43. (G) Neonatal rat ventricular cardiomyocytes were transfected with Vezf1 siRNA (100 nM) or control siRNA and 2 days later cells were treated with Iso (1 µM) for 24 h. Shown is qRT-PCR analysis for expression of Vezf1, β-MHC (myosin heavy chain beta-subunit), Ska (skeletal alpha actin), ANP (atrial natriuretic peptide) and β-MHC versus Ska ratio. Results are normalized to expression of 18S (18S ribosomal RNA). n = 4. (H-I) Neonatal rat ventricular cardiomyocytes were transfected with Vezf1 siRNA (100 nM) or control siRNA (100 nM) and 3 days later cells were treated with either vehicle, phenylephrine (PE, 100 µM) or basic fibroblast growth factor (FGF, 20 ng/ml) for 48 h. (H) Shown is immunoblot analysis for β-MHC, ANP, GAPDH, Ska and Vinculin. (I) Shown is β-MHC versus Ska ratio from immunoblot quantification. n = 4. *P < 0.05, **P < 0.01, ***P < 0.001. Data are presented as mean ± SD.
To study the biological function of Vezf1 in cardiomyocytes, we depleted Vezf1 in adult rat ventricular cardiomyocytes by RNAi, which resulted in 70% decrease in Vezf1 mRNA level compared to the control siRNA treated cells (Fig. 5C, p < 0.0001). Microscopy analysis of cardiomyocyte size shows that depletion of Vezf1 by two different siRNAs significantly attenuates isoprenaline-induced increase in cardiomyocyte size (Fig. 5D). Vezf1 siRNA treated cardiomyocytes also display significantly shorter sarcomere length compared to control cells after 24 h of isoprenaline treatment (Fig. 5E). We then analyzed the effect of Vezf1 depletion on Ca2+ handling in isolated cardiomyocytes. Similar to findings in zebrafish in vivo, Vezf1 knockdown in adult rat cardiomyocytes has no effect on Ca2+ transient kinetics (Fig. 5F). However, Vezf1 knockdown significantly attenuated cardiomyocyte shortening (Fig. 5F). While Vezf1 knockdown in zebrafish did not induce apoptosis at baseline or following hemodynamic stress, we addressed the role of Vezf1 in regulating cardiomyocyte viability under hypoxic stress. We found that Vezf1 depletion has no effect on cardiomyocyte viability in normoxic conditions and analysis for cardiomyocyte death immediately after 4 h of hypoxia, or after 5 h or 24 h reperfusion shows no difference between the Vezf1 siRNA and control siRNA treated cells (Fig. S4).
3.6. Vezf1 regulates cardiomyopathy related genes
To identify the molecular targets of Vezf1 in cardiomyocytes, we performed RNA sequencing analysis of samples from Vezf1 siRNA treated NRVMs. We found that Vezf1 silencing resulted in significant regulation of 1144 transcripts (FDR < 0.05, ArrayExpress accession E-MTAB-7124). 28 transcripts were upregulated and 53 transcripts downregulated by more than 2-fold (FDR < 0.05, Table S2). KEGG pathway analysis of the differentially expressed genes identified 14 gene functions including cardiac muscle contraction and dilated cardiomyopathy. The only pathway significantly enriched in Reactome pathway analysis was muscle contraction. Among the genes most significantly regulated by Vezf1, were cardiac muscle contraction and cardiomyopathy related genes Myh7 (β-MHC), Atp1a2, and Acta1 (Ska) (Table S2). qPCR analysis of RNA samples from NRVMs confirmed that transcription of Myh7 and Atp1a2 were decreased and Ska mRNA levels were increased in response to Vezf1 knockdown (Fig. S5). RNA sequencing further showed that a number of MCAT element containing genes were significantly regulated by Vezf1 in rat cardiomyocytes, including Myh7, Atp1a2, Tcap, Myh11, Cacng7, Acta1, Tnnc2, MYH6, cardiac troponin T and cardiac troponin C (Table S2, ArrayExpress accession E-MTAB-7124).
To investigate if Vezf1 knockdown affects stress-induced expression of cardiomyopathy related genes, we treated NRVMs with isoprenaline for 24 h. We found that knockdown of Vezf1 in NRVMs prevented both basal and isoprenaline –induced expression of β-MHC (Fig. 5G). Vezf1 silencing enhanced isoprenaline-induced skeletal alpha actin (Acta1, Ska) expression, but had no effect on either basal or isoprenaline-induced expression of ANP (Fig. 5G). Consequently, the β-MHC/Ska mRNA expression ratio was profoundly decreased in Vezf1 depleted cardiomyocytes (Fig. 5G). To investigate whether Vezf1 depletion similarly affects protein levels of its target genes, NRVMs were transfected with an siRNA targeting a distinct site in Vezf1 mRNA, and the cells were stimulated with phenylephrine or basic fibroblast growth factor for 48 h. Western blot analysis of protein samples shows that Vezf1 siRNA decreased both basal and hypertrophic stimuli-induced β-MHC protein levels (Fig. 5H). In agreement with the mRNA data, Vezf1 knockdown resulted in an increase in Ska protein levels, but only had a modest effect on basal or hypertrophic agonist-induced ANP levels (Fig. 5H). Similar to mRNA data, Vezf1 silencing resulted in a significant reduction in β-MHC/Ska ratio at protein level (Fig. 5I).
To investigate whether Vezf1 targets the same genes in zebrafish, qPCR analysis of zebrafish samples at 1 dpf showed that Vezf1 knockdown decreased expression of vMHC (zebrafish orthologue for β-MHC) by 92% compared to controls (Fig. S6). Expression of Ska, on the other hand, was significantly increased in Vezf1 KD zebrafish hearts (Fig. S6). Consequently, the vMHC/Ska mRNA ratio was diminished in hearts of Vezf1 KD zebrafish (Fig. S6).
3.7. Vezf1 regulates β-MHC promoter luciferase reporter
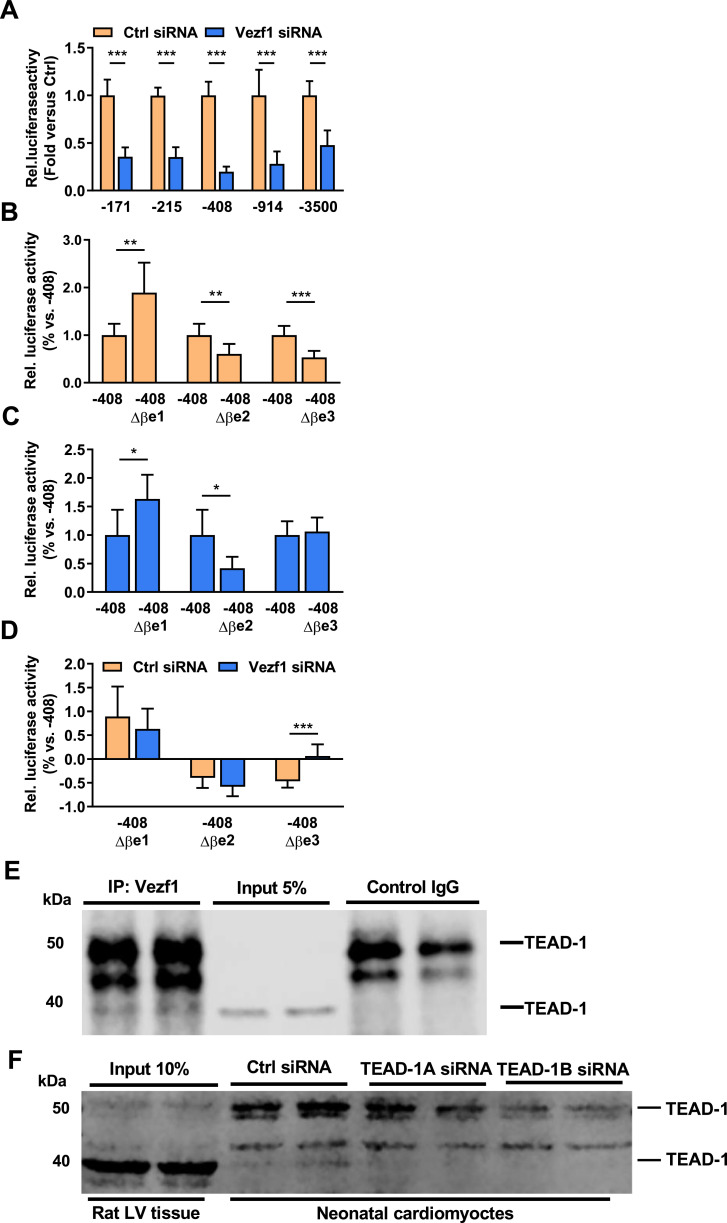
To evaluate if Vezf1 regulates β-MHC promoter activity, NRVMs were co-transfected with five different β-MHC promoter luciferase reporter constructs (−171 to −3500 bp) together with Vezf1 siRNA or control siRNA. We found that Vezf1 siRNA treatment decreases the relative luciferase activity of β-MHC promoters by 52–80% with the most pronounced effect in the −408 β-MHC reporter construct (Fig. 6A). The five β-MHC promoter luciferase reporter constructs (−171 to −3500 bp) were then co-transfected with mouse Vezf1 plasmids or empty pGL3basic plasmids. We found that overexpression of Vezf1 leads to a significant increase in the activity of the −914 β-MHC and −3500 β-MHC promoter luciferase constructs (Fig. S7). Overexpression of Vezf1 also results in an increase in cardiomyocyte size and augments β-MHC expression, but that did not reach significance (Fig. S7).
Fig. 6.
Vezf1 targets an MCAT binding site within β-MHC promoter and interacts with TEAD-1. (A) Neonatal rat ventricular cardiomyocytes (NRVMs) were transfected with β-MHC promoter (−171 to −3500 bp) luciferase reporter constructs together with Vezf1 siRNA (100 nM) or control siRNA (100 nM). 3 days later, relative luciferase activities were analyzed. Firefly luciferase activities were normalized to renilla luciferase activity and expressed as relative to the activity of respective β-MHC promoter luciferase construct treated with control siRNA. n = 4. (B–D) NRVMs were transfected with an intact −408 β-MHC promoter construct or with −408 β-MHC promoter construct containing either Δβe1, Δβe2 or Δβe3 mutation together with (B) control siRNA (100 nM) (n = 6–8) or (C) Vezf1 siRNA (100 nM) (n = 3–7). 3 days later, relative luciferase activities of the different lengths of β-MHC promoter constructs were analyzed. Firefly luciferase activities were normalized to renilla luciferase and the data are expressed as relative to that of the wild-type −408 β-MHC promoter construct activity. (D) Comparison of relative change in luciferase activity of −408 β-MHC Δβe1, Δβe2 or Δβe3 promoter construct to that of wild-type −408 β-MHC promoter luciferase construct in control siRNA and Vezf1 siRNA treated cells. n = 3–8. (E) Adult rat left ventricular (LV) tissue samples were immunoprecipitated with Vezf1 antibody or control IgG. Shown is immunoblot analysis for Transcription enhancer factor-1 (TEAD-1) in immunoprecipitates and LV lysates. (F) NRVMs were transfected with two distinct TEAD-1 siRNAs (100 nM) or control siRNA (100 nM) and 3 days later protein samples were collected. Shown is immunoblot analysis for TEAD-1 in rat LV lysates and in NRVMs. *P < 0.05, **P < 0.01, ***P < 0.001. Data are presented as mean ± SD.
3.8. Vezf1 targets an MACT binding site within the β-MHC promoter and interacts with TEAD-1
To investigate the mechanism by which Vezf1 regulates β-MHC gene expression, we performed mutational analysis of the −408 β-MHC promoter construct. As RNA sequencing identified a number of genes with MCAT binding sites regulated by Vezf1, we utilized two −408 β-MHC promoter constructs with MCAT site mutations (Δβe2 and Δβe3) [26,27]. In addition, a −408 β-MHC promoter construct with mutation of a constitutive repressive site was utilized (Δβe1) [27]. The activities of the mutated −408 β-MHC promoter constructs were then investigated in control siRNA and Vezf1 siRNA treated NRVMs. In control siRNA treated NRVMs, the Δβe1 construct shows 90% increase in activity compared to the wild-type −408 β-MHC promoter construct (Fig. 6B), whereas the Δβe2 and Δβe3 constructs show 40–50% decrease in luciferase activity. In Vezf1 siRNA treated NRVMs, responses to Δβe1 and Δβe2 mutations are similar to those of control siRNA treated cells (Fig. 6C). However, unlike control siRNA treated cells, the mutation of an MCAT binding site at −211 to −189 (Δβe3) has no effect in Vezf1 silenced cells. These data thus indicate that Vezf1 targets the βe3 site of the β-MHC promoter. Comparisons of the relative responses to the −408 β-MHC promoter mutations in control siRNA and Vezf1 siRNA treated cells are shown in Fig. 6D. MCAT motifs in the Myh7 gene are binding sites for transcriptional enhancer factors (TEADs/TEFs) [28]. To investigate whether Vezf1 regulation of Myh7 involves binding of Vezf1 to TEFs, we immunoprecipitated rat LV lysates with Vezf1 antibody and analyzed for co-immunoprecipitation of TEF factors. We found that Vezf1 binds TEAD-1 in hearts in vivo (Fig. 6E). The specificity of the 40 kD TEAD-1 band was further confirmed by knockdown of TEAD-1 with two different siRNAs in neonatal rat cardiomyocytes (Fig. 6F).
4. Discussion
Vezf1 has previously been associated with regulation of endothelial cell function. However, the role of Vezf1 in cardiac biology remains unknown. In the current study, we found that Vezf1 expression is decreased in LV samples of patients with idiopathic and ischemic cardiomyopathy, and in mice subjected to experimental MI. Studies in zebrafish show that Vezf1 is necessary for isoprenaline-induced compensatory ventricular hypertrophy and increase in ventricular systolic function. Analysis of cardiac tissues showed that Vezf1 knockdown decreases cardiomyocyte size, but does not affect cardiomyocyte viability. In agreement with findings in zebrafish, Vezf1 depletion in isolated cardiomyocytes has no effect on cardiomyocyte viability, but decreases cardiomyocyte shortening and cardiomyocyte growth in response to isoprenaline. The defective contractile response to isoprenaline stimuli in zebrafish prompted us to investigate if Vezf1 knockdown affects Ca2+ cycling. However, Vezf1 knockdown either in zebrafish or in isolated cardiomyocytes did not affect Ca2+ transient kinetics.
Analysis for Vezf1 target genes identified Myh7 as a key regulatory target in cardiomyocytes and this was further confirmed with a notable reduction in Myh7/β-MHC expression at the protein level. Strikingly, analysis for zebrafish ventricular MHC (vMHC) expression at 1 dpf (when Vezf1 deletion had not yet induced a cardiac phenotype) showed 92% reduction in vMHC expression. In zebrafish, vMHC is the likely orthologue of the mammalian β-MHC/Myh7, with its similar ventricle-specific expression pattern [29]. Myosin is the main component of the sarcomere thick filament and the intrinsic ATPase activity of the myosin heavy chains transduces chemical energy into mechanical energy resulting in cell contraction. The two major cardiac MHC genes, α-MHC (Myh6) and β-MHC, have been shown to be reciprocally regulated during development and during cardiac pathologies [30]. Notably, in the current study the robust decrease in Myh7 expression in response to Vezf1 knockdown in cardiomyocytes was not associated with upregulation of Myh6 expression that could rescue the total MHC levels. Instead, we found that expression of thin filament skeletal α-actin (Ska) was increased in Vezf1 KD hearts and Vezf1 silenced cardiomyocytes resulting in a robust decrease in the myosin/actin ratio. Normal myosin/actin ratio in the myocardium is 1.0/4.1 [31] and maintenance of the structure and function of cardiac muscle requires precise control of the synthesis, assembly, and turnover of the sarcomere proteins [32]. Previously, reduction in myosin/actin ratio has been observed in pulmonary artery hypertension patients, who have reduced LV ejection fraction and reduced LV cardiomyocyte size [33]. The decrease in myosin/actin ratio did not result in ventricular dysfunction in unstressed Vezf1 KD hearts, which is likely due to substantial contractile reserve (the difference between myocardial contractility in response to stress compared to contractility at rest) of the myocardium. However, the disturbed protein balance in the cardiomyocyte contractile unit provides a potential mechanism for the defective contractile response of Vezf1 KD hearts during hemodynamic stress.
RNA sequencing analysis revealed that the highest regulated genes were ones containing a muscle-specific MCAT element in their regulatory regions, such as β-MHC (Myh7), Atp1a2, Tcap, Myh11 and Ska (Acta1) that were all regulated by more than log 2 fold. Prior data shows that MCAT motifs (binding sites for TEAD/TEF transcription factors) in the Myh7 gene are essential for Myh7 gene expression [28]. By using mutational analysis, we demonstrate that an MCAT element in the Myh7 promoter at least partially mediates the transcriptional response to Vezf1 knockdown. However, the proximal promoter region of Myh7 does not contain putative binding sites for Vezf1. Interestingly, cardiomyocyte knockdown of TEAD-1 in neonatal mice in vivo has been shown to profoundly augment Ska expression and decrease Myh7 expression [34]. Tamoxifen inducible cardiomyocyte-specific knockdown of TEAD-1 in the adult mice resulted in dilated cardiomyopathy and significant reduction in Myh7 expression [35]. Moreover, overexpression of TEAD-1 was sufficient to enhance Myh7 expression [36]. In the current study, analysis for Vezf1 binding partners showed that Vezf1 co-immunoprecipitates with TEAD-1 in hearts in vivo. In addition, Vezf1 shares common downstream target genes with TEAD-1. However, differently from TEAD-1 overexpression [36], Vezf1 overexpression is not sufficient to drive Myh7 expression.
Previous data have shown that Vezf1 has a central role in vasculogenesis and angiogenesis [10,12]. In agreement, we observed that Vezf1 knockdown in zebrafish resulted in altered formation of the two major trunk axial vessels and increased distance between intersegmental vessels following β-adrenergic stress. Endothelial cell Vezf1 is a positive regulator of endothelin-1 (ET-1) [11], which is known to induce cardiac hypertrophy, enhance cardiac contractile function, and regulate cardiomyocyte Ca2+ homeostasis [37], [38], [39]. We cannot thus rule out that Vezf1-mediated regulation of endothelial cell -derived factors, such as ET-1, may have had effect on the observed cardiac phenotype, such as reduced cardiac growth in the zebrafish [40]. However, we found that Vezf1 is expressed in adult cardiomyocytes in levels comparable to that in endothelial cells, and directly regulates expression of cardiac genes. Lack of targeting Vezf1 in the adult heart is a limitation of the current study. In future studies, it would be of interest to determine the role of Vezf1 in the stressed myocardium by targeting Vezf1 in cardiomyocytes. Interestingly, a different set of genes was regulated by Vezf1 in cardiomyocytes compared to that previously described in endothelial cells [12]. Interaction of Vezf1 with TEAD-1 actually indicates that cell type-specific transcriptional cofactors participate in Vezf1-mediated transcriptional regulation.
In summary, our data shows that in addition to regulating angiogenesis, Vezf1 is expressed in cardiomyocytes and directly regulates genes involved in cardiac contractile function and dilated cardiomyopathy. We further find that zebrafish Vezf1 is necessary for hemodynamic stress-induced increase in cardiac contractile function and loss of Vezf1 results in greater propensity towards heart failure. Molecular analysis for Vezf1 function in cardiomyocytes identifies TEAD-1 as a binding partner of Vezf1. These data thus identify previously unappreciated biological functions for Vezf1 in cardiomyocytes. However, further studies are needed to evaluate the significance of decreased Vezf1 expression in diseased human myocardium and to understand cell type-specific effects of Vezf1 in the adult heart.
Funding source
This work was supported by research funding of the Helsinki-Uusimaa Hospital District (state funding for university-level health research) (to I.T.) and by grants from the Academy of Finland (grant N:o to 256908 J.U., grant N:o 268505 to J.M. and grant N:o 297094 to R.K.), Jane and Aatos Erkko Foundation (to R.K and H.H.), Aarne Koskelo Foundation (to J.P., S.N. and I.T.), the Finnish Foundation for Cardiovascular Research (to J.P., J.U., T.A., I.T. and R.K.), the Finnish Cultural Foundation (to T.A., S.N. and H.W.), the Finnish Foundation for Laboratory Medicine (to P.L.), the Finska Läkaresällskapet (to I.T.) and the Liv och Hälsa Foundation (to I.T.). The funders played no role in study design, data collection or interpretation, decision to publish or preparation of the manuscript.
Author contributions
J.P., J.U., I.T. and R.K. designed the study. J.P., T.A., J.U., T.K., J.S., S.P., S.N., H.W., R.L., K.P., K.I. and J.M. performed acquisition of data. J.P., J.U., T.A. T.K., J.S., S.P., J.J., H.H., K.I., P.L, J.M., I.T. and R.K. analyzed and interpreted the data. J.P., T.A., T.K. and R.K. drafted the manuscript. J.M. and I.T. made critical revision to the manuscript.
Declaration of Competing Interest
The authors have nothing to disclose.
Acknowledgments
We thank Marja Arbelius, Kirsi Salo; Ilkka Miinalainen and staff of the BCO Electron Microscopy Core; and staff of the Zebrafish Unit and the Biomedicum Imaging Unit for excellent technical assistance.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.102608.
Appendix. Supplementary materials
References
- 1.Roth G.A., Johnson C., Abajobir A. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol. 2017;70(1):1–25. doi: 10.1016/j.jacc.2017.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Berlo J.H., Maillet M., Molkentin J.D. Signaling effectors underlying pathologic growth and remodeling of the heart. J Clin Invest. 2013;123(1):37–45. doi: 10.1172/JCI62839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oka T., Akazawa H., Naito A.T., Komuro I. Angiogenesis and cardiac hypertrophy: maintenance of cardiac function and causative roles in heart failure. Circ Res. 2014;114(3):565–571. doi: 10.1161/CIRCRESAHA.114.300507. [DOI] [PubMed] [Google Scholar]
- 4.Pikkarainen S., Tokola H., Kerkela R., Ruskoaho H. GATA transcription factors in the developing and adult heart. CardiovascRes. 2004;63(2):196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 5.Akazawa H., Komuro I. Cardiac transcription factor Csx/Nkx2-5: its role in cardiac development and diseases. Pharmacol Ther. 2005;107(2):252–268. doi: 10.1016/j.pharmthera.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Oka T., Maillet M., Watt A.J. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98(6):837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 7.Heineke J., Auger-Messier M., Xu J. Cardiomyocyte GATA4 functions as a stress-responsive regulator of angiogenesis in the murine heart. JClinInvest. 2007;117(11):3198–3210. doi: 10.1172/JCI32573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong J.W., Leahy A., Lee H.H., Stuhlmann H. Vezf1: a zn finger transcription factor restricted to endothelial cells and their precursors. DevBiol. 1999;206(2):123–141. doi: 10.1006/dbio.1998.9144. [DOI] [PubMed] [Google Scholar]
- 9.Kuhnert F., Campagnolo L., Xiong J.W. Dosage-dependent requirement for mouse Vezf1 in vascular system development. DevBiol. 2005;283(1):140–156. doi: 10.1016/j.ydbio.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou Z., Ocaya P.A., Sun H., Kuhnert F., Stuhlmann H. Targeted Vezf1-null mutation impairs vascular structure formation during embryonic stem cell differentiation. ArteriosclerThrombVascBiol. 2010;30(7):1378–1388. doi: 10.1161/ATVBAHA.109.200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aitsebaomo J., Kingsley-Kallesen M.L., Wu Y., Quertermous T., Patterson C. Vezf1/DB1 is an endothelial cell-specific transcription factor that regulates expression of the endothelin-1 promoter. J Biol Chem. 2001;276(42):39197–39205. doi: 10.1074/jbc.M105166200. [DOI] [PubMed] [Google Scholar]
- 12.Gerald D., Adini I., Shechter S. RhoB controls coordination of adult angiogenesis and lymphangiogenesis following injury by regulating VEZF1-mediated transcription. NatCommun. 2013;4:2824. doi: 10.1038/ncomms3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gowher H., Stuhlmann H., Felsenfeld G. Vezf1 regulates genomic DNA methylation through its effects on expression of DNA methyltransferase Dnmt3b. Genes Dev. 2008;22(15):2075–2084. doi: 10.1101/gad.1658408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waits E.R., Nebert D.W. Genetic architecture of susceptibility to PCB126-Induced developmental cardiotoxicity in zebrafish. ToxicolSci. 2011;122(2):466–475. doi: 10.1093/toxsci/kfr136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mickelson E.C., Robinson W.P., Hrynchak M.A., Lewis M.E. Novel case of del(17)(q23.1q23.3) further highlights a recognizable phenotype involving deletions of chromosome (17)(q21q24) Am J Med Genet. 1997;71(3):275–279. doi: 10.1002/(sici)1096-8628(19970822)71:3<275::aid-ajmg5>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 16.Gut P., Reischauer S., Stainier D.Y.R., Arnaout R. Little fish, big data: zebrafish as a model for cardiovascular and metabolic disease. Physiol Rev. 2017;97(3):889–938. doi: 10.1152/physrev.00038.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hookana E., Junttila M.J., Kaikkonen K.S. Comparison of family history of sudden cardiac death in nonischemic and ischemic heart disease. Circ Arrhythm Electrophysiol. 2012;5(4):757–761. doi: 10.1161/CIRCEP.112.971465. [DOI] [PubMed] [Google Scholar]
- 18.Sundvik M., Kudo H., Toivonen P., Rozov S., Chen Y.C., Panula P. The histaminergic system regulates wakefulness and orexin/hypocretin neuron development via histamine receptor H1 in zebrafish. FASEB J. 2011;25(12):4338–4347. doi: 10.1096/fj.11-188268. [DOI] [PubMed] [Google Scholar]
- 19.Kaslin J., Panula P. Comparative anatomy of the histaminergic and other aminergic systems in zebrafish (Danio rerio) J Comp Neurol. 2001;440(4):342–377. doi: 10.1002/cne.1390. [DOI] [PubMed] [Google Scholar]
- 20.Westerfield M. 4th editor. University of Oregon Press; Eugene: 2000. The zebrafish book A guide for the laboratory use of zebrafish (Danio rerio) [Google Scholar]
- 21.Dash S.N., Narumanchi S., Paavola J. Sept7b is required for the subcellular organization of cardiomyocytes and cardiac function in zebrafish. Am J Physiol Heart Circ Physiol. 2017;312(5):H1085–H1H95. doi: 10.1152/ajpheart.00394.2016. [DOI] [PubMed] [Google Scholar]
- 22.Kerkela R., Karsikas S., Szabo Z. Activation of hypoxia response in endothelial cells contributes to ischemic cardioprotection. MolCell Biol. 2013;33(16):3321–3329. doi: 10.1128/MCB.00432-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magga J., Vainio L., Kilpio T. Systemic blockade of ACVR2B ligands protects myocardium from acute ischemia-reperfusion injury. Mol Ther. 2019;27(3):600–610. doi: 10.1016/j.ymthe.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaikkonen L., Magga J., Ronkainen V.P. p38alpha regulates SERCA2a function. J Mol Cell Cardiol. 2014;67:86–93. doi: 10.1016/j.yjmcc.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gore A.V., Monzo K., Cha Y.R., Pan W., Weinstein B.M. Vascular development in the zebrafish. Cold Spring Harb Perspect Med. 2012;2(5) doi: 10.1101/cshperspect.a006684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knotts S., Rindt H., Neumann J., Robbins J. In vivo regulation of the mouse beta myosin heavy chain gene. J Biol Chem. 1994;269(49):31275–31282. [PubMed] [Google Scholar]
- 27.Huey K.A., Roy R.R., Haddad F., Edgerton V.R., Baldwin K.M. Transcriptional regulation of the type I myosin heavy chain promoter in inactive rat soleus. Am J Physiol Cell Physiol. 2002;282(3):C528–C537. doi: 10.1152/ajpcell.00355.2001. [DOI] [PubMed] [Google Scholar]
- 28.Rindt H., Gulick J., Knotts S., Neumann J., Robbins J. In vivo analysis of the murine beta-myosin heavy chain gene promoter. J Biol Chem. 1993;268(7):5332–5338. [PubMed] [Google Scholar]
- 29.Jin D., Ni T.T., Hou J., Rellinger E., Zhong T.P. Promoter analysis of ventricular myosin heavy chain (vmhc) in zebrafish embryos. Dev Dyn. 2009;238(7):1760–1767. doi: 10.1002/dvdy.22000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadal-Ginard B., Mahdavi V. Molecular basis of cardiac performance. plasticity of the myocardium generated through protein isoform switches. J ClinInvest. 1989;84(6):1693–1700. doi: 10.1172/JCI114351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami U., Uchida K. Contents of myofibrillar proteins in cardiac, skeletal, and smooth muscles. J Biochem. 1985;98(1):187–197. doi: 10.1093/oxfordjournals.jbchem.a135257. [DOI] [PubMed] [Google Scholar]
- 32.Wang X. Heart failure and protein quality control. Circ Res. 2006;99(12):1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 33.Manders E., Bogaard H.J., Handoko M.L. Contractile dysfunction of left ventricular cardiomyocytes in patients with pulmonary arterial hypertension. J Am Coll Cardiol. 2014;64(1):28–37. doi: 10.1016/j.jacc.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Liu R., Jagannathan R., Li F. Tead1 is required for perinatal cardiomyocyte proliferation. PLoS ONE. 2019;14(2) doi: 10.1371/journal.pone.0212017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu R., Lee J., Kim B.S. Tead1 is required for maintaining adult cardiomyocyte function, and its loss results in lethal dilated cardiomyopathy. JCI Insight. 2017;2(17) doi: 10.1172/jci.insight.93343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsika R.W., Ma L., Kehat I. TEAD-1 overexpression in the mouse heart promotes an age-dependent heart dysfunction. J Biol Chem. 2010;285(18):13721–13735. doi: 10.1074/jbc.M109.063057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito H., Hirata Y., Hiroe M. Endothelin-1 induces hypertrophy with enhanced expression of muscle- specific genes in cultured neonatal rat cardiomyocytes. Circ Res. 1991;69(1):209–215. doi: 10.1161/01.res.69.1.209. [DOI] [PubMed] [Google Scholar]
- 38.Shohet R.V., Kisanuki Y.Y., Zhao X.S., Siddiquee Z., Franco F., Yanagisawa M. Mice with cardiomyocyte-specific disruption of the endothelin-1 gene are resistant to hyperthyroid cardiac hypertrophy. Proc Natl Acad Sci U S A. 2004;101(7):2088–2093. doi: 10.1073/pnas.0307159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szokodi I., Kerkela R., Kubin A.M. Functionally opposing roles of extracellular signal-regulated kinase 1/2 and p38 mitogen-activated protein kinase in the regulation of cardiac contractility. Circulation. 2008;118(16):1651–1658. doi: 10.1161/CIRCULATIONAHA.107.758623. [DOI] [PubMed] [Google Scholar]
- 40.Brutsaert DL. Cardiac endothelial-myocardial signaling: its role in cardiac growth, contractile performance, and rhythmicity. Physiol Rev. 2003;83(1):59–115. doi: 10.1152/physrev.00017.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.