Summary
Three-dimensional documentation of the axonal pathways connecting grey matter components of the human brain has wide-ranging scientific and clinical applications. Recent attempts to map human structural connectomes have concentrated on using tractography results derived from diffusion-weighted imaging data, but tractography is an indirect method with numerous limitations. Advances in holographic visualization platforms provide a new medium to integrate anatomical data, as well as a novel working environment for collaborative interaction between neuroanatomists and brain imaging scientists. Therefore, we developed the first holographic interface for building axonal pathways, populated it with human histological and structural MRI data, and assembled world expert neuroanatomists to interactively define axonal trajectories of the cortical, basal ganglia, and cerebellar systems. This blending of advanced visualization hardware, software development, and neuroanatomy data enabled the translation of decades of amassed knowledge into a human axonal pathway atlas that can be applied to educational, scientific, or clinical investigations.
Keywords: Basal Ganglia, Subthalamic Nucleus, Hyperdirect Pathway, Deep Brain Stimulation
eTOC blurb:
Petersen et al. use group-based holographic visualization to construct axonal pathway trajectories in the human brain via interactive collaboration by world expert neuroanatomists. They blend advanced visualization hardware, software development, and neuroanatomy data to overcome the limitations of tractography.
Introduction
Visualizing and understanding the axonal connections of the human brain has immense value in applications that range from training medical students to planning neurosurgical procedures. As such, neuroanatomists have used traditional histologic techniques for nearly a century to describe the white matter connections of the human brain [Agrawal et al., 2011]. Human anatomical results are ideally coupled with more detailed experiments performed in the non-human primate brain to delineate details of the specific connections and somatotopic organization of individual axonal pathways [Van Essen and Glasser, 2018]. However, direct translation of the complex axonal trajectories and detailed connectomic information available from monkey studies is often difficult to scale and render within the context of the human brain. In addition, assumptions must be made on the cross-species homolog of each pathway, and its exact anatomical location in the human brain.
The advent of diffusion-weighted imaging (DWI) [Basser et al., 1994], and the application of tractography algorithms to that data [Mori et al., 1999], have provided unique opportunities to create in vivo simulations of structural connectivity in the human brain. Tractography-based structural connectomes have subsequently become a driving force in human brain imaging research [Setsompop et al., 2013; Yeh et al., 2018], and are beginning to play a role in patient-specific neurosurgical procedure planning [Essayed et al., 2017; Sammartino and Hodaie, 2018]. However, tractography suffers from inherent limitations in reconstructing white matter pathways [Jones et al., 2013; Thomas et al., 2014], and decades of tractography validation studies have repeatedly called for alternative strategies to more accurately map the axonal pathways of the human brain [Maier-Hein et al., 2017; Schilling et al., 2019].
The goal of this study was to employ new holographic visualization technologies to facilitate data integration and collaborative interaction for the generation of human axonal pathway models. Our specific focus was the creation of detailed representations of the axonal pathways in the subthalamic region, a brain area considered highly problematic for tractographic reconstructions, but a common surgical target for deep brain stimulation (DBS) therapies. While it is generally accepted that tractography can generate reasonable estimates of large pathways, such as the corpus callosum or corticospinal tract, it has known difficulties with smaller pathways. This is especially true if those smaller pathways also intersect large pathways, such as the connections between the basal ganglia and diencephalon. In addition, anatomical reconstructions of large pathways typically show a somatotopic organization of the fibers within the tract, but tractography alone is not always capable of simulating that somatotopy. Therefore, we set out to collate decades of anatomical knowledge about the cortical and subcortical axonal pathways coursing through the subthalamic region and integrate that information into a model of the human brain.
Generating an anatomically accurate representation of the axonal pathways in the human subthalamic region is a challenging 3D task. Many different pathways are crossing each other and following tortuous paths. In addition, the detailed anatomical knowledge on the specific features of each pathway is spread across multiple experts, each with their own unique understanding of the system. To address this challenge, we employed group-based holographic visualization to help us perform complex 3D tasks as collaborative interactions. Our holographic visualization system integrated human histological and structural MRI data with estimated trajectories for each pathway. Each of these trajectories could be interactively manipulated with a series of control points. The control points could be placed and scaled within the holographic brain model based on structural and/or histological landmarks available in the data. We then assembled world expert neuroanatomists to discuss, define, and finalize the axonal trajectories to the best of their collective knowledge, all within the holographic visualization environment. The outcomes of this project were a first of its kind holographic visualization program for developing anatomical priors for use in tractography research, as well as the first anatomically realistic model of the major axonal pathways in the human subthalamic region. We propose that both of these advances have applications in medical education and neurosurgical research.
Results
This project generated three types of results. First, we assembled a human brain model that served as the foundation for creating our axonal pathway representations. Second, we designed a holographic visualization platform for analyzing and interacting with brain imaging data. Third, we used the human brain model and holographic visualization to create axonal pathway reconstructions that connect the cortical, basal ganglia, and cerebellar systems.
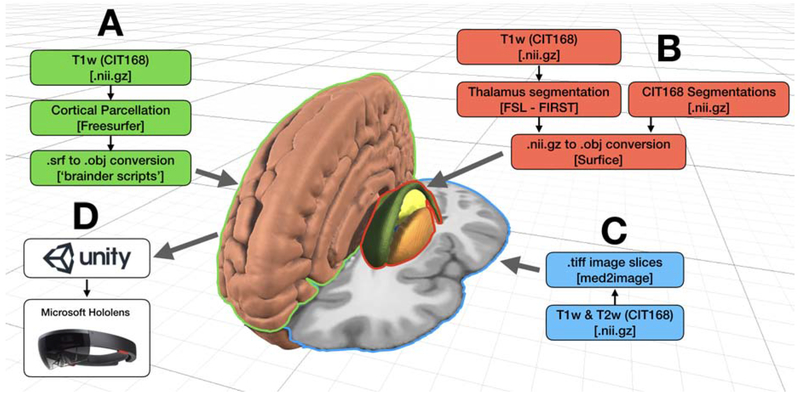
The CIT168 brain atlas [Pauli et al., 2018] provided the foundation for our human brain model (Figure 1). This brain atlas was originally constructed using diffeomorphic registration of T1- and T2-weighted structural magnetic resonance images from 168 typical adults drawn from the Human Connectome Project [Van Essen et al., 2013]. 3D volumes representing cortical parcellations and subcortical nuclei were rendered and integrated with 2D image slices associated with the axial, coronal, and sagittal views of the CIT168 MRI data. These triangulated surface models and image slices were then ported to the Unity programming environment, so the results could be simulated as holograms by the Microsoft HoloLens platform (see Methods) (Figure 1).
Figure 1.
Building a holographic brain model. A) Cortical surface models converted to object formats (.obj) compatible with Unity. B) Subcortical segmentations converted to .obj. C) MRI data converted from nifti to .tiff. D) Unity-compatible data from brain structures and segmentations are visualized in the Microsoft HoloLens system. Beige volume – cortical surface, green volume – caudate, orange volume – putamen, yellow volume – thalamus.
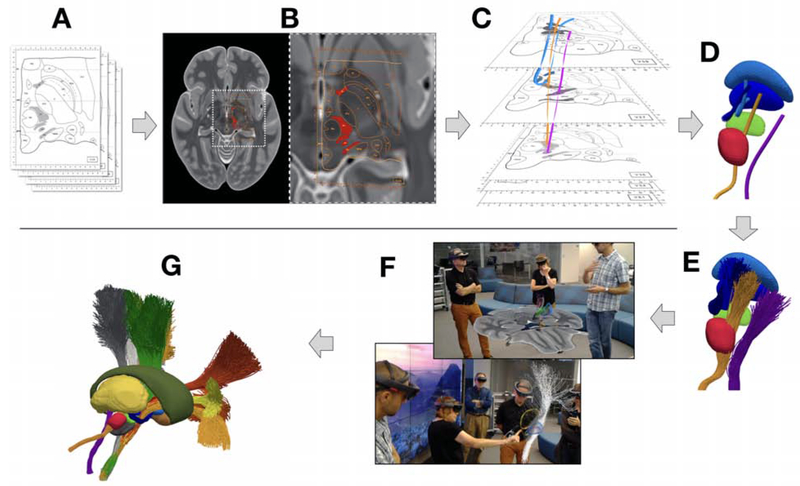
Histological results of from the Morel stereotactic atlas of the human thalamus and basal ganglia [Morel, 2007; Gallay et al., 2008] were co-registered with the CIT168 MRI slices (see Methods) (Figure 2). These results provided myelin stain based descriptions of the cerebellothalamic, pallidothalamic, and medial lemniscus axonal pathways in axial and sagittal 2D slices (Figure 2A,B). We then assembled 3D trajectories for these pathways and generated streamline approximations to simulate the distribution of fibers within the anatomy (Figure 2C,D,E).
Figure 2.
Pathway generation process. A) Histological data from the Morel atlas [Morel, 2007; Gallay et al., 2008]. B) Histological data fitted to the CIT 168 brain atlas [Pauli et al., 2018]. C) Preliminary pathway trajectories were generated using information from the MRI, histology, and previous literature. D) Mean trajectories were generated for each pathway. E) Preliminary streamline bundles were generated. F) Using the HoloLens system, these pathways were visualized, discussed, and manually edited via holographic interactions with the neuroanatomists. G) Finalized pathways.
The general process of importing histological results and developing streamline approximations for each pathway was repeated for the posterior limb of internal capsule [Morecraft et al., 2017] (Figure 3A), anterior limb of internal capsule [Safadi et al., 2018] (Figure 3B), and hyperdirect pathway [Haynes and Haber, 2013; Coude et al., 2018] (Figure 4). However, the available anatomical results for these pathways were derived from the non-human primate experiments, so direct co-registration with the CIT168 MRI slices was not explicit, but inferred from expert knowledge. Similarly, detailed anatomical reconstructions are available for the axonal trajectories connecting the subthalamic nucleus (STN) and globus pallidus (GP) in the monkey brain [Sato et al., 2000a,b]. As such, that information was used to guide our representation of those pathways in the human brain.
Figure 3.
Somatotopic organization of the internal capsule. A) Axial view of the posterior limb at descending levels, showing (i) pathway organization (ii) cooresponding illustration from Morecraft et al. [2017] macaque histology. B) Coronal view of the anterior limb, showing (i) replicated organization of five prefrontal bundles (ii) 3D reconstruction of the same bundles from Safadi et al. [2018] macaque histology.
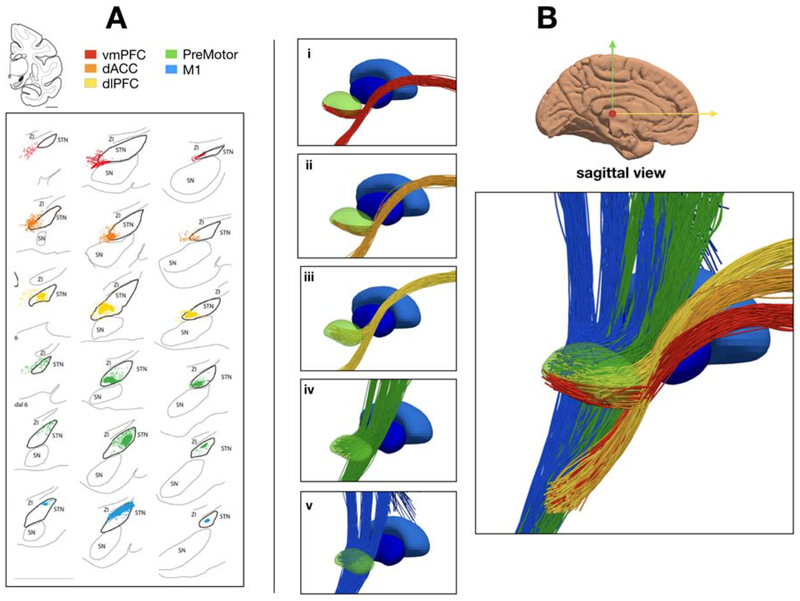
Figure 4.
Hyperdirect Pathway. A) Five termination regions of the STN from Haynes & Haber [2013] macaque histology. B) Detailed view of hyperdirect streamlines from (i) vmPFC (ii) dACC (iii) dlPFC (iv) Premotor (v) M1, color-coded based on the legend in A. Green volume – subthalamic nucleus, dark blue volume – globus pallidus internus, light blue volume – globus pallidus exturnus.
Attempting to visualize, and render in one’s mind, the complex 3D neuroanatomy of the subthalamic region can be difficult using traditional 2D computer screens. This task becomes even more complicated when attempting to verify the anatomical accuracy and continuity of various axonal pathways with respect to all of the other surrounding structures. Therefore, we turned to holographic visualization to help us refine the pathway descriptions (Figure 2F) (see Methods). The most valuable component of the holographic visualization experience was the ability to perform group-based interactions with the data. We assembled collections of world expert neuroanatomists on multiple occasions for interactive pathway editing sessions (see Methods). During these sessions, we could all see the same hologram that was fixed to a common space in the room, toggle different components of the brain model on/off, and interactively manipulate the control points describing each pathway trajectory (see Methods) (Figure 2F). Most importantly, all of these technical aspects of the model were available while maintaining eye contact with each other in the real world, which facilitated the communication of ideas. The supplementary material provides some short video clips demonstrating the interactions. In between holographic group sessions, the engineering team used the anatomist input to update the pathway descriptions and refine components of the human brain model (see Methods).
During our pathway editing process, histological data was considered paramount when defining the axonal trajectories, but many gaps needed to be filled with collective knowledge that was uniquely accessed via the holographic group sessions. In such situations, the anatomist input was coupled with structural landmarks available in the CIT168 MRI data to provide the constraints necessary to define anatomically realistic trajectories for the pathways. The result of this iterative process was our collective definition of anatomical priors that are representative of 12 cortical projections (Figure 3), as well as their corresponding hyperdirect innervations of the STN (Figure 4), and 6 pathways associated with the basal ganglia or thalamus (Figure 5). The streamlines associated with these pathways, and their quantitative coordinates in CIT168 space, are available for download as VTK files in the Open Science Framework (https://osf.io/mhd4z). In addition, a web-based Unity program is provided for interactive visualization of the entire human brain model (https://bit.ly/2V8XywA).
Figure 5.
Atlas pathways. Top row shows the surface ROIs used to generate the cortical pathways. Following rows shows the motor, prefrontal, thalamus, and basal ganglia pathways, respectively. Streamlines color-coded based on legend. Yellow volume – thalamus, green volume – subthalamic nucleus, dark blue volume – globus pallidus internus, light blue volume – globus pallidus exturnus, dark green volume – caudate, red volume – red nucleus.
Discussion
The educational desire, and clinical need, for an anatomically accurate pathway atlas for the human subthalamic region has existed for decades, especially following the therapeutic success of subthalamic DBS for the treatment of Parkinson’s disease symptoms. In theory, most of the anatomical data necessary to assemble a subthalamic pathway atlas has been available via the combination of results from many different studies, but never formally collated into a single coordinate system, or translated into the human space. This missing step can be attributed to the substantial technical challenges associated with integrating the various datasets, and then visualizing that complex 3D information in a way that would be amenable to atlas generation. However, the recent advent of holographic visualization systems that facilitate group-based interaction has changed the equation. We recognized that any attempt to assemble an axonal pathway atlas from histological results and structural MRI data would require extensive neuroanatomist expertise to fill the inherent gaps in the available information. However, no single person has all of the anatomical knowledge and experience necessary to achieve that goal for every pathway of interest. Therefore, the unique opportunity we leveraged was the combination of advanced holographic visualization of integrated datasets with simultaneous interactive inputs from collections of world expert neuroanatomists. We propose that this general framework can be repeated throughout the brain to develop the anatomical priors necessary for the evolution of human connectomic analyses.
Countless human tractography studies have been performed to simulate the connections between cortex, basal ganglia, thalamus, and cerebellum [e.g. Meola et al., 2016; Plantinga et al., 2018]. However, tractography alone is an inherently limited approach that currently requires alternative or combinatorial strategies to accurately map the axonal pathways of the human brain [Schilling et al., 2019]. This issue is compounded by the lack of anatomical axonal pathway “gold standards” in the human brain to help verify or refute the accuracy of tractographic simulations. Our results provide an independently generated pathway atlas that explicitly integrates anatomical detail in the subthalamic region that far exceeds the capabilities of any current tractography algorithm or dataset. As such, our data represents a test case for the evaluation of novel tractography strategies moving forward.
Human Connectome Project (HCP) datasets represent a standard for high quality DWI data, and to further improve the robustness and applicability of those results, group connectomes have recently been constructed [Yeh et al., 2018]. Therefore, we performed a simple comparison of our anatomically defined pathways with the streamlines available in a group connectome specifically created to study DBS of the subthalamic region [Horn et al., 2019] (Figure 6). The substantial discrepancies between the anatomist-defined atlas pathways and the group connectome streamlines are easily apparent. Further, nearly all of the connecting streamlines predicted by the group connectome appear to be false positive trajectories. Therein lies the danger of using anatomically unconstrained methods to infer axonal pathway information from a water diffusion displacement profile in an underdetermined inverse problem [Thomas et al., 2014]. The simulations in Figure 6 highlight the need to use detailed anatomical foundations, or anatomical priors, when attempting to create connectomic datasets, even when using high quality DWI data.
Figure 6.
Pathway atlas vs. group connectome. A) Cerebello-thalamic pathway. B) Pallido-thalamic pathway (ansa lenticularis and lenticular fasiculus). C) Subthalamo-pallidal pathway. First column shows the ROIs used to extract streamlines. Second Column shows the reference pathways. Final three columns show streamlines extracted from the group connectome data. First, streamlines that terminate within the defined ROIs (passing through the SCP ROI in top row). Second, streamlines that pass through ROIs. Third, streamlines passing through resampled into a Track Density Image (TDI).
While the pathway models generated in this study represent the most anatomically detailed 3D representations of human subthalamic axonal trajectories ever created, they still suffer from numerous limitations. First, no anatomical standards currently exist for defining these pathways in the human brain. As such, an important validation experiment for our holographic reconstruction methods would be to recreate established pathways in the macaque brain to more explicitly demonstrate the legitimacy of this new methodology. Nonetheless, our goal was to address the pressing need for something that could serve as an initial standard for human analyses, based on the collective knowledge of our anatomy experts, as well as the latest histologic and MRI data. Clearly, our pathway models will need to evolve with time as additional anatomical information becomes available and new anatomical mapping techniques are developed. For example, HCP data [Yeh et al., 2018], 7T MRI data [Duchin et al., 2018], and 3D polarized light imaging (3D-PLI) [Schmitz et al., 2018] all provide augmentative information that could be used to evolve the human pathway models in the future. The second major limitation of this study was that many of the details included in the human pathway atlas were derived from scientific results generated in the macaque brain. This required inter-species assumptions on the maintenance of somatotopic organizations of axonal trajectories within pathways, as well as their respective termination points in the basal ganglia targets. For example, previous tractographic analyses suggest that the human internal capsule is somatotopically organized in the same fashion as the macaque [e.g. Archer et al., 2018; Safadi et al., 2018], but real anatomical documentation of these kinds of details are lacking in the human. The third major limitation of our atlas, is that it is an atlas, an amalgamation of many different anatomical features that may not be specifically representative of any single individual. Nonetheless, advances in non-linear warping algorithms do provide opportunities to translate the idealized anatomical information provided in our atlas into the context of MRI datasets from a real person [Avants et al., 2014] (Figure 6). Supplementary Figures S1 and S2 demonstrate that process with a small, medium, and large sized brain selected from the HCP database.
The primary applications of the pathway atlas are in medical education and connectomic research. Detailed anatomical information about the human brain and its axonal connections provides greater opportunities to facilitate medical training experiences that highlight the role of brain connectivity on behavioral function, as well as the subsequent loss of function with disease (e.g. multiple sclerosis or stroke). Such training can also leverage the group-based holographic visualization capabilities, as is being done with gross anatomy education [Wish-Baratz et al., 2019]. There are also many potential benefits of augmented reality technology in neurosurgery, and with it, the need for prospective comparison studies with traditional visualization strategies on 2D computer screens [Meola et al., 2017]. Along that line, neurosurgical procedures are increasingly considering axonal connections in their planning, but limited opportunities currently exist for neurosurgeons to visualize (or learn about) the specific trajectories of anatomically verified pathways in the context of the human brain. Tractography has long attempted to serve that goal, but the limitations of clinical-grade DWI datasets are typically considered inadequate for creating accurate pathway reconstructions [Maier-Hein et al., 2017]. However, the convergence of tractography algorithms with anatomically guided constraints represents a growing trend in DWI analyses. Such steps toward anatomical realism may be especially relevant for DBS research, where the use of tractography is prevalent, but the applicability of unconstrained tractography methods is questionable [Gunalan et al., 2018] (Figure 6).
In summary, this study developed the first holographic interface for building axonal pathways, populated that system with human histological and structural MRI data, and assembled world expert neuroanatomists to interactively define axonal trajectories of the cortical, basal ganglia, and cerebellar networks. This blending of advanced visualization hardware, software development, and neuroanatomy data enabled the translation of decades of amassed knowledge into a human axonal pathway atlas that has both educational and research applications. We propose that this general framework can be repeated throughout the brain to develop the anatomical priors necessary for the evolution of human connectomic analyses.
STAR Methods
LEAD CONTACT and MATERIALS AVAILABILITY
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Cameron McIntyre (ccm4@case.edu). The streamlines associated with the pathways, and their quantitative coordinates in CIT168 space, are available for download as VTK files in the Open Science Framework (https://osf.io/mhd4z). In addition, a web-based Unity program is provided for interactive visualization of the entire human brain model (https://bit.ly/2V8XywA). The pathway atlas is the property of CWRU and may not be used for commercial purposes without the written consent of CWRU.
EXPERIMENTAL MODEL
Not Applicable.
METHOD DETAILS
Building the holographic brain model
The holographic brain model was generated using the CIT168 brain atlas as the reference coordinate system [Pauli et al., 2018] (Figure 1). This atlas contains high-resolution segmentations of the basal ganglia structures, and the included T1w and T2w image templates provide sufficient anatomical detail for identifying reference landmarks and computing non-linear transformations to patient data. To visualize MRI data (slices, segmentations, and streamlines) using the Microsoft HoloLens platform, we first converted the CIT168 images into formats that are compatible with the Unity development framework (https://unity.com/). The T1w and T2w CIT168 datasets were converted from 3D niftis to a stack of 2D axial, coronal and sagittal slices in .tiff format, using the Python toolbox, ‘med2image’ (https://github.com/FNNDSC/med2image). To generate a segmentation of the thalamus, we processed the CIT168 T1w image using FSL’s automated segmentation tool (FIRST) [Patenaude et al., 2011]. The probabilistic basal ganglia segmentations (4D niftis) were first split into separate nifti files for each subcortical brain structure and then converted to 3D surface meshes (in Wavefront OBJ (.obj) format)) using the Surfice toolbox (https://www.nitrc.org/projects/surfice/). We applied a threshold 0.33 to the probabilisitic segmentations. Each mesh file was then loaded in the 3D modelling software ‘Blender’ (https://www.blender.org/) and manually separated into left and right models. All subcortical meshes were imported as .objs into Unity (Figure 1B). Next, we processed the T1w template data using the Freesurfer software package [Fischl, 2012] to generate 3D meshes of the cortical and cerebellar surfaces, as well as a model of the white matter skeleton. Individual cortical regions were also parcellated in Freesurfer using the desikan-killiany anatomical atlas. All Freesurfer meshes were converted to Unity-compatible .obj files (https://brainder.org).
Integrating the Morel atlas
To provide anatomical references of the thalamic nuclei, as well as thalamic pathways, we integrated histological data from the ‘Stereotactic Atlas of the Human Thalamus and Basal Ganglia’ [Morel, 2007; Gallay et al., 2008]. The Morel atlas provides diagrams along the axial, sagittal and coronal stereotactic planes (AC-PC reference system) annotated to highlight thalamic and basal ganglia structures and key basal ganglia pathways (ansa lenticularis, lentifular fasciculus, thalamic fasciculus, medial lemniscus and cerebellothalamic pathway). These diagrams were imported into the Histolozee toolbox (http://picsl.upenn.edu/software/histolozee/). We used the slice position and spacing as annotated relative to the AC-PC reference system to co-register the histologic atlas slices to the CIT168 brain atlas. Each histological diagram was manually adjusted using in-plane transformations (translation, rotation, scale and skew) to fit the anatomical reference points visible in the T1w and T2w CIT168 datasets. This resulted in three sets (axial, coronal, and sagittal) of 2D diagrams fitted to the left hemisphere of the CIT168 brain atlas. These fitted diagrams were exported as nifti images (axial: 0.043 x 0.043 x 0.2, coronal: 0.040 x 0.2 x 0.040, sagittal: 0.2 x 0.071 x 0.071). Using custom in-house developed Unity scripts, these slice stacks were then integrated and overlaid on the CIT168 .tiff image data already imported into Unity.
Building the pathways
The fitted Morel atlas diagrams were overlaid onto the CIT168 T1w and T2w image data and used to plot control points delineating a single mean trajectory for each pathway of interest. These trajectories represented the course of the ansa lenticularis, lenticular fasciculus, medial lemniscus and cerebellothalamic pathways. A preliminary pathway bundle was then defined with 500 streamlines that were generated by defining a start and end region (Figure 7). A quadratic spline was fitted to each streamline, generating a smooth trajectory. Two outputs were then available to import into Unity: the preliminary pathway bundles, and the set of mean trajectory control points.
Figure 7.
Example of HoloLens-facilitated pathway refinement. A) Preliminary mean trajectory control points for the ansa lenticularis were edited based on interactive discussion with the anatomical team, shifting from (i) to (ii). The shape of individual control points were also refined to provide details about the span and spread of the pathway as it passed through a given brain region (iii). The refined bundle (iv) was then generated from the edited control points. B) Axial plane showing the histological diagram overlaid onto the CIT168 T2w image. This shows the relationship between generated reference pathways (clipped to plane) and pathways delineated using histological data.
The pallido-subthalamic and subthalamo-pallidal pathways were initially constructed by first segmenting the basal ganglia structures (STN, GPi and GPe) into two divisions along each structure’s principal component axis. This provided basic ventro-medial and dorso-lateral regions, roughly designed to represent associative and somatosensory divisions of the basal ganglia structures. We then constructed pathways connecting the STN and GPi, and the STN and GPe, by connecting two randomized points within the somatosensory and associative divisions, respectively. For the pallido-subthalamic connections from GPe to STN, we duplicated the STN to GPe set of streamlines.
Construction of the cortical projections into the subthalamic region began with using Freesurfer to define the white matter skeleton of the CIT168 brain atlas. We manually defined cortical regions corresponding to primary motor cortex, supplementary motor area (SMA) and premotor cortex (Figure 5, top panel). The primary motor cortex was then further segmented into three major regions representing the face-neck, the upper-extremities, and the lower-extremities. This resulted in five motor cortex regions of interest. We next defined a series of descending control points (each control point with an associated radius) to guide streamlines from the each of the five motor cortex regions through the internal capsule, all of which were arbitrarily truncated at the brainstem. The control point locations were selected such that there was a somatopic organization of the projections from primary motor cortex (face, upper ex., lower ex.) (Figure 3A) [Morecraft et al., 2017]. In addition, we imposed an anterior-posterior organization within the internal capsule of first the SMA fibers, then the premotor fibers, and finally the primary motor cortex projections (Figure 3A) [Morecraft et al., 2017].
Reconstructions of the cortical projections from the prefrontal cortex (PFC) followed the same general strategy as performed for the motor projections. We manually defined cortical regions corresponding to the dorsolateral-PFC (dlPFC), dorsomedial-PFC (dmPFC), ventrolateral-PFC (vlPFC), ventromedial-PFC (vmPFC) and anterior cingulate cortex (ACC) within the CIT168 brain atlas (Figure 5, top panel). Streamlines from each of these cortical regions were then guided through the anterior limb of the internal capsule (ALIC) and subthalamic region using control points, replicating the organizational patterns found in recent anatomical studies [Lehman et al., 2013; Safadi et al., 2018] (Figure 3B). We then arbitrarily truncated these trajectories at the pons.
With the cortical projections within the internal capsule in place, we then turned our attention to definition of hyperdirect axon collaterals to the STN. We constructed sets of branching axon collaterals from the primary motor cortex (face, upper-ex., lower-ex.), premotor, dlPFC, vmPFC and ACC projections that went into the STN, as described in Haynes and Haber [2013] and Coude et al. [2018] (Figure 4). This was accomplished by selecting 100 random streamlines from each cortical projection bundle. Along each of these streamlines, we selected a collateral starting point on the main corticofugal trajectory that was 4-6 mm (randomly selected) above the STN center-of-gravity [Coude et al., 2018]. From there, collaterals were projected into five pre-specified regions of the STN (Figure 4A), which were defined from the results of Haynes and Haber [2013].
Holographic editing sessions with anatomists
After generating preliminary pathway representations, we integrated them into the Unity platform. We then created a HoloLens application that allowed us to simulatenously visualize (and toggle on/off) the MRI data, histology data, 3D segmentations of basal ganglia structures, and pathway bundles (Figure 1). In addition, the HoloLens application allowed us to manually refine the control points that formed the basis of each of the pathways. Potential manual refinements included changing the position of each control point, as well as its 3D shape and size, which we used as a proxy for the spatial spread of the pathway at that point in the anatomy (Figure 7).
PLS, SNH, MP, YS, were introduced to the holographic brain model as part of two separate daylong sessions held at the Interactive Commons on the campus of CWRU (July 24, 2018 and December 20, 2018). They provided the expert anatomical knowledge to manually refine the preliminary pathways. The sessions involved visual inspections of the pathways, and extended discussions around the neuroanatomical plausibility of the preliminary delineations. The output from these discussions were both quantitative (e.g. move this control point 3-4 mm) and qualitative (e.g. the trajectory should bend more here) comments that were used to refine the preliminary pathways. A goal of the group discussions was to develop a consensus definition for each pathway that represented the collective understanding of the expert anatomists. As such, we discussed one pathway at a time, and then ended the days with a collective view of the entire system for final thoughts. Following each editing session with the anatomists, the engineering team regenerated the detailed streamline renderings of the pathways with the new/updated control point information.
We found that one of the anatomists would gravitate toward controlling the discussion on their respective “favorite” pathway(s) and the other anatomists would then take a more passive role until their “favorite” pathway was up for discussion. As such, inter-rater variability and/or disagreement was not a major issue in the workflow. However, as the leading anatomist was working, the other anatomists would chime in on a regular basis, asking questions or looking for clarification on why the leader was making their specific comments/changes for the model. It was those questions that then drove the engineering team to integrate new data or pull up different views for visualization during the discussion. For example, during our 1st group working day, the model was focused on using MRI data to guide the pathway development process. However, the main insight from those 1st day discussions was that we needed to transition our focus toward histological data because the anatomists wanted something they felt represented a more precise standard for us to base our decisions. Therefore, the 2nd meeting was focused on “validating” that the pathways precisely aligned with the available histology data.
During each day with the anatomists, we did 2-3 hours of interactive holographic work in the morning, and 2-3 hours of holographic work in the afternoon, which was followed by ~1 hour summarizing our conclusions from the day. We worked through each pathway on both meeting days, but some pathways received more time/effort than others. For example, the hyperdirect collaterals and STN innervation, ansa lenticularis, and cerebellothalamic pathways were the most complicated and took the most time. However, each pathway received at least 4 different iterations of interactive group discussion, where each day with the anatomists used a 2-iteration process. The 1st iteration in the morning focused on the pathway, independent of other pathways, and 2nd iteration in the afternoon, was primarily concerned with its place relative to the surrounding pathways/structures.
Warping pathways to a de novo subject
We randomly selected individual subject datasets from the human connectome project and used the ANTs toolbox (http://stnava.github.io/ANTs/) to estimate a nonlinear transformation from the CIT168 brain atlas (T1w image) to the subject’s T1w image (see Supplemental Figures S1 and S2). Using this transformation, we warped our reference pathways to the subject space. Next, we extracted a ‘group connectome’ dataset from the Lead-DBS Toolbox [Horn et al., 2019] and estimated the nonlinear transformation to the CIT168 brain atlas. Then, applying both warps, we transformed the group connectome to subject space, where we extracted streamlines corresponding to three pathways (Figure 6). We then visually compared the group connectome pathways to those predicted by our subthalamic pathway atlas.
QUANTIFICATION and STATISTICAL ANALYSIS
Not Applicable.
DATA and CODE AVAILABILITY
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and Algorithms | ||
| CIT168 | Pauli et al., 2018; http://doi.org/10.17605/OSF.IO/JKZWP | |
| HoloLens | https://www.microsoft.com/en-us/hololens | |
| med2image | https://github.com/FNNDSC/med2image | |
| FIRST | Patenaude et al., 2011; https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FIRST/UserGuide | |
| Surfice | https://www.nitrc.org/projects/surfice/ | |
| Blender | https://www.blender.org/ | |
| Freesurfer | Fischl, 2012; https://surfer.nmr.mgh.harvard.edu/ | |
| Brainder | https://brainder.org | |
| Histolozee | http://picsl.upenn.edu/software/histolozee/ | |
| ANTs | http://stnava.github.io/ANTs/ | |
| Lead-DBS | Horn et al., 2019; https://www.lead-dbs.org/ | |
| Pathways | This paper; https://osf.io/mhd4z | |
| Brain Model | This paper; https://bit.ly/2V8XywA | |
Highlights:
Developed the first holographic interface for building axonal pathways.
Group-based interactive definition of axonal trajectories by neuroanatomists.
Translation of decades of amassed knowledge into a 3D human axonal pathway atlas.
Acknowledgements
This work was supported by the National Institutes of Health (R01 NS105690).
Conflict of Interest Statement
CCM is a paid consultant for Boston Scientific Neuromodulation, receives royalties from Hologram Consultants, Neuros Medical, Qr8 Health, and is a shareholder in the following companies: Hologram Consultants, Surgical Information Sciences, Autonomic Technologies, Cardionomic, Enspire DBS, Cortics. MAG’s laboratory receives research support from Siemens Healthineers. MAG receives royalties from Siemens and GE. MVP, MAG, and CCM have patent and patent applications related to this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Material
Supplemental Video
References
- Agrawal A, Kapfhammer JP, Kress A, Wichers H, Deep A, Feindel W, Sonntag VK, Spetzler RF, Preul MC. Josef Klingler’s models of white matter tracts: influences on neuroanatomy, neurosurgery, and neuroimaging. Neurosurgery. 2011. August;69(2):238–52. [DOI] [PubMed] [Google Scholar]
- Archer DB, Vaillancourt DE, Coombes SA. A Template and Probabilistic Atlas of the Human Sensorimotor Tracts using Diffusion MRI. Cereb Cortex. 2018. May 1;28(5):1685–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC. The Insight ToolKit image registration framework. Front Neuroinform. 2014. April 28;8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994. January;66(1):259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudé D, Parent A, Parent M. Single-axon tracing of the corticosubthalamic hyperdirect pathway in primates. Brain Struct Funct. 2018. December;223(9):3959–3973. [DOI] [PubMed] [Google Scholar]
- Duchin Y, Shamir RR, Patriat R, Kim J, Vitek JL, Sapiro G, Harel N. Patient-specific anatomical model for deep brain stimulation based on 7 Tesla MRI. PLoS One. 2018. August 22;13(8):e0201469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essayed WI, Zhang F, Unadkat P, Cosgrove GR, Golby AJ, O’Donnell LJ. White matter tractography for neurosurgical planning: A topography-based review of the current state of the art. Neuroimage Clin. 2017. June 15;15:659–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B FreeSurfer. Neuroimage. 2012. August 15;62(2):774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallay MN, Jeanmonod D, Liu J, Morel A. Human pallidothalamic and cerebellothalamic tracts: anatomical basis for functional stereotactic neurosurgery. Brain Struct Funct. 2008. August;212(6):443–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunalan K, Howell B, McIntyre CC. Quantifying axonal responses in patient-specific models of subthalamic deep brain stimulation. Neuroimage. 2018. May 15;172:263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes WI, Haber SN. The organization of prefrontal-subthalamic inputs in primates provides an anatomical substrate for both functional specificity and integration: implications for Basal Ganglia models and deep brain stimulation. J Neurosci. 2013. March 13;33(11):4804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn A, Li N, Dembek TA, Kappel A, Boulay C, Ewert S, Tietze A, Husch A, Perera T, Neumann WJ, Reisert M, Si H, Oostenveld R, Rorden C, Yeh FC, Fang Q, Herrington TM, Vorwerk J, Kuhn AA. Lead-DBS v2: Towards a comprehensive pipeline for deep brain stimulation imaging. Neuroimage. 2019. January 1;184:293–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, Knösche TR, Turner R. White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion MRI. Neuroimage. 2013. June;73:239–54. [DOI] [PubMed] [Google Scholar]
- Lehman JF, Greenberg BD, McIntyre CC, Rasmussen SA, Haber SN. Rules ventral prefrontal cortical axons use to reach their targets: implications for diffusion tensor imaging tractography and deep brain stimulation for psychiatric illness. J Neurosci. 2011. July 13;31(28):10392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier-Hein KH, Neher PF, Houde JC, Côté MA, Garyfallidis E, Zhong J, Chamberland M, Yeh FC, Lin YC, Ji Q, Reddick WE, Glass JO, Chen DQ, Feng Y, Gao C, Wu Y, Ma J, Renjie H, Li Q, Westin CF, Deslauriers-Gauthier S, González JOO, Paquette M, St-Jean S, Girard G, Rheault F, Sidhu J, Tax CMW, Guo F, Mesri HY, Dávid S, Froeling M, Heemskerk AM, Leemans A, Boré A, Pinsard B, Bedetti C, Desrosiers M, Brambati S, Doyon J, Sarica A, Vasta R, Cerasa A, Quattrone A, Yeatman J, Khan AR, Hodges W, Alexander S, Romascano D, Barakovic M, Auría A, Esteban O, Lemkaddem A, Thiran JP, Cetingul HE, Odry BL, Mailhe B, Nadar MS, Pizzagalli F, Prasad G, Villalon-Reina JE, Galvis J, Thompson PM, Requejo FS, Laguna PL, Lacerda LM, Barrett R, Dell’Acqua F, Catani M, Petit L, Caruyer E, Daducci A, Dyrby TB, Holland-Letz T, Hilgetag CC, Stieltjes B, Descoteaux M The challenge of mapping the human connectome based on diffusion tractography. Nat Commun. 2017. November 7;8(1):1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola A, Yeh FC, Fellows-Mayle W, Weed J, Fernandez-Miranda JC. Human Connectome-Based Tractographic Atlas of the Brainstem Connections and Surgical Approaches. Neurosurgery. 2016. September;79(3):437–55. [DOI] [PubMed] [Google Scholar]
- Meola A, Cutolo F, Carbone M, Cagnazzo F, Ferrari M, Ferrari V. Augmented reality in neurosurgery: a systematic review. Neurosurg Rev. 2017. October;40(4):537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morecraft RJ, Binneboese A, Stilwell-Morecraft KS, Ge J. Localization of orofacial representation in the corona radiata, internal capsule and cerebral peduncle in Macaca mulatta. J Comp Neurol. 2017. November 1;525(16):3429–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel A Stereotactic Atlas of the Human Thalamus and Basal Ganglia. Informa Healthcare USA; 2007. [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 1999. February;45(2):265–9. [DOI] [PubMed] [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011. June 1;56(3):907–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli WM, Nili AN, Tyszka JM. A high-resolution probabilistic in vivo atlas of human subcortical brain nuclei. Sci Data. 2018. April 17;5:180063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantinga BR, Temel Y, Duchin Y, Uludağ K, Patriat R, Roebroeck A, Kuijf M, Jahanshahi A, Ter Haar Romenij B, Vitek J, Harel N. Individualized parcellation of the subthalamic nucleus in patients with Parkinson’s disease with 7T MRI. Neuroimage. 2018. March;168:403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safadi Z, Grisot G, Jbabdi S, Behrens TE, Heilbronner SR, McLaughlin NCR, Mandeville J, Versace A, Phillips ML, Lehman JF, Yendiki A, Haber SN. Functional Segmentation of the Anterior Limb of the Internal Capsule: Linking White Matter Abnormalities to Specific Connections. J Neurosci. 2018. February 21;38(8):2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammartino F, Hodaie M. Diffusion Tensor Imaging of the Basal Ganglia for Functional Neurosurgery Applications. Prog Neurol Surg. 2018;33:62–79. [DOI] [PubMed] [Google Scholar]
- Sato F, Lavallée P, Lévesque M, Parent A. Single-axon tracing study of neurons of the external segment of the globus pallidus in primate. J Comp Neurol. 2000a. January 31;417(1):17–31. [PubMed] [Google Scholar]
- Sato F, Parent M, Levesque M, Parent A. Axonal branching pattern of neurons of the subthalamic nucleus in primates. J Comp Neurol. 2000b. August 14;424(1):142–52. [DOI] [PubMed] [Google Scholar]
- Schilling KG, Nath V, Hansen C, Parvathaneni P, Blaber J, Gao Y, Neher P, Aydogan DB, Shi Y, Ocampo-Pineda M, Schiavi S, Daducci A, Girard G, Barakovic M, Rafael-Patino J, Romascano D, Rensonnet G, Pizzolato M, Bates A, Fischi E, Thiran JP, Canales-Rodríguez EJ, Huang C, Zhu H, Zhong L, Cabeen R, Toga AW, Rheault F, Theaud G, Houde JC, Sidhu J, Chamberland M, Westin CF, Dyrby TB, Verma R, Rathi Y, Irfanoglu MO, Thomas C, Pierpaoli C, Descoteaux M, Anderson AW, Landman BA. Limits to anatomical accuracy of diffusion tractography using modern approaches. Neuroimage. 2019. January 15; 185:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz D, Muenzing SEA, Schober M, Schubert N, Minnerop M, Lippert T, Amunts K, Axer M. Derivation of Fiber Orientations From Oblique Views Through Human Brain Sections in 3D-Polarized Light Imaging. Front Neuroanat. 2018. September 27;12:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Kimmlingen R, Eberlein E, Witzel T, Cohen-Adad J, McNab JA, Keil B, Tisdall MD, Hoecht P, Dietz P, Cauley SF, Tountcheva V, Matschl V, Lenz VH, Heberlein K, Potthast A, Thein H, Van Horn J, Toga A, Schmitt F, Lehne D, Rosen BR, Wedeen V, Wald LL. Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. Neuroimage. 2013. October 15;80:220–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, Pierpaoli C. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci U S A. 2014. November 18;111(46):16574–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF. Parcellating Cerebral Cortex: How Invasive Animal Studies Inform Noninvasive Mapmaking in Humans. Neuron. 2018. August 22;99(4):640–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K; WU-Minn HCP Consortium. The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013. October 15;80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wish-Baratz S, Gubatina AP, Enterline R, Griswold MA. A new supplement to gross anatomy dissection: HoloAnatomy. Med Educ. 2019. May;53(5):522–523. [DOI] [PubMed] [Google Scholar]
- Yeh FC, Panesar S, Fernandes D, Meola A, Yoshino M, Fernandez-Miranda JC, Vettel JM, Verstynen T. Population-averaged atlas of the macroscale human structural connectome and its network topology. Neuroimage. 2018. September;178:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.