Abstract
Hypotheses
The drive to survive is a biological universal. Intelligent behaviour is usually recognized when individual organisms including plants, in the face of fiercely competitive or adverse, real-world circumstances, change their behaviour to improve their probability of survival.
Scope
This article explains the potential relationship of intelligence to adaptability and emphasizes the need to recognize individual variation in intelligence showing it to be goal directed and thus being purposeful. Intelligent behaviour in single cells and microbes is frequently reported. Individual variation might be underpinned by a novel learning mechanism, described here in detail. The requirements for real-world circumstances are outlined, and the relationship to organic selection is indicated together with niche construction as a good example of intentional behaviour that should improve survival. Adaptability is important in crop development but the term may be complex incorporating numerous behavioural traits some of which are indicated.
Conclusion
There is real biological benefit to regarding plants as intelligent both from the fundamental issue of understanding plant life but also from providing a direction for fundamental future research and in crop breeding.
Keywords: Intelligence, adaptability, learning, real-world circumstances, selection, systems biology
INTRODUCTION
The idea that plants are intelligent has been controversial since it was first described (Trewavas, 2003). There have been three published criticisms (Firn, 2004; Alpi et al., 2007; Chamowitz, 2018). The first two have already been answered in print although readers must judge how well (Trewavas, 2004a, 2007). The third will be considered later. The trio of critiques each raise different issues, suggesting that they arise from differing perceptions of plant behaviour and plant abilities.
Three issues seem to be at the root of the criticism. They are all answered in greater detail in this article. First is the idea that nervous systems are required for intelligence. The counter argument is that intelligent behaviour is reported in single cells and bacteria. Such organisms obviously do not have a nervous system but do use electrical information and Ca2 signalling as do higher plants.
Second is the influence of laboratory experience on perceptions of plant behaviour. The need to examine plant behaviour and intelligence in wild (real-world) circumstances was indicated in Trewavas (2003). Darwinian over-production of seed ensures that virtually all plants die before completing the life cycle. The plant environment contains predators and grazers of all kinds, threats of disease and stringent competition. The inbuilt driving forces of individual survival and thence to reproduction are fundamental to life of all kinds. In these unpredictable and varying circumstances the aim of intelligence in all individuals is to modify behaviour to improve the probability of survival. The single environment of the laboratory with individual plants grown under ideal, non-threatening conditions disguises the reality of wild plant life and real behaviour. The reason for intelligent behaviour is, then, not obviously apparent and is easily dismissed as not being relevant (Chamowitz, 2018). Better knowledge of the ecological literature is an effective counter argument.
The third issue is the relationship of adaptability to intelligence. We have used the term ‘adaptability’ throughout to refer to the individual plant, with the consequences that follow from individuality. Darwin always believed that selection started with the individual and later evolutionary writing put changes in behaviour as the first step (Mayr, 2001). Without recognizing the individual nature of adaptability (and physiology has usually ignored it) leads to a lack of recognition that its characteristics are surely selectable.
Where this is most important is in crop production where the emphasis on yield and stability may have inevitably and unsurprisingly diminished crop adaptability. More seriously the present gene pool for crops may have largely eliminated a necessity for crops that are grown in a variety of field environments and which require the ability to adapt. If it is the case that present crops are the result of selection, driving the genome along an irreversible branch, then the only solution is to start again with wild ancestors but with a better understanding of what agriculture in the future requires. This may be the only route to really identifying the essential genomic traits of adaptability. We also consider that adaptability may be a generic term that actually covers a range of behavioural traits, and showing this does now need investigation. Recognition of the presence of intelligent capabilities that incorporate adaptability will improve appreciation and research.
UNDERSTANDING THE BASIS OF INTELLIGENCE
The general requirement for intelligent behaviour throughout life
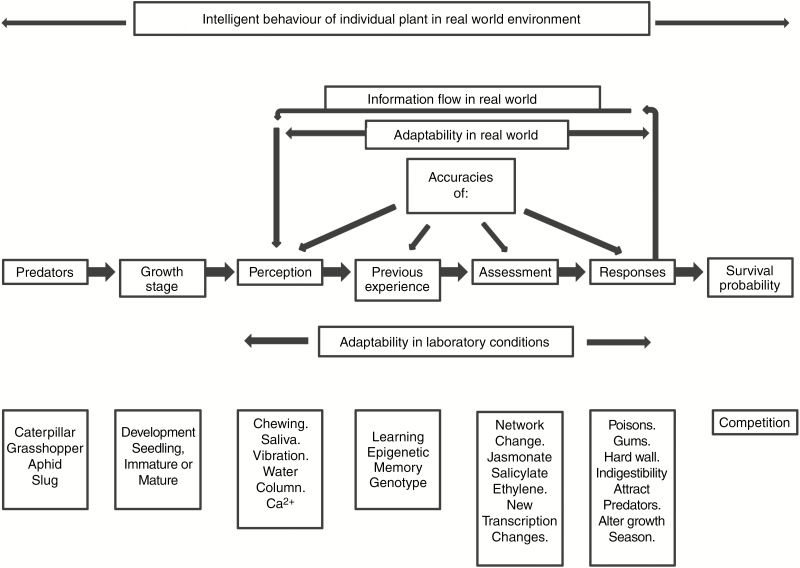
Behaviour is generally recognized as intelligent when an individual organism in fiercely competitive or threatening circumstances modifies its behaviour to improve its chances of survival. Such circumstances are experienced by plants and all other organisms that live in wild, real-world environments and in which survival is a very common uncertainty (Gilbert, 2001; Trewavas, 2003). Figure 1 indicates the features of intelligent behaviour for an individual plant using recognized predator–prey relationships. This figure shows the probable sequences of perception through to responses and survival probability. The features above this sequence (accuracies of perception, experience, assessment and responses) and feedback information to increase or decrease the strength of the ultimate response are only relevant to real-world circumstances but are variables that inevitably impact on survival probability and on subsequent individual competition. They probably remain unrecognized in laboratory research because of the common methodology of aggregating and averaging data amongst a number of individuals and use of just one environment. However, the accuracies of perception, experience, assessment and response will be important variables in individual wild plants. It needs to be emphasized here that adaptability will be different in laboratory conditions compared to real-world situations. The blocks below the main perception line in Figure 1 are some of the known contributors on each of the sub-characteristics of transduction. They are indicants, not an exhaustive list.
Fig. 1.
Intelligence schedule of predator–prey relationships in individual plants. The figure summarizes a main sequence of predation through to survival probability. In laboratory conditions, adaptability involves only the sequence perception to responses. Above the main sequence are real-world conditions with the additional inputs of accuracies of perception, previous experiences, assessment and responses which undoubtedly vary between individuals. There is also a feedback system of information flow from responses back to perception to modulate and assess the continued predation and subsequent response. The blocks below the main sequence are suggested constituents of each step. These lists are indicants only and are not exhaustive. Intelligent behaviour covers the range from predators to survival probability and does need real-world circumstances for its demonstration of improved survival probability amongst competition.
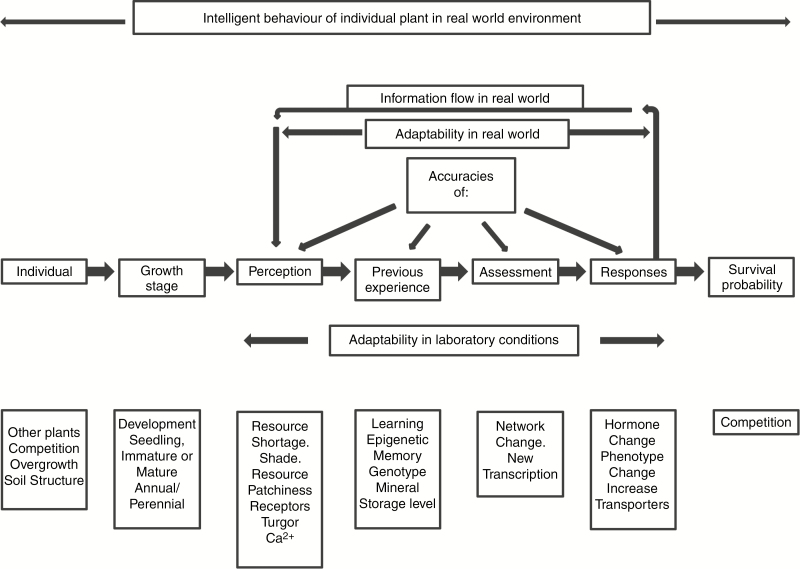
Figure 2 indicates a similar structure and conclusions for resource acquisition, which can be likened to the plant acting as predator and the prey as resource. The visible response here is phenotypic plasticity, either increasing the size of organs or increasing their number (Trewavas, 2003; Sultan, 2015), and at the same time diminishing those that function less well (Trewavas, 2014). The same overall transduction response sequence is maintained, as are the requirements for accuracies in real-world circumstances for individual plants.
Fig. 2.
Intelligence schedule of resource acquisition in individual plants. The figure summarizes the main sequence from individual to survival probability. In laboratory conditions adaptability covers only the range of perception to responses. Above the main sequence are real-world conditions only, with the additional complications of individual variation in accuracies of perception, previous experience, assessment and responses. The feedback system of information from response to perception should modulate the extent of response and ensure it is controlled. The blocks below each main step are suggested and the lists are not intended to be exhaustive. Intelligent behaviour covers the whole range from individual plant to survival probability and competition.
Different combinations of responses and different combinations of proteins will be selected and used (Figures 1 and 2). Some combinations will work well, others less so but this will be crucially dependent on the specific biological and abiotic environment experienced at the time. As this environment changes, it can be expected that previous failures in response now become useful. Additional options include speed or size of response, quick learning, and accessibility of and relevance of epigenetic memory amongst others. Numerous observations indicate that individuals vary in the extent and the speed with which they respond to external signals (e.g. Liptay and Davidson, 1971; Bazzaz, 1996).
The most comprehensive definition of intelligence highlights the importance of the individual
Two psychologists, Legg and Hunter (2007), collected all the definitions of intelligence they could find, 70 in total, and provided a consensus of what they rightly termed ‘universal intelligence’. Below we have replaced the word ‘agent’ with ‘plant’ in their definition:
‘(1). Intelligence is a property that an individual plant has as it interacts with its environment. (2). Intelligence is related to the plants ability to succeed or profit with respect to some goal or objective. (3). Intelligence depends on how able the plant is to adapt to different objectives and environments’ (Legg and Hunter, 2007, p. 5). This definition emphasizes the individual.
Section 1 describes plant behaviour. Changes in visible responses to resource fluctuation often lead to phenotypic plasticity as in Figure 2. Resistance to herbivory, disease and developed resistance to some extreme abiotic signals (high and low temperature, drought, etc.) are commonly identified only with molecular changes. Comparison of resistant and sensitive individuals reveals an eventual phenotypic difference.
Section 2. The goal is the maximal number of viable siblings (fitness); the equally prominent objective through the life cycle is survival, accumulation of resources and the optimal positioning of a maximal number of flowers. Intelligent differences between individuals should make a material difference but optimal intelligence in any one individual is probably limited to particular environments.
Section 3 emphasizes that intelligence relates to the skill with which adaptability can be used in different environmental circumstances. Reliance here is placed on either the accuracies of perception, previous experience, assessment and continuing change in response (Figure 1) and the size and nature of the threatening circumstance (e.g. herbivory), or the identity of the competition. Learning about the environment will contribute to the skill required but learning is adaptive behaviour (Plotkin, 1988)
Intelligence of the individual organism can then be broken down into three separate but complex systems: behaviour–adaptability–environments. These three systems integrate the individual plant into a very complex emergent property that we call intelligence. Intelligence also incorporates negative feedback adjusting behaviour and adaptability as time and response progress (Figure 1). However, note that adaptability is different between laboratory environments (one circumstance) and numerous real-world circumstances which involve survival and attempted completion of the life cycle (Figures 1 and 2).
Intention and purpose
Active behaviour is most easily defined as purposeful or intentional when it is goal orientated (Rosenblueth et al., 1943; Russell, 1946; Turner, 2007). Romanes (1884) acquired 40 years-worth of information and cuttings from Darwin on intelligence before writing the first book on animal intelligence covering protozoa to primates but excluding humans. He concluded that: ‘Intelligence is the faculty which is concerned in the intentional adaptability of means to ends’. The ‘means’ are what the individual plant has to hand, usually changes in meristem behaviour; the ends are the goals or objectives: ‘When we find that an individual profits by individual experience and acts on its perception, it sounds, less unusual, to perceive it as displaying intelligence’ (Romanes, 1884, p. 17). Stenhouse (1974) indicates intelligence to be ‘adaptively variable behaviour during the lifetime of the individual’.
Sternberg (1985) summarized human intelligence as ‘purposive adaptation to real world contexts’ and emphasized practical intelligence (Sternberg and Wagner, 1986; Beer, 1990). He regarded human circumstances as real world because of the variety of environments in which individuals grow and develop.
The goal underpinning intention and purposeful intelligence in individual plants is, of course, survival; maximal numbers of viable siblings usually requires other plants of the same species for fertilization.
Attitudes about human intelligence mislead as to its general nature
Human intelligence is a psychological and educational subject, used initially to try and identify individuals with special educational needs (Binet and Simon, 1916). Despite some claims to the contrary (Chamowitz, 2018), there is general agreement amongst psychologists as to what human intelligence is (reasoning, numeracy and language skills) and that it can be accurately measured, as for example with IQ (Deary, 2001; see Box 1).
BOX 1: DO PSYCHOLOGISTS DISAGREE ABOUT HUMAN INTELLIGENCE?
Chamowitz (2018) stated that the concept of human intelligence is vague and subjective, and that there is no agreement on its meaning. Is that claim supported by the literature?
An analysis of 40 short articles on intelligence found problem-solving and adaptation to be the commonest descriptors of human intelligence (Sternberg and Berg, 1986). Ninety-seven per cent of +660 psychologists surveyed for their opinion on human intelligence identified problem-solving, an ability to acquire knowledge (learn) and reasoning as the important aspects of human intelligence, and also agreed it could be measured accurately (Snyderman and Rothman, 1988). Gottfredson and 52 others (Gottfredson, 1997): ‘Human intelligence is a very general mental capability that involves the ability to reason, plan, solve problems, learn quickly and learn from experience.’ ‘Intelligence can be measured and intelligence tests (IQ) do it well.’ ‘They are among the most accurate (in technical terms, reliable and valid) of all psychological tests and assessments.’ The first IQ test was constructed by Alfred Binet, Theodore Simon and Lewis Terman at Stanford in the early 1900s and was used to identify children with special educational needs (Binet and Simon, 1916). It is still commonly used today along with one constructed by Weschler (Deary, 2001). Spearman’s (1904) important general intelligence or g, the most thoroughly established factor of human intelligence supported by over 400 publications, is considered later.
A task force of eminent psychologists was set up in 1996 to state the nature of human intelligence and direction for research findings. ‘Individuals differ from one another in their ability to understand complex ideas, to adapt effectively to their environment, to learn from experience’ (Neisser, et al., 1996). A further implied claim was that Sternberg and Binet disagreed about the meaning of intelligence (Chamowitz, 2018). Sternberg’s (2006) assessment of Binet and Simon’s view of intelligent thought is that ‘direction and adaptation certainly fits with contemporary views of intelligence and Binet’s notion of criticism actually seems prescient considering the current appreciation of metacognitive processes as a key aspect of intelligence’ (Sternberg, 2006, p. 488). There will always be individuals who disagree with the views propounded by over 95 % of psychologists on human intelligence but Sternberg and Binet are not in that category. Sternberg (1986) sees intelligence as occurring throughout the natural world.
Because we are acutely aware of our own intelligence and human intelligence is so common by experience and discussion, we are reluctant to allow that other organisms can be intelligent too. We impose our own animal view on all other organisms, which leads to expectations of visible movement as expressions of intelligence that in plants cannot be fulfilled. Modern medicine, agriculture and social structures protect against the vagaries of environmental and disease problems. A good biological reason for human intelligence is not apparent because survival is no longer a consideration. When the word intelligence is used to describe the behavioural qualities of other organisms, its necessary relationship to survival is thus not recognized.
The mechanisms of intelligent behaviour between plants and animals are entirely different. Both rely on communication using electrical and chemical means in differing proportions. The words hormones, behaviour, disease, reproduction, stem cells, male, female, growth, development, vascular tissue, circulation and many others are all used to describe analogous processes between higher animals and plants but whose mechanisms are, unsurprisingly, very different. Learning, memory and intelligence fall into the same category.
During evolution, plant and animal progenitors separated when single-celled. Complex movement and a nervous system probably evolved in animals from the positive feedback inherent in predator/prey relationships in which speed to observe and speed to catch, or not be caught, were improved by a proto-nervous system that connected sensory and motor systems (Jekely et al., 2015; Keijzer, 2017). Photosynthetic plants needed a wall, to constrain osmotically active materials, but which constrained movement. In multicellular plants it provided the skeletal structure. However, crucially both plants and animals retained adaptability. Alan Turing (1947) identified the relationship of intelligence with adaptability.
INTELLIGENT BEHAVIOUR IS REPORTED IN SINGLE CELLS AND BACTERIA
A common anthropomorphic view of the natural world is constructed by placing organisms on an evolutionary tree according to how near they reflect the potentialities of humans (Lovejoy, 1936). Vertebrates are considered more advanced than invertebrates and mammals more advanced than birds, and any multicellular animal is considered more advanced than single cells or bacteria. Plants, on this basis, if they are considered at all, are evolutionary simpletons. They do not fulfil the prime assumption that without obvious movement in our time frame, they cannot be intelligent. However, the real world is full of challenging problems and environmental variations for all organisms that need solution if they are to survive.
The biogenic route inverts anthropomorphism (Lyon, 2006). It starts by asking what capabilities can be identified in single cells and then looks at increasingly complex multicellular organisms, identifying how these capabilities are changed during evolution. In 1897, the originator of IQ measurements, Alfred Binet (see Box 1) published a short book of his observations on protozoan behaviour which he described as behaving intelligently. Jennings (1906) detailed his investigations on behaviour in Stentor and Amoeba. In identifying their behaviour as intelligent, he stated (p. 334) that intelligence is ‘held to consist in the modification of behaviour in accordance with experience’. ‘Thus it seems possible to trace forward from the simpler organisms some of the phenomena which we know from objective evidence to exist in ourselves.’ The single-cell organisms that authors have identified as expressing intelligent behaviour are Stentor, Didinium, Amoeba, Spirostomum, Paramecium and Physarum in experiments that demonstrate learning, memory, speed versus accuracy, diagnosis of error and correction, cheating during reproduction, choice, assessment, and environment and food discrimination (Binet, 1897; Jennings, 1906; Smith, 1908; Gelber, 1952; Jensen, 1957; Hinkle and Wood, 1994; Armus et al., 2006; Clark, 2010, 2013; Trewavas, 2014; and see list of Physarum references in Trewavas, 2017). Because evolution tinkers with material already to hand, intelligent behaviour in more complex organisms did not appear de novo, but was present before the major evolutionary divisions into plants, fungi and animals (Clark, 2010, 2013). Nervous systems have elaborated what was already present in single-cell progenitors.
Bacterial swimming represents one aspect of behaviour for about half of bacterial species. Decisions that lead to intelligent swimming towards food or away from toxins are modified by assessments of costs and benefits, present internal state (whether ‘fed or hungry’), probable future conditions, strength of various gradients, error perception and correction, and no doubt more (Adler and Tso, 1974; Allmann, 1999; Lyon, 2006; Westerhoff et al., 2014). Bacteria learn and also express self-recognition, memory, associative learning, anticipation, adaptation and reflection, and make decisions when given choices (Allmann, 1999; Hoffer et al., 2001; Gibbs et al., 2008; Westerhoff et al., 2014). EMBO organized meetings on neural networks in bacteria (Golden, 2003; Armitage et al., 2005). ‘Mind may be the result of interacting cells. Mind and body perceiving are equally self-referring, self-reflexive processes already present in the earliest bacteria’ (Margulis and Sagan, 1995, p. 32). Even phages, bacterial viruses, have been shown to have a remarkable social life. Using peptide communication they decide whether to lie low in the host cell, or when to replicate and burst out in search of new hosts (Dolgin, 2019).
The capabilities here reflect the need to deal with the varying, uncertain world as it is. Multicellular plants evolved from archaea via single cells. Would any of these valuable behavioural capabilities have been discarded when the self-same situations of a challenging uncertain environment needed to be solved?
INVESTIGATIVE BENEFITS TO IDENTIFYING PLANTS AS INTELLIGENT
We consider that viewing plant behaviour through the lens of intelligence generates numerous benefits. Specifically, it provides for:
(1) A better understanding of plant behaviour and in particular the necessary requirements which need experimental investigation (such as decision-making, error perception, anticipation, speed versus accuracy, etc., see later) for intelligent, adaptive responses.
(2) An important focus for investigations of systems biology that can improve understanding by enabling analysis of system network structure involved in adaptability.
(3) A common base to adaptability and intelligence in all organisms? High-throughput molecular techniques (DNA and RNA sequencing and translation, methylome, phosphorylome and interactome approaches) are now sufficiently advanced in this regard.
(4) Better understanding of the molecular and genetic basis of adaptability, which should help breeding through genetic markers and adaptation to a greater range of farm environments, given that many present crop species are regarded as having been selected for aspects of adaptability (Matsuo, 1975).
(5) Improvement of crop management by new techniques of plant hardening-inspired plant learning skills, and optimizing plant–plant networking in integrative crop systems with different species, supporting a more sustainable agriculture (Bruce, 2010).
(6) A better appreciation of its evolutionary implications, specifically the relevance of intelligent behaviour to understanding of selection and fitness. The psychologist Jonathan Schull (1990), in two articles on species intelligence, commented that: ‘As I understand it, the intelligence of a species of plant, might exceed that of a species of intelligent animals’ (p. 104). That can now be investigated. Darwin (1871) stated that: ‘Intelligence is based on how efficient a species becomes at doing the things they need to survive.’
(7) By considering plants as self-referring organisms, a better understanding of the plant–plant interactions and communication and, as a consequence, a deeper comprehension of ecological relationships, such as cooperation and altruism among plants, rather than the pure Darwinian struggle of life (Dudley, 2015).
(8) A better understanding of the implications of McClintock’s challenge to plant scientists by this Nobel prize-winning plant scientist which clearly starts by recognizing plants as intelligent organisms. ‘A goal for the future would be to determine the extent of knowledge the cell has of itself and how it uses that knowledge in a thoughtful manner when challenged’ (McClintock, 1984).
(9) A fuller understanding of what a plant actually is. If the intelligent aspects of plant life fit most concepts and factual evidence supports it, then we must describe plants as intelligent beings, not because we want to or arbitrarily choose to, but because they are.
PLANTS ARE COMPLEX ADAPTIVE SYSTEMS
The individual plant and its constituent cells are described as complex because they exhibit four characteristics: connection, interdependence, diversity and adaptation (Holland, 1995; Miller and Page, 2007). Emergence is a property of complexity (Anderson, 1972; Boogerd et al., 2005), exemplified by plant self-organization (Trewavas, 2014). A basic trade-off in a complex system pits exploration against exploitation. Exploration is the search for optimal solutions; exploitation uses the best-discovered solution. In any system exploration and exploitation should be in reasonable balance.
The plant and cell environment is learnt and is cognitive
Each seedling learns and remembers the variations in space and time in its environment. Those for light and gravity were known from Darwin’s time; intermittent signalling led to memories lasting hours. Optimal functioning can only be expected in wild environments in which the individual species has evolved. That constraint applies to all organisms.
Some coupling between environmental factors is common (e.g. light, daylength and temperature) reducing the perceived complexity, but this may be variable among species (Souza et al., 2015). The ability to predict environmental coupling should provide a fitness advantage. Even simple microbes can learn these relationships and anticipate future environmental change (Baliga, 2008; Tagkopoulos et al., 2008). The important conclusion from these two articles is bacterial cognition. The environment is internally mapped and the constructed controlling networks simulate the perceived environmental structure. Plant learning is likely to be similar.
Learning involves protein interactions conditional on ‘if/then’ interactions
Experience guides adaptable changes in an organism’s structure (molecular and physical) so that as time passes the organism makes better use of its environment for its own ends; it profits from experience (Holland, 1995; Miller and Page, 2007). If there is no adaptation or learning, the parts of the system follow simple rules and remain at equilibrium. Small amounts of learning or adaptation allow the system components to work out how to interact with one another to form a complex whole: adaptation generates self-organization (Trewavas, 2014; De la Fuente, 2015; Wegner and Lüttge, 2019).
Cells contain large numbers of proteins that act simultaneously as signals through interactions with each other. The familiar global properties of synthesis and degradation cascades, energy generation, secretion and electrical changes are thus constructed. These (often) non-linear interactions require tight coordination and use tags, sequence signals that enable accurate recognition of other individual proteins or complexes during interaction as signals in cycles or cascades. The actions of all proteins depend on the signals they receive although it is conditional: they have an ‘if/then’ structure. If the signal vector is present then the consequence of interaction proceeds (Holland, 1995; Miller and Page, 2007). Many times, it is likely that the sequence will abort because the next protein (the next step) is not present. Transduction chains (bit-strings) may be constrained to four steps; more steps increase the chances of error (Lestas et al., 2010). The consequence may also be complex: feedback initiation of cellular global consequence, such as initiation of division or change in growth direction.
Learning requires molecular construction, reconstruction and competition
Complexes of signal and developmental-processing proteins construct modules with defined functions, rules or programmes. Davidson (2010) has described the structure of a number of these modules and has illustrated the potential functions of each. There may well be thousands varying in size, and this size distribution is perhaps represented as a power law, varying from complex clusters of perhaps 50–100 or more different proteins as expected for the cell cycle, to those involving four or five proteins for more common metabolic requirements (Barabasi and Oltvait, 2004; Milo et al., 2004; Ma and Gao, 2012).
The number of proteins in Arabidopsis is more than the 24 000 or so recognized genes because of isoforms, post-translational modifications, genetic variants, sometimes extra sequence copies and splice variants (Gan et al., 2011; Reddy et al., 2013). These additional variants increase the numbers of potential modules available. There is already considerable sequence variation amongst different Arabidopsis accessions and substantial epigenomic modifications too (Kawakatsu et al., 2015; The 1001 Genomes Consortium, 2016). These data argue against uniform mechanisms of learning.
The modules described above act as building blocks (Holland, 1995). When new situations emerge or as development proceeds, various modules are combined together in different combinations providing a potential route through to: (1) either different adaptive responses, or (2) a changing environment, or (3) to the next stage in development or (4) even to unusual or novel situations. The only overall requirement for the precise mixtures of modules is that of downward causation; they must provide for cellular survival. However, numerous combinations will form and these can be regarded as varying in capability to satisfy the need for continued development and adaptation: the precise molecular routes between cells, and between individuals, will be different. There will be competition amongst these combinations and the better ones will win and the cell will remember and use them preferentially in the future (De la Fuente, 2015; Demongeot et al., 2019). Those that generate inadequate learning schedules will probably fail, a common end for many seedlings.
On this proposed mechanism the learnt molecular route is probably different in each individual
The situation is like an auction with various bids offered and the best succeed in providing the better functions or rules. The strength assigned to any combination(s) of modules will depend on its overall contribution to the complex adaptive system (Holland, 2006). This is trial and error learning, a learning method first described by Thorndike (1911). Adaptation changes with time and in the direction of improvement, because the spectrum of synthesized proteins changes in response to the memory of previous conditions and developmental age. Exploration is gradually replaced by exploitation.
Developmental modification (phenotypic plasticity) is progressively constructed or reconstructed (Oyama et al., 2001). However, this is unlikely to be straightforward plasticity but instead irregular and partial. The cell system or individual plant will learn and identify more productive combinations and in due course forward better fitness combinations.
While current views merely emphasize that genomic variation is the reason for individual seedling variation, the view propounded here places this variation instead as primarily learnt, coupled with the lottery of what networks are finally constructed and integrated with the specifics of environmental variation that each experiences. The developing plant cell is like a table in which decision-makers debate a question and respond collectively to the information put to them with answers that can vary to a degree with each cell or individual plant (Levy et al., 2010). The presence of plant somatic mosaics is one result (Watahiki and Trewavas, 2019).
There are a variety of leverage points
All adaptive systems have leverage points where a simple intervention has a lasting effect (Holland, 2006). In plants these may be hormones (which act to help synchronize the inevitable cell variability of a tissue to a common goal; Bradford and Trewavas, 1994) or any of the sRNAs, proteins, mRNA and ions that circulate throughout.
Sparse coding of sensory inputs
When individual plant tissues perceive particular local environments, do they construct an image of the distributions of light, mineral distribution, soil structure or selective regions of herbivore damage? The presence of somatic mosaics, in which individual cells in a single plant tissue differ in response to each other from the same signal, suggests the potential for pattern formation rather than a uniform image (Watahiki and Trewavas, 2019). The pattern formed could be learnt and remembered, improving the response to a subsequent signal. If different signals construct different patterns, those are likely to be learnt and remembered too. In addition, the tissue could construct an integrated response when the pattern is recognized.
Sparse coding relies on using minimal information to store and memorize such potential patterns (Willshaw et al., 1969). The benefit is a reduction of interference or competition between a number of experienced different patterns, making memory formation easier and more reliable (Olshausen and Field, 2004)
One potential example might be in root cap cells in Arabidopsis. The root cap is a developing tissue of only several hundred cells in which cell replacement is continuous as outer cells are sloughed off. The gravity-sensing capability of the individual cell, or very small clusters of root cap cells, varies substantially, thus indicating it is a somatic mosaic (Blancaflor et al., 1998). The gravity signal constructs a pattern that is then interpreted holistically. The cap is also sensitive to at least eight different signals when imposed singly (Trewavas, 2017). Each of these can be expected to construct a different pattern of sensing and sparse coding eases the specific pattern memories required. Because numerous signals are present at any one time, sparse coding helps to resolve the issue of which is the most critical to respond to. When a root impacts a stone in the soil, a kind of dog-leg structure of the root is formed and maintained despite continued cap cell formation. Touch and gravity produce the immediate patterns that are constructed and remembered, enabling the root to slide over the obstruction (Massa and Gilroy, 2009).
Leaf epidermal ocelli can focus light onto the epidermal basal membrane that can sense changing light patterns and thus act as a sensory epithelium (Haberlandt, 1914, p. 626; Baluška and Mancuso 2016; Gianoli 2016). Leaves are somatic mosaics in their cellular response to red light and potentially other signals (Watahiki and Trewavas, 2019). Different patterns of light impact can be constructed and the memory used to sensitively modify petiole or pulvinus movement.
REAL-WORLD CIRCUMSTANCES, INTELLIGENCE AND INDIVIDUALITY IN EVOLUTION
When chimpanzees were taught sign language, they acquired some 300 words over two years and the trainers involved rated them as equivalent to 2-year-old children (Gardner and Gardner, 1969). However, when placed in a new troop, an individual animal knows in a few seconds its place in the pecking order. How long would a tribe of 2-year-old children last in the wild? Intelligence of any organism has to be judged in the framework of the environment in which it evolved. Chimpanzees did not evolve in a human-constructed environment but in real-world circumstances in jungle conditions. They recognize by learning all the necessary and critical factors in their environment and do not need sign language to appreciate them; but recognition of pecking order is essential to individual survival. Learning sign language in a laboratory is not.
The requirement for real-world circumstances for expression of plant intelligence
Ecological developmental biology (eco-devo) represents the meeting of development with the real world (Gilbert, 2001; Sultan, 2015). Scott F. Gilbert (2001), who introduced the term ‘eco-devo’, emphasized the necessity of using the ‘real world’ in studying development. Many environmental factors experienced together interact with each other and modify animal embryological development; as they do in the developing plant phenotype. The system structure of any individual wild plant is a composite of the individual organism together with the complexity of its external environment, which is rarely still. The known number of individual environmental signals to which plants have been reported to sense and which elicit phenotypic change is over 60 (Trewavas, 2014, p. 70). And some of these, such as water, wind and light, can vary on a minute-to-minute or even second-to-second basis.
Predation and disease represent other uncertain and variable contingencies
Providing the environment does not kill the organism prematurely (common enough in seed germination), there will be a compatibility or congruence that forms a unity between the structure of its environment and that of the individual. As long as this compatibility exists, the environment and individual act as mutual sources of perturbation, changing the internal state of the individual in a form of structural coupling (Varela, 1979; Maturana and Varela, 1980, 1987).
The consequence of seeing plants only in laboratory conditions can encourage the view that plants have an overall plan which is simply fulfilled during germination, growth and flowering. Plans are of poor value in an unpredictable real-world circumstance. Given the quantitative and qualitative environmental variations that occur naturally, real plant environments must be in the many thousands. In contrast, laboratory and growth room circumstances are few or can be regarded as just one.
Real-world circumstances are the environments in which plants have evolved and the circumstances in which intelligence and adaptability will optimally contribute to survival. It is not genes (with a few exceptions), nor particular phenotypes found in certain environments, but instead the capacity to deal with environmental challenge, uncertainty and change during the life cycle that is the major focus of selection (McNamara and Houston, 1996; Schlichting and Pigliucci, 1998).
Investigating the ‘real world’ environmental effects using Arabidopsis
Schilchting and Pigliucci (1998) cautioned against using real-world circumstances for investigation. Their concern was that there may be difficulty in reproducibility. However, a pioneering paper by Richards et al. (2012) accumulated direct information on gene expression throughout a ‘real-world’ Arabidopsis life cycle. Features of environmental change to specific genetic modifications were recorded. We regard this important approach as productive and surely an important future direction. Darwin’s (1880) hugely productive output on plant behaviour merely used his weakly (or un-)controlled front room. And his two important books on variation (Darwin, 1868) record information accumulated without any controlled conditions.
The largely ignored importance of individuality
Darwin considered that natural selection operated at the level of the individual. Yet the individual plant does not receive the attention in plant research that perhaps it ought (Watahiki and Trewavas, 2019). The presentation of published data is commonly expressed as averages or means with some statistical estimate of variation. However, the average does not exist (Williams, 1956; Weiss, 1973). That has substantial consequences for mechanisms that are commonly deduced from the average and unfortunately simplify what may be considerably more complex.
In real-world circumstances, fierce competition can threaten survival and requires internal assessment
The common phenotypic changes in leaf, stem and root plasticity induced by various signals suggest that a fierce fight for resources is a very common and expected experience. The behaviour of any individual is probably dependent on the behaviour of those other plants that surround it (good examples of these are illustrated in Bazzaz,1996, pp. 112–114). Perceiving the potential identity of competitive neighbours and thus responding beneficially to them comes from a variety of information: the changing intensity, quality and direction of light, direction and concentrations of volatiles, root secreted chemicals, direct touch and information through mycorrhizal networks (Novoplansky, 2009; Trewavas, 2016b). Several different kinds of competitive response are recognized, according to the information gained: avoidance, confrontation and tolerance (Novoplansky, 2009). The information gained needs intelligent assessment. Self-competition in both shoot and root must be minimized. In addition, the expenditure (costs) involved in plasticity needs to be minimized and benefit maximized. Without an internal assessment that indicates the optimal future changes, it is difficult to see how this response to competition can be accomplished.
Niche construction requires cognition and intentionality
Both shoots and roots modify their local environment. Modification requires the continual input of a changing adaptability as development progresses (Trewavas, 2009). Cognitive mapping is commonly connected to organisms such as plants or microbes that modify their environment to a particular end (Turner, 2018). Intention describes the evident drive in development towards a future goal and is present in any individual plant.
The goal in niche construction is the production of an equable soil environment for root growth, root function and soil exploitation. In plant roots growing in soil, intentional actions are the variable secretions of: (1) enzymes, organic and inorganic acids to mobilize phosphate; (2) mucilage to improve soil structure and lateral root penetration; (3) strigolactone to attract mycorrhizal symbionts – a whole hyphal network can convey information on disease and herbivory in adjacent plants, provide additional phosphate and iron and improve resistance to disease; (4) a variety of other organic chemicals, to attract microbes that live both inside and outside the root and improve disease resistance; and (5) the easier detection of competitive neighbours and gradients of water and N (Kloepper et al., 2004; Gorzelak et al., 2015; Santhanam et al., 2015; Song et al., 2015; Novoplansky, 2019).
Each of these events is controlled through a changing conversation with the external soil circumstance and other organisms, which indicates intention (Trewavas, 2009). When roots proliferate abundantly as a result of competition, the intention is to occupy soil space, deny soil resources to competitors and to act territorially, plausibly another cognitive capability (Robinson, 1996; Schenk et al., 1999; Trewavas, 2014).
Natural selection relies on individual adaptability
Organic selection was first clearly identified by discussions between Baldwin, Osborn and Lloyd Morgan (Baldwin, 1896; Osborn, 1897). The clearest statement of this mechanism was provided by Osborn (1897, p. 946): ‘Ontogenetic adaptation (phenotypic plasticity, intelligent behaviour) is of a very profound character, it enables animals and plants to survive very critical changes in their environment. Thus all individuals of a race are similarly modified over such long periods of time that very gradually congenital variations, which happen to coincide with the ontogenetic adaptive modifications, are collected and become phylogenic. Thus, there would result an apparent but not real transmission of acquired characters.’ Baldwin recognizes these ontogenetic adaptations as critical in plants: ‘these adaptations are seen in a remarkable way in plants, in unicellular organisms and in very young children’. ‘There seems to be a readiness and capacity to rise to the occasion as it were and make gain out of the circumstances of its life’ (p. 443). ‘The most plastic individuals will be preserved to do the advantageous things for which their variations show them to be the most fit’. ‘The future development of each stage of a species development, must be in the direction thus ratified by intelligence’ (Baldwin, 1896, pp. 447–448).
Organic selection was designed to explain how some organisms seem so well adapted to their environment (birds and wings for example; Corning, 2003). The process was omitted by the so-called modern synthesis of evolution that developed in the 1930s, based as it was only on mutations and strict genetic heritability. Mayr (2001) regarded the first step in selection as a change in behavior, thus placing adaptability at its forefront.
Organic selection is a distinct form of individual selection that speeds up evolution and has been regarded as modificatory steering. After environmental shifts, plants with greater plasticity adapt more quickly, and may arise with higher probability or with lower cost (Bateson, 1963). Unless there were modifications in all aspects of the phenotype, selection of one phenotypic character might become limited by others that do not respond in the same way. Organic selection using adaptability can clarify the evolutionary trade-offs in natural selection between exploration and exploitation.
CROP ADAPTABILITY INDICATES WHY INTELLIGENT PLANT BEHAVIOURS ARE FRAMED IN TERMS OF FITNESS
The future need to increase crop yield is well understood and has been the target of research for centuries. Adaptable behaviours have been recognized in many such species (rice, corn, wheat, barley and, of course, Arabiodopsis), but early studies were framed in terms of seed yield comparisons amongst many varieties of the same crop or plant grown in different agricultural conditions (Finlay and Wilkinson, 1963; Matsuo, 1975). The aim was of course to locate the varieties and environmental conditions that by interaction or synergism might provide for maximum seed crop production; the interaction is commonly summarized as genotype × environment (G×E). A farm is a complex, integrated system, with interactions through many of its parts and the overall network structure determined by the characteristics of the farmer him/herself (Trewavas, 2004b).
Maximum seed yield is also a proxy for fitness in wild plants, but there are two primary differences between farm and wildness. First, while many crops are derivatives of wild plants, millennia of selection, breeding and use of mutants has effectively severed their connection. The morphology is often substantially different. Much of this difference has been originally at the level of development and thus probably reflects in turn genomic manipulation. Second, the cultivated field is an environment. Even Lamarck (1809, translated 1914) recognized the role of cultivation itself: ‘All botanists know that plants translated from their natal spot (the wild) into gardens gradually undergo changes which in the end make them unrecognisable’ (1914, p. 215). Is this epigenetic change?
Present crop species do not seem to survive in wild conditions. In the 19th century, Broadbalk experiments (Rothamsted) indicated that wheat disappeared within 2 years in a fallow field and corn is similar (Beadle, 1980). Crops retain some features of adaptability but others have been eliminated. Crop plants are then chimeric constructs in intelligence terms: part human, part plant.
Detailed analysis of corn has led to the conclusion that the genomic regions used for adaptation of corn to North America have limited its ability to adapt to different natural environments (Gage et al., 2017). Breeding has emphasized stability rather than plasticity in many different characteristics, although sufficient phenotypic plasticity may remain to improve corn yields (Kusmec et al., 2018). Corn does respond to abiotic stresses and different cultivars exhibit differential gene expression (Waters et al., 2017). There may also be a cost to plasticity, and it may be disfavoured under other circumstances, so its elimination may increase yield, although the literature is still uncertain on this issue (Schlichting and Pigliucci, 1998; Sultan and Spencer, 2002; Auld et al., 2010; Palacio-Lopez et al., 2015). Abiotic stress of different varieties of corn indicates substantial variation in cis and regulatory features (Waters et al., 2017). Such research emphasizes the need for clarity on the genomic nature of adaptability.
However, it is the familiarity of plant scientists with the manipulation of plants to produce crop species that undermines an ability to see intelligence in operation. There is no goal of fitness, merely farming yield. It is a problem that besets all domesticated organisms coupled with poor awareness of wild behaviour. When wild plants that are placed in shade generate leaves that have a larger surface area, or when a climbing plant offered a poor support (glass rod) unwinds and searches for a better one, or when Simmondsia turns its leaves at midday in a vertical direction to that of the sun, the words ‘smart’, ‘clever’ and ‘intelligent’ come obviously to mind (Trewavas, 2014; Sultan, 2015). No matter what adaptability remains in a crop it will be under our control, our intelligence, not that of the plant independently. That is the critical distinction.
IS THERE AN EQUIVALENT IN PLANT ADAPTABILITY TO GENERAL HUMAN INTELLIGENCE, G?
General human intelligence or g
General intelligence (g) was first introduced by Spearman in 1904. His contention was that people with good verbal comprehension, or processing speed, for example, tended also to have good working memory and perceptual organization and reasoning. Over 400 papers have clearly established the issue; correlation factors are between 0.6 and 0.8 (Deary, 2001). Detterman (1982, 1986) identified general human intelligence, ‘g’, as part of an intelligence hierarchical system, now indicated in textbooks on human intelligence (Cianciolo and Sternberg, 2004; Sternberg, 2006). There is a strong heritable character to g based on studies of separate identical twins. This suggests there could be an underlying discrete but similar mechanism of adaptability that is coupled to human IQ characteristics. Is there an equivalent in plant adaptability or plasticity?
A specific basis for plant adaptability?
The Holy Grail for crop breeding would be to identify a simple set of genes that change a non-adaptable plant into an adaptable one. Some evidence suggests that this may be possible. Adaptation circumstances commonly modify many phenotypic characteristics in concert (Schlichting, 1986, 1989, Schlichting and Levin, 1990; see also diagram in West-Eberhard, 2002, p. 297). Plasticity (adaptability, intelligence) exhibits some heritable characteristics (Jain, 1978; Jain and Martins, 1979; Schlichting, 1986, 1989; Schlichting and Pigliucci, 1998; Grenier et al., 2016). Distinct generalist and specialist congeners of Polygonum species clearly differ in adaptability to water availability (Sultan et al., 2009). Seed from Nicotiana rustica, isolated from individuals experiencing good environments, exhibited greater plasticity in height than those in poor environments (Jinks and Pooni, 1982). In yeast, a set of 900 genes responds similarly to a diverse array of environmental stresses and share common regulatory themes (Gasch et al., 2000; Causton et al., 2001; Brooks et al., 2011). A plant stress gene database has been constructed (Borkotoky et al., 2013). However, there are molecular differences between stressful and milder circumstances in water deprivation and probably other stresses (Baerenfaller et al., 2012; Fleta-Soriano and Munne-Bosch, 2016).
One common event following numerous, but different, environmental signals and stressful conditions is rapid cytosolic Ca2+ transients (Trewavas, 2011). Microtubules and microfilaments connect the outer membrane with structural and molecular aspects of the cytoplasm (Kolling et al., 2019). Such Ca2+ transients may simply disaggregate the present microfilaments and microtubules (effectively wiping the cell slate clean) so cells can accommodate to the new environmental situation now experienced. The specific events known to follow these transients then arise from activation of Ca2+- and Ca2+-calmodulin-activated kinases, CDPKs, others that act in downstream interpretation and enable the construction of a new cellular network.
Evidence that adaptability is instead a complicated molecular and genetic process
The real difficulty in trying to disentangle the nature of adaptability comes from trying to assess what measure of adaptability can actually be used for genetic investigations. This difficulty is clearly outlined by Laitinen and Nikoloski (2019), who also state that the genetic basis of adaptability is not understood. Box 2 indicates briefly some of the known information on the genetic basis of adaptability.
BOX 2. MOLECULAR INVESTIGATIONS OF QUANTITATIVE ADAPTABILITY.
Two important points were established about systems behaviour from investigations of control theory of metabolic systems in vivo (Flint et al., 1981; Fell, 1997). While most control of any metabolic sequence was in the sequence itself, substantial amounts of control were found outside through connections with the greater metabolic network. To substantially increase the flux of material through any pathway required increasing the amounts of all pathway enzymes.
Genetic investigations of abiotic stresses indicate numerous contributing quantitative trait loci (QTLs), many of which have only marginal effects (Des Marais et al., 2013). From a network perspective these marginal QTLs could be secondary connections of the primary interpretative network to other less critical processes or changes. Expression QTLs (eQTLs) and transcriptomics are more informative. Very large numbers of genes have their expression increased under drought stress for example, but to very different extents compared to water deficit adaptation (Baerenfaller et al., 2012; Rasheed et al., 2016; Rymaszewski et al., 2017). Water deprivation modifies root anatomy and morphology and in rice 76 loci and 233 candidate genes were predicted to be responsible (Kadam et al., 2017).
Although many transcripts appear to be controlled by locally present cis-eQTLs, as much as 70 % of eQTLs in maize, rice and Brassica rapa are trans-acting loci clustered into genomic hotspots and influencing many thousands of genes (Hansen et al., 2008; Des Marais et al., 2013). QTLs are associated in some animal cells with chromatin modification, transcription factor binding, histone modification, gene expression and DNA methylation (Banovich et al., 2014). One potential for some plant trans-eQTLs is that they act like ‘enhancers’ which activate genes and promoter regions at least a megabase away (Weber et al., 2016). One additional way to increase flux through pathways is using feedforward mechanisms implicit in much protein phosphorylation. With over 1000 protein kinases and one-third of proteins phosphorylated, there is certainly potential for investigation, but this is technologically constrained by the difficulties of identifying which substrates are phosphorylated by which kinases (Sopko and Andrews, 2008; Cheng et al., 2014; Bhaskara et al., 2017). Yeast provides a model of what can clearly be achieved using protein chips, or mass spectrometry, to construct the dynamic phosphorylome (Cheng et al., 2015).
There is a benefit here to describing plant adaptability as an aspect of intelligence because there is a commonality of problem with human intelligence too. It is common to segregate any trait as being due to genotype [H], environment [E] and any interaction as [H] × [E] measured by ANOVA (Laitenen and Nikoloski, 2019). If [H] × [E] is not statistically significant then the effects of H and E are simply additive. In that case a single group of genes [H] ‘could be’ responsible for all forms of adaptability. Wahlstein (1990) demonstrates the fallacy of this approach, because ANOVA is in many cases insufficiently sensitive to demonstrate a lack of interaction. This difficulty first appeared in human intelligence studies with unwarranted emphasis on a supposed lack of any environmental interaction and thus on genetic variance only. However, a norm of reaction for human intelligence has been illustrated covering the known range from IQ measurements (Platt and Sanislow, 1988).
When plants are considered as complex adaptive systems (as described earlier), the learning process results from interactions between environmental signals and the molecular pathways that interpret them. Later environmental changes or the experience of novel conditions requiring adaptive modification are again learnt. The memory, generated as a result of signals, can be understood as having an epigenetic basis via DNA or mRNA modification (Covelo-Molares et al., 2018; Ginsburg and Jablonka, 2010). Thus, the genetic input in adaptability is, to an extent, environment-specific. In animals, intelligent responses to new situations depend on previous rearing conditions (Wahlstein, 1990) and have also been demonstrated clearly for plant growing conditions (Turkington, 1983).
An alternative approach seeks to understand the nature of the cellular networks that underpin adaptability and other developmental issues. These networks are formed from the interactions between the thousands of cellular proteins present at any one developmental stage. Network changes are fundamental to understanding the molecular nature of adaptability. Box 3 summarizes some of the current information. In Arabidopsis, post-translational modifications of various kinds and other misinterpretations probably more than double the proteins expressed from 24 000 identified genes (Watahiki and Trewavas, 2019). Describing the complete, plant cellular network will be challenging. However, given the progress in yeast and bacteria this could be accomplished if large numbers of laboratories are prepared to collaborate.
BOX 3: NETWORK ANALYSIS NECESSARY TO INVESTIGATE ADAPTABILITY.
The current activity using high-throughput methodology has accelerated the development of databases that store information on how genes, proteins and metabolites interact with each other. The interactions construct networks and the biological networks have similar structures (proteins, genes, mRNA, transcription factors etc.) that interact to form nodes connected to other proteins or agents by edges and thus an interactome. Such interactions generate emergent properties (Johnson, 2001; Boogerd et al., 2005). Nodes or hubs are recognized as having a large number of connecting edges (commonly called degrees) with the distribution of connections recognized as a power law (Barabasi and Oltvait, 2004; Milo et al., 2004; Zhu et al., 2007; Ma and Gao, 2012). Power law relations indicate a scale-free structure (Bak and Snedden, 1993; Bak, 1999; He, 2014). A variety of methods are available that enable visualization of the network (Baryshnikova, 2016). Nodes of high degree are considered as probably being essential, in that their loss by mutation is generally lethal or extremely damaging (Zotenko et al., 2008; Jalili et al., 2016), although Ahmed et al. (2018) provide alternative information and detailed analysis of a plant cell surface interactome.
A variety of methods have evolved to analyse an interactome network and these include betweeness (the number of shortest paths between two nodes, Zhu et al., 2009), influence of any particular node using weighted k-shell decomposition (Pei et al., 2014; Wei et al., 2015) and bottlenecks (i.e. network nodes that have many shortest paths going through them). These are considered analogous to bridges or tunnels and are very likely central and essential (Yu et al., 2007).
The connection strength between nodes can change, allowing plastic adaptable behaviour (Zhu et al., 2007). While more connected networks (higher connectance) support fine-tuning regulation, fewer tight links (lower connectance) improve flexibility. However, both strategies are interchangeable, increasing the arsenal of phenotypic plasticity that enables plant stability (Bertolli et al., 2013). Lower connectance is common under equable growth conditions but increases under stress, enabling faster responses, the so-called ‘stability-complexity hypothesis’ (Souza et al., 2005). However, there are alternatives via different pathways of connections of edges and knots in networks. There is no one-sidedness of either positive or negative interactions and in the effects on stability. It is a basic feature of the organization of networks that they always comprise positive and negative feedbacks (Souza and Lüttge, 2015). Connectance plasticity may be species-specific (Souza et al., 2009; Souza and Lüttge, 2015). Phosphorylation is one way in which connection strength can be altered in cellular networks as required by adaptability. Future approaches are needed to assess weighted values to connection strength in cellular networks.
Synaptic plasticity (connection strength) in neural systems is thought to be altered by the frequency of action potential movement (Hebb, 1949). Initiation of mechanically induced Ca2+ transients in plant cells also increases the synthesis of proteins (Ca2+-dependent kinases, calmodulin and others) that in turn increase information flow through this Ca2+-initiated pathway (Trewavas, 1999). This is a Hebbian type of control although slower than used in nervous systems.
Ma et al. (2009) have defined network topologies that enable adaptation using the two requirements of sensitivity and precision. Their analysis indicates minimal requirements involve feedback and feedforward circuitry and with modification provide for robust adaptation. Such a structure exists for perhaps the simplest of intelligence – chemotaxis by Escherichia coli, in toxin or nutrient gradients (Hoffer et al., 2001). Network resilience (the maintenance of function despite errors, failures and environmental challenges) depends on network density, heterogeneity and symmetry (Gao et al. 2016). The relationship to adaptability is obvious.
THE CRITICAL TRAITS THAT CONSTRUCT ADAPTIBLE INTELLIGENT BEHAVIOUR
Adaptability, despite seemingly a simple term, is one of considerable complexity. Numerous forms of animal behaviour are assumed by some to require a neural investment; and yet, even animals, with a central nervous system (CNS), engage in behaviours and make decisions without the CNS (e.g. immune systems). Furthermore, single-celled organisms and bacteria (summarized earlier in this article) clearly behave in a variety of ways. Eisenstein (1975) argues that interpolation of neural cells in multicellular animals elaborates but does not fundamentally change the basic behaviours (learning, memory, etc.) already exhibited by single-celled organisms. While increasing numbers of connected nerve cells could generate emergent properties, even three of four neurons connected together exhibit memory, error correction, time sequence retention and a capacity for solving optimization problems (Hopfield, 1982). Plants do use forms of electrical connection (Calvo et al., 2017) and these abilities can be seen as derived from unicellular ancestors along with learning and memory that are present in motile single plant cells.
Systems biology is a productive approach to unpick adaptability
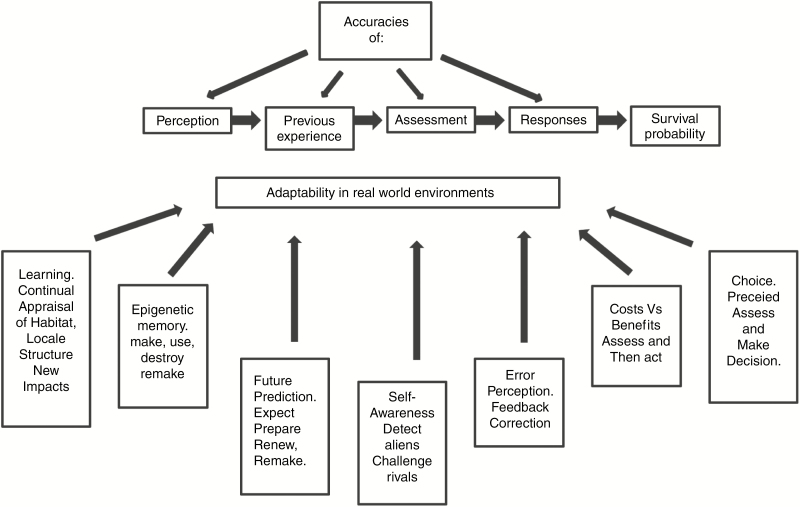
The traits described below (Fig. 3) we regard as essential constituents of plant intelligence and also adaptability. Understanding their molecular basis can be investigated using the wealth of technologies in present-day systems biology. High-throughput methods of genetic analysis, transcriptional and translational change, DNA, chromatin and mRNA modifications, interactome and phosphorylome analyses converge on providing the essential information (Gutierrez et al., 2005; Joyard and McCormick, 2010; Boogerd et al., 2013; Sheth and Thaker, 2014).
Fig. 3.
Potential behavioural traits that can influence individual adaptability in real world conditions. This figure summarizes some of the behavioural traits indicated in the section ‘The critical traits that construct adaptible intelligent behaviour’.
Learning and memory as part of adaptability
Habituation
Habituation is the response decrement to an intermittent but repetitive stimulus. It is a form of ubiquitous learning, present in single cells, mammals and plants (Eisenstein et al., 1980; Gagliano et al., 2014, 2018). Habituation as learning can be recognized by using a similar but non-identical stimulus whose response remains unchanged during the habituation process. Habituation was convincingly identified in the drop response of Mimosa (Gagliano et al., 2014). Eisenstein et al. (1980) proposed that in single aneural cells, habituation resulted from a progressive reduction in the size of cytosolic Ca2+ transients. Repetitive wind stimuli applied to tobacco seedlings led to progressive reduction in cytosolic Ca2+ transients whilst cold shock-induced Ca2+ transients remained unaffected (Knight et al., 1992). This is also an example of habituation that supports the proposed mechanism of Eisenstein et al. (1980). The adaptive function of habituation provides for a rapid maximization of the organism’s overall readiness to cope with novel stimuli and to minimize unnecessary costs: the so-called ‘behavioural homeostasis theory’ (Turner, 2007; Eisenstein et al., 2012). All forms of habituation require the frequency and number of stimuli to be remembered and are (for the most part) adaptive responses.
Direct experimental evidence of associative learning has been published (Gagliano et al., 2016). A neutral cue was used to demonstrate its potential during foraging for resources and is the first example of an important plant learning capability. Learning about increasing nutrient supply (trajectory sensitivity) can lead to anticipation and enhanced root branching (Shemesh et al., 2010). Systems analysis should help with appreciation of its molecular basis.
Sensitization
Sensitization occurs when overall responsiveness is increased by the subsequent signal (Eisenstein et al., 2012). Tendril curling is sensitized when tendrils are touched in darkness and then briefly exposed to blue light (Jaffe and Shotwell, 2006). The first few light exposures increase curling rates before a decline. Abiotic and herbivore signals also can be placed in this class of behaviour. Brief treatments lead to an enhanced response on a second or third stimulation (Bruce et al., 2007; Frost et al., 2008).
Memory, an important adaptive response that has to be learnt
The presence of a memory of a previous stimulus is recognized because subsequent stimuli of the same kind now exhibit an altered molecular or phenotypic response. There are numerous examples of memory in plants (Trewavas, 2009). Herbivory can be regarded as a predator–prey relationship, and plant memory here (priming) was first identified in predation (Baldwin and Schmelz, 1996; Ruuhola et al., 2007). Plants that have been attacked previously respond more quickly and to a greater extent (Frost et al., 2008). Disease attacks and abiotic stresses lead to the establishment of a memory of the challenge that is used in defence from subsequent episodes (e.g. Bruce et al., 2007; Fleta-Seriano and Munne-Bosch, 2016). Some memories such as in Dionea are clearly electrical and short lived. Others involve longer term changes in gene expression and longer again in chromatin structural alterations that can last for years (Probst and Scheid, 2015). There are also well-established reports of the effects of moderate changes in temperature, soil mineral changes and effects of various chemicals or physical treatments that can last for 5–12 generations (e.g. Cullis, 2005; Highkin, 1958; Hill, 1965; Moss and Mullett, 1982). These events are probably epigenetic in origin, as described for memory in non-neural cells (Ginsburg and Jablonka, 2009).
Similar variation in memory lengths to the above exist in the human brain. Short-term memory in the brain depends on glutamate-sensitive Ca2+ channels. Such channels have been identified in plant cells and contribute to long-distance communication as well as herbivory resistance (Toyota et al., 2018). Long-term neural memory requires protein synthesis and also involves epigenetic chromatin changes in nerve cells (Jarome and Lubin, 2014).
Error perception and correction in adaptability
In real-world circumstances, errors in development or in signal response are likely to be common. Error diagnosis and correction is found in single-celled organisms (Clark, 2010, 2013). Error perception and correction involves mechano-sensitive Ca2+ channels acting in Hebbian-like mode (Clark, 2010, 2013). Such mechano-sensitive channels are present in plants (Basu and Haswell, 2017)
Visible error correction was reported by Darwin (1875) and von Sachs (1887). In climbing plants provided with an unsuitable support such as a glass rod, curling starts, stops, unwinds and then searches elsewhere, an indication of adaptability and obvious intelligence. If an etiolated plant is exposed to a brief flash of light, it will start to phototropically bend and then straighten after 1–2 h. Presumably such plants use checkpoints for curling, bending, etc. Systems biology might be able to identify them.
Speed versus accuracy
If changes in environmental conditions are fast, then this increases the likelihood of adaptive errors in response. Furthermore, new learning will be necessary unless the condition has been previously experienced. Comparisons of abiotic stress of sudden change, for example, to water depletion or heat shock temperature (e.g. conditions requiring massive chaperonin synthesis), compared to slower manipulation to the same level, will reveal probable errors and also errors in adaptability.
Decisions and discrimination between choices
Decisions are an essential part of growth and development and their adaptive, intelligent modifications. Its costs and benefits, the potential trade-offs involved and an assessment of present internal circumstances would seem essential along with assessment of any that are anticipated in the near future. The internal circumstances will be encoded in the networks both inside and outside cells within tissues and the whole plant; anticipated futures probably result from past experience, which is remembered and expectation of repetition. Decisions do require crossing a threshold in development, and in somatic mosaic tissues some form of quorum sensing seems appropriate (Trewavas, 2012). Feedforward mechanisms using bi-stable switches are likely to be involved so that the size of change necessary to cross the threshold will be very small. Systems analysis is needed to describe both the threshold and how it is exceeded. When provided with several different environments for growth, plants choose the environment which best increases growth rates (Trewavas, 2014). How these decisions are made requires an examination of learning pathways, memories and future assessment.
Costs and benefits
Costs are again probably estimated from prior experiences and memory of them as do benefits. They form a crucial part of adaptive assessments and responses. Herbivore resistance (or resource limitation) provides good examples of costs and benefits (Bloom et al., 1985; Cipollini et al., 2014; Zust and Agrawal, 2017). The primary costs of herbivory come from the use of basic resources to synthesize numerous natural pesticides, which thus reduce those for growth and then life cycle completion, the ultimate driver. A suggested very simple interaction structure for resistance has been indicated but needs more details on molecular interactions and network structures (Zust and Agrawal, 2017).
Resource limitations of fixed carbon (C) can increase shoot growth whilst N and P deficiency increases root growth. The cost is overall growth reduction and the benefit of adaptability is to best retain the target of the life cycle but with a likely reduction in stored resources for seed production. Bloom et al. (1985) creatively used numerous economic terms to describe resource limitation. Using this analogy now requires these economic terms to be replaced by molecular network formations described by Davidson (2010). Economies are also complex adaptive structures and a further approach is to use the ECHO model generated by Holland (1996) based on adaptive behaviour. This model is also applicable to herbivory.
Self-awareness and self-recognition
Numerous investigations have indicated that plants are directly aware of neighbours and take competitive, adaptive action (Schenk et al., 1999; Falik et al., 2003, 2006, 2011; Gruntmann and Novoplansky, 2004; Herben and Novoplansky, 2008; Gagliano et al., 2012; Gagliano and Renton, 2013; Novoplansky, 2019). One good possibility for root recognition is secretion of peptides, which can be detected by cell-surface-resident, receptor-like kinases that respond to peptides (Ma et al., 2016). Kin recognition and separated vegetative clones could well operate in a similar manner. Secreted peptide sequences could be intentionally changed with time, thus destroying original kin or clone recognition and leading to recognition as aliens (Gruntmann and Novoplansky, 2004; Dudley and File, 2007; Pennisi, 2019). Recognizing aliens implies recognition of self too (although for other options, see Novoplansky, 2009).
Anticipation of future environmental change
Anticipation of future environmental change is known to occur and improves adaptability (Aphalo and Ballare, 1995; Gagliano et al., 2016; Novoplansky, 2016; Calvo and Friston, 2017). Long-term memory of environmental change with network structures remaining from previous encounters is one possibility. Certainly, the presence of a memory of any event will alter assumptions of future environmental change. Environmental influences experienced by the mother plant can survive into siblings (Trewavas, 2014).
Cognition and consciousness
Cognition is sometimes confused with consciousness, which leads to its rejection as being present in plants (Segundo-Ortin and Calvo, 2019). Cognition results from detection of environmental variables and enables a mapping process to indicate what is present and in many cases where (Calvo, 2007; Gagliano, 2015). This has already been mentioned several times in this article. With regard to consciousness, ‘Not just animals are conscious but every organic being every autopoietic cell is conscious. In the simplest sense, consciousness is an awareness of the outside world’ (Margulis and Sagan, 1995, p.122). On this basis plants have at least a simple consciousness (Calvo, 2017, 2018; Calvo et al., 2017; Gagliano, 2017).
CONCLUSION
This article has highlighted adaptability as a critical property that incorporates learning and memory, and is involved in evolution and individual survival of wild plants. Intriguingly, exploration and exploitation (competition and selection) are involved in all these processes. The ability to sense the extent of the environment for any individual wild plant is, along with its genomic, learnt and memory structures, a critical issue for fitness. The complexity involved is substantial and we consider that framing this capability as intelligent rightly ascribes the recognition of adaptability as a critical element to the life cycle of any individual. ‘The phenomena of irritability both in the vegetable and animal kingdom must in the main be purposeful. All those adaptations in the organism are purposeful which contribute to its maintenance and insure its existence’ (von Sachs, 1882, p. 601). The goal of understanding plant intelligence is very unlikely ever to be achieved by looking at one environmental feature at a time in laboratory conditions as suggested by Chamowitz (2018). It will be necessary instead to use real-world conditions, examining multiple interactions and with experiments spread over time.
FUNDING
P.C. is supported by the Office of Naval Research Global (Award No. N62909-19-1-2015). M.G. is supported by the Templeton World Charity Foundation (TWCF) under the Diverse Intelligences Initiative. G.M.S. is supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq grant 302715/2018-5).
ACKNOWLEDGEMENTS
We are very grateful to Professor Ariel Novoplansky for a critical reading of the manuscript and numerous suggestions for improvement. He is not responsible for the views expressed within. The authors declare no conflicts of interest.
LITERATURE CITED
- Adler J, Tao W-W. 1974. Decision making in bacteria: chemotactic response of Escherichia coli to conflicting stimuli. Science 184: 1292–1294. [DOI] [PubMed] [Google Scholar]
- Ahmed H, Howton TC, Sun Y, Weinberger N, Belkhadir Y, Mukhtar MS. 2018. Network biology discovers pathogen contact points in host protein–protein interactomes. Nature Communications 19: 23121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmann J. 1999. Evolving brains. Scientific American Library. [Google Scholar]
- Alpi A, Amrhein N, Bert A, et al. 2007. Plant neurobiology: no brain, no gain. Trends in Plant Science 12: 135–136. [DOI] [PubMed] [Google Scholar]
- Anderson PW. 1972. More is different. Science 177: 393–396. [DOI] [PubMed] [Google Scholar]
- Aphalo PJ, Ballare CL. 1995. On the importance of information acquiring systems in plant–plant interactions. Functional Ecology 9: 5–14. [Google Scholar]
- Armitage JI, Holland IB, Jenai U, Kenny B. 2005. “Neural networks” in bacteria: making connections. Journal of Bacteriology 187: 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armus HL, Montgomery AR, Jellison JL. 2006. Discrimination learning in Paramecia. Psychological Record 56: 489–498. [DOI] [PubMed] [Google Scholar]
- Auld JR, Agrawal AA, Relyea RA. 2010. Re-evaluating the costs and limits of adaptive phenotypic plasticity. Proceedings of the Royal Society London Series B 277: 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Massonnet C, Walsh S, et al. 2012. Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Molecular Systems Biology 8: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak P. 1999. How nature works. The science of self-organised criticality. New York: Springer. [Google Scholar]
- Bak P, Snedden K. 1993. Punctuated equilibrium and criticality in a simple model of evolution. Physical Review Letters 71: 4083–4086. [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Schmelz EA. 1996. Immunological “memory” in the induced accumulation of nicotine in wild tobacco. Ecology 77: 236–246. [Google Scholar]
- Baldwin JM. 1896. A new factor in evolution. American Naturalist 30: 441–451; 536–553. [Google Scholar]
- Baliga NS. 2008. The scale of prediction. Science 320: 1297–1299. [DOI] [PubMed] [Google Scholar]
- Baluška F, Mancuso S. 2016. Vision in plants via plant-specific ocelli? Trends in Plant Science 21: 727–730. [DOI] [PubMed] [Google Scholar]
- Banovich NE, Lan X, McVicker G, et al. 2014. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications and gene expression levels. PLoS Genetics 10: e1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi A-L, Oltvai ZN. 2004. Network biology: understanding the cells functional organisation. Nature Reviews Genetics 5: 101–115. [DOI] [PubMed] [Google Scholar]
- Baryshnikova A. 2016. Exploratory analysis of biological networks through visualization, clustering, and functional annotation in cytoscape. Cold Spring Harbor Protocols 2016: pdb.prot077644. [DOI] [PubMed] [Google Scholar]
- Basu D, Haswell ES. 2017. Plant mechano-sensitive ion channels: an ocean of possibilities. Current Opinion in Plant Biology 40: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzaz FA. 1996. Plants in changing environments. Cambridge: Cambridge University Press. [Google Scholar]
- Bateson G. 1963. The role of somatic change in evolution. Evolution 17: 529–539. [Google Scholar]
- Beadle GW. 1980. The ancestry of corn. Scientific American 242: 112–119. [Google Scholar]
- Beer RD. 1990. Intelligence as adaptive behaviour; an experiment in computational ethology. San Diego: Academic Press. [Google Scholar]
- Bertolli S, Vítolo H, Souza G. 2013. Network connectance analysis as a tool to understand homeostasis of plants under environmental changes. Plants 2: 473–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara GB, Wen T-N, Nguyen TT, Verslues PE. 2017. Protein phosphatase 2Cs and microtubule-associated stress protein 1 control microtubule stability, plant growth, and drought response. The Plant Cell 29: 169–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet A. 1897. The psychic life of micro-organisms. Chicago: Open Court Publishing. [Google Scholar]
- Binet A, Simon T. 1916. The development of intelligence in children (The Binet-Simon Scale) (Kite ES, Trans.). Baltimore: Williams & Wilkins Co. [Google Scholar]
- Blancaflor EB, Fasano JM, Gilroy S. 1998. Mapping the functional roles of cap cells in the response of Arabidopsis primary root to gravity. Plant Physiology 116: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom AJ, Chapin FS III, Mooney HA. 1985. Resource limitation in plants – an economic analogy. Annual Review of Ecology and Systematics 16: 363–392. [Google Scholar]
- Boogerd FC, Bruggemena FJ, Richardson RC. 2013. Mechanistic explanations and models in molecular systems biology. Foundations of Science 18: 725–744. [Google Scholar]
- Boogerd FC, Brugemann FJ, Richardson RC, Stephan A, Westerhoff HV. 2005. Emergence and its place in nature: a case study of biochemical networks. Synthese 145: 131–164. [Google Scholar]
- Borkotoky S, Saravanan V, Jaiswal A, et al. 2013. The Arabidopsis stress gene responsive database. International Journal of Plant Genomics 949564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford K, Trewavas AJ. 1994. Sensitivity thresholds and variable time scales in plant hormone action. Plant Physiology 105: 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Turkarsian S, Beer KD, Lo FY, Baliga NS. 2011. Adaptation of cells to new environments. Wiley Interdisciplinary Reviews of Systems Biology and Medicine 3: 544–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce TJA. 2010. Exploiting plant signals in sustainable agriculture. In: Baluska F, Ninkovic V, eds. Plant communication from an ecological perspective. Berlin: Springer-Verlag, 215–228. [Google Scholar]
- Bruce TJA, Matthes MA, Napier JC, Pickett JA. 2007. Stressful “memories” of plants: evidence and possible mechanisms. Plant Science 173: 603–607. [Google Scholar]
- Calvo P. 2007. The quest for cognition in plant neurobiology. Plant Signaling and Behavior 2: 208–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo P. 2017. What is it like to be a plant? Journal of Consciousness Studies 24: 205–227. [Google Scholar]
- Calvo P. 2018. Caterpillar/basil-plant tandems. Animal Sentience 11: 100. [Google Scholar]
- Calvo P, Friston K. 2017. Predicting green: really radical (plant) predictive processing. Journal of the Royal Society Interface 14: 20170096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo P, Sahi VP, Trewavas AJ. 2017. Are plants sentient? Plant, Cell & Environment 40: 2858–2869. [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, et al. 2001. Remodelling of the yeast genome expression in response to environmental changes. Molecular Biology of the Cell 12: 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamowitz D. 2018. Plants are intelligent – now what. Nature Plants 4: 622–623. [DOI] [PubMed] [Google Scholar]
- Cheng C, Anderes E, Yan K-K, Ung M, Wang D, Gerstein M. 2015. An approach for determining and measuring network hierarchy applied to comparing the phosphorylome and the regulome. Genome Biology 16: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Deng W, Wang Y, Ren J, Liu Z, Xue Y. 2014. dbPPT: a comprehensive database of protein phosphorylation in plants. Database bau121: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciolo AT, Sternberg RJ. 2004. Intelligence, a brief history. Oxford: Blackwells. [Google Scholar]
- Cipollini D, Walters D, Voelckel C. 2014. Costs of resistance in plants: from theory to evidence. Annual Plant Reviews 47: 263–307. [Google Scholar]
- Clark KB. 2010. Origins of learned reciprocity in solitary ciliates searching grouped ‘courting’ assurances at quantum efficiencies. Biosystems 99: 27–41 [DOI] [PubMed] [Google Scholar]
- Clark KB. 2013. Ciliates learn to diagnose and correct classical error syndromes in mating strategies. Frontiers in Microbiology 4: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corning P. 2003. Natures magic. Synergy in evolution and the fate of humankind. Cambridge: Cambridge University Press. [Google Scholar]
- Covelo-Molares H, Bartosovic M, Vanacova S. 2018. RNA methylation in nuclear pre-mRNA processing. Wiley Interdisciplinary Reviews RNA 9: e1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis CA. 2005. Mechanism and control of rapid genomic changes in flax. Annals of Botany 95: 204–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1868. The variation of animals and plants under domestication. Volumes 1 and 2 London: John Murray. [Google Scholar]
- Darwin C. 1871The descent of man. London: John Murray. [Google Scholar]
- Darwin C. 1875. On the movements and habits of climbing plants. London: John Murray. [Google Scholar]
- Darwin C, Darwin F. 1880. The power of movements in plants. London: John Murray. [Google Scholar]
- Davidson EH. 2010. Emerging properties of animal gene regulatory networks. Nature 468: 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ. 2001. Intelligence. A very short introduction. Oxford: Oxford University Press. [Google Scholar]
- De la Fuente IM. 2014. Metabolic dissipative structures. In: Aon MA, Saks V, Schlattner U, eds. Systems biology of metabolic and signaling networks: energy, mass and information transfer. New York: Springer Books, 179–212. [Google Scholar]
- De la Fuente I. 2015. Elements of the cellular metabolic structure. Frontiers in Molecular Biosciences 2. doi: 10.3389/fmolb.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, Hernandez KM, Juenger TE. 2013. Genotype-by-environment interaction and plasticity: exploring genomic responses of plants to the abiotic environment. Annual Review of Ecology and Systematics 44: 5–29. [Google Scholar]
- Demongeot J, Hasgui H, Thellier M. 2019. Memory in plants: Boolean modeling of the learning and store/recall memory functions in response to environmental stimuli. Journal of Theoretical Biology 467:123–133. [DOI] [PubMed] [Google Scholar]
- Detterman DK. 1982. Does “g” exist? Intelligence 6: 99–108. [Google Scholar]
- Detterman DK. 1986. Human intelligence is a complex system of separate processes. In: Sternberg RJ, Detterman DK, eds. What is intelligence? Norwood: Ablex Publishing, 57–63. [Google Scholar]
- Dolgin E. 2019. The secret social lives of viruses. Nature 570, 290–292. [DOI] [PubMed] [Google Scholar]
- Dudley SA. 2015. Plant cooperation. AoB Plants 7plv113. doi: 10.1093/aobpla/plv113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley SA, File AL. 2007. Kin recognition in an annual plant. Biology Letters 3: 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein EM, Brunder DG, Blair HJ. 1980. Habituation and sensitisation in an aneural cell: some comparative and theoretical considerations. Neuroscience and Behavioral Reviews 6: 183–194. [DOI] [PubMed] [Google Scholar]
- Eisenstein EM. 1975. Aneural organisms in neurobiology. New York: Plenum. [Google Scholar]
- Eisenstein EM, Eisenstein DL, Sarma JSM, Knapp H, Smith JC. 2012. Some speculative ides about the behaviour homeostasis theory as to how the simple learned behaviours of habituation and sensitisation improve organism’s survival throughout phylogeny. Communicative and Integrative Biology 5: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falik O, de Kroon H, Novoplansky A. 2006. Physiologically-mediated self/non-self, discrimination in Trifolium repens has mixed effects on plant performance. Plant Signalling and Behaviour 1: 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falik O, Mordoch Y, Quansah L, Fait A, Novoplansky A. 2011. Rumor has it …: relay communication of stress cues in plants. PLoS One 6: e23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falik O, Reides P, Gersani M, Novoplansky A. 2003. Self/ non-self discrimination in roots. Journal of Ecology 91: 525–531. [Google Scholar]
- Fell D. 1997. Understanding the control of metabolism. London: Portland Press. [Google Scholar]
- Finlay KW, Wilkinson GN. 1963. The analysis of adaptation in a plant-breeding programme. Australian Journal of Agricultural Research 14: 742–754. [Google Scholar]
- Firn R. 2004. Plant intelligence: an alternative viewpoint. Annals of Botany 93: 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleta-Seriano E, Munne-Bosch S. 2016. Stress memory and the inevitable effects of drought: a physiological perspective. Frontiers in Plant Science 7: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Tateson RW, Bartelmess IB, Porteous DJ, Donachie WD, Kacser H. 1981. Control of the flux of the arginine pathway in Neurospora crassa. Biochemical Journal 200: 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost CJ, Mescher MC, Carlson JE, de Moraes C. 2008. Plant defence priming against herbivores: getting ready for a different battle. Plant Physiology 146: 118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage JL, Jarquin D, Romay C, et al. 2017. The effect of artificial selection on phenotypic plasticity in maize. Nature Communications 8: 1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliano M. 2015. In a green frame of mind: perspectives on the behavioural ecology and cognitive nature of plants. AoB Plants 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliano M. 2017. The mind of plants: thinking the unthinkable. Communicative and Integrative Biology 10: e1288333. [Google Scholar]
- Gagliano M, Abramson C, Depczynski M. 2018. Plants learn and remember, let’s get used to it. Oecologia 186: 29–31. [DOI] [PubMed] [Google Scholar]
- Gagliano M, Renton M. 2013. Love thy neighbour: facilitation through an alternative signalling modality in plants. BMC Ecology 13: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliano M, Renton M, Depczynski M, Mancuso S. 2014. Experience teaches plants to learn faster and forget slower in environments where it matters. Oecologia 175: 63–72. [DOI] [PubMed] [Google Scholar]
- Gagliano M, Renton M, Duvdevani N, Timmins M, Mancuso S. 2012. Out of sight but not out of mind: alternative means of communication in plants. PLoS One 7: e37382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliano M, Vyazovsky VV, Borbely AA, Grimonprez M, Depczynski M. 2016. Learning by association in plants. Scientific Reports 6: 38427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Stegele O, Behr J, et al. 2011. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Barzel B, Barabasi A-L. 2016. Universal resilience patterns in complex networks. Nature 530: 307–312. [DOI] [PubMed] [Google Scholar]
- Gardner RA, Gardner BT. 1969. Teaching sign language to a chimpanzee. Science 165: 664–672. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman DT, Kao CM, et al. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Molecular Biology of the Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelber B. 1952. Investigations of the behaviour of Paramecium aurelia. Journal of Comparative Physiology and Psychology 45: 58–65. [DOI] [PubMed] [Google Scholar]
- Gianoli E. 2016. Eyes in the chameleon vine? Trends in Plant Science 22: 4–5. [DOI] [PubMed] [Google Scholar]
- Gibbs KA, Urbanowski ML, Greenberg EP. 2008. Genetic determinants of self-identity and social recognition. Science 321: 256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF. 2001. Ecological development biology: developmental biology meets the real world. Developmental Biology 233: 1–12. [DOI] [PubMed] [Google Scholar]
- Ginsburg S, Jablonka E. 2009. Epigenetic learning in non-neural organisms. Journal of Bioscience 34: 633–646. [DOI] [PubMed] [Google Scholar]
- Golden SS. 2003. Think like a bacterium. EMBO Reports 4: 15–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzelak MA, Asay AK, Pickles BJ, Simard SW. 2015. Inter-plant communication through mycorrhizal networks mediates complex adaptive behaviour in plant communities. AoB Plants 7: 050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfredson LS. 1997. Mainstream science on intelligence, history and bibliography: an editorial with 52 signatories. Intelligence 24: 13–23. [Google Scholar]
- Grenier S, Barre P, Litrico I. 2016. Phenotypic plasticity and selection: nonexclusive mechanisms of adaptation. Scientifica 2016: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntmann M, Novoplansky A. 2004. Physiologically-mediated self/non-self discrimination in roots. Proceedings of the National Academy of Sciences USA 101: 3863–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Shasha DE, Coruzzi GM. 2005. System biology for the virtual plant. Plant Physiology 138: 550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberlandt G. 1914. Physiological plant anatomy (translated by Drummond M.). London: Macmillan and Co. Ltd. [Google Scholar]
- Hansen BG, Halkier BA, Kliebenstein DJ. 2008. Identifying the molecular basis of QTLs: eQTLs add a new dimension. Trends in Plant Science 13: 72–77. [DOI] [PubMed] [Google Scholar]
- He BJ. 2014. Scale-free brain activity: past, present and future. Trends in Cognitive Sciences 18: 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebb D. 1949. The organisation of behaviour. A neuropsychological theory. New York: Wiley and Sons. [Google Scholar]
- Herben T, Novoplansky A. 2008. Implications of self/non-self discrimination for spatial patterning of clonal plants. Evolutionary Ecology 22: 337–350. [Google Scholar]
- Highkin HR. 1958. Temperature-induced variability in peas. American Journal of Botany 45: 626–631. [Google Scholar]
- Hill J. 1965. Environmental induction of heritable changes in Nicotiana. Nature 207: 732–734. [Google Scholar]
- Hinkle DJ, Wood DC. 1994. Is tube escape learning by protozoa, associative learning? Behavioural Neuroscience 108: 94–99. [DOI] [PubMed] [Google Scholar]
- Hoffer SM, Westerhoff HV, Hellingwerf KJ, Postma PW, Tommassen J. 2001. Autoamplification of two component regulatory system results in learning behaviour. Journal of Bacteriology 183: 4914–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JH. 1995. Hidden order. How adaptation builds complexity. New York: Perseus Books. [Google Scholar]
- Holland JH. 2006. Studying complex adaptive systems. Journal of Systems Science and Complexity 19: 1–8. [Google Scholar]
- Hopfield JJ. 1982. Neural networks and physical systems with emergent collective, computational abilities. Proceedings of the National Academy of Sciences USA 79: 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe M, Shotwell M. 2006. Physiological studies on tendrils. XI. Storage of tactile information prior to the blue light activation effect. Physiologia Plantarum 50: 78–82. [Google Scholar]
- Jain SK. 1978. Inheritance of phenotypic plasticity in soft chess, Bromus mobilis. Experientia 34: 835–836. [Google Scholar]
- Jain SK, Martins PS. 1979. Ecological genetics of the colonizing ability of rose clover (Trifolium hirtum All.). American Journal of Botany 66: 361–366. [Google Scholar]
- Jalili M, Salehzadeh-Yazdi Y, Gupta S, et al. 2016. Evolution of centrality measurements for the detection of essential proteins in biological networks. Frontiers in Physiology 7: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Lubin FD. 2014. Epigenetic mechanisms of memory formation and reconsolidation. Neurobiology, Learning and Memory 115: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jékely G, Keijzer F, Godfrey-Smith P. 2015. An option space for early neural evolution. Philosophical Transactions of the Royal Society of London Series B. Biological Sciences 370: 1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings HS. 1906. Behaviour of the lower organisms. New York: Columbia University Press. [Google Scholar]
- Jensen DD. 1957. Experiments on learning in Paramecia. Science 125: 191–192. [DOI] [PubMed] [Google Scholar]
- Jinks JL, Pooni HS. 1982. Determination of the environmental sensitivity of selection lines of Nicotiana rustica by the selection environment. Heredity 49: 291–294. [DOI] [PubMed] [Google Scholar]
- Johnson SB. 2001. Emergence: the connected lives of ants, brains, cities and software. New York: Scribner. [Google Scholar]
- Joyard J, McCormick S. 2010. Plant systems biology. Plant Physiology 152: 401. [Google Scholar]
- Kadam NN, Tamilselvan A, Lawas LMF, et al. 2017. Genetic control of plasticity in root morphology and anatomy of rice in response to water deficit. Plant Physiology 174: 2302–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Huang S-SC, Jupe F, et al. 2015. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzer FA. 2017. Evolutionary convergence and biologically embodied cognition. Interface Focus 7: 20160123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper JW, Ryu C-M, Zhang S. 2004. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94: 1259–1266. [DOI] [PubMed] [Google Scholar]
- Knight MR, Smith SM, Trewavas AJ. 1992. Wind-induced plant motion immediately increases cytosolic calcium. Proceedings of the National Academy of Sciences USA 89: 4967–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolling M, Kumari P, Burstenbinder K. 2019. Calcium- and calmodulin-regulated microtubule-associated proteins as signal-integration hubs at the plasma membrane-cytoskeleton nexus. Journal of Experimental Botany 70: 387–396. [DOI] [PubMed] [Google Scholar]
- Kusmec A, Deleon N, Schnable PS. 2018. Harnessing phenotypic plasticity to improve maize yields. Frontiers in Plant Science 9: 1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen RAE, Nikoloski Z. 2019. Genetic basis of plasticity in plants. Journal of Experimental Botany 70: 739–745. [DOI] [PubMed] [Google Scholar]
- Lamarck J. 1809. Zoological philosophy. Translation 1914 by Elliott H. London: MacMillan. [Google Scholar]
- Legg S, Hunter M. 2007. A collection of definitions of intelligence. In: Goertzel B, Wang P, eds. Advances in artificial general intelligence: concepts, architectures and algorithms. Frontiers in Artificial Intelligence and Applications. Amsterdam: IOS Press, 17–24. [Google Scholar]
- Lestas I, Vinnicombe G, Paulsson J. 2010.Fundamental limits on the suppression of molecular fluctuations. Nature 467: 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy ED, Landry CR, Michnik SW. 2010. Signalling through cooperation. Science 328: 983–984. [DOI] [PubMed] [Google Scholar]
- Liptay A, Davidson D. 1971. Coleoptile growth: variation in elongation patterns of individual coleoptiles. Annals of Botany 35: 991–1002. [Google Scholar]
- Lovejoy AO. 1936.The great chain of being. New York: Harper and Row. [Google Scholar]
- Lyon P. 2006. The biogenic approach to cognition. Cognitive Processes 7: 11–29. [DOI] [PubMed] [Google Scholar]
- Ma W, Trusina A, El-Samad H, Lim WA, Tang C. 2009. Defining network topologies that can achieve biochemical adaptation. Cell 138: 760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Gao L. 2012. Biological network analysis: insights into structure and function. Briefings in Functional Genomics 11: 434–442. [DOI] [PubMed] [Google Scholar]
- Ma X, Xu G, He P, Shan L. 2016. SERKing coreceptors for receptors. Trends in Plant Science 21: 1017–1033. [DOI] [PubMed] [Google Scholar]
- Margulis L, Sagan D. 1995. What is life? London: Weidenfield and Nicolson Ltd. [Google Scholar]
- Massa G, Gilroy S. 2009. Touch modulates gravity sensing to regulate the growth of primary roots of Arabidopsis thaliana. Plant Journal 33: 435–445. [DOI] [PubMed] [Google Scholar]
- Matsuo T. 1975. Adaptability in plants – with special reference to crop yield. JIBP Synthesis 6: 1–217. [Google Scholar]
- Maturana HR, Varela FG. 1980. Autopoiesis and cognition: the realization of the living. Dordrecht: Reidel. [Google Scholar]
- Maturana HR, Varela FG. 1987. The tree of knowledge. Boston: Shambhala. [Google Scholar]
- Mayr E. 2001. What evolution is. New York: Basic Books. [Google Scholar]
- McClintock B. 1984. The significance of responses of the genome to challenge. Science 226: 792–801. [DOI] [PubMed] [Google Scholar]
- McNamara JM, Houston AI. 1996. State dependent life histories. Nature 380: 215–221. [DOI] [PubMed] [Google Scholar]
- Miller JH, Page SE. 2007. Complex adaptive systems. Princeton: Princeton University Press. [Google Scholar]
- Milo R, Itzkovitz S, Kashtan N, et al. 2004. Super-families of evolved and designed networks. Science 303: 1538–1542. [DOI] [PubMed] [Google Scholar]
- Moss GI, Mullett JH. 1982. Potassium release and seed vigour in germinating bean seed as influenced by temperature over the previous five generations. Journal of Experimental Botany 33: 1147–1160. [Google Scholar]
- Neisser U, Boodoo G, Bouchard TJ, Brody N, Ceci SJ. 1996. Intelligence knowns and unknowns. American Psychologist 51: 77–101. [Google Scholar]
- Novoplansky A. 2009. Picking battles wisely: plant behaviour under competition. Plant, Cell and Environment 32: 726–741. [DOI] [PubMed] [Google Scholar]
- Novoplansky A. 2016. Future perception in plants. In: Nadin M, ed. Anticipation across disciplines. Cognitive Systems Monographs 29: 57–69. [Google Scholar]
- Novoplansky A. 2019. What plant roots know? Seminars in Cell and Developmental Biology 92: 126–133. [DOI] [PubMed] [Google Scholar]
- Olshausen BA, Field DJ. 2004. Sparse coding of sensory inputs. Current Opinion in Neurobiology 14: 481–487. [DOI] [PubMed] [Google Scholar]
- Osborn HF. 1897. The limits of organic selection. American Naturalist 31: 944–951. [Google Scholar]
- Oyama S, Griffiths PE, Gray RD. 2001. Introduction: what is developmental systems theory. In: Oyama S, Griffiths PE, Gray RD, eds. Cycles of contingency. Developmental systems and evolution. Cambridge: MIT Press, 1–13. [Google Scholar]
- Palacio-Lopez K, Beckage B, Scheiner S, Molofsky J. 2015. The ubiquity of phenotypic plasticity in plants: as synthesis. Ecology and Evolution 5: 3389–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei S, Muchnik L, Amdrade JS, Zheng Z, Makse HA. 2014. Searching for superspreaders of information in real world social media. Scientific Reports 4: 5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. 2019. Do plants favour their kin? Science 363: 15–16. [DOI] [PubMed] [Google Scholar]
- Platt SA, Sanislow CA. 1988. Norm-of-reaction: definition and misinterpretation of animal research. Journal of Comparative Psychology 102: 254–261. [DOI] [PubMed] [Google Scholar]
- Plotkin HC. 1988. Learning and evolution. In: Plotkin HC, ed. The role of behaviour in evolution. Cambridge: MIT Press, 133–165. [Google Scholar]
- Probst AV, Scheid OM. 2015. Stress-induced structural changes in plant chromatin. Current Opinion in Plant Biology 27: 8–16. [DOI] [PubMed] [Google Scholar]
- Rasheed S, Bashir K, Matsui A, Tanaka M, Seki M. 2016. Transcriptomic analysis of soil-grown Arabidopsis thaliana roots and shoots in response to drought stress. Frontiers in Plant Science 7: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Marquez Y, Kalyna M, Barta A. 2013. Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CL, Rosas U, Banta J, Bhambra N, Puruggan MD. 2012. Genome wide patterns of Arabidopsis gene expression in Nature. PLoS Genetics 8: e1002662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. 1996. Resource capture by localised root proliferation: why do plants bother? Annals of Botany 77: 179–185. [Google Scholar]
- Romanes GJ. 1884. Animal intelligence. London: Kegan Paul, Trench, Trubner & Co. Ltd. [Google Scholar]
- Rosenblueth A, Weiner N, Bigelow J. 1943. Behaviour, purpose and teleology. Philosophy of Science 19:18–24. [Google Scholar]
- Russell ES. 1946. The directiveness of organic activities. Cambridge: Cambridge University Press. [Google Scholar]
- Ruuhola T, Salminesin J-T, Haviola S, Yang S, Rantal MJ. 2007. Immunological memory of mountain birches: effects of phenolics on performance of the autumnal moth depend on herbivory history of trees. Journal of Chemical Ecology 35: 1160–1175. [DOI] [PubMed] [Google Scholar]
- Rymaszewski W, Vile D, Bediee A, et al. 2017. Stress-related gene expression reflects morpho-physiological responses to water deficit. Plant Physiology 174: 1913–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Sachs J. 1887. Lectures on the physiology of plants. Oxford: Clarendon Press. [Google Scholar]
- Shemesh H, Ovadia O, Novoplansky A. 2010. Anticipating future conditions via trajectory sensitivity. Plant Signalling and Behavior 5: 1501–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam R, Luu VT, Weinhold A, Goldberg J, Oh Y, Baldwin IT. 2015. Native root associated bacteria, rescue a plant from sudden-wilt disease that emerged during continuous cropping. Proceedings of the National Academy of Sciences USA 112: E5013–E5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk HJ, Callaway RM, Mahall BE. 1999. Spatial root segregation: are plants territorial? Advances in Ecological Research 28: 145–180. [Google Scholar]
- Schlichting CD. 1986. The evolution of phenotypic plasticity in plants. Annual Review of Ecology and Systematics 17: 667–693. [Google Scholar]
- Schlichting CD. 1989. Phenotypic plasticity in Phlox. II. Plasticity of character correlation. Oecologia 78: 496–501. [DOI] [PubMed] [Google Scholar]
- Schlichting CD, Levin DA. 1990. Phenotypic plasticity in Phlox. III. Variation among natural populations of Phlox drummondii. Journal of Evolutionary Biology 3: 411–428. [Google Scholar]
- Schlichting CD, Pigliucci M. 1998. Phenotypic evolution. A reaction norm perspective. Sunderland: Sinauer Associates. [Google Scholar]
- Schull JA. 1990. Are species intelligent? Behavioural and Brain Sciences 13: 63–108. [Google Scholar]
- Scott-Kelso TA. 1995. Dynamic patterns: the self-organisation of brains and behaviour. Cambridge: MIT Press. [Google Scholar]
- Segundo-Ortin M, Calvo P. 2019. Are plants cognitive? A reply to Adams. Studies in History and Philosophy of Science Part A 73: 64–71. [DOI] [PubMed] [Google Scholar]
- Sheth BP, Thaker VS. 2014. Plant systems biology. Insight, advances and challenges. Planta 240: 35–54. [DOI] [PubMed] [Google Scholar]
- Smith S. 1908. The limits of educability of Paramecium. Journal of Comparative Neurology and Psychology 18: 499–510. [Google Scholar]
- Snyderman M, Rothman S. 1988. The IQ controversy, the media and public policy. Piscataway: Transaction Books. [Google Scholar]
- Song YY, Simard SW, Carroll A, Mohn WW, Zeng RS. 2015. Defoliation of interior douglas elicits carbon transfer and stress signalling to Ponderosa pine neighbours through ectomycorrhizal networks. Scientific Reports 5: 8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Andrews BJ. 2008. Linking the kinome and phosphorylome – a comprehensive review of approaches to kinase targets. Molecular Biosystems 4: 920–933. [DOI] [PubMed] [Google Scholar]
- Souza GM, Pincus SM, Monteiro JAF. 2005. The complexity-stability hypothesis in plant gas exchange under water deficit. Brazilian Journal of Plant Physiology 17: 273–281. [Google Scholar]
- Souza GM, Ribeiro RV, Prado CHBS, Damineli DSC, Sato AM, Oliveira MS. 2009. Using network connectance and autonomy analyses to uncover patterns of photosynthetic responses in tropical woody species. Ecological Complexity 6: 15–26. [Google Scholar]
- Souza GM, Lüttge U. 2015. Stability as a phenomenon emergent from plasticity, complexity and diversity in eco-physiology. Progress in Botany 76: 211–239. [Google Scholar]
- Spearman C. 1904. “General intelligence” objectively determined and measured. American Journal of Psychology 15: 201–293. [Google Scholar]
- Stenhouse D. 1974. The evolution of intelligence. A general theory and some of its implications. London: George Allen and Unwin. [Google Scholar]
- Sternberg RJ. 1985. Beyond IQ: a triarchic theory of human intelligence. Cambridge: Cambridge University Press. [Google Scholar]
- Sternberg RJ. 1986. A framework for understanding conceptions of intelligence. In: Sternberg RJ, Detterman DK, eds. What is intelligence? Norwood: Ablex Publishing, 3–19. [Google Scholar]
- Sternberg RJ. 2006. Cognitive psychology. Califonia: Thompson and Wadworth. [Google Scholar]
- Sternberg RJ, Berg CA. 1986. Quantitative integration. Definitions of intelligence: a comparison of the 1921 and 1986 symposia. In: Sternberg RJ, Detterman DK, eds. What is intelligence? Norwood: Ablex Publishing, 155–163. [Google Scholar]
- Sternberg RJ, Wagner RK. 1986. Practical intelligence. Cambridge: Cambridge University Press. [Google Scholar]
- Sultan SE. 2015. Organism and environment: ecological development, niche construction, and adaptation. Oxford: Oxford University Press. [Google Scholar]
- Sultan SE, Barton K, Wilczek AM. 2009. Contrasting patterns of transgenerational plasticity in ecologically distinct congeners. Ecology 90: 1831–1839. [DOI] [PubMed] [Google Scholar]
- Sultan SE, Spencer HG. 2002. Metapopulation structure favors plasticity over local adaptation. The American Naturalist 160: 271–283 [DOI] [PubMed] [Google Scholar]
- Tagkopoulos I, Liu Y-C, Tavazole S. 2008. Predictive behaviour within microbial genetic networks. Science 320: 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1001 Genomes Consortium 2016. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorndike EL. 1911. Animal intelligence: Experimental studies. New York: Macmillan. [Google Scholar]
- Toyota M, Spenser D, Sawai-Toyota S, et al. 2018. Glutamate triggers long-distance, calcium-based plant defense signalling. Science 361: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Trewavas AJ. 1999. Le Calcium c’est la Vie, calcium makes waves. Plant Physiology 120: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas AJ. 2003. Aspects of plant intelligence. Annals of Botany 9: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas AJ. 2004a Aspects of plant intelligence-an answer to Firn. Annals of Botany 93: 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas AJ. 2004b A critical assessment of organic farming and food assertions with particular respect to the UK and the potential environmental benefits of no-till agriculture. Crop Protection 23: 757–781. [Google Scholar]
- Trewavas AJ. 2007. Response to Alpi et al.: Plant neurobiology – all metaphors have value. Trends in Plant Science 12: 231–233. [DOI] [PubMed] [Google Scholar]
- Trewavas AJ. 2009. What is plant behaviour? Plant, Cell and Environment 32: 606–616. [DOI] [PubMed] [Google Scholar]
- Trewavas AJ. 2011. Plant cell calcium, past and future. In: Luan S, ed. Coding and decoding of calcium signals in plants. Berlin: Springer-Verlag, 1–7. [Google Scholar]
- Trewavas AJ. 2012. Information, noise and communication: thresholds as controlling elements in development. In: Witzany G, Baluska F, eds. Biocommunication of plants. Signalling and communication in plants, 14. Berlin: Springer-Verlag, 11–35. [Google Scholar]
- Trewavas AJ. 2014. Plant behaviour and intelligence. Oxford: Oxford University Press. [Google Scholar]
- Trewavas AJ. 2016a Intelligence, cognition and language of green plants. Frontiers in Plant Science 7: 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas AJ. 2016b Plant intelligence: an overview. Bioscience 66: 542–551. [Google Scholar]
- Trewavas AJ. 2017. The foundations of plant intelligence. Interface Focus 7: 20160098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turing AM. 1947. Lecture to the London Mathematical Society on 20 February 1947. In: Carpenter BE, Doran RW, eds. A.M.Turing’s ACE Report of 1946 and Other Papers. Cambridge: MIT Press. [Google Scholar]
- Turkington R. 1983. Plasticity in growth of dry matter distributions of two genotypes of Trifolium repens grown in different environments of neighbours. Canadian Journal of Botany 61: 2186–2194. [Google Scholar]
- Turner JS. 2007. The tinkerers accomplice – how design emerges from life itself. London: Harvard University Press. [Google Scholar]
- Turner JS. 2018. Purpose and desire. What makes something alive and why modern Darwinism has failed to explain it. New York: Harper Collins Publishers. [Google Scholar]
- Varela F. 1979. Principles of biological autonomy. New York: Elsevier-North Holland. [Google Scholar]
- Wahlstein D. 1990. Insensitivity of the analysis of variance to heredity–environment interaction. Behavioural and Brain Sciences 13: 109–161. [Google Scholar]
- Watahiki M, Trewavas AJ. 2019. Systems, variation, individuality and plant hormones. Progress in Biophysics and Molecular Biology 146: 3–22. [DOI] [PubMed] [Google Scholar]
- Waters AJ, Makaarevitch I, et al. 2017. Natural variation for gene expression responses to stress in maize. Plant Journal 89: 706–717. [DOI] [PubMed] [Google Scholar]
- Weber B, Zicola J, Oka R, Stam M. 2016. Enhancers; a call for discovery. Trends in Plant Science 21: 974–987. [DOI] [PubMed] [Google Scholar]
- Wegner LH, Lüttge U, eds.2019. Emergence and modularity in life sciences. Berlin: Springer-Nature. [Google Scholar]
- Wei B, Liu J, Wei D, Gao C, Deng Y. 2014. Weighted k-shell decomposition for complex networks based on potential edge weights. Physica A 420: 277–283. [Google Scholar]
- Weiss PA. 1973. The science of life: the living system- a system for living. New York:Futura Publishing Co. [Google Scholar]
- West-Eberhard MJ. 2002. Developmental plasticity and evolution. Oxford: Oxford University Press. [Google Scholar]
- Westerhoff HV, Brooks AN, Simeonidis E, et al. 2014. Macromolecular networks and intelligence in micro-organisms. Frontiers in Microbiology 5: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. 1956. Biochemical individuality. The basis of the genetotrophic concept. New York: John Wiley &Sons. [Google Scholar]
- Willshaw DJ, Buneman OP, Longuet-Higgins HC. 1969. Non-holographic associative memory. Nature 222: 960–962. [DOI] [PubMed] [Google Scholar]
- Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein MI. 2007. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Computational Biology 3: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Gerstein M, Snyder M. 2007. Getting connected: analysis and principles of biological networks. Genes and Development 21: 1010–1024. [DOI] [PubMed] [Google Scholar]
- Zotenko E, Mestre J, O’Leary DP, Przytycka TM. 2008. Why do hubs in the yeast protein interaction network tend to be essential: re-examining the connections between the network. topology and essentiality. PLoS Computational Biology 4: e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust T, Agrawal AA. 2017. Trade-offs between plant growth and defense against insect herbivory: an emerging mechanistic synthesis. Annual Review of Plant Biology 68: 513–534. [DOI] [PubMed] [Google Scholar]