Abstract
Background and Aims
The tremendously unbalanced distribution of species richness across clades in the tree of life is often interpreted as the result of variation in the rates of diversification, which may themselves respond to trait evolution. Even though this is likely a widespread pattern, not all diverse groups of organisms exhibit heterogeneity in their dynamics of diversification. Testing and characterizing the processes driving the evolution of clades with steady rates of diversification over long periods of time are of importance in order to have a full understanding of the build-up of biodiversity through time.
Methods
We studied the macroevolutionary history of the species-rich tree fern family Cyatheaceae and inferred a time-calibrated phylogeny of the family including extinct and extant species using the recently developed fossilized birth–death method. We tested whether the high diversity of Cyatheaceae is the result of episodes of rapid diversification associated with phenotypic and ecological differentiation or driven by stable but low rates of diversification. We compared the rates of diversification across clades, modelled the evolution of body size and climatic preferences and tested for trait-dependent diversification.
Key Results
This ancient group diversified at a low and constant rate during its long evolutionary history. Morphological and climatic niche evolution were found to be overall highly conserved, although we detected several shifts in the rates of evolution of climatic preferences, linked to changes in elevation. The diversification of the family occurred gradually, within limited phenotypic and ecological boundaries, and yet resulted in a remarkable species richness.
Conclusions
Our study indicates that Cyatheaceae is a diverse clade which slowly accumulated morphological, ecological and taxonomic diversity over a long evolutionary period and provides a compelling example of the tropics as a museum of biodiversity.
Keywords: Diversification, macroevolution, tree ferns, Cyatheaceae, phylogeny, target sequencing, climatic preferences, fossilized birth–death, phenotypic evolution, gradual evolution, species richness, divergence times
INTRODUCTION
The distribution of biodiversity is strikingly unbalanced when comparing species richness among lineages and one of the main challenges in evolutionary biology is to understand the underlying factors that are responsible for this uneven distribution (Wiens, 2017). This challenge is commonly tackled by comparing the patterns of diversification among regions and taxonomic groups (Antonelli et al., 2015; Eiserhardt et al., 2017), but this requires comparing large numbers of species to have a good understanding of the potential processes involved. Fortunately, the increasing availability of DNA sequence data coupled with advances in phylogenetic comparative methods over recent decades provides us with powerful tools to study the assembly of Earth’s biodiversity through time and space.
The heterogeneity of species richness across clades is likely the result of variations in the rates of speciation or extinction (Wiens, 2017). Changes in diversification rates, measured as the difference between the estimated rates of speciation and extinction, have been attributed to a wide range of factors. Intrinsic traits such as the tank habit in bromeliads (Silvestro et al., 2014) or heterostyly in primroses (de Vos et al., 2014) have been linked with accelerated diversification. Ecological interactions, for instance clownfish mutualism with sea anemones (Litsios et al., 2012) or plant interactions with pollinators (Breitkopf et al., 2015; Serrano-Serrano et al., 2017), have also been hypothesized to play a role in species diversification. Finally, the ecological niche can also affect the diversity between lineages, as shown in the large fern family Polypodiaceae, where lineage diversification is positively associated with the colonization of a wider elevation range (Sundue et al., 2015).
Clade age has also been proposed as an explanation for differences in species richness between groups of organisms through the ‘clade-age’ hypothesis, which argues that older clades tend to have more species simply because they have had more time to accumulate diversity (Wiens, 2017). In this case, species richness results from the steady accumulation of lineages through time rather than from shifts in the rates of diversification. Stable evolutionary dynamics have been reported, for example, in Neotropical Troidini butterflies (Condamine et al., 2012) and several plant groups, such as figs (Bruun-Lund et al., 2018), malagasy Angraecum orchids (Andriananjamanantsoa et al., 2016), Annonaceae (Couvreur et al., 2011b), Australian Proteaceae (Cardillo and Pratt, 2013) and liverworts (Wilson et al., 2007). The clade-age hypothesis also provides a possible scenario to explain geographical variation in species richness, whereby more diverse regions, for instance the tropics, would be the result of early colonizations rather than variation in the rate of clade diversification among regions (Brown, 2014). In contrast to the clade-age hypothesis, a recent study has proposed that diversification is a time-dependent process whereby younger clades diversify more rapidly than older clades (Henao Diaz et al., 2019). This time dependency could potentially disrupt a correlation between the age of a clade and its diversity.
Alternatively, instead of invoking a single underlying factor, some studies have proposed a more nuanced model of evolutionary dynamics where both time and rates of diversification contribute to the explanation of diversity patterns. For instance, the palm family (Arecaceae) exhibited a constant rate of diversification during most of its long evolutionary history, but several increases in the rate of diversification have punctuated the recent evolution of this group (Couvreur et al., 2011a; Baker and Couvreur, 2013). Similarly, the assembly of fern diversity appears to have been shaped by a strong turnover of clades resulting from the combination of both diversity-dependent speciation and climate-driven extinction (Lehtonen et al., 2017).
In this context, the long-term drivers of the diversification history of old and species-rich plant clades can give important insights into the build-up of biodiversity through time. We focused here on the scaly tree fern family Cyatheaceae, an iconic plant group of humid forests comprising about 660 known species distributed worldwide in tropical and subtropical regions. This species richness is remarkable compared with the seven other families of the Cyatheales, which contain from a single species in the Thyrsopteridaceae to ~35 species in the Dicksoniaceae (PPG, 2016). Phylogenetic studies have estimated the divergence between the Cyatheaceae and its sister family the Dicksoniaceae around 145 Ma and have shown that both Gondwanan vicariance and long-distance dispersal contributed to generating the current distribution of scaly tree ferns (Korall and Pryer, 2014). Significant among-clade variations in the rate of diversification have been reported for the family (Janssen et al., 2008; Ramírez-Barahona et al., 2016), and a recent study also found a positive association between the rates of morphological and climatic niche evolution and the rates of diversification, suggesting that the evolutionary dynamics of Cyatheaceae followed those of an adaptive radiation (Ramírez-Barahona et al., 2016). However, only about 10 % of the total number of Cyatheaceae species were represented in these studies. Such a low taxonomic sampling weakens phylogenetic comparative analyses (Heath et al., 2008; Silvestro et al., 2011) and calls into question our understanding of the evolutionary history of the group.
Here, we combined molecular data at an unprecedented level of taxonomic sampling with fossil information to reconstruct a new dated phylogenetic tree of the Cyatheaceae. The origin of the group dates back to the Jurassic and recent theory suggests that diversification in such an old group could be the result of a steady but slow accumulation of species through time (Henao Diaz et al., 2019). Alternatively, the high diversity of the family could be associated with the evolution of intrinsic or ecological traits in specific lineages driving species diversification (Ramírez-Barahona et al., 2016). We used macroevolutionary models to test several hypotheses related to species and trait diversification of the Cyatheaceae. We first tested whether the build-up of species diversity in this group followed the expectation of the clade-age hypothesis, which would imply that species richness in this family increased at a steady rate through its long evolutionary history. As an alternative hypothesis, we tested instead whether their great diversity originated from variations in the rate of diversification between lineages. Second, we investigated the role played by morphological and ecological traits during the evolution of the group by reconstructing the evolutionary dynamics of these traits through time and testing their impact on the rates of diversification. Our findings show that, unlike many species-rich groups, the family Cyatheaceae is an old lineage that diversified at an exceptionally low and homogeneous pace without undergoing extensive morphological evolution. Although the rate of evolution of climatic preferences accelerated in several clades, likely in response to dispersals to higher elevation habitats, this was decoupled from the diversification of the group.
MATERIALS AND METHODS
Taxonomic sampling
Our dataset includes 323 species of Cyatheaceae representing ~49 % of the total extant diversity of the group. All genera were included, with sampling fractions of 49 % in Alsophila, 47 % in Cyathea, 72 % in Gymnosphaera and 49 % in Sphaeropteris. Sixty-six accessions were previously published and we took them directly from GenBank. The rest of the sampled material (257) was collected and identified personally (by one of the authors); voucher material for published sequences (Korall et al., 2007) was also revised (by one of the authors). For taxonomy we follow the recent reinstatement of Gymnosphaera as a separate genus from Alsophila (Dong and Zuo, 2018). As outgroup, we included 12 species from three genera (Calochlaena, Dicksonia and Lophosoria) of the sister family Dicksoniaceae.
DNA extraction, amplification and sequencing
DNA extraction was performed with the NucleoSpin® Plant II (Macherey Nagel, Düren, Germany) kit following the manufacturer’s protocol. We generated sequence data for three chloroplast markers: trnL-trnF [including the trnL group I intron (g1i) and the trnL-trnF intergenic spacer (IGS)], trnG-trnR [including the trnG group II intron (g2i) and the trnG-trnR IGS) and rpl16 (rps3-rpl16, including parts of the rps3 exon, as well as the rps3-rpl16 IGS and the rpl16 g2i]. For PCR and sequencing reactions of trnG-R, the primer set and PCR program described by Nagalingum et al. (2007) were used. The PCR and sequencing reactions of trnL-F and rpl16 followed the protocol by Noben et al. (2017). Reactions were performed either on a Biometra TProfessional TRIO thermocycler (Biometra, Göttingen, Germany) or on an Eppendorf Mastercycler EPGradient S (Eppendorf, Hamburg, Germany). For sequencing, the services of Macrogen Europe (Amsterdam, Netherlands) and GATC Biotech (Konstanz, Germany) were used. All the sequences newly generated in this study are deposited in GenBank (GenBank numbers will be added upon acceptance).
Phylogenetic analyses
All sequences were aligned using the MAFFT (Katoh et al., 2002) plug-in in Geneious 6.1.8 (Biomatters). Alignments were visually inspected and ambiguous regions were excluded. The three markers were concatenated into a single alignment using SequenceMatrix 1.7.8 (Vaidya et al., 2011). We partitioned the alignment by genes and the best model of nucleotide substitution for each region was selected based on the Akaike information criterion using jModelTest 2.1.7 (Darriba et al., 2012). Although additional chloroplast genes are available with relatively good species sampling in GenBank, we did not use these sequences for our final analyses since they increased the proportion of missing data and adding them to our own molecular dataset resulted in a similar topology (see Supplementary Data for details), similar branch length estimates (Supplementary Data Fig. S1) and did not improve substantially the resolution of the estimated phylogenetic tree (Supplementary Data Figs S2 and S3). Phylogenetic inference and divergence time estimates were made using the fossilized birth–death (FBD) model (Heath et al., 2014) implemented in BEAST 2.4.7 (Bouckaert et al., 2014). Unlike traditional node calibration, FBD analysis allows the use of all available fossil evidence and not only the oldest unequivocal fossil for a given clade. Hence, we selected a set of 41 fossils of Cyatheales, 13 of which belonged to the Cyatheaceae, and constrained their placement using taxonomic information (Supplementary Data Table S1). Importantly, we differ from previous phylogenetic studies of tree ferns regarding the placement of the fossil Kuylisporites mirabilis (93.9–100.5 Myr; Mohr and Lazarus, 1994), which has been used to calibrate the crown node of Cyatheaceae based on its resemblance to spores of extant species of Cyathea and Alsophila. However, this interpretation was due to a misleading taxonomic treatment (the species Cyathea decurrens used to be treated as Alsophila decurrens) and the Kuylisporites spore type is actually only found in extant species of Cyathea and not in any other genus. Therefore, in our FBD analysis this fossil was only allowed to be placed in the Cyathea lineage and we used the age intervals of the fossils to indicate their respective age. The first appearance of the fossil genus Cyathocaulis (Upper Jurassic, 145–165.5 Ma) was used to constrained the origin of the Cyatheaceae (see Supplementary Data for details; Korall et al., 2014). We performed two runs of 500 million Markov chain Monte Carlo (MCMC) generations in BEAST 2.4.7, sampling every 50 000 generations. Convergence and effective sample size (ESS) were checked in Tracer 1.5.0 (Rambaut and Drummond, 2009). The two posterior distributions of trees were combined after removal of a burn-in of 10 % using LogCombiner. We used TreeAnnotator to obtain the maximum clade credibility (mcc) tree after removing the fossil taxa from the combined posterior distributions of trees. In all subsequent macroevolutionary analyses, outgroup species were removed from the phylogenetic tree. Additionally, for comparison we performed divergence time estimation using traditional node dating using four fossil calibrations, including K. mirabilis to calibrate the stem node of Cyathea (see Supplementary Data for details). We also assessed the effect of using single versus multiple occurrences for each fossil during the FBD analyses (Supplementary Data Figs S5 and S6).
Diversification rate analyses
We used a variety of approaches to estimate net diversification rates across the Cyatheaceae family and to assess the presence of rate heterogeneity. First, we used BAMM 2.5 (Rabosky, 2014) to infer lineage-specific diversification rates on the mcc tree from BEAST. We specified clade-specific sampling fractions to account for non-random incomplete taxon sampling. Given the recent debate on prior sensitivity of the BAMM model (Moore et al., 2016; Rabosky et al., 2017), we ran the analysis under different priors for the number of expected shifts (expectedNumberofShifts = 0.1; 1; 5; 10; 50) to assess the robustness of our result to prior parameterization. Each analysis was run for 20 million generations, sampling every 2000 generations. Convergence and ESS values were checked using the R package coda (Plummer et al., 2006) and outputs were analysed in the R package BAMMtools (Rabosky et al., 2014). Second, we used BayesRate (Silvestro et al., 2011) to test for different scenarios of diversification. We accounted for phylogenetic uncertainty by running the analysis over a sample of the posterior distribution of trees. Unlike BAMM, BayesRate does not estimate per-branch rate and require the specification of a priori branches with putatively varying rates of diversification. As we did not have strong assumptions about rate variation in the family, we implemented (1) a four-rate model in which each genus had its own rate and (2) a two-rate model with equal speciation and extinction rates for the sister genera Cyathea and Alsophila and a second set of rates for the remaining genera – Gymnosphaera and Sphaeropteris. We tested these two models against a constant-rate model by computing the marginal likelihoods of the three alternative scenarios. We then estimated speciation, extinction and net diversification rates under the best model by performing an MCMC analysis on a set of 100 trees randomly sampled from the posterior distribution of BEAST, running 10 million generations per tree. Finally, we computed the rates of net diversification (i.e. speciation minus extinction) per clade (for clade names see Supplementary Data Fig. S4) using the method of moments (Magallón and Sanderson, 2001; Meyer and Wiens, 2018). Net diversification rates were computed with both stem and crown ages and under zero extinction as well as high extinction (ε = 0.9; Magallón and Sanderson, 2001).
Morphological data
We scored a matrix of three quantitative morphological traits (trunk height, lamina length and petiole length) for the species included in the phylogeny. All these variables relate to plant height and crown size, which vary with light availability (Arens and Sanchez Baracaldo, 2000). Published taxonomic revisions for the Neotropics (Windisch, 1977, 1978; Proctor, 1989; Rojas-Alvarado, 2001; Christenhusz, 2009; Lehnert, 2009, 2011, 2012, 2014, 2016; Lehnert and Weigand, 2013, 2017; Maciel, et al., 2017), Africa (Holttum, 1981; Janssen and Rakotondrainibe, 2007, 2008) and Australasia (Holttum, 1959, 1963, 1964, 1965; Lehnert et al., 2013) were consulted for character coding, which also drew from our own field observations. We computed overall body size, defined as the sum of trunk height, petiole length and lamina length. All values were then log-transformed. Body size has been shown by Ramírez-Barahona et al. (2016) to be one of the main drivers of Cyatheaceae diversification, although in their study low taxonomic sampling prevented any precise modelling of the evolution of this trait through time. We therefore investigated the dynamics of morphological change through time, by modelling body size evolution on our newly inferred time-calibrated phylogenetic tree under a relaxed Brownian motion process, using reversible-jump MCMC (Eastman et al., 2011) as implemented in the function rjmcmc.bm of the R package geiger 2.0.6 (Harmon et al., 2008). This Bayesian method, which is analogous to the phenotypic evolutionary rate analysis of BAMM, quantifies the heterogeneity in the rate of evolution of a continuous character across branches in the phylogeny by estimating the number and placement of rate shifts. We did not test multiple models of trait evolution but only focused on Brownian motion because our goal here was to investigate the variation in the rate of evolution of traits through time and between lineages. We performed an analysis of 10 million MCMC generations, sampling every 1000 generations. Convergence was checked using the R package coda (Plummer et al., 2006). To make sure that phylogenetic uncertainty did not bias our estimates, we repeated the analysis on 100 trees randomly extracted from the posterior distribution of BEAST, each time running 5 million MCMC generations and sampling every 1000 generations. We then summarized the mean number of shifts and the mean rate per run. Finally, we re-evaluated the hypothesis that the rates of diversification in the Cyatheaceae were positively correlated with the rate of evolution of body size using structured rate permutations (STRAPP; Rabosky and Huang, 2016) as implemented in the R package BAMMtools (Rabosky et al., 2014).
Ecological data
We collected occurrence data for all species included in our phylogeny from nine online herbaria (E, GBIF, K, NY, SING, SpeciesLink, Tropicos, US, W) as well as three personal databases from our collaborators (Marcus Lehnert, Rodrigo Cámara Leret, Wilson D. Rodríguez Duque). All records were checked for (1) taxonomic correctness, using The Plant List (2013), and (2) regional GPS precision to country/state borders, using the World Administrative Borders from GADM (http://www.gadm.org/version2). We corrected the synonyms present in the databases with their accepted names and removed duplicates. Environmental data were extracted from CHELSA version 1.2 (Karger et al., 2017), recalculated to 2 × 2 km resolution in R 3.4.3 (R Core Team, 2016). We used the extract function from the raster package 2.6–7 (Hijmans and van Etten, 2014) and interpolated values between the four nearest raster cells (method = bilinear) to account for local GPS errors at the scale of a few kilometres. For each occurrence, we extracted the values of annual precipitation and maximum temperature of the warmest month, i.e. the two climatic variables that have been shown to have the strongest limiting effect on tree fern distributions (Bystriakova et al., 2011), and calculated their mean value for each species. The evolution of climatic preferences, along with morphological evolution, has been shown to be linked with increased diversification in the Cyatheaceae family (Ramírez-Barahona et al., 2016). Thus, we analysed the two climatic variables following the same procedure as for body size, to re-assess the evolution of climatic preferences within the family and its impact on species diversification. We tested for heterogeneity in the rate of evolution of ecological traits by modelling the evolution of precipitation and temperature along the phylogenetic tree. We chose this single-variable approach because the use of principal component scores from multivariate analyses such as PCA or OMI can potentially biased the results of macroevolutionary analyses (Uyeda et al., 2015). We performed a reversible-jump MCMC analysis of 10 million MCMC generations, sampling every 1000 generations, using the rjmcmc.bm function in R, and repeated the analysis on 100 phylogenetic trees from the posterior distribution. We then tested for trait-dependent diversification using STRAPP (Rabosky and Huang, 2016) as implemented in the R package BAMMtools (Rabosky et al., 2014). Additionally, we recorded the mean elevation for the species included in the phylogeny using the same published taxonomic revisions that we consulted to score morphological data (see above) and visualized their elevational distribution by making the histogram of mean elevation and boxplots of the maximum elevation at different latitudes.
RESULTS
Phylogenetic reconstruction
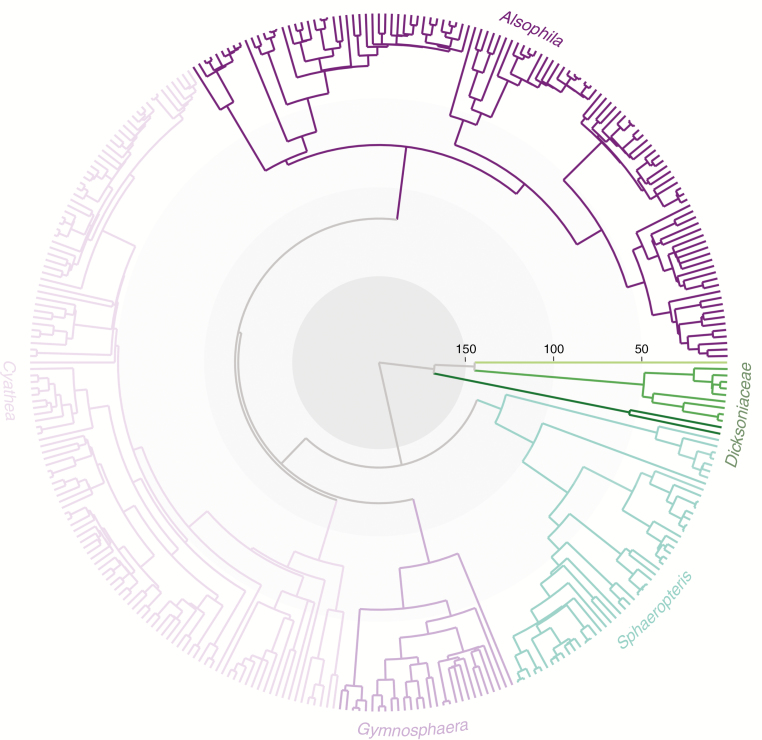
Our concatenated alignment of the three chloroplast genes was composed of 335 individuals and had a length of 3002 bp. For each DNA region, the best model of nucleotide substitution was GTR+G. The two runs of the FBD analysis in BEAST2 gave congruent results and all parameters had effective sample size values above 200. The topology of the maximum clade credibility tree supported the monophyly of the four genera (Fig. 1). The topology of the marginate-scaled clade was recovered with Gymnosphaera sister to a clade formed by Alsophila and Cyathea, but this relationship had a posterior probability of 0.33 and was therefore not supported (support values are provided in Supplementary Data Fig. S3). Relationships among closely related species were also poorly resolved, as indicated by low node posterior probabilities, especially within Cyathea and Alsophila. The Neotropical species of Alsophila formed a monophyletic clade. Divergence time estimates showed that Cyatheaceae diverged from their sister group, the Dicksoniaceae family, around 200 Ma [95 % HPD (highest posterior density) of stem node, 184.74–217.74] and the most recent common ancestor of all extant species is 140 Myr old (95 % HPD of crown node. 108.63–170.86). Sphaeropteris had a crown age of ~125 Myr and the remaining three genera had crown ages between 75 and 80 Myr. Overall, the node-dating analysis resulted in a similar topology with constantly younger age estimates, especially in the deeper nodes of the tree (Table 1).
Fig. 1.
Time-calibrated phylogeny of the Cyatheaceae. Numbers indicate million years.
Table 1.
Clade age estimates from the node-dating (ND) and FBD analyses
| NDmin | NDmax | NDmean | FBDmin | FBDmax | FBDmean | |
|---|---|---|---|---|---|---|
| Stem Cyatheaceae | 147.48 | 172.40 | 157.75 | 184.74 | 217.74 | 199.47 |
| Crown Cyatheaceae | 101.06 | 136.83 | 117.80 | 108.63 | 170.86 | 139.42 |
| Crown Alsophila | 55.41 | 86.86 | 71.02 | 52.32 | 99.08 | 75.15 |
| A. abbottii | 25.41 | 47.56 | 40.04 | 18.32 | 42.81 | 30.08 |
| A. cuspidata | 15.63 | 35.05 | 26.01 | 18.48 | 47.31 | 34.60 |
| A. australis | 42.01 | 76.82 | 60.28 | 39.61 | 85.98 | 62.84 |
| A. humilis | 11.20 | 32.00 | 20.66 | 9.99 | 32.00 | 20.30 |
| A. manniana | 11.19 | 51.15 | 29.47 | 10.42 | 52.12 | 28.94 |
| A. smithii | 36.30 | 65.77 | 50.78 | 33.83 | 73.20 | 52.78 |
| Crown Cyathea | 57.56 | 85.42 | 71.45 | 60.34 | 105.78 | 81.30 |
| C. cnemidaria | 19.99 | 41.69 | 30.51 | 18.96 | 45.59 | 31.69 |
| C. armata | 36.85 | 64.63 | 50.46 | 39.26 | 77.19 | 57.70 |
| C. decurrens | 33.26 | 70.88 | 51.71 | 30.65 | 80.23 | 55.42 |
| C. divergens | 32.13 | 54.68 | 44.74 | 29.41 | 59.08 | 48.44 |
| C. gibbosa | 26.64 | 45.34 | 35.76 | 29.14 | 57.03 | 37.56 |
| Crown Gymnosphaera | 54.79 | 91.34 | 73.18 | 53.63 | 110.77 | 81.35 |
| Gymnosphaera ‘Asian-clade’ | 33.29 | 60.83 | 46.51 | 31.95 | 68.28 | 48.79 |
| Gymnosphaera ‘Malagasy-clade’ | 4.37 | 31.66 | 16.04 | 2.94 | 28.04 | 14.06 |
| Crown Sphaeropteris | 81.45 | 126.48 | 106.72 | 89.13 | 160.38 | 124.53 |
| S. horrida | 10.97 | 35.11 | 22.58 | 10.59 | 37.27 | 22.76 |
| Sphaeropteris ‘Fourniera-clade’ | 16.27 | 56.28 | 35.18 | 15.34 | 60.81 | 35.35 |
| S. glauca | 29.08 | 56.79 | 49.33 | 27.97 | 62.17 | 50.82 |
| Sphaeropteris ‘Sarcopholis-clade’ | 17.27 | 42.57 | 29.39 | 15.98 | 44.65 | 29.77 |
| Sphaeropteris ‘Schizocaena-clade’ | 7.90 | 30.03 | 18.38 | 7.08 | 30.13 | 18.07 |
| Crown Dicksoniaceae | 117.83 | 151.95 | 136.93 | 128.38 | 201.51 | 167.10 |
| Crown Dicksonia | 40.01 | 72.26 | 55.03 | 23.76 | 72.24 | 47.46 |
Diversification rates
The results of the BAMM analyses with different priors on the number of rate shifts were similar, indicating that the analyses were robust to prior influence. Therefore, we present only the results of the analysis with a prior rate shift of 1. The rate of diversification slowly increased through time [mean net diversification rate 0.039 (95 % HPD 0.004–0.055) lineages per Myr at the root and 0.070 (95 % HPD 0.055–0.078) lineages per Myr at the tips] and BAMM detected no shift in the rate of diversification across the Cyatheaceae (Fig. 2). In BayesRate, although the model with the highest marginal likelihood was the two-rate model (Supplementary Data Table S2), with higher posterior net diversification rate for Alsophila and Cyathea (0.0608 lineages per Myr; 95 % HPD 0.040–0.084) than for Gymnosphaera and Sphaeropteris (0.0242 lineages per Myr; 95 % HPD 0.006–0.042), neither speciation nor extinction rates were actually significantly different in the two groups (Supplementary Data Fig. S7). Per-clade rates of net diversification estimated with the methods of moments varied between 0.025 and 0.142 lineages per Myr under no extinction and between 0.009 and 0.072 lineages per Myr when extinction was set to 0.9 (Supplementary Data Table S3). Despite some differences among rate estimates, all methods concurred in finding low diversification rates, with little evidence for strong rate heterogeneity across branches.
Fig. 2.
Results of BAMM analysis. Best configuration (left) and net diversification rate through time (right).
Morphological evolution
The analysis of body size evolution showed a constant background rate of 0.0038 and a single shift with a posterior probability of 0.48 towards an increased rate detected in a small clade of four species of Cyathea (Supplementary Data Fig. S8). This result was robust to phylogenetic uncertainty, as shown by the distributions of the mean number of shifts and mean rate across the 100 additional runs (Supplementary Data Fig. S10). The result of the STRAPP analysis indicated no significant correlation between body size and net rates of diversification (Table 2).
Table 2.
Correlation coefficients from STRAPP analyses
| Body size | Max temperature | Annual precipitation | |
|---|---|---|---|
| R | −0.069 | 0.032 | 0.002 |
| P value | 0.904 | 0.940 | 0.928 |
Climatic niche evolution
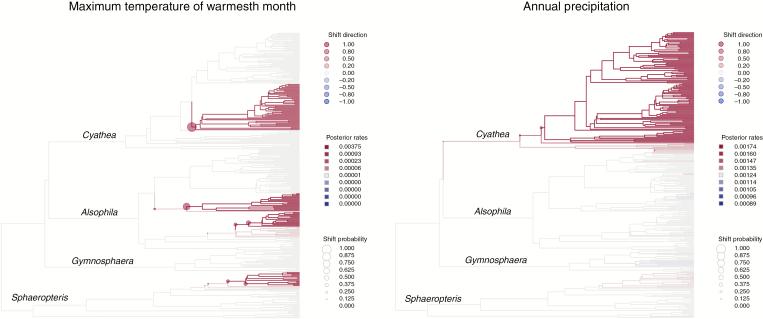
The analysis of trait evolution recovered a heterogeneous process for the evolution of temperature preferences, with a median posterior rate of 0.00001 and four shifts towards a rate of 0.00375 with posterior probabilities between 0.39 and 0.99 (Fig. 3), in the clades exhibiting the highest mean elevation and the widest elevational range. For annual precipitation, the median posterior rate was 0.00124 and rate acceleration was detected for the entire Neotropical Cyathea clade, which showed a rate of 0.00174 (Fig. 3). However, the posterior probability of this shift was only 0.14, indicating poor support (Fig. 3). The 100 replicated MCMC analyses showed that topological variation did not significantly impact the estimated rates and number of shifts of these two climatic variables (Supplementary Data Fig. S7). The STRAPP analysis showed that neither maximum temperature nor annual precipitation was correlated with net rates of diversification (Table 2).
Fig. 3.
Posterior rates of the evolution of maximum temperature (left) and precipitation (right) modelled under a relaxed Brownian motion.
DISCUSSION
In this study, we evaluated several hypotheses related to the evolutionary history of a diverse clade of tree ferns and investigated the impact of morphological and climatic traits on the diversification of the group. We asked whether the build-up of species diversity in this group occurred through a steady accumulation of species through time or whether their great diversity originated instead from variations in the rate of diversification between lineages. We then investigated the role played by morphological and ecological traits in their evolution and tested the impact of these traits on the rates of diversification in the group.
Based on an unprecedented sampling of nearly half of extant species diversity and integrating data from the rich fossil record of tree ferns, we estimated divergence times using the FBD model. In agreement with published phylogenies (Janssen et al., 2008; Korall and Pryer 2014; Ramírez-Barahona et al., 2016), we found support for four main clades within Cyatheaceae, which are here referred to as the genera Alsophila, Cyathea, Gymnosphaera and Sphaeropteris. We also recovered a monophyletic clade for the Neotropical species of Alsophila, which were split between two different clades in a previous phylogeny with lower sampling (Korall and Pryer, 2014). As in most previous studies (Korall et al., 2007; Janssen et al., 2008; Korall and Pryer, 2014; Ramírez-Barahona et al., 2016), we found low support for the phylogenetic relationships among Alsophila, Cyathea and Gymnosphaera, which is not surprising given that our molecular dataset is restricted to three chloroplast loci and that Cyatheaceae have a notably low rate of molecular evolution (Korall et al., 2010). Since additional plastid sequences available in public repositories did not appear to improve substantially the resolution of the phylogeny, it is likely that only the addition of nuclear genes could yield a more robust resolution of the deep and shallow phylogenetic splits in this slowing evolving group. Alternatively, the low amount of divergence at the molecular level between Alsophila, Cyathea and Gymnosphaera (Fig. 1) may result from a high level of incomplete lineage sorting resulting in a ‘hard’ polytomy in this part of the phylogenetic tree.
Whether we used the FBD process or node dating, the recovered age estimates point to an older origin of the family than in most of the previous estimates (Janssen et al., 2008; Korall and Pryer, 2014; Ramírez-Barahona et al., 2016; Testo and Sundue, 2016; Lehtonen et al., 2017). These differences in dating can be explained by several factors, including the different taxonomic assignment of the fossil K. mirabilis and the better taxon sampling and more realistic evolutionary models used in our phylogenetic analyses. The inclusion of old fossils representing stem lineages in the FBD analyses probably explains why the age estimates recovered by this method are older than those obtained with node calibration (Saladin et al., 2017).
Results from our diversification analyses using different methods are coherent, and consistently indicate that the rate of diversification in Cyatheaceae was low and underwent very little variation throughout their long evolutionary history. This finding is consistent with a near-stable sampled diversity in the fossil record of Cyatheales (Lehtonen et al., 2017), but contrasts strongly with previous reports of dramatic heterogeneity in the rates of diversification among Cyatheaceae (Janssen et al., 2008; Ramírez-Barahona et al., 2016). Previous studies were based on just about 10 % of the extant species and we believe that the 5-fold increase in taxon sampling in our analysis and advances in molecular dating methods are the basis of the divergent conclusions regarding the tempo of diversification in the group. Our estimates of the net diversification rates are 10–40 times slower than those of the fastest plant radiations reported to date, such as in Dianthus (Valente et al., 2010), Lupinus (Hughes and Eastwood, 2006) or Andean Campanulaceae (Lagomarsino et al., 2016). The estimates obtained for the Cyatheaceae are nevertheless among the intermediate values for fern families, where net diversification rates range from 0.001 in Thyrsopteridaceae to 0.132 in Athyriaceae (Testo and Sundue, 2018). However, apart from the Hymenophyllaceae, fern families with more than 600 species are generally younger clades that diversified at higher rates than the Cyatheaceae, whereas many of the early branching fern clades are characterized by much lower current diversity (Testo and Sundue, 2016, 2018). The Cyatheaceae therefore exhibit an uncommon pattern of an old lineage that has successfully diversified by accumulating species at a very low rate through time without any significant among-clade variation. The exact factors that have allowed the Cyatheaceae to diversify so extensively when compared with other tree fern families remain to be investigated, but this will require a much denser taxonomic sampling in the other families than what we have in our current study.
Our analyses have also shown that the modes of evolution differed substantially between morphological and climatic traits. The rate of body size evolution was remarkably low and constant across the phylogeny. This may seem surprising given that tree ferns differ considerably in size, ranging from minute rock-hugging species with leaves 15 cm long to very large species 20 m tall with 5-m long leaves. However, dwarf species are found only in Neotropical species of Cyathea, where dwarf growth has repeatedly evolved as an adaptation to exposed rocky outcrop habitats. This is most conspicuous in the Hymenophyllopsis group, with 11 species (Maciel et al., 2017), but also concerns about 25 other species, which, based on morphology, should be distributed in several other clades. Due to the inaccessibility of these habitats, many of these species have only recently been described (Lehnert, 2006; Tejedor and Calatayud, 2017; Acuña-Tarazona et al., 2018), and their diversity may be underestimated. Acceleration in the rate of body size evolution may thus be expected in dwarf Cyathea, but the shift detected in a clade of four of these species was not strongly supported (Supplementary Data Fig. S8), which may be due to the limited sampling of these groups. Although undersampling of dwarf species may underestimate morphological evolution in Neotropical Cyathea, it is unlikely to affect the overall result of low and nearly constant rate of body size evolution in the family. Korall et al. (2010) reported exceptionally slow rates of molecular evolution in tree ferns in comparison with other fern lineages, while a recent study found a negative correlation between substitution rates and body size in tree ferns (Barrera-Redondo et al., 2018), but it is unclear whether this could actually be the underlying factor limiting the rate of phenotypic evolution. Given this result and the homogeneous rate of diversification estimated, it is thus not surprising that we did not detect any association between body size and diversification.
In contrast, the evolution of climatic preferences followed a more heterogeneous process. For precipitation, a small rate increase was detected in the clade of Neotropical Cyathea, whereas for temperature several shifts occurred independently within four clades in three different genera (Fig. 3). These four clades correspond to a primarily Andean clade in Cyathea and primarily New Guinean clades in Alsophila and Sphaeropteris. We interpret these patterns as the results of the topo-climatic heterogeneity of these mountains: the Andes and New Guinea are the most extensive and the highest tropical mountain systems, allowing niche shifts along the elevational gradient. However, whereas New Guinea mostly has humid climatic conditions in the mountains, the Andes have a much wider range of precipitation regimes, ranging from xeric to hyper-humid, which probably explains why only a single shift was detected for precipitation in the Neotropical clade of Cyathea. However, these shifts in the evolution of climatic preferences likely triggered by the colonization of mountain ranges were decoupled from species diversification, as shown by the lack of association between climatic traits and the rates of net diversification. This result was unexpected given that tropical mountains probably played an important role during the evolutionary history of Cyatheaceae. Indeed, these ecosystems harbour most of the global fern diversity, and colonization of montane habitats has been hypothesized to be the primary driver of diversification in several fern lineages (Kessler et al., 2016). For example, in Polypodiaceae it has been shown that shifts in elevation, but not in leaf area, were positively associated with the rates of diversification, supporting the idea that exploration of new habitats rather than morphological innovation drove the diversification of the group (Sundue et al., 2015). Environmental factors were also associated with diversification in a study using palaeontological data to investigate the drivers of diversification in all ferns (Lehtonen et al., 2017), but the effect was correlated with extinction rates and not origination rates, which were largely diversity-dependent. In Cyatheaceae, tropical mountains likely provided new suitable habitats in which topographical complexity triggered allopatric speciation. Indeed, geographical isolation favoured by geological events has been suggested as an important factor in promoting differentiation in tree ferns (Ramírez-Barahona and Luna-Vega, 2015). Although the distribution of Cyatheaceae reaches high latitudes, their altitudinal range is greatest closer to the Equator and decreases towards higher latitudes (Supplementary Data Fig. S9). The appearance of montane habitats in tropical regions may have compensated the range contraction that Cyatheaceae underwent during their evolutionary history, as shown by the latitudinal range of their fossil record exceeding their extant distribution (Collinson, 2001). For example, Late Cretaceous fossil spores (Kuylisporites) similar to the spores of extant Cyathea are found in Siberia. Records of Cyatheaceae are also known from Antarctica in the Eocene, but are restricted to Central America by the Pliocene (Collinson, 2001). Therefore, rather than triggering a sharp biome shift that could increase net diversification rates, dispersals into higher elevations in the tropics may have simply represented an opportunity for Cyatheaceae to track or slightly expand their preferred climatic conditions. This is coherent with the fact that tree ferns are restricted to warm and humid environments with low seasonality and exhibit strong niche conservatism (Bystriakova et al., 2011; Sosa et al., 2016).
Our findings regarding morphological and climatic niche evolution disagree with the results of Ramírez-Barahona et al. (2016), who reported a positive correlation between the rates of body size and climatic niche evolution and the rates of diversification. Although our phylogeny is neither complete nor fully supported, we argue that our modelling of trait evolution, which took into account phylogenetic uncertainty, is more realistic than the approach used by Ramírez-Barahona et al. (2016), who circumvented a poor taxonomic sampling by modelling trait evolution on simulated trees, a procedure that has been shown to yield biased estimates (Rabosky, 2015). Conversely, and in line with our own results, a recent study across all ferns found a lack of association between the rates of diversification and the rates of body size evolution, a result that held true when looking at the Cyatheaceae in particular (Testo and Sundue, 2018).
Conclusions
Our study provides a new perspective on the evolution of one of the most conspicuous vegetation elements of tropical montane forests. Rather than interpreting the present-day diversity of the Cyatheaceae as the result of adaptive bursts of diversification, as previously suggested (Ramírez-Barahona et al., 2016), our findings indicate that their diversity steadily accumulated over time. Colonization of mountain habitats seems to have triggered a small acceleration of niche evolution but it was not followed by rapid radiation, unlike what has been observed in several plant groups, particularly in the Andes (Hughes and Atchison, 2015; Uribe-Convers and Tank, 2015; Pérez-Escobar et al., 2017; Pouchon et al., 2018; but see Wagner et al., 2013; Salariato et al., 2016). Many empirical examples support the view that changes in diversification rates linked to trait evolution are the primary driver of species diversity (Matuszak et al., 2016; Onstein et al., 2017), but counter-examples have shown that it is not a ubiquitous pattern (Vamosi and Vamosi, 2011; Rabosky et al., 2012). The present study provides further evidence that a species-rich clade can result from the slow and steady accumulation of species over long evolutionary times and that rapid morphological differentiation, the evolution of key innovations and niche divergence are not a prerequisite for a clade to thrive for hundreds of millions of years.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: correlation of branch length of the two RaxML trees. Figure S2: boxplots of bootstrap values for the two phylogenetic trees. Figure S3: clades used to compute net diversification rates with the method of moments. Figure S4: maximum clade credibility tree from the BEAST2 analyses under the FBD process. Figure S5: comparison of parameter estimates from the FBD run with several fossils per taxon and a single fossil per taxon. Figure S6: correlation of clade age estimates between the FBD run with a single fossil per taxon and the FBD run with several fossils per taxon. Figure S7: differences between the posterior rates of the two groups for the best model in the BayesRates analysis. Figure S8: posterior rates of body size evolution modelled under relaxed Brownian motion. Figure S9: elevational distribution of the Cyatheaceae. Figure S10: histograms of the mean rate and mean number of shifts across the 100-replicate rjmcmc analysis for body size, annual precipitation and maximum temperature of warmest month. Table S1: fossils included in the FBD analysis. Table S2: model comparison in BayesRates and parameter estimation for the best model with two rates. Table S3: net diversification rate estimates from the method of moments.
FUNDING
M.L. received funding from the German Research Foundation (LE 1826/3 and LE 1826/4) for conducting field work and sequencing, and from SYNTHESYS (GB-TAF-4927 and GB-TAF-6305) for herbarium studies. N.S. and M.K. received funding from the Swiss National Science Foundation (CRSII3-147630) and N.S. from the University of Lausanne.
ACKNOWLEDGEMENTS
For help during field work and providing samples, we thank G. Calatayud, L. F. Giraldo G., W. D. Rodríguez D. and A. Tejedor. We thank the Vital-IT facilities of the Swiss Institute of Bioinformatics for the use of their HPC infrastructure. M.K., N.S. and O.L. designed the study. M.L. collected and identified the samples and compiled morphological data and elevation data. M.L., A.W. and S.N. carried out the molecular laboratory work. A.W. compiled GPS occurrences and extracted climatic variables. O.L. performed phylogenetics and macroevolutionary analyses with the help of J.R., D.S. and N.S. O.L. led the writing with significant contributions from all authors. The authors declare no conflict of interest. DNA sequences will be deposited in GenBank. Phylogenetic trees, matrices of morphological and ecological data will be available in Dryad. Genbank accessions and the detailed species list is available at the following doi:10.5281/zenodo.3483489.
LITERATURE CITED
- Acuña-Tarazona M, Huamán-Melo E, Toledo-Aceves T, Mehltreter K. 2018. Cyathea leoniae (Cyatheaceae), a new pinnate-pinnatifid tree fern species from Northern Peru. Phytotaxa 344: 191–197. [Google Scholar]
- Andriananjamanantsoa HN, Engberg S, Louis EE, Brouillet L. 2016. Diversification of Angraecum (Orchidaceae, Vandeae) in Madagascar: revised phylogeny reveals species accumulation through time rather than rapid radiation. PLoS ONE 11: e0163194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli A, Zizka A, Silvestro D, Scharn R, Cascales-Miñana B, Bacon CD. 2015. An engine for global plant diversity: highest evolutionary turnover and emigration in the American tropics. Frontiers in Genetics 6: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arens NC, Sanchez Baracaldo P. 2000. Variation in tree fern stipe length with canopy height: tracking preferred habitat through morphological change. American Fern Journal 90: 1–15. [Google Scholar]
- Baker WJ, Couvreur TLP. 2013. Global biogeography and diversification of palms sheds light on the evolution of tropical lineages. II. Diversification history and origin of regional assemblages. Journal of Biogeography 40: 286–298. [Google Scholar]
- Barrera-Redondo J, Ramírez-Barahona S, Eguiarte LE. 2018. Rates of molecular evolution in tree ferns are associated with body size, environmental temperature and biological productivity. Evolution 72: 1050–1062. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, et al. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkopf H, Onstein RE, Cafasso D, Schlüter PM, Cozzolino S. 2015. Multiple shifts to different pollinators fuelled rapid diversification in sexually deceptive Ophrys orchids. New Phytologist 207: 377–389. [DOI] [PubMed] [Google Scholar]
- Brown JH. 2014. Why are there so many species in the tropics? Journal of Biogeography 41: 8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun-Lund S, Verstraete B, Kjellberg F, Rønsted N. 2018. Rush hour at the museum – diversification patterns provide new clues for the success of figs (Ficus L., Moraceae). Acta Oecologica 90: 4–11. [Google Scholar]
- Bystriakova N, Schneider H, Coomes D. 2011. Evolution of the climatic niche in scaly tree ferns (Cyatheaceae, Polypodiopsida). Botanical Journal of the Linnean Society 165: 1–19. [Google Scholar]
- Cardillo M, Pratt R. 2013. Evolution of a hotspot genus: geographic variation in speciation and extinction rates in Banksia (Proteaceae). BMC Evolutionary Biology 13: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenhusz MJM. 2009. New combinations and an overview of Cyathea subg Hymenophyllopsis (Cyatheaceae). Phytotaxa 1: 37–42. [Google Scholar]
- Collinson ME. 2001. Cainozoic ferns and their distribution. Brittonia 53: 173–235. [Google Scholar]
- Condamine FL, Silva-Brandão KL, Kergoat GJ, Sperling FAH. 2012. Biogeographic and diversification patterns of Neotropical Troidini butterflies (Papilionidae) support a museum model of diversity dynamics for Amazonia. BMC Evolutionary Biology 12: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur TLP, Forest F, Baker WJ. 2011a Origin and global diversification patterns of tropical rain forests: inferences from a complete genus-level phylogeny of palms. BMC Biology 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur TLP, Pirie MD, Chatrou LW, et al. 2011. b Early evolutionary history of the flowering plant family Annonaceae: steady diversification and boreotropical geodispersal. Journal of Biogeography 38: 664–680. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz LFH, Harmon LJ, Sugawara MT, Miller ET, Pennell MW. 2019. Macroevolutionary diversification rates show time dependency. Proceedings of the National Academy of Sciences 116: 7403–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong SY, Zuo ZY. 2018. On the recognition of Gymnosphaera as a distinct genus in Cyatheaceae. Annals of the Missouri Botanical Garden 103: 1–24. [Google Scholar]
- Eastman JM, Alfaro ME, Joyce P, Hipp AL, Harmon LJ. 2011. A novel comparative method for identifying shifts in the rate of character evolution on trees. Evolution 65: 3578–3589. [DOI] [PubMed] [Google Scholar]
- Eiserhardt WL, Couvreur TLP, Baker WJ. 2017. Plant phylogeny as a window on the evolution of hyperdiversity in the tropical rainforest biome. New Phytologist 214: 1408–1422. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24: 129–131. [DOI] [PubMed] [Google Scholar]
- Heath TA, Hedtke SM, Hillis DM. 2008. Taxon sampling and the accuracy of phylogenetic analyses. Journal of Systematics and Evolution 46: 239–257. [Google Scholar]
- Heath TA, Huelsenbeck JP, Stadler T. 2014. The fossilized birth-death process for coherent calibration of divergence-time estimates. Proceedings of the National Academy of Sciences of the USA 111: E2957–E2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao Diaz LF, Harmon LJ, Sugawara MTC, Pennell MW. 2019. Macroevolutionary diversification rates show time-dependency. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ. 2015. Raster: geographic data analysis and modeling. R package version 2.4-15. http://CRAN.R-project.org/package=raster [Google Scholar]
- Hijmans RJ, van Etten J. 2014. raster: Geographic data analysis and modeling. R package version 2. [Google Scholar]
- Holttum RE. 1959. Pteridophyta. Flora Malesiana Series 2 1: 1–64. [Google Scholar]
- Holttum RE. 1963. Cyatheaceae. Flora Malesiana Series 2 1: 65–176. [Google Scholar]
- Holttum RE. 1964. The tree ferns of the genus Cyathea in Australasia and the Pacific. Blumea 12: 241–274. [Google Scholar]
- Holttum RE. 1965. Tree ferns of the genus Cyathea Sm. in Asia (excluding Malaysia). Kew Bulletin 19: 463–487. [Google Scholar]
- Holttum RE. 1981. The tree ferns of Africa. Kew Bulletin 36: 463–482. [Google Scholar]
- Hughes CE, Atchison GW. 2015. The ubiquity of alpine plant radiations: from the Andes to the Hengduan Mountains. New Phytologist 207: 275–282. [DOI] [PubMed] [Google Scholar]
- Hughes C, Eastwood R. 2006. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proceedings of the National Academy of Sciences of the USA 103: 10334–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen T, Rakotondrainibe F. 2007. An update of the revision of Cyathea subgen. Alsophila sect. Gymnosphaera (Cyatheaceae) in Madagascar and the Comoros including a discussion of putative hybridization events. Adansonia 29: 195–213. [Google Scholar]
- Janssen T, Rakotondrainibe F. 2008. A revision of the indusiate scaly tree ferns (Cyatheaceae, Cyathea subgen. Alsophila sect. Alsophila) in Madagascar, the Comoros and the Seychelles. Adansonia 30: 221–376. [Google Scholar]
- Janssen T, Bystriakova N, Rakotondrainibe F, Coomes D, Labat J-N, Schneider H. 2008. Neoendemism in Madagascan scaly tree ferns results from recent, coincident diversification bursts. Evolution 62: 1876–1889. [DOI] [PubMed] [Google Scholar]
- Karger DN, Conrad O, Böhner J, et al. 2017. Climatologies at high resolution for the earth’s land surface areas. Scientific Data 4: 170122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M, Karger DN, Kluge J. 2016. Elevational diversity patterns as an example for evolutionary and ecological dynamics in ferns and lycophytes. Journal of Systematics and Evolution 54: 617–625. [Google Scholar]
- Korall P, Pryer KM. 2014. Global biogeography of scaly tree ferns (Cyatheaceae): evidence for Gondwanan vicariance and limited transoceanic dispersal. Journal of Biogeography 41: 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korall P, Conant DS, Metzgar JS, Schneider H, Pryer KM. 2007. A molecular phylogeny of scaly tree ferns (Cyatheaceae). American Journal of Botany 94: 873–886. [DOI] [PubMed] [Google Scholar]
- Korall P, Schuettpelz E, Pryer KM. 2010. Abrupt deceleration of molecular evolution linked to the origin of arborescence in ferns. Evolution 64: 2786–2792. [DOI] [PubMed] [Google Scholar]
- Lagomarsino LP, Condamine FL, Antonelli A, Mulch A, Davis CC. 2016. The abiotic and biotic drivers of rapid diversification in Andean bellflowers (Campanulaceae). New Phytologist 210: 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert M. 2006. New species and records of tree ferns (Cyatheaceae, Pteridophyta) from the northern Andes. Organisms Diversity and Evolution 6: 321–322. [Google Scholar]
- Lehnert M. 2009. Resolving the Cyathea caracasana complex (Polypodiopsida: Cyatheaceae). Stuttgarter Beiträge zur Naturkunde A, Neue Serie 2: 409–445. [Google Scholar]
- Lehnert M. 2011. Species of Cyathea in America related to the western Pacific species C. decurrens. Phytotaxa 26: 39–59. [Google Scholar]
- Lehnert M. 2012. A synopsis of the species of Cyathea (Cyatheaceae-Polypodiopsida) with pinnate to pinnate-pinnatifid frond. Phytotaxa 61: 17–36. [Google Scholar]
- Lehnert M. 2014. Do you know Cyathea divergens (Cyatheaceae-Polypodiopsida)? Phytotaxa 161: 1–42. [Google Scholar]
- Lehnert M. 2016. A synopsis of the exindusiate species of Cyathea (Cyatheaceae-Polypodiopsida) with bipinnate-pinnatifid or more complex fronds, with a revision of the C. lasiosora complex. Phytotaxa 243: 1–53. [Google Scholar]
- Lehnert M, Weigand A. 2013. A proposal to distinguish several taxa in the Brazilian tree fern Cyathea corcovadensis (Cyatheaceae). Phytotaxa 155: 35–49. [Google Scholar]
- Lehnert M, Weigand A. 2017. A synopsis of the Neotropical species of Cyathea (Cyatheaceae; Polypodiopsida) with bipinnate fronds. Brittonia 69: 71–90. [Google Scholar]
- Lehnert M, Coritico FP, Darnaedi D, et al. 2013. Taxonomic and ecological notes on the Alsophila hornei complex (Cyatheaceae-Polypodiopsida), with the description of the new species A. phlebodes from New Guinea. Systematic Botany 38: 875–886. [Google Scholar]
- Lehtonen S, Silvestro D, Karger DN, et al. 2017. Environmentally driven extinction and opportunistic origination explain fern diversification patterns. Scientific Reports 7: 4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsios G, Sims CA, Wüest RO, Pearman PB, Zimmermann NE, Salamin N. 2012. Mutualism with sea anemones triggered the adaptive radiation of clownfishes. BMC Evolutionary Biology 12: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciel S, Lehnert M, Hirai RY, Prado J. 2017. Three new species of the Cyathea “Hymenophyllopsis” clade (Cyatheaceae) from Venezuela and Brazil. Phytotaxa 329: 159–166. [Google Scholar]
- Magallón S, Sanderson MJ. 2001. Absolute diversification rates in angiosperm clades. Evolution 55: 1762–1780. [DOI] [PubMed] [Google Scholar]
- Matuszak S, Favre A, Schnitzler J, Muellner-Riehl AN. 2016. Key innovations and climatic niche divergence as drivers of diversification in subtropical Gentianinae in southeastern and eastern Asia. American Journal of Botany 103: 899–911. [DOI] [PubMed] [Google Scholar]
- Meyer ALS, Wiens JJ. 2018. Estimating diversification rates for higher taxa: BAMM can give problematic estimates of rates and rate shifts. Evolution 72: 39–53. [DOI] [PubMed] [Google Scholar]
- Mohr BAR, Lazarus DB. 1994. Paleobiogeographic distribution of Kuylisporites and its possible relationship to the extant fern genus Cnemidaria (Cyatheaceae). Annals of the Missouri Botanical Garden 81: 758–767. [Google Scholar]
- Moore BR, Höhna S, May MR, Rannala B, Huelsenbeck JP. 2016. Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proceedings of the National Academy of Sciences of the USA 113: 9569–9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalingum NS, Schneider H, Pryer KM. 2007. Molecular phylogenetic relationships and morphological evolution in the heterosporous fern genus Marsilea. Systematic Botany 32: 16–25. [Google Scholar]
- Noben S, Kessler M, Quandt D, et al. 2017. Biogeography of the Gondwanan tree fern family Dicksoniaceae – a tale of vicariance, dispersal and extinction. Journal of Biogeography 44: 2648–2659. [Google Scholar]
- Onstein RE, Baker WJ, Couvreur TLP, Faurby S, Svenning JC, Kissling WD. 2017. Frugivory-related traits promote speciation of tropical palms. Nature Ecology and Evolution 1: 1903–1911. [DOI] [PubMed] [Google Scholar]
- Pérez-Escobar OA, Chomicki G, Condamine FL, et al. 2017. Recent origin and rapid speciation of Neotropical orchids in the world’s richest plant biodiversity hotspot. New Phytologist 215: 891–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer M, Best N, Cowles K, Vines K. 2006. CODA: convergence diagnosis and output analysis for MCMC. R News 6: 7–11. [Google Scholar]
- Pouchon C, Fernández A, Nassar JM, et al. 2018. Phylogenomic analysis of the explosive adaptive radiation of the Espeletia complex (Asteraceae) in the tropical Andes. Systematic Biology 67: 1041–1060. [DOI] [PubMed] [Google Scholar]
- PPG I. 2016. A community-derived classification for extant lycophytes and ferns. Journal of Systematics and Evolution 54: 563–603. [Google Scholar]
- Proctor GR. 1989. Ferns of Puerto Rico and the Virgin Islands. Memoirs of the New York Botanical Garden, Vol. 53. New York: New York Botanical Garden Press. [Google Scholar]
- R Core Team. 2016. R: A language and environment for statistical computing. Versión 3.4.3. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rabosky DL. 2014. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE 9: e89543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL. 2015. No substitute for real data: a cautionary note on the use of phylogenies from birth-death polytomy resolvers for downstream comparative analyses. Evolution 69: 3207–3216. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Huang H. 2016. A robust semi-parametric test for detecting trait-dependent diversification. Systematic Biology 65: 181–193. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, Slater GJ, Alfaro ME. 2012. Clade age and species richness are decoupled across the eukaryotic tree of life. PLoS Biology 10: e1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, Grundler M, Anderson C, et al. 2014. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods in Ecology and Evolution 5: 701–707. [Google Scholar]
- Rabosky DL, Mitchell JS, Chang J. 2017. Is BAMM flawed? Theoretical and practical concerns in the analysis of multi-rate diversification models. Systematic Biology 66: 477–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. 2009. Tracer: MCMC trace analysis tool, version 1.5. http://tree.bio.ed.ac.uk/software/tracer. [Google Scholar]
- Ramírez-Barahona S, Luna-Vega I. 2015. Geographic differentiation of tree ferns (Cyatheales) in tropical America. American Fern Journal 105: 73–85. [Google Scholar]
- Ramírez-Barahona S, Barrera-Redondo J, Eguiarte LE. 2016. Rates of ecological divergence and body size evolution are correlated with species diversification in scaly tree ferns. Proceedings of the Royal Society B: Biological Sciences 283: 20161098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Alvarado AF. 2001. Nuevas especies, nombres nuevamente utilizados y nuevas distribuciones en los helechos arborescentes (Filicales: Cyatheaceae) para el Neotrópico. Revista de Biología Tropical 49: 453–466. [PubMed] [Google Scholar]
- Saladin B, Leslie AB, Wüest RO, et al. 2017. Fossils matter: improved estimates of divergence times in Pinus reveal older diversification. BMC Evolutionary Biology 17: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salariato DL, Zuloaga FO, Franzke A, Mummenhoff K, Al-Shehbaz IA. 2016. Diversification patterns in the CES clade (Brassicaceae tribes Cremolobeae, Eudemeae, Schizopetaleae) in Andean South America. Botanical Journal of the Linnean Society 181: 543–566. [Google Scholar]
- Serrano-Serrano ML, Rolland J, Clark JL, Salamin N, Perret M. 2017. Hummingbird pollination and the diversification of angiosperms: an old and successful association in Gesneriaceae. Proceedings of the Royal Society B: Biological Sciences 284: 20162816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestro D, Schnitzler J, Zizka G. 2011. A Bayesian framework to estimate diversification rates and their variation through time and space. BMC Evolutionary Biology 11: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestro D, Zizka G, Schulte K. 2014. Disentangling the effects of key innovations on the diversification of Bromelioideae (Bromeliaceae). Evolution 68: 163–175. [DOI] [PubMed] [Google Scholar]
- Sosa V, Ornelas JF, Ramírez-Barahona S, Gándara E. 2016. Historical reconstruction of climatic and elevation preferences and the evolution of cloud forest-adapted tree ferns in Mesoamerica. PeerJ 4: e2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundue MA, Testo WL, Ranker TA. 2015. Morphological innovation, ecological opportunity, and the radiation of a major vascular epiphyte lineage. Evolution 69: 2482–2495. [DOI] [PubMed] [Google Scholar]
- Tejedor A, Calatayud G. 2017. Eleven new scaly tree ferns (Cyathea: Cyatheaceae) from Peru. American Fern Journal 107: 156–191. [Google Scholar]
- Testo W, Sundue M. 2016. A 4000-species dataset provides new insight into the evolution of ferns. Molecular Phylogenetics and Evolution 105: 200–211. [DOI] [PubMed] [Google Scholar]
- Testo WL, Sundue MA. 2018. Are rates of species diversification and body size evolution coupled in the ferns? American Journal of Botany 105: 525–535. [DOI] [PubMed] [Google Scholar]
- The Plant List 2013. http://www.theplantlist.org/.
- Uribe-Convers S, Tank DC. 2015. Shifts in diversification rates linked to biogeographic movement into new areas: an example of a recent radiation in the Andes. American Journal of Botany 102: 1854–1869. [DOI] [PubMed] [Google Scholar]
- Uyeda JC, Caetano DS, Pennell MW. 2015. Comparative analysis of principal components can be misleading. Systematic Biology 64: 677–689. [DOI] [PubMed] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180. [DOI] [PubMed] [Google Scholar]
- Valente LM, Savolainen V, Vargas P. 2010. Unparalleled rates of species diversification in Europe. Proceedings of the Royal Society B: Biological Sciences 277: 1489–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamosi JC, Vamosi SM. 2011. Factors influencing diversification in angiosperms: at the crossroads of intrinsic and extrinsic traits. American Journal of Botany 98: 460–471. [DOI] [PubMed] [Google Scholar]
- de Vos JM, Hughes CE, Schneeweiss GM, Moore BR, Conti E. 2014. Heterostyly accelerates diversification via reduced extinction in primroses. Proceedings of the Royal Society B: Biological Sciences 281: 20140075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner N, Silvestro D, Brie D, et al. 2013. Spatio-temporal evolution of Fosterella (Bromeliaceae) in the Central Andean biodiversity hotspot. Journal of Biogeography 40: 869–880. [Google Scholar]
- Wiens JJ. 2017. What explains patterns of biodiversity across the Tree of Life? New research is revealing the causes of the dramatic variation in species numbers across branches of the Tree of Life. BioEssays 39: 1600128. [DOI] [PubMed] [Google Scholar]
- Wilson R, Heinrichs J, Hentschel J, Gradstein SR, Schneider H. 2007. Steady diversification of derived liverworts under Tertiary climatic fluctuations. Biology Letters 3: 566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windisch PG. 1977. Synopsis of the genus Sphaeropteris with a revision of the neotropical exindusiate species. Botanische Jahrbücher der Systematik 92: 176–198. [Google Scholar]
- Windisch PG. 1978. The systematics of the group of Sphaeropteris hirsuta (Cyatheaceae). In: Maguire B, ed. The botany of the Guayana Highland. Memoirs of the New York Botanical Garden, Vol. 29 New York: New York Botanical Garden Press, 2–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.