Abstract
Ischemia-reperfusion (I/R) injury occurs during cardiac surgery and is the major factor leading to heart dysfunction and heart failure. Our previous study showed that gene and microRNA expression profiles are altered in heart grafts with extended I/R injury. In this study, we, for the first time, demonstrated that I/R injury upregulates the expression of Polo-like kinase 2 (Plk2) but decreases miR-128 expression in heart cells both in vitro and in vivo. Silencing Plk2 using small interfering RNA (siRNA) protects cells from Antimycin A-induced cell apoptosis/death. Silencing Plk2 also decreases phosphorylated p65 expression but increases Angiopoietin 1 expression. In addition, Plk2 is negatively regulated by miR-128. miR-128 exerts a protective effect on cell apoptosis similar to Plk2 siRNA in response to I/R stress. Methylation inhibitor 5-azacytidine (5-AZ) increases the expression of miR-128 and subsequently reduces Plk2 expression and cell apoptosis. In conclusion, this study demonstrated that Plk2 regulated by miR-128 induces cell apoptosis/death in response to I/R stress through activation of the nuclear factor κB (NF-κB) signal pathway. miR-128 and Plk2 are new targets for preventing cardiac I/R injury or oxidative stress-mediated injury.
Keywords: Plk2, miR-128, I/R, apoptosis, Antimycin A, heart, H9c2 cell
Introduction
Ischemic disease is characterized by a decrease in blood supply to an organ, causing tissues to be deprived of oxygen and nutrients, which eventually leads to necrosis. Reperfusion is the only way to resuscitate ischemic tissue. Unfortunately, reperfusion itself has shown to further exacerbate damage to cardiac tissue. Ischemia-reperfusion (I/R) injury impairs recovery and hinders the effectiveness of treatments against cardiac ischemic diseases. I/R injury is also encountered during some medical interventions such as heart transplantation and coronary artery bypass grafting. This decreases the therapeutic potential and long-term patient survival of these treatments. There is no known therapy to reduce injuries due to I/R. It is vital to determine the mechanistic pathway of I/R injury so that therapies can be developed to effectively reduce damage to the myocardium.
Polo-like kinase 2 (Plk2) is a member of the polo-box family of serine/threonine kinases and plays a role in cell division. Plk2, initially identified in the early 1990s as an early response gene in serum-starved cells and originally named serum-inducible kinase (Snk), is the least characterized of the polo-like kinases.1, 2, 3, 4 Literature has reported that oxidative stress, reactive oxygen species (ROS), and calcium may induce Plk2 expression in mitochondrial dysfunction cells.5 Plk2 has recently been discovered as a tumor suppressor associated with apoptosis.6, 7, 8 Our cDNA microarray assay previously showed that Plk2 is overexpressed in transplanted heart grafts with extended cold I/R injury.9 In this study, we aimed to investigate the role of Plk2 in cardiac cells in response to I/R injury using an in vitro cell culture model and to dissect the underlying molecular mechanism by which Plk2 induces I/R injury.
Results
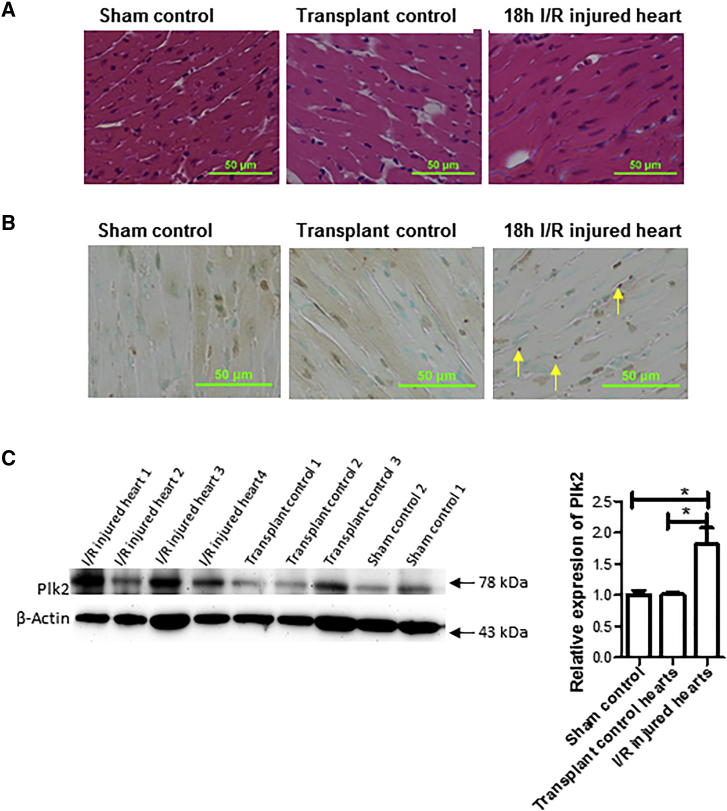
Plk2 Is Upregulated in Prolonged Cold I/R Injured Hearts
To explore new mechanistic signal pathways, we previously performed a cDNA microarray assay for I/R injured heart tissues. The microarray data showed that the expression of Plk2 was increased by 2-fold, as compared with transplant control hearts without extended I/R.9 To confirm the microarray results, we performed western blotting to detect Plk2 expression at the protein level. We excised heart organs from donor mice and stored them in University of Wisconsin (UW) solution at 4°C for 18 h to induce cold ischemia prior to transplantation. After 18 h of preservation, donor hearts were implanted into syngeneic recipient C57BL/6 mice. Twenty-four hours after transplantation, the heart grafts were harvested for histopathological analysis and western blotting. As shown in Figures 1A and 1B, 18 h of cold I/R increased histopathological changes and cell apoptosis and necrosis in the heart grafts. Plk2 was upregulated at the protein level by prolonged 18-h cold I/R as compared with the grafts without extended 18-h cold ischemia or sham controls, which were not transplanted (Figure 1C).
Figure 1.
Plk2 Expression Was Upregulated in I/R Injured Hearts In Vivo
Donor hearts were isolated from C57BL/6 mice and preserved in UW solution at 4°C for 18 h. After 18-h preservation, donor hearts (I/R injured hearts) were implanted into syngeneic recipient C57BL/6 mice. Some hearts were transplanted immediately after retrieval from donors without 18-h preservation at 4°C and used as a transplant control. Some hearts collected from mice that underwent abdominal open/closure surgery but without heart transplantation were included as a sham control. Twenty-four hours after transplantation, the heart grafts were harvested and subjected to H&E staining, TUNEL assay, and gene expression by western blotting. (A) H&E staining. (B) TUNEL assay for apoptosis. (C) Plk2 expression. Left: representative images from western blot results. Right: relative expression of Plk2 at the protein levels. Band densities were measured by the ImageJ program. The ratio of Plk2/actin density was calculated and normalized with the sham controls. Data were presented as mean ± SD. n = 3 for sham control and n = 5/group for transplant hearts. *p < 0.05.
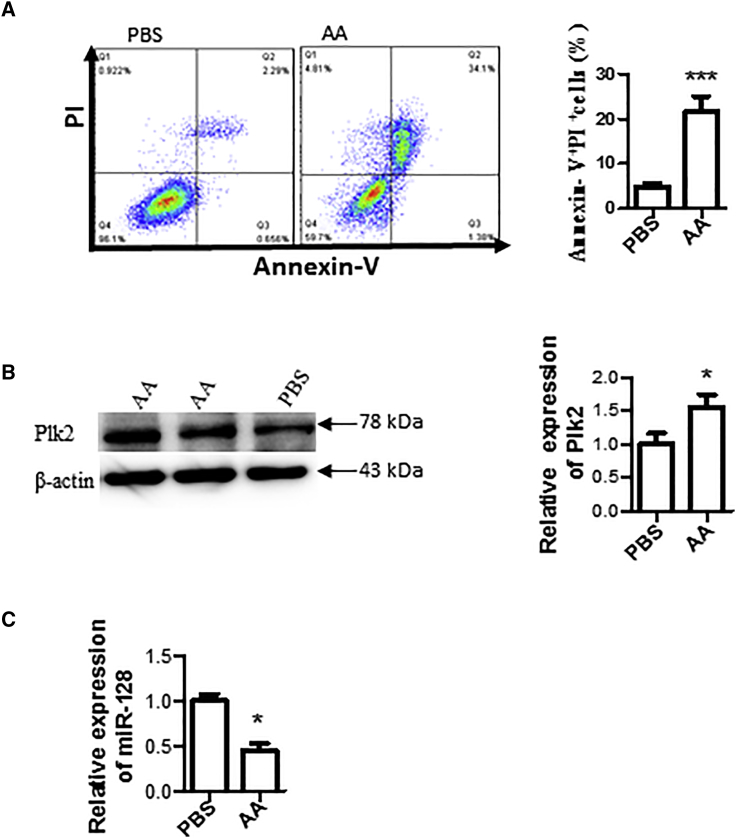
In Vitro I/R Stimulated with AA Induces Cardiac Cell Apoptosis, Increases Plk2 Expression, and Reduces miR-128
Antimycin A (AA), an inhibitor of complex III, produces ROS and can induce oxidative stress.10 AA is commonly used to simulate I/R injury in vitro.11 Therefore, we used an in vitro AA simulated I/R model in this study. H9c2 cells were cultured and treated with 20 μM AA for 3 h followed by 3 h of reperfusion with complete medium. AA treatment not only caused I/R injury with increased cell apoptosis and death, determined by double staining with Annexin V and propidium iodide (PI) and flow cytometry analysis, compared with the PBS control without AA treatment (Figure 2A), but also upregulated the expression of Plk2 in the cells as measured by western blotting (Figure 2B). Moreover, AA treatment reduced the expression of miR-128 (Figure 2C).
Figure 2.
In Vitro I/R Stimulated with AA Induced Cardiac Cell Apoptosis/Death, Increased Plk2 Expression, and Reduced miR-128
(A) Cell apoptosis. H9c2 cells were cultured and treated with 20 μM AA or PBS (as a control) for 3 h, followed by 3-h reperfusion with complete medium. Cells were double stained with FITC-Annexin V and PI, followed by flow cytometry analysis. Left panel: representative images from flow cytometry result; right panel: summarized results of flow cytometry. n = 5; ***p < 0.001. (B) Plk2 expression. Total proteins were extracted from the above cells, and Plk2 expression was detected by western blotting. Left panel: representative images from three independent western blotting experiments; right panel: semiquantitative results of western blotting. Data were normalized with PBS control. *p < 0.05. (C) miR-128 expression. miRNA was extracted, and the expression of miR-128 was determined by qRT-PCR. SNORD61.1 was used an internal loading control, and data were normalized with the PBS control. n = 3; *p < 0.05.
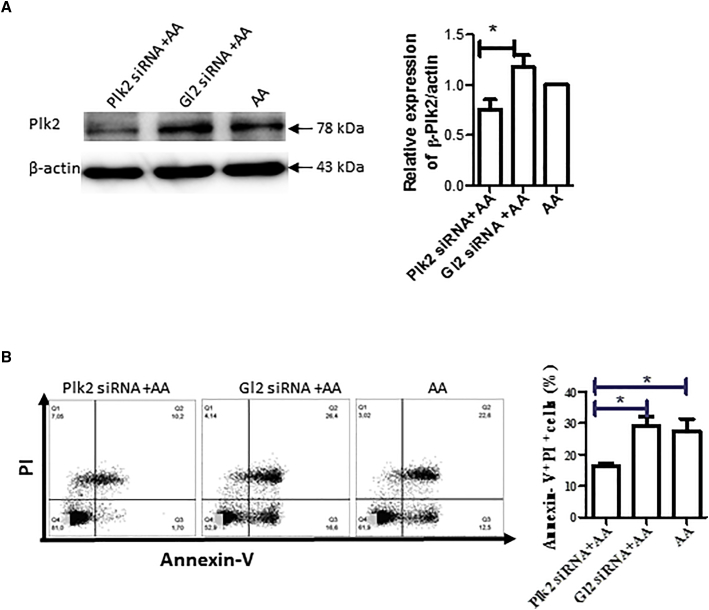
Knockdown of Plk2 Using siRNA Reduces Apoptosis Induced by AA
To understand the role of Plk2 in cardiac I/R injury, we knocked down the Plk2 gene using small interfering RNA (siRNA). H9C2 cells were cultured, transfected with Plk2 siRNA, and then treated with AA. The expression of Plk2 was remarkably decreased in cells transfected with Plk2 siRNA as compared with control Gl2 siRNA and AA control (Figure 3A). Transfection with Plk2 siRNA significantly reduced cell apoptosis and death as seen by reduced Annexin V+PI+ cells (Figure 3B).
Figure 3.
Plk2 siRNA Knocked Down Plk2 and Reduced Cell Apoptosis Induced by I/R
(A) Gene silencing of Plk2. H9c2 cells were transfected with Plk2 siRNA or control Gl2 siRNA. Forty-eight hours after transfection, cells were subjected to AA treatment followed by 3-h reperfusion. Proteins were extracted from the cells, and Plk2 expression was detected by western blotting. Left panel: representative images from three independent experiments; right panel: semiquantitative results of western blotting. The relative expression of Plk2 was normalized with the AA control. n = 3; *p < 0.05. (B) Knockdown of Plk2 reduces apoptosis. Twenty-four hours after transfection, cells were subjected to AA. Three hours after reperfusion, cell apoptosis was detected by staining with FITC-Annexin V and PI, followed by flow cytometry. Left panel: representative images from four independent experiments; right panel: summarized results of flow cytometry. n = 3; *p < 0.05.
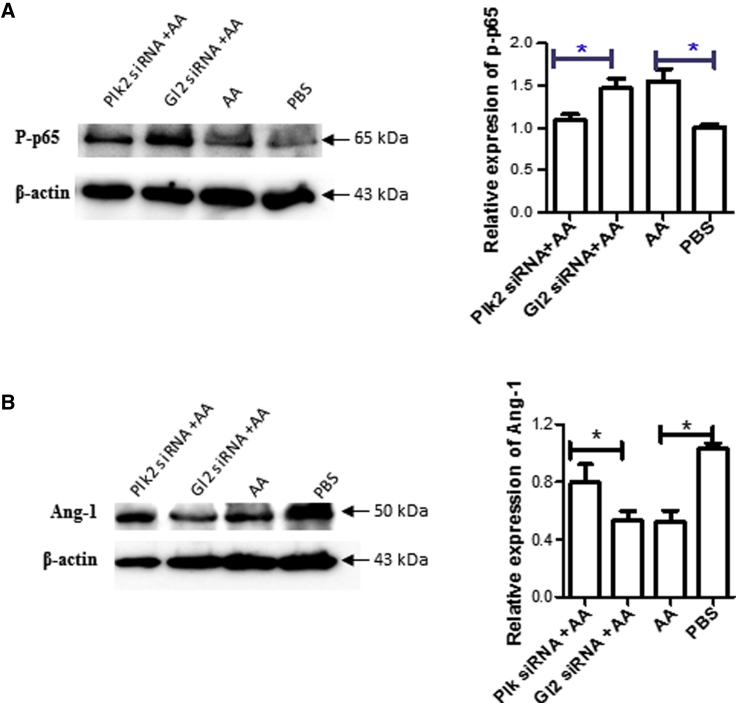
Knockdown of Plk2 Inactivates the NF-κB Pathway and Reverses the Loss of Angiopoientin-1
Literature has reported that the nuclear factor κB (NF-κB) pathway is activated by I/R,12 and that angiopoietin (Ang-1) plays a protective role in I/R injury in heart transplantation.13 Accordingly, we detected the expression of phosphorylated p65 (p-p65) and Ang-1 to understand how Plk2 protects cells from I/R injury. We found that AA treatment increased p-p65 (Figure 4A), whereas it decreased Ang-1 (Figure 4B) as compared with the control. Transfection with Plk2 siRNA reduced the expression of p-p65 (Figure 4A) and increased Ang-1 expression (Figure 4B) as compared with control siRNA, indicating that Plk2 siRNA reversed the effect of AA on the expression of these genes.
Figure 4.
Knockdown of Plk2 Reduced the Expression of p-p65 but Increased Ang-1
H9c2 cells were transfected with Plk2 siRNA, and total proteins were extracted as described in Figure 3. (A and B) The expression of p-p65 (A) and Ang-1 (B) was detected by western blotting. The relative expression of Plk2 was semiquantified by measuring the density of western blotting bands, and normalized with the PBS control. β-Actin was used as an internal loading control. Left panel: representative images from experiments (n = 3); right panel: semiquantitative results of western blotting (n = 3). *p < 0.05.
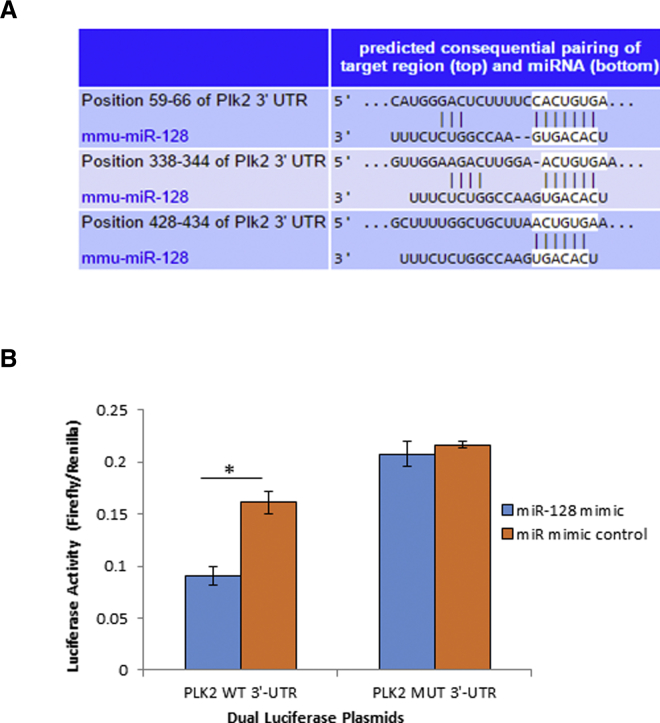
Plk2 Was Negatively Regulated by miR-128
To understand how Plk2 is regulated, we conducted computational analysis using bioinformatic microRNA (miRNA) target prediction tools. Bioinformatic analysis revealed that the 3′ untranslated region (UTR) of the Plk2 gene contains three putative conserved binding sites of miR-128 (Figure 5A), which was reportedly downregulated in I/R injured heart cells in our previous microarray study9 and in AA-treated heart cells. To identify and confirm whether miR-128 targets Plk2, we performed a luciferase reporter assay.14 We cloned both the wild-type (WT) 3′ UTR of Plk2-expressing luciferase reporter vector and mutant (MUT) 3′ UTR vector in which the seed region nucleotides of miR-128 were MUT. We then co-transfected the luciferase reporter construct harboring the WT 3′ UTR or MUT of Plk2 with miR-128. Forty-eight hours after co-transfection, firefly and Renilla luciferase activities were measured separately, and the ratio of firefly/Renilla luciferase activity was calculated. The luciferase reporter assay showed a significant decrease in the ratio of firefly/Renilla luciferase activity in cells co-transfected with the dual-luciferase reporter plasmids containing the WT Plk2 3′ UTR and the miR-128 mimic when compared with the miR mimic control (p < 0.05; Figure 5B). However, there was no significant difference in the ratio of firefly/Renilla luciferase activity when the MUT Plk2 3′ UTR dual-luciferase vector was co-transfected with either the miR-128 mimic or miR mimic control (Figure 5B). This indicates that miR-128 binds to the 3′ UTR of Plk2 mRNA, and that Plk2 is a target of miR-128.
Figure 5.
Plk2 Was Negatively Regulated by miR-128
(A) Putative binding sites of miR-128 at the 3′ UTR of the Plk2 gene predicated by TargetScan. (B) miR-128 binds to 3′ UTR of Plk2. A fragment of Plk2 3′ UTR containing the sequence of the putative binding sites to miR-128 was inserted into the luciferase reporter vector pMIR-Reporter producing a construct named PLK2 WT 3′ UTR. A mutant Plk2 3′ UTR plasmid (PLK2-MUT 3′ UTR) was created by replacing wild-type miR-128 targeting site sequence (ACUGUG) with GAGGCG. Cells were co-transfected with the above constructed WT or MUT pmir-Plk2-UTR and miR-128 mimic, or mimic control. Forty-eight hours after co-transfection, luciferase activity was detected using a dual-luciferase activity kit. The ratio of firefly/Renilla luciferase activity was calculated. n = 3; *p < 0.05.
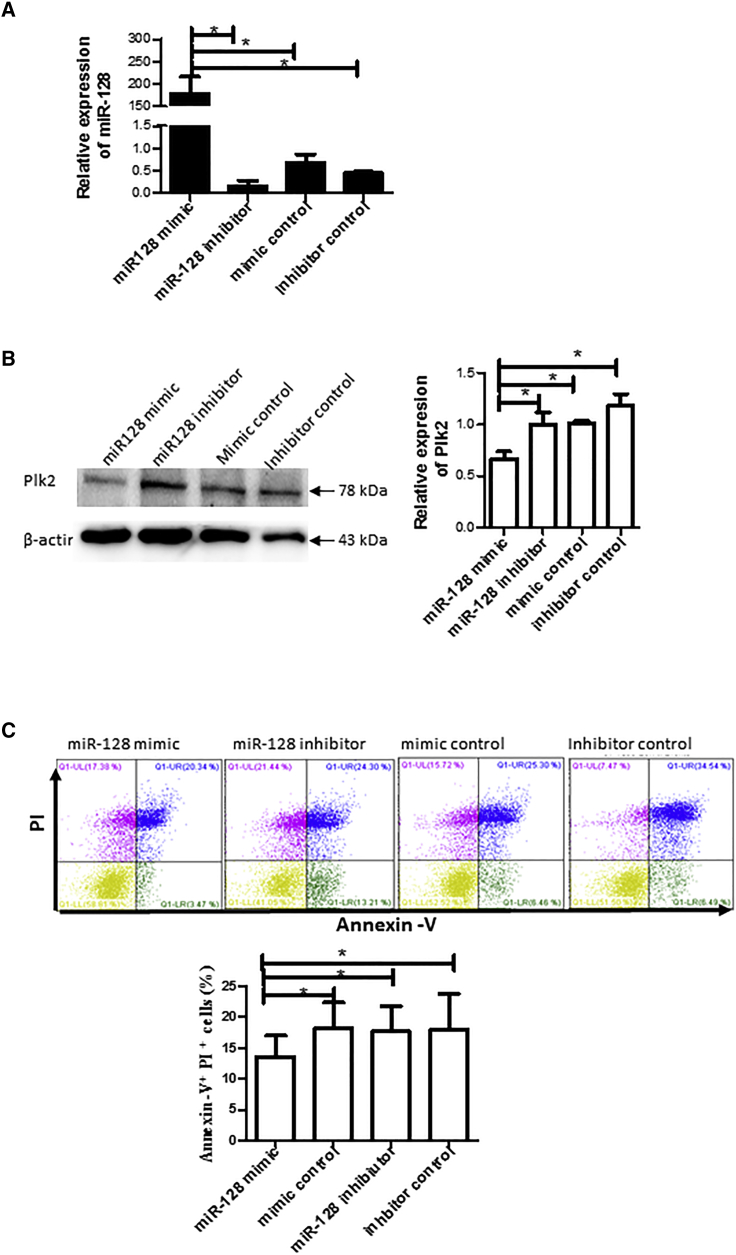
Introduction of Exogenous miR-128 Mimic Inhibits Apoptosis/Death and Reduces Plk2 Expression in Response to AA
To further validate the regulation of miR-128 on Plk2, we transfected cells with miR-128 mimic, miR-128 inhibitor, or their controls for 24 h prior to exposure to AA. We confirmed that transfection with miR-128 mimic dramatically increased miR-128 levels in the cells, whereas miR-128 inhibitor decreased miR-128 (Figure 6A). We also found that transfection with miR-128 mimics decreased the expression of Plk2 (Figure 6B) while increasing cell apoptosis (Figure 6C).
Figure 6.
Transfection of miR-28 Mimic Reduced Plk2 Expression and Cell Apoptosis/Death
(A) miR-128 expression. H9c2 cells were plated in a six-well plate 1 day prior to transfection. The cells were transfected with 1 μg miR-128 mimic, mimic control, miR-128 inhibitor, or inhibitor control with 2 μL Lipofectamine 2000. Twenty-four hours after transfection, total RNA was extracted, and miR-128 expression was detected by qRT-PCR using SYBR Green qPCR kits, normalized to control cells transfected with control mimic. n = 3; *p < 0.05. (B) Plk2 expression. Proteins were extracted from the above transfected cells 48 h after transfection. Plk2 expression was detected by western blotting with Plk2 antibodies. Left panel: representative images of western blotting; right panel: semiquantitative results of western blotting. n = 3; *p < 0.05. (C) mimR-128 reduced cell apoptosis. The above transfected cells were subjected to AA, and cell apoptosis was determined by Annexin V and PI staining and flow cytometry. Upper panel: representative image of flow cytometry; bottom panel: statistical results of flow cytometry. n = 3; *p < 0.05.
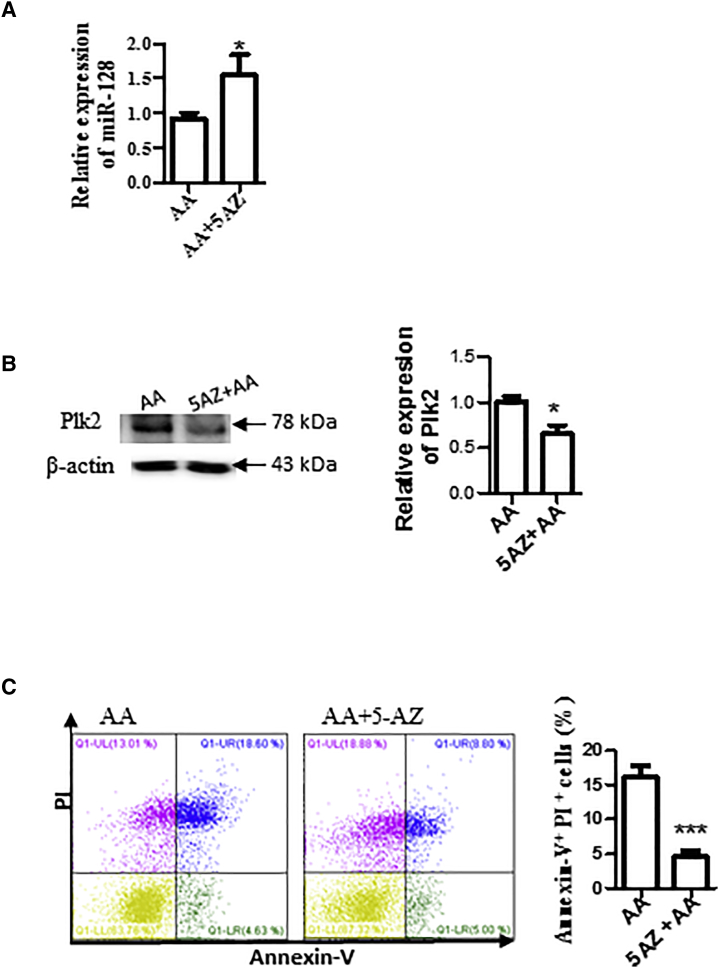
Methylation Inhibits miR-128 Expression
Hypoxia-induced methylation and hypermethylation were observed during I/R.15 Many miRNAs are associated with CpG islands and regulated by DNA methylation.16,17 We investigated whether the downregulation of miR-128 was caused by methylation. To do this, we treated H9c2 cells with methylation inhibitor 5-azacytidine (5-AZ) and then detected miR-128 expression by qRT-PCR. As shown in Figure 7A, 5-AZ treatment increased miR-128 expression. We also found that treatment with 5′−AZ reduced cell apoptosis/death induced by AA (Figure 7B) and the expression of Plk2 (Figure 7C).
Figure 7.
Inhibition of Methylation by 5-AZ Increased miR-128 Expression
(A) 5-AZ increased miR-128 expression. H9c2 cells were treated with 5 μmol/L methylation inhibitor 5-AZ and subjected to AA treatment. miR-128 expression was then detected by qRT-PCR. (B) 5-AZ reduced Plk2 expression. Plk2 expression was detected by western blotting. Left panel: representative image of western blotting; right panel: semiquantitative results of western blotting. n = 3; *p < 0.05. (C) 5-AZ attenuated cell apoptosis/death induced by AA. Cells were collected and stained with Annexin V-FITC and PI, and then analyzed by flow cytometry. Left panel: representative images of flow cytometry results; right panel: statistical analysis of cell apoptosis. n = 3; ***p < 0.001.
Discussion
In this study, we, for the first time, demonstrated that I/R injury upregulated Plk2 in heart cells in vitro and in vivo. The increased Plk2 plays a detrimental role in heart cells in response to I/R stress. Plk2 was negatively regulated by miR-128. Knockdown of Plk2 by siRNA or miR-128 mimic reduced I/R-mediated cell apoptosis/death by regulating the NF-κB signal pathway and Ang-1.
Plk2 is one of the PLK family members and is the least characterized of the polo-like kinases.1, 2, 3, 4 The Plk2 gene, located in chromosome 5 at 5q12.1–q13.2,18 is a target of p53.19 Plk2 is intermediately expressed in the heart2 and participates in proliferation and early lineage commitment of cardiac progenitor cells.20 Plk2 is an early response gene to stresses, and its expression is immediately elevated under various stresses.3,4 Most reported studies of Plk2 are focused on cancer and neurodegenerative diseases.19,21,22 Zou et al.23 have recently reported that PLK2 is upregulated in kidneys from diabetic kidney disease patients and podocytes treated with high D-glucose. The authors demonstrated that knockdown of Plk2 reduces high D-glucose-induced apoptosis and proinflammatory cytokines such as tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), and IL-1β.23 In this study, we first verified Plk2 overexpression in heart cells under I/R stress in vitro and in vivo. We demonstrated that knockdown of Plk2 with siRNA reduced cell apoptosis under I/R stress in vitro, indicating that Plk2 plays a causative role in I/R injury. We also found that Plk2 siRNA reduced the expression of p-p65, which was upregulated by I/R, but increased Ang-1 expression. Literature has reported that the NF-κB pathway is activated during I/R, and activation of this pathway causes cell death.12 We and other groups have previously reported that I/R injury results in loss of Ang-1 expression in heart tissues and cells.9,24 Ang-1 plays a cardioprotective effect against I/R, and overexpression of Ang-1 prevents cardiac I/R injury.12,13,24 Thus, we propose that Plk2 siRNA protects heart cell apoptosis/death from I/R injury through inactivation of the NF-κB pathway and preserving Ang-1 protein.
miRNAs are single-stranded RNAs of 18–24 nt in length and are generated from an endogenous transcript that consists of a hairpin structure.25, 26, 27, 28, 29 miRNA is a major negative gene regulator by partially base-pairing with the 3′ UTR of the target, leading to translational repression.30, 31, 32, 33, 34 Approximately one-third of genes are regulated by miRNAs.35 It has been reported that miR-12636,37 and miR-27a38 regulate Plk2 expression. Our previous studies have showed that I/R altered the expression of miRNAs, including miR-128, in the hearts.9 In this study, we investigated whether and how Plk2 was regulated by miR-128 through a series of experiments. We confirmed that I/R downregulated miR-128 in H9c2 cells in which Plk2 was upregulated. Computational analyses showed that, in the Plk2 gene, there are binding sites of miR-128. A luciferase activity reporter assay further confirmed that miR-128 binds to the 3′ UTR of the Plk2 gene. Transfection of miR-128 mimic reduced Plk2 expression, indicating that Plk2 is negatively regulated by miR-128. We also observed that overexpression of miR-128 reduced cell death, similar to Plk2 siRNA effect, further indicating that miR-128 negatively regulates Plk2. In support, a more recent study also reported that miR-128 negatively regulates Plk2 in liver cancer cells.21 Furthermore, it was reported in the literature that miR-128 upregulated the anti-apoptotic factor Bcl-2 in neuroblastoma cells,39 and that introducing Blc-2 in a mouse I/R model reduced injury,40 which aligns with our results that overexpression of miR-128 reduced cell apoptosis/death. However, Zeng et al.41 reported that inhibition of miR-128 reduced cell apoptosis through targeting activation of peroxisome proliferator-activated receptor gamma (PPARG) in a rabbit coronary artery ligation model with 60 min of ischemia. Chen et al.42 recently showed that miR-128-3p increased human cardiomyocyte cell apoptosis in an 18-h hypoxia/2-h reperfusion in vitro model. The discrepancy of these two studies might be because miRNA expression is spatial and temporal, and the effects of miRNA are cell, tissue, and model specific.43,44
Epigenetic regulation, such as DNA-methylation, affects miRNA expression.45 In this study, we also investigated the effect of methylation on miR-128 expression. We found that treatment with a methyltransferase inhibitor 5-AZ increased miR-128 expression and deceased Plk2 expression induced by AA, resulting in a reduction in cell apoptosis. It shows that methylation is involved in miR-128 expression and in the miR-128-mediated protective effect.
We recognize that there are several limitations to the present study that need to be considered when interpreting these results. The effect of miR-128/Plk2 was tested in one cell line H9c2 in vitro, and we used an AA model. This is a simulated model and may not reflect in vivo and clinical scenarios. The function of this axis needs to be investigated in other cardiomyocytes and cardiac fibroblast cell lines, especially primary cardiomyocytes and primary cardiac fibroblasts in future studies. In vivo animal studies should be conducted in future studies as well.
In conclusion, this study demonstrated that Plk2 induced cell apoptosis/death in response to I/R stress through activation of the NF-κB signal pathway. Furthermore, Plk2 was negatively regulated by miR-128 which plays a protective role in heart cells in response to AA stress. The increased understanding of the upstream and downstream regulatory pathways of Plk2 and miRNAs would open up the possibility of targeting them with novel therapeutic interventions to decrease I/R injury.
Materials and Methods
Animals
C57B/6 mice were obtained from Charles River Laboratories (Canada). All animal experiments in the study were conducted in accordance with the guidelines established by the Canadian Council of Animal Care and were approved by the Animal Care Committee of Western University.
Cell Culture
H9c2 cells (Rattus norvegicus heart myoblast cell line) were purchased from ATCC (Burlington, ON, Canada) and cultured with Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Mississauga, ON, Canada) supplemented with 10% GIBCO fetal bovine serum (FBS; Thermo Fisher Scientific) and 100 U/mL penicillin-streptomycin (Thermo Fisher Scientific), at conditions of 37°C and 5% CO2 until 80%–90% confluence for subculturing.
In Vitro I/R Model Induced by AA
H9c2 cells were plated in a six-well plate with a density of 9 × 104 cells/well and grown at 37°C for 24 h prior to starting the I/R protocol. Hypoxic stress was induced by washing the cells with PBS and replacing the media with 1 mL of 20 μM AA (Sigma-Aldrich, Oakville, ON, Canada) in Ca2+/Mg2+ PBS (Thermo Fisher Scientific, Burlington, ON, Canada) and incubating them at 37°C for 3 h. After 3 h of ischemic stress, the cells were washed with PBS, and reperfusion was induced by replacing the media with 1 mL 10% FBS-supplemented DMEM. The cells were cultured at 37°C for another 3 h (mimicking reperfusion phase). This is a chemical method of stimulating I/R injury.11
In Vivo Cardiac I/R Model in Heart Transplant
A syngeneic murine heart transplantation with a donor heart that underwent extended cold I/R was performed as described previously.9,46 In brief, healthy C57BL/6 mice at 8 weeks of age were anesthetized with ketamine/xylene, and the hearts were retrieved and preserved with UW solution at 4°C for 18 h, then implanted into syngeneic C57BL/6 recipient mice. Twenty-four hours after revascularization, animals were sacrificed for further experiment. Donor hearts immediately implanted into recipients without 18-h cold ischemia were used as a transplant control. Hearts from sham surgery (sham control) were also included.
Cell Transfection
H9c2 cells (80,000 cells/well) were plated in a six-well plate and cultured in complete DMEM medium (CM) 1 day before transfection. Cells were transfected with plk2 siRNA (Thermo Fisher Scientific), miR-128 mimics, miR-128 inhibitors, or their controls (GenePharma, Shanghai, China) at a final concentration of 30 nM using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. A 1:2 ratio of siRNA or miRNA/Lipofectamine 2000 was used. After the transfection solution was added, cells were incubated for 4 h at 37°C and 5% CO2, and then DMEM supplemented with 20% FBS by volume was added to each transfection well. Cells were incubated overnight (18 h) at 37°C and 5% CO2 before being subjected to simulated cold I/R injury.
Apoptosis Analysis by Flow Cytometry
H9c2 cells were plated in six-well plates and then subjected to transfection and simulated cold I/R injury as described above. After 3 h of simulated reperfusion, cells were trypsinized using Trypsin-EDTA (Thermo Fisher Scientific) and washed with PBS. Cells were double stained with fluorescein isothiocyanate (FITC)-labeled Annexin V and PI using the Annexin V Apoptosis Detection Kit (BD Biosciences, Mississauga, ON, Canada) according to the manufacturer’s instructions. Cells were then analyzed by flow cytometry using a CytoFLEX system (Beckman Coulter, Mississauga, ON, Canada) and the CytExpert software.
miRNA Isolation and qRT-PCR Analysis
Total RNA, including miRNA, was extracted from cells using the miRNeasy Mini Kit (QIAGEN, Mississauga, ON, Canada). cDNA for gene expression was synthesized using oligo-dT and reverse transcriptase (Thermo Fisher Scientific). cDNA of miRNA-128 was synthesized using the qScript microRNA cDNA synthesis kit (Quanta Biosciences, San Francisco, CA, USA) or miScript II RT Kit (QIAGEN) according to the manufacturers’ instructions. The cDNA was then subjected to quantitative PCR (qPCR) with respect to miRNA-128 expression using the qScript One-Step SYBR Green qRT-PCR kit (Quanta Biosciences) or miScript SYBR Green PCR Kit (QIAGEN). qPCR was performed using the CFX Connect Real-Time PCR Detection System (Bio-Rad, Mississauga, ON, Canada). SNORD61 (QIAGEN) was used as an internal loading reference. PCR cycling conditions for qScript One-Step SYBR Green qRT-PCR kit were as follows: 95°C for 2 min followed by 40 cycles of 95°C for 5 s, 60°C for 30 s, and 72°C for 30 s, or 95°C for 15 min followed by 40 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 30 s for the miScript SYBR Green PCR Kit.
Luciferase Assay to Demonstrate miR-128 Binding to 3′ UTR of PLK2
Dual-luciferase reporter assay systems were purchased from Promega (Madison, WI, USA). Dual-luciferase reporters containing the WT and MUT 3′ UTR of PLK2 in the multiple cloning sites were constructed by Norclone (London, ON, Canada). The MUT 3′ UTR of PLK2 contained the same sequence as the WT sequence except the three potential binding sequences of “ACUGUG” (predicted by TargetScan) were replaced with “GAGGCG” (five-base mutation). H9c2 cells were plated onto a 12-well plate (80,000 cells/well) containing 1 mL of CM/well and incubated for 18 h at 37°C and 5% CO2. Wells (n = 3 for each dual transfection group) were each transfected with one of the following co-transfections: miR-128 mimic and dual-luciferase reporter plasmid containing the WT 3′ UTR of PLK2; miR-128 mimic and dual-luciferase reporter plasmid containing the MUT 3′ UTR of PLK2; miRNA mimic control and dual-luciferase reporter plasmid containing the WT 3′ UTR of PLK2; and miRNA mimic control and dual-luciferase reporter plasmid containing the MUT 3′ UTR of PLK2. The co-transfection of miRNA and the dual-luciferase vector was performed using miRNA to a final concentration of 30 nM and the dual-luciferase vector at 0.10 μg. Transfection was performed according to the manufacturer’s instructions using Lipofectamine 2000 and Opti-MEM reduced serum medium (Thermo Fisher Scientific). A 1:2 ratio of miRNA/Lipofectamine 2000 was used. Cells were transfected at 37°C and 5% CO2 for 48 h. The luciferase assay was performed according to the manufacturer’s instructions. Firefly (Photinus pyralis) luciferase activity and Renilla (Renilla reniformis) luciferase activity were measured at 560 and 480 nm, respectively, using a VICTOR Multilabel Plate Reader (PerkinElmer, Woodbridge, ON, Canada). The relative expression of firefly luciferase was calculated as Renilla luciferase activity was used as a normalizer.
Western Blotting
Heart tissues or cells were harvested, washed with PBS, and then lysed with radioimmunoprecipitation assay (RIPA) buffer containing 1 mM protease inhibitor phenylmethylsulfonyl fluoride (PMSF; Thermo Fisher Scientific) to extract total protein. Cell lysate was centrifuged for 20 min at 15,000 rpm, and supernatant was collected. The concentration of protein was measured using a Bradford assay (Thermo Fisher Scientific). A total of 25 μg total protein was loaded on 12% polyacrylamide gels and run for 60–80 min at 100 voltage. Separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes. Transferred membranes were blocked with 5% fat-free milk powder in Tris-buffered saline with 0.25% Tween 20 (TBST) for 30 min at room temperature and then blotted with the primary antibodies against mouse Plk2 (1:1,000 dilution; Santa Cruz Biotechnology, Dallas, TX, USA), phosphorylated Rel A p65 (1:1,000 dilution; Cell Signaling Technology), Ang-1 (1:1,000 dilution; Abcam, Boston, MA, USA), and β-actin (1:4,000 dilution; Santa Cruz Biotechnologies) at 4°C overnight. The blotted membranes were washed with TBST containing 0.25% Tween 20 for 10 min at room temperature and repeatedly washed for three times. Washed membranes were blotted with appropriated secondary antibodies (Santa Cruz Biotechnologies) for 60 min at room temperature. Proteins were developed with ECL kits (Bio-Rad) and visualized by FluorChem M system (ProteinSimple, San Jose, CA, USA). The density of bands was quantified using the ImageJ program (https://imagej.nih.gov/ij/).
Histological Analysis
Heart grafts were collected from mice, and tissue slices were fixed in 10% formalin and processed for histology examination using standard techniques. Formalin tissue was embedded in paraffin, and 5-μm sections were stained with hematoxylin and eosin (H&E). Histological changes of heart grafts were assessed by a pathologist in a blind manner for damage of epicardium, myocardium, endocardium, intimal thickness, infarction, neutrophil infiltration, and fibrosis.
Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick-End Labeling (TUNEL) Assay
Cell apoptosis in heart grafts was detected by the TUNEL assay using paraffin-embedded tissue sections and an in situ cell death detection kit according to the manufacturer’s instructions (Roche, Mississauga, ON, Canada).
Statistical Analyses
Data were presented as means ± standard error; n ≥ 3. Paired t tests were performed for comparisons between two groups. A one-way analysis of variance (ANOVA) followed by the Newman-Keuls test was used for comparisons between more than two groups. A p value <0.05 indicated a statistically significant difference. All analyses were performed using GraphPad Prism, version 7 (La Jolla, CA, USA).
Author Contributions
D.Z., E.S., F.L., Q.L., B.W., Q.Z., C.Z., and H.Z. executed the experiments and data analysis, and interpreted the results. A.W. participated in manuscript writing. K.L. and X.Z. contributed to financial support, project design, and manuscript writing.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (CIHR), the Natural Sciences and Engineering Research, and the Bureau of Science and Technology of Jilin Province, China (20160414054GH).
Contributor Information
Kexiang Liu, Email: kxliu64@hotmail.com.
Xiufen Zheng, Email: xzheng26@uwo.ca.
References
- 1.Ma S., Liu M.A., Yuan Y.L., Erikson R.L. The serum-inducible protein kinase Snk is a G1 phase polo-like kinase that is inhibited by the calcium- and integrin-binding protein CIB. Mol. Cancer Res. 2003;1:376–384. [PubMed] [Google Scholar]
- 2.Simmons D.L., Neel B.G., Stevens R., Evett G., Erikson R.L. Identification of an early-growth-response gene encoding a novel putative protein kinase. Mol. Cell. Biol. 1992;12:4164–4169. doi: 10.1128/mcb.12.9.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anger M., Kues W.A., Klima J., Mielenz M., Kubelka M., Motlik J., Esner M., Dvorak P., Carnwath J.W., Niemann H. Cell cycle dependent expression of Plk1 in synchronized porcine fetal fibroblasts. Mol. Reprod. Dev. 2003;65:245–253. doi: 10.1002/mrd.10289. [DOI] [PubMed] [Google Scholar]
- 4.Liby K., Wu H., Ouyang B., Wu S., Chen J., Dai W. Identification of the human homologue of the early-growth response gene snk, encoding a serum-inducible kinase. DNA Seq. 2001;11:527–533. doi: 10.3109/10425170109041337. [DOI] [PubMed] [Google Scholar]
- 5.Li J., Ma W., Wang P.Y., Hurley P.J., Bunz F., Hwang P.M. Polo-like kinase 2 activates an antioxidant pathway to promote the survival of cells with mitochondrial dysfunction. Free Radic. Biol. Med. 2014;73:270–277. doi: 10.1016/j.freeradbiomed.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coley H.M., Hatzimichael E., Blagden S., McNeish I., Thompson A., Crook T., Syed N. Polo Like Kinase 2 Tumour Suppressor and cancer biomarker: new perspectives on drug sensitivity/resistance in ovarian cancer. Oncotarget. 2012;3:78–83. doi: 10.18632/oncotarget.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kis E., Szatmári T., Keszei M., Farkas R., Esik O., Lumniczky K., Falus A., Sáfrány G. Microarray analysis of radiation response genes in primary human fibroblasts. Int. J. Radiat. Oncol. Biol. Phys. 2006;66:1506–1514. doi: 10.1016/j.ijrobp.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Kim J.H., Ku B., Lee K.S., Kim S.J. Structural analysis of the polo-box domain of human Polo-like kinase 2. Proteins. 2015;83:1201–1208. doi: 10.1002/prot.24804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou L., Zang G., Zhang G., Wang H., Zhang X., Johnston N., Min W., Luke P., Jevnikar A., Haig A., Zheng X. MicroRNA and mRNA signatures in ischemia reperfusion injury in heart transplantation. PLoS ONE. 2013;8:e79805. doi: 10.1371/journal.pone.0079805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madungwe N.B., Zilberstein N.F., Feng Y., Bopassa J.C. Critical role of mitochondrial ROS is dependent on their site of production on the electron transport chain in ischemic heart. Am. J. Cardiovasc. Dis. 2016;6:93–108. [PMC free article] [PubMed] [Google Scholar]
- 11.Kurian G.A., Pemaih B. Standardization of in vitro cell-based model for renal ischemia and reperfusion injury. Indian J. Pharm. Sci. 2014;76:348–353. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J.R., Yu H.L. Effect of NF-κB signaling pathway mediated by miR-711 on the apoptosis of H9c2 cardiomyocytes in myocardial ischemia reperfusion. Eur. Rev. Med. Pharmacol. Sci. 2017;21:5781–5788. doi: 10.26355/eurrev_201712_14025. [DOI] [PubMed] [Google Scholar]
- 13.Lee S.W., Won J.Y., Lee H.Y., Lee H.J., Youn S.W., Lee J.Y., Cho C.H., Cho H.J., Oh S., Chae I.H., Kim H.S. Angiopoietin-1 protects heart against ischemia/reperfusion injury through VE-cadherin dephosphorylation and myocardiac integrin-β1/ERK/caspase-9 phosphorylation cascade. Mol. Med. 2011;17:1095–1106. doi: 10.2119/molmed.2011.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hullinger T.G., Montgomery R.L., Seto A.G., Dickinson B.A., Semus H.M., Lynch J.M., Dalby C.M., Robinson K., Stack C., Latimer P.A. Inhibition of miR-15 protects against cardiac ischemic injury. Circ. Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heylen L., Thienpont B., Naesens M., Busschaert P., Depreeuw J., Smeets D., Jochmans I., Monbaliu D., Pirenne J., Lerut E. Ischemia-induced DNA hypermethylation during kidney transplant predicts chronic allograft injury. J. Am. Soc. Nephrol. 2018;29:1566–1576. doi: 10.1681/ASN.2017091027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong L., Wang F., Huang X., Liu Z.H., Zhao T., Wu L.Y., Wu K., Ding X., Liu S., Wu Y. DNA demethylation regulates the expression of miR-210 in neural progenitor cells subjected to hypoxia. FEBS J. 2012;279:4318–4326. doi: 10.1111/febs.12021. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y., Wang L.L., Li Y.H., Gao X.N., Liu Y., Yu L. Effect of CpG island methylation on microRNA expression in the k-562 cell line. Biochem. Genet. 2012;50:122–134. doi: 10.1007/s10528-011-9478-9. [DOI] [PubMed] [Google Scholar]
- 18.Thierry-Mieg D., Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthew E.M., Hart L.S., Astrinidis A., Navaraj A., Dolloff N.G., Dicker D.T., Henske E.P., El-Deiry W.S. The p53 target Plk2 interacts with TSC proteins impacting mTOR signaling, tumor growth and chemosensitivity under hypoxic conditions. Cell Cycle. 2009;8:4168–4175. doi: 10.4161/cc.8.24.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mochizuki M., Lorenz V., Ivanek R., Della Verde G., Gaudiello E., Marsano A., Pfister O., Kuster G.M. Polo-like kinase 2 is dynamically regulated to coordinate proliferation and early lineage specification downstream of yes-associated protein 1 in cardiac progenitor cells. J. Am. Heart Assoc. 2017;6:e005920. doi: 10.1161/JAHA.117.005920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joo M.S., Shin S.B., Kim E.J., Koo J.H., Yim H., Kim S.G. Nrf2-lncRNA controls cell fate by modulating p53-dependent Nrf2 activation as an miRNA sponge for Plk2 and p21cip1. FASEB J. 2019;33:7953–7969. doi: 10.1096/fj.201802744R. [DOI] [PubMed] [Google Scholar]
- 22.Wang R., Wang Y., Qu L., Chen B., Jiang H., Song N., Xie J. Iron-induced oxidative stress contributes to α-synuclein phosphorylation and up-regulation via polo-like kinase 2 and casein kinase 2. Neurochem. Int. 2019;125:127–135. doi: 10.1016/j.neuint.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Zou H.H., Yang P.P., Huang T.L., Zheng X.X., Xu G.S. Plk2 plays an essential role in high d-glucose-induced apoptosis, ros generation and inflammation in podocytes. Sci. Rep. 2017;7:4261. doi: 10.1038/s41598-017-00686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Syrjälä S.O., Nykänen A.I., Tuuminen R., Raissadati A., Keränen M.A., Arnaudova R., Krebs R., Koh G.Y., Alitalo K., Lemström K.B. Donor heart treatment with comp-ang1 limits ischemia-reperfusion injury and rejection of cardiac allografts. Am. J. Transplant. 2015;15:2075–2084. doi: 10.1111/ajt.13296. [DOI] [PubMed] [Google Scholar]
- 25.Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutvágner G., Zamore P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 27.Lim L.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 28.Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 29.Piubelli C., Meraviglia V., Pompilio G., D’Alessandra Y., Colombo G.I., Rossini A. Micrornas and cardiac cell fate. Cells. 2014;3:802–823. doi: 10.3390/cells3030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayr C., Hemann M.T., Bartel D.P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seitz H., Youngson N., Lin S.P., Dalbert S., Paulsen M., Bachellerie J.P., Ferguson-Smith A.C., Cavaillé J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 32.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 33.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 34.Leaman D., Chen P.Y., Fak J., Yalcin A., Pearce M., Unnerstall U., Marks D.S., Sander C., Tuschl T., Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 35.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Liu L.Y., Wang W., Zhao L.Y., Guo B., Yang J., Zhao X.G., Hou N., Ni L., Wang A.Y., Song T.S. Mir-126 inhibits growth of SGC-7901 cells by synergistically targeting the oncogenes PI3KR2 and Crk, and the tumor suppressor PLK2. Int. J. Oncol. 2014;45:1257–1265. doi: 10.3892/ijo.2014.2516. [DOI] [PubMed] [Google Scholar]
- 37.Deng S., Wang H., Jia C., Zhu S., Chu X., Ma Q., Wei J., Chen E., Zhu W., Macon C.J. Microrna-146a induces lineage-negative bone marrow cell apoptosis and senescence by targeting polo-like kinase 2 expression. Arterioscler. Thromb. Vasc. Biol. 2017;37:280–290. doi: 10.1161/ATVBAHA.116.308378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian Y., Fu S., Qiu G.B., Xu Z.M., Liu N., Zhang X.W., Chen S., Wang Y., Sun K.L., Fu W.N. MicroRNA-27a promotes proliferation and suppresses apoptosis by targeting PLK2 in laryngeal carcinoma. BMC Cancer. 2014;14:678. doi: 10.1186/1471-2407-14-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guidi M., Muiños-Gimeno M., Kagerbauer B., Martí E., Estivill X., Espinosa-Parrilla Y. Overexpression of miR-128 specifically inhibits the truncated isoform of NTRK3 and upregulates BCL2 in SH-SY5Y neuroblastoma cells. BMC Mol. Biol. 2010;11:95. doi: 10.1186/1471-2199-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brocheriou V., Hagège A.A., Oubenaïssa A., Lambert M., Mallet V.O., Duriez M., Wassef M., Kahn A., Menasché P., Gilgenkrantz H. Cardiac functional improvement by a human Bcl-2 transgene in a mouse model of ischemia/reperfusion injury. J. Gene Med. 2000;2:326–333. doi: 10.1002/1521-2254(200009/10)2:5<326::AID-JGM133>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 41.Zeng X.C., Li L., Wen H., Bi Q. MicroRNA-128 inhibition attenuates myocardial ischemia/reperfusion injury-induced cardiomyocyte apoptosis by the targeted activation of peroxisome proliferator-activated receptor gamma. Mol. Med. Rep. 2016;14:129–136. doi: 10.3892/mmr.2016.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen G.H., Xu C.S., Zhang J., Li Q., Cui H.H., Li X.D., Chang L.P., Tang R.J., Xu J.Y., Tian X.Q. Inhibition of miR-128-3p by Tongxinluo Protects Human Cardiomyocytes from Ischemia/reperfusion Injury via Upregulation of p70s6k1/p-p70s6k1. Front. Pharmacol. 2017;8:775. doi: 10.3389/fphar.2017.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sempere L.F., Sokol N.S., Dubrovsky E.B., Berger E.M., Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev. Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 44.Zhu W., Yang L., Du Z. MicroRNA regulation and tissue-specific protein interaction network. PLoS ONE. 2011;6:e25394. doi: 10.1371/journal.pone.0025394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xin L., Liu L., Liu C., Zhou L.Q., Zhou Q., Yuan Y.W., Li S.H., Zhang H.T. DNA-methylation-mediated silencing of miR-7-5p promotes gastric cancer stem cell invasion via increasing Smo and Hes1. J. Cell. Physiol. 2020;235:2643–2654. doi: 10.1002/jcp.29168. [DOI] [PubMed] [Google Scholar]
- 46.Zheng X., Lian D., Wong A., Bygrave M., Ichim T.E., Khoshniat M., Zhang X., Sun H., De Zordo T., Lacefield J.C. Novel small interfering rna-containing solution protecting donor organs in heart transplantation. Circulation. 2009;120:1099–1107. doi: 10.1161/CIRCULATIONAHA.108.787390. 1 p following 1107. [DOI] [PubMed] [Google Scholar]